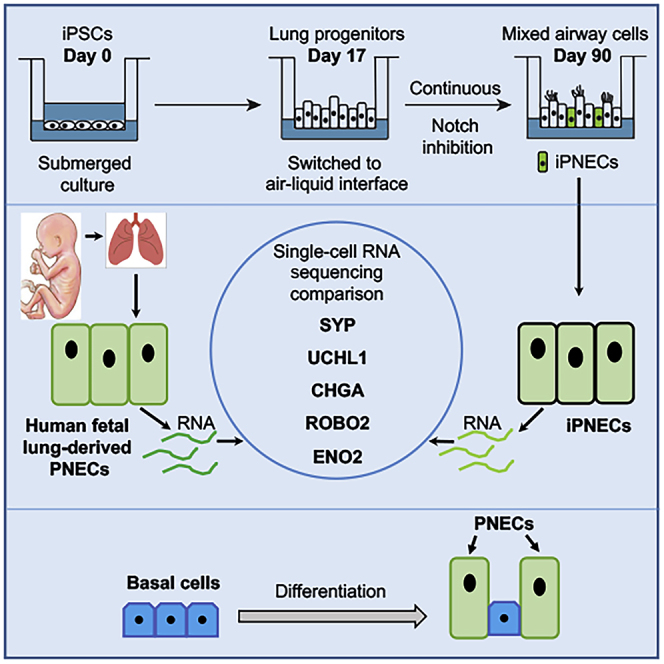
Summary
Expansion of pulmonary neuroendocrine cells (PNECs) is a pathological feature of many human lung diseases. Human PNECs are inherently difficult to study due to their rarity (<1% of total lung cells) and a lack of established protocols for their isolation. We used induced pluripotent stem cells (iPSCs) to generate induced PNECs (iPNECs), which express core PNEC markers, including ROBO receptors, and secrete major neuropeptides, recapitulating known functions of primary PNECs. Furthermore, we demonstrate that differentiation efficiency is increased in the presence of an air-liquid interface and inhibition of Notch signaling. Single-cell RNA sequencing (scRNA-seq) revealed a PNEC-associated gene expression profile that is concordant between iPNECs and human fetal PNECs. In addition, pseudotime analysis of scRNA-seq results suggests a basal cell origin of human iPNECs. In conclusion, our model has the potential to provide an unlimited source of human iPNECs to explore PNEC pathophysiology associated with several lung diseases.
Subject Areas: Cell Biology, Stem Cells Research, Transcriptomics
Graphical Abstract
Highlights
-
•
PNECs can be efficiently generated from induced pluripotent stem cells (iPSCs)
-
•
Induced PNECs express key PNEC markers and express and secrete all major neuropeptides
-
•
Induced PNECs resemble the transcriptomic profile of human primary fetal PNECs
-
•
Basal cells are able to differentiate into PNECs
Cell Biology; Stem Cells Research; Transcriptomics
Introduction
Pulmonary neuroendocrine cells (PNECs) are endogenous chemosensory cells that form around gestational week 8 in humans, making them the first differentiated cell type to appear in fetal lung (Cutz et al., 1985). Innervated clusters of PNECs in the human airway epithelium are referred to as “neuro-epithelial bodies or NEBs” (Lauweryns and Peuskens, 1972, Lauweryns and Van Ranst, 1988). PNECs are multifunctional cells with known roles in oxygen sensing, redistribution and regulation of blood flow especially during hypoxia, modulation of immune responses, and stimulation of intraepithelial vagosensory nerve fibers (Linnoila, 2006, Sunday, 1996). In addition, PNECs have been reported to serve as a stem cell niche during severe lung injury, with the potential to give rise to club and ciliated cells (Ouadah et al., 2019, Song et al., 2012). PNEC ontogeny, however, is not entirely clear. A recent lineage tracing study in mice showed that a small population of cells expressing the basal cell marker, tumor protein 63+ (TP63+), and labeled at embryonic day (E) 9.5, gave rise to calcitonin gene-related polypeptide alpha (CGRP)-expressing PNECs, that can be detected at E18.5 by immunostaining (Yang et al., 2018). However, whether basal cells have the capacity to give rise to PNECs in the human fetal or adult lung remains to be determined.
A spectrum of lung disorders including neuroendocrine hyperplasia of infancy (NEHI), chronic obstructive pulmonary disease (COPD), cystic fibrosis, asthma, bronchopulmonary dysplasia (BPD), and pulmonary hypertension are associated with expansion of PNECs in the airway epithelium (Cutz, 2015, Cutz et al., 2007, Gu et al., 2014, Johnson et al., 1985, Johnson and Georgieff, 1989, Pan et al., 2006, Schindler et al., 1995). The pathophysiological consequences of increased PNEC abundance in these diseases is not clear. As PNECs comprise <1% of all respiratory cell types, primary PNECs are inherently difficult to isolate in sufficient quantities for detailed characterization. We, therefore, addressed an unmet need to develop a model system for efficiently generating human PNECs to study their physiological and pathological functions using induced pluripotent stem cells (iPSCs).
Within the lung milieu, activation of pro-neural transcription factor ASCL1 (Achaete-Scute Family BHLH Transcription Factor 1) is required for cells to form the pulmonary neuroendocrine lineage (Linnoila, 2006). The Notch-HES1/HEY1 (Hairy/Enhancer-Of-Split related BHLH transcription factor family) pathway regulates the non-neuroendocrine fate of lung endoderm by repressing pro-neural genes like ASCL1 (Henke et al., 2009, Nelson et al., 2009). Recently, it has been shown that inhibition of Notch can increase PNEC production (Chen et al., 2019). These studies, however, focused specifically on modeling small cell lung carcinoma (SCLC) using human embryonic stem cell-derived PNECs, or on generating proximal airway epithelial spheroids from human pluripotent cells (Chen et al., 2019, Konishi et al., 2016). In-depth characterization of iPNECs or comparison at the transcriptional level with primary PNECs was not performed. In this article, we report the differentiation of iPSCs to human iPNECs with a gene expression profile similar to that of primary fetal PNECs that could be used in future studies of pathophysiological changes in diseases such as NEHI or BPD.
Results
Directed Differentiation of iPSCs to iPNECs
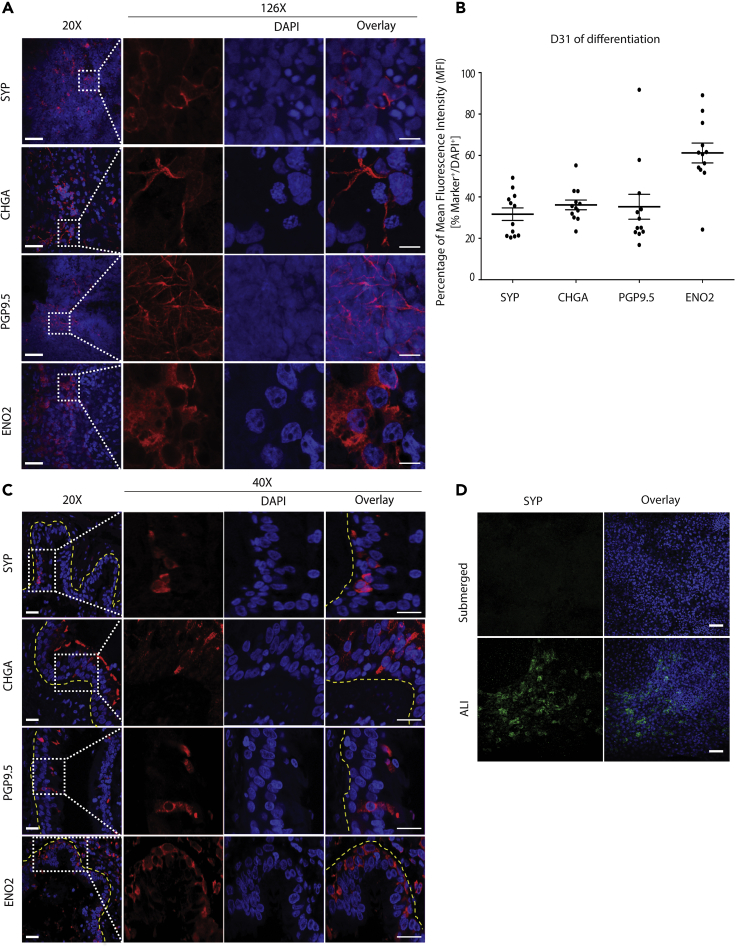
We adapted our previously published differentiation protocol to create airway epithelium from human iPSCs, recapitulating the key stages of embryonic lung development (Firth et al., 2014). iPSCs were differentiated in culture without sorting, resulting in a mixed population of mesoderm and epithelium comprising the proximal airways. To validate the presence of PNECs in our directed differentiation, cultures were stained for a panel of genes known to be expressed in primary human PNECs (Linnoila, 2006, Sunday, 1996), including synaptophysin (SYP), chromogranin A (CHGA), PGP9.5 (expressed by the gene UCHL1), and neuron-specific enolase (ENO2). After 31 days of differentiation, including culture at an air-liquid interface (ALI) from Day 17, cells expressing these PNEC markers were observed in cultures generated from three independent iPSC lines (Figures 1A and S1A). Expression of SYP, CHGA, PGP9.5, and ENO2 was quantified as a percentage of the mean fluorescence intensity (MFI) of the respective marker normalized to the expression of nuclear marker 4′,6-diamidino-2-phenylindole (DAPI). The distribution of these markers ranged from 31.64% ± 3.01% MFI for SYP to 61.20% ± 4.78% MFI for ENO2 (Figure 1B). Orthogonal projection of the differentiated cultures revealed that iPNECs are present in closer proximity to the apical-air interface (Figure S1B). Analysis of key PNEC markers, including ENO2 and SYP, throughout the differentiation time course from iPSC indicated that ENO2 protein was robustly expressed from Day 13 (Figure S2A), whereas SYP expression was increased gradually after exposure at the ALI, from Day 17 onward. (Figure S2B). Expression of these PNEC markers was additionally validated in explanted human adult lung tissue sections (Figure 1C). Cells expressing SYP, CHGA, and PGP9.5 were identified within the epithelium of the distal airways. In support of observations in our iPNEC cultures, ENO2 expression was also more widespread than the other PNEC markers in the human lung sections (Figure 1C), consistent with ENO2 being expressed in other lung cells in addition to PNECs. Indeed, our data indicated that KRT5+ basal cells co-express ENO2 in adult human lung tissues (Figure S2C). This is also supported by data deposited in the LungMAP database (www.lungmap.net). Gastrin-releasing protein (GRP), also known as bombesin, is another key marker of PNECs whose expression peaks during human fetal lung development around week 20–23 (Linnoila, 2006, Sunday, 1996, Sunday et al., 1991). GRP mRNA expression peaked during specification of the primordial lung progenitor cells from ventralized anterior foregut endoderm, Days 13–17 of our iPSC differentiation protocol (Figure S2D), and then decreased.
Figure 1.
Directed Differentiation of Human iPSCs into iPNECs
(A) Representative immunofluorescence (IF) images of iPNECs at Day 31 of differentiation at the ALI show expression of SYP, CHGA, PGP9.5, and ENO2 (all red). Left panels (20×) represent low-power images (scale bars, 50 μm), and right panels (40×) show magnified views (scale bars, 10 μm).
(B) Quantification of PNEC marker expression at Day 31 of differentiation expressed as percentage MFI of each marker in relation to total DAPI fluorescence. Data are expressed as mean ± SEM from 3 independent experiments.
(C) Representative IF images of primary PNECs in adult human distal lung show expression of SYP, CHGA, PGP9.5, and ENO2 (all red). Yellow dotted lines demarcate the airways. Left panels (20×) represent low-power images (scale bars, 100 μm), and right panels (40×) show magnified views of the marked areas (scale bars, 50 μm).
(D) Representative IF images of iPNECs in submerged and ALI cultures stained for SYP (green). Scale bars, 200 μm. Nuclei are counterstained with DAPI (blue).
See also, Figures S1 and S2.
Induction of PNECs Is Augmented by Air-Liquid Interface Culture during Directed Differentiation
Based on our observations that SYP expression increased after exposure of the differentiation cultures to an ALI, we evaluated whether the air interface was an important factor in PNEC induction. We continued the culture of primordial lung progenitors at Day 17 of differentiation (Firth et al., 2014), in both submerged and ALI conditions. Owing to consistency in staining across experimental replicates, the relative MFI of SYP:DAPI was compared (Figure 1B). After 14 additional days of differentiation through to experimental Day 31, SYP+ iPNECs were only detected in the presence of an ALI (Figure 1D, bottom panel). No SYP-expressing cells were observed in continually submerged cultures (Figure 1D, top panel), indicating that air exposure provides a microenvironment suited to the continued development of PNECs.
Characterization of Human iPNECs
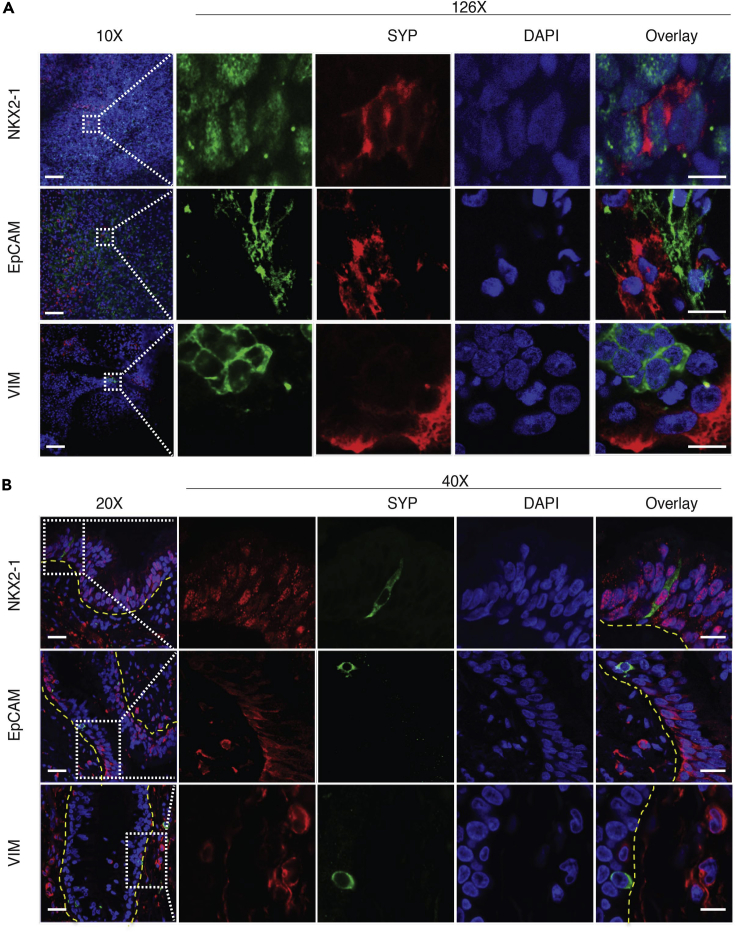
To comprehensively characterize and validate iPNECs, we co-labeled the differentiating cultures with NKX2.1, a primordial lung endoderm transcription factor; EPCAM, a pan-epithelial marker; and vimentin, VIM, a mesenchymal marker, in combination with the PNEC marker SYP. SYP+ iPNECs co-express NKX2.1 supporting a lung endodermal origin, but do not express VIM or EPCAM (Figures 2A and 2B). Human lung tissue sections were used to validate this co-staining pattern: primary PNECs within these sections also co-express NKX2.1, but not EPCAM or VIM, supporting our iPNEC data (Figure 2B). NKX2.1 also identifies neuronal cells developing in the forebrain (Germain et al., 2013, Goulburn et al., 2011); therefore, to exclude the possibility that our differentiation was creating forebrain neurons we stained for additional neuronal-specific marker, microtubule-associated protein 2 (MAP2), at Days 31 and 60 of differentiation. No MAP2 expression was evident at either of these time points (data not shown).
Figure 2.
Characterization of Human iPNECs
(A) Representative immunofluorescence (IF) images of ALI cultures at Day 31 of directed differentiation: SYP+ iPNECs (red), NKX2.1, EPCAM, and vimentin (VIM) (all green). Left panels (20×) represent low-power images (scale bars, 200 μm), and right panels (40×) show magnified views of the marked areas (scale bars, 10 μm).
(B) Representative IF images of adult human distal lung sections: SYP+ iPNECs (green), NKX2.1, EPCAM, and VIM (all red). Yellow dotted lines demarcate the airways. Left panels (20X) represent low-power images, and right panels (40×) show magnified views of the marked areas (scale bars, 50 μm). Nuclei are counterstained with DAPI (blue).
NOTCH Inhibition Augments iPNEC Specification and Phenotypic Complexity
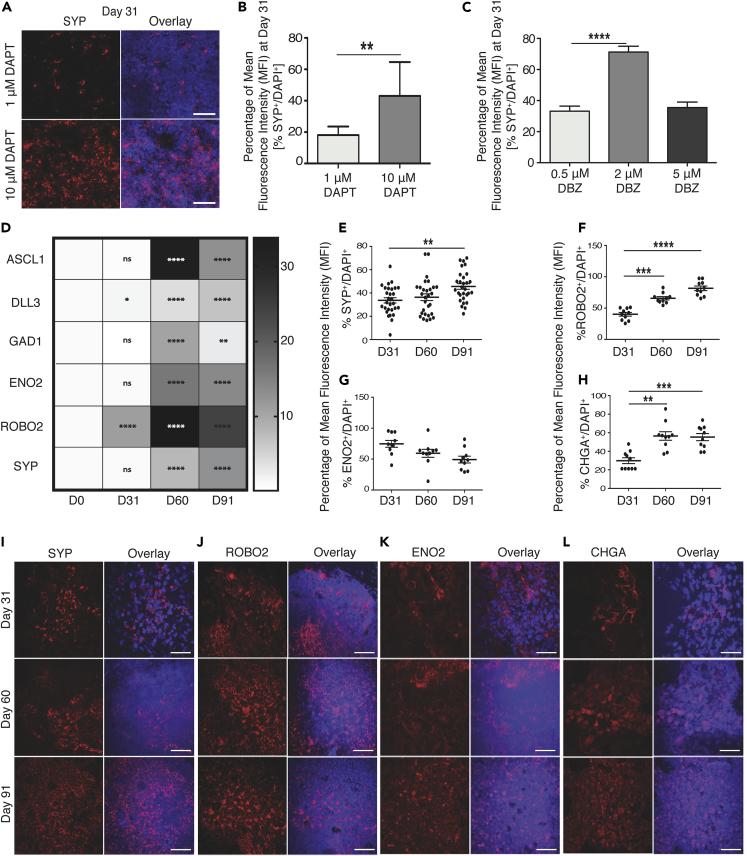
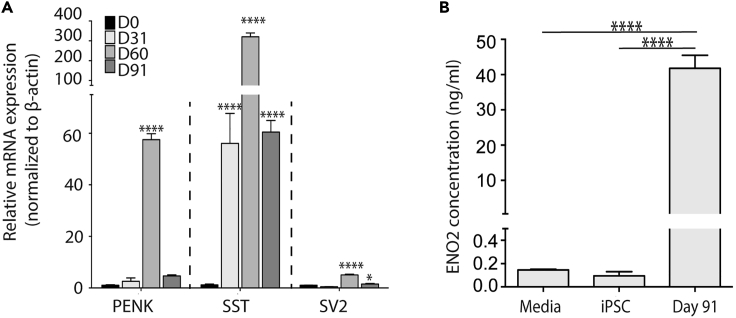
A transcriptional program regulated by transcription factor ASCL1 is required for activating the neuroendocrine lineage in developing lung to generate PNECs (Linnoila, 2006). ASCL1 expression is known to be repressed by NOTCH signaling, which supports expansion and differentiation of lung basal and secretory cells, respectively. During iPSC differentiation, ASCL1 mRNA is detectable from Day 10 (Figure S2E), preceding the appearance of SYP+ cells from Day 13 (Figure S2B). We also observe that expression of HEY1/HES1, downstream targets of the NOTCH pathway, peaks between Days 10 and 17 of differentiation (Figures S2F and S2G), whereas ASCL1 peaks at Day 31 (Figure S2E). This suggests that in our differentiation protocol, much like during development, there is an inverse correlation between activity of the Notch signaling pathway and ASCL1 expression. To evaluate the impact of Notch inhibition on neuroendocrine differentiation and expansion during our differentiation protocol, we performed a dose-response to γ-secretase/Notch inhibitor, 3tert-Butyl(2S)-2-[[(2S)-2-[[2-(3,5-difluorophenyl) acetyl] amino] propanoyl] amino]-2-phenylacetate (DAPT). As before, marker expression was quantified as a percentage of the MFI for the respective marker normalized to the MFI of nuclear marker DAPI. Continuous addition of 1, 10, and 20 μM DAPT to cultures from Day 17 onward resulted in a dose-dependent increase in the relative MFI of SYP at Day 31 (Figures S3A, 3A, and 3B). The effect of Notch inhibition was validated using a second Notch signaling inhibitor, dibenzazepine (DBZ), at 0.5, 2, and 5 μM. A 2-fold increase in relative MFI of SYP was observed when increasing the concentration of DBZ from 0.5 to 2 μM (Figures 3C and S3B). However, no further increase in the MFI ratio of SYP was observed with an increase to 5 μM DBZ treatment. These findings support inhibition of Notch signaling as an important mechanism for augmenting PNEC differentiation and expansion. To deduce the optimal concentrations of Notch inhibitors, DAPT and DBZ, required for iPNEC differentiation, we also co-labeled Day 31 cultures with cleaved caspase 3, an apoptotic marker to evaluate whether high concentrations of Notch inhibitors induced toxicity and cell death. Based on immunofluorescent staining, an increase in the caspase 3+ relative MFI percentage was observed at the highest concentration (20 μM DAPT and 5 μM DBZ) of the Notch inhibitors (Figures S3A and S3B), indicating that 10 μM DAPT or 2 μM DBZ were optimal for induction of iPNECs. These concentrations were used for all subsequent experiments. Extended culture of iPNECs with continuous addition of DAPT/DBZ for up to 90 days resulted in a significant increase in mRNA expression for representative PNEC genes ASCL1, DLL3, GAD1/GAD67, ENO2, ROBO2, and SYP (Figure 3D). In addition, there was a significant increase in SYP, ROBO2, and CHGA staining (Figures 3E, 3F, and 3H). ENO2 was robustly expressed by Day 31 with no further increase at Day 91 (Figure 3G). These data support a role for continuous repression of Notch signaling in augmenting iPNEC expansion and homeostasis. Interestingly, SYP, ROBO2, and ENO2 staining patterns show an intricate arborized network formation over time in culture. Using Sholl analysis (Figure S4A), we quantified this arborization as a change in the number of intersections (Figures S4B, S4D, and S4F) and branching density (Figures S4C, S4E, and S4G) of SYP+, ROBO2+, and ENO2+ iPNECs (Binley et al., 2014, Bird and Cuntz, 2019, Sholl, 1953, Stanko and Fenton, 2017, Ferreira et al., 2014). Such changes in the cellular complexity of the iPNECs likely reflect phenotypic maturation similar to that observed in the NEBs of adult lungs (Pan et al., 2004, Pan et al., 2006).
Figure 3.
Extended Notch Inhibition Increases iPNEC Specification and Phenotypic Complexity
(A–C) (A) Representative immunofluorescence (IF) images and quantification of SYP+ iPNECs (red) at Day 31 of differentiation in the presence of DAPT (1 μM and 10 μM). Quantification of SYP expression (MFI expressed as a percentage of total DAPI fluorescence) at Day 31 of differentiation in the presence of (B) DAPT (1 μM and 10 μM) (paired t test) and (C) DBZ (0.5 μM, 2 μM and 5 μM) (one-way ANOVA). Data are expressed as mean ± SEM from 3 independent experiments. ∗∗p < 0.01, ∗∗∗∗p < 0.0001.
(D–L) (D) Quantitative RT-PCR showing PNEC marker expression after 91 days of differentiation. Data are normalized to internal control β-actin and expressed as comparisons among Days 31, 60, and 91 with respect to Day 0. N = 3 experimental replicates each with 3 technical replicates. One-way ANOVA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001. MFI expressed as percentage of DAPI fluorescence for cells expressing SYP+ (E), ROBO2+ (F), ENO2+ (G), and CHGA+ (H) at Days 31, 60, and 91 of differentiation. One-way ANOVA. ∗∗p < 0.01, ∗∗∗∗p < 0.0001. Scale bars, 100 μm. Nuclei are counterstained with DAPI (blue). (E) Representative IF images show SYP+ (I), ROBO2+ (J), ENO2+ (K), and CHGA+ (L) cells at Days 31, 60, and 91 of differentiation (all red).
See also Figures S3 and S4.
Transcriptomic Characterization of iPNECs
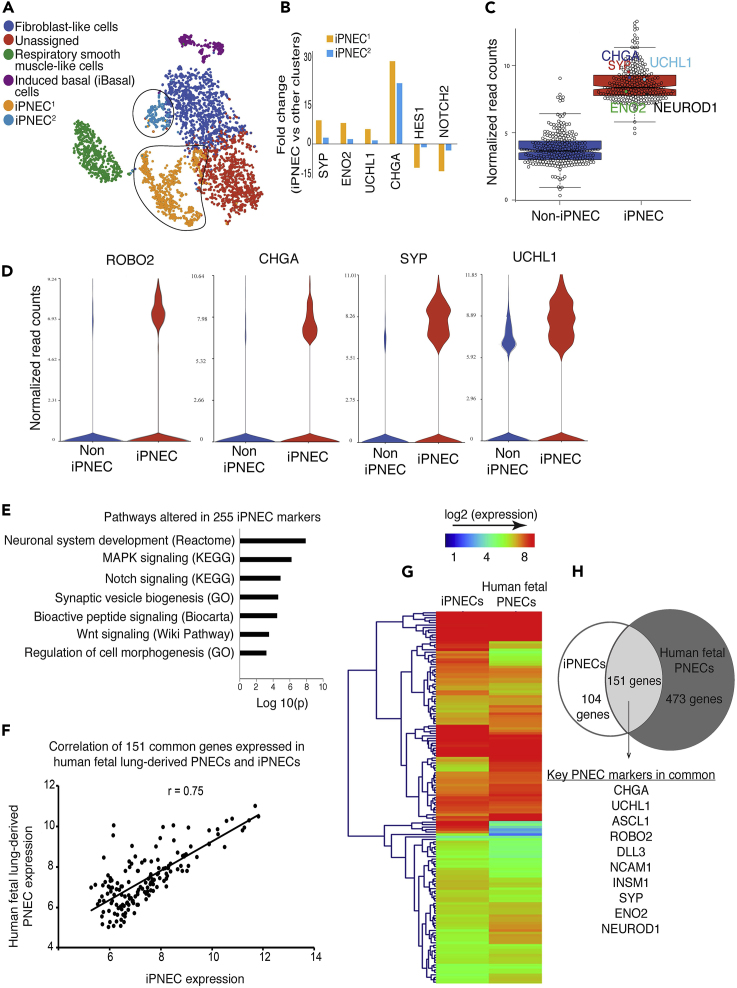
To evaluate the transcriptomic profile of iPNECs, we performed single-cell RNA sequencing (scRNA-seq) of cultures maintained with continuous treatment with 10 μM DAPT at Day 91 of iPSC differentiation (for schematic see Figure S5A). Two independent biological replicates were sequenced and analyzed. Based on the quality parameters as detailed in Transparent Methods, we selected a set of 1,705 and 1,502 cells from replicates 1 and 2, respectively, for analysis. The two replicates were corrected for any batch effect before further analysis. A subset of 1,171 most variable genes that showed a high level of concordance between the two replicates was selected for further downstream analysis (Pearson correlation coefficient, r = 0.94, Figure S5B). A flowchart describing the bioinformatics analysis pipeline for the scRNA-seq data is shown in Figure S5C. Unsupervised graph-based clustering (false discovery rate, FDR p < 0.05) and non-linear dimensionality reduction distributes the cells into six independent cell populations on Day 91 of differentiation (Figure 4A). Two of these clusters, labeled iPNEC1 and iPNEC2, representing >30% of the cells captured, were defined by a transcriptomic signature to reflect a neuroendocrine lineage determined by the expression of known PNEC genes including, but not limited to, SYP, CHGA, PGP9.5, ENO2, and ROBO2. Further in-depth analysis revealed that these two clusters are separated from the other cell clusters by differential expression (fold change, FC) of key PNEC marker genes SYP, ENO2, UCHL1, and CHGA, and Notch pathway genes, HES1 and NOTCH2 (Figure 4B). The larger cluster, iPNEC1 (yellow dots), has a higher degree of Notch repression and a higher FC of SYP, ENO2, UCHL1, and CHGA than the smaller cluster, iPNEC2 (blue dots). For all subsequent analyses the two clusters, iPNEC1 and iPNEC2 were grouped together as iPNECs. A set of 255 differentially enriched genes (FDR p < 0.05; FC ≥ 1.5) was identified using gene set enrichment analysis against a compendium of 350 different human tissues and cell types; we refer to this set of genes as iPNEC-associated genes. Figure S5C shows a flowchart describing the process of identifying 255 iPNEC-associated genes. In a genome-wide comparison, these 255 genes are significantly enriched (FDR p < 0.5, FC ≥ 1.5) in the putative iPNEC cell population compared with other populations (Figure 4C) and includes established PNEC marker genes including SYP, CHGA, and ROBO2 (Figure 4D). UCHL1, although highly enriched within the iPNEC cluster, is also expressed in the non-iPNEC population. According to the LungMAP database, UCHL1 is also expressed to some extent by lung mesenchyme (www.lungmap.net). We observed co-differentiation of mesenchymal-like clusters in our Day 91 cultures (VIM+ fibroblast-like and ACTA2+ smooth muscle cells in Figure 4A), which could contribute to UCHL1 expression within the non-iPNEC population. PNECs receive stimulation from neurotransmitters/neurosecretions innervating the lung and respond to such stimuli by releasing hormones (neuropeptides and amines). PNECs, therefore, have important functions in assimilating characteristics of both the nervous and endocrine systems. Accordingly, analysis of the pathways most enriched in the 255 gene set includes neuronal system development, synaptic vesicle biogenesis, and bioactive peptide signaling (Figure 4E), consistent with some of the known functions of PNECs. We also observe Notch signaling as one of the most differentially altered pathways (Figure 4G), reflecting the fact that continued Notch inhibition was administered to induce a neuroendocrine fate.
Figure 4.
Transcriptomic Characterization of iPNECs
(A) tSNE plot identifies six cell clusters in two replicates of 91-day old iPSC differentiated cells.
(B) Expression of representative genes between iPNECs1 and iPNECs2. Fold change represents differential expression between iPNEC1/2 clusters versus other cells.
(C) Box whisker plot comparing 255 iPNEC-associated genes within iPNEC and non-iPNEC populations. Key PNEC markers are shown (CHGA, SYP, UCHL1, ENO2, and NEUROD1).
(D) Violin plots showing expression of individual PNEC marker genes (ROBO2, CHGA, SYP, and UCHL1) within iPNEC and non-iPNEC populations.
(E) Signaling pathways altered within 255 iPNEC-associated genes.
(F) Scatterplot showing concordance of expression of 151 common genes (Pearson correlation coefficient, r = 0.75) in Day 91 iPNECs and human fetal lung-derived primary PNECs pooled from gestational weeks 15, 18, and 21.
(G) Heatmap comparing the gene expression signature between iPNECs and human fetal lung-derived primary PNECs pooled from gestational weeks 15, 18, and 21.
(H) Venn diagram showing the number of genes overlapping between iPNECs and human fetal lung-derived primary PNECs and the known key PNEC markers that are common between the two samples.
See also Figures S5 and S6.
Given the limited number of PNECs in human lung, performing RNA sequencing on primary PNECs is challenging. We, therefore, procured the only available scRNA-seq dataset of human fetal lung explants pooled from gestational weeks 15, 18, and 21 (EMBL-EBI ArrayExpress Database: E-MTAB-8221) (Miller et al., 2018) and compared the neuroendocrine cluster (220 cells) with the iPNEC cluster within our scRNA-seq dataset (GEO accession number: GSE146990). Comparative analysis revealed a significant correlation coefficient (r = 0.75) with a high concordance of 151 genes found to be commonly expressed between iPNECs and fetal PNECs (Figure 4F). One-way hierarchical clustering indicates an overall similar gene expression pattern between iPNECs and human primary fetal PNECs (Figure 4G) including expression of all known major PNEC genes such as SYP, CHGA, PGP9.5, ROBO2, ASCL1, NCAM1, ENO2, DLL3, and NEUROD1 (Figure 4H and Tables S1, S2, and S3), indicating considerable overlap between the transcriptomes of iPNECs and primary human fetal PNECs.
PNECs are considered neuroepithelial in nature; however, we did not observe co-staining of iPNECs or primary PNECs in human lung tissue sections stained with epithelial marker EPCAM (Figures 2A and 2B). To further clarify the whether EPCAM is expressed in PNEC we further analyzed the scRNA-seq data. In fetal PNECs, Miller et al., identified a neuroendocrine cell population within a computationally extracted epithelial cluster based on high EPCAM expression (Miller et al., 2018). Detailed analysis of the human fetal lung scRNA-seq data enabled a differentiation of two subsets of fetal PNECs based on EPCAM expression: of the 220 fetal PNECs 77 were EPCAM+ and 143 EPCAM−. Analysis of our 255 PNEC-associated genes in EPCAM− fetal PNECs indicated a significant concordance of 243 genes between both datasets (read count ≥2, r = 0.6) (Figure S5D). We, therefore, demonstrate that (1) a majority of human fetal PNECs do not express EPCAM and (2) iPNECs have significant gene expression concordance with human fetal PNECs.
iPNECs Express and Secrete Major Neuropeptides
PNECs contain vesicles filled with neuropeptides, amines, and neurotransmitters that are secreted upon stimulation. Recently, neuropeptides proenkephalin (PENK), somatostatin (SST), and neurotransmitter synaptic vesicle protein 2 (SV2) have been shown to be secreted by primary PNECs (Branchfield et al., 2016). Differentiation cultures at days 31, 60, and 91 containing greater than 30% iPNECs (Figures 3 and 4) express PENK, SST, and SV2 at the mRNA level (Figure 5A). ENO2 secretion has been widely used as a biochemical marker to monitor the progression of PNEC-associated disorders, namely SCLC and pulmonary neuroendocrine tumors (Anastasiades et al., 1987, Kasprzak et al., 2007). Immunostaining of our cultures at Day 91 confirms that the majority of SYP+ cells co-express ENO2 (Figure S6B). We, therefore, proceeded to perform an ELISA to evaluate whether SYP+ iPNECs also secrete ENO2. Media were collected from the basolateral compartment of Day 91 ALI cultures used for the scRNA-seq experiment using basal media and iPSC-conditioned media as negative controls and recombinant ENO2 as a positive control. Day 91 differentiation cultures, containing >30% iPNECs (Figures 3 and 4), expressed ENO2 at mRNA (Figures 3D and S6A) and protein levels (Figure 5B) in the absence of external stimulation, showing similarity in terms of their gene expression profile to primary PNECs as previously shown (Cutz et al., 2008). It is important to note that scRNA-seq analysis indicates that ENO2 expression is only found in the defined iPNEC populations (Figure S6A) supporting selective release of ENO2 from iPNEC in our Day 91 cultures.
Figure 5.
Neuropeptide and Marker Expression Profile of iPNECs
(A) qRT-PCR showing the neuropeptide profile of cultures at Day 0, 31, 60, and 91 of differentiation. Data are normalized to internal control β-actin and expressed as comparisons among Days 31, 60, and 91 with respect to Day 0. N = 3 experimental replicates each with 3 technical replicates. One-way ANOVA. ∗p < 0.05, ∗∗∗∗p < 0.0001.
(B) Enzyme-linked immunosorbent assay (ELISA) data showing secreted levels of enzyme ENO2 in the culture media. N = 4 experimental replicates. One-way ANOVA, ∗∗∗∗p < 0.0001.
See also Figure S6.
Human Basal Cells Are Capable of Differentiating into PNECs
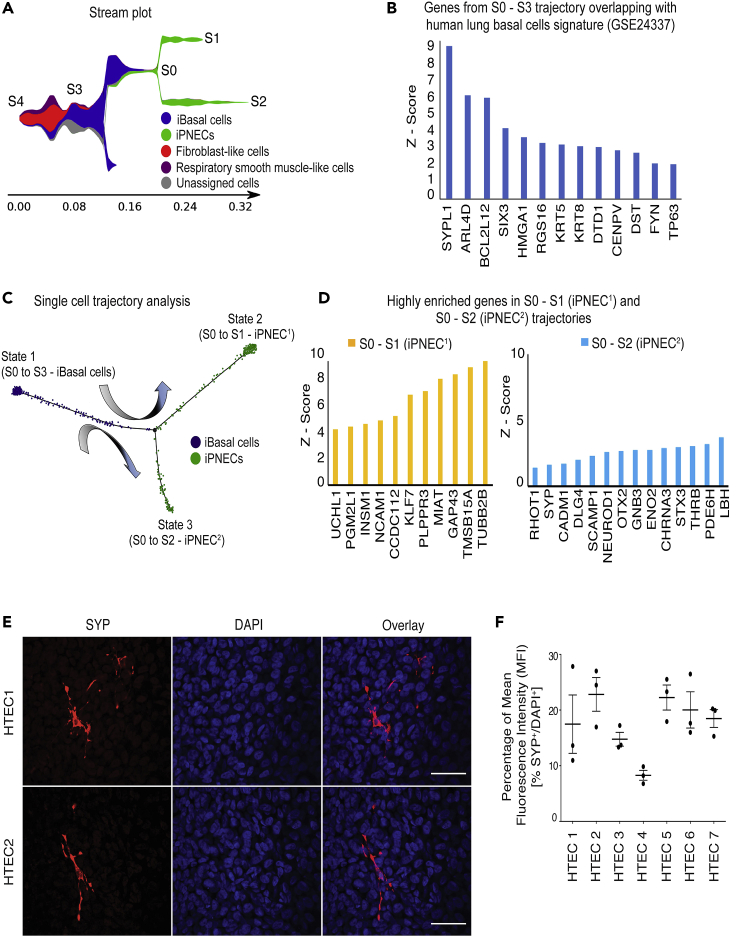
Lineage tracing studies in mice have indicated that TP63+ basal cells can give rise to PNECs (Yang et al., 2018). To determine whether human basal cells are also able to differentiate into PNECs, we performed two experiments: (1) pseudotime analysis of our human differentiation Day 91 iPNEC scRNA-seq data and (2) ALI differentiation of purified human tracheal epithelial cells (HTECs). We analyzed the scRNA-seq data of Day 91 differentiation cultures to track cell population changes across the differentiation trajectory (Figure 4). A stream plot was generated using Stream and Monocle3 for easy visualization of the different cell types in our cultures projected onto the differentiation trajectory “tree” according to their pseudotime locations and the distances from their assigned branches (Figure 6A). Based on the cell state during the reprograming each cell type/population is assigned to a distinct “branch” of the “tree” representing a specific cell fate choice/developmental lineage that proceeds down a distinct path. Cells on the same or nearby branches are predicted to be hierarchically related. The stream plot (Figure 6A) indicates that several different cell types are present in our Day 91 differentiation cultures including basal cells, respiratory smooth muscle-like cells, fibroblast-like cells, and iPNECs. We demonstrate that the key genes expressed in the S3 – S0 branch overlap with the published human lung basal cell database (GSE24337), confirming that iBasal cells in this culture possess a transcriptomic signature resembling that or primary human basal cells (Figure 6B). We tracked six key PNEC marker genes, INSM1, UCHL1, NCAM1, SYP, NEUROD1, and ENO2 across the branches where a significant increase in their expression was detected as the cells transition from the iBasal cell state into the two iPNEC trajectories S0 – S1 or S0 – S2 (p < 0.001, Figures S7A–S7F). To further confirm the differentiation of iBasal cells to iPNECs, we used the reverse graph embedding tool in Moncole3 to reconstruct an unbiased lineage trajectory and assigned iBasal cells as the starting cell lineage (Figure 6C). Cells start at the root (State 1) and progress to one of the two alternative reprogramming outcomes, denoted by State 2 or State 3. Monocole3 trajectories support that iBasal cells differentiate into two states, State 2 and State 3, which correspond to the two different states of iPNECs, iPNEC1 and iPNEC2 respectively. Therefore, it is possible that the two populations observed in graph-based clustering in Figure 4A represent two different physiological states of iPNECs, as observed by Monocle3 (Trapnell, 2020, Trapnell et al., 2014a, Trapnell et al., 2014b). Analysis of the differentially expressed genes between State 2 (iPNEC1) and State 3 (iPNEC2) indicates a difference in the level of the PNEC marker gene expression. UCHL1, NCAM1, and INSM1 are high in iPNEC1, whereas SYP and ENO2 are high in iPNEC2 (Figure 6D). Overall, the pseudotime trajectory analysis supports an iBasal cell predating iPNECs in the differentiation protocol described in this article. Purification and expansion of iPSC-derived basal cells is currently challenging. We therefore used HTECs to corroborate our in silico finding. HTECs, consisting almost exclusively of a self-renewing population of KRT5+/TP63+ basal cells (Figure S8A), were cultured at the ALI and stained for SYP. After more than 75 days of differentiation at the ALI, the distribution of SYP staining in the cultures was estimated to be 17.71% ± 2.49% based on the relative MFI percentage of SYP:DAPI (Figures 6E, 6F, and S8B). Using two independent approaches our data strongly suggest that human basal cells, both iPSC-derived and primary, are capable of giving rise to PNECs.
Figure 6.
Human Basal Cells Differentiate into PNECs
(A) Stream plot of scRNA-seq data showing lineage differentiation trajectories of iPSC-derived Day 91 cultures. Cells are assigned a distinct color based on cell types. Cells on the same branch are predicted to be hierarchically related.
(B) Z-score of highly enriched genes across S3 – S0 trajectory that overlaps with human lung basal cell microarray dataset GSE24337). Relative gene expression level across each trajectory was calculated as a Z-score of log2 (CPM+1) where CPM denotes counts per million. A positive Z-score indicates a score that is higher than the mean.
(C) Monocle-based lineage reconstruction of pseudotemporal trajectories superimposing both cell fate and cell states during differentiation from iBasal cells to iPNECs. Cells can differentiate from State1 (iBasal cells) to State 2 (iPNEC1) or State 3 (iPNEC2).
(D) Z score of highly enriched genes across trajectories S0 – S1 and S0 – S2 corresponding to two states of iPNECs, iPNEC1 and iPNEC2 respectively.
(E) Representative immunofluorescence images showing SYP+ PNECs (red) in >78 days ALI cultures from primary HTEC. Scale bars, 25 μm.
(F) Quantification of SYP+ PNECs in HTEC cultures expressed as MFI percentage of SYP fluorescence relative to total DAPI fluorescence.
Data are expressed as mean ± SEM. N = 7 experimental replicates. See also Figures S7 and S8.
Discussion
We have established a directed differentiation strategy for deriving iPNECs from human iPSCs that are transcriptomically similar to human fetal primary PNECs. Generation of these cells from a renewable and patient-specific cellular source overcomes limitations with regard to accessibility to sufficient primary human PNECs and enhances our capacity to model human diseases, like NEHI. Recent reports have demonstrated the generation of PNEC-like cells from human pluripotent stem cells; these studies, however, focused primarily on their sporadic presence in spheroid cultures or on their potential as lung cancer models (Chen et al., 2019, Konishi et al., 2016) (17, 18). To date, there has been no in-depth phenotypic and transcriptional characterization of these cells to validate their similarity to their primary counterparts. The iPNEC differentiation protocol described in this article is adapted from our previously established airway epithelial differentiation protocol to specifically promote differentiation toward the neuroendocrine lineage (Firth et al., 2014). Using this model we are able to generate iPNECs expressing all major PNEC marker genes including SYP, CHGA, PGP9.5, ROBO2, and ENO2, consistent with primary human and rodent PNECs (Branchfield et al., 2016, Linnoila, 2006, Sunday, 1996). By specifically adapting the culture conditions to favor expansion of PNECs we are able to generate cultures with an increased efficiency of generating iPNECs over currently published protocols (Chen et al., 2019). Our differentiation is robust across multiple iPSC lines despite inherent variability between stem cells lines/clones with regard to their differentiation efficiency. Furthermore, differentiation protocols, such as those developed in this study, rely on the sequential application of growth factors and cytokines directing differentiation toward lung epithelium: at any given point cells may be captured at varied stages of differentiation and maturation, again contributing to heterogeneity. It should be noted that our iPNEC protocol does not utilize any purification steps and thus an endpoint culture composed of mesenchymal (niche) cells and epithelium is generated with the iPNECs. Isolation of iPSC-derived lung basal cells, akin to primary HTEC, and differentiation toward PNECs may reduce this variability; however, loss of putative niche cells may also impact efficiency. Such factors will be evaluated in a subsequent study (Ryan laboratory, unpublished data).
Interestingly, we found that exposure to an air interface was important for the continued development of iPNECs. This condition typically mimics the microenvironment of the postnatal and adult lung (Van Haute et al., 2009, Wong et al., 2015, Wong et al., 2012, Yamaya et al., 1992). This observation has not previously been reported, and although PNECs first appear in the embryonic lung when the lung is filled with amniotic fluid (Hooper and Harding, 1995, Morrisey and Hogan, 2010), it is in the adult lung, when the lung is exposed to air/oxygen, that PNECs are known to function as airway chemosensors (Garg et al., 2019, Linnoila, 2006, Nikolić et al., 2018, Sunday, 1996). Our protocol mimics this trajectory, with iPNECs being detected for the first time around Day 13 of differentiation, correlating with the early specification of NKX2.1-expressing anterior foregut endoderm cells (Figure S1B); at this stage the cultures are still submerged. Persistent culture in submerged growth conditions after specification of the primordial lung progenitor results in complete loss of the SYP-expressing cells suggesting that this microenvironment is not conducive to their continued growth and proliferation. There are a number of factors that may influence this including exposure to a more hypoxic environment upregulating hypoxia response genes such as HIF-1α and HIF-2α causing an upregulation of Notch signaling, as previously reported (Gerovac et al., 2014). Notch upregulation, in turn, is known to be antagonistic toward PNEC fate induction (Yao et al., 2018). Existing literature has not specifically indicated a requirement for an ALI during differentiation to generate PNEC-like cells (Chen et al., 2019, Konishi et al., 2016). Other iPSC-derived PNECs have been generated in submerged cultures by formation of spheroids/organoids perhaps accounting for differences in iPNEC yield observed between these protocols and our current protocol. The continued and strong suppression of Notch signaling favors PNEC specification in our protocol. We did not observe significant numbers of ciliated cells under these conditions in contrast to our original protocol. We speculate that this may be due to continuous and increased (10-fold higher) levels of Notch suppression. These cultures were observed for over 3 months, and a substantial increase in both the number and the phenotypic complexity of iPNECs was evident. Although it is currently not possible to determine the precise maturity of iPNECs due to a lack of adult human isolated PNECs for direct comparison, our data suggest that the increase in branched network formation in iPNECs may correlate with their cellular maturation over time.
PNECs are considered neuroepithelial in nature (Cutz et al., 2013, Verckist et al., 2017) as they arise from the undifferentiated epithelial progenitors located at the Id2+ (inhibitor of DNA binding 2) distal tips of the branching epithelium along with all other epithelial cell lineages within the lung (Rawlins et al., 2009, Song et al., 2012). However, neither primary nor induced SYP+ PNECs were observed to co-express the epithelial marker, EPCAM. This finding is in contrast to a recent study in mouse (Montoro et al., 2018), where analysis of scRNA-seq on purified EPCAM+ tracheal epithelial cells identified a specific cell cluster as neuroendocrine. It should be noted that no analysis of EPCAM-negative cells was performed. There are also no published studies that show co-staining of human PNECs with EPCAM. Furthermore, we found that 65% of human fetal PNECs analyzed in the dataset from Miller et al. is EPCAM− and shows a high level of gene expression concordance with our iPNECs (Miller et al., 2018). Interestingly, we did not observe CGRP expression in our iPNECs. CGRP is often used as a PNEC marker; however, its expression and distribution in PNECs appears to vary among species. CGRP expression in humans, for example, has been observed to correlate to a cancer phenotype (Edbrooke et al., 1985) and observed to only be present in ∼20% of primary lung PNECs (Weichselbaum et al., 2005). In human lungs, bombesin/GRP is more robust and widely accepted as a marker of fetal-stage PNECs (Linnoila, 2006, Sunday, 1996, Sunday et al., 1991). Consistent with the literature, we also observe GRP expression peaking during the earlier stages of iPNEC differentiation (Days 13 and 17).
Primary PNECs are known to contain neuropeptides that are stored in dense core vesicles in the cytoplasm and are secreted in response to external stimuli such as hypoxia, nicotine, or tobacco smoke (Branchfield et al., 2016, Cutz et al., 2013, Linnoila, 2006). The iPNECs generated in this study exhibit similar secretion of ENO2 and expression of neuropeptides. ROBO genes that help form sensory organoid clusters in vivo via a Slit-ligand-dependent process are also a known feature of mouse PNECs (Branchfield et al., 2016).
Due to a lack of gene expression data focusing on PNECs only a small list of genes is available to define and study PNEC. We hypothesize that our 255 iPNEC-associated genes could serve as a more comprehensive gene set for PNECs and acknowledge that it will be important to validate the iPNEC-associated gene signature in adult human PNECs. Interestingly, our scRNA-seq data indicated two distinct iPNEC clusters from both the t-distributed Stochastic Neighbor Embedding (tSNE) and stream plot analyses, with additional analysis of cell trajectory predicting that the two clusters correspond to two individual physiological states. In a recent publication, rare Notch2+ NEstem and intermediate transit amplifying Hes1+ Notch-active NE cells that contribute to epithelial repair during lung injury in mice were identified (Ouadah et al., 2019). Despite maintaining our 90-day iPSC PNEC differentiation in the continued presence of Notch inhibition we are able to observe two individual iPNEC populations with differential levels of Notch2 and Hes1 expression. We hypothesize that these two distinct subpopulations, iPNEC11 (low Notch) and iPNEC2 (high Notch), may resemble the PNEC heterogeneity observed by Ouadah and colleagues (Ouadah et al., 2019); however, this will require further validation and development of a similar injury model using human iPSC-derived PNECs.
In conclusion, we have successfully derived PNECs from human iPSCs that resemble the known phenotype of human fetal and adult primary PNECs at the cellular and molecular levels. This represents an important step toward creating human cellular models for investigation of lung diseases characterized by PNEC hyperplasia, such as NEHI.
Limitations of the Study
In this study, we undertook the directed differentiation of human iPSCs into PNECs and performed robust characterization of these iPNECs at the cellular and molecular levels. Although we were able to demonstrate significant gene expression concordance with human fetal lung PNECs, it will be important to also make comparisons to the transcriptomes of human adult lung PNECs as these data become available to determine the maturation state of our iPNECs. With <1% of the cellular composition of the adult lung being PNECs, it remains challenging to capture sufficient cells to profile. Currently, methods for the specific isolation/enrichment of PNECs do not exist, making such comparisons extremely challenging. Although we have included experiments that support a functional profile for the iPNECs such as the expression and secretion of major neuropeptides (Figure 5), additional experiments will be important to further establish their functional competency. Such experiments might include measurement of neuropeptide release under hypoxic conditions and mechanical stress or in response to allergic stimulants, which would also be beneficial for disease modeling. Although analysis of our scRNA-seq datasets indicates the presence of two PNEC populations, in the current article the functional differences between these two populations were not addressed. In future experiments it would be interesting to determine whether these are two independent PNEC populations or whether they are cells at different stages along the differentiation trajectory. It also remains unclear whether primary or induced PNECs can be expanded ex vivo. This would require an extensive series of experiments to optimize both the purification and culture of both PNEC and iPNECs; however, this would be incredibly valuable to further study the biology of PNECs. Finally, to definitively confirm a basal cell origin of PNECs, lineage tracing will need to be performed in both primary HTECs and iPSC-derived cultures requiring the generation of iPSC reporter lines.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We acknowledge the use of the services at the USC Optical Imaging Facility, Genomics Core in the Department of Biomedical Sciences at the Cedars-Sinai Medical Center, and the LungMAP Consortium. This work was supported by NIH grants R35HL135747, R00NS077435, and R01NS097850; the New York Stem Cell Foundation; the USC Broad Innovation Award; and the Hastings Foundation. J.K.I. is a New York Stem Cell Foundation-Robertson Investigator. Z.B. is Ralph Edgington Chair in Medicine and Hastings Professor of Medicine. The LungMAP Consortium (U01HL122642) and the LungMAP Data Coordinating Center (1U01HL122638) are funded by the National Heart, Lung, and Blood Institute (NHLBI).
Author Contributions
Conceptualization, P.H., A.L.R., and Z.B.; Methodology, P.H., J.K.I., A.L.R., and Z.B.; Investigation, P.H., V.P., S.L.B, J.R.S., B.A.C., B.R.S., A.C., A.J.M., and J.R.S.; Writing – Original Draft, P.H., A.L.R.; Writing – Review & Editing, P.H., A.L.R., J.K.I., and Z.B.; Funding Acquisition, J.K.I. and Z.B.; Resources, J.K.I., A.L.R., and Z.B.; Supervision, J.K.I., A.L.R., and Z.B.
Declaration of Interests
J.K.I. is a co-founder of AcuraStem Incorporated. J.K.I. declares that he is bound by confidentiality agreements that prevent him from disclosing details of his financial interests in this work. All data needed to evaluate the conclusions in the article are present in the article paper and/or the Supplemental Information. Additional data related to this article may be requested from the authors.
Published: May 22, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101083.
Contributor Information
Justin K. Ichida, Email: ichida@usc.edu.
Amy L. Ryan (Firth), Email: amy.firth@med.usc.edu.
Zea Borok, Email: zborok@med.usc.edu.
Data and Code Availability
We have uploaded and made available our single-cell RNA sequencing data generated from our Day 91 differentiation cultures on Gene Expression Omnibus (GEO) database under accession number: GSE146990.
Supplemental Information
List of genes for human fetal PNECs that did not overlap with iPNECs.
References
- Anastasiades K.D., Mullins R.E., Conn R.B. Neuron-specific enolase. Assessment by ELISA in patients with small cell carcinoma of the lung. Am. J. Clin. Pathol. 1987;87:245–249. doi: 10.1093/ajcp/87.2.245. [DOI] [PubMed] [Google Scholar]
- Binley K.E., Ng W.S., Tribble J.R., Song B., Morgan J.E. Sholl analysis: a quantitative comparison of semi-automated methods. J. Neurosci. Methods. 2014;225:65–70. doi: 10.1016/j.jneumeth.2014.01.017. [DOI] [PubMed] [Google Scholar]
- Bird A.D., Cuntz H. Dissecting Sholl analysis into its functional components. Cell Rep. 2019;27:3081–3096.e5. doi: 10.1016/j.celrep.2019.04.097. [DOI] [PubMed] [Google Scholar]
- Branchfield K., Nantie L., Verheyden J.M., Sui P., Wienhold M.D., Sun X. Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science. 2016;351:707–710. doi: 10.1126/science.aad7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.J., Poran A., Unni A.M., Huang S.X., Elemento O., Snoeck H.-W., Varmus H. Generation of pulmonary neuroendocrine cells and SCLC-like tumors from human embryonic stem cells. J. Exp. Med. 2019;216:674–687. doi: 10.1084/jem.20181155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutz E. Hyperplasia of pulmonary neuroendocrine cells in infancy and childhood. Semin. Diagn. Pathol. 2015;32:420–437. doi: 10.1053/j.semdp.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Cutz E., Gillan J.E., Bryan A.C. Neuroendocrine cells in the developing human lung: morphologic and functional considerations. Pediatr. Pulmonol. 1985;1:S21–S29. [PubMed] [Google Scholar]
- Cutz E., Pan J., Yeger H., Domnik N.J., Fisher J.T. Recent advances and controversies on the role of pulmonary neuroepithelial bodies as airway sensors. Semin. Cell Dev. Biol. 2013;24:40–50. doi: 10.1016/j.semcdb.2012.09.003. [DOI] [PubMed] [Google Scholar]
- Cutz E., Yeger H., Pan J., Ito T. Pulmonary neuroendocrine cell system in health and disease. Curr. Respir. Med. Rev. 2008 http://www.eurekaselect.com/83021/article [Google Scholar]
- Cutz E., Yeger H., Pan J. Pulmonary neuroendocrine cell system in pediatric lung disease-recent advances. Pediatr. Dev. Pathol. 2007;10:419–435. doi: 10.2350/07-04-0267.1. [DOI] [PubMed] [Google Scholar]
- Edbrooke M.R., Parker D., McVey J.H., Riley J.H., Sorenson G.D., Pettengill O.S., Craig R.K. Expression of the human calcitonin/CGRP gene in lung and thyroid carcinoma. EMBO J. 1985;4:715–724. doi: 10.1002/j.1460-2075.1985.tb03688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira T.A., Blackman A.V., Oyrer J., Jayabal S., Watt A.J., Sjostrom P.J., van Meyel A.J. Neuronal morphometry directly from bitmap images. Nature Methods. 2014;11:982–984. doi: 10.1038/nmeth.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.L., Dargitz C.T., Qualls S.J., Menon T., Wright R., Singer O., Gage F.H., Khanna A., Verma I.M. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2014;111:E1723–E1730. doi: 10.1073/pnas.1403470111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A., Sui P., Verheyden J.M., Young L.R., Sun X. Chapter Three - consider the lung as a sensory organ: a tip from pulmonary neuroendocrine cells. In: Wellik D.M., editor. Current Topics in Developmental Biology, Organ Development. Academic Press; 2019. pp. 67–89. [DOI] [PubMed] [Google Scholar]
- Germain N.D., Banda E.C., Becker S., Naegele J.R., Grabel L.B. Derivation and isolation of nkx2.1-positive basal forebrain progenitors from human embryonic stem cells. Stem Cells Dev. 2013;22:1477–1489. doi: 10.1089/scd.2012.0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerovac B.J., Valencia M., Baumlin N., Salathe M., Conner G.E., Fregien N.L. Submersion and hypoxia inhibit ciliated cell differentiation in a Notch-dependent manner. Am. J. Respir. Cell Mol. Biol. 2014;51:516–525. doi: 10.1165/rcmb.2013-0237OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulburn A.L., Alden D., Davis R.P., Micallef S.J., Ng E.S., Yu Q.C., Lim S.M., Soh C.-L., Elliott D.A., Hatzistavrou T. A targeted NKX2.1 human embryonic stem cell reporter line enables identification of human basal forebrain derivatives. Stem Cells. 2011;29:462–473. doi: 10.1002/stem.587. [DOI] [PubMed] [Google Scholar]
- Gu X., Karp P.H., Brody S.L., Pierce R.A., Welsh M.J., Holtzman M.J., Ben-Shahar Y. Chemosensory functions for pulmonary neuroendocrine cells. Am. J. Respir. Cell Mol. Biol. 2014;50:637–646. doi: 10.1165/rcmb.2013-0199OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henke R.M., Meredith D.M., Borromeo M.D., Savage T.K., Johnson J.E. Ascl1 and Neurog2 form novel complexes and regulate Delta-like3 (Dll3) expression in the neural tube. Dev. Biol. 2009;328:529–540. doi: 10.1016/j.ydbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper S.B., Harding R. Fetal lung liquid: a major determinant of the growth and functional development of the fetal lung. Clin. Exp. Pharmacol. Physiol. 1995;22:235–247. doi: 10.1111/j.1440-1681.1995.tb01988.x. [DOI] [PubMed] [Google Scholar]
- Johnson D.E., Georgieff M.K. Pulmonary neuroendocrine cells: their secretory products and their potential roles in health and chronic lung disease in infancy. Am. Rev. Respir. Dis. 1989;140:1807–1812. doi: 10.1164/ajrccm/140.6.1807. [DOI] [PubMed] [Google Scholar]
- Johnson D.E., Kulik T.J., Lock J.E., Elde R.P., Thompson T.R. Bombesin-, calcitonin-, and serotonin-immunoreactive pulmonary neuroendocrine cells in acute and chronic neonatal lung disease. Pediatr. Pulmonol. 1985;1:S13–S20. [PubMed] [Google Scholar]
- Kasprzak A., Zabel M., Biczysko W. Selected markers (chromogranin A, neuron-specific enolase, synaptophysin, protein gene product 9.5) in diagnosis and prognosis of neuroendocrine pulmonary tumours. Pol. J. Pathol. 2007;58:23–33. [PubMed] [Google Scholar]
- Konishi S., Gotoh S., Tateishi K., Yamamoto Y., Korogi Y., Nagasaki T., Matsumoto H., Muro S., Hirai T., Ito I. Directed induction of functional multi-ciliated cells in proximal airway epithelial spheroids from human pluripotent stem cells. Stem Cell Rep. 2016;6:18–25. doi: 10.1016/j.stemcr.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauweryns J.M., Peuskens J.C. Neuro-epithelial bodies (neuroreceptor or secretory organs?) in human infant bronchial and bronchiolar epithelium. Anatomical Rec. 1972;172:471–481. doi: 10.1002/ar.1091720301. [DOI] [PubMed] [Google Scholar]
- Lauweryns J.M., Van Ranst L. Protein gene product 9.5 expression in the lungs of humans and other mammals. Immunocytochemical detection in neuroepithelial bodies, neuroendocrine cells and nerves. Neurosci. Lett. 1988;85:311–316. doi: 10.1016/0304-3940(88)90584-8. [DOI] [PubMed] [Google Scholar]
- Linnoila R.I. Functional facets of the pulmonary neuroendocrine system. Lab. Invest. 2006;86:425–444. doi: 10.1038/labinvest.3700412. [DOI] [PubMed] [Google Scholar]
- Miller A.J., Yu Q., Czerwinski M., Tsai Y.-H., Conway R.F., Wu A., Holloway E.M., Walker T., Glass I.A., Treutlein B. Basal stem cell fate specification is mediated by SMAD signaling in the developing human lung (preprint) Dev. Biol. 2018 doi: 10.1101/461103. [DOI] [Google Scholar]
- Montoro D.T., Haber A.L., Biton M., Vinarsky V., Lin B., Birket S.E., Yuan F., Chen S., Leung H.M., Villoria J. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisey E.E., Hogan B.L.M. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev. Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson B.R., Hartman B.H., Ray C.A., Hayashi T., Bermingham-McDonogh O., Reh T.A. Acheate-scute like 1 (Ascl1) is required for normal Delta-like (Dll) gene expression and Notch signaling during retinal development. Dev. Dyn. 2009;238:2163–2178. doi: 10.1002/dvdy.21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolić M.Z., Sun D., Rawlins E.L. Human lung development: recent progress and new challenges. Development. 2018;145 doi: 10.1242/dev.163485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouadah Y., Rojas E.R., Riordan D.P., Capostagno S., Kuo C.S., Krasnow M.A. Rare pulmonary neuroendocrine cells are stem cells regulated by Rb, p53, and Notch. Cell. 2019;179:403–416.e23. doi: 10.1016/j.cell.2019.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Luk C., Kent G., Cutz E., Yeger H. Pulmonary neuroendocrine cells, airway innervation, and smooth muscle are altered in Cftr null mice. Am. J. Respir. Cell Mol. Biol. 2006;35:320–326. doi: 10.1165/rcmb.2005-0468OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Yeger H., Cutz E. Innervation of pulmonary neuroendocrine cells and neuroepithelial bodies in developing rabbit lung. J. Histochem. Cytochem. 2004;52:379–389. doi: 10.1177/002215540405200309. [DOI] [PubMed] [Google Scholar]
- Rawlins E.L., Clark C.P., Xue Y., Hogan B.L.M. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development. 2009;136:3741–3745. doi: 10.1242/dev.037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler M.B., Bohn D.J., Bryan A.C., Cutz E., Rabinovitch M. Increased respiratory system resistance and bronchial smooth muscle hypertrophy in children with acute postoperative pulmonary hypertension. Am. J. Respir. Crit. Care Med. 1995;152:1347–1352. doi: 10.1164/ajrccm.152.4.7551393. [DOI] [PubMed] [Google Scholar]
- Sholl D.A. Dendritic organization in the neurons of the visual and motor cortices of the cat. J. Anat. 1953;87:387–406.1. [PMC free article] [PubMed] [Google Scholar]
- Song H., Yao E., Lin C., Gacayan R., Chen M.-H., Chuang P.-T. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc. Natl. Acad. Sci. U S A. 2012;109:17531–17536. doi: 10.1073/pnas.1207238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanko J.P., Fenton S.E. Quantifying branching density in rat mammary gland whole-mounts using the sholl analysis method. J. Vis. Exp. 2017:e55789. doi: 10.3791/55789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunday M.E. Pulmonary neuroendocrine cells and lung development. Endocr. Pathol. 1996;7:173–201. doi: 10.1007/BF02739921. [DOI] [PubMed] [Google Scholar]
- Sunday M.E., Hua J., Torday J. Bombesin may play a role in fetal lung growth and maturation in utero and in lung organ culture. Chest. 1991;99:21S. doi: 10.1378/chest.99.3_supplement.21s. [DOI] [PubMed] [Google Scholar]
- Trapnell C. monocle: clustering, differential expression, and trajectory analysis for single- cell RNA-Seq. Bioconductor version: release (3.10) 2020. [DOI]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. Pseudo-temporal ordering of individual cells reveals dynamics and regulators of cell fate decisions. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Haute L., De Block G., Liebaers I., Sermon K., De Rycke M. Generation of lung epithelial-like tissue from human embryonic stem cells. Respir. Res. 2009;10:105. doi: 10.1186/1465-9921-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verckist L., Lembrechts R., Thys S., Pintelon I., Timmermans J.-P., Brouns I., Adriaensen D. Selective gene expression analysis of the neuroepithelial body microenvironment in postnatal lungs with special interest for potential stem cell characteristics. Respir. Res. 2017;18:87. doi: 10.1186/s12931-017-0571-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaum M., Sparrow M.P., Hamilton E.J., Thompson P.J., Knight D.A. A confocal microscopic study of solitary pulmonary neuroendocrine cells in human airway epithelium. Respir. Res. 2005;6:115. doi: 10.1186/1465-9921-6-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Bear C.E., Chin S., Pasceri P., Thompson T.O., Huan L.-J., Ratjen F., Ellis J., Rossant J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012;30:876–882. doi: 10.1038/nbt.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong A.P., Chin S., Xia S., Garner J., Bear C.E., Rossant J. Efficient generation of functional CFTR-expressing airway epithelial cells from human pluripotent stem cells. Nat. Protoc. 2015;10:363–381. doi: 10.1038/nprot.2015.021. [DOI] [PubMed] [Google Scholar]
- Yamaya M., Finkbeiner W.E., Chun S.Y., Widdicombe J.H. Differentiated structure and function of cultures from human tracheal epithelium. Am. J. Physiol. 1992;262:L713–L724. doi: 10.1152/ajplung.1992.262.6.L713. [DOI] [PubMed] [Google Scholar]
- Yang Y., Riccio P., Schotsaert M., Mori M., Lu J., Lee D.-K., García-Sastre A., Xu J., Cardoso W.V. Spatial-temporal lineage restrictions of embryonic p63+ progenitors establish distinct stem cell pools in adult airways. Dev. Cell. 2018;44:752–761.e4. doi: 10.1016/j.devcel.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao E., Lin C., Wu Q., Zhang K., Song H., Chuang P.T. Notch signaling controls transdifferentiation of pulmonary neuroendocrine cells in response to lung injury. Stem Cells. 2018;36:377–391. doi: 10.1002/stem.2744. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of genes for human fetal PNECs that did not overlap with iPNECs.
Data Availability Statement
We have uploaded and made available our single-cell RNA sequencing data generated from our Day 91 differentiation cultures on Gene Expression Omnibus (GEO) database under accession number: GSE146990.