Abstract
Naked mole-rats (NMRs) have amongst the longest lifespans relative to body size of any known, non-volant mammalian species. They also display an enhanced stress resistance phenotype, negligible senescence and very rarely are they burdened with chronic age-related diseases. Alternative splicing (AS) dysregulation is emerging as a potential driver of senescence and ageing. We hypothesised that the expression of splicing factors, important regulators of patterns of AS, may differ in NMRs when compared to other species with relatively shorter lifespans. We designed assays specific to NMR splicing regulatory factors and also to a panel of pre-selected brain-expressed genes known to demonstrate senescence-related alterations in AS in other species, and measured age-related changes in the transcript expression levels of these using embryonic and neonatal developmental stages through to extreme old age in NMR brain samples. We also compared splicing factor expression in both young mouse and NMR spleen and brain samples. Both NMR tissues showed approximately double the expression levels observed in tissues from similarly sized mice. Furthermore, contrary to observations in other species, following a brief period of labile expression in early life stages, adult NMR splicing factors and patterns of AS for functionally relevant brain genes remained remarkably stable for at least two decades. These findings are consistent with a model whereby the conservation of splicing regulation and stable patterns of AS may contribute to better molecular stress responses and the avoidance of senescence in NMRs, contributing to their exceptional lifespan and prolonged healthspan.
Electronic supplementary material
The online version of this article (10.1007/s11357-019-00150-7) contains supplementary material, which is available to authorized users.
Keywords: Naked mole-rat, Heterocephalus glaber, Ageing, Splicing factors, hnRNPs, Srsfs, Klotho, CDKN2A, Cognitive function, DNA repair
Introduction
Naked mole-rats (NMRs; Heterocephalus glaber) are eusocial animals which live in subterranean colonies of up to 300 individuals with a single breeding female, in a manner analogous to bees or other social insects (Jarvis 1981; Brett 1991). Similarly sized rodents such as mice have a lifespan limited to 3–4 years, whereas NMRs typically survive to over 20 years of age, with a maximum recorded lifespan of 37 years in a still extant male. This is approximately 5-fold greater than that predicted from their small body size (Edrey et al. 2011). This extraordinary lifespan is accompanied by extended “healthspan”, with virtually no increase in Gompertzian mortality risk with age (Buffenstein et al. 2012; Ruby et al. 2018). Both fertility and neurogenesis persist for decades (Holmes 2016; Lewis and Buffenstein 2016a, b). NMRs are resistant to both spontaneous and experimentally induced cancer (Liang et al. 2010; Delaney et al. 2013; Deweerdt 2014). Moreover, NMRs demonstrate few age-related metabolic, cardiac or vascular changes (O'Connor et al. 2002; Csiszar et al. 2007; Buffenstein 2008; Grimes et al. 2014; Triplett et al. 2015a, b; Delaney et al. 2016). Indeed, considerable evidence suggests they maintain an overall youthful phenotype throughout their long lives (Buffenstein et al. 2019), for example they retain foetal haemoglobin throughout their lives, facilitating more efficient oxygen transport and clearly an evolved adaption to the hypoxic challenges of their colonial, subterranean habitation. Having resided in this harsh milieu since the early Miocene, they have evolved numerous adaptations to deal with the many conditions imposed by their subterranean habitation (e.g. hypercapnia, hypoxia, exposure to heavy metals, and plant secondary defence mechanisms) for review see (Lewis and Buffenstein 2016a, b). For example, NMRs can survive 18 min in pure nitrogen atmospheres and under these anoxic conditions switch from glucose-based anaerobic metabolism to that of fructose and appear to have several neuroprotective mechanisms to prevent neuronal death under hypoxia (Park et al. 2017; Munro et al. 2019). These long living, resilient animals therefore present an interesting paradox in ageing biology and an opportunity to examine fundamental mechanisms of ageing.
Several molecular explanations for the enhanced longevity and negligible levels of senescence (Buffenstein 2008) of NMRs have been proposed, most of which align to the established “hallmarks of ageing” (Lopez-Otin et al. 2013; Dammann 2017). NMRs have a low metabolic rate approximately 70% of that of a laboratory mouse (Yahav and Buffenstein 1991), enhanced expression of DNA repair genes (MacRae et al. 2015), resistance to oxidative stress (Lewis et al. 2013), well-maintained proteostasis (Perez et al. 2008; Pride et al. 2015), enhanced autophagy (Zhao et al. 2014), improved telomere maintenance (Evfratov et al. 2014), a stable epigenome (Tan et al. 2017), a well-maintained circulating and hepatic metabolome consistent with methionine and dietary restriction (Heinze et al. 2018; Lewis et al. 2018; Viltard et al. 2019) and concomitant high levels of cystathionine β-synthase (CBS) activation (Olecka et al. 2018), a novel immune system (Hilton et al. 2019), and show few signs of cellular senescence both in vitro (Zhao et al. 2018) and in vivo (Buffenstein 2008). The lack of senescence is of particular interest since targeted ablation of p16INK-4a positive senescent cells in genetically altered mouse models is able to extend lifespan and bring about improvements in many of the features of old age (Baker et al. 2011; Baker et al. 2016; Baar et al. 2017; Baar et al. 2018; Baker and Petersen 2018). Senescent cells are thought to drive ageing phenotypes by a number of mechanisms, not least the secretion of the senescence-associated secretory phenotype (SASP) and transcriptional changes brought about through chromatin remodelling (van Deursen 2014). Senotherapeutic interventions in man are in their infancy, but have been reported to have some beneficial effects in patients with idiopathic pulmonary fibrosis or chronic kidney disease (Hickson et al. 2019; Justice et al. 2019).
Cellular senescence can arise because of failure in molecular stress response as may occur during oncogenic insults (de Magalhaes and Passos 2018) and during development. A key component of this is the ability to maintain an active and plastic transcriptome. In response to internal and external stimuli, cells must be able to effectively reprogram their gene expression (Guzikowski et al. 2019). Alternative splicing (AS), one mechanism by which genes can produce a variety of functionally different isoforms from a single coding unit, is a key component of transcriptomic plasticity, and is especially responsive to stress (Nevo et al. 2012; Pai and Luca 2019). Unlike constitutive splicing, which occurs for all multi-exon genes, AS brings about the expression of a repertoire of multiple isoforms, which may be temporally or spatially regulated, or differ in response to stimuli. Alternative isoforms commonly have variant or antagonistic functions (Stamm et al. 2005; Villate et al. 2008). AS events may comprise alternative (cassette exons) exons, alternative 5′ or 3′ splice site usage, intron inclusion or deletions of parts of exons. More than one of these phenomena may be active in any given gene (Pai and Luca 2019). AS decisions are made by the binding of serine-arginine rich (SR) splicing activator proteins (SRSFs) and heterogeneous nuclear ribonucleoprotein particle (hnRNP) splicing inhibitor proteins to exon and intron splicing enhancer or silencer sequences around the splice sites (Cartegni et al. 2002). Each splice site is regulated by the combinational and competitive binding of a unique combination of activators and inhibitors (Smith and Valcarcel 2000). These activator and inhibitor proteins are collectively termed splicing factors. AS has previously been reported in NMRs, as in other higher organisms, including for important senescence-associated loci such as CDKN2A (Tian et al. 2015).
Correct regulation of AS is critical to ageing; genes encoding components of the splicing regulatory machinery are amongst the most dysregulated by age in human populations and in senescent human fibroblasts, endothelial cells, cardiomyocytes and astrocytes. Splicing factor expression is mainly downregulated in senescent cells of these subtypes, but tissue-specific differences in both the identity of affected splicing factors and directionality do exist (Harries et al. 2011; Holly et al. 2013; Latorre et al. 2017; Latorre et al. 2018a; Latorre et al. 2018b; Lye et al. 2019). The expression of splicing regulatory factor genes are also associated with lifespan and dietary restriction in mice and other species (Heintz et al. 2016; Lee et al. 2016; Lee et al. 2019a), and predictively linked with human ageing phenotypes in population studies (Lee et al. 2019b; Lye et al. 2019). Splicing factors expression is tightly connected to control of cell proliferation with splicing factors frequently mutated in cancer (Seiler et al. 2018). Finally, restoration of splicing factor expression using small molecules or targeted genetic interventions is able to reverse multiple features of senescence in aged human primary cells in vitro (Latorre et al. 2017; Latorre et al. 2018a, c).
We hypothesised that given the lack of visible signs of senescence in ageing NMRs and the importance of splicing factor regulation in the context of senescence, that splicing factor dysregulation and the consequent changes to the splicing patterns in ageing cells and tissues (Harries et al. 2011; Latorre et al. 2018b; Lye et al. 2019) may not be a feature of NMR ageing. We aimed to characterise the abundance of an a priori panel of 20 splicing factors known to be important in ageing and senescence from our previous work (Holly et al. 2013; Latorre et al. 2017; Latorre et al. 2018a, b, c), in a series of whole brain samples originating from embryonic NMRs all the way up to extreme old age. Brain expression levels of a panel of senescence-related genes, and AS patterns of a candidate series of functionally relevant brain isoforms previously identified to be altered in replicatively senescent human astrocytes (Lye et al. 2019) were also assessed as a functional output of splicing factor expression. We determined that although changes are evident in the brain expression of splicing factor genes between embryonic and adult states as expected, splicing factor expression trajectories remain static over the course of NMR ageing, as do the expression patterns of important alternatively spliced brain function genes. Furthermore we identified no increase in the expression of important molecular markers of cellular senescence (including isoforms of the Cdkn1a Tp53 and Cdkn2a genes), in accordance with the negligible senescence phenotype in these animals. Our data are supportive of a hypothesis that the extraordinary longevity of NMRs may arise at least in part from maintained transcriptomic molecular stress responses, conservation of regulated alternative splicing leading to avoidance of cellular senescence, a post-transcription mechanism of gene expression regulation.
Methods
Animal characteristics and husbandry
The NMRs used in this study were part of the well characterised Calico colony. The progenitors of these animals were collected in Kenya. All animals are microchipped at 90 days of age, providing each individual with a unique nine-digit identifying number. NMRs were housed in multi-chambered plexiglass burrow systems in animal rooms maintained at 28–30 °C and 30–50% relative humidity, in attempts to simulate climatic conditions in their native equatorial habitat. The animals were fed ad libitum with fruit and vegetables (bananas, apples, oranges, butternut squash, red bell pepper, romaine lettuce, cucumber, green beans, corn, carrots and red garnet yams) and supplemented with a high protein and vitamin enriched cereal (Pronutro, South Africa).
Animals, ranging from neonates to 22 years old, as well as pregnant females were euthanised using isoflurane and killed by cardiac exsanguination. Four to eight animals of both sexes were used in each age cohort (Table 1). Tissues were harvested from the embryos and animals of different ages and immediately flash frozen in liquid nitrogen and stored at -80 °C until analysed. Brain and spleen samples were used in these studies. We chose spleen because, as a lymphoid organ, it consists of large numbers of white blood cells. Most of the transcripts extracted from spleen will arise from B cells, T cells and mononuclear phagocytes and may allow a window on “inflammaging”, thought to be a prominent ageing mechanism (Zuo et al. 2019). Brain was chosen as splicing patterns are especially complex and functionally important in this organ (Karlsson and Linnarsson 2017), and dysregulated splicing is a feature of many neurodegenerative and neurodevelopmental diseases in brain (Nik and Bowman 2019). All animal use and experiments were approved by the Buck Institute Institutional Animal Care and Use Committee (IACUC) protocol number A10173.
Table 1.
Details of animals used in the study. Shown here are the numbers and age ranges of naked mole rats included in each age category in the current study
| Sample subset | Age range (days) | n |
|---|---|---|
| Foetus | – | 5 |
| 1 day | 1 | 6 |
| 2 weeks | 14 | 4 |
| 3–4 years | 1095–1793 | 8 |
| 5–6 years | 2163–2478 | 4 |
| 7–8 Years | 2631–2920 | 4 |
| 9–12 years | 3306–4264 | 6 |
| 13–15 years | 4745–5326 | 5 |
| 17–20 years | 6264–7300 | 5 |
| 21–22 years | 7842–8159 | 4 |
Candidate genes selected for analysis
An a priori list of splicing factor candidate genes were chosen based on associations with ageing or senescence in multiple human ageing cohorts and in senescent primary human cell lines (Harries et al. 2011; Holly et al. 2013; Latorre et al. 2017; Latorre et al. 2018b). Some of the splicing factors in this list have also been shown to associate with lifespan in both mice and humans (Lee et al. 2016) and cognitive dysfunction in human populations (Lee et al. 2019b), or to be involved in molecular responses to dietary restriction (Lee et al. 2019a). The list of genes included the negative regulatory splicing factors Hnrnpa0, Hnrnpa1, Hnrnpa2b1, Hnrnpd, Hnrnph3, Hnrnpk, Hnrnpm, Hnrnpul2, the positive regulatory splicing enhancers Pnisr, Srsf1, Srsf2, Srsf3, Srsf6, Tra2b and the core components of the spliceosome Sf1 and Sf3b1. A further panel of alternatively spliced candidate genes where specific isoforms have known links to brain function/dysfunction (Lye et al. 2019) and/or have been demonstrated to be dysregulated in senescent astrocytes (Lye et al. 2019) or linked with cognitive dysfunction in human populations (Lye et al. 2019), and/or neurodegenerative disease were chosen for analysis. These were Aph1a, App, Aqp4, Gfap, Klotho, Mapt, Psen1 and Psen2. An additional panel of transcripts known to be associated with senescence were also used: Atm, Cdkn1a, Cdkn2a, Cdkn2b and Tp53. Table 2 lists all genes/isoforms tested in this panel, rationale for their inclusion and references to supporting literature. Isoform structures are given in Supplementary figure S1. Six endogenous control genes were also selected on the basis of stability in ageing human populations (Harries et al. 2011): Hprt1, Idh3b, Polr2a, Ppia, Tbp and Ywhaz, however assays to Hprt1 and Ywhaz were subsequently excluded due to poor assay performance.
Table 2.
Cognition and senescence-related transcripts and isoforms used for expression analysis. Shown here are the transcripts and isoforms selected for analysis and a brief description of their function. Details of the type of AS event for each isoform, and the exons involved are provided in the references indicated
| Gene | Transcripts/isoforms | Function | References | |
|---|---|---|---|---|
| Cognition-related transcripts | Aph1a | Aph1aL | Subunit of the gamma-secretase complex involved in β-amyloid processing | (Ma et al. 2005), (Serneels et al. 2005) |
| Aph1aS | ||||
| App | App695 | Precursor to β-amyloid protein | (Rohan de Silva et al. 1997), (Zhang et al. 2011) | |
| App714 | ||||
| App751 | ||||
| AppALL | ||||
| Aqp4 | Aqp4-M1 | Pore-forming intrinsic membrane protein | (De Bellis et al. 2014), (Jung et al. 1994), (Mader and Brimberg 2019) | |
| Aqp4-M23 | ||||
| Gfap | Gfapα | Astrocyte intermediate filament protein | (Kamphuis et al. 2012) | |
| Gfapδ | ||||
| Mapt | Mapt-3R | Microtubule-associated protein tau involved in neurofibrillary tangles | (Goedert et al. 1989), (Lacovich et al. 2017) | |
| Mapt-4R | ||||
| Klotho | mKlotho | Endocrine factor which improves cognitive performance in ageing | (Masso et al. 2015) | |
| sKlotho | ||||
| Psen1 | Psen1 | Subunit of the gamma-secretase complex involved in β-amyloid processing | (De Jonghe et al. 1999), (Janssen et al. 2000) | |
| Psen1 (insTAC) | ||||
| Psen1 (VRSQ) | ||||
| Psen2 | Psen2 | Subunit of the gamma-secretase complex involved in β-amyloid processing | (Moussavi Nik et al. 2015), (Sato et al. 1999) | |
| Psen2 (Δexon5) | ||||
| Senescence-related transcripts | Atm | Atm | DNA damage repair | (Marechal and Zou 2013) |
| Cdkn1a | Cdkn1a (p21a) | Inhibitory to proliferation | (Kaija et al. 2012), (Nozell and Chen 2002) | |
| Cdkn2a | Cdkn2a (p14ARF) | p53 pathway to cell cycle arrest | (Tian et al. 2015), (Kim et al. 2011) | |
| Cdkn2a (p16INK4a) | RB1 pathway to cell cycle arrest | |||
| Cdkn2b | Cdkn2b (p15INK4b) | TGFβ pathway to cell cycle arrest | (Tian et al. 2015) | |
| Tp53 | Tp53 | Cell cycle regulation | (Vousden and Lane 2007) |
RNA extraction
Snap-frozen tissues were first treated with RNAlater™-ICE Frozen Tissue Transition Solution (ThermoFisher, Waltham, MA, USA) according to the manufacturer’s instructions, to allow handling of tissue without RNA degradation occurring due to thawing of sample. Tissue sections were placed in 1 mL TRI Reagent® Solution (ThermoFisher, Waltham, MA, USA) supplemented with the addition of 10 mM MgCl2 to aid recovery of microRNAs (Kim et al. 2012). Samples were completely homogenised in a bead mill (Retsch Technology GmbH, Haan, Germany) at a frequency of 30 cycles per second for 15 min. Phase separation was carried out using chloroform. Total RNA was precipitated from the aqueous phase by means of an overnight incubation at − 20 °C with isopropanol. A total of 1.2 μl Invitrogen™ GlycoBlue™ Coprecipitant (ThermoFisher, Waltham, MA, USA) was added prior to incubation to aid pellet recovery. RNA pellets were ethanol-washed twice and re-suspended in 1× TE buffer, pH 8.0. RNA quality and concentration were assessed by NanoDrop spectrophotometry (NanoDrop, Wilmington, DE, USA).
Reverse transcription
For RTPCR and Sanger sequencing as described below, a pool of 1000 ng of total RNA taken from 10 samples (one sample chosen at random from each NMR age group) was reverse transcribed using EvoScript Universal cDNA Master kit (Roche LifeScience, Burgess Hill, West Sussex, UK) in single 20 μl reactions, according to the manufacturer’s instructions, with the exception of a change to the extension phase of the reaction: a step of 30 min at 65 °C was used instead of 15 min at 65 °C. Resulting cDNA samples were diluted to a final volume of 75 μl with dH2O, and repeated as necessary to provide sufficient template for all RTPCR and sequencing reactions carried out. For assay validation by standard curve, reactions as described above were carried out in quadruplicate and pooled prior to a 6-step 1:2 dilution series. For qPCR, 1500 ng of total RNA was reverse transcribed using SuperScript® VILO™ cDNA Synthesis Kit (ThermoFisher, Waltham, MA, USA) in 20 μl reactions, according to the manufacturer’s instructions (Thermofisher n.d.a, b). Resulting cDNA samples were diluted to a final volume of 150 μl with dH2O to ensure sufficient volume for all subsequent qPCR reactions.
Assay design
Quantitative real-time reverse transcriptase PCR (qPCR) assays to mouse splicing factor assays were obtained from Thermo Fisher (ThermoFisher, Waltham, MA, USA). Assay Ids are available on request. Assays to NMR splicing factors and alternatively expressed isoforms were custom designed to determine expression levels of all candidate mRNAs. Given that the current NMR genome is not completely annotated, this required a four-step design process, as follows:
“In silico” sequence alignment: Where genes were not being tested for alternate isoform expression, the genome sequences of genes in question were obtained from the UCSC Genome Browser (Kent et al. 2002) for the following species: NMR, Guinea pig, Rat, Mouse and Human (see Supplementary Table S1 for assemblies used). Intronic sequences were removed and resulting “in silico-spliced” sequences manually aligned across all 5 species to identify areas of highest homology. Where feasible, exon boundaries within the regions of high homology were then selected as target areas for design of qPCR assays. Where alternate isoforms exist and were required to be assessed, evidence from previous literature was sought to identify the isoforms in question, with particular focus on differential function of specific isoforms (the nature of each splicing change and the precise exons involved are shown in Supplementary Figure S1, and referenced in Table 2). Dependent upon the species in which previous work had been carried out, one or more sequences were obtained from the UCSC Genome Browser (Kent et al. 2002) along with the NMR sequence. Once again, “in silico splicing” and manual alignment was carried out as described in the previous paragraph, however in these cases the areas of alternative splicing were selected as targets for assay design.
Validation of exon junction sequences: Prior to design of qPCR assays, sequence verification was carried out by conventional RTPCR amplification of the predicted fragments to confirm identity. In all cases, primers were designed to the NMR sequence, with the aim of amplifying the target areas as defined above and confirming amplicon identity based on expected size. Where alternative spliced isoforms were to be amplified (and where isoform structure allowed) multiple primer sets were designed to amplify each isoform individually. In the cases of Aph1a, App and Mapt, this was not possible therefore one set of primers was designed to amplify all possible isoforms along with a set of nested primers to isolate individual isoforms using gel electrophoresis and band-stab PCR (Harries et al. 2004). Primer sequences are given in Supplementary Table S2. Template cDNA for the RTPCR was created from a pool of mRNA from all NMR age groups. This pool was reverse transcribed as described above and used in PCR reactions using either Microzone Megamix Royal (Clent Life Science, Stourbridge, UK) or Platinum™ II Hot-Start Green PCR Master Mix (ThermoFisher, Waltham, MA, USA). Reaction conditions were set according to manufacturer’s instructions except in some cases where annealing temperatures and cycle numbers used were specific to each amplicon. These are given in Supplementary Table S2. Amplicons were then checked using gel electrophoresis, and those which matched the expected size were taken forward for the next steps. Of the genes chosen, App and the p21b isoform of Cdkn1a proved refractory to RTPCR amplification and size-based verification. In the case of App, the predicted sequence was used for the assay design. For p21b, sequence homology between the known (human/mouse) variants and the NMR genome assembly was too low to design an assay with any degree of confidence, so this isoform was omitted at this stage.
Sanger sequencing: Once amplicons had been verified by size, each was then sequenced to absolutely confirm the in silico-predicted spliced mRNA sequence. Sanger sequencing was carried out on the ABI 3730 platform using standard protocols. PCR primers were designed to incorporate M13 forward and reverse tag sequences to enable use of a common sequencing primer for all reactions (see Supplementary Table S2). Sequence verification was successful in all cases, however the NMR Psen1 sequence showed no evidence of either the 3 base insertion (insTAC) or the 4 amino acid (VRSQ) splice variants previously reported in human studies (De Jonghe et al. 1999; Janssen et al. 2000), while the NMR Psen2 sequence did not contain the exon 5 skipped (PS2V) splice variant seen in human and animal studies (Sato et al. 1999; Moussavi Nik et al. 2015). Consequently, both genes were omitted from further analysis.
Assay design and verification: Following sequence confirmation, TaqMan® assays were designed to target regions as defined above. Forward primer, reverse primer and reporter sequences of all assays designed are given in Supplementary Table S3. To determine assay efficiency and linearity, standard curves were created from qPCR data generated using a serial dilution of NMR cDNA (see “Reverse transcription” section above). Assay efficiencies and r2 values are given in Supplementary Table S3.
Quantitative reverse transcriptase real-time PCR
A total of 1.0 μl cDNA (reverse transcribed as indicated above) was added to a 5 μl qRTPCR reaction including 2.5 μl TaqMan® Universal Master Mix II, no UNG (ThermoFisher, Waltham, MA, USA) and 0.25 μl custom TaqMan® probe and primer mix (corresponding to 900 nM each primer and 250 nM probe). Reactions were run in triplicate on 384-well plates using the QuantStudio 12 K Flex Real-Time PCR System (ThermoFisher, Waltham, MA, USA). Amplification conditions were a single cycle of 95 °C for 10 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. We first compared levels of splicing factor transcripts in spleen and brain tissue from young NMR (12 months) and young mice (3–4 months), followed by assessment of splicing factor expression and alternative splicing patterns of key senescence or brain-related genes in an extended series of NMR brain tissues from foetal stages to extreme old age (21 to 22 years).
Data preparation
EDS files were uploaded to the ThermoFisher Cloud (ThermoFisher, Waltham, MA, USA) and analysed using the Relative Quantification qPCR App within the software (ThermoFishera). To ensure we are looking at real difference and not efficiency differences, assays were designed to the same splice boundary and are as conserved as possible. Relative threshold data were calculated using the Crt method employed by the ThermoFisher analytical software (ThermoFisherb) along with correction for assay efficiencies (Supplementary Table S3) to produce relative threshold data and then imported into Excel (Microsoft, Redmond, WA, USA) and used for analysis using the 2−ΔΔCrt method. Separate analyses were carried out for the splicing factor species comparison, splicing factor brain series and the isoform/senescence brain series datasets. First, Crt data from all transcripts measured, housekeeping endogenous controls, calculated arithmetic and geometric means of these controls along with calculated “global” arithmetic and geometric means across all genes measured for each dataset were then uploaded to the RefFinder webtool (Xie et al. 2012) to establish the most stable transcript(s). For both the species comparison and the splicing factor brain series, the geometric mean of the 4 housekeeping endogenous controls (Idh3b, Ppia, Polr2a and Tbp) was most stable, while in the isoform/senescence brain series the geometric mean across all genes measured was the most stable, thus these were used for the respective ΔCrt normalisation steps. For the ΔΔCrt normalisation, in the case of the species comparison the arithmetic mean expression value of all transcripts across all mouse samples from both tissues was used as the comparator. In the splicing factor brain series, the arithmetic mean expression level in the 3–4 year old animals for each transcript was used for ΔΔCrt normalisation, as this represents a mature (i.e. non-developmental), but comparatively young time point for comparison. For the alternatively spliced isoform/senescence brain series, the arithmetic mean of the expression levels in the 3–4-year-old animals for each isoform was used (as above) for the statistical analyses as this enabled us to test expression differences between time points longitudinally for each isoform, however for visualisation purposes (see Figs. 4 and 5) and to allow for illustration of the relative expression levels of each isoform, a separate analysis was carried out using a median value across all isoforms for the ΔΔCrt normalisation. Following these ΔΔCrt normalisation steps, fold-changes were calculated using the 2−ΔΔCrt method. To ensure normal distribution and for ease of visualisation, data were subsequently log transformed; in the case of the species comparison, a log10 transformation was used due to the effect sizes, while for the splicing factor brain series and isoform/senescence brain series, a natural log was employed.
Fig. 4.
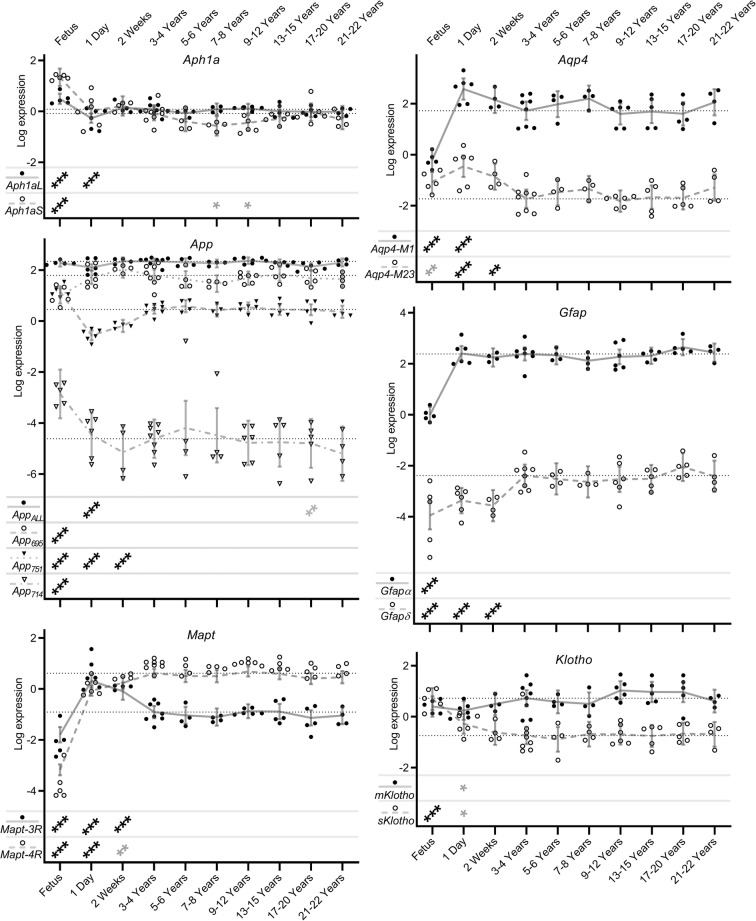
Cognition-related alternatively spliced isoform transcript expression levels in naked mole-rat brain during development and ageing. Shown here are transcript expression levels of cognition-related alternatively spliced isoform transcripts in NMR brain during development and ageing from foetus through to 20–22 years old. In order to show levels of individual isoform transcripts, expression levels are plotted relative to the median expression value of all isoforms across all time points within each gene. Data points represent β-coefficients of log fold-change in expression levels for each transcript in each animal relative to the mean expression of each isoform in the 3–4 year old animals (null point for each isoform shown as a light dotted line). Grey lines denote the mean values with error bars at the 95% confidence intervals. Isoform identities are shown in the legend at the lower left of each graph. Transcripts showing significant differences from the 3–4 year old comparator group are denoted with stars: * = p < 0.05, ** = p < 0.01, *** = p < 0.001. Associations which remain significant after FDR correction (p < 0.002) are indicated in black, while those in grey represent nominal associations (p < 0.05).
Fig. 5.
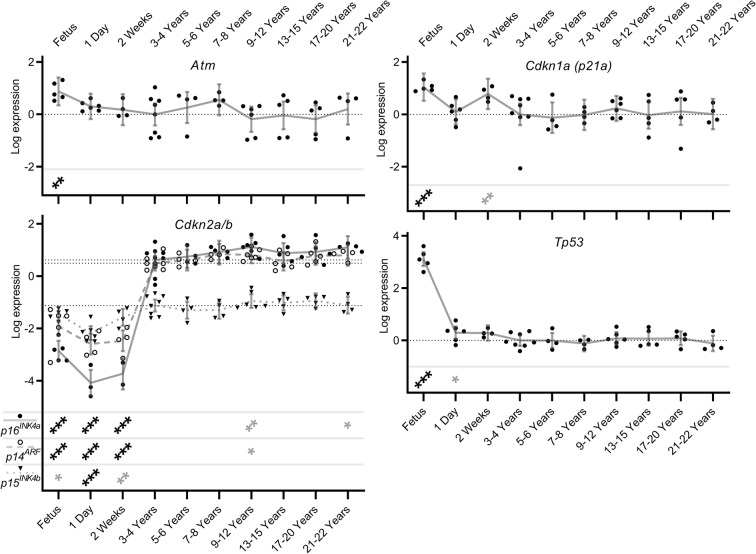
Senescence-related alternatively spliced isoform transcript expression levels in naked mole-rat brain during development and ageing. Shown here are transcript expression levels of senescence-related alternatively spliced isoform transcripts in NMR brain during development and ageing from foetus through to 20–22 years old. In order to show levels of individual isoform transcripts, expression levels are plotted relative to the median expression value of all isoforms across all time points within each gene. Data points represent β-coefficients of log fold-change in expression levels for each transcript in each animal relative to the mean expression of each isoform in the 3–4-year-old animals (null point for each isoform shown as a light dotted line). Grey lines denote the mean values with error bars at the 95% confidence intervals. Where applicable, isoform identities are shown in the legend at the lower left of the graph. Transcripts showing significant differences from the 3–4 year old comparator group are denoted with stars: * = p < 0.05, ** = p < 0.01, *** = p < 0.001. Associations which remain significant after FDR correction (p < 0.002) are indicated in black, while those in grey represent nominal associations (p < 0.05).
Statistical analysis
Differences in gene expression were tested using Student’s t test for the species comparison dataset and linear regressions for both the splicing factor brain series and isoform/senescence brain series datasets. Student’s t test were carried out in SPSS v15.0 (IBM, Armonk, NY, USA) and linear regressions in STATA v15.1 (StataCorp, College Station, TX, USA). Benjamini, Krieger and Yekutieli false discovery rate (FDR) calculations (Benjamini et al. 2006) were then performed to account for multiple testing using GraphPad Prism 8.2.0 (GraphPad Software, San Diego, CA, USA), with the q value set at 1%.
Results
Splicing factors are generally expressed at higher levels in young NMR tissues compared to young mouse tissues
We compared splicing factor expression in the spleen and brain of young mice and naked mole-rats. We used spleen and brain because of the known role of inflammation in the ageing process and the known complexity and prevalence in age-related diseases linked to brain splicing patterns. Looking at splicing factor expression as a whole, relative levels across all transcripts measured are on average 1.6× and 2.1× higher in young NMR brain and spleen respectively compared with young mice. In spleen, all splicing factors tested demonstrated altered expression in NMRs compared with mice. Most genes were upregulated. In brain, although slightly fewer splicing factors demonstrated altered expression compared to mice, major differences in expression were apparent, particularly for the Hnrnpa0, Hnrnph3, Hnrnpk, Hnrnpul2, Srsf1, Srsf2, Srsf3 and Srsf6 genes (p = < 0.001 for each). There was a striking similarity in the overall pattern of species differences in splicing factor expression in both tissues, with 13 out of the 16 splicing factors measured sharing directionality and approximate effect size across the two tissues (Fig. 1, Supplementary Table S4).
Fig. 1.
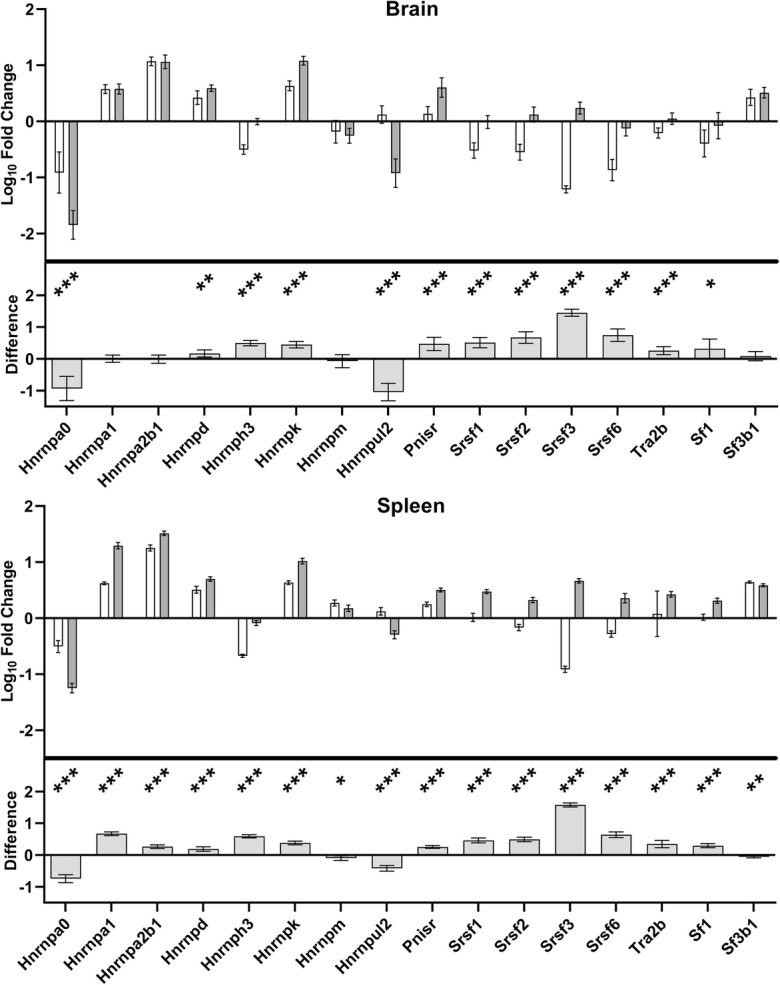
Comparison of splicing factor transcript expression levels between mouse and naked mole-rat (NMR) in brain and spleen. The upper panel of each graph shows log10 fold-change in expression levels, relative to the mean expression across all transcripts in both tissues in mice, with open bars denoting expression in mouse and grey bars showing expression in the NMR. The lower panel of each graph shows the difference between the species. Significant differences between species are denoted with stars: * = p < 0.05, ** = p < 0.01, *** = p < 0.001. All significant differences noted on the graphs met the FDR correction criteria at p < 0.046.
Splicing factor transcript expression remains stable during NMR ageing
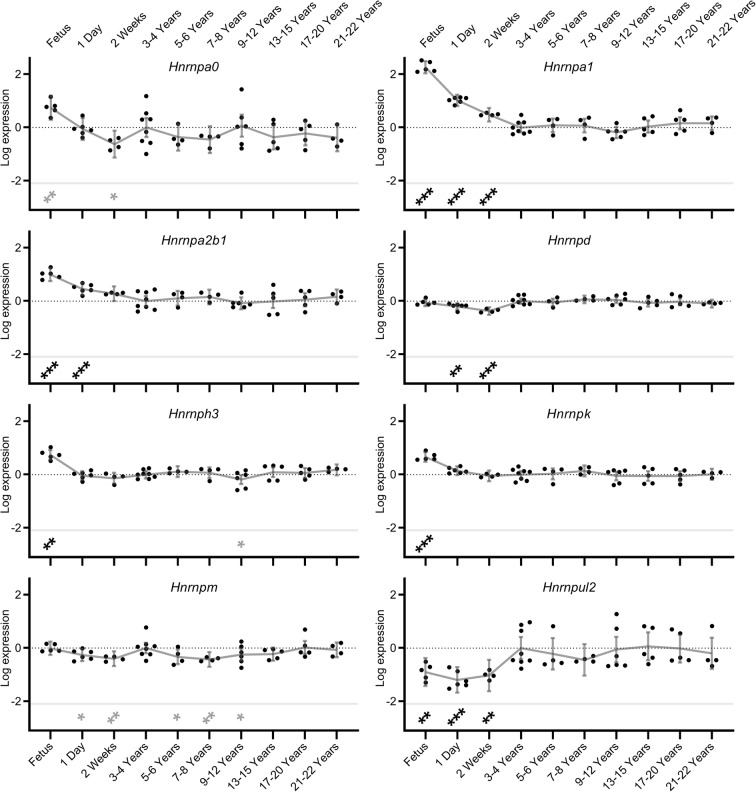
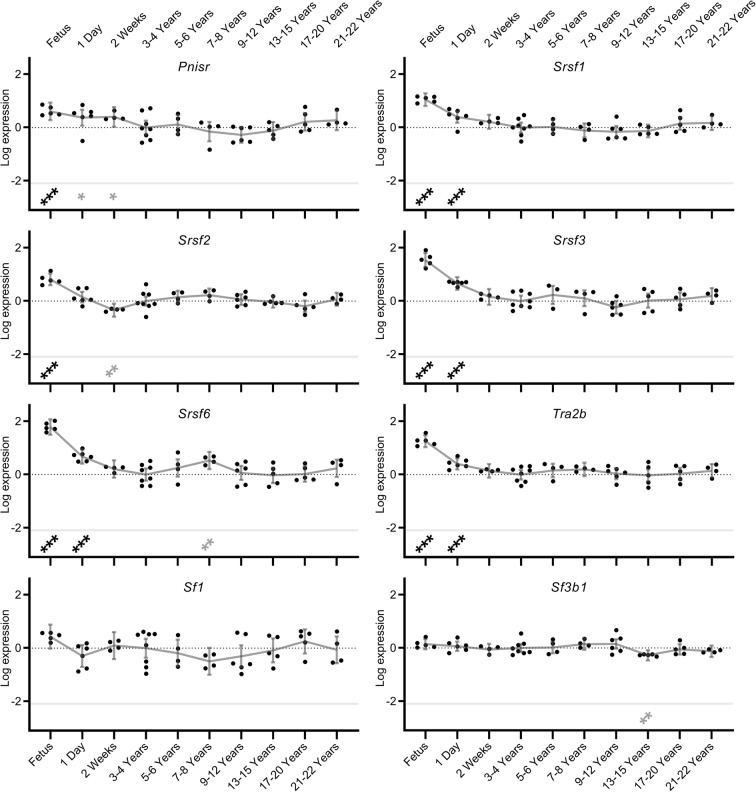
To investigate changes in splicing factor expression across the lifecourse, splicing factor expression was measured from developmental stages (foetus, 1-day-old or 2-week-old animals) until old age (21–22 years). For all regulatory splicing factor transcripts measured, significant expression changes were noted in the brain tissue during at least one of the developmental phases (foetus, 1-day-old or 2-week-old). Once the NMRs were older than 2 weeks of age, for both splicing activators and inhibitor transcripts, in most cases splicing factor expression declined markedly (p < 0.001) and stabilised at these low levels for more than two decades. This was most marked for Hnrnpa1, Hnrnpa2b1, Hnrnph3 and Hnrnpk splicing inhibitors (beta coefficients of expression change between foetal and 3/4 years were: 2.264, 0.997, 0.743 and 0.664 respectively; all p = <0.001) and Srsf1, Srsf2, Srsf3, Srsf6 and Tra2b splicing activators (beta coefficients: 1.050, 0.812, 1.541, 1.786 and 1.257 respectively; all p = <0.001; Figs. 2 and 3; Supplementary Table S5). However in the case of Hnrnpul2, levels were lower in developmental states (up to 2 weeks of age) and were higher in the older age cohorts (beta coefficient: − 0.898; p = 0.001). During progression from mature adults (3–4 years old) to old age (21 to 22 years), almost no alterations in splicing factor expression were noted. Only 3 of 16 regulatory splicing factors (Hnrnph3, Hnrnpm and Srsf6) showed slight alterations in splicing factor expression during ageing in comparison to the 3–4-year-old animals (beta coefficients: − 0.181, − 0.245 and 0.529 respectively p = 0.036, 0.036 and 0.002; Figs. 2 and 3, Supplementary Table S5). Similarly, the core spliceosome element Sf3b1 did not show any alterations in brain expression levels either during developmental phases or during ageing, with the exception of a single nominally significant reduction in expression of Sf3b1 in the 13–15-year-old group (beta coefficient: − 0.275; p = 0.006; Supplementary table S6). This is contrary to previous observations from our group in other species, where dysregulation of splicing factor expression was noted in senescent astrocytes (Supplementary figure S2a; data from Lye et al. 2019).
Fig. 2.
Splicing inhibitor expression levels in naked mole-rat (NMR) brain during development and ageing. Transcript expression levels of Hnrnp splicing inhibitors in NMR brain from foetus through to 20–22 years old are shown. Data points represent β-coefficients of log fold-change in expression levels for each transcript in each animal relative to the mean expression in the 3–4 year old animals. The null point for each transcript is shown as a light dotted line. Grey lines denote the mean values with error bars at the 95% confidence intervals. Transcripts showing significant differences from the 3–4 year old comparator group are denoted with stars: * = p < 0.05, ** = p < 0.01, *** = p < 0.001. Associations which remain significant after FDR correction (p < 0.002) are indicated in black, while those in grey represent nominal associations (p < 0.05).
Fig. 3.
Splicing activator and core spliceosome component expression levels in naked mole-rat brain during development and ageing. Transcript expression levels of Srsf splicing activators in NMR brain from foetus through to 20–22 years old are shown. Data points represent β-coefficients of log fold-change in expression levels for each transcript in each animal relative to the mean expression in the 3–4-year-old animals. The null point for each transcript is shown as a light dotted line. Grey lines denote the mean values with error bars at the 95% confidence intervals. Transcripts showing significant differences from the 3–4 year old comparator group are denoted with stars: * = p < 0.05, ** = p < 0.01, *** = p < 0.001. Associations which remain significant after FDR correction (p < 0.002) are indicated in black, while those in grey represent nominal associations (p < 0.05).
Cognition-related alternatively spliced isoform transcripts remain stable during NMR ageing
To demonstrate the functional consequences of splicing factor changes during NMR development and ageing, we assessed changes in the isoform repertoire of a panel of genes that we have previously demonstrated to be differentially expressed in human astrocyte senescence; some of which are also associated with cognitive decline in human populations (Lye et al. 2019). Sanger sequencing of the relevant isoform-specific exon boundaries demonstrated that the majority of the isoforms selected for assessment were also present in NMR. This included the p14Arf, p15 and p16INK-4a isoforms of the CDKN2A gene; the mole-rat p16INK-4a transcript has poor sequence similarity to that of the mouse, and reportedly presents with two early stop codons, predicting a truncated protein and while the functional domains appear to be partially preserved, the function of this cellular senescence pathway may be already compromised (Kim et al. 2011). The only cases where we did not find evidence for a NMR isoform corresponding to the human and mouse isoforms was the p21b isoform of Cdkn1a. Similar to the pattern seen in the splicing factors, we identified few changes in the expression levels of specific isoforms of cognition-related genes in adult animals. Levels of Aph1a isoforms demonstrated elevated expression in the foetal period (beta coefficients: 0.411 and 1.423, both p = <0.001; Fig. 4; Supplementary table S6) as did the expression of 2 of the 4 isoforms (App714 and App751) expressed from the App gene (beta coefficients: 1.758, and 0.699; both p = <0.001; Fig. 4; Supplementary table S6). Conversely one App isoform, App695 demonstrated decreased expression in the foetal period (beta coefficient: − 0.809; p = <0.001). Similarly, levels of both isoforms of Mapt, both isoforms of Gfap and the M1 isoform of Aqp4 also demonstrated decreased expression in the foetal period (beta coefficients: − 1.096, − 4.661, − 2.393, − 1.563 and − 1.941; all p = <0.001; Fig. 4; Supplementary table S6). In most cases, isoforms were regulated independently of other isoforms of the same gene, indicating that these represent splicing changes and not changes in pre-mRNA transcription per se. Conversely, few changes were noted after 3–4 years old, with isoform ratios remaining consistent throughout the NMR ageing process. (Fig. 4; Supplementary table S6). This is contrary to observations from our group in other species, where dysregulation of this isoform set was reported in senescent astrocytes (supplementary figure S2b; data from Lye et al. 2019).
Senescence-related transcripts remain stable during NMR ageing
The negligible senescence noted in NMR (Buffenstein 2008; Stenvinkel and Shiels 2019) led us to evaluate the expression of known markers of senescence across the lifecourse in our samples. As seen for the splicing factor genes, expression levels of Atm, Cdkn1a, and Tp53 genes were elevated in the foetal state (beta coefficients: 0.883, 1.040 and 3.111; p = 0.002, < 0.001 and < 0.001 respectively; Fig. 5; Supplementary table S7). Conversely, all 3 isoforms of the Cdkn2a/b locus (p14ARF, p16INK4a and p15INK4b) demonstrated reduced expression in foetal animals compared with the mature animals (beta coefficients − 2.414, − 3.472 and − 0.336; p = <0.001, < 0.001 and 0.024 respectively; Fig. 5; Supplementary table S7). Subsequent trajectory for all the molecular markers of senescence remained remarkably static from young adult to old age (Fig. 5; Supplementary Table S7). This is in striking contrast to our observations in replicatively senescent human astrocytes, where marked dysregulation of this isoform set was observed (Supplementary Figure S1b; data from Lye et al. 2019).
Discussion
Here we present evidence that, contrary to observations in both man and mouse, and more in keeping with their negligible senescence phenotype (Buffenstein 2008), adult NMRs maintain splicing regulatory capacity and conservation of regulated alternative splicing with advancing age. Splicing factor expression is higher in young adult NMRs than in young adult mice. We have also identified that contrary to observations from ourselves and others in humans (Harries et al. 2011; Holly et al. 2013; Wang et al. 2018) and in human senescent cells of several different lineages (Holly et al. 2013; Latorre et al. 2017; Latorre et al. 2018b; Lye et al. 2019), splicing factor expression and patterns of isoform expression in NMRs do not appear to alter with age. Splicing factor expression has been demonstrated to correlate with median strain lifespan in mice (Lee et al. 2016), and to have potential mechanistic roles in response to dietary restriction in mice and invertebrates (Heintz et al. 2016; Lee et al. 2019a) and may also contribute to the superior stress resilience observed in NMRs (Lewis et al. 2012).
Superior stress response is a well-established characteristic of NMRs. NMRs have several enhanced stress resistance mechanisms including enhanced mitochondrial antioxidant defences, enhanced proteostasis and improved maintenance of genomic stability than do other shorter lived rodents (Perez et al. 2009; Petruseva et al. 2017; Munro et al. 2019). AS is another fundamental cellular stress resilience mechanism. The ability to adjust transcriptomic output has been associated with resistance to several endogenous and exogenous cellular stressors in plants and animals (Mastrangelo et al. 2012; Pai and Luca 2019). AS is governed by the combinatorial control of a series of splicing activators (SR proteins) and inhibitors (hnRNPs) (Smith and Valcarcel 2000). These regulators are dynamically expressed and responsive to multiple cellular stressors (Biamonti and Caceres 2009; Yamamoto et al. 2016; Kim Guisbert and Guisbert 2017; Jeffery et al. 2019). In humans and other species, splicing factor expression has been demonstrated to display altered expression in ageing and ageing phenotypes (Harries et al. 2011; Lee et al. 2016; Lee et al. 2019b; Lye et al. 2019). Conceptually, one might expect to see elevated expression of splicing enhancers and decreased expression of splicing silencers in species with conserved splicing regulation. However, each splice event is regulated by the combinatorial binding of a unique battery of splicing activators; inclusion of an exon therefore may rely upon the activation of one splice site, as well as the inhibition of a competitor site. For these reasons, we do not expect to see directionality changes at the level of the whole transcriptome. The data presented here suggest that not only do NMRs start with elevated splicing factor expression compared to similarly sized rodents, but their splicing regulatory capacity is well-maintained in old age.
Of all the organs, the brain is known for its diverse splicing patterns and requirement for correctly regulated splicing (Karlsson and Linnarsson 2017). This contributes to many aspects of brain function such as activity patterns and behavioural state (Que et al. 2019), correct configuration of neural circuits (Sudhof 2017) and neuronal differentiation, function and plasticity (Furlanis and Scheiffele 2018). Accordingly, dysregulation of splicing patterns is characteristic of many neurodegenerative and neurodevelopmental diseases in humans such as Alzheimer’s disease, Parkinson’s disease and Autism (Brudek et al. 2016; Parikshak et al. 2016; Raj et al. 2018). The reduced tolerance for splicing dysregulation in brain makes it an ideal organ to study the long term trajectory of splicing factor regulation. Dysregulated splicing factor and isoform expression is a feature of senescent astrocytes (Lye et al. 2019), as well as senescent skin and lung fibroblasts, endothelial cells and cardiomyocytes (Holly et al. 2013; Latorre et al. 2017; Latorre et al. 2018b). In other tissues, dysregulated splicing factor expression has also been suggested to be a driver of cellular senescence; targeted knockdown of the splicing activator SRSF2 or the splicing inhibitor HNRNPD in early passage human primary endothelial cells is sufficient to induce senescence (Latorre et al. 2018c), and restoration of splicing factor expression using small molecule or genetic means in human primary skin and lung fibroblasts and endothelial cells rescues multiple aspects of the senescent cell phenotype (Latorre et al. 2017; Latorre et al. 2018a; Latorre et al. 2018c). The accumulation of senescent cells, whilst known to confer early life longevity benefits in terms of guarding against malignancy, contributing to wound repair and embryonic development (Attaallah et al. 2019) is known to be linked to ageing phenotypes. Targeted removal of such cells is able to bring about a remarkable rejuvenation of multiple aspects of age-related pathology in genetically modified animals (Baker et al. 2011; Baker et al. 2016; Baar et al. 2017; Baar et al. 2018; Baker and Petersen 2018). Newly emerging markers of senescence such as the chromatin remodelling component HMGB2 are elevated in senescent endothelial cells and demonstrably higher in the circulation of people on adverse ageing trajectories (Lawrence et al. 2018). These observations suggest that the accumulation of senescent cells may be at least partly causal to age-related phenotypes, and that new approaches such as Ramen microspectroscopy that are able to detect their accumulation in vivo (Liendl et al. 2019) may provide useful biomarkers to assess the efficacy of senotherapeutic interventions in the future.
The data presented here are consistent with a model by which the extreme longevity and conserved healthspan characteristic of NMRs may arise from conservation of alternative splicing regulation and maintenance of patterns of AS. This maintained transcriptomic plasticity could then ensure a more robust response to internal and external environmental stressors and avoidance of cellular senescence. These data add weight to the hypothesis that splicing regulation is a key feature in the avoidance of cellular senescence, and provide a potential explanation for the extreme longevity seen in NMRs. In the future, the genes and pathways responsible for maintenance of splicing regulation and molecular stress response may be promising future therapeutic targets for healthspan and lifespan interventions.
Electronic supplementary material
(DOCX 637 kb)
Acknowledgements
Calico Life Sciences LLC kindly provided the funding for this study. We thank the animal care staff of Calico for their diligent and dedicated care of the naked mole-rat colony.
Author contributions
BPL designed and carried out all in vitro experiments. MS collected the samples and cared for and aged the animals used. RB was responsible for suggesting this study, experimental study design, provision of tissue samples for analysis, funding the study and drafting components of the manuscript. LWH was responsible for experimental study design, day to day project management and drafting the manuscript. All authors edited and approved the final version of this manuscript.
Compliance with ethical standards
Conflict of interest
RB and MS are employees of Calico Life Sciences LLC and have no other competing financial interests to declare. LWH is a trustee of the British Society for Research on Ageing (BSRA).
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
R. Buffenstein, Email: rbuffen@calicolabs.com
L. W. Harries, Email: L.W.Harries@exeter.ac.uk
References
- Attaallah A, Lenzi M, Marchionni S, Bincoletto G, Cocchi V, Croco E, Hrelia P, Hrelia S, Sell C, Lorenzini A (2019) A pro longevity role for cellular senescence. Geroscience [DOI] [PMC free article] [PubMed]
- Baar M, Brandt R, Putavet D, Klein J, Derks K, Bourgeois B, Stryeck S, Rijksen Y, van Willigenburg H, Feijtel D, van der Pluijm I, Essers J, Van Cappellen WA, van Jcksen W, I, Houtsmuller AB, Pothof J, De Bruin RW, Madi T, Hoeijmakers JG, Campisi J, de Keizer P (2017) Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169:133–147 [DOI] [PMC free article] [PubMed]
- Baar MP, Perdiguero E, Munoz-Canoves P, de Keizer PL. Musculoskeletal senescence: a moving target ready to be eliminated. Curr Opin Pharmacol. 2018;40:147–155. doi: 10.1016/j.coph.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Petersen RC. Cellular senescence in brain aging and neurodegenerative diseases: evidence and perspectives. J Clin Invest. 2018;128(4):1208–1216. doi: 10.1172/JCI95145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, Kirkland JL, van Deursen JM. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Childs BG, Durik M, Wijers ME, Sieben CJ, Zhong J, Saltness RA, Jeganathan KB, Verzosa GC, Pezeshki A, Khazaie K, Miller JD, van Deursen JM. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93(3):491–507. [Google Scholar]
- Biamonti G, Caceres JF. Cellular stress and RNA splicing. Trends Biochem Sci. 2009;34(3):146–153. doi: 10.1016/j.tibs.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Brett R. The population structure of naked mole-rat colonies. The Biology of the Naked Mole-Rat. J. J. A. R. Sherman PW. Princeton: Princeton University Press; 1991. pp. 97–136. [Google Scholar]
- Brudek T, Winge K, Rasmussen NB, Bahl JM, Tanassi J, Agander TK, Hyde TM, Pakkenberg B. Altered alpha-synuclein, parkin, and synphilin isoform levels in multiple system atrophy brains. J Neurochem. 2016;136(1):172–185. doi: 10.1111/jnc.13392. [DOI] [PubMed] [Google Scholar]
- Buffenstein R. Negligible senescence in the longest living rodent, the naked mole-rat: insights from a successfully aging species. J Comp Physiol B. 2008;178(4):439–445. doi: 10.1007/s00360-007-0237-5. [DOI] [PubMed] [Google Scholar]
- Buffenstein R, Park T, Hanes M, Artwohl J (2012) The laboratory rabbit, guinea pig, hamster, and other rodents. Elsevier
- Buffenstein R, Lewis KN, Gibney PN, Narayan V, Grimes KM, Smith M, Lin T, Brown-Borg HM (2019) Probing Paedomorphy and Prolonged Lifespan in the Naked Mole-rat and Dwarf Mouse. Physiology in press [DOI] [PubMed]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3(4):285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Csiszar A, Labinskyy N, Orosz Z, Buffenstein R, Ungvari Z. Vascular aging in the longest-living rodent, the naked mole-rat. FASEB J. 2007;21(6):A851–A851. doi: 10.1152/ajpheart.01287.2006. [DOI] [PubMed] [Google Scholar]
- Dammann P. Slow aging in mammals-lessons from African mole-rats and bats. Semin Cell Dev Biol. 2017;70:154–163. doi: 10.1016/j.semcdb.2017.07.006. [DOI] [PubMed] [Google Scholar]
- De Bellis M, Pisani F, Mola MG, Basco D, Catalano F, Nicchia GP, Svelto M, Frigeri A. A novel human aquaporin-4 splice variant exhibits a dominant-negative activity: a new mechanism to regulate water permeability. Mol Biol Cell. 2014;25(4):470–480. doi: 10.1091/mbc.E13-06-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonghe C, Cruts M, Rogaeva EA, Tysoe C, Singleton A, Vanderstichele H, Meschino W, Dermaut B, Vanderhoeven I, Backhovens H, Vanmechelen E, Morris CM, Hardy J, Rubinsztein DC, St George-Hyslop PH, Van Broeckhoven C. Aberrant splicing in the presenilin-1 intron 4 mutation causes presenile Alzheimer’s disease by increased Abeta42 secretion. Hum Mol Genet. 1999;8(8):1529–1540. doi: 10.1093/hmg/8.8.1529. [DOI] [PubMed] [Google Scholar]
- de Magalhaes JP, Passos JF. Stress, cell senescence and organismal ageing. Mech Ageing Dev. 2018;170:2–9. doi: 10.1016/j.mad.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Delaney MA, Nagy L, Kinsel MJ, Treuting PM. Spontaneous histologic lesions of the adult naked mole rat (Heterocephalus glaber): a retrospective survey of lesions in a zoo population. Vet Pathol. 2013;50(4):607–621. doi: 10.1177/0300985812471543. [DOI] [PubMed] [Google Scholar]
- Delaney MA, Kinsel MJ, Treuting PM. Renal pathology in a nontraditional aging model: the naked mole-rat (Heterocephalus glaber) Vet Pathol. 2016;53(2):493–503. doi: 10.1177/0300985815612557. [DOI] [PubMed] [Google Scholar]
- Deweerdt S. Comparative biology: naked ambition. Nature. 2014;509(7502):S60–S61. doi: 10.1038/509S60a. [DOI] [PubMed] [Google Scholar]
- Edrey YH, Hanes M, Pinto M, Mele J, Buffenstein R. Successful aging and sustained good health in the naked mole rat: a long-lived mammalian model for biogerontology and biomedical research. ILAR J. 2011;52(1):41–53. doi: 10.1093/ilar.52.1.41. [DOI] [PubMed] [Google Scholar]
- Evfratov SA, Smekalova EM, Golovin AV, Logvina NA, Zvereva MI, Dontsova OA. Structural features of the telomerase RNA gene in the naked mole rat Heterocephalus glaber. Acta Nat. 2014;6(2):41–47. [PMC free article] [PubMed] [Google Scholar]
- Furlanis E, Scheiffele P. Regulation of neuronal differentiation, function, and plasticity by alternative splicing. Annu Rev Cell Dev Biol. 2018;34:451–469. doi: 10.1146/annurev-cellbio-100617-062826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M, Spillantini MG, Potier MC, Ulrich J, Crowther RA. Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 1989;8(2):393–399. doi: 10.1002/j.1460-2075.1989.tb03390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes KM, Reddy AK, Lindsey ML, Buffenstein R. And the beat goes on: maintained cardiovascular function during aging in the longest-lived rodent, the naked mole-rat. Am J Physiol Heart Circ Physiol. 2014;307(3):H284–H291. doi: 10.1152/ajpheart.00305.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzikowski AR, Chen YS, Zid BM. Stress-induced mRNP granules: form and function of processing bodies and stress granules. Wiley Interdiscip Rev RNA. 2019;10(3):e1524. doi: 10.1002/wrna.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harries LW, Ellard S, Jones RW, Hattersley AT, Bingham C. Abnormal splicing of hepatocyte nuclear factor-1 beta in the renal cysts and diabetes syndrome. Diabetologia. 2004;47(5):937–942. doi: 10.1007/s00125-004-1383-x. [DOI] [PubMed] [Google Scholar]
- Harries LW, Hernandez D, Henley W, Wood AR, Holly AC, Bradley-Smith RM, Yaghootkar H, Dutta A, Murray A, Frayling TM, Guralnik JM, Bandinelli S, Singleton A, Ferrucci L, Melzer D. Human aging is characterized by focused changes in gene expression and deregulation of alternative splicing. Aging Cell. 2011;10(5):868–878. doi: 10.1111/j.1474-9726.2011.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz C, Doktor TK, Lanjuin A, Escoubas CC, Zhang Y, Weir HJ, Dutta S, Silva-Garcia CG, Bruun GH, Morantte I, Hoxhaj G, Manning BD, Andresen BS, Mair WB. Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans. Nature. 2016;541(7635):102–106. doi: 10.1038/nature20789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinze I, Bens M, Calzia E, Holtze S, Dakhovnik O, Sahm A, Kirkpatrick JM, Szafranski K, Romanov N, Sama SN, Holzer K, Singer S, Ermolaeva M, Platzer M, Hildebrandt T, Ori A. Species comparison of liver proteomes reveals links to naked mole-rat longevity and human aging. BMC Biol. 2018;16(1):82. doi: 10.1186/s12915-018-0547-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickson LJ, Langhi Prata LGP, Bobart SA, Evans TK, Giorgadze N, Hashmi SK, Herrmann SM, Jensen MD, Jia Q, Jordan KL, Kellogg TA, Khosla S, Koerber DM, Lagnado AB, Lawson DK, LeBrasseur NK, Lerman LO, McDonald KM, McKenzie TJ, Passos JF, Pignolo RJ, Pirtskhalava T, Saadiq IM, Schaefer KK, Textor SC, Victorelli SG, Volkman TL, Xue A, Wentworth MA, Wissler Gerdes EO, Zhu Y, Tchkonia T, Kirkland JL. Senolytics decrease senescent cells in humans: preliminary report from a clinical trial of dasatinib plus quercetin in individuals with diabetic kidney disease. EBioMedicine. 2019;47:446–456. doi: 10.1016/j.ebiom.2019.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton HG, Rubinstein ND, Janki P, Ireland AT, Bernstein N, Fong NL, Wright KM, Smith M, Finkle D, Martin-McNulty B, Roy M, Imai DM, Jojic V, Buffenstein R. Single-cell transcriptomics of the naked mole-rat reveals unexpected features of mammalian immunity. PLoS Biol. 2019;17(11):e3000528. doi: 10.1371/journal.pbio.3000528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly AC, Melzer D, Pilling LC, Fellows AC, Tanaka T, Ferrucci L, Harries LW. Changes in splicing factor expression are associated with advancing age in man. Mech Ageing Dev. 2013;134(9):356–366. doi: 10.1016/j.mad.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MM. Social regulation of adult neurogenesis: a comparative approach. Front Neuroendocrinol. 2016;41:59–70. doi: 10.1016/j.yfrne.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Janssen JC, Hall M, Fox NC, Harvey RJ, Beck J, Dickinson A, Campbell T, Collinge J, Lantos PL, Cipolotti L, Stevens JM, Rossor MN. Alzheimer’s disease due to an intronic presenilin-1 (PSEN1 intron 4) mutation: a clinicopathological study. Brain. 2000;123(Pt 5):894–907. doi: 10.1093/brain/123.5.894. [DOI] [PubMed] [Google Scholar]
- Jarvis JU. Eusociality in a mammal: cooperative breeding in naked mole-rat colonies. Science. 1981;212(4494):571–573. doi: 10.1126/science.7209555. [DOI] [PubMed] [Google Scholar]
- Jeffery N, Richardson S, Chambers D, Morgan NG, Harries LW (2019) Cellular stressors may alter islet hormone cell proportions by moderation of alternative splicing patterns. Hum Mol Genet [DOI] [PMC free article] [PubMed]
- Jung JS, Bhat RV, Preston GM, Guggino WB, Baraban JM, Agre P. Molecular characterization of an aquaporin cDNA from brain: candidate osmoreceptor and regulator of water balance. Proc Natl Acad Sci U S A. 1994;91(26):13052–13056. doi: 10.1073/pnas.91.26.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice JN, Nambiar AM, Tchkonia T, LeBrasseur NK, Pascual R, Hashmi SK, Prata L, Masternak MM, Kritchevsky SB, Musi N, Kirkland JL (2019) Senolytics in idiopathic pulmonary fibrosis: results from a first-in-human, open-label, pilot study. EBioMedicine [DOI] [PMC free article] [PubMed]
- Kaija HM, Sarkioja T, Kortelainen ML, Vuoristo JT, Huikuri HV, Porvari KS. Stress-specific responses of p21 expression: implication of transcript variant p21 alt-a in long-term hypoxia. J Cell Biochem. 2012;113(2):544–552. doi: 10.1002/jcb.23377. [DOI] [PubMed] [Google Scholar]
- Kamphuis W, Mamber C, Moeton M, Kooijman L, Sluijs JA, Jansen AH, Verveer M, de Groot LR, Smith VD, Rangarajan S, Rodriguez JJ, Orre M, Hol EM. GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS One. 2012;7(8):e42823. doi: 10.1371/journal.pone.0042823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson K, Linnarsson S. Single-cell mRNA isoform diversity in the mouse brain. BMC Genomics. 2017;18(1):126. doi: 10.1186/s12864-017-3528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Guisbert KS, Guisbert E. SF3B1 is a stress-sensitive splicing factor that regulates both HSF1 concentration and activity. PLoS One. 2017;12(4):e0176382. doi: 10.1371/journal.pone.0176382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EB, Fang X, Fushan AA, Huang Z, Lobanov AV, Han L, Marino SM, Sun X, Turanov AA, Yang P, Yim SH, Zhao X, Kasaikina MV, Stoletzki N, Peng C, Polak P, Xiong Z, Kiezun A, Zhu Y, Chen Y, Kryukov GV, Zhang Q, Peshkin L, Yang L, Bronson RT, Buffenstein R, Wang B, Han C, Li Q, Chen L, Zhao W, Sunyaev SR, Park TJ, Zhang G, Wang J, Gladyshev VN. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479(7372):223–227. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Yeo J, Kim B, Ha M, Kim VN. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol Cell. 2012;46(6):893–895. doi: 10.1016/j.molcel.2012.05.036. [DOI] [PubMed] [Google Scholar]
- Lacovich V, Espindola SL, Alloatti M, Pozo Devoto V, Cromberg LE, Carna ME, Forte G, Gallo JM, Bruno L, Stokin GB, Avale ME, Falzone TL. Tau isoforms imbalance impairs the axonal transport of the amyloid precursor protein in human neurons. J Neurosci. 2017;37(1):58–69. doi: 10.1523/JNEUROSCI.2305-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E, Birar VC, Sheerin AN, Jeynes JCC, Hooper A, Dawe HR, Melzer D, Cox LS, Faragher RGA, Ostler EL, Harries LW. Small molecule modulation of splicing factor expression is associated with rescue from cellular senescence. BMC Cell Biol. 2017;18(1):31. doi: 10.1186/s12860-017-0147-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E, Ostler EO, Faragher RGA, Harries LW. FOXO1 and ETV6 genes may represent novel regulators of splicing factor expression in cellular senescence. FASEB J. 2018;33:1086–1097. doi: 10.1096/fj.201801154R. [DOI] [PubMed] [Google Scholar]
- Latorre E, Pilling LC, Lee BP, Bandinelli S, Melzer D, Ferrucci L, Harries LW. The VEGFA156b isoform is dysregulated in senescent endothelial cells and may be associated with prevalent and incident coronary heart disease. Clin Sci (Lond) 2018;132(3):313–325. doi: 10.1042/CS20171556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latorre E, Torregrossa R, Wood ME, Whiteman M, Harries LW. Mitochondria-targeted hydrogen sulfide attenuates endothelial senescence by selective induction of splicing factors HNRNPD and SRSF2. Aging (Albany NY) 2018;10(7):1666–1681. doi: 10.18632/aging.101500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence I, Bene M, Nacarelli T, Azar A, Cohen JZ, Torres C, Johannes G, Sell C. Correlations between age, functional status, and the senescence-associated proteins HMGB2 and p16(INK4a) Geroscience. 2018;40(2):193–199. doi: 10.1007/s11357-018-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BP, Pilling LC, Emond F, Flurkey K, Harrison DE, Yuan R, Peters LL, Kuchel GA, Ferrucci L, Melzer D, Harries LW. Changes in the expression of splicing factor transcripts and variations in alternative splicing are associated with lifespan in mice and humans. Aging Cell. 2016;15(5):903–913. doi: 10.1111/acel.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BP, Mulvey L, Barr G, Garratt J, Goodman E, Selman C, Harries LW (2019a) Dietary restriction in ILSXISS mice is associated with widespread changes in splicing regulatory factor expression levels. Exp Gerontol:110736 [DOI] [PubMed]
- Lee BP, Pilling LC, Bandinelli S, Ferrucci L, Melzer D, Harries LW. The transcript expression levels of HNRNPM, HNRNPA0 and AKAP17A splicing factors may be predictively associated with ageing phenotypes in human peripheral blood. Biogerontology. 2019;20(5):649–663. doi: 10.1007/s10522-019-09819-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K, Buffenstein R. The naked mole-rat: a resilieint rodent model of aging, longevity and healthspan. In: M. a. M. Kaeberlein, G, editor. Handbook of the biology of aging. London: Academic press; 2016. [Google Scholar]
- Lewis KN, Buffenstein R. The naked mole-rat: a resilient rodent model of aging, longevity and healthspan. Handbook of the biology of aging. M.R. Kaeberlein and G. Martin: London, Academic Press; 2016. [Google Scholar]
- Lewis KN, Mele J, Hornsby PJ, Buffenstein R. Stress resistance in the naked mole-rat: the bare essentials - a mini-review. Gerontology. 2012;58(5):453–462. doi: 10.1159/000335966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KN, Andziak B, Yang T, Buffenstein R. The naked mole-rat response to oxidative stress: just deal with it. Antioxid Redox Signal. 2013;19(12):1388–1399. doi: 10.1089/ars.2012.4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis KN, Rubinstein ND, Buffenstein R. A window into extreme longevity; the circulating metabolomic signature of the naked mole-rat, a mammal that shows negligible senescence. Geroscience. 2018;40(2):105–121. doi: 10.1007/s11357-018-0014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Mele J, Wu Y, Buffenstein R, Hornsby PJ. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9(4):626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liendl L, Grillari J, Schosserer M (2019) Raman fingerprints as promising markers of cellular senescence and aging. Geroscience [DOI] [PMC free article] [PubMed]
- Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye J, Latorre E, Lee BP, Bandinelli S, Holley JE, Gutowski NJ, Ferrucci L, Harries L. Astrocyte senescence may drive alterations in GFAPa, CDKN2A p14ARF and TAU3 transcript expression and contribute to cognitive decline. Geroscience. 2019;42:1–13. doi: 10.1007/s11357-019-00100-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G, Li T, Price DL, Wong PC. APH-1a is the principal mammalian APH-1 isoform present in gamma-secretase complexes during embryonic development. J Neurosci. 2005;25(1):192–198. doi: 10.1523/JNEUROSCI.3814-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae, S. L., M. M. Croken, R. B. Calder, A. Aliper, B. Milholland, R. R. White, A. Zhavoronkov, V. N. Gladyshev, A. Seluanov, V. Gorbunova, Z. D. Zhang and J. Vijg (2015). DNA repair in species with extreme lifespan differences. Aging (Albany NY) 7(12): 1171–1184 [DOI] [PMC free article] [PubMed]
- Mader S, Brimberg L (2019) Aquaporin-4 water channel in the brain and its implication for health and disease. Cells 8(2) [DOI] [PMC free article] [PubMed]
- Marechal A, Zou L (2013) DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb Perspect Biol 5(9) [DOI] [PMC free article] [PubMed]
- Masso A, Sanchez A, Gimenez-Llort L, Lizcano JM, Canete M, Garcia B, Torres-Lista V, Puig M, Bosch A, Chillon M. Secreted and transmembrane alphaKlotho isoforms have different spatio-temporal profiles in the brain during aging and Alzheimer’s disease progression. PLoS One. 2015;10(11):e0143623. doi: 10.1371/journal.pone.0143623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrangelo AM, Marone D, Laido G, De Leonardis AM, De Vita P. Alternative splicing: enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012;185-186:40–49. doi: 10.1016/j.plantsci.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Moussavi Nik SH, Newman M, Wilson L, Ebrahimie E, Wells S, Musgrave I, Verdile G, Martins RN, Lardelli M. Alzheimer’s disease-related peptide PS2V plays ancient, conserved roles in suppression of the unfolded protein response under hypoxia and stimulation of gamma-secretase activity. Hum Mol Genet. 2015;24(13):3662–3678. doi: 10.1093/hmg/ddv110. [DOI] [PubMed] [Google Scholar]
- Munro D, Baldy C, Pamenter ME, Treberg JR. The exceptional longevity of the naked mole-rat may be explained by mitochondrial antioxidant defenses. Aging Cell. 2019;18(3):e12916. doi: 10.1111/acel.12916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo Y, Kamhi E, Jacob-Hirsch J, Amariglio N, Rechavi G, Sperling J, Sperling R. Genome-wide activation of latent donor splice sites in stress and disease. Nucleic Acids Res. 2012;40(21):10980–10994. doi: 10.1093/nar/gks834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nik S, Bowman TV. Splicing and neurodegeneration: insights and mechanisms. Wiley Interdiscip Rev RNA. 2019;10(4):e1532. doi: 10.1002/wrna.1532. [DOI] [PubMed] [Google Scholar]
- Nozell S, Chen X. p21B, a variant of p21(Waf1/Cip1), is induced by the p53 family. Oncogene. 2002;21(8):1285–1294. doi: 10.1038/sj.onc.1205191. [DOI] [PubMed] [Google Scholar]
- O'Connor TP, Lee A, Jarvis JU, Buffenstein R. Prolonged longevity in naked mole-rats: age-related changes in metabolism, body composition and gastrointestinal function. Comp Biochem Physiol A Mol Integr Physiol. 2002;133(3):835–842. doi: 10.1016/s1095-6433(02)00198-8. [DOI] [PubMed] [Google Scholar]
- Olecka M, Huse K, Platzer M. The high degree of cystathionine beta-synthase (CBS) activation by S-adenosylmethionine (SAM) may explain naked mole-rat's distinct methionine metabolite profile compared to mouse. Geroscience. 2018;40(4):359–360. doi: 10.1007/s11357-018-0035-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai AA, Luca F. Environmental influences on RNA processing: biochemical, molecular and genetic regulators of cellular response. Wiley Interdiscip Rev RNA. 2019;10(1):e1503. doi: 10.1002/wrna.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, Hartl C, Leppa V, Ubieta LT, Huang J, Lowe JK, Blencowe BJ, Horvath S, Geschwind DH. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540(7633):423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park TJ, Reznick J, Peterson BL, Blass G, Omerbasic D, Bennett NC, Kuich P, Zasada C, Browe BM, Hamann W, Applegate DT, Radke MH, Kosten T, Lutermann H, Gavaghan V, Eigenbrod O, Begay V, Amoroso VG, Govind V, Minshall RD, Smith ESJ, Larson J, Gotthardt M, Kempa S, Lewin GR. Fructose-driven glycolysis supports anoxia resistance in the naked mole-rat. Science. 2017;356(6335):307–311. doi: 10.1126/science.aab3896. [DOI] [PubMed] [Google Scholar]
- Perez VI, Buffenstein R, Masamsetti P, Salmon AB, Mele J, Friguet B, Ward W, Richardson A, Chaudhuri A. Maintenance of protein stability and resistance to oxidative stress during aging in the longest living rodent, the naked mole-rat; are these determinants of longevity? Free Radic Biol Med. 2008;45:S99–S99. [Google Scholar]
- Perez VI, Buffenstein R, Masamsetti V, Leonard S, Salmon AB, Mele J, Andziak B, Yang T, Edrey Y, Friguet B, Ward W, Richardson A, Chaudhuri A. Protein stability and resistance to oxidative stress are determinants of longevity in the longest-living rodent, the naked mole-rat. Proc Natl Acad Sci U S A. 2009;106(9):3059–3064. doi: 10.1073/pnas.0809620106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruseva IO, Evdokimov AN, Lavrik OI. Genome stability maintenance in naked mole-rat. Acta Nat. 2017;9(4):31–41. [PMC free article] [PubMed] [Google Scholar]
- Pride H, Yu Z, Sunchu B, Mochnick J, Coles A, Zhang Y, Buffenstein R, Hornsby PJ, Austad SN, Perez VI. Long-lived species have improved proteostasis compared to phylogenetically-related shorter-lived species. Biochem Biophys Res Commun. 2015;457(4):669–675. doi: 10.1016/j.bbrc.2015.01.046. [DOI] [PubMed] [Google Scholar]
- Que L, Winterer J, Foldy C. Deep survey of GABAergic interneurons: emerging insights from gene-isoform transcriptomics. Front Mol Neurosci. 2019;12:115. doi: 10.3389/fnmol.2019.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Li YI, Wong G, Humphrey J, Wang M, Ramdhani S, Wang YC, Ng B, Gupta I, Haroutunian V, Schadt EE, Young-Pearse T, Mostafavi S, Zhang B, Sklar P, Bennett DA, De Jager PL. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat Genet. 2018;50(11):1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohan de Silva HA, Jen A, Wickenden C, Jen LS, Wilkinson SL, Patel AJ. Cell-specific expression of beta-amyloid precursor protein isoform mRNAs and proteins in neurons and astrocytes. Brain Res Mol Brain Res. 1997;47(1–2):147–156. doi: 10.1016/s0169-328x(97)00045-4. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Smith M, Buffenstein R (2018) Naked Mole-Rat mortality rates defy gompertzian laws by not increasing with age. Elife 7 [DOI] [PMC free article] [PubMed]
- Sato N, Hori O, Yamaguchi A, Lambert JC, Chartier-Harlin MC, Robinson PA, Delacourte A, Schmidt AM, Furuyama T, Imaizumi K, Tohyama M, Takagi T. A novel presenilin-2 splice variant in human Alzheimer’s disease brain tissue. J Neurochem. 1999;72(6):2498–2505. doi: 10.1046/j.1471-4159.1999.0722498.x. [DOI] [PubMed] [Google Scholar]
- Seiler, M., S. Peng, A. A. Agrawal, J. Palacino, T. Teng, P. Zhu, P. G. Smith, N. Cancer Genome Atlas Research, S. Buonamici and L. Yu (2018). Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep 23(1): 282–296 e284 [DOI] [PMC free article] [PubMed]
- Serneels L, Dejaegere T, Craessaerts K, Horre K, Jorissen E, Tousseyn T, Hebert S, Coolen M, Martens G, Zwijsen A, Annaert W, Hartmann D, De Strooper B. Differential contribution of the three Aph1 genes to gamma-secretase activity in vivo. Proc Natl Acad Sci U S A. 2005;102(5):1719–1724. doi: 10.1073/pnas.0408901102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25(8):381–388. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Stenvinkel P, Shiels PG. Long-lived animals with negligible senescence: clues for ageing research. Biochem Soc Trans. 2019;47(4):1157–1164. doi: 10.1042/BST20190105. [DOI] [PubMed] [Google Scholar]
- Sudhof TC. Synaptic neurexin complexes: a molecular code for the logic of neural circuits. Cell. 2017;171(4):745–769. doi: 10.1016/j.cell.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Ke Z, Tombline G, Macoretta N, Hayes K, Tian X, Lv R, Ablaeva J, Gilbert M, Bhanu NV, Yuan ZF, Garcia BA, Shi YG, Shi Y, Seluanov A, Gorbunova V. Naked mole rat cells have a stable Epigenome that resists iPSC reprogramming. Stem Cell Reports. 2017;9(5):1721–1734. doi: 10.1016/j.stemcr.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ThermoFisher (n.d.a) Cloud Connect. from https://www.thermofisher.com/uk/en/home/cloud.html
- ThermoFisher (n.d.b) Crt, a relative threshold method for qPCR data analysis on the QuantStudio™ 12K Flex system with OpenArray® technology. from https://assets.thermofisher.com/TFS-Assets/LSG/brochures/CO28730-Crt-Tech-note_FLR.pdf
- Tian X, Azpurua J, Ke Z, Augereau A, Zhang ZD, Vijg J, Gladyshev VN, Gorbunova V, Seluanov A. INK4 locus of the tumor-resistant rodent, the naked mole rat, expresses a functional p15/p16 hybrid isoform. Proc Natl Acad Sci U S A. 2015;112(4):1053–1058. doi: 10.1073/pnas.1418203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triplett JC, Swomley A, Kirk J, Lewis K, Orr M, Rodriguez K, Cai J, Klein JB, Buffenstein R, Butterfield DA. Metabolic clues to salubrious longevity in the brain of the longest-lived rodent: the naked mole-rat. J Neurochem. 2015;134(3):538–550. doi: 10.1111/jnc.13149. [DOI] [PubMed] [Google Scholar]
- Triplett JC, Tramutola A, Swomley A, Kirk J, Grimes K, Lewis K, Orr M, Rodriguez K, Cai J, Klein JB, Perluigi M, Buffenstein R, Butterfield DA. Age-related changes in the proteostasis network in the brain of the naked mole-rat: implications promoting healthy longevity. Biochim Biophys Acta. 2015;1852(10 Pt A):2213–2224. doi: 10.1016/j.bbadis.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen JM. The role of senescent cells in ageing. Nature. 2014;509(7501):439–446. doi: 10.1038/nature13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villate O, Rastrojo A, Lopez-Diez R, Hernandez-Torres F, Aguado B. Differential splicing, disease and drug targets. Infect Disord Drug Targets. 2008;8(4):241–251. doi: 10.2174/187152608786734188. [DOI] [PubMed] [Google Scholar]
- Viltard, M., S. Durand, M. Perez-Lanzon, F. Aprahamian, D. Lefevre, C. Leroy, F. Madeo, G. Kroemer and G. Friedlander (2019). The metabolomic signature of extreme longevity: naked mole rats versus mice. Aging (Albany NY) 11(14): 4783–4800 [DOI] [PMC free article] [PubMed]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Wang K, Wu D, Zhang H, Das A, Basu M, Malin J, Cao K, Hannenhalli S. Comprehensive map of age-associated splicing changes across human tissues and their contributions to age-associated diseases. Sci Rep. 2018;8(1):10929. doi: 10.1038/s41598-018-29086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol: Biol; 2012. [DOI] [PubMed] [Google Scholar]
- Yahav S, Buffenstein R. Huddling behavior facilitates homeothermy in the naked mole rat Heterocephalus-Glaber. Physiol Zool. 1991;64(3):871–884. [Google Scholar]
- Yamamoto K, Furukawa MT, Fukumura K, Kawamura A, Yamada T, Suzuki H, Hirose T, Sakamoto H, Inoue K. Control of the heat stress-induced alternative splicing of a subset of genes by hnRNP K. Genes Cells. 2016;21(9):1006–1014. doi: 10.1111/gtc.12400. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer’s disease. Mol Brain. 2011;4:3. doi: 10.1186/1756-6606-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Lin L, Kan G, Xu C, Tang Q, Yu C, Sun W, Cai L, Xu C, Cui S. High autophagy in the naked mole rat may play a significant role in maintaining good health. Cell Physiol Biochem. 2014;33(2):321–332. doi: 10.1159/000356672. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tyshkovskiy A, Munoz-Espin D, Tian X, Serrano M, de Magalhaes JP, Nevo E, Gladyshev VN, Seluanov A, Gorbunova V. Naked mole rats can undergo developmental, oncogene-induced and DNA damage-induced cellular senescence. Proc Natl Acad Sci U S A. 2018;115(8):1801–1806. doi: 10.1073/pnas.1721160115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo L, Prather ER, Stetskiv M, Garrison DE, Meade JR, Peace TI, Zhou T (2019) Inflammaging and oxidative stress in human diseases: from molecular mechanisms to novel treatments. Int J Mol Sci 20(18) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 637 kb)