Abstract
In the current spread of novel coronavirus (SARS-CoV-2), antiviral drug discovery is of great importance. AutoDock Vina was used to screen potential drugs by molecular docking with the structural protein and non-structural protein sites of new coronavirus. Ribavirin, a common antiviral drug, remdesivir, chloroquine and luteolin were studied. Honeysuckle is generally believed to have antiviral effects in traditional Chinese medicine. In this study, luteolin (the main flavonoid in honeysuckle) was found to bind with a high affinity to the same sites of the main protease of SARS-CoV-2 as the control molecule. Chloroquine has been proved clinically effective and can bind to the main protease; this may be the antiviral mechanism of this drug. The study was restricted to molecular docking without validation by molecular dynamics simulations. Interactions with the main protease may play a key role in fighting against viruses. Luteolin is a potential antiviral molecule worthy of attention.
Keywords: 2019-nCoV, AutoDock Vina, chloroquine, remdesivir, ribavirin, luteolin
1. Introduction
The novel coronavirus, 2019-nCoV, known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1], emerged recently in Hubei province, P.R. China [2, 3]. The whole-genome sequence of 2019-nCoV was first released on January 10, 2020 [4]. 2019-nCoV has a wide range of infection in mammals, including humans. This characteristic of transmission leads to the possibility of transmission from animals to humans. The 2019-nCoV is highly transmissible and can lead to mild to severe respiratory tract infections [5]. The spread of 2019-nCoV has drawn great attention and created concern worldwide. There have been two coronavirus-related crises in humans since 2003 [6]. Severe acute respiratory syndrome coronavirus (SARS-CoV) broke out in 2003 and the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) emerged in the Arabian Peninsula in 2012 with a fatality rate of 35% [7].
Coronaviruses (CoVs) encode replicase complex (ORF1ab), expressed in the form of polyproteins (pp), which synthesize non-structural proteins (nsp) and four structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins [8], during proteolytic processing [9]. The main protease, 3CL protease (3CLpro) is a key enzyme in the processing of polyproteins pp1a and pp1ab. ORF1a and ORF1ab are cleaved by papain-like protease (PLpro, nsp3) and 3C-like protease (3CLpro, nsp5) to produce the nsp [10]. The SARS-CoV 3CLpro has an important function and is considered an active target for antiviral drugs. Many 3CLpro inhibitors have been reported over the past decade [11] and a variety of inhibitors have been found through screening and structure-based design [12]. PLpro is an indispensable enzyme in virus replication and infection of the host and is an important target for coronavirus inhibitors.
A recent study showed that 2019-nCoV uses angiotensin-converting enzyme 2 (ACE2) as the entry receptor into host cells [13]. S protein, a type I glycoprotein on the surface of the virus, plays a crucial role during virus entry into the host cells [14]. S protein can assist viral binding to the host acceptor, which has attracted great attention because of its function in receptor binding. The receptor binding domain (RBD) of S protein binds to the host cell. A total of 72% of the amino acid sequences in the RBDs from SARS-CoV and 2019-nCoV are identical. However, in 2019-nCoV, the rigid prolyl residues are replaced with a distinct loop with flexible glycyl residues [15]. A unique phenylalanine in the loop (F486) can penetrate into the hydrophobic pocket of ACE2 [16] and may play a key role in acceptor recognition. Nsp12 is a viral RNA-dependent RNA polymerase (RdRp) with co-factors nsp7 and nsp8 and possesses high polymerase activity. Four functional proteins in 2019-nCoV, 3CLpro (the main protease), RdRp, PLpro, and S, were studied as potential drug targets. There is no approved antiviral drug for treatment of COVID-19. The fastest way to find anti-2019-nCoV drugs is to screen drugs that are commonly used in the clinic.
Chloroquine is an antimalarial drug made by Bayer in Germany in 1934 to replace natural antimalarial drugs. This drug was found to be efficacious in the treatment of patients infected with SARS-CoV-2 [17, 18, 19]. Chloroquine inhibited quinone reductase 2, which is structurally similar to UDP-N-acetylglucosamine 2-epimerase, an enzyme involved in the biosynthesis of sialic acids. The possible interference by chloroquine of sialic acid biosynthesis could account for the broad antiviral spectrum of this drug [19]. Chloroquine can also impair early stage virus replication by interfering with the pH-dependent endosome-mediated viral entry of enveloped viruses as well as the post-translational modification of viral proteins. However, the mechanism of antiviral action of chloroquine against 2019-nCoV is not clear.
Ribavirin is a traditional antiviral drug widely used in the clinic for treating a variety of viral infections but it has no significant effect on SARS-CoV-2 [20]. In contrast, the new antiviral drug, remdesivir was found to be effective in preventing replication of this virus [21] and is a possible therapeutic option for COVID-19 [22,23].
Lianhuaqingwen (LH) is a traditional Chinese medicine (TCM) preparation that has been shown to have broad-spectrum antiviral effects on a series of influenza viruses. LH also showed anti-2019nCoV activity [24]. Honeysuckle and forsythia are commonly used as antiviral ingredients in TCM and are included in LH. The type and quantity of active components in the TCM formula are vital for the function of the preparation. Forsythia has long been used as an antipyretic, anti-inflammatory and anti-infectious agent in East Asia [25]. The compounds in forsythia showed antiviral activity against H1N1 virus and respiratory syncytial virus (RSV) [26], [27], [28], [29]. The main ingredients in honeysuckle exhibit antibacterial and antiviral activities [30, 31, 32]. Luteolin is the main flavonoid in honeysuckle [33]. TCM may be an effective alternative treatment for respiratory infections. In 2003, TCM was widely used to treat severe acute respiratory syndrome (SARS). The high similarity in genomic and structural characterization between 2019-nCoV and SARS indicates that TCM may have potential use in the current outbreak [34]. In this study, the binding of chloroquine, remdesivir, ribavirin and luteolin with the main proteins of 2019-nCoV (3CLpro, PLpro, RdRp, and S) were carried out by computational methods.
2. Methods and materials
2.1. Sequence alignment and modeling
The newly-emerged SARS-CoV-2 nucleotide gene (NC_045512.2) was retrieved from the National Center for Biotechnology Information (NCBI) nucleotide database. The important antivirus drug target proteins like 3CLpro, PLpro, and RdRp are highly conserved between SARS-CoV and 2019-nCoV, particularly in terms of functional sites.
The model of the COVID-19 main protease was downloaded from the Protein Data Bank (www.rcsb.org). The crystal structure of the COVID-19 main protease in complex with an inhibitor N3 (PDB ID: 6LU7, chain A) [35] was chosen as the model of 3CLpro, and the inhibitor N3 was removed. The ligand N3 (N-[(5-methylisoxazol-3yl)carbonyl]alanyl-l-valyl-n~1~-((1R,2Z)-4-(benzyloxy)-4-oxo-1-{[(3R)-2-oxopyrrolidin-3-yl]methyl}but-2-enyl)-l-leucinamide) was used as a control. Recent studies highlighted that SARS-CoV-2 genes share <80% nucleotide identity and 89.10% nucleotide similarity with SARS-CoV genes [36, 37]. The X-ray structure of PLpro of human coronavirus papain-like proteases was used as the model of 2019-nCoV PLpro (PDB ID: 4OVZ, chain A) [38]. The ligand P85 (N-[(4-fluorophenyl)methyl]-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide) was used as a control. The SARS-HCoV solved structure (PDB ID: 6NUS, chain A) was used for binding since it was the most sequelogous solved structure (97.08% sequence identity) to SARS-CoV-2 RdRp. 6NUS is a SARS-HCoV non-structural protein 12 (nsp12) solved structure (cryo-electron microscopy) with 3.5 Å resolution, which was deposited in the protein data bank in 2019 [39]. Remdesivir, an antagonist of 2019-nCOV RdRp, was chosen as a control. The model of 2019-nCoV S glycoprotein (PDB ID: 6VSB, chains A, B, C) [40] was used as the model for molecular docking. 6VSB was S protein prefusion with a single receptor-binding domain solved structure (cryo-electron microscopy) with 3.46 Å resolution. The ligand NAG (N-acetyl-D-glucosamine) of 6VSB was used as a control.
2.2. Molecular docking
AutoDock Vina software was utilized in all the docking experiments, with the optimized model as the docking target. The screening method is restricted to molecular docking, and molecular dynamics simulation has not been carried out. Remdesivir and ribavirin are RdRp antagonists. Remdesivir may be effective against the new coronavirus; however, ribavirin is invalid. Molecular docking was used to study the binding difference between the two molecules. Chloroquine has been proved to be clinically effective, and its molecular target should be further studied. In the context of TCM in the treatment of new coronavirus, luteolin, the main flavonoid of honeysuckle (the main antiviral component in LH), was selected for molecular docking to further study the drug. Prior to testing the ligands against SARS-CoV-2 target proteins, the structures of the small molecules were optimized using the classical MM2 force field; the active site aspartates of targets were treated as rigid. Potential antiviral drugs were shown by the effective relationship between molecules and receptor targets. Grid box (60 Å × 60 Å × 60 Å) centered at (-28.059, 9.486, 61.528) Å, for the 3CLpro, grid box (60 Å × 60 Å × 60 Å) centered at (-13.953, 51.391, -31.750) Å, for the PLpro, grid box (126 Å × 126 Å × 126 Å) centered at (251.541, 330.170, 152.338) Å, for the S protein, and grid box (126Å × 126 Å × 126 Å) centered at (152.179, 167.664, 166.985) Å, for the RdRp, were used in the docking experiments by utilizing the AutoDock tools.
3. Results and discussion
3.1. 3CL protease
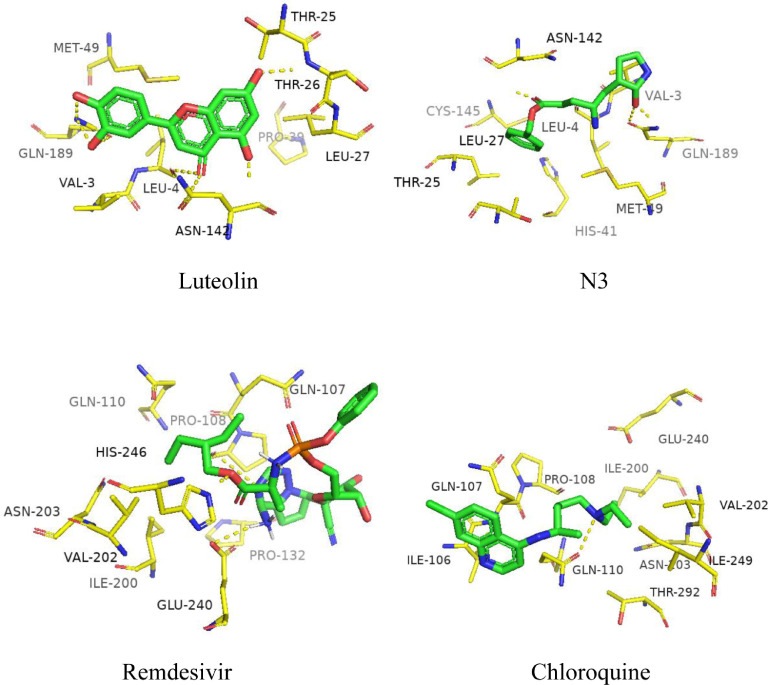
N3 molecule is the ligand molecule isolated from the crystal structure of 3CLpro, which was used as the binding site control. According to the analysis of docking results (Fig. 1 ), the interactions between luteolin and binding sites are highly consistent with that of N3. Luteolin formed five hydrogen bonds with GLN-189, LEU-4, ASN-142 and THR-26, respectively. MET-49 and VAL-3 formed hydrophobic interaction with luteolin. With regard to N3, 3-H bonds were formed with CYS-145 and GLN189. N3 formed a π-cation contact with HIS-41, and hydrophobic reactions with THR-25, LEU-27, LEU-4, VAL-3 and ASN-142. There is a hydrogen bond interaction between chloroquine and GLN-110 at the binding site, hydrophobic reactions with ILE 249, ILE-200, and a π-cation contact with GLN-107. Remdesivir formed 3-H bonds with GLU-240, HIS-246 and PRO-108, hydrophobic reactions with PRO-132 and ILE-200, and a π-cation contact with GLN-107.
Fig. 1.
Binding region of 3CL.
3.2. PLpro
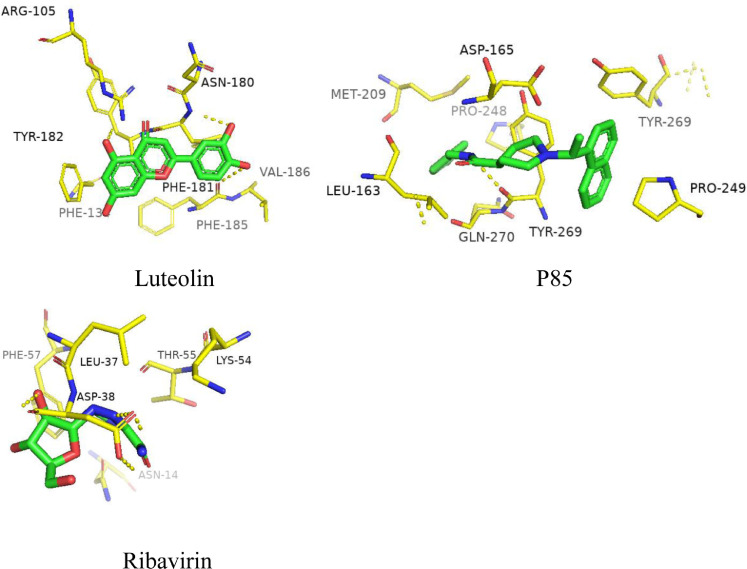
The ligand P85 of PLpro was used as a control. Fig. 2 shows P85 formed hydrogen bonds with TYR-269. Luteolin formed hydrogen bonds with ASN-180, ARG-105 and PHE-185 in the sites. The interactions between ribavirin and ASP-38 were hydrogen bonds. The binding sites of luteolin and ribavirin are inconsistent with those of the control molecule.
Fig. 2.
Binding region of PLpro.
3.3. RdRp
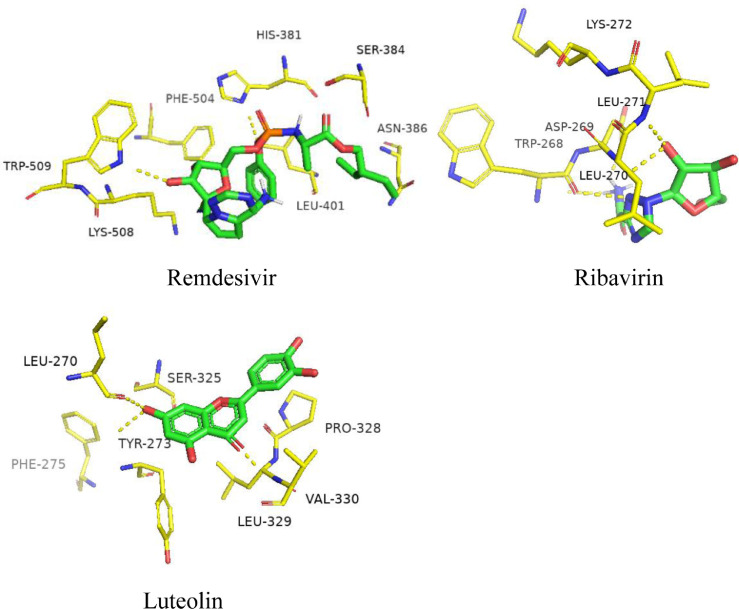
Fig. 3 shows remdesivir formed 2-H bonds with TRP-509 and HIS-381, π-cation contacts with PHE-504, and hydrophobic reactions with LYS-508, LEU-401, ASN-386, and SER-384. Ribavirin formed 5-H bonds with LEU-271, LEU-270, ASP-269 and TRP-268. Luteolin formed 3-H bonds with TYR-273, LEU-270 and VAL-330, two π-cation reactions with SER-325 and PHE-275, and two hydrophobic reactions with LEU-329 and PRO-328. The binding sites of luteolin and ribavirin are inconsistent with those of remdesivir; however, the binding sites of ribavirin and luteolin are close, indicating another active site.
Fig. 3.
Binding region of RdRp.
3.4. Spike protein
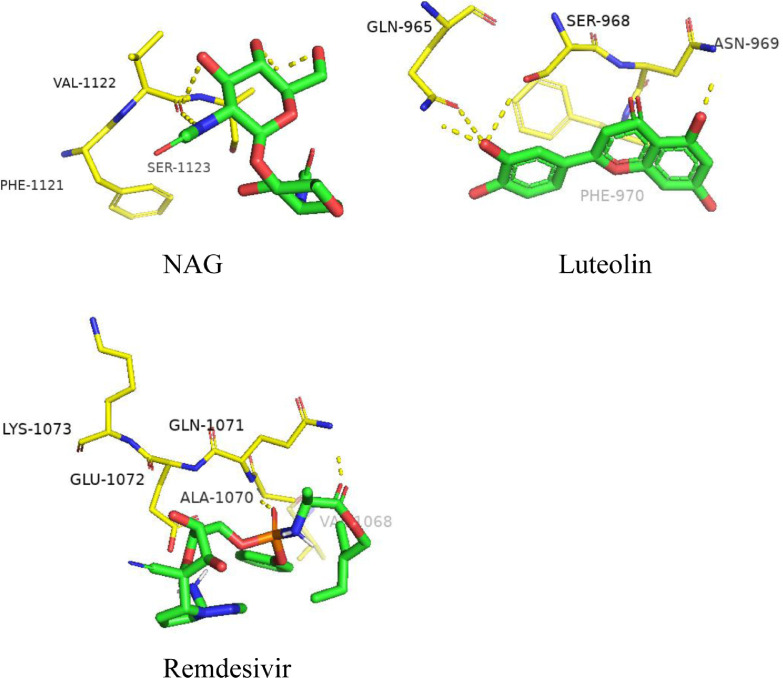
Fig. 4 shows the interactions formed between the ligands NAG, luteolin and remdesivir against the S protein of 2019-nCOV after docking. NAG was found to form 3-H bonds with residues VAL-1122 and SER-1123. In addition, NAG built π-cation contacts with PHE-1121. In luteolin, 4H-bonds were formed with the residues GLN-965, SER-968 and ASN-969. π-cation interaction was formed between luteolin and PHE-970. Remdesivir formed 2-H bonds with ALA-1070 and GLN-1071, and hydrophobic interaction with GLU-1072, as seen in Fig. 4. The binding site of remdesivir is close to that of the control molecule NAG.
Fig. 4.
Binding region of spike protein.
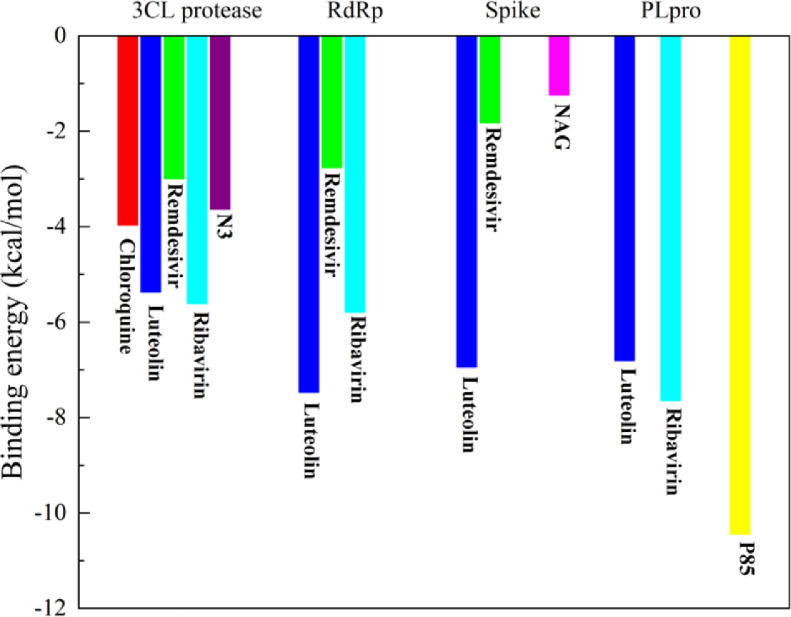
The binding energy of luteolin to the main protease was -5.37 kcal/mol, and that of N3 was -3.63 kcal/mol (Fig. 5). The binding energy of luteolin to the active site is even smaller than that of the control molecule, indicating that luteolin has a higher binding activity. With regard to RdRp and S protein, luteolin also has a smaller binding energy. From the point of view of binding energy, luteolin shows strong interactions with the targets of the new coronavirus.
Fig. 5.
Binding energy.
4. Conclusion
LH may be effective against the new coronavirus. Honeysuckle and forsythia are the main antiviral drugs in the formula. Luteolin (the main flavonoid of honeysuckle) can bind tightly to the main protease of the new coronavirus. Luteolin binds to the active sites of the main protease of 2019-nCOV with a lower binding energy than the ligand of the crystal structure, which implies a possible strong antiviral activity. This study indicates the potential of TCM in the treatment of the new coronavirus. Chloroquine has been shown to be a potentially effective treatment for the new coronavirus; binding with the main protease may be a supplement to its mechanisms of action. Luteolin may be useful in the selection of new compounds that bind specifically to the SARS-CoV-2 main protease sites.
Declarations
Funding: The authors wish to thank the general projects of scientific research plan of the Beijing Municipal Commission of Education for financial support under the project KM202010858002.
Competing Interests: None
Ethical Approval: Not required.
Editor: Jean-Marc Rolain
References
- 1.Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–440. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupia T, Scabini S, Mornese Pinna S, Di Perri G, De Rosa FG, Corcione S. 2019 novel coronavirus (2019-nCoV) outbreak: A new challenge. J Global Antimicrob Resis. 2020;21:22–27. doi: 10.1016/j.jgar.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Tawfiq JA. Asymptomatic coronavirus infection: MERS-CoV and SARS-CoV-2 (COVID-19). Travel Med Infect Dis2020:101608. [DOI] [PMC free article] [PubMed]
- 4.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Zowalaty ME, Järhult JD. From SARS to COVID-19: A previously unknown SARS-related coronavirus (SARS-CoV-2) of pandemic potential infecting humans – Call for a One Health approach. One Health. 2020;9 doi: 10.1016/j.onehlt.2020.100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020 Feb 27 doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S-W, Lin C-W. Human coronaviruses: Clinical features and phylogenetic analysis. BioMedicine. 2013;3:43–50. doi: 10.1016/j.biomed.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Konno H, Onuma T, Nitanai I, Wakabayashi M, Yano S, Teruya K, et al. Synthesis and evaluation of phenylisoserine derivatives for the SARS-CoV 3CL protease inhibitor. Bioorg Med Chem Lett. 2017;27:2746–2751. doi: 10.1016/j.bmcl.2017.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V, Shin JS, Shie J-J, Ku KB, Kim C, Go YY, et al. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLPro inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luan J, Lu Y, Jin X, Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Comm. 2020;526(1):165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ibrahim IM, Abdelmalek DH, Elshahat ME, Elfiky AA. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun J, He W-T, Wang L, Lai A, Ji X, Zhai X, et al. COVID-19: Epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med. 2020;26(5):483–495. doi: 10.1016/j.molmed.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Guo Y, Pan Y, Zhao ZJ. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Comm. 2020;525:135–140. doi: 10.1016/j.bbrc.2020.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao J, Tian Z, Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. BioSci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 18.Gautret P, Lagier J-C, Parola P, Hoang VT, Meddeb L, Mailhe M, et al. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Devaux CA, Rolain J-M, Colson P, Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pillaiyar T, Meenakshisundaram S, Manickam M. Recent discovery and development of inhibitors targeting coronaviruses. Drug Discov Today. 2020;25(4):668–688. doi: 10.1016/j.drudis.2020.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colson P, Rolain J-M, Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ribavirin Elfiky AA. remdesivir, sofosbuvir, galidesivir, and tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sciences. 2020 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Tawfiq JA, Al-Homoud AH, Memish ZA. Remdesivir as a possible therapeutic option for the COVID-19. Travel Med Infect Dis. 2020 doi: 10.1016/j.tmaid.2020.101615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Runfeng L, Yunlong H, Jicheng H, Weiqi P, Qinhai M, Yongxia S, et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharm Res. 2020 doi: 10.1016/j.phrs.2020.104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu H-F, Hsiao P-C, Kuo T-C, Chiang S-T, Chen S-L, Chiou S-J, et al. Antioxidant and anti-inflammatory activities of Lonicera japonica Thunb. var. sempervillosa Hayata flower bud extracts prepared by water, ethanol and supercritical fluid extraction techniques. Ind Crops Prod. 2016;89:543–549. doi: 10.1016/j.indcrop.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang K-L, Liu R-X, Zhao L, Xie Z-P, Zhang S-M, Dai S-J. Labdane diterpenoids from Forsythia suspensa with anti-inflammatory and anti-viral activities. Phytochemistry. 2020;173 doi: 10.1016/j.phytochem.2020.112298. [DOI] [PubMed] [Google Scholar]
- 27.Zhao L, Xiang K-L, Liu R-X, Xie Z-P, Zhang S-M, Dai S-J. Anti-inflammatory and anti-viral labdane diterpenoids from the fruits of Forsythia suspensa. Bioorg Chem. 2020;96 doi: 10.1016/j.bioorg.2020.103651. [DOI] [PubMed] [Google Scholar]
- 28.Lee JJ, Kim KH, Kim EJ, Choi J-Y, Kim S-J, Jeong S-I, et al. Anti-inflammatory activity of the decoction of Forsythia suspensa (Thunb.) Vahl is related to Nrf2 and A20. J Ethnopharmacol. 2018;227:97–104. doi: 10.1016/j.jep.2018.08.027. [DOI] [PubMed] [Google Scholar]
- 29.Du J, Hu Z, Yu Z, Li H, Pan J, Zhao D, et al. Antibacterial activity of a novel Forsythia suspensa fruit mediated green silver nanoparticles against food-borne pathogens and mechanisms investigation. Mater Sci Eng C Mater Biol Appl. 2019;102:247–253. doi: 10.1016/j.msec.2019.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Zhang B, Nan T-G, Zhan Z-L, Kang L-P, Yang J, Lai C-J-S, et al. A monoclonal antibody-based enzyme-linked immunosorbent assay for the determination of chlorogenic acid in honeysuckle. J Pharm Biomed Anal. 2018;148:1–5. doi: 10.1016/j.jpba.2017.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Li C, Zheng Y, Li Y, Peng G. Study on the anaphylactoid of three phenolic acids in honeysuckle. J Ethnopharmacol. 2015;170:1–7. doi: 10.1016/j.jep.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 32.Liu M, Yu Q, Yi Y, Xiao H, Putra DF, Ke K, et al. Antiviral activities of Lonicera japonica Thunb. Components against grouper iridovirus in vitro and in vivo. Aquaculture. 2020;519 [Google Scholar]
- 33.Li C, Zang C, Nie Q, Yang B, Zhang B, Duan S. Simultaneous determination of seven flavonoids, two phenolic acids and two cholesterines in Tanreqing injection by UHPLC-MS/MS. J Pharm Biomed Anal. 2019;163:105–112. doi: 10.1016/j.jpba.2018.08.058. [DOI] [PubMed] [Google Scholar]
- 34.Ling C-Q. Traditional Chinese medicine is a resource for drug discovery against 2019 novel coronavirus (SARS-CoV-2) J Integr Med. 2020;18:87–88. doi: 10.1016/j.joim.2020.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M(pro) from COVID-19 virus and discovery of its inhibitors. Nature. 2020 Apr 9 doi: 10.1038/s41586-020-2223-y. [DOI] [Google Scholar]
- 36.Ul Qamar MT, Alqahtani SM, Alamri MA, Chen L-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J Pharm Analysis. 2020 Mar 26 doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Yang X-L, Wang X-G, Hu B, Zhang L, Zhang W, et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. bioRxiv 2020:2020.01.22.914952.
- 38.Baez-Santos YM, Barraza SJ, Wilson MW, Agius MP, Mielech AM, Davis NM, et al. X-ray structural and biological evaluation of a series of potent and highly selective inhibitors of human coronavirus papain-like proteases. J Med Chem. 2014;57:2393–2412. doi: 10.1021/jm401712t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10:2342. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]