Summary
Rpn13/Adrm1 is recruited to the proteasome by PSMD1/Rpn2, where it serves as a substrate receptor that binds preferentially to K48-linked ubiquitin chains, an established signal for protein proteolysis. Here, we use NMR to solve the structure of hRpn13 Pru:hRpn2 (940–953):K48-diubiquitin. Surprisingly, hRpn2-bound hRpn13 selects a dynamic, extended conformation of K48-diubiquitin that is unique from previously determined structures. NMR experiments on free K48-diubiquitin demonstrate the presence of the reported ‘closed’ conformation observed by crystallography, but also this more extended state, in which the hRpn13-binding surface is exposed. This extended K48-diubiquitin conformation is defined by interactions between L73 from G76-linked (distal) ubiquitin and a Y59-centered surface of K48-linked (proximal) ubiquitin. Furthermore, hRpn13 exchanges between the two ubiquitins within 100 ms, although prefers the proximal ubiquitin due to interactions with the K48 linker region. Altogether, these data lead to a revised model of how ubiquitinated substrates interact with the proteasome.
Graphical Abstract
eTOC Blurb
hRpn13 is a ubiquitin receptor assembled into the proteasome by hRpn2. Lu et al. used NMR to solve the structure of hRpn2-bound hRpn13 with K48-diubiquitin to discover the chemical basis of hRpn13 preference for the K48 linkage and a dynamic and extended conformation for K48-diubiquitin.
Introduction
The 26S proteasome, composed of a 20S catalytic core particle (CP) capped at either end with a 19S regulatory particle (RP), performs regulated protein degradation in cells. The RP binds and processes ubiquitinated substrates to ultimately passage them to the CP for hydrolysis into short peptides, reviewed in (Ehlinger and Walters, 2013; Finley et al., 2016). Substrate receptors Rpn1/S2/PSMD2, Rpn10/S5a/PSMD4 and Rpn13/Adrm1 in the RP capture ubiquitinated substrates by recognition of ubiquitin and/or shuttle factors bound to ubiquitinated substrates (Chen et al., 2019; Chen et al., 2016; Hiyama et al., 1999; Husnjak et al., 2008; Schreiner et al., 2008; Shi et al., 2016; Walters et al., 2002; Young et al., 1998; Zhang et al., 2009). These three receptors additionally contribute ubiquitin processing enzymes to the proteasome, deubiquitinating enzymes Usp14 (Borodovsky et al., 2001; Leggett et al., 2002; Verma et al., 2000) and Uch37 (Lam et al., 1997) for Rpn1 and Rpn13 respectively, and ubiquitin E3 ligase E6AP for Rpn10 (Buel et al., 2019).
hRpn13 has emerged as a therapeutic target (Anchoori et al., 2018; Anchoori et al., 2013; Kisselev, 2013; Lu et al., 2017; Randles et al., 2016; Song et al., 2019; Song et al., 2016; Trader et al., 2015) with synergy to current proteasome inhibitors of the CP that are used to treat hematological cancers (Song et al., 2016; Trader et al., 2015). An N-terminal Pru (pleckstrin-like receptor for ubiquitin) domain in hRpn13 binds ubiquitin (Husnjak et al., 2008; Schreiner et al., 2008) and the proteasome (Hamazaki et al., 2006; Jorgensen et al., 2006; Lu et al., 2015; Qiu et al., 2006; Yao et al., 2006) while a C-terminal DEUBAD (DEUBiquitinase ADaptor) domain recruits and activates deubiquitinating enzyme (DUB) Uch37 (Hamazaki et al., 2006; Qiu et al., 2006; Yao et al., 2006), one of three DUBs that remove ubiquitin from substrates prior to entry into the CP catalytic chamber.
To bind the proteasome, the hRpn13 Pru domain forms extensive hydrophobic interactions with a 14-residue intrinsically disordered region at the extreme C-terminal end of hRpn2 (Lu et al., 2015; Lu et al., 2017; VanderLinden et al., 2017). A model generated by using HADDOCK and sparse NMR data as well as an experimental crystal structure have defined three loops in the hRpn13 Pru domain to bind the L8-I44-V70 hydrophobic patch of monoubiquitin (Schreiner et al., 2008; VanderLinden et al., 2017). Recently, a structure of hRpn13 Pru with K48-diubiquitin has also been reported based on NMR data, concluding that hRpn13 selectively enriches a preexisting compact state of K48-diubiquitin (Liu et al., 2019). Our data herein suggest an alternative interaction mechanism and structure for hRpn13 binding to K48-diubiquitin. We use NMR to solve the structure of hRpn2 (940–953)-bound hRpn13 Pru with K48-diubiquitin. We find hRpn13 to bind dynamically to each ubiquitin of K48-diubiquitin, with preference for the ubiquitin linked by K48, named proximal for its free G76, which could in principle be linked to a substrate. In contrast to the earlier study, we find an extended conformation for K48-diubiquitin that is unique from previously determined structures and is selected for binding by hRpn2-bound hRpn13.
Results
Structure of hRpn2-bound hRpn13 complexed with K48-diubiquitin
We previously found hRpn13 to bind preferentially to K48-linked ubiquitin chains over all other linkage types (Chen et al., 2016) and for the extreme C-terminal 14 amino acids of hRpn2 to be sufficient for hRpn13 binding (Lu et al., 2015; Lu et al., 2017; VanderLinden et al., 2017). In an effort to define mechanistically the binding interactions of hRpn2-bound hRpn13 with K48-diubiquitin, we prepared two differentially labeled samples for NMR experiments. Each sample contained 13C labeled hRpn13 Pru domain and unlabeled 14-amino acid, hRpn13-binding hRpn2 (940–953). K48-diubiquitin was also present in each sample with either the proximal (Ternary-13C-P) or distal (Ternary-13C-D) ubiquitin 13C labeled (Figure 1A). In each case, 1.2-fold molar excess K48-diubiquitin was added to premixed equimolar hRpn13 Pru and hRpn2 (940–953). For each sample, 13C-dispersed NOESY spectra recorded intramolecular interactions for the 13C-labeled protein as well as intermolecular interactions among all components and 13C-half-filtered NOESY spectra (Chen and Walters, 2012; Lee et al., 1994; Walters et al., 2001; Wider et al., 1991) were used to select for interactions between the 13C- and 12C-labeled components (Figures 1B and S1). Similar intermolecular interactions were observed between hRpn13 and the proximal (Figures 1B left panel and S1A) or distal (Figures 1B right panel and S1B) ubiquitin moiety, indicating that hRpn13 binds to both ubiquitins and does so with a common binding mechanism. The NOEs to distal ubiquitin were weaker compared to proximal ubiquitin (compare right and left panels in Figure 1B and Figures S1A–S1B), indicating hRpn13 preference for the K48-linked proximal ubiquitin, as previously observed for experiments performed without hRpn2 present (Schreiner et al., 2008). In total, we identified 72 or 56 intermolecular interactions between hRpn13 and the proximal or distal ubiquitin moieties, respectively (Tables 1 and 2). 722 NOE interactions were observed within hRpn13 and 104 between hRpn13 and hRpn2 that were identical compared to data collected without K48-diubiquitin present (Figures S2A–S2B) (Lu et al., 2017), indicating that the hRpn13 structure and interaction with hRpn2 was not altered by binding to K48-diubiquitin. We combined all experimental data as described in STAR Methods and listed in Tables 1 and 2 to calculate the structure of hRpn2-bound hRpn13 at either ubiquitin of K48-diubiquitin.
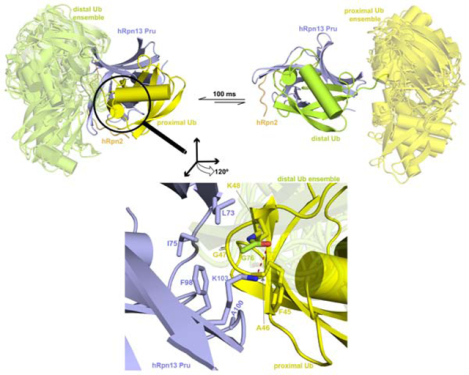
Figure 1. Structure of hRpn2-bound hRpn13 with K48-diubiquitin.
(A) Schematic representation of interactions (arrows) detected in 13C-edited NOESY spectra recorded on two differentially labeled samples at 25°C and pH 6.5; Ternary- 13C-P (0.6 mM 13C-hRpn13 Pru (purple), 0.6 mM hRpn2 (940–953, orange) and 0.72 mM K48-diubiquitin with 13C-proximal ubiquitin (yellow)) and Ternary-13C-D (0.6 mM 13C-hRpn13 Pru, 0.6 mM hRpn2 (940–953) and 0.72 mM K48-diubiquitin with 13C-distal ubiquitin (green)). The ubiquitin linker region is displayed as a black connecting curve. 13C-labeled constituents are indicated with “13C” in red.
(B) Selected regions highlighting intermolecular NOE interactions detected in 13C-half-filtered NOESY experiments (100 ms mixing time) acquired on sample Ternary-13C-D (left) or Ternary-13C-P (right), as indicated in (A). Breakthrough signals at the diagonal are labeled by the letter D (red) while assignments for hRpn13 Pru (purple), hRpn2 (orange), proximal ubiquitin (gray) or distal ubiquitin (green) are provided.
(C and D) Ribbon diagram representation for the 15 lowest energy structures without violations for Ternary-P (C) or Ternary-D (D) colored as in (A) with hRpn13 Pru, hRpn2, and the bound ubiquitin superimposed.
(E and F) Ribbon diagram of a representative structure for Ternary-P (E) or Ternary-D (F) selected from the bundles shown in (C) or (D) respectively retaining for the unbound ubiquitin the full conformational ensemble present in (C) and (D) respectively. Secondary structural elements are labeled and the unbound ubiquitins are displayed in a transparent view. See also Figures S1 and S2.
Table 1.
Structural statistics for the structure of hRpn2-bound hRpn13 at proximal ubiquitin of K48-diubiquitin
| Complex | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance restraints | |
| Total NOE | 5,475 |
| Intra-residue | 1,591 |
| Inter-residue | 3,884 |
| Sequential (|i-j| = 1) | 1,022 |
| Non-sequential (|i-j| > 1) | 2,542 |
| Intermolecular NOEs | 320 |
| hRpn13:hRpn2 | 224 |
| hRpn13:Proximal Ub | 72 |
| Proximal Ub:Distal Ub | 24 |
| Hydrogen bonds | 100 |
| Intramolecular | 97 |
| Intermolecular | 3 |
| hRpn13:hRpn2 | 2 |
| hRpn13:Proximal Ub | 1 |
| Total dihedral angle restraints | 604 |
| Phi | 302 |
| Psi | 302 |
| Structure statistics | |
| Violations (mean and SD) | |
| Distance constraints (Å) | 0.051 ± 0.001 |
| Dihedral angle constraints (°) | 0.370 ± 0.034 |
| Max. dihedral angle violation (> 5 °) | 0 |
| Max. distance constraint violation (> 0.5 Å) | 0 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (°) | 0.494 ± 0.014 |
| Impropers (°) | 0.350 ± 0.010 |
| Average pairwise root-mean-square deviation** (Å) | |
| Heavy atoms | 1.00 ± 0.13 |
| Backbone | 0.48 ± 0.09 |
Statistics for 15 lowest energy structures without violations for hRpn13 Pru (K21-N130), bound proximal ubiquitin (M1-L71), and hRpn2 (942–951).
Table 2.
Structural statistics for the structure of hRpn2-bound hRpn13 at distal ubiquitin of K48-diubiquitin
| complex | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance restraints | |
| Total NOE | 5,459 |
| Intra-residue | 1,591 |
| Inter-residue | 3,868 |
| Sequential (|i-j| = 1) | 1,022 |
| Non-sequential (|i-j| > 1) | 2,542 |
| Intermolecular NOEs | 304 |
| hRpn13:hRpn2 | 224 |
| hRpn13: Distal Ub | 56 |
| Proximal Ub:Distal Ub | 24 |
| Hydrogen bonds | 99 |
| Intramolecular | 97 |
| Intermolecular | 2 |
| Total dihedral angle restraints | 604 |
| Phi | 302 |
| Psi | 302 |
| Structure statistics | |
| Violations (mean and SD) | |
| Distance constraints (Å) | 0.051 ± 0.001 |
| Dihedral angle constraints (°) | 0.371 ± 0.052 |
| Max. dihedral angle violation (> 5 °) | 0 |
| Max. distance constraint violation (> 0.5 Å) | 0 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (°) | 0.494 ± 0.013 |
| Impropers (°) | 0.348 ± 0.007 |
| Average pairwise root-mean-square deviation** (Å) | |
| Heavy atoms | 1.07 ± 0.16 |
| Backbone | 0.59 ± 0.14 |
Statistics for 15 lowest energy structures without violations for hRpn13 Pru (K21-N130), bound distal ubiquitin (M1-L71), and hRpn2 (942–951).
Excluding the unbound ubiquitin moiety, the 15 lowest energy structures without violations converged to a backbone root mean square deviation (RMSD) of 0.48 Å or 0.59 Å respectively for the complex with hRpn13 at proximal (Ternary-P, Figure 1C) or distal (Ternary-D, Figure 1D) ubiquitin. For clarity, a representative ribbon diagram for Ternary-P (Figure 1E) or Ternary-D (Figure 1F) is included with a transparent view of the conformational ensemble possible for the unbound ubiquitin. In both cases, hRpn2 (940–953) extends across hRpn13 Pru where it interacts with β1, β2, β7, β8 and connecting loops β1-β2, β6-β7, β8-α1 (Figures 1E–1F), similar to the interactions observed without ubiquitin present (Lu et al., 2017) (Figure S2C). In addition, the structure of each ubiquitin moiety is unchanged compared to free monoubiquitin (PDB code 1UBQ) (Vijay-Kumar et al., 1987), with an RMSD for the secondary structural elements of 0.60 ± 0.14 Å (Figure S2D).
hRpn2-bound hRpn13 interacts dynamically with ubiquitins of K48-diubiquitin
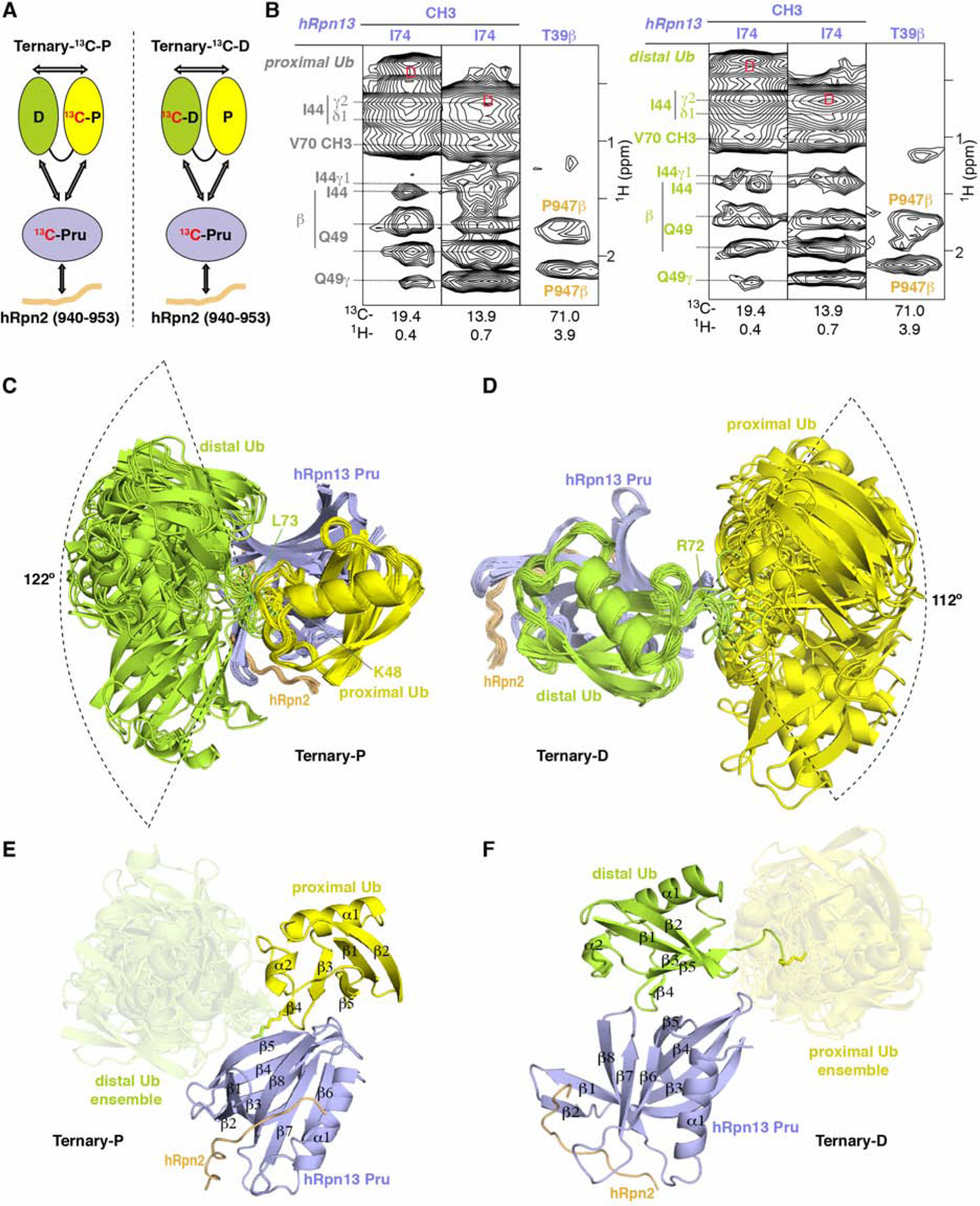
As described above, two distinct binding states were observed, differentiated by hRpn2-bound hRpn13 interaction with either the proximal or distal ubiquitin of K48-diubiquitin (Figure 1). Our NMR experiments further indicated that hRpn2-bound hRpn13 exchanges dynamically between these two states. More specifically, the 13C-dispersed NOESY spectra for the differentially labeled hRpn2:hRpn13:K48-diubiquitin samples (Figure 1A) were recorded with a 100 ms mixing time, during which NOE interactions were transferred between the two states. Characteristic of the hRpn13 being at either ubiquitin, we observed two sets of proximal and distal ubiquitin signals for amino acids at the binding surface, including the A46 Hα and methyl groups (Figure 2A). One set of signals was not shifted by hRpn2:hRpn13 addition, indicating an unbound state – either due to hRpn13 binding at the other ubiquitin or to the slight molar excess of K48-diubiquitin compared to hRpn2:hRpn13 (Figure 2B). Indicative of exchange between the two observed states, NOEs were detected between the A46 unbound-state and bound-state methyl signals, as well as to and between the A46 Hα signals. Furthermore, intermolecular NOEs involving hRpn13 F98 and K103 were transferred to the unbound states of A46 (Figure 2A). These transferred NOEs from the bound to unbound state indicate that hRpn13 interacts dynamically with K48-diubiquitin such that during 100 ms, an hRpn13-bound ubiquitin moiety is released.
Figure 2. hRpn2-bound hRpn13 interacts dynamically with ubiquitins of K48-diubiquitin.
(A) Expanded regions for ubiquitin A46 from a 13C-edited NOESY spectrum (100 ms mixing time) on sample Ternary-13C-P (Figure 1A, left panel) or Ternary-13C-D (Figure 1A, right panel). NOEs between A46 signals from the free and bound states are labeled and indicated with dashed black lines whereas intermolecular NOEs to hRpn13 are labeled and indicated with a solid purple line. (B) Schematic representation of exchangeable states presence in sample Ternary-13C-P or Ternary-13C-D with coloring as in Figure 1A.
Binding mechanisms of hRpn13 for K48-diubiquitin
Ubiquitin receptors commonly bind a hydrophobic ubiquitin surface centered on L8, I44, and V70 (Randles and Walters, 2012), which are located in ubiquitin loop β1-β2, β3, and β5, respectively, and part of the hRpn13 recognition surface (Figures 3A–3B). hRpn13 binds this ubiquitin surface at a location remote from where hRpn2 binds and formed by β4, β8 and loops connecting β3-β4, β5-β6, and β7-β8 (Figures 1E–1F and 3A). Arginine substitution of hRpn13 F76, located in the β5-β6 loop, abrogates hRpn13 binding to ubiquitin (Schreiner et al., 2008) and we find this amino acid to interact with the distal and proximal L8-I44-V70 ubiquitin patch of K48-diubiquitin similarly (Figures 3A–3B), as defined by our experimental data (Figure S3). At either side of F76, hRpn13 L56 and I74 form hydrophobic contacts to ubiquitin R42, I44, Q49 and V70 (Figure 3B), as indicated by NOE interactions (Figure S1).
Figure 3. Binding mechanisms of hRpn13 for K48-diubiquitin.
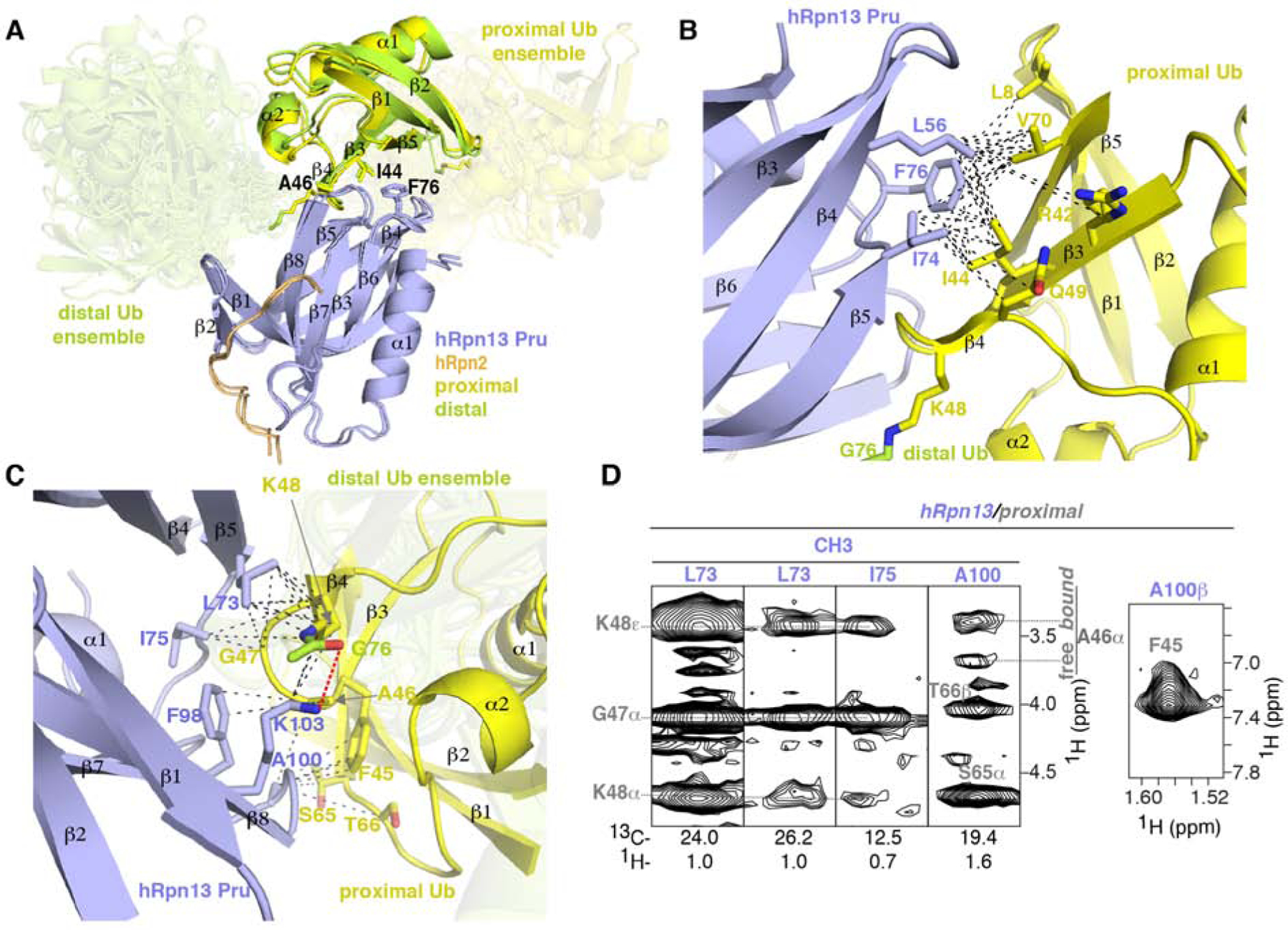
(A) Superposition of Ternary-P (displayed as in Figure 1E) and Ternary-D (displayed as in Figure 1F) overlaying the hRpn13-bound ubiquitin moieties. Key interacting amino acid sidechains are shown, including hRpn13 F76 and ubiquitin I44 and A46.
(B and C) Enlarged view of the interactions between hRpn13 and proximal ubiquitin for Ternary-P, shown as in (A). Black dashed lines represent intermolecular NOE interactions involving hRpn13 L56, I74 or F76 and proximal ubiquitin L8, R42, I44, K48, Q49 or V70 (B) as well as hRpn13 L73, I75, F98, A100 or K103 and proximal ubiquitin F45-K48 or S65-T66 (C). In (C), a hydrogen bond between the hRpn13 K103 ε-ammonium group and the carbonyl of isopeptide bonded ubiquitin G76 is represented by a red dashed line. Nitrogen and oxygen atoms at the interaction surface are colored in blue and red, respectively.
(D) Selected regions from a 13C-half-filtered NOESY experiment (100 ms mixing time) acquired on sample Ternary-13C-D (Figure 1A) highlighting interactions displayed in (C) between hRpn13 (labeled purple) and proximal ubiquitin (labeled gray) of K48-diubiquitin. Two sets of signals are observed for ubiquitin A46 and labeled according to chemical shift position overlapping with free ubiquitin (free) or shifted by hRpn13 binding (bound). See also Figures S1 and S3.
In the region at ubiquitin A46, hRpn13 F98 also contributes to ubiquitin binding, interacting with this residue, while further interactions are made by hRpn13 A100 to ubiquitin F45, S65 and T66 (Figure 3C). These contacts are indicated by NOEs (Figures 2A, 3D and S1), some of which transferred to signals from unbound ubiquitin (Figures 3D and S1). As discussed above, the presence of these transferred NOEs indicated dynamic exchange between the hRpn13-bound and hRpn13-free states for the ubiquitin moieties of K48-diubiquitin.
Furthermore, hRpn13 L73, I75 and K103 form extensive interactions with ubiquitin G47 and K48 (Figure 3C), which for the proximal ubiquitin is adjacent to or at the ubiquitin linker region. Loss of the charge for the proximal ubiquitin K48 sidechain enables closer contacts with surrounding hydrophobic groups from these hRpn13 amino acids (Figure 3C). In addition, a hydrogen bond is formed between the hRpn13 K103 ε-ammonium group and the carbonyl oxygen of the isopeptide bonded ubiquitin G76 (Figure 3C). These interactions provide a rationale for hRpn13 preference for K48-linked ubiquitin.
Unbound ubiquitin is only partially constrained in complex with hRpn2-bound hRpn13
24 NOEs were detected between the two ubiquitin moieties involving distal ubiquitin L73 and proximal ubiquitin amino acids, including Q49, E51, R54, D58, and Y59 (Figures 4A–4B, Tables 1 and 2). These interactions were detected for both sample Ternary-13C-P and Ternary-13C-D (Figures 1A, 4A–4B, S4A–S4B) and are derived by the sidechain of L73 from distal ubiquitin being buried by hydrophobic groups from proximal ubiquitin amino acids at or near the linker region centered around Y59 (Figures 4C–4D). Only K48-linked ubiquitin chains can form linkage-dependent interactions with the Y59-centered surface, as it is spatially close to the K48 sidechain and remote from all other amino acids used to form ubiquitin chains (Figure S4C). At either side of this surface are the sidechain γ and δ groups of R54 and the linked K48 sidechain, thus placing L73 between K48 and R54, a location ideal for interaction with the aromatic sidechain of Y59 (Figures 4B–4D).
Figure 4. Unbound ubiquitin is only partially constrained in complex with hRpn2:hRpn13.
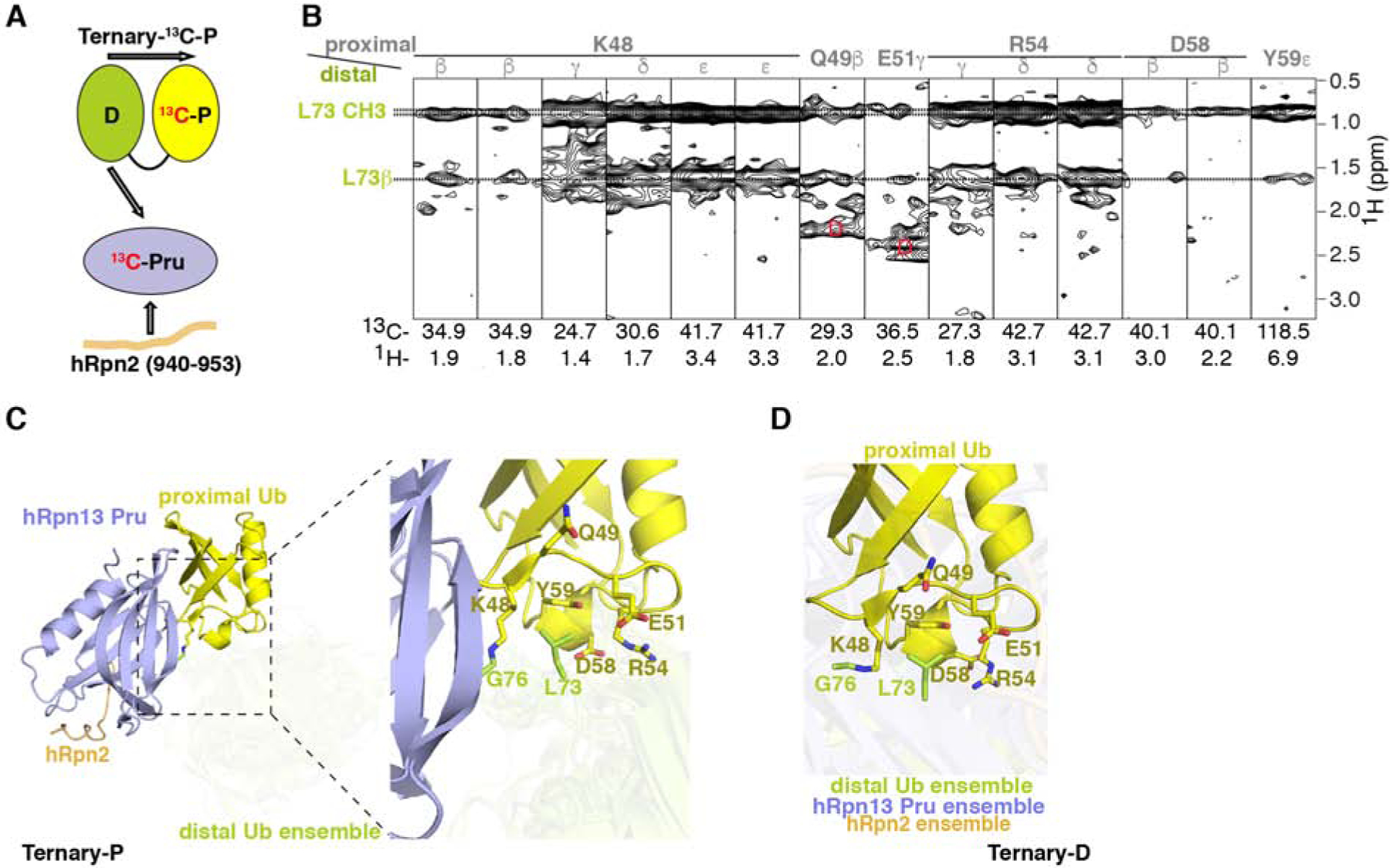
(A) Schematic representation of interactions in Ternary-13C-P detectable by the 13C-half-filtered NOESY experiment (indicated by arrows) with the coloring of Figure 1A.
(B) Selected regions from a 13C-half-filtered NOESY experiment (100 ms mixing time) acquired on sample Ternary-13C-P (A) highlighting interactions between the two ubiquitin moieties, with proximal ubiquitin assignments in gray and distal ubiquitin assignments in green. Breakthrough signals at the diagonal are labeled by the letter D (red).
(C and D) Representative Ternary-P (C, displayed as in Figure 3C) and Ternary-D (D) structures illustrating interactions between distal ubiquitin L73 and proximal ubiquitin K48, Q49, E51, R54, D58 and Y59. The region expanded to the right in (C) is indicated by a dashed rectangle. In (D), a representative structure is displayed for the superposition of proximal ubiquitin when hRpn2 (940–953):hRpn13 Pru is bound to distal ubiquitin with the conformational ensemble for the components not superimposed displayed in a transparent view. See also Figure S4.
The interactions between distal ubiquitin L73 and the proximal ubiquitin Y59-centered surface as well as the isopeptide bond that links the two ubiquitins partially constrain their relative orientation. However, distal ubiquitin L73 is not rigidly constrained relative to neighboring R72 (Figures 1C–1D). This feature, together with the lack of additional interactions involving the unbound ubiquitin, enables conformational heterogeneity such that relative to the linker region, distal ubiquitin can rotate about an angle of ~122° when hRpn13 is present at proximal u biquitin (Figure 1C), and proximal ubiquitin rotates about an angle of ~112° when hRpn 13 binds distal ubiquitin (Figure 1D). Thus, a striking commonality between the two K48-diubiquitin binding states of hRpn2:hRpn13 is the geometric freedom of the unbound ubiquitin, inducing flexibility for the ubiquitin moieties neighboring that bound by hRpn13.
hRpn2-bound hRpn13 binds to an extended conformational state of K48-diubiquitin
Two distinct conformational states have been resolved for free K48-diubiquitin, a ‘closed’ form (Figure 5A) in which the ubiquitins pack against each other (Cook et al., 1992; Trempe et al., 2010) and an ‘opened’ form (Figure 5B) with the ubiquitins further apart (Hirano et al., 2011; Lai et al., 2012). Distal ubiquitin L73 is spatially close to proximal ubiquitin K48 in the ‘closed’ but not ‘opened’ form. However, in both conformational states of K48-diubiquitin, proximal ubiquitin Q49, E51, R54, D58 and Y59 are too far for the observed NOE interactions with distal ubiquitin L73 (this study, Figure 5C). The detected inter-ubiquitin NOEs thus suggest the presence of a K48-diubiquitin conformational state that is unique from those previously discovered.
Figure 5. hRpn2-bound hRpn13 binds to an extended conformational of K48-diubiquitin.
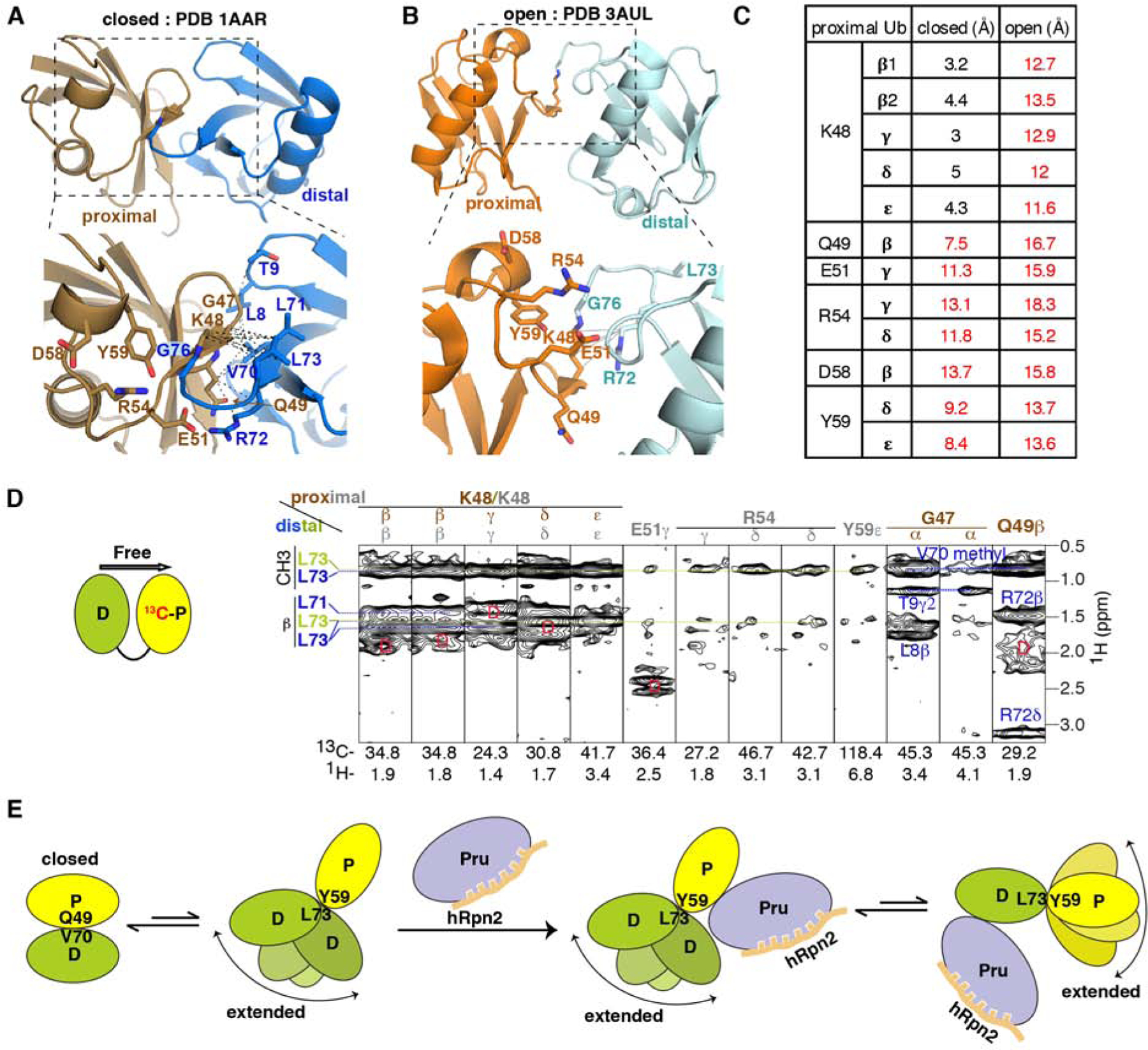
(A and B) Ribbon diagram of free K48-diubiquitin structures solved by x-ray crystallography with an expansion (dashed rectangles) in the lower panels. Two conformational states have been reported and named ‘closed’ (PDB 1AAR, A, proximal in brown, distal in blue) or ‘open’ (PDB 3AUL, B, proximal in orange and distal in cyan). Sidechains of interacting ubiquitin residues are displayed (bottom panels), with nitrogen and oxygen in blue and red, respectively. Black dashed lines represent inter-ubiquitin NOE interactions detected in the experiment of (D), involving proximal ubiquitin G47-Q49 and distal ubiquitin L8-T9 or V70-L73 (A). Gray solid lines represent interactions close enough for expected NOE interactions between proximal ubiquitin K48 and distal ubiquitin R72 (B); these were not detected in our 13C-half-filtered NOESY experiment.
(C) Measured distances between proximal ubiquitin residues and distal ubiquitin L73 for the ‘closed’ (Figure 5A) and ‘open’ (Figure 5B) crystal structures of free K48-diubiquitin. Distances too large for NOE interactions are indicated in red.
(D) Selected regions from a 13C-half-filtered NOESY experiment (100 ms mixing time, right panel) acquired on 0.3 mM K48-diubiquitin with 13C-labeled proximal (left panel, yellow) and unlabeled distal (left panel, green) ubiquitin at 25°C and pH 6.5 highlighting NOE interactions between the two ubiquitin moieties. NOE interactions expected from the ‘closed’ conformation are labeled brown for proximal ubiquitin and blue for distal ubiquitin whereas those consistent with the structure used to bind hRpn13 (Figures 1C–1F) are labeled gray for proximal ubiquitin and green for distal ubiquitin. Breakthrough diagonal signals are indicated with a red ‘D’.
(E) Cartoon depicting the dynamics of free K48-diubiquitin and following interaction with hRpn2-bound hRpn13. When free, K48-diubiquitin exchanges between the ‘closed’ conformation resolved by x-ray crystallography (A) and an ensemble of extended conformations, as detected in this study. The ‘closed’ conformation is dominant for free K48-diubiquitin. The extended conformation is selected by hRpn2-bound hRpn13, which binds either ubiquitin with preference for the proximal moiety. See also Figure S4.
We tested whether these interactions exist in free K48-diubiquitin or alternatively, are induced by binding to hRpn13 by recording a 13C-half-filtered NOESY spectrum on free K48-diubiquitin with the proximal ubiquitin 13C-labeled (Figure 5D, left panel). Interactions expected for the ‘closed’ K48-diubiquitin conformation were represented strongly in this experiment. In particular, NOEs were detected between distal ubiquitin L71 and L73 and proximal ubiquitin K48 as well as between distal ubiquitin L8, T9, V70 or R72 and proximal ubiquitin G47 or Q49 (Figure 5D, right panel). This second set of NOE interactions are exclusive to the ‘closed’ conformation (Figure 5A) and not present in the spectra recorded with hRpn2-bound hRpn13 present (Figure 4B), suggesting that this conformational state is not productive for binding to hRpn13. We were not able to find interactions between the two ubiquitins characteristic of the ‘opened’ crystal structure, such as between proximal ubiquitin K48 and distal ubiquitin R72 (Figures 5B and 5D), consistent with a previous report (Lai et al., 2012).
Similar to the spectra recorded with hRpn2-bound hRpn13 present, we detected NOE interactions between distal ubiquitin L73 and proximal ubiquitin R54 or Y59 for the free K48-diubiquitin sample (Figure 5D, right panel). These NOEs were weaker compared to when hRpn2-bound hRpn13 was present (Figure 5D, right panel versus 4B) and also compared to the NOEs representative of the K48-diubiquitin ‘closed’ conformation (Figure 5D).
Altogether, our data indicate that free K48-diubiquitin exists in solution predominately in the ‘closed’ conformation, but also in an extended conformation that orients distal L73 towards a proximal ubiquitin surface at R54 and Y59 and that this latter conformational state is selected for by hRpn2-bound hRpn13 (Figure 5E); for simplicity, we henceforth refer to this state as ‘extended’. The extended K48-diubiquitin state is dynamic but does not include the ‘open’ conformation observed by x-ray crystallography (Figure S4D), which does not orient distal L73 towards the Y59-centered proximal ubiquitin surface.
Comparison with previous structures of hRpn13 complexes with ubiquitin
hRpn2-bound hRpn13 Pru complexed with monoubiquitin has been crystallized to have two conformational states per asymmetric unit, which superimpose onto hRpn2-hRpn13 Pru bound to the proximal ubiquitin with an RMSD of 0.968 Å (Figure 6A, dark red) or 0.963 Å (Figure 6A, gray) (VanderLinden et al., 2017). Thus, the binding interactions in these complexes are similar. Some subtle differences exist involving L8 and as expected, K48. In the crystal structures, ubiquitin L8 is spatially close to hRpn13 F76 and P77 (for PDB 5V1Y-1) or F76, P77 and P131 (for PDB 5V1Y-2) (Figure 6B, red and gray). We detected intermolecular NOE interactions from proximal or distal ubiquitin L8 to hRpn13 F76 (Figure S3) but not to P77 or P131. Both proximal and distal ubiquitin I44 and V70 also have NOEs to hRpn13 F76 (Figure S3). Thus, in Ternary-P and Ternary-D, proximal L8 is close to hRpn13 F76, but further away from P77 and P131 than in the crystal structures with monoubiquitin (Figure 6B).
Figure 6. Comparison with previous structures of hRpn13 complexed with ubiquitin.
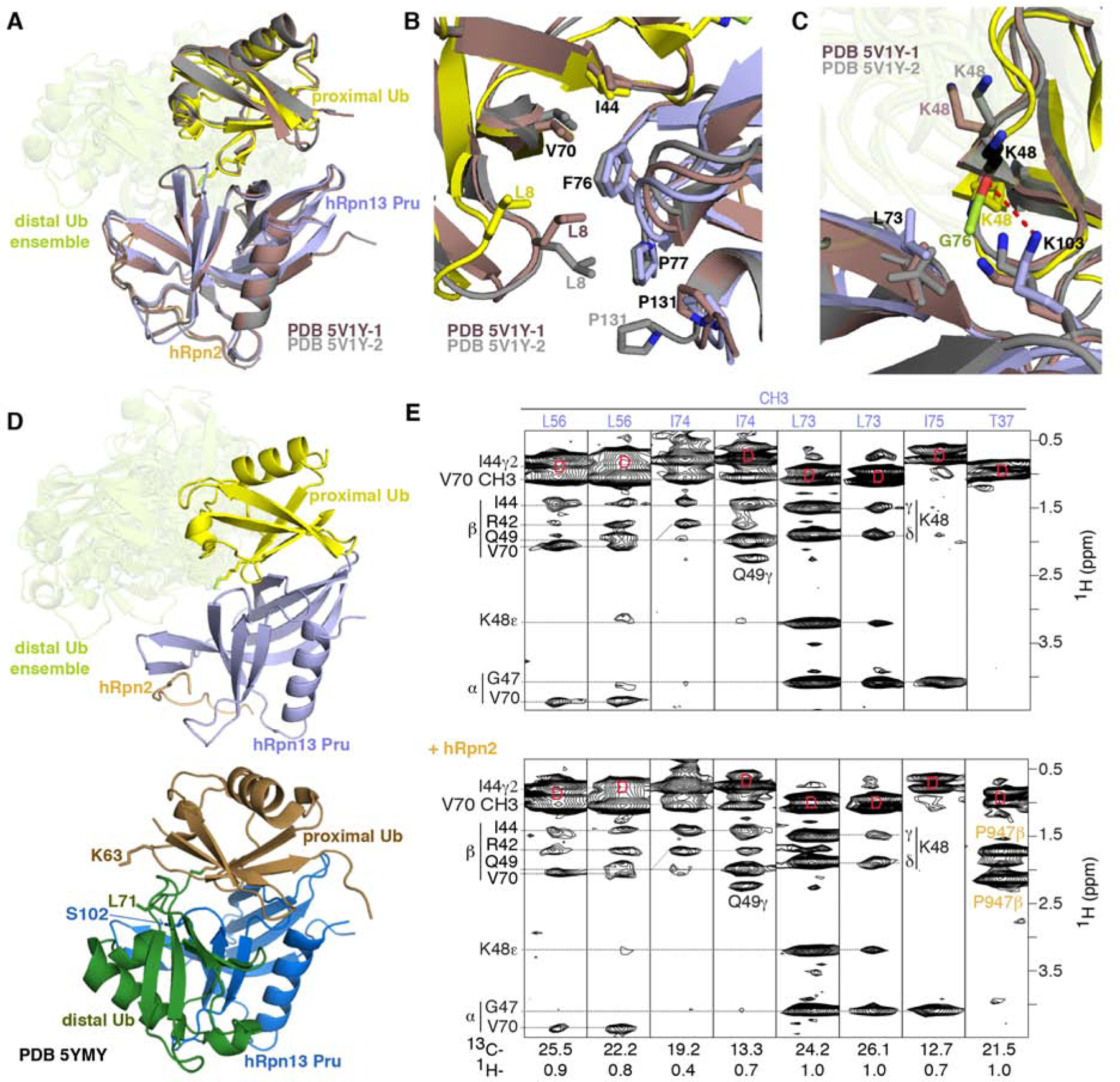
(A, B and C) Superposition of hRpn13 Pru:hRpn2 (940–953):ubiquitin for the two available crystal forms (PDB 5V1Y-1 in dark red and PDB 5V1Y-2 in gray) and our Ternary-P structure presented as in Figure 1E. Expanded regions centered on ubiquitin L8-I44-V70 (B) and ubiquitin K48 (C) are included. Interactions to hRpn13 F76, P77, and P131 are highlighted in (B) demonstrating similarity among the three structures for hRpn13 F76 but divergence for ubiquitin L8 and hRpn13 P131. In the crystal structures with monoubiquitin, K48 adopts two configurations (displayed in grey and black) for PDB 5V1Y-2 and one conformation for PDB 5V1Y-1 (dark red). The isopeptide bonded ubiquitin K48 is directed closer to L73 compared to in the structures with monoubiquitin (C). From this location, a hydrogen bond (red dashed line) forms between the hRpn13 K103 ε-ammonium group and the carbonyl of isopeptide bonded ubiquitin G76. Nitrogen and oxygen atoms are displayed in blue and red, respectively.
(D) Structural comparison for Ternary-P (top panel, displayed as in A) and structure PDB-5YMY of hRpn13 Pru (blue):K48-diubiquitin (bottom panel, proximal or distal ubiquitin in brown or dark green, respectively) with identical orientation for hRpn13 Pru. In PDB-5YMY, hRpn13 S102, proximal ubiquitin K63 and distal ubiquitin L71 are displayed and labeled.
(E) Selected regions of a 13C-half-filtered NOESY experiment (mixing time 100 ms) acquired on a mixture of 0.25 mM 13C-labeled hRpn13 Pru with 1.2-fold molar excess unlabeled monoubiquitin without (top panel) or with 1.2-fold molar excess hRpn2 (940–953) (bottom panel) at 25°C and pH 6.5. Diagonal br eakthrough signals are labeled by ‘D’ (red) and assignments for hRpn13 Pru (purple), ubiquitin (black) and hRpn2 (orange) included. See also Figure S3.
In the crystal structures with monoubiquitin, K48 adopts multiple conformations (Figure 6C, red, gray or black). When linked to G76 however K48 converges (Figure 1C) to interact with hRpn13 L73 and K103 (Figure 6C). These interactions are indicated by myriad NOEs between these residues (Figures 3D and S1). The isopeptide bonded K48 sidechain is closer to hRpn13 L73 methyl groups compared to in the structures with monoubiquitin (Figure 6C), a conformational change likely favored by loss of the positive charge. In addition, the aforementioned hydrogen bond between the hRpn13 K103 ε-ammonium group and the carbonyl oxygen of the isopeptide bonded ubiquitin G76 is enabled by the shifting of proximal ubiquitin K48 towards hRpn13 K103 (Figure 6C).
Previously, a structure determined by NMR was reported for hRpn13 Pru complexed with K48-diubiquitin (PDB-5YMY, (Liu et al., 2019)). In contrast to our structure, this structure reports only one configuration for hRpn13 binding to K48-diubiquitin with both ubiquitins binding to hRpn13 at the same time (Figure 6D, bottom panel). The differences between our structure and 5YMY are not caused by hRpn2, as 13C-half-filtered NOESY spectra recorded on a mixture of 0.25 mM 13C-labeled hRpn13 Pru with 1.2-fold molar excess unlabeled monoubiquitin without and with hRpn2 (940–953) detected identical NOE interactions (Figure 6E, top versus bottom panel). This finding is consistent with our previous demonstration that hRpn2 has no effect on ubiquitin binding for the single domain hRpn13 Pru fragment; however, hRpn2 does activate ubiquitin binding for full length hRpn13 by abolishing intramolecular interactions between the hRpn13 Pru and DEUBAD domains (Chen et al., 2010).
Our NOE data are not consistent with the 5YMY structure, and furthermore, only one intermolecular NOE distance constraint between hRpn13 Pru and distal ubiquitin of K48-diubiquitin was listed for 5YMY. This interaction was between hRpn13 Pru S102 Hα and the distal ubiquitin L71 methyl groups (Figure 6D, bottom panel). We were unable to observe an NOE corresponding to or supporting this interaction in our NMR spectra. In addition, we were unable to find other intermolecular NOEs that would be expected from PDB-5YMY, such as proximal ubiquitin K63 interaction with distal ubiquitin L71 methyl groups (Figure 6D, bottom panel).
Discussion
Unexpectedly, we found an inherently dynamic, extended conformational state for free K48-diubiquitin. We propose that only K48-linked ubiquitin chains are able to adopt this conformational state, with distal ubiquitin L73 buried by proximal ubiquitin K48, R54 and Y59, as these amino acids are remote from distal ubiquitin G76 for all other linkage types (Figure S4C). As discussed above for free K48-diubiquitin, inspection of structures deposited for free K48-tetraubiquitin (PDB: 1F9J (Phillips et al., 2001), 2O6V (Eddins et al., 2007), 3ALB (Satoh et al., 2010), and 1TBE (Cook et al., 1994)) as well as other complexed structures available for K48-diubiquitin with other binding partners, including deubiquitinase MINDY-1 MIU2 (PDB: 5MN9 (Kristariyanto et al., 2017)), ubiquitin receptors Rpn1 (PDB: 2N3V, 2N3W (Shi et al., 2016)) and Rpn10 (PDB: 2KDE, 2KDF (Zhang et al., 2009)), E2 Ube2K (PDB: 6IF1 (Lee et al., 2018)), and gp78 E3 ligase CUE domain (PDB: 2LVP, 2LVQ (Liu et al., 2012)), revealed that this dynamic extended conformational state that we discover herein was not observed in previous studies.
The hRpn13-binding surface is exposed in the extended configuration of K48-linked chains and this surface is also used to bind many other ubiquitin receptors, including the other proteasome substrate receptors Rpn1 and Rpn10 (Shi et al., 2016; Wang et al., 2005; Zhang et al., 2009). We also observed the ‘closed’ K48-diubiquitin conformation; this state buries the L8-I44-V70 binding surface of both ubiquitins (Cook et al., 1992; Trempe et al., 2010). We propose that it is the dynamic, extended conformation of K48-linked chains, rather than the ‘closed’ state, from which binding to receptors is most frequently initiated.
Even in our samples with excess K48-diubiquitin compared to hRpn2-bound hRpn13, the ‘closed’ state was not observed, suggesting that dynamic binding to hRpn13 is faster than the time needed to reform the ‘closed’ state. Thus, the presence of hRpn13 changes the time scale for dynamic reorientation of two ubiquitin moieties, which in free K48-diubiquitin is reported to occur on a time scale of 9–30 ns (Ryabov and Fushman, 2007). Substrates are degraded at the proteasome in timescales of seconds with substrate unfolding being the rate limiting step (Bard et al., 2019). We propose that the ability of hRpn13 to maintain ubiquitin chains in the extended conformation with exposed receptor binding sites over hundreds of milliseconds would aid binding to other ubiquitin receptors in the proteasome over the required duration for substrate unfolding, especially weaker binding hRpn1 (Shi et al., 2016).
Contribution of dynamic binding to ubiquitin chains at the proteasome
Substrates for the proteasome are diverse, as is the ubiquitin signal that directs them there, which can be attached at multiple sites and/or with a variety of linkage configurations (Dikic et al., 2009; Komander and Rape, 2012; Liu and Walters, 2010). How the proteasome handles this multi-level diversity remains elusive. Following binding to the proteasome, ubiquitin chains on substrates must be removed by deubiquitinating enzymes while the substrates themselves engage the ATPase ring. Flexible, unstructured initiation sites within the substrate, either naturally occurring or generated by interaction with Cdc48/p97 (Olszewski et al., 2019), promote proteolysis by productive interactions with the translocation mechanisms of the proteasome ATPase ring (Bard et al., 2019; Fishbain et al., 2015; Prakash et al., 2004). These events occur at fairly restrictive locations in the proteasome RP (de la Pena et al., 2018; Dong et al., 2019), suggesting that substrate and ubiquitin chain orientation would be important determinants of degradation efficiency.
The dynamic interactions between hRpn2-bound hRpn13 and K48-diubiquitin similarly occur with the other major substrate receptors in the proteasome, Rpn1 and Rpn10 (Shi et al., 2016; Zhang et al., 2009). In our previous studies, we found each of these proteins to interact dynamically with either ubiquitin of a K48-linked chain, in these cases through two ubiquitin-binding sites (Shi et al., 2016; Zhang et al., 2009). We propose that the dynamic nature of receptor binding to the ubiquitin chains aids in orienting substrates at the proteasome for downstream processing by the deubiquitinating enzymes and ATPase ring. In this model (Figure 7), the receptors stochastically sample different locations of ubiquitin chains to induce movement of the attached substrate until a binding configuration is obtained that is conducive for processive deubiquitination and/or substrate interaction with the proteasome ATPase ring.
Figure 7. . Model illustrating how dynamic interactions between ubiquitin chains and proteasome receptors may stochastically drive substrate proteolysis.
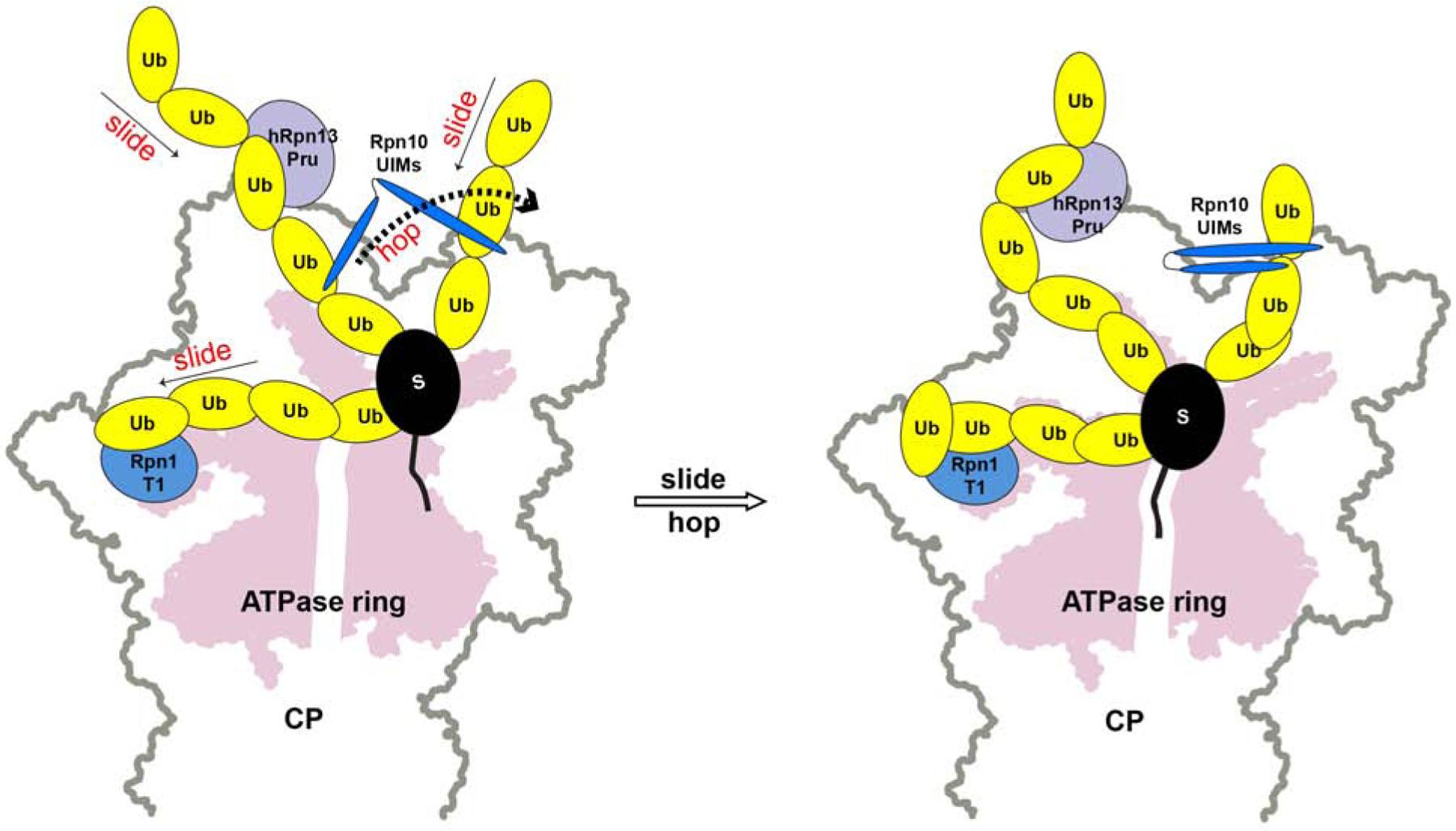
Based on our previous and current NMR data, we propose that the three substrate receptors hRpn1 (blue), hRpn10 (dark blue) and hRpn13 (purple) bind dynamically to ubiquitin chains (yellow) attached to substrate (black), sliding within a chain and hopping between chains, causing motion for the substrate (left panel) until a productive orientation is formed whereby the substrate engages the ATPase ring (pink, right panel).
Such dynamic interactions with ubiquitin chains may be used broadly by ubiquitin receptor proteins in signaling pathways. For example, Rap80 contains two ubiquitin-binding regions with spacing within a helix optimal for binding to neighboring ubiquitins of K63-linked chains (Sato et al., 2009; Walters and Chen, 2009). When interacting with longer ubiquitin chains, Rap80 similarly binds dynamically (Markin et al., 2010).
In summary, our findings highlight the dynamic interactions that occur between proteasome receptors and K48-linked ubiquitin chains. We expect these dynamic interactions to also occur with other ubiquitin chain linkage types and to be a natural necessity of the substrate diversity at the proteasome.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kylie J. Walters (kylie.walters@nih.gov). This study did not generate new unique reagents.
Experimental Model and Subject Details
E. coli strains
Escherichia coli BL21(DE3)pLysS or BL21 (DE3) cells (Invitrogen) were grown in Fernbach flasks at 37°C in a shaker at 250 rpm unti l induction in either Luria-Bertani broth or M9 minimal media with 13C glucose as the sole carbon source. In all cases, antibiotic selection was used at 100 μg/mL ampicillin and 34 μg/mL chloramphenicol. Following induction with isopropyl-β-D-thiogalactoside (0.4 mM), the temperature was reduced to 17°C for hRpn13 Pru and ubiquitin.
Method Details
NMR sample preparation
hRpn13 Pru (1–150), hRpn2 (940–953), and K48-diubiquitin were produced from Escherichia coli BL21(DE3)pLysS or BL21 (DE3) cells (Invitrogen) as previously described (Cook et al., 1992; Lu et al., 2017). Affinity chromatography was used for hRpn13 and hRpn2 respectively by using His and glutathione S-transferase tags at the N-terminus followed by a PreScission protease cleavage site. K48-diubiquitin was made by including additional amino acid D77 in the proximal ubiquitin (Ub-D77) and substituting K48 with arginine (Ub-K48R) in the distal ubiquitin. Protein expression was induced by isopropyl-β-D-thiogalactoside (0.4 mM) for 4 h at 37 °C or 20 h at 17°C following growth at 37 °C to an OD 600 value of 0.6. The cells were collected by centrifugation at 4,550g for 30 min, lysed by sonication and cell debris removed by centrifugation at 31,000g for 30 min. The lysates for hRpn2 (940–953) or hRpn13 Pru were incubated with Glutathione S-sepharose 4B (GE Healthcare Life Sciences) for 3 h or Talon Metal Affinity resin (Clontech) for 1 h, respectively, and the resin washed extensively with buffer A (20 mM sodium phosphate, 300 mM NaCl, 10 mM βME, pH 6.5). hRpn2 (940–953) or hRpn13 Pru (1–150) was eluted from the resin by overnight incubation with 50 units per mL PreScission protease (GE Healthcare Life Sciences) in buffer B (20 mM sodium phosphate, 50 mM NaCl, 2 mM DTT, pH 6.5). The eluent was subjected to size exclusion chromatography with a Superdex75 column on an FPLC system equilibrated with buffer B for further purification. The lysates for Ub-D77 or Ub-K48R in buffer C (50 mM Tris, 1mM PMSF, protease inhibitor cocktail, 1mM DTT, pH 7.6) were titrated with 70% perchloric acid to a final concentration of 2% (v/v) perchloric acid to precipitate impurities and centrifugated at 31,000g for 30 min. The supernatant was dialyzed into buffer D (50 mM Ammonium Acetate, pH 4.5), applied to a cation exchange column (SP-Sepharose), and eluted by a gradient of buffer D and E (50 mM Ammonium Acetate, 600 mM NaCl, pH 4.5).13C glucose was used for isotopic labelling.
0.88 mM 13C-labeled Ub-D77(proximal) or Ub-K48R (distal) with 1.2-fold molar excess unlabeled Ub-K48R or Ub-D77 respectively were incubated at 37 °C for 4 h in reaction buffer (50 mM Tris, 5 mM MgCl2, 10 mM creatine phosphate, 0.6 U/mL inorganic pyrophosphatase, 0.6 U/mL creatine kinase, 2 mM ATP) with 20 μM E2–25K and 0.1 μM E1. 10 μM E2–25K and 0.05 μM E1 was next added and incubation continued at 37 °C overnight, after which the reaction was quenched by addition of 5 mM DTT and 1 mM EDTA. The pH was lowered to 4.5 and the product purified by using a Mono S column. A mixture of hRpn13 Pru with 1.2-fold molar excess hRpn2 (940–953) was prepared from the separately purified proteins and then passed over the Superdex75 column equilibrated with buffer B. The complex of hRpn13 Pru with hRpn2 (940–953) was added to 1.2-fold molar excess K48-diubiquitin to assemble the ternary complexes in buffer B.
NMR experiments
All NMR experiments were conducted at 25 °C and pH 6.5 on Bruker Avance 850 or 900 MHz spectrometers equipped with cryogenically cooled probes. Intramolecular or intermolecular NOE distance constraints for structure calculations were obtained by using 13C-edited NOESY spectra (100 ms mixing time) or 13C-half-filtered NOESY spectra (100 ms mixing time) on mixtures of 0.6 mM 13C-labeled hRpn13 Pru, equimolar unlabeled hRpn2 (940–953) and 1.2-fold molar excess K48-diubiquitin with either ubiquitin 13C-labeled. Intermolecular NOEs for hRpn13 with monoubiquitin were determined by using a 13C-half-filtered NOESY spectrum recorded with 100 ms mixing time on a mixture of 0.25 mM 13C-labeled hRpn13 Pru with 1.2-fold molar excess unlabeled monoubiquitin or a mixture of 0.25 mM 13C-labeled hRpn13 Pru with 1.2-fold molar excess unlabeled monoubiquitin and hRpn2 (940–953). Another 13C-half-filtered NOESY spectrum was recorded with 100 ms mixing time on 0.3 mM K48-diubiquitin with 13C-labeled proximal ubiquitin. The 13C-edited and 13C-half-filtered NOESY spectra were acquired on samples dissolved in D2O or 75% D2O. NMRPipe (Delaglio et al., 1995) was used to process data and XEASY (Bartels et al., 1995) was used to visualize and analyze spectra.
Structure determination
Assignment of intramolecular NOE interactions in the 13C-dispersed NOESY spectra was aided by previous spectra recorded on hRpn2-bound hRpn13 (Lu et al., 2017). The NOE interactions observed within hRpn13 and between hRpn13 and hRpn2 were preserved in the complexes with K48-diubiquitin, indicating that the hRpn13 structure and interaction with hRpn2 was not altered by binding to K48-diubiquitin. The quality of 13C-dispersed NOESY spectra for hRpn2-bound hRpn13 with K48-diubiquitin was poorer than that without K48-diubiquitin; therefore, intramolecular NOE-derived distance constraints for hRpn13 were obtained from 15N-dispersed NOESY spectra and 13C-dispersed NOESY spectra recorded on 15N, 13C labeled hRpn13 with hRpn2. We also obtained additional intermolecular NOE-derived distance constraints by using 13C-half-filtered NOESY, 15N NOESY, and 2D 1H, 1H NOESY experiments on the hRpn13:hRpn2 complex without K48-diubiquitin. Intramolecular backbone φ and ψ torsion angles as well as hydrogen bonds were also derived from the data recorded without K48-diubiquitin. In addition, we included intramolecular hRpn2 distances based on our experimental 15N-dispersed and 13C-dispersed NOESY spectra recorded on 15N, 13C labeled hRpn2 with hRpn13 (Lu et al., 2017). Experimental intramolecular NOE-derived distance constraints were used for proximal and distal ubiquitin recorded on hRpn2-bound hRpn13 mixed with K48-diubiquitin. In addition, intramolecular backbone φ and ψ torsion angles as well as hydrogen bonds were included based on a previous structure of monoubiquitin (PDB 1D3Z) (Cornilescu et al., 1998). These constraints were all combined (Tables 1 and 2) to calculate the structure of hRpn2-bound hRpn13 Pru at proximal or distal ubiquitin of K48-diubiquitin by using simulated annealing algorithms in XPLOR-NIH 2.50 (http://nmr.cit.nih.gov/xplor-nih/) (Schwieters et al., 2003). In the first iteration, 20 linear starting structures were subjected to 19,400 simulated annealing and cooling steps of 0.005 ps. The lowest energy structure was then used as the starting structure for a second iteration of simulated annealing to generate 100 structures. The 15 lowest energy structures without violations for Ternary-P were analyzed and a hydrogen bond between hRpn13 K103 ε-ammonium group and the carbonyl oxygen of isopeptide bonded ubiquitin G76 was found in 5 structures. This hydrogen bond was therefore included in a new iteration of Ternary-P structure calculations. The 15 lowest energy structures without violations were finally selected for visualization and statistical analyses. Structure evaluation was performed with the program PROCHECK-NMR (Laskowski et al., 1996); the percentage of residues for Ternary-P in the most favored, additionally allowed, generously allowed and disallowed regions is 89.6, 9.9, 0.5 and 0.0, respectively and for Ternary-D 89.2, 10.3, 0.6 and 0.0, respectively. Visualization was performed with MOLMOL (Koradi et al., 1996) and PyMOL (PyMOL Molecular Graphics System, http://www.pymol.org).
Quantification and Statistical Analysis
The number of violations and values for average and one standard deviation (SD) from average reported in Tables 1 and 2 were calculated by XPLOR-NIH. Values for average pairwise root-mean-square deviation and one SD were calculated by MOLMOL.
Data and Code Availability
The structural coordinates and chemical shift data for Ternary-P and Ternary-D have been deposited into the Protein Data Bank (PDB) and Biological Magnetic Resonance Data Bank (BMRB) with accession codes 6UYI (Ternary-P), 6UYJ (Ternary-D) and 28042.
Supplementary Material
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli BL21(DE3) | Invitrogen | C600003 |
| Escherichia coli BL21(DE3)pLysS | Invitrogen | C606003 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| D2O | Sigma-Aldrich | 191701 |
| 13C glucose | Cambridge Isotope Laboratories, Inc. | CLM-1396–5 |
| PreScission protease | GE Healthcare Life Sciences | GE27-0843-01 |
| protease inhibitor cocktail | Roche | 04693159001 |
| inorganic pyrophosphatase | Sigma-Aldrich | I1643–100UN |
| creatine kinase | Sigma-Aldrich | C3755–1KU |
| ATP | Sigma-Aldrich | A2383–5G |
| E1 | Boston Biochem, Inc. | E-306 |
| Isopropyl- β-D-thiogalactoside | UBPBio | P1010–10 |
| Glutathione S-sepharose 4B | GE Healthcare Life Sciences | 17-0756-05 |
| SP-Sepharose | GE Healthcare Life Sciences | 17-0729-10 |
| Talon Metal Affinity resin | Clontech Laboratories, Inc. | 635502 |
| E2–25K | This paper | N/A |
| hRpn13 Pru (1–150) | This paper | N/A |
| hRpn2 (940–953) | This paper | N/A |
| Ub-D77 | This paper | N/A |
| Ub-K48R | This paper | N/A |
| K48-diubiquitin | This paper | N/A |
| Deposited Data | ||
| Crystal structure of K48-tetraubiquitin | (Phillips et al., 2001) | PDB: 1F9J |
| Crystal structure of K48-tetraubiquitin | (Eddins et al., 2007) | PDB: 2O6V |
| Crystal structure of cyclic K48-tetraubiquitin | (Satoh et al., 2010) | PDB: 3ALB |
| Crystal structure of K48-tetraubiquitin | (Cook et al., 1994) | PDB: 1TBE |
| Crystal structure of Ube2K with K48-diubiquitin | (Lee et al., 2018) | PDB: 6IF1 |
| NMR structure of gp78 E3 ligase CUE domain with K48-diubiquitin | (Liu et al., 2012) | PDB: 2LVP, 2LVQ |
| Crystal structure of MINDY-1 tMIU with K48-diubiquitin | (Kristariyanto et al., 2017) | PDB: 5MN9 |
| NMR structure of Rpn1 with K48-diubiquitin | (Shi et al., 2016) | PDB: 2N3V, 2N3W |
| NMR structure of hRpn10 with K48-diubiquitin | (Zhang et al., 2009) | PDB: 2KDE, 2KDF |
| NMR solution structure of monoubiquitin | (Cornilescu et al., 1998) | PDB: 1D3Z |
| Crystal structure of monoubiquitin | (Vijay-Kumar et al., 1987) | PDB: 1UBQ |
| Crystal structure of K48-diubiquitin in the “closed” conformation | (Cook et al., 1992) | PDB: 1AAR |
| Crystal structure of K48-diubiquitin in the “open” conformation | (Hirano et al., 2011) | PDB: 3AUL |
| NMR solution structure of hRpn13 Pru:hRpn2 | (Lu et al., 2017) | PDB: 6CO4 |
| Crystal structure of hRpn13 Pru:hRpn2:monoubiquitin | (VanderLinden et al., 2017) | PDB: 5V1Y |
| NMR solution structure of hRpn13 Pru:K48-diubiquitin | (Liu et al., 2019) | PDB: 5YMY |
| Chemical shift data for Ternary-P and Ternary-D | This paper | BMRB: 28042 |
| NMR structure of Ternary-P | This paper | PDB: 6UYI |
| NMR structure of Ternary-D | This paper | PDB: 6UYJ |
| Software and Algorithms | ||
| XPLOR-NIH | (Schwieters et al., 2003) | http://nmr.cit.nih.gov/xplor-nih/ |
| XEASY | (Bartels et al., 1995) | N/A |
| NMRPipe | (Delaglio et al., 1995) | https://www.ibbr.umd.edu/nmrpipe/install.html |
| PROCHECK-NMR | (Laskowski et al., 1996) | https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/ |
| PyMOL | N/A | http://www.pymol.org |
| MOLMOL | (Koradi et al., 1996) | https://sourceforge.net/projects/molmol/ |
Highlights.
Chemical origin of Rpn13 preference for K48-linked Ub chains revealed
NMR demonstrates highly dynamic interactions between hRpn2:hRpn13 and K48-diUb
K48-diUb adopts a dynamic, extended conformation that hRpn13 selects
Structure of hRpn2:Rpn13 bound to K48-diUb described
Acknowledgements
This work was supported by the Intramural Research Program of the CCR, NCI, NIH (ZIA BC011490). We gratefully thank Janusz Koscielniak and Jinfa Ying for their maintenance of the NMR spectrometers and Xiang Chen for useful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Anchoori RK, Jiang R, Peng S, Soong RS, Algethami A, Rudek MA, Anders N, Hung CF, Chen X, Lu X, et al. (2018). Covalent Rpn13-Binding Inhibitors for the Treatment of Ovarian Cancer. ACS Omega 3, 11917–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, et al. (2013). A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24, 791–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard JAM, Bashore C, Dong KC, and Martin A (2019). The 26S Proteasome Utilizes a Kinetic Gateway to Prioritize Substrate Degradation. Cell 177, 286–298 e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Xia TH, Billeter M, Guntert P, and Wuthrich K (1995). The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6, 1–10. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, and Ploegh HL (2001). A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J 20, 5187–5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buel GR, Chen X, Chari R, O’Neill MJ, Ebelle DL, Jenkins C, Sridharan V, Tarasov SG, Tarasova NI, Andresson T, et al. (2019). Structure of E3 ligase E6AP with a novel proteasome-binding site provided by substrate receptor hRpn10. Nat Commun, accepted. [DOI] [PMC free article] [PubMed]
- Chen X, Ebelle DL, Wright BJ, Sridharan V, Hooper E, and Walters KJ (2019). Structure of hRpn10 Bound to UBQLN2 UBL Illustrates Basis for Complementarity between Shuttle Factors and Substrates at the Proteasome. J Mol Biol 431, 939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lee BH, Finley D, and Walters KJ (2010). Structure of proteasome ubiquitin receptor hRpn13 and its activation by the scaffolding protein hRpn2. Mol Cell 38, 404–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Randles L, Shi K, Tarasov SG, Aihara H, and Walters KJ (2016). Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure 24, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, and Walters KJ (2012). Identifying and studying ubiquitin receptors by NMR. Methods Mol Biol 832, 279–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook WJ, Jeffrey LC, Carson M, Chen Z, and Pickart CM (1992). Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). J Biol Chem 267, 16467–16471. [DOI] [PubMed] [Google Scholar]
- Cook WJ, Jeffrey LC, Kasperek E, and Pickart CM (1994). Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol 236, 601–609. [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Marquardt JL, Ottiger M, and Bax A (1998). Validation of protein structure from anisotropic carbonyl chemical shifts in a dilute liquid crystalline phase. Journal of the American Chemical Society 120, 6836–6837. [Google Scholar]
- de la Pena AH, Goodall EA, Gates SN, Lander GC, and Martin A (2018). Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, and Bax A (1995). NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–293. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, and Walters KJ (2009). Ubiquitin-binding domains - from structures to functions. Nat Rev Mol Cell Biol 10, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, and Mao Y (2019). Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddins MJ, Varadan R, Fushman D, Pickart CM, and Wolberger C (2007). Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol 367, 204–211. [DOI] [PubMed] [Google Scholar]
- Ehlinger A, and Walters KJ (2013). Structural insights into proteasome activation by the 19S regulatory particle. Biochemistry 52, 3618–3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D, Chen X, and Walters KJ (2016). Gates, Channels, and Switches: Elements of the Proteasome Machine. Trends Biochem Sci 41, 77–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbain S, Inobe T, Israeli E, Chavali S, Yu H, Kago G, Babu MM, and Matouschek A (2015). Sequence composition of disordered regions fine-tunes protein half-life. Nat Struct Mol Biol 22, 214–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, and Murata S (2006). A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J 25, 4524–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Serve O, Yagi-Utsumi M, Takemoto E, Hiromoto T, Satoh T, Mizushima T, and Kato K (2011). Conformational dynamics of wild-type Lys-48-linked diubiquitin in solution. J Biol Chem 286, 37496–37502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, and Hanaoka F (1999). Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem 274, 28019–28025. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, and Dikic I (2008). Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453, 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JP, Lauridsen AM, Kristensen P, Dissing K, Johnsen AH, Hendil KB, and Hartmann-Petersen R (2006). Adrm1, a putative cell adhesion regulating protein, is a novel proteasome-associated factor. J Mol Biol 360, 1043–1052. [DOI] [PubMed] [Google Scholar]
- Kisselev AF (2013). A novel bullet hits the proteasome. Cancer Cell 24, 691–693. [DOI] [PubMed] [Google Scholar]
- Komander D, and Rape M (2012). The ubiquitin code. Annu Rev Biochem 81, 203–229. [DOI] [PubMed] [Google Scholar]
- Koradi R, Billeter M, and Wuthrich K (1996). MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14, 51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- Kristariyanto YA, Abdul Rehman SA, Weidlich S, Knebel A, and Kulathu Y (2017). A single MIU motif of MINDY-1 recognizes K48-linked polyubiquitin chains. EMBO Rep 18, 392–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MY, Zhang D, Laronde-Leblanc N, and Fushman D (2012). Structural and biochemical studies of the open state of Lys48-linked diubiquitin. Biochim Biophys Acta 1823, 2046–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Xu W, DeMartino GN, and Cohen RE (1997). Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385, 737–740. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, and Thornton JM (1996). AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8, 477–486. [DOI] [PubMed] [Google Scholar]
- Lee JG, Youn HS, Kang JY, Park SY, Kidera A, Yoo YJ, and Eom SH (2018). Crystal structure of the Ube2K/E2–25K and K48-linked di-ubiquitin complex provides structural insight into the mechanism of K48-specific ubiquitin chain synthesis. Biochem Biophys Res Commun 506, 102–107. [DOI] [PubMed] [Google Scholar]
- Lee W, Revington MJ, Arrowsmith C, and Kay LE (1994). A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett 350, 87–90. [DOI] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, and Finley D (2002). Multiple associated proteins regulate proteasome structure and function. Mol Cell 10, 495–507. [DOI] [PubMed] [Google Scholar]
- Liu F, and Walters KJ (2010). Multitasking with ubiquitin through multivalent interactions. Trends Biochem Sci 35, 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Chen Y, Li J, Huang T, Tarasov S, King A, Weissman AM, Byrd RA, and Das R (2012). Promiscuous interactions of gp78 E3 ligase CUE domain with polyubiquitin chains. Structure 20, 2138–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Dong X, Yi HW, Yang J, Gong Z, Wang Y, Liu K, Zhang WP, and Tang C (2019). Structural basis for the recognition of K48-linked Ub chain by proteasomal receptor Rpn13. Cell Discov 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Liu F, Durham SE, Tarasov SG, and Walters KJ (2015). A High Affinity hRpn2-Derived Peptide That Displaces Human Rpn13 from Proteasome in 293T Cells. PLoS One 10, e0140518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Nowicka U, Sridharan V, Liu F, Randles L, Hymel D, Dyba M, Tarasov SG, Tarasova NI, Zhao XZ, et al. (2017). Structure of the Rpn13-Rpn2 complex provides insights for Rpn13 and Uch37 as anticancer targets. Nat Commun 8, 15540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markin CJ, Xiao W, and Spyracopoulos L (2010). Mechanism for recognition of polyubiquitin chains: balancing affinity through interplay between multivalent binding and dynamics. J Am Chem Soc 132, 11247–11258. [DOI] [PubMed] [Google Scholar]
- Olszewski MM, Williams C, Dong KC, and Martin A (2019). The Cdc48 unfoldase prepares well-folded protein substrates for degradation by the 26S proteasome. Commun Biol 2, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips CL, Thrower J, Pickart CM, and Hill CP (2001). Structure of a new crystal form of tetraubiquitin. Acta Crystallogr D Biol Crystallogr 57, 341–344. [DOI] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, and Matouschek A (2004). An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 11, 830–837. [DOI] [PubMed] [Google Scholar]
- Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, and Goldberg AL (2006). hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J 25, 5742–5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles L, Anchoori RK, Roden RB, and Walters KJ (2016). The Proteasome Ubiquitin Receptor hRpn13 and Its Interacting Deubiquitinating Enzyme Uch37 Are Required for Proper Cell Cycle Progression. J Biol Chem 291, 8773–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles L, and Walters KJ (2012). Ubiquitin and its binding domains. Front Biosci (Landmark Ed) 17, 2140–2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryabov YE, and Fushman D (2007). A model of interdomain mobility in a multidomain protein. J Am Chem Soc 129, 3315–3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Mimura H, Yamashita M, Yamagata A, and Fukai S (2009). Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by tandem UIMs of RAP80. EMBO J 28, 2461–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Sakata E, Yamamoto S, Yamaguchi Y, Sumiyoshi A, Wakatsuki S, and Kato K (2010). Crystal structure of cyclic Lys48-linked tetraubiquitin. Biochem Biophys Res Commun 400, 329–333. [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, and Groll M (2008). Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453, 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, and Clore GM (2003). The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160, 65–73. [DOI] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F, et al. (2016). Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Park PMC, Wu L, Ray A, Picaud S, Li D, Wimalasena VK, Du T, Filippakopoulos P, Anderson KC, et al. (2019). Development and preclinical validation of a novel covalent ubiquitin receptor Rpn13 degrader in multiple myeloma. Leukemia. [DOI] [PMC free article] [PubMed]
- Song Y, Ray A, Li S, Das DS, Tai YT, Carrasco RD, Chauhan D, and Anderson KC (2016). Targeting proteasome ubiquitin receptor Rpn13 in multiple myeloma. Leukemia 30, 1877–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trader DJ, Simanski S, and Kodadek T (2015). A reversible and highly selective inhibitor of the proteasomal ubiquitin receptor rpn13 is toxic to multiple myeloma cells. J Am Chem Soc 137, 6312–6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Brown NR, Noble ME, and Endicott JA (2010). A new crystal form of Lys48-linked diubiquitin. Acta Crystallogr Sect F Struct Biol Cryst Commun 66, 994–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderLinden RT, Hemmis CW, Yao T, Robinson H, and Hill CP (2017). Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism. J Biol Chem 292, 9493–9504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, and Deshaies RJ (2000). Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11, 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S, Bugg CE, and Cook WJ (1987). Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol 194, 531–544. [DOI] [PubMed] [Google Scholar]
- Walters KJ, and Chen X (2009). Measuring ubiquitin chain linkage: Rap80 uses a molecular ruler mechanism for ubiquitin linkage specificity. EMBO J 28, 2307–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters KJ, Ferentz AE, Hare BJ, Hidalgo P, Jasanoff A, Matsuo H, and Wagner G (2001). Characterizing protein-protein complexes and oligomers by nuclear magnetic resonance spectroscopy. Methods Enzymol 339, 238–258. [DOI] [PubMed] [Google Scholar]
- Walters KJ, Kleijnen MF, Goh AM, Wagner G, and Howley PM (2002). Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry 41, 1767–1777. [DOI] [PubMed] [Google Scholar]
- Wang Q, Young P, and Walters KJ (2005). Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol 348, 727–739. [DOI] [PubMed] [Google Scholar]
- Wider G, Weber C, and Wuethrich K (1991). Proton-Proton Overhauser Effects of Receptor-Bound Cyclosporin A Observed with the Use of a Heteronuclear-Resolved Half-Filter Experiment. J Am Chem Soc 113, 4676–4678. [Google Scholar]
- Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, and Cohen RE (2006). Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol 8, 994–1002. [DOI] [PubMed] [Google Scholar]
- Young P, Deveraux Q, Beal RE, Pickart CM, and Rechsteiner M (1998). Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem 273, 5461–5467. [DOI] [PubMed] [Google Scholar]
- Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, Haririnia A, Storaska AJ, Cole JL, Fushman D, et al. (2009). Structure of the s5a:k48-linked diubiquitin complex and its interactions with rpn13. Mol Cell 35, 280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structural coordinates and chemical shift data for Ternary-P and Ternary-D have been deposited into the Protein Data Bank (PDB) and Biological Magnetic Resonance Data Bank (BMRB) with accession codes 6UYI (Ternary-P), 6UYJ (Ternary-D) and 28042.


