Abstract
Ubiquitination is a highly conserved post-translational modification in eukaryotes, well known for targeting proteins for degradation by the 26S proteasome. Proteins destined for proteasomal degradation are selected by E3 ubiquitin ligases. Cullin-RING E3 ubiquitin ligases (CRLs) are the largest superfamily of E3 ubiquitin ligases, with over 400 members known in mammals. These modular complexes are tightly regulated in the cell. In this chapter, we highlight recent structural and biochemical advances shedding light on the assembly and architecture of cullin- RING ligases, their dynamic regulation by a variety of host factors, and their manipulation by viral pathogens and small molecules.
Keywords: Ubiquitin, Ubiquitination, Ubiquitylation, E3 ligase, Cullin, Cullin-RING ligase, NEDD8, Neddylation, Protein-protein interaction, Viral hijacking
12.1. Introduction: Overview and Function of CRLs
Ubiquitin (Ub) is a small, 8.5 kDa protein that is conserved across all eukaryotes and that regulates important cellular processes (Hershko and Ciechanover 1998). Indeed ubiquitous, it is expressed at very high concentrations in vivo, at approximately 85 μM in human embryonic kidney (HEK) cells for example (Kaiser et al. 2011). The covalent attachment of Ub to a target protein, termed ubiquitylation or ubiquitination, has a variety of effects on the regulation and fate of the target protein. Ub-mediated protein degradation by the 26S proteasome was discovered in the late 1970s and early 1980s. This work led to the receiving of the 2004 Nobel Prize in Chemistry by Aaron Ciechanover, Avram Hershko, and Irwin Rose (Ciechanover et al. 2004). Besides its role in proteasomal degradation, ubiquitination has been shown to mediate many other processes, such as protein-protein interactions, subcellular localization, and DNA repair. This process requires a cascade of three enzymes: E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes, and E3 ubiquitin ligases (reviewed in Pickart 2001). E1 activating enzymes transfer Ub in an ATP-dependent manner to the E2 conjugating enzyme. The E3 Ub ligase bridges the E2 and the target protein (the substrate) to facilitate the transfer of Ub to a lysine or the N-terminus of the target.
E3 ligases are responsible for selecting substrates to be ubiquitinated. While humans only have two E1 activating enzymes, UBA1 and UBA6, and ~30 different E2 conjugating enzymes, there are over 600 different E3 ligases (van Wijk and Timmers 2010). E3 ligases are divided into three main families: the homologous to E6AP carboxyl terminus (HECT), the really interesting new gene and U box (RING and U box), and the RING-between-RING (RBR) families (Berndsen and Wolberger 2014). The HECT and RBR families catalyze ubiquitination in two steps. First, Ub is catalytically transferred from the E2 to the E3, and then from the E3 to the substrate. In contrast, the RING family mediates direct transfer of ubiquitin from the E2 to the substrate, bypassing an E3-Ub intermediate. While not catalyzing a reaction, the RING family is still considered an enzyme (Bulatov and Ciulli 2015). The majority of E3 ligases belong to the RING family (Deshaies and Joazeiro 2009). Within that family, cullin-RING E3 ligases (CRLs) are the largest superfamily, with over 200 members and are responsible for ~20% of all ubiquitination in cells (Soucy et al. 2009; Petroski and Deshaies 2005). The core CRL consists of four components: a cullin protein that serves to scaffold the CRL, a RING finger protein that binds to an E2 ubiquitin conjugating enzyme, a substrate receptor that recognize the target protein, and adaptor proteins that bridge the substrate receptor to the cullin (Fig. 12.1a).
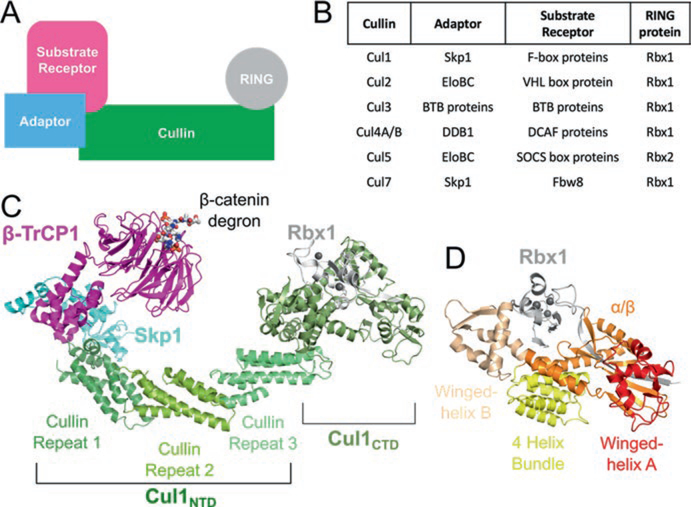
Fig. 12.1.
Architecture of cullin-RING ligases. (a) Schematic representation of the components of a CRL. (b) Specific protein factors for each CRL family. (c) Structural architecture of a CRL1 in ribbon representation generated from PDB IDs 1LDK and 1P22. (d) Organization of Cul1 CTD–Rbx1. The domains of Cul1 are labeled in (c) and (d). The Zn2+ ions in (c) and (d) are shown as gray spheres
Mammals express seven canonical cullin proteins (Cul1, Cul2, Cul3, Cul4A, Cul4B, Cul5, and Cul7) that form modular, multisubunit CRLs, designated CRL1–7. Cullins are named because they “cull” (originally meaning to selectively slaughter a herd of wild animals) proteins for proteasomal degradation (Sarikas et al. 2011). Bioinformatic analysis suggests that the canonical eukaryotic cullins arose from three ancestral genes (Culα, Culβ, Culγ) where Cul1, Cul2, Cul5, and Cul7 descend from Culα, Cul3 from Culβ, and Cul4A/Cul4B from Culγ (Marin 2009). Cul4A and Cul4B mainly differ by an additional 149 amino acids at the Cul4B N-terminus, a portion of which harbors a nuclear localization sequence (Zou et al. 2009). Cul7 is an atypical cullin as it is much larger than the average cullin (~1700 amino acids compared to ~760 amino acids). Mammals also encode two other proteins that contain cullin homology domains, Cul9/PARkin-like cytoplasmic protein (PARC), and subunit 2 of the anaphase promoting complex/cyclosome (APC2), but these two proteins are overall highly divergent from canonical cullins as they contain additional domains (Skaar et al. 2007; Zachariae et al. 1998).
Within a CRL, the cullin protein serves as the central scaffold of the E3 Ub ligase (Fig. 12.1a). The C-terminal domain of a cullin anchors the RING protein, either Rbx1 or Rbx2, which recruit the Ub-charged E2 enzymes. At the opposite end of the cullin, the N-terminal domain interacts with the adaptor component of the CRL. Adaptors share one of two common folds that are used to interact with cullins. Skp1, Elongin C, and Bric-a-brac, Tramtrack and Broad Complex (BTB) proteins share a common α-helical BTB fold to interact with their CRL, while the DDB1– Cul4-associated factor (DCAF) family uses a WD40 β–propeller fold.
The adaptor component of a CRL helps link the cullin to a family of substrate receptors (Fig. 12.1b). Typically, each substrate receptor family contains anywhere from approximately 30 to 70 proteins (Sarikas et al. 2011). For example, the Skp1 adaptor protein connects the F-box substrate receptor protein family, with over 60 representatives, to a CRL1 (Skaar et al. 2013). However, the atypical Cul7, to date, has only one identified substrate receptor protein, Fbw8 (Dias et al. 2002). A specific CRL complex can be denoted with its substrate receptor, such as CRL7Fbw8. The substrate receptor has an additional domain that is responsible for recognizing a substrate of the CRL through a ‘degron’ (Fig. 12.1c), a specific amino acid sequence that is often post-translationally modified (Bulatov and Ciulli 2015; Lydeard et al. 2013). Thus, the substrate receptor component is responsible for dictating the specificity of a particular CRL towards its cellular target proteins. Since various substrate receptors can target a repertoire of substrates, CRLs are involved in a diverse array of biological processes and are associated with a multitude of diseases (Table 12.1).
Table 12.1.
List of some diseases associated with cullin-RING ligases
| CRL | Cellular function | Related diseases |
|---|---|---|
| CRL1β-TrCP1 | Cell adhesion and signaling | Gastric cancer; split hand-split foot malformation |
| CRL1Skp2 | Cell division | Lung cancer; squamous cell carcinoma |
| CRL1Atrogin1 | Muscle differentiation | Muscle wasting; dilated cardiomyopathy |
| CRL2LRR-1 | Cell migration | |
| CRL2VHL | Hypoxic response | von Hippel-Lindau disease; renal cell carcinomas |
| CRL3KLHL9 | Cytokinesis | Early onset distal myopathy |
| CRL3SPOP | Cell growth and development | |
| CRL3Keap1 | Oxidative stress response | Pulmonary papillary; adenocarcinoma |
| CRL4DDB2 | DNA damage response | Xeroderma pigmentosum; cockayne syndrome |
| CRL4Cdt2 | Cell division | |
| CRL5SOCS3 | Cytokine signaling | Diabetes |
| CRL5ASB3 | T-cell signaling regulation |
12.1.1. CRL1 Defines the Prototypical CRL
The prototypical and best-characterized CRL is CRL1 or Skp1–Cul1–F-box (SCF) E3 ligase. Much of our understanding of CRLs comes from CRL1. The structure of CRL1Skp2, containing Skp1, the F-box domain of Skp2 (F-boxSkp2), Cul1, and Rbx1 (Zheng et al. 2002) was the first to provide insight toward the overall CRL arrangement. The complex has an arch or banana-shaped architecture, with the curved Cul1 at the center, scaffolding Skp1 and Skp2 at its N-terminus and Rbx1 at its C-terminus (Fig. 12.1c). The spatial organization and overall architecture of CRL1 is representative for all CRLs.
Cul1 is composed of a helical N-terminal domain (NTD) and a globular C-terminal domain (CTD) (Fig. 12.1c). The N-terminus can be divided into three cullin repeat domains, each composed of 5-helix bundles, with the first cullin repeat containing an additional helix inserted after helix α4 (Fig. 12.1c). The first cullin repeat interacts with Skp1 and F-boxSkp2 via hydrophobic and electrostatic interactions with helices α2 and α5, which is a conserved mechanism across all CRLs. The F-box motif is a three-helix bundle that packs along the helices of Skp1 (Fig. 12.1b) (Schulman et al. 2000). The Cul1 C-terminus is globular, containing a 4-helix bundle, an α/β domain, and two winged-helix domains, A and B (Fig. 12.1d). The CTD creates a V-shaped binding-pocket for Rbx1. Rbx1 contains three segments: an N-terminal β-strand that inserts into the Cul1 α/β domain, a flexible linker region, and a C-terminal RING finger domain. The Rbx1 RING domain contains a RING motif, which binds two zinc ions, but with an additional 20-residue insertion, forming an additional zinc ion coordinating site (Zheng et al. 2002). In the crystal structure, Rbx1 packs against the winged-helix B of Cul1, in an inactive conformation (discussed in Sect. 12.2.1). The Cul1 NTD and CTD are rigid with respect to each other, acting as a stiff platform for the proteins at each end.
The two domains of Cul1 can be expressed in vitro independently of one another. With solubility-enhancing mutations at the interfaces, either the Cul1 NTD or the Cul1 CTD with Rbx1 can be expressed and purified from E. coli. This technique has been utilized for the expression and crystallization of fragments of Cul3 and Cul5 (Errington et al. 2012; Canning et al. 2013; Muniz et al. 2013). The two wild-type halves can be co-expressed together to reconstitute a ‘full-length’ Cul1 in a ‘split-n- coexpress’ method (Zheng et al. 2002).
12.1.2. CRL2 and CRL5: The Same Adaptor, Different Substrate Receptors
Among the CRL families, CRL2 and CRL5 are the most similar to CRL1 (Fig. 12.2a). While Skp1 is the adaptor protein for CRL1, the obligate heterodimer of Elongin B and Elongin C (EloBC) is the adaptor component for CRL2 and CRL5. Even with the same adaptor, Cul2 and Cul5 recruit different substrate receptors. VHL box substrate receptors interact with Cul2/EloBC while SOCS box proteins bind Cul5/EloBC. The VHL box motif is similar in sequence and structure to the SOCS box motif, making it difficult to predict their assembly with either Cul2 or Cul5 (Fig. 12.2b) (Mahrour et al. 2008). Both the VHL box and the SOCS box motifs are composed of a BC box and a cullin box. The BC box mediates the association with EloBC while the cullin box is important for Cul2/Cul5 specificity. Swapping of cullin boxes can switch the substrate receptor preference for Cul2 or Cul5 (Kamura et al. 2004; Mahrour et al. 2008).
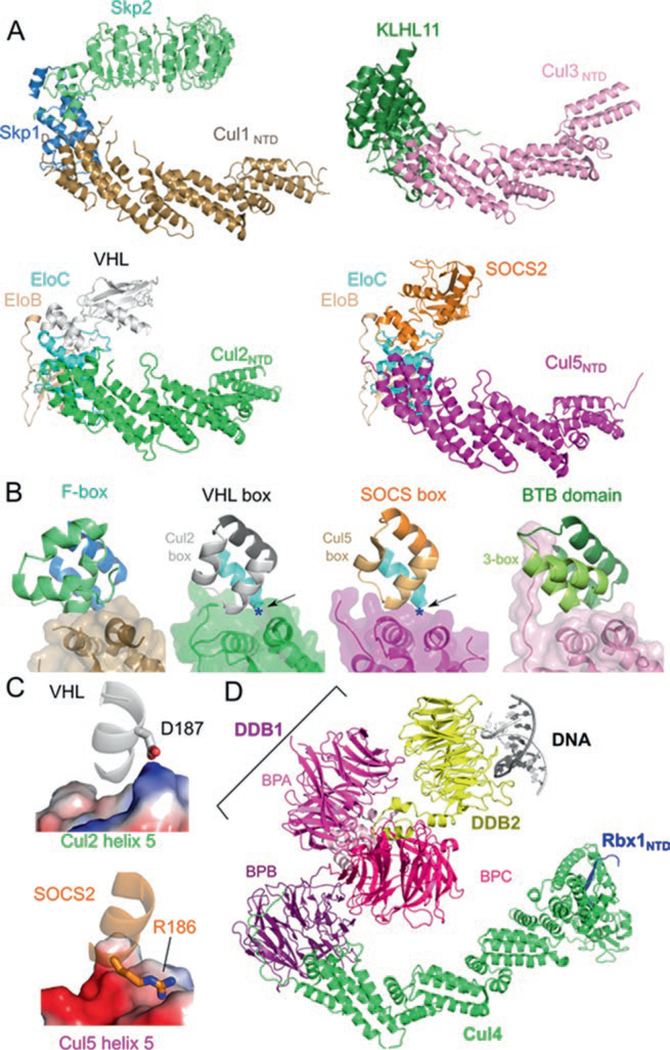
Fig. 12.2.
Similar architecture of cullin-RING ligases. (a) Structures of VHL–EloBC–Cul2NTD (PDB ID: 4WQO), SOCS2–EloBC–Cul5NTD (PDB ID: 4JGH), Skp2–Skp1–Cul1NTD (PDB ID: 1LDK), and KLHL11-Cul3NTD (PDB ID: 4AP2) show a common spatial organization of components. Cullin repeats 2 and 3 of Cul2 were modeled from Cul1. (b) The cullin–adaptor–substrate receptor binding regions for each CRL shown in (a) are in ribbon representations with semitrans-parent surfaces shown for the cullins. The Cul2 box, the Cul5 box, and the 3-box are marked. (c) Charge complementary at the binary cullin–substrate receptor interface between VHL and Cul2 helix α5 (top) or between SOCS2 and Cul5 helix α5 (bottom). Cullins are shown in electrostatic surface representation (Blue: positive; Red: negative). Each region is marked in (b) with a blue asterisk and with the point of view depicted by the arrow. (d) Structure of the CRL4DDB2–UV-damaged DNA complex (PDB ID: 4A0K) in ribbon representation. The domains of DDB1 are marked
Charge complementary helps dictate substrate receptor preference for Cul2 or Cul5. The structures of CRL2VHL and CRL5SOCS2 reveal that the interactions between substrate receptor and cullin are centered around cullin helices α2 and α5 (Guo et al. 2014; Kim et al. 2013; Nguyen et al. 2015). The surface charge polarity between VHL and Cul2 helix α5 and between SOCS2 and Cul5 helix α5 are opposite, with the Cul2 helix α5 interface being basic and the Cul5 counterpart being acidic (Fig. 12.2c). In addition, the CRL2VHL structure revealed that the determinants for Cul2 selection can lie outside of the cullin box (Nguyen et al.2015). The residues used by the substrate receptors to recruit the respective cullin are not conserved across the other VHL or SOCS box proteins. Therefore, even with the knowledge of charge complementarity at the substrate receptor-cullin interface, a more explicit rule to predict Cul2/5 preference is lacking and will require additional CRL2/5 structures.
12.1.3. CRL3: Integrating Adaptors and Substrate Receptors to Target One or Two Copies of a Substrate
CRL3 fuses the functions of the substrate receptor and adaptor into a single polypeptide, a BTB protein. BTB proteins contain the BTB domain, which is responsible for binding to Cul3, and a protein-protein interaction domain to target a substrate. The BTB domain is structurally homologous to the adaptors Skp1 and EloC in CRL1 and CRL2/5, respectively (Fig. 12.2b). However, not all proteins containing BTB domains bind to Cul3 (Genschik et al. 2013). Only those that contain an additional motif, termed the 3-box, are recruited to CRL3. The 3-box is analogous to the other cullin-binding motifs such as the F-box, VHL box, and SOCS box, as it aids in the binding of Cul3 (Zhuang et al. 2009). The 3-box creates a hydrophobic groove into which the 22 amino acid N-terminal extension of Cul3 is inserted. Deletion of the Cul3 N-terminal extension results in a 30-fold loss of binding (Canning et al. 2013). Overall, assembly of CRL3s is similar in architecture to CRL1/2/5 (Fig. 12.2a) (Canning et al. 2013; Ji and Prive 2013; Errington et al. 2012).
BTB proteins can dimerize to control substrate ubiquitination. This leads to an assembled CRL3 that has two substrate receptors and two catalytic RING domains, which can either act independently and target two substrates total (one for each Rbx1), or the dimer can act to target a single protein. The BTB protein Keap1 falls into the latter category and is thought to recognize two distinct degrons present in the same target substrate, Nrf2 (Tong et al. 2006, 2007). In addition, multimeric CRL3 assemblies can be formed through BTB proteins containing the BACK domains, such as SPOP. CRL3SPOP has a much higher E3 ligase activity than that of a dimerization-deficient mutant, which could be due to either enhanced avidity or increased local concentration of CRLs (Errington et al. 2012). While some CRL1 F-box proteins, such as Fbw7, have been reported to dimerize (Welcker and Clurman 2007), the ability to form dimeric CRL is a hallmark characteristic of the CRL3 family.
12.1.4. CRL4: Propellers Drive Assembly
CRL4s are highly distinct from the other CRLs as they use the adaptor protein damaged DNA binding protein 1 (DDB1), which does not have the BTB fold found in the adaptors of CRL1/2/3/5. Instead, DDB1 consists of three WD40 β-propeller domains (BPA, BPB, BPC) and a helical C-terminal domain (Fig. 12.2d). Only the BPB domain interacts with Cul4, and while it is structurally distinct from the other adaptors, it binds to Cul4 in an equivalent manner as other adaptor-cullins interactions. BPB recognizes Cul4 helices α2 and α5 of the first cullin repeat and is wrapped around by a long N-terminal extension of Cul4. BPA and BPC form a cleft that is the binding site for the DCAF substrate receptors. The DDB1 BPA-BPC double propeller and the BPB propeller arrangement can be quite flexible, with a 150° range of rotational freedom (Angers et al. 2006; Zimmerman et al. 2010). This flexibility may allow binding of substrates of various sizes and shapes or to scan a large area centered around the DCAF–substrate complex (Scrima et al. 2008). DCAF proteins utilize a short helix, termed the H-box motif, or two conserved Asp-x-Arg motifs located at the bottom surface of the WD40 propeller (WDXR motif) to interact with DDB1 (Angers et al. 2006; Jin et al. 2006; Li et al. 2010). While CRL4 exhibits vast structural differences in its subunit composition, the overall architecture is surprisingly similar to the other CRLs.
CRL4s have been shown to recognize non-canonical substrates. They are the only CRLs known to date where the substrate may not be a protein, but DNA. CRL4DDB2 detects UV-induced lesions in DNA and ubiquitinates nearby proteins to help direct nucleotide excision repair (Chen et al. 2001; Fischer et al. 2011; Scrima et al. 2008). Small molecules can also modulate the substrate specificity of a CRL. The immune modulatory drugs thalidomide and its derivatives lenalidomide and pomalidomide bind to the CRL4 substrate receptor Cereblon, prevent targeting of its native substrates, and subvert it to target casein kinase 1α for polyubiquitination (Petzold et al. 2016). These examples demonstrate that CRL can recognize non- protein substrates and that they can be altered to target non-canonical proteins (discussed in Sects. 12.3 and 12.4).
12.2. Regulation of CRLs
The active state of CRLs must be tightly regulated to ensure proper cellular fitness. This is achieved by post-translational modifications of cullins and controlled alterations to the subunit composition of the complexes. These are highly dynamic processes that require the interplay of many other proteins. Here we describe how neddylation affects CRLs, how CAND1 allows for remodeling of substrate receptors on CRLs, and how E3 activity can be downregulated by the protein Glomulin.
12.2.1. Neddylation Activates CRLs Through Conformational Change at the Cullin-RING Interface
All cullins are post-translationally modified by covalent attachment of the protein Neural precursor cell Expressed Developmentally Down-regulated protein 8 (NEDD8) to activate ligase activity (Hori et al. 1999; Pan et al. 2004). The attachment of NEDD8, termed neddylation, requires a cascade of enzymes similar to those required for ubiquitination. NAE, a heterodimer of NAE1 and UBA3, acts as the E1 for neddylation while UBC12 and UBE2F act as the NEDD8 E2s. Interestingly and distinct from ubiquitination, a NEDD8 E3 and a ‘co-E3’ are required for neddylation. The NEDD8 E3 is the RING protein of the CRLs, either Rbx1 or Rbx2, while the ‘co-E3’ is the protein Defective in Cullin Neddylation 1 (DCN1) (Kurz et al. 2005). Both Rbx1/Rbx2 and DCN1 are required for the NEDD8-charged E2 to neddylate a cullin. DCN1 helps recruit the E2 by using its Potentiating Neddylation (PONY) domain to clasp onto both the cullin and the acetylated N-terminal helix of the E2 UBC12, while the RING correctly positions the E2 via contacts with both UBC12 and NEDD8 (Scott et al. 2014).
Neddylation occurs on the cullin CTD (Lys720 in the winged-helix B motif of Cul1) and leads to a conformation change of the cullin winged helix B motif and the cullin–RING interface (Fig. 12.3a). NEDD8 interacts with the winged-helix B motif in the cullin CTD, leading to the displacement of Rbx1 from the cullin. In this conformation, Rbx1 can sample multiple conformations to bring the E2 closer to the target substrate tethered at the other end of the cullin by the substrate receptor (Fig. 12.3a). Small angle X-ray scattering and biochemical studies validated that NEDD8-induced conformational change results in the catalytically active CRL state (Duda et al. 2008).
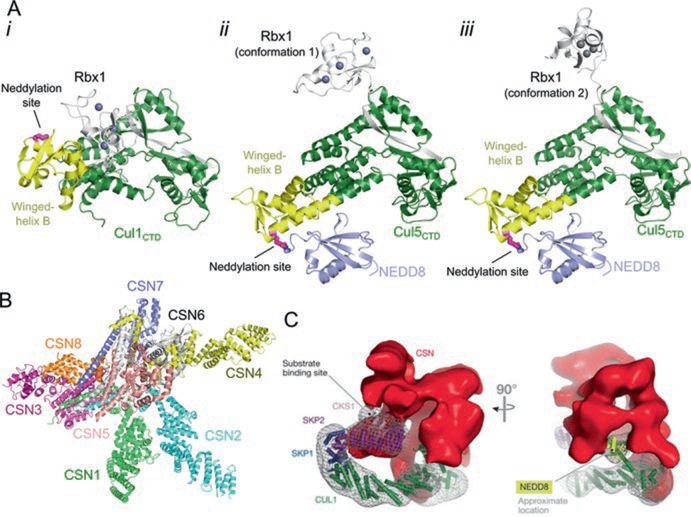
Fig. 12.3.
Neddylation activates cullin-RING ligases. (a) i, Structure of unneddylated, inactive Cul1CTD–Rbx1 (PDB ID: 1LDK). Lys720, the neddylation site, is shown in magenta as sticks. ii and iii, Neddylation of CulCTD causes a change in orientation of the winged-helix B motif and rearrangement of Rbx1 (PDB ID: 4P5O). Shown are two conformations of Rbx1 observed in the crystal structure. The isopeptide bond between Cul5 and NEDD8 is shown as sticks. (b) Crystal structure of the CSN (PDB ID: 4D10). (c) Negative staining electron microscopy reconstruction of CSN bound to CRL1Skp2 (EMD-2173) (Reprinted from Lydeard et al. 2013)
Deneddylation is regulated by the COP9 Signalosome (CSN), which is a ~ 350 kDa, eight-subunit complex that harbors similarity to the 19S lid of the proteasome (Pick et al. 2009; Lyapina et al. 2001). It is composed of CSN1–8 where CSN5 is the catalytic subunit, a zinc metalloproteinase that cleaves NEDD8. The CRL-free CSN is catalytically inactive and has three organizational centers: a bottom base, a central box, and a cap. The base is a hand-like structure that is composed of CSN1–4 and CSN7–8. The central box sits at the palm of the hand and is a large helical bundle containing the C-terminal helices of every CSN subunit. The cap sits at the top of the box and consists of a CSN5–CSN6 heterodimer (Fig. 12.3b) (Deshaies 2014). CSN4 senses binding of a neddylated CRL and leads to a conformational rearrangement of the CSN that activates CSN5 to cleave NEDD8 (Lingaraju et al. 2014).
Besides deneddylation, CSN has additional regulatory roles that are not well understood. in vivo, 10–20% of CRLs are associated with CSN at steady-state level, with only a two-fold decrease when neddylation is inhibited, revealing that the association is NEDD8-independent (Bennett et al. 2010). This is supported by experiments in vitro, as electron microscopy structures of CSN bound to either CRL1Skp2 or CRL4DDB2 revealed that CSN interacts with unneddylated CRLs and blocks the binding sites for both the substrate receptor and the E2 (Fig. 12.3c) (Enchev et al. 2012; Cavadini et al. 2016). In addition, deneddylation does not always occur even when CSN binds neddylated CRLs (Bennett et al. 2010; Emberley et al. 2012). This suggests that either binding of CSN to CRL itself is not sufficient for deneddylation or another regulatory step is required. However, these studies could be biased by the epitope-tagged CSN that may lack the catalytic subunit (Lydeard et al. 2013). Moreover, when preformed CSN–CRL complexes are incubated with a substrate in vitro, CSN dissociates from CRL while the substrate associates (Bennett et al. 2010; Fischer et al. 2011; Emberley et al. 2012). This suggests a mode of CRL regulation in which high substrate levels out-compete CSN for CRL. Binding between CSN and CRL can also be regulated by inositol hexakisphosphate (Scherer et al. 2016). Further experiments are needed to completely delineate the deneddylase-independent roles of CSN in regulating CRLs.
12.2.2. CAND1 Acts as an Adaptor Protein Exchange Factor
Cullin-associated NEDD8-dissociated factor 1 (CAND1) was first identified as an inhibitor of CRL activity, but further studies revealed that it promotes the dissociation of the bound substrate receptor from a cullin to allow the binding of a new substrate receptor. CAND1 is a ~ 136 kDa HEAT (huntingtin, elongation factor 3, protein phosphatase 2A, TOR) – repeat protein that reversibly binds to deneddylated CRL by wrapping around both the NTD and CTD of the cullin (Goldenberg et al. 2004). CAND1 binding occludes both the substrate receptor binding site and the NEDD8 acceptor lysine site to inhibit neddylation, supporting its role as a negative regulator of CRLs (Fig. 12.4a) (Goldenberg et al. 2004). This is in contrast to genetic studies that showed it as a positive regulator, promoting CRL activity (Zhang et al. 2008). These conflicting views were resolved by a Förster resonance energy transfer (FRET) study, which revealed that CAND1 significantly enhances (by õne million fold) the dissociation of incorporated substrate receptors from CRLs. The addition of CAND1 to a mixture of pre-assembled CRL1β-TRCP and a free F-box protein-adaptor, Skp1–FBXW7, led to the removal of β-TRCP and the exchange of FBXW7 onto CRL1 (Pierce et al. 2013). CAND1 seems to behave like a substrate exchange factor, analogous to guanine nucleotide exchange factors that promote exchange of GTP for GDP in GTPases.
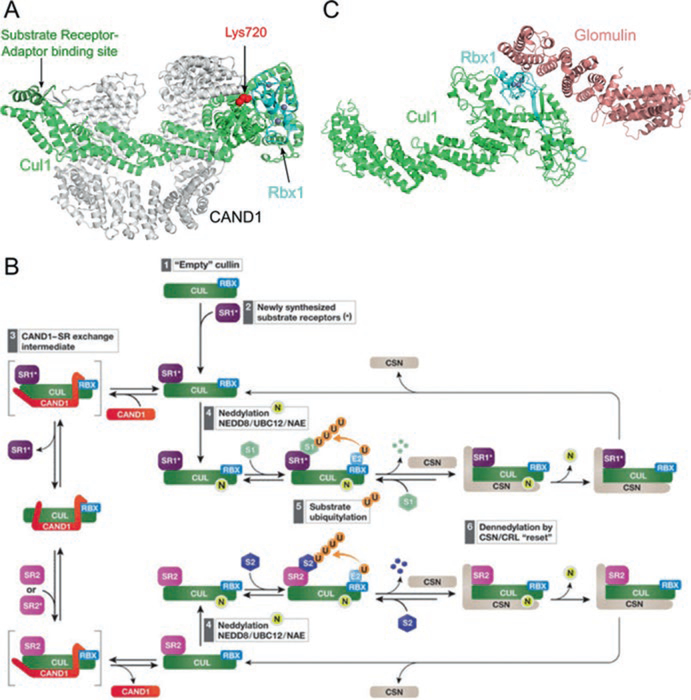
Fig. 12.4.
Regulation of cullin-RING ligases by CAND1 and Glomulin. (a) Structure of Cul1–Rbx1 inhibited by CAND1 (PDB ID: 1U6G) in ribbon representation. CAND1 sterically blocks the neddylation site on the cullin (shown in red spheres) and the substrate receptor binding site at the cullin NTD (highlighted in dark green). (b) Model of remodeling of CRL composition by CAND1 and CSN (Reprinted from Lydeard et al. 2013. See main text for description). (c) Structural inhibition of Rbx1 by glomulin (PDB ID: 4F52) show in ribbon representation. Glomulin inhibits CRL activity by binding to Rbx1 and blocking the E2 interacting site. The Zn2+ ions in (a) and (c) are shown as gray spheres
Figure 12.4b summarizes a model for CAND1-mediated substrate receptor exchange (Lydeard et al. 2013). A cullin-RING binary complex (step 1) engages the first substrate receptor–adaptor, SR1 (step 2). This assembled complex could either encounter CAND1 to replace SR1 with a different substrate receptor–adaptor, SR2, (step 3) or could become neddylated, recruit substrates, and start ubiquitin-mediated degradation (steps 4 and 5). As the substrate concentration decreases, CSN deneddylates the CRL and then dissociates (step 6), freeing the CRL complex to repeat the cycle in step 2. CAND1 exchanges SR1 with SR2 through a proposed intermediate where CAND1 and SR2 bind simultaneously, but transiently, with the cullin–RING binary complex (step 3). Exchange of SR1 for SR2 would change which substrates are targeted in the cell.
12.2.3. Glomulin Sterically Blocks Rbx1-E2 Interactions
The Rbx1 utilizing activity of CRLs can be negatively regulated by another HEAT-repeat protein called Glomulin, which specifically interacts with Rbx1, but not Rbx2 (Tron et al. 2012). This interaction inhibits CRLs by obscuring the E2-binding site, preventing both ubiquitination and neddylation (Fig. 12.4c). This interaction is independent of the neddylation state of the cullin or the presence of CAND1 in vitro (Duda et al. 2012). However, the in vivo role of Glomulin is less understood as it only affects CRL1Fbxw7 activity, yet its deletion leads to decreased cellular levels of Cul1, Rbx1, and Fbxw7 (Tron et al. 2012). Genetic analysis reveals that Glomulin mutations cause glomuvenous malformation disease, characterized by cutaneous venous lesions (Brouillard et al. 2002). The mechanism of this Glomulin-associated disease is currently unknown.
12.3. Viral Hijacking of CRLs: Turning the Cellular Machinery Against Itself
Viruses often hijack host E3 ubiquitin ligases by mimicking the substrate receptors to evade the immune system and promote replication (Barry and Früh 2006) (Fig. 12.5a). For example, Epstein-Barr virus expresses BZLF1 (also known as EB1, Zta, or ZEBRA) to target p53 for degradation via both CRL2 and CRL5, promoting viral proliferation during the lytic phase (Sato et al. 2009). Similarly, paramyxoviruses, Hepatitis B virus (HBV), and human immunodeficiency virus (HIV) have evolved various accessory proteins to target host factors for polyubiquitination and degradation (Barry and Früh 2006; Jia et al. 2015). For the purpose of this review, we will focus on some recent examples of structural studies on viral hijacking of CRLs.
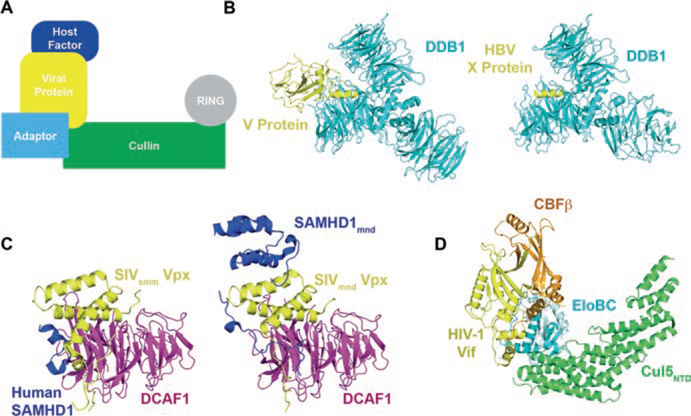
Fig. 12.5.
Viral hijacking of cullin-RING ligases. (a) Schematic of how viral proteins recruit host factors to CRL for ubiquitination and degradation. (b) Paramyxoviruses V protein (PDB ID: 2B5L) and HBV X protein (PDB ID: 3I7H) bind to DDB1 using a short helix, the H-Box. (c) SIV Vpx- DCAF1 complexes interact with the human SAMHD1 C-terminus (PDB ID: 4CC9) and the mandrill SAMHD1 N-terminus (PDB ID: 5AJA) using similar binding modes. (d) HIV-1 Vif interacts with Cul5 with the help of CBFβ (PDB ID 4N9F)
12.3.1. Paramyxoviruses and Hepatitus B Virus Utilizes CRL4 for Successfully Infection
Paramyxoviruses V protein and HBV X protein function as substrate receptors to target host factors to CRL4 for polyubiquitination and degradation. Paramyxoviruses V protein targets the host Signal Transducer and Activator of Transcription (STAT) protein, which is involved in the interferon signaling pathway, thus overcoming the host interferon antiviral responses (Ulane and Horvath 2002; Precious et al. 2005). In HBV, the X protein (HBx), which is essential for virus replication in vivo (Bouchard and Schneider 2004), also interacts with DDB1 (Sitterlin et al. 1997). It has recently been reported that HBx functions by targeting the structural maintenance of chromosomes (Smc) complex Smc5/6 to CRL4 for degradation (Decorsière et al. 2016).
The paramyxoviruses V protein and HBx directly interact with the CRL4 adaptor protein DDB1 and they do so in a very similar manner (Fig. 12.5b) (Angers et al. 2006; Li et al. 2006, 2010). Paramyxoviruses V protein and HBx both present a 3-turn helix, which docks into the binding pocket enclosed by BPA and BPC. The helix mainly contacts the top of BPC, using hydrophobic residues on one helical face and forming hydrogen bonds with BPC residues. Although there are no common residues shared between paramyxoviruses V protein and HBx helices, their DBB1 interaction modes are remarkably similar and mimic the interaction between DBB1 and its cellular substrate receptors (e.g. DCAF9, DDB2, etc) (Li et al. 2010). This short helical domain was therefore named the H-box.
Paramyxoviruses V protein makes additional interactions with DDB1 outside the H-box (Li et al. 2006), like other DBB1 substrate receptors. It has been suggested that the DDB1 and H-box interaction alone is not sufficient for productive CRL formation. The CRLs are activated when additional interactions between the substrate receptors (such as V protein, DCAF9 and DDB2) and DDB1 are formed. This bipartite interaction may allow CRL4 to switch between productive and nonproductive forms without disassembling the complex (Li et al. 2010).
12.3.2. SIVs Employ Vpx to Suppress SAMHD1 Via CRL4
Simian immunodeficiency viruses (SIVs) express the accessory protein Vpx to hijack CRL4 to evade the host antiviral protein SAMHD1 (Hrecka et al. 2011; Laguette et al. 2011). SAMHD1 represses lentiviral infection in non-dividing cells by depleting cellular dNTP levels, inhibiting reverse transcription (Kim et al. 2012; St Gelais et al. 2012). Interestingly,Vpx from different lineages of SIVs targets different regions of SAMHD1 to a CRL4. Vpx from HIV-2 and SIVsmm (SIV infecting sooty mangabey monkey) lineages interact with the SAMHD1 C-terminus, whereas Vpx from SIVmnd-2 (mandrill-infecting) lineage targets the N-terminus of SAMHD1. When expressed in human cell lines, Vpx of SIVsmm can recognize the C-terminal tail of human SAMHD1 for degradation (Schwefel et al. 2014).
Although Vpx proteins from different species recognize distinct regions of SAMHD1, they interact with the substrate receptor DCAF1 in a remarkably similar manner (Fig. 12.5c) (Schwefel et al. 2014, 2015). Vpx consists of a flexible N-terminal region and a 3-helix globular domain, which interact with the side and the top regions of DCAF1 respectively. The C-terminal tail of human SAMHD1 is located at the SIVsmmVpx–DCAF1 interaction interface, making contacts with both components. In contrast, the non-structured N-terminal region of mandrill SAMHD1 is sandwiched between SIVmnd-2Vpx N-terminus and DCAF1, while the SAM domain of mandrill SAMHD1 makes additional contacts with Vpx helices to stabilize the complex.
The two variable regions (VR1 and VR2) of Vpx are responsible for the ability of the architecturally similar Vpx–DCAF1 complexes to bind different regions of SAMHD1. The Vpx of HIV-2 and SIVsmm feature a GEET-motif in VR1 located just N-terminal of helix 1. In contrast, the SIVnmd-2Vpx VR1 comprises a proline followed by several other conserved residues, P(x)GAG[D/E]V/A, which is conserved among SIV Vpx that recognize SAMHD1 N-terminus (Schwefel et al. 2015). Moreover, the VR2 of Vpx has two conserved tyrosine residues, facilitating the complex formation with SAMHD1 and DCAF1. On the SAMHD1 side, two critical residues Phe52 and Phe15 also control the interaction between mandrill SAMHD1 N-terminus and Vpx-DCAF1 complex.
HIV-1 encodes a Vpx homolog protein in its genome, Vpr, which induces host G2/M cell cycle arrest, but does not target human SAMHD1 for degradation. Presumably, this is due to Vpr’s role in targeting an unknown factor for degradation by CRL4. Recently, the crystal structure of HIV-1 Vpr bound to DDB1–DCAF1 and a substrate, uracil DNA glycosylase 2 was solved. This revealed that Vpr engages DCAF1 in a similar mode as Vpx does, but Vpr uses different structural regions to recruit its substrate (Wu et al. 2016).
12.3.3. HIV-1 Vif Downregulates the Immune Response by Targeting APOBEC3 Proteins to a CRL5
HIV express virion infectivity factor (Vif) to target the host antiviral proteins APOBEC3G (A3G) and APOBEC3F (A3F) to a CRL5 for polyubiquitination and subsequent proteasomal degradation (Sheehy et al. 2002; Yu et al. 2003). A3G and A3F are cytidine deaminases that lethally hypermutate the viral genome to suppress HIV infection (Harris et al. 2003; Lecossier et al. 2003; Mangeat et al. 2003). Unlike any other known CRL, cellular or viral, Vif recruits an additional cofactor, core binding factor β (CBFβ), to form the CRL5 (Jager et al. 2012; Zhang et al. 2012). CBFβ is a transcription factor that normally interacts with the RUNX family of proteins to promote cell differentiation (de Bruijn and Speck 2004; Ito 2008). In CRL5, CBFβ acts as a molecular chaperone to stabilize the Vif structure and allow it to interact with Cul5 (Fribourgh et al. 2014).
The crystal structure of the Vif–CBFβ–EloBC–Cul5NTD complex reveals how Vif recruits components of the E3 ligase (Fig. 12.5d) (Guo et al. 2014). Vif is at the center of the complex, buttressing CBFβ, EloC, and Cul5NTD. Vif is composed of two domains, a larger α/β and a smaller α-helical domain with a Zn2+ binding motif serving to organize the two domains. The α/β domain has no known structural homology to other proteins. CBFβ makes extensive interactions with the α/β domain as well as the Zn2+ binding motif, serving as a molecular chaperone to help Vif fold into a proper conformation. The Vif α-helical domain interacts with EloBC through the conserved BC box motif as shown previously (Stanley et al. 2008), but uses a non-canonical motif to engage Cul5. Interestingly, Vif still interacts with the same general interface of Cul5 as other substrate receptors, making the overall assembly of CRL5Vif similar to the other CRLs.
While HIV-1 Vif was originally found to recruit CRL5, lentiviral Vifs have shown some promiscuity or ambiguity on the exact CRL they recruit. HIV-1 Vif has also been shown to recruit CRL2 depending on the cell line (Jager et al. 2012; Kane et al. 2015). Likewise, Vif from bovine immunodeficiency virus (BIV) and maedi-visna virus (MVV), a sheep-specific lentivirus, have been shown to associate with both CRL2 and CRL5 (Zhang et al. 2014; Kane et al. 2015). The composition of these CRLs also differs from that of HIV-1 Vif CRL5, as BIV Vif does not require an additional cellular cofactor while MVV Vif recruits cyclophilin A instead of CBFβ (Kane et al. 2015). Given the similarities between CRL2 and CRL5, it could be to the advantage of the virus to recruit both CRLs to achieve its goal.
12.4. Therapeutic Targeting of CRL
The misregulation of protein levels by the ubiquitin-proteasome system (UPS) causes a large number of disorders, including cancer, diabetes, and neurodegenerative diseases (Table 12.1). While some proteasome inhibitors are effective at treating certain diseases, such as Bortezomib treatment for multiple myeloma and mantle cell lymphoma, these drugs are broadly cytotoxic due to nonspecific blocking of all UPS-dependent protein degradation (Orlowski and Kuhn 2008). Interfering with CRL functions represents an alternative strategy to modulate the UPS in a more specific way. While small-molecule inhibitors of specific substrate receptors have been reviewed (Bulatov and Ciulli 2015), we will focus on two more general strategies targeting CRLs.
12.4.1. Inactivation of Neddylation by MLN4924
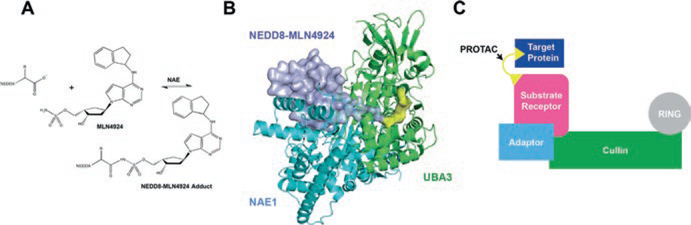
Currently MLN4924 (also known as pevonedistat or TAK-924) is the only general, small-molecule inhibitor of CRLs, targeting the neddylation reaction (Soucy et al. 2009). It is a sulfamate analog of AMP and is a mechanism-based inhibitor of NAE, the E1 for neddylation (Fig. 12.6a). Specifically, MLN4924 binds to the E1 nucleotide binding pocket on the NAE1 subunit of NAE, which then catalyzes the addition of MLN4924 to the C-terminus of NEDD8, forming a stable NEDD8–MLN4924 adduct (Fig. 12.6b). The modified NEDD8 adduct remains tightly bound to NAE. The crystal structure of the NEDD8–MLN4924 adduct with NAE reveals that NEDD8–MLN4924 mimics the charged NEDD8–AMP intermediate, but cannot be processed further by enzymes in the NEDD8 pathway (Brownell et al. 2010; Petroski 2010; Huang et al. 2007; Walden et al. 2003). Thus, MLN4924, through the actions of NAE, generates a null NEDD8 that cannot be conjugated onto cullins. This leads to an overall decrease in active CRLs and an accumulation of substrates that would normally be targeted for degradation.
Fig. 12.6.
Therapeutic manipulation of the cullin-RING ligase activity. (a) MLN4924 is covalently linked to the C-terminus of NEDD8 through catalysis by NAE. (b) The NAE heterodimer, composed of NAE1 and UBA3, forms a tight complex with the NEDD8–MLN4924 adduct. NEDD8–MLN4924 is shown in surface representation, with MLN4924 in yellow (PDB ID: 3GZN). (c) A PROTAC bridges target protein to CRL for ubiquitination and degradation
This general CRL inhibition has been shown to be an effective treatment for some cancers. In HCT-116 cells, a human colon cancer cell line, treatment with MLN4924 leads to S-phase arrest and eventually apoptosis (Soucy et al. 2009). While the MLN4923-triggered apoptosis is not completely understood, these cells accumulate multiple copies of DNA without undergoing mitosis, termed rereplication. This leads to an accumulation of DNA damage (Archambault et al. 2005), but it is unknown whether the DNA damage or accumulation of CRL substrates is responsible for induction of apoptosis. This phenotype is also observed in vivo, as MLN4924 suppressed the growth of xenografted tumors in various mouse models (Kuo et al. 2015; Milhollen et al. 2010; Soucy et al. 2009). It is unclear why MLN4924 affects tumors more strongly than normal cells. Nonetheless, it is undergoing Phase I clinical trials in patients suffering from advanced nonhematologic malignancies, relapsed/refractory lymphoma or multiple myeloma (Shah et al. 2016; Sarantopoulos et al. 2015).
12.4.2. Small-Molecule Subversion of CRL Targeting by PROTACs
By taking advantage of knowledge of substrate receptor binding sites, PROTACs (proteolysis targeting chimeras) have been developed to modulate specific protein levels by CRLs. PROTACs are bifunctional small molecules that simultaneously bind a target protein and the substrate receptor of a ubiquitin ligase, thus leading to the ubiquitination and degradation of the target (Fig. 12.6c). In essence, they bridge novel proteins to CRLs for polyubiquitination and degradation. The first PROTAC, Protac-1, was composed of a phosphopeptide fragment of IκBα, a ligand for the F-box protein β-TRCP, that was fused via a linker to the small molecule ovalicin, which binds to methionine aminopeptidase-2 (MetAP-2), which is involved in G1 phase cell cycle arrest and is not normally targeted to any CRL. in vitro, Protac-1 has been shown to target MetAP-2 to the CRL1β-TRCP for polyubiquitination (Sakamoto et al. 2001). Furthermore, in Xenopus egg extracts, Protac-1 led to the degradation of MetAP-2 within 30 min (Sakamoto et al. 2001). However, Protac-1 is not cell permeable, so it cannot act in vivo, limiting its therapeutic potential. Recently, many PROTACs have been developed with improved pharmacological properties (Toure and Crews 2016). Many of these take advantage of small molecules in place of phosphopeptides to recruit either the substrate receptors VHL or Cereblon to target oncogenic proteins for degradation and are functional in vivo (Bondeson et al. 2015; Winter et al. 2015). Unlike traditional small-molecule inhibitors, PROTACs can act catalytically to induce multiple rounds of targeting polyubiquitination and degradation (Bondeson et al. 2015), allowing for lower doses to be effective. While no PROTACs have made it to clinical trials yet, there is great potential for the PROTAC technology as a viable therapeutic pathway to reprogram a CRL to target any protein of interest.
12.5. Future Perspectives
CRLs are the largest superfamily of E3 ubiquitin ligase and play critical roles in a diverse range of cellular processes. The extensive studies discussed above have shed enormous insight into the architecture and regulatory mechanisms of these macromolecular complexes. Yet while the general architecture of CRLs are known, much remain to be answered regarding their assembly. While we know surface charge complementarity contribute to the selection of VHL box or SOCS box proteins in CRL2/5s, there is no explicit rule to define the preference of a VHL box or SOCS box protein for Cul2 or Cul5. In addition, even with the knowledge of assembled and NEDD8-activated CRLs, it is unknown how an E2 enzyme targets specific lysines on the substrate or how an E2 accommodates the initial target site and then elongates the ubiquitin chain during polyubiquitination. In the complex case of the dimeric CRL3Keap1, with two binding sites for a single substrate, it will be interesting to see how the two E2s are positioned relative to the single Nrf2 substrate. Structural biology, in particular the rapidly developing field of cryo-electron microscopy (cryo-EM), will likely play a major role in answering these questions.
The regulation of CRL assembly and activity is very dynamic and more complex than previously thought. A recent study revealed that the RBR E3 ARIH1 and CRL1/2/3s work in concert to polyubiquitinate substrates. ARIH1 binds to neddylated CRLs to add the first Ub to the substrate while the CRL and its E2 elongate the Ub chain (Scott et al. 2016). However, it will be important to determine the dynamics of the ARIH1-CRL interactions with respect to ARIH1 monoubiquitin kinetics (Kleiger and Deshaies 2016). While the kinetics of CAND1 association with CRL and its effect on substrate receptor exchange have been revealed, the kinetics of CSN association with CRL and the subsequent deneddylation are unknown. Of particular interest is the interplay of these regulatory factors in vivo. How the combined action of CSN, CAND1, substrates, and substrate receptors, at relevant in vivo protein concentrations, affects CRL assembly needs to be addressed. To address these questions, fluorescence microscopy of endogenous proteins and single molecule techniques may be necessary.
Finally, with the structural knowledge of how viruses hijack CRLs for their own survival, the road to rational drug design at the viral protein-CRL interface is now open. An alternative avenue focusing on CRL regulation, PROTACs or other approaches to manipulate CRLs can be used to combat viral hijacking of CRLs or other CRL related diseases. The development of these new therapeutics will likely depend on the collaboration between continued research in basic biology and efforts in pharmaceutical sciences.
Acknowledgements
We thank Olga Buzovetsky, Xiaofei Jia, Kirsten Knecht, April Rose, Sarah Smaga, and Samantha Ziegler for critical comments and discussion. This work was supported in part by NIH grants AI078831 and AI16313 to Y.X.
Contributor Information
Henry C. Nguyen, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511, USA Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94158, USA.
Wei Wang, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511, USA.
Yong Xiong, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511, USA.
References
- Angers S, Li T, Yi XH, MacCoss MJ, Moon RT, Zheng N (2006) Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery. Nature 443(7111):590–593. doi: 10.1038/Nature05175 [DOI] [PubMed] [Google Scholar]
- Archambault V, Ikui AE, Drapkin BJ, Cross FR (2005) Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol Cell Biol 25(15):6707–6721. doi: 10.1128/MCB.25.15.6707-6721.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry M, Früh K (2006) Viral modulators of cullin RING ubiquitin ligases: culling the host defense. Sci Signal 2006(335):pe21–pe21. doi: 10.1126/stke.3352006pe21 [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Rush J, Gygi SP, Harper JW (2010) Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143(6):951–965. doi: 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndsen CE, Wolberger C (2014) New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21(4):301–307. doi: 10.1038/nsmb.2780 [DOI] [PubMed] [Google Scholar]
- Bondeson DP, Mares A, Smith IED, Ko E, Campos S, Miah AH, Mulholland KE, Routly N, Buckley DL, Gustafson JL, Zinn N, Grandi P, Shimamura S, Bergamini G, Faelth-Savitski M, Bantscheff M, Cox C, Gordon DA, Willard RR, Flanagan JJ, Casillas LN, Votta BJ, den Besten W, Famm K, Kruidenier L, Carter PS, Harling JD, Churcher I, Crews CM (2015) Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat Chem Biol 11(8):611–U120. doi: 10.1038/Nchembio.1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard MJ, Schneider RJ (2004) The enigmatic X Gene of hepatitis B virus. J Virol 78(23):12725–12734. doi: 10.1128/JVI.78.23.12725-12734.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouillard P, Boon LM, Mulliken JB, Enjolras O, Ghassibe M, Warman ML, Tan OT, Olsen BR, Vikkula M (2002) Mutations in a novel factor, glomulin, are responsible for glomuvenous malformations (“glomangiomas”). Am J Hum Genet 70(4):866–874. doi: 10.1086/339492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR (2010) Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell 37(1):102–111. doi: 10.1016/j.molcel.2009.12.024 [DOI] [PubMed] [Google Scholar]
- Bulatov E, Ciulli A (2015) Targeting Cullin-RING E3 ubiquitin ligases for drug discovery: structure, assembly and small-molecule modulation. Biochem J 467:365–386. doi: 10.1042/Bj20141450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Ayinampudi V, Marsden BD, Gileadi O, Knapp S, von Delft F, Bullock AN (2013) Structural basis for Cul3 protein assembly with the BTB-Kelch family of E3 ubiquitin ligases (vol 288, pg 7803, 2013). J Biol Chem 288(39):28304–28304. doi: 10.1074/Jbc.A112.437996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavadini S, Fischer ES, Bunker RD, Potenza A, Lingaraju GM, Goldie KN, Mohamed WI, Faty M, Petzold G, Beckwith RE, Tichkule RB, Hassiepen U, Abdulrahman W, Pantelic RS, Matsumoto S, Sugasawa K, Stahlberg H, Thoma NH (2016) Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature 531(7596):598–603. doi: 10.1038/nature17416 [DOI] [PubMed] [Google Scholar]
- Chen X, Zhang Y, Douglas L, Zhou P (2001) UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J Biol Chem 276(51):48175–48182. doi: 10.1074/jbc.M106808200 [DOI] [PubMed] [Google Scholar]
- Ciechanover A, Hershko A, Rose I (2004) The Nobel Prize in Chemistry 2004. Nobel Media AB; http://www.nobelprize.org/nobel_prizes/chemistry/laureates/2004/ Accessed 15 Mar 2016 [Google Scholar]
- de Bruijn MF, Speck NA (2004) Core-binding factors in hematopoiesis and immune function. Oncogene 23(24):4238–4248. doi: 10.1038/sj.onc.1207763 [DOI] [PubMed] [Google Scholar]
- Decorsière A, Mueller H, van Breugel PC, Abdul F, Gerossier L, Beran RK, Livingston CM, Niu C, Fletcher SP, Hantz O, Strubin M (2016) Hepatitis B virus X protein identifies the Smc5/6 complex as a host restriction factor. Nature 531(7594):386–380. doi: 10.1038/nature17170 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ (2014) Structural biology: corralling a protein-degradation regulator. Nature 512(7513):145–146. doi: 10.1038/nature13644 [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CAP (2009) RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434. doi: 10.1146/Annurev.Biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Dias DC, Dolios G, Wang R, Pan ZQ (2002) CUL7: a DOC domain-containing cullin selectively binds Skp1.Fbx29 to form an SCF-like complex. Proc Natl Acad Sci U S A 99(26):16601–16606. doi: 10.1073/pnas.252646399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA (2008) Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134(6):995–1006. doi: 10.1016/j.cell.2008.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Olszewski JL, Tron AE, Hammel M, Lambert LJ, Waddell MB, Mittag T, DeCaprio JA, Schulman BA (2012) Structure of a glomulin-RBX1-CUL1 complex: inhibition of a RING E3 ligase through masking of its E2-binding surface. Mol Cell 47(3):371–382. doi: 10.1016/j.molcel.2012.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emberley ED, Mosadeghi R, Deshaies RJ (2012) Deconjugation of Nedd8 from Cul1 Is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J Biol Chem 287(35):29679–29689. doi: 10.1074/jbc.M112.352484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enchev RI, Scott DC, da Fonseca PCA, Schreiber A, Monda JK, Schulman BA, Peter M, Morris EP (2012) Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep 2(3):616–627. doi: 10.1016/J.Celrep.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington WJ, Khan MQ, Bueler SA, Rubinstein JL, Chakrabartty A, Prive GG (2012) Adaptor protein self-assembly drives the control of a cullin-RING ubiquitin ligase. Structure 20(7):1141–1153. doi: 10.1016/J.Str.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Fischer ES, Scrima A, Bohm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, Iwai S, Gut H, Sugasawa K, Thoma NH (2011) The molecular basis of CRL4(DDB2/CSA) ubiquitin ligase architecture, targeting, and activation. Cell 147 (5):1024–1039. doi: 10.1016/J.Cell.2011.10.035 [DOI] [PubMed] [Google Scholar]
- Fribourgh JL, Nguyen HC, Wolfe LS, Dewitt DC, Zhang W, Yu XF, Rhoades E, Xiong Y (2014) Core binding factor beta plays a critical role by facilitating the assembly of the Vif-cullin 5 E3 ubiquitin ligase. J Virol 88(6):3309–3319. doi: 10.1128/JVI.03824-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genschik P, Sumara I, Lechner E (2013) The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J 32(17):2307–2320. doi: 10.1038/emboj.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N (2004) Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 119(4):517–528. doi: 10.1016/j.cell.2004.10.019 [DOI] [PubMed] [Google Scholar]
- Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z (2014) Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505(7482):229–233. doi: 10.1038/nature12884 [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113(6):803–809. doi: 10.1016/s0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479. doi: 10.1146/annurev.biochem.67.1.425 [DOI] [PubMed] [Google Scholar]
- Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K (1999) Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18(48):6829–6834. doi: 10.1038/sj.onc.1203093 [DOI] [PubMed] [Google Scholar]
- Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J (2011) Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474(7353):658–661. doi: 10.1038/nature10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DT, Hunt HW, Zhuang M, Ohi MD, Holton JM, Schulman BA (2007) Basis for a ubiquitin-like protein thioester switch toggling E1-E2 affinity. Nature 445(7126):394–398. doi: 10.1038/nature05490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y (2008) RUNX genes in development and cancer: regulation of viral gene expression and the discovery of RUNX family genes. Adv Cancer Res 99:33–76. doi: 10.1016/S0065-230X(07)99002-8 [DOI] [PubMed] [Google Scholar]
- Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, Clarke SC, Shales M, Mercenne G, Pache L, Li K, Hernandez H, Jang GM, Roth SL, Akiva E, Marlett J, Stephens M, D’Orso I, Fernandes J, Fahey M, Mahon C, O’Donoghue AJ, Todorovic A, Morris JH, Maltby DA, Alber T, Cagney G, Bushman FD, Young JA, Chanda SK, Sundquist WI, Kortemme T, Hernandez RD, Craik CS, Burlingame A, Sali A, Frankel AD, Krogan NJ (2012) Global landscape of HIV-human protein complexes. Nature 481(7381):365–370. doi: 10.1038/nature10719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji AX, Prive GG (2013) Crystal structure of KLHL3 in complex with Cullin3. PLoS One 8(4):e60445. doi: 10.1371/journal.pone.0060445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Zhao Q, Xiong Y (2015) HIV suppression by host restriction factors and viral immune evasion. Curr Opin Struct Biol 31:106–114. doi: 10.1016/j.sbi.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin JP, Arias EE, Chen J, Harper JW, Walter JC (2006) A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell 23(5):709–721. doi: 10.1016/j.molcel.2006.08.010 [DOI] [PubMed] [Google Scholar]
- Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR (2011) Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat Methods 8(8):691–696. doi: 10.1038/nmeth.1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI (2004) VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 18(24):3055–3065. doi: 10.1101/gad.1252404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JR, Stanley DJ, Hultquist JF, Johnson JR, Mietrach N, Binning JM, Jonsson SR, Barelier S, Newton BW, Johnson TL, Franks-Skiba KE, Li M, Brown WL, Gunnarsson HI, Adalbjornsdottir A, Fraser JS, Harris RS, Andresdottir V, Gross JD, Krogan NJ (2015) Lineage-specific viral hijacking of non-canonical E3 ubiquitin ligase cofactors in the evolution of Vif Anti-APOBEC3 activity. Cell Rep 11(8):1236–1250. doi: 10.1016/j.celrep.2015.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA (2012) Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem 287(26):21570–21574. doi: 10.1074/jbc.C112.374843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kwak MJ, Ku B, Suh HY, Joo K, Lee J, Jung JU, Oh BH (2013) Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes. Acta Crystallogr D Biol Crystallogr 69(Pt 8):1587–1597. doi: 10.1107/S0907444913011220 [DOI] [PubMed] [Google Scholar]
- Kleiger G, Deshaies R (2016) Tag team ubiquitin ligases. Cell 166(5):1080–1081. doi: 10.1016/j.cell.2016.08.014 [DOI] [PubMed] [Google Scholar]
- Kuo KL, Ho IL, Shi CS, Wu JT, Lin WC, Tsai YC, Chang HC, Chou CT, Hsu CH, Hsieh JT, Chang SC, Pu YS, Huang KH (2015) MLN4924, a novel protein neddylation inhibitor, suppresses proliferation and migration of human urothelial carcinoma: in vitro and in vivo studies. Cancer Lett 363(2):127–136. doi: 10.1016/j.canlet.2015.01.015 [DOI] [PubMed] [Google Scholar]
- Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M (2005) The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature 435(7046):1257–1261. doi: 10.1038/nature03662 [DOI] [PubMed] [Google Scholar]
- Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M (2011) SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474(7353):654–657. doi: 10.1038/nature10117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ (2003) Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300 (5622):1112–1112. doi: 10.1126/Science.1083338 [DOI] [PubMed] [Google Scholar]
- Li T, Chen X, Garbutt KC, Zhou P, Zheng N (2006) Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124(1):105–117. doi: 10.1016/j.cell.2005.10.033 [DOI] [PubMed] [Google Scholar]
- Li T, Robert EI, van Breugel PC, Strubin M, Zheng N (2010) A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. Nat Struct Mol Biol 17(1):105–111. doi: 10.1038/nsmb.1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, Fischer ES, Thoma NH (2014) Crystal structure of the human COP9 signalosome. Nature 512(7513):161–165. doi: 10.1038/nature13566 [DOI] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ (2001) Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292(5520):1382–1385. doi: 10.1126/science.1059780 [DOI] [PubMed] [Google Scholar]
- Lydeard JR, Schulman BA, Harper JW (2013) Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep 14(12):1050–1061. doi: 10.1038/embor.2013.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW (2008) Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to elongin BC-based ubiquitin ligases. J Biol Chem 283(12):8005–8013. doi: 10.1074/Jbc.M706987200 [DOI] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D (2003) Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424(6944):99–103. doi: 10.1038/Nature01709 [DOI] [PubMed] [Google Scholar]
- Marin I (2009) Diversification of the cullin family. BMC Evol Biol 9:267. doi: 10.1186/1471-2148-9-267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG (2010) MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-{kappa} B-dependent lymphoma. Blood 116(9):1515–1523. doi: 10.1182/blood-2010-03-272567 [DOI] [PubMed] [Google Scholar]
- Muniz JR, Guo K, Kershaw NJ, Ayinampudi V, von Delft F, Babon JJ, Bullock AN (2013) Molecular architecture of the ankyrin SOCS box family of Cul5-dependent E3 ubiquitin ligases. J Mol Biol 425(17):3166–3177. doi: 10.1016/j.jmb.2013.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HC, Yang H, Fribourgh JL, Wolfe LS, Xiong Y (2015) Insights into cullin-RING E3 ubiquitin ligase recruitment: structure of the VHL-EloBC-Cul2 complex. Structure. doi: 10.1016/j.str.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ, Kuhn DJ (2008) Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin Cancer Res 14(6):1649–1657. doi: 10.1158/1078-0432.Ccr-07-2218 [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K (2004) Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23(11):1985–1997. doi: 10.1038/sj.onc.1207414 [DOI] [PubMed] [Google Scholar]
- Petroski MD (2010) Mechanism-based neddylation inhibitor. Chem Biol 17(1):6–8. doi: 10.1016/j.chembiol.2010.01.002 [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of Cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6(1):9–20. doi: 10.1038/Nrm1547 [DOI] [PubMed] [Google Scholar]
- Petzold G, Fischer ES, Thoma NH (2016) Structural basis of lenalidomide-induced CK1alpha degradation by the CRL4 ubiquitin ligase. Nature. doi: 10.1038/nature16979 [DOI] [PubMed] [Google Scholar]
- Pick E, Hofmann K, Glickman MH (2009) PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol Cell 35(3):260–264. doi: 10.1016/j.molcel.2009.07.009 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533. doi: 10.1146/Annurev.Biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RLJ, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan SO, Deshaies RJ (2013) Cand1 promotes assembly of new SCF complexes through dynamic exchange of F Box proteins. Cell 153 (1):206–215. doi: 10.1016/J.Cell.2013.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Precious B, Childs K, Fitzpatrick-Swallow V, Goodbourn S, Randall RE (2005) Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J Virol 79(21):13434–13441. doi: 10.1128/jvi.79.21.13434-13441.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ (2001) Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A 98(15):8554–8559. doi: 10.1073/pnas.141230798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarantopoulos J, Shapiro GI, Cohen RB, Clark JW, Kauh JS, Weiss GJ, Cleary JM, Mahalingam D, Pickard MD, Faessel HM, Berger AJ, Burke K, Mulligan G, Dezube BJ, Harvey RD (2015) Phase I study of the investigational NEDD8-activating enzyme inhibitor Pevonedistat (TAK-924/MLN4924) in patients with advanced solid tumors. Clin Cancer Res. doi: 10.1158/1078-0432.CCR-15-1338 [DOI] [PubMed] [Google Scholar]
- Sarikas A, Hartmann T, Pan ZQ (2011) The cullin protein family. Genome Biol 12(4). doi: 10.1186/Gb-2011-12-4-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Kamura T, Shirata N, Murata T, Kudoh A, Iwahori S, Nakayama S, Isomura H, Nishiyama Y, Tsurumi T (2009) Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Path 5(7). doi: 10.1371/Journal.Ppat.1000530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer PC, Ding Y, Liu Z, Xu J, Mao H, Barrow JC, Wei N, Zheng N, Snyder SH, Rao F (2016) Inositol hexakisphosphate (IP6) generated by IP5K mediates cullin-COP9 signalosome interactions and CRL function. Proc Natl Acad Sci U S A 113(13):3503–3508. doi: 10.1073/pnas.1525580113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ERE, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavietich NP (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408(6810):381–386. doi: 10.1038/35042620 [DOI] [PubMed] [Google Scholar]
- Schwefel D, Groom HCT, Boucherit VC, Christodoulou E, Walker PA, Stoye JP, Bishop KN, Taylor IA (2014) Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505(7482):234–238. doi: 10.1038/nature12815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwefel D, Boucherit Virginie C, Christodoulou E, Walker Philip A, Stoye Jonathan P, Bishop Kate N, Taylor Ian A (2015) Molecular determinants for recognition of divergent SAMHD1 proteins by the lentiviral accessory protein Vpx. Cell Host Microbe 17(4):489–499. doi: 10.1016/j.chom.2015.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Rhee DY, Duda DM, Kelsall IR, Olszewski JL, Paulo JA, de Jong A, Ovaa H, Alpi AF, Harper JW, Schulman BA (2016) Two distinct types of E3 ligases work in unison to regulate substrate ubiquitylation. Cell 166(5):1198–1214 e1124. doi: 10.1016/j.cell.2016.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA (2014) Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157(7):1671–1684. doi: 10.1016/j.cell.2014.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH (2008) Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135(7):1213–1223. doi: 10.1016/j.cell.2008.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JJ, Jakubowiak AJ, O’Connor OA, Orlowski RZ, Harvey RD, Smith MR, Lebovic D, Diefenbach C, Kelly K, Hua Z, Berger AJ, Mulligan G, Faessel HM, Tirrell S, Dezube BJ, Lonial S (2016) Phase I study of the novel investigational NEDD8-activating enzyme inhibitor pevonedistat (MLN4924) in patients with relapsed/refractory multiple myeloma or lymphoma. Clin Cancer Res 22(1):34–43. doi: 10.1158/1078-0432.CCR-15-1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418(6898):646–650. doi: 10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Sitterlin D, Lee TH, Prigent S, Tiollais P, Butel JS, Transy C (1997) Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J Virol 71(8):6194–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, Florens L, Tsutsumi T, Arai T, Tron A, Swanson SK, Washburn MP, DeCaprio JA (2007) PARC and CUL7 form atypical cullin RING ligase complexes. Cancer Res 67(5):2006–2014. doi: 10.1158/0008-5472.CAN-06-3241 [DOI] [PubMed] [Google Scholar]
- Skaar JR, Pagan JK, Pagano M (2013) Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14(6):369–381. doi: 10.1038/nrm3582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458(7239):732–736. doi: 10.1038/nature07884 [DOI] [PubMed] [Google Scholar]
- Stanley BJ, Ehrlich ES, Short L, Yu Y, Xiao Z, Yu XF, Xiong Y (2008) Structural insight into the human immunodeficiency virus Vif SOCS box and its role in human E3 ubiquitin ligase assembly. J Virol 82(17):8656–8663. doi: 10.1128/JVI.00767-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L (2012) SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4 + T-lymphocytes cannot be upregulated by interferons. Retrovirology 9(1):1–15. doi: 10.1186/1742-4690-9-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Katoh Y, Kusunoki H, Itoh K, Tanaka T, Yamamoto M (2006) Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol 26(8):2887–2900. doi: 10.1128/MCB.26.8.2887-2900.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M (2007) Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 27(21):7511–7521. doi: 10.1128/MCB.00753-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toure M, Crews CM (2016) Small-molecule PROTACS: new approaches to protein degradation. Angew Chem Int Ed Eng 55(6):1966–1973. doi: 10.1002/anie.201507978 [DOI] [PubMed] [Google Scholar]
- Tron AE, Arai T, Duda DM, Kuwabara H, Olszewski JL, Fujiwara Y, Bahamon BN, Signoretti S, Schulman BA, DeCaprio JA (2012) The glomuvenous malformation protein Glomulin binds Rbx1 and regulates cullin RING ligase-mediated turnover of Fbw7. Mol Cell 46(1):67–78. doi: 10.1016/j.molcel.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulane CM, Horvath CM (2002) Paramyxoviruses SV5 and HPIV2 assemble STAT protein ubiquitin ligase complexes from cellular components. Virology 304(2):160–166. doi: 10.1006/viro.2002.1773 [DOI] [PubMed] [Google Scholar]
- van Wijk SJL, Timmers HTM (2010) The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J 24(4):981–993. doi: 10.1096/fj.09-136259 [DOI] [PubMed] [Google Scholar]
- Walden H, Podgorski MS, Huang DT, Miller DW, Howard RJ, Minor DL, Holton JM, Schulman BA (2003) The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1. Mol Cell 12(6):1427–1437. doi: 10.1016/S1097-2765(03)00452-0 [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE (2007) Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div 2:7. doi: 10.1186/1747-1028-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, Bradner JE (2015) Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 348(6241):1376–1381. doi: 10.1126/science.aab1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhou X, Barnes CO, DeLucia M, Cohen AE, Gronenborn AM, Ahn J, Calero G (2016) The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat Struct Mol Biol 23(10):933–940 doi: 10.1038/nsmb.3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF (2003) Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302(5647):1056–1060. doi: 10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- Zachariae W, Shevchenko A, Andrews PD, Ciosk R, Galova M, Stark MJ, Mann M, Nasmyth K (1998) Mass spectrometric analysis of the anaphase-promoting complex from yeast: identification of a subunit related to cullins. Science 279(5354):1216–1219. doi: 10.1126/science.279.5354.1216 [DOI] [PubMed] [Google Scholar]
- Zhang W, Ito H, Quint M, Huang H, Noel LD, Gray WM (2008) Genetic analysis of CAND1-CUL1 interactions in Arabidopsis supports a role for CAND1-mediated cycling of the SCFTIR1 complex. Proc Natl Acad Sci U S A 105(24):8470–8475. doi: 10.1073/pnas.0804144105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Du J, Evans SL, Yu Y, Yu XF (2012) T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481(7381):376–379. doi: 10.1038/nature10718 [DOI] [PubMed] [Google Scholar]
- Zhang WY, Wang H, Li ZL, Liu X, Liu GC, Harris RS, Yu XF (2014) Cellular requirements for bovine immunodeficiency virus Vif-mediated inactivation of bovine APOBEC3 proteins. J Virol 88(21):12528–12540. doi: 10.1128/Jvi.02072-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song LZ, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP (2002) Structure of the Cul1- Rbx1- Skp1-F box(Skp2) SCF ubiquitin ligase complex. Nature 416(6882):703–709. doi: 10.1038/416703a [DOI] [PubMed] [Google Scholar]
- Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, Hoggard T, Harper JW, White KP, Schulman BA (2009) Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell 36(1):39–50. doi: 10.1016/j.molcel.2009.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman ES, Schulman BA, Zheng N (2010) Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol 20(6):714–721. doi: 10.1016/J.Sbi.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Mi J, Cui J, Lu D, Zhang X, Guo C, Gao G, Liu Q, Chen B, Shao C, Gong Y (2009) Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J Biol Chem 284(48):33320–33332. doi: 10.1074/jbc.M109.050427 [DOI] [PMC free article] [PubMed] [Google Scholar]