Abstract
Adherent-invasive Escherichia coli (AIEC) strains have been extensively related to Crohn’s disease (CD) etiopathogenesis. Higher AIEC prevalence in CD patients versus controls has been reported, and its mechanisms of pathogenicity have been linked to CD physiopathology. In CD, the therapeutic armamentarium remains limited and non-curative; hence, the necessity to better understand AIEC as a putative instigator or propagator of the disease is certain. Nonetheless, AIEC identification is currently challenging because it relies on phenotypic assays based on infected cell cultures which are highly time-consuming, laborious and non-standardizable. To address this issue, AIEC molecular mechanisms and virulence genes have been studied; however, a specific and widely distributed genetic AIEC marker is still missing. The finding of molecular tools to easily identify AIEC could be useful in the identification of AIEC carriers who could profit from personalized treatment. Also, it would significantly promote AIEC epidemiological studies. Here, we reviewed the existing data regarding AIEC genetics and presented those molecular markers that could assist with AIEC identification. Finally, we highlighted the problems behind the discovery of exclusive AIEC biomarkers and proposed strategies to facilitate the search of AIEC signature sequences.
Keywords: Crohn’s disease, Adherent-invasive Escherichia coli, Molecular markers, Genetics, Inflammatory bowel disease, Signature sequences
Core tip: In this review, we thoroughly review the approaches for deciphering adherent-invasive Escherichia coli (AIEC) genetics. The characteristics of putative AIEC molecular markers that could assist in AIEC identification are described. We then discuss several aspects that could explain the difficulty behind the discovery of suitable biomarkers and highlight the importance of standardizing AIEC protocols in order to increase the probability of finding these biomarkers. Finally, we point out new approaches for looking for signature sequences that need to take into account the AIEC phylogenetic origin and strain virulence under particular experimental conditions.
INTRODUCTION
Non-pathogenic Escherichia coli (E. coli) strains are common colonizers of the mucus layer of the intestinal tract and have a mutualistic relationship with their hosts. However, some E. coli strains have evolved virulent behavior. Among those, strains belonging to the adherent-invasive E. coli (AIEC) pathovar are suggested to be of particular concern. AIEC isolates lack typical E. coli virulence factors but are phenotypically characterized by their capability to adhere to and invade intestinal epithelial cells (IECs), in addition to surviving and replicating inside macrophages without inducing host-cell death[1]. Using in vitro and in vivo studies, AIEC interactions with IECs have been described to occur through its binding to host receptors, which in turn, promotes intestinal epithelial permeability[2-5]. Additionally, in animal studies, induction of high levels of cytokine secretion and exacerbation of intestinal inflammation in susceptible hosts due to AIEC presence has been reported[6-8]. Since a high prevalence of AIEC has been depicted in the mucosa of Crohn’s disease (CD) patients[1,9-16] and molecular mechanisms of AIEC virulence have been associated with disease pathogenesis[2,4-6,8,17-25], AIEC has been pointed out to take part in the complex multifactorial aetiology of CD.
It is of paramount importance to further decipher the role of AIEC in CD (such as disease specificity or association with active disease), AIEC host range and transmission paths in order to define measures of contamination risk and prevention and/or to provide personalized treatments for AIEC carriers. One reason for the lack of information in these aspects is due to the absence of an AIEC molecular biomarker. Its identification relies on phenotypic traits undergoing cell-culture infection assays, which are extremely time-consuming and hard to standardize. In this review, we aimed to provide a description of AIEC genetics based on the knowledge obtained by different approaches. Moreover, putative genetic/phenotypic markers for rapid AIEC identification have been gathered. We also researched putative reasons why finding AIEC molecular genetic signatures is challenging and discussed new strategies that could shed light on this field.
APPROACHES FOLLOWED TO DECIPHER AIEC GENETICS
Once Darfeuille-Michaud et al[1] defined the AIEC pathotype in 2004, a search for unique genes that could explain its phenotype began. Several approaches have been followed for deciphering AIEC genetics (gene prevalence, point mutations and gene expression) in which both known and novel genes have been studied.
First studies based on polymerase chain reaction (PCR)-based gene prevalence[11,26] indicated that AIEC strains did not harbor any particular genetic trait that could distinguish them from commensals and they did not commonly present virulence genes previously described in other E. coli pathotypes. In line with this observation, the first genome sequencing studies[27-30] together with the most recent genomic studies[31-36] demonstrated again that there was no gene strictly associated with the AIEC phenotype of the strains. Even though PCR-based and genomic studies focusing on gene content reported some genes to be more prevalent in AIEC versus non-AIEC strains (Invasion-related genes: malX[37], pic[38]; Capsule formation-related genes: kpsMTII[37]; Adhesion-related genes: lpfA[31], papGII/III[38]; Resistance-related genes: gipA[39], ibeA[40], iss[38]; Iron scavenging-related genes: chuA[13], pduC[31]; Toxin-related genes: colV[40], vat[38].). However, a low difference in AIEC/non-AIEC gene prevalence was reported for these genes (18%-34%; Table 1)[41,42], and only the vat gene was found to be differently distributed between AIEC and non-AIEC strains in other strain collections[32,43]. No confirmation of the findings in other studies has been obtained for 10/12 of these genes (malX[26,44], kpsMTII[16,26], lpfA and gipA[13], chuA[31,38], pduC[44], ibeA[37,38], colV[31], pic[37], papGII/III[16,37] and iss[31]). As a previous study pointed out[34], it is likely that the associations described are phylogenetic in nature and do not reflect the pathogenic potential of the strains.
Table 1.
Phenotype, phylogroup and prevalence of virulence genes found to be more frequent in adherent-invasive Escherichia coli than non-adherent-invasive Escherichia coli strains in PCR-based and genomic studies
| Virulence gene |
Group of study (n) |
Phylogroup (n)1 |
Prevalence (%) |
||||||
| AIEC | non-AIEC | A | B1 | B2 | D | Others | AIEC | non-AIEC | |
| malX[37] | 49 | 1342 | 39 | 19 | 98 | 19 | 8 | 71 | 47 |
| kpsMTII[37] | 49 | 1342 | 39 | 19 | 98 | 19 | 8 | 71 | 52 |
| pduC[31] | 24 | 25 | 14 | 16 | 10 | 9 | 0 | 50 | 20 |
| lpfA[31] | 24 | 25 | 14 | 16 | 10 | 9 | 0 | 71 | 20 |
| lpfA + gipA[39] | 35 | 103 | Undetermined | 31 | 0 | ||||
| chuA[13] | 15 | 37 | 11 | 5 | 18 | 18 | 0 | 93 | 59 |
| ibeA[40] | 19 | 57 | Undetermined | 37 | 3 | ||||
| colV[40] | 19 | 57 | Undetermined | 42 | 16 | ||||
| vat[38] | 22 | 37 | 9 | 8 | 29 | 12 | 1 | 59 | 30 |
| pic[38] | 22 | 37 | 9 | 8 | 29 | 12 | 1 | 41 | 16 |
| papGII/III[38] | 22 | 37 | 9 | 8 | 29 | 12 | 1 | 18 | 0 |
| iss[38] | 22 | 37 | 9 | 8 | 29 | 12 | 1 | 32 | 11 |
Based on house-keeping genes identified by triplex PCR[41] or by structure analysis included in the multilocus sequence typing analyses[42].
Nineteen intestinal pathogenic Escherichia coli strains and 78 ExPEC strains isolated from animals and 37 human mucosal-associated non-AIEC strains. AIEC: Adherent-invasive Escherichia coli.
Controversial results on gene frequency may be explained by differential strain collections (origin of isolation, host and phylogenetic origin) and the amount of strains considered (Table 1 and Table 2). Our strain collection[37,38] is mainly composed of B2 strains, but for example, the collection of a previous study[31] is enriched in A and B1. As a consequence, the results of studies comparing unequal strains could be questioned. Such is the case of Desilets et al[32] who reported that B2-strains harbored three genomic regions that were absent in non-AIEC strains, but in the last group, all were non-clinical isolates, and only two B2 strains were considered. Since the AIEC pathotype is genetically highly diverse by phylogroup and invasive determinants, cross-validation of observations in a strain collection is strongly recommended.
Table 2.
Review of studies in which the prevalence of particular virulence genes has been examined according to the adherent-invasive Escherichia coli pathotype
| Ref. | AIEC | Non-AIEC | Genes studied |
| Darfeuille-Michaud et al[1], 2004 | 26 | 0 | afaD, eae, ipaC, tia |
| Martinez-Medina et al[11], 2009 | 22 | 38 | afa/draBC, bfpA, cdtB,cnf1, eae, eltA, est, fimAvMT78, fimH, hlyA, ibeA, ipaH, iucD, neuC, papC, pCDV432, sfa/focDE, stx1, stx2 |
| Martinez-Medina et al[26], 2009 | 27 | 59 | afa/draBC, bfpA, bmaE, cdtB, cnf1, cvaC, eae, eltA, est, fimA, fimAvMT78, fimH, focG, gafD, hlyA, ibeA ,ipaH, iroN, iucD, kpsMII, kpsMIII, malX, neuC, papC, papGI, papGII, papGIII alleles,pCDV432, sat, sfa/focDE, sfaS, stx1, stx2, traT, usp |
| Martinez-Medina et al[37], 2011 | 49 | 134 | afa/draBC, astA, bmaE, chuA, cnf, csgA, cvaB, cvaC, eaI, eitA, eitC, etsB, etsC, fimC, focG, fyuA, gafD, gimB, hlyA, hlyF, hra, ibeA, iha, ireA, iroN, irp2, iss, iucD, iutA, kpsMTII, malX, mat, neuC, nfaE, ompA, ompT, papC, papEF, papGI, papGII, papGII/III, papGIII, pic, pks, sat, sfa/foc, sfaS, sitA, sitD (chr.), sitD (epis.), tia, traT, tsh, vat |
| Chassaing et al[5], 2011 | 249 | lpfA | |
| Conte et al[16], 2014 | 27 | 0 | afa/draBC, aggR, cnf1, cvaC, fimH, focG, fyuA, gafD, hlyA, ibeA, iutA, kpsMT1, kpsMT5, kpsMTII, kpsMTIII, nfaE, pAA, PAI1, papA, papC, papEF, papG alleles, sfa/focDE, traT |
| Vazeille et al[39], 2016 | 35 | 103 | lpfA + gipA |
| Céspedes et al[13], 2017 | 15 | 37 | afa/draBC, aufA, cdtB, chuA, cnf1, cvaC, eaaA, eatA, ecNA144, espC, espP, fhuD, fimAvMT78 , fimH, gipA, hlyA, ibeA, irp2, neuC, papC, pet, pic, ratA, sat, sepA, sfa/focDE, sigA, tsh, vat |
| Dogan et al[40], 2018 | 19 | 57 | afaC, chuA, cnf1, colV, focG, fyuA, gsp, hcp, ibeA, iss, kpsMII, lpfA, malX, papC, pduC, pmt1, ratA, sfaDE, traC |
| Camprubí-Font et al[38], 2019 | 48 | 56 | afa/draBC, bmaE, csgA, fimC, focG, gafD, hra, iha, mat, nfaE, papC, papEF, papGII/III, papGI, papGII, papGIII, sfa/foc, sfaS, tsh, chuA, eitA, eitC, fyuA, ireA, iroN, irp2, iucD, iutA, sitA, sitD, (epis.), sitD (chr.), iss, neuC, kpsMTII, ompA, ompT, traT, astA, cnf, sat, vat, hlyA, hlyF, ibeA, gimB, tia, malX, pic, pks, eaI, cvaB, cvaC, etsB, etsC, lpfA141, lpfA154, fimH, chiA, astA, cnf, sat, vat |
| Camprubí-Font et al[46], 2019 | 13 | 30 | ompA, ompC, ompF |
Genes associated with pathotype or origin of isolation are highlighted in bold.
Pathogenicity island described in a virulent uropathogen. AIEC: Adherent-invasive Escherichia coli; CD: Crohn’s disease patients; UC: Ulcerative colitis patients.
Besides, it has been suggested that variations in the sequence of particular genes (fimH, chiA and ompA) may uncover AIEC virulence abilities[2,3,18]. For FimH, previous studies have found some polymorphisms conferring higher adhesion ability but they have not detected a variant more prevalent in AIEC than in non-AIEC iso-lates[13,32,34,38,44,45], yet one has hypothesised that gene expression might explain the phenotype[45]. Regarding OmpA, five amino acid variants (V114I, F131V, D132Y, T228N, and A276G) were described when AIEC reference strain LF82 and the commensal K-12 protein sequence were compared. In this study, Rolhion et al[3] suggested that the amino acid substitutions present in the LF82 protein sequence favors invasion. Likewise, for ChiA, five amino acid changes (Q362K, E370K, V378A, V388E, and E548V) were found located in a chitin binding domain of AIEC strain LF82 in comparison with K-12[18]. These differences in the amino acid sequence were thought to be responsible for the capability of the strain to adhere and invade IECs, as well as, to be a putative AIEC identification marker. Therefore, one of the studies conducted by our research group consisted of the examination of the protein sequences of ChiA, OmpA, OmpC, and OmpF in a large collection of strains[38,46]. In general, no relevant differences in the pathoadaptative mutations according to pathotype were reported; instead, most of them were related to phylogroup. Only one amino acid substitution in OmpA (A200V) and three in OmpC (S89N, V220I, and W231D) were associated with pathotype, but these genetic traits presented low specificity and sensibility as markers for AIEC screening. Despite no particular mutations in ChiA were associated with AIEC pathotype, we found that the LF82 ChiA sequence variant was mainly shared by AIEC strains. Nonetheless, it only comprised 35.5% of all AIEC strains. Thus, at this point, given that neither prevalence nor point mutations of the already described virulence genes (VGs) could uncover the basis of AIEC phenotype, identification of new genetic elements and application of novel techniques are required.
In 2010, the first AIEC genomes were sequenced, and since then many comparative genomics studies have been conducted in the attempt to elucidate the characteristics of the AIEC genome and to identify a genetic biomarker (Table 3)[47]. However, no gene or sequence exclusive to the AIEC pathotype has yet been identified. As a consequence, analysis of single nucleotide polymorphisms (SNPs) in the whole genome has attracted attention since it provided a novel approach to look for AIEC genetic markers. The first study using this methodology took place in 2015 in which only B2 strains were included[33]. Twenty-nine SNPs that could differentiate four AIEC together with 51 ExPEC strains from the commensal and other ExPEC strains were identified but no specific characteristic capable of distinguishing the AIEC pathotype was found[33]. This observation was in concordance with results from a study by O’Brien et al[34], who analyzed differences in base composition of genes among AIEC and non-AIEC strains from the same sequence type. No clustering of AIEC strains was observed. In contrast, the comparative genomics study of AIEC/non-AIEC strain pairs[36] revealed three SNPs [E3-E4_4.3(2), E3-E4_4.4 and E5-E6_3.16 = 3.22(2)] that resulted in differential nucleotide distribution between AIEC and non-AIEC strains in a larger strain collection (22 AIEC and 28 non-AIEC strains). However, there was no nucleotide only present in AIEC strains and absent in non-AIEC. Thus, this study corroborated the absence of AIEC-specific genetic markers widely distributed across all AIEC strains. In fact, the results obtained by analyzing gene prevalence and point mutations reinforce the idea that no particular VGs or pathoadaptative mutations described so far are specifically linked with the AIEC pathotype although, diverse genetic traits could lead to the same phenotype. However, studies reinforcing this hypothesis are absent, and a specific signature sequence of these strains remains to be elucidated.
Table 3.
Summary of the comparative genomics studies conducted in adherent-invasive Escherichia coli to date
| Ref. | AIEC | Non-AIEC | Phylogroup | AIEC origin of isolation |
| Miquel et al[27], 2010 | 1 | 211 | AIEC: B2; Commensals: 4A, 2B1, 1B2; ExPEC: 2B1, 6B2, 3D, 3E | From an I-CD patient |
| Nash et al[28], 2010 | 2 | 101 | AIEC: B2; Commensals: 2A; ExPEC: 7B2, 1E | From I-CD patients |
| Dogan et al[31], 2014 | 24 | 25 | 14 strains from A phylogroup, 16 B1, 10 B2 and 9 D2 | From I-CD patients and controls |
| Desilets et al[32], 2015 | 143 | 6 | AIEC: A: 1; B1: 1; B2: 10; D: 1; F: 1. non-AIEC: A: 2; B1: 2; B2: 2 | From CD and UC patients[47] |
| Zhang et al[35], 2015 | 13 | 11 | AIEC: 1A, 1B1, 4B2, 1D, 5 Unknown. non-AIEC: 3A, 8 Unknown | From CD and UC patients and non-CD subjects |
| Deshpande et al[33], 2015 | 4 | 13071 | All B2 | From CD patients |
| O’Brien et al[34], 2015 | 11 | 30 | All B2, ST95 | From IBD patients and controls |
| Camprubí-Font et al[36], 2018 | 3 | 3 | AIEC: 1 B1, 1 B2 and 1 D. Non-AIEC: 1 B1, 1 B2 and 1 D | From CD patients and controls |
The strain collection examined according to pathotype and phylogroup is depicted. Adherent-invasive Escherichia coli origin of isolation and study observations are also presented.
Include commensals and ExPEC.
Human AIEC: 1A, 1B1, 1B2 and 1D; Murine AIEC: 1B1 and 1 B2; Dog AIEC: 2 B2; Human non-AIEC A phylogroup.
Apart from LF82, UM146 and NRG857c the other strains were only assessed for intramacrophage replication in J774 cells. AIEC: Adherent-invasive Escherichia coli; CD: Crohn’s disease; UC: Ulcerative colitis.
In spite of the advances in the understanding of AIEC genetics, AIEC/non-AIEC differential gene expression has been scarcely studied[21,35,48]. Indeed, three earlier studies examined only LF82 against HS or K-12 gene expression. Furthermore, they studied only one gene during intramacrophage bacterial replication[21], seven genes in the presence of bile salts[48] or comparative transcriptomics while growing in Luria broth medium[35]. Our research contributed to these findings by studying outer membrane proteins (OMPs) gene expression in a collection of AIEC/non-AIEC strains[46]. We analyzed gene expression during bacterial intestinal epithelial cells (IEC) invasion. An increase in OMPs expression was reported in non-AIEC strains during IECs infection in comparison to the expression during growth in the supernatant of cell cultures, while AIEC strains only presented differences between conditions for ompA gene expression. Consequently, it is suggested that OMPs expression may participate in bacterial adhesion to IECs and intracellular persistence. Future work is required to confirm the implication of the differential expression in the AIEC phenotype by performing expression mutants and deciphering whether the differential expression is a trait common to all AIEC strains by studying the gene expression in a larger strain collection.
PUTATIVE BIOMARKERS TO ASSIST AIEC IDENTIFICATION
To date, eight genetic elements have been suggested as putative AIEC molecular markers (Table 4), however most of them presented either present low sensitivity or have been studied in a reduced number of strains. The putative biomarkers presented by Dogan et al[31] and Vazeille et al[39] were more prevalent in AIEC than in non-AIEC strains, nonetheless they were also present in non-AIEC strains (pduC and lpfA) (although in low percentages), or found only in a reduced number of AIEC strains (lpfA + gipA). As a consequence, the specificity values remained high, but the sensitivity values were low. The opposite occurred for the chuA gene[13]; in this case, it was present in nearly all of the AIEC strains and in more than 50% of non-AIEC strains and yielded a high sensitivity and high probability of false-positives (low specificity). Deshpande et al[33] discovered 29 SNPs that could differentiate a group of AIEC strains from a group of ExPEC and commensal strains (all from the B2 phylogroup), but they only studied four AIEC strains. Moreover, the three genomic regions described by Desilets et al[32] also raised interest. Nevertheless, it should be noted that only six non-AIEC strains were included, and AIEC strains were classified based only in the capacity to replicate within macrophages. Likewise, as only B2 strains were studied, the general utility of this approach for any putative AIEC strain remains to be determined.
Table 4.
Genetic elements more frequently found in strains from the adherent-invasive Escherichia coli pathotype and suggested as putative adherent-invasive Escherichia coli molecular markers
| Marker |
Group of study (n) |
Prevalence (%) |
Sensitivity (%) | Specificity (%) | Accuracy (%) | ||
| AIEC | non-AIEC | AIEC | non-AIEC | ||||
| pduC[31]1 | 24 | 25 | 50 | 20 | 50 | 80 | 65 |
| lpfA[31]1 | 24 | 25 | 71 | 20 | 71 | 80 | 75 |
| 29 SNPs[33]2 | 4 | 1307 | 100 | 4 | - | - | - |
| lpfA + gipA[39] | 35 | 103 | 31 | 0 | 31 | 100 | 83 |
| 3 genomic regions[32]3 | 14 | 6 | 79 | 0 | 79 | 100 | 85 |
| chuA[13]4 | 15 | 37 | 93 | 59 | 93 | 41 | 56 |
| SNP algorithm[36] | 22 | 29 | - | - | 82 | 86 | 84 |
| pic + ampR[38] | 22 | 27 | 86 | 33 | 86 | 67 | 75 |
This strain collection was mainly formed by strains from A and B1 phylogroup (14 A, 16 B1, 10 B2 and 9D).
Only B2 strains were included. In this case, the non-AIEC group included commensal and ExPEC strains.
Only present in B2 AIEC strains. The strains’ phylogroup were: AIEC: 1 A, 1 B1, 10 B2, 1 D and 1 F; non-AIEC: 2 A, 2 B1 and 2 B2.
Strain collection with mainly B2 and D strains (11 A, 5 B1, 18 B2 and 18 D). AIEC: Adherent-invasive Escherichia coli.
Along this line, two additional markers that present either higher sensitivity or have been studied in a larger strain collection than the previous ones were presented[36,38]. On one hand, in a recent study we have deeply characterized genetically and phenotypically a collection of AIEC and non-AIEC strains isolated from the intestinal mucosa of humans[38]. Therein, AIEC screening could be assisted by the evaluation of two traits (the presence of pic gene and ampicillin resistance). Although these traits are not specific and widely distributed across the pathotype, E. coli strains that have resistance to ampicillin and harbor the pic gene present an 82% probability of being AIEC. Its major problem was a high rate of false-positives; thus, it could only be used as an initial screening tool, and AIEC strains predicted by this method should be further tested phenotypically. Besides, this marker has been only studied in a particular strain collection; therefore, further validation in external collections would be required. On the other hand, in contrast to previous studies seeking to find AIEC genetic markers, the genome of three strain pairs that could be considered clones but that differed in phenotype were compared[36]. Using this methodological approach, the combination of three point mutations (E3-E4_4.4, E5-E6_3.16 = 3.22(2), and E5-E6_3.12) resulted in the prediction of AIEC phenotype with a sensitivity of 82%, a specificity of 86%, and an accuracy of 84%. So far, to our knowledge, this method is the best one out of the available methods. However, before drawing conclusions on whether a molecular marker is adequate to identify AIEC strains, we recommend performing additional analyses to confirm the specificity, sensitivity, and accuracy of this method. First, the results should be verified using a larger set of strains, including AIEC and non-AIEC strains from other geographical origins. Second, since AIEC strains present similar genetic traits as ExPEC strains[1,9,11], determining the specificity of the method with other E. coli pathotypes, in particular ExPEC strains, would also be required. Finally, if the results of the previously mentioned analysis confirmed the usefulness of the purposed method, testing the utility of the tool in clinical specimens (both fecal and tissue biopsies) should be considered.
POSSIBLE REASONS WHY THE SEARCH FOR AIEC MOLECULAR MARKERS IS CHALLENGING
Failure to detect a molecular property strictly associated with AIEC so far could be explained by: How AIEC might be emerged or the fact that the approaches used so far are not enough appropriate. Moreover, the lack of a standardized method for AIEC phenotypic characterization can add confusion in the search for distinctive traits and/or in its validation in external strains collections.
AIEC isolates by no means represent uniform populations[32,34,35]. This pathotype is highly diverse based on genetic and phenotypic characteristics such as virulence gene carriage or serotype. Even though most of them belong to the B2 phylogroup, they can comprise all the principal phylogenetic groups (A, B1, B2, D, and others)[9-11,49,50]. Moreover, they present genetic similarities with ExPEC strains[1,9,11]. Therefore, the AIEC phenotype might be driven by the combination of various virulence genes that do not necessarily need to be the same for each AIEC strain. Since different mechanisms are involved in the colonization of the epithelium by AIEC, the hypothesis considers that there is no key determinant in common for all the AIEC strains, and different ones can lead to the same phenotype gains plausibility. One study recently described that the genetics of one particular AIEC strain changes during host-to-host transmissions[51], resulting in strains with different phenotypes that compete with the parental strain and present a mobile element that is only maintained in specific conditions. Therefore, these finding indicate that this strain can easily adapt to specific environmental pressures genetically, making the search for biomarkers even more complex.
Moreover, differential gene expression may determine the phenotypic characteristics of AIEC strains. Indeed, this finding could explain why previous works have not found a gene or a point mutation that is widely distributed and specific to AIEC. So far, only two studies have described the transcriptome of AIEC[35,48]. A total of only three AIEC strains have been studied, and the selected experimental designs did not allow the best picture of the real expression profiles during AIEC gut colonization to be obtained. New experimental approaches directed at examining these elements under particular conditions in which AIEC isolates behave differently from other strains may help in finding molecular markers for AIEC detection that will probably be useful for clinical samples. Modulation of gene expression might be determined in various ways, such as DNA methylation or transposable elements. DNA methylation has been described to occur in bacteria in a manner that clonal bacterial populations can be split by switching among alternative DNA methylation patterns[52]. For instance, as studied in an uropathogenic E. coli strain, the Pap pilin causes variations in the phase by a mechanism which involves methylation[53]. Likewise, in terms of transposable elements, one study previously demonstrated that through constant macrophage exposure, a commensal E. coli strain can evolve into a pathogenic strain (such as being able to survive inside macrophages or escape) by the acquisition of transposable element insertion[54]. On the whole, epigenetics and transposable elements are unexplored in AIEC research and should be considered once looking for AIEC characteristic elements.
Finally, regardless of the above-mentioned possible reasons, once looking for AIEC biomarkers, the first question the scientists face is the standardization of the current AIEC identification method. The lack of uniformity among laboratories is very problematic since it can result in different classification assays, which may lead to incorrect associations between genetic and phenotypic features. The vast majority of studies have classified an isolate as AIEC by analyzing all of its phenotypic characteristics in vitro; nonetheless, some discrepancies exist in the protocols (Table 5) and the selected cell lines (Figure 1)[55-62]. Variances in the multiplicity of infection (MOI) and time of infection, in addition to incubation conditions occurred. In terms of invasion assays, while most analyses were performed at a MOI of 10 with an infection time of 3 h and subsequent 1-h incubation with gentamicin (100 µg/mL), others assessed the invasive capacity with a higher MOI (20 or 100), less time of infection (30 min or, 1 or 2 h) and different antibiotic concentrations (50 µg/mL or 3 mg/mL). Additionally, there is even more variability with the protocols used to determine the capacity of the strains to survive and replicate inside macrophages. In these cases, the highest discrepancy occurred with respect to the infection conditions since some performed a centrifugation step to facilitate bacterial intramacrophage uptake, whereas others did not. After this time of infection, non-phagocytosed bacteria were treated with antibiotics at different concentrations and different incubation times. The most common procedure included a first step of 1 h with higher antibiotic concentration (100 µg/mL) followed by a second step of 24 h incubation with reduction in antibiotic concentration (15, 20, or 50 µg/mL) even though other studies performed only one incubation step that consisted of 1 or 24 h steps with the same concentration of antibiotic (20, 50, or 100 µg/mL or 3 mg/mL).
Table 5.
Comparison of the principal experimental conditions of the protocols used to assess bacterial invasion to intestinal epithelial cells and survival and replication inside macrophages
|
Invasion assays | |||
| MOI | Infection conditions | Incubation conditions | Ref. |
| 10 | 30 min | 3 h with amikacin 100 µg/mL | [13] |
| 10 | 1 h | 2 h with gentamicin 100 µg/mL | [70] |
| 10 or 20 | 3 h | 1 h with gentamicin 100 µg/mL | [1,9,11,14,16,31,34,47,56-60,63-65,71] |
| 10 | 3 h | 1 h with gentamicin 3 mg/mL | [72] |
| 100 | 2 h | 1 h with gentamicin 50 µg/mL | [54] |
| 100 | 3 h | 1 h with gentamicin 50 µg/mL | [62] |
| Survival and replication assays | |||
| 10 | 20 min | Media replacement with gentamicin 100 µg/mL for 40 min and media replacement with gentamicin 50 µg/mL for 24 h | [16] |
| 10 | 2 h | Media replacement with amikacin 100 µg/mL for 3 and 24 h | [13] |
| 10 | 2 h | Media replacement with gentamicin 100 µg/mL for 1 h and media replacement with gentamicin 20 µg/mL for 24 h | [1,34,58] |
| 10 or 100 | Centrifugation 10 min at 1000 g and incubation 10 min | Media replacement with gentamicin 100 µg/mL for 40 min and media replacement with gentamicin 20 µg/mL for 24 h | [11,47,66] |
| 10 | Centrifugation 5 min at 500 g and incubation 30 min | Media replacement with gentamicin 100 µg/mL for 2 h and media replacement with gentamicin 15 µg/mL for 24 h | [70] |
| 20 | 2 h | Media replacement with gentamicin 100 µg/mL for 1 h and media replacement with gentamicin 20 µg/mL for 24 h | [9,31] |
| 20 | 2 h | Media replacement with gentamicin 100 µg/mL for 1 and 24 h | [14] |
| 20 | 2 h | Media replacement with gentamicin 3 mg/mL for 1 and 24 h | [72] |
| 100 | Centrifugation 10 min at 1000 g and incubation 10 min | Media replacement with gentamicin 20 µg/mL for 1 and 24 h | [21,59] |
| 100 | 2 h | Media replacement with gentamicin 50 µg/mL for 1 and 24 h | [54] |
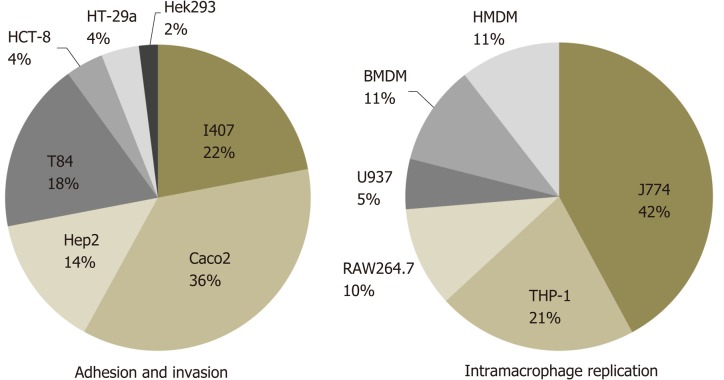
Figure 1.
Review of cell lines used for adherent-invasive Escherichia coli identification. Analysis of the cell lines used for adhesion and invasion assays are based on 29 previously published works[1,9,11,13,14,16,18,21,31,34,39,43-45,55-62,64,66-68,70-72,74] while for intramacrophage replication 17 studies were considered[1,9,11,13,14,16,21,31,34,39,44,55,57,62,66-68,74].
Moreover, the cell lines used to date (Figure 1) might not be the most appropriate considering that for instance, I-407 and Hep-2 originate from cervical and epithelial carcinomas of unknown origin, respectively, and both result from HeLa contamination. As an exception, Caco-2 and T84 are derived from colorectal carcinomas, but it is poorly defined how applicable they are for AIEC identification based on CD pathogenesis. Similarly, for intramacrophage survival, the cell lines mostly used are J774 which is derived from murine origin[1,9-13,15,16]. Some studies have started to use human macrophage-derived monocytes, THP-1 cells, but bacterial intramacrophage survival methods differ among them[34,62,63].
In view of the lack of standardization, adhesion and invasion indices in addition to the replication index of the strains are highly variable among research groups. Taking into account the indices of the LF82 AIEC strain, which is commonly used as control in these procedures, the adhesion index fluctuates between 4.8 and 62.8 bacteria/cell[1,18,34,39,45,62,64-66], the invasion index varies from 0.12% to 12.2%[1,9,16,34,39,62,65-73] and the intramacrophage survival and replication index ranges between 223.0% and 580.0%[1,9,16,21,34,62,66,74]. This finding is of particular concern, especially for those strains that present low indices, which are close to the threshold value that classifies the strain as adherent/invasive. In this case, one strain in one laboratory may be considered AIEC, while in another may be classified as non-AIEC. Therefore, there is the need to solve this discrepancy in order to regulate AIEC strain classification. Without consistency in the actual screening method, it is difficult to search for AIEC genetic differences as we might be using inaccurate isolates. Although hypothesis can be obtained in a particular strain collection, then in the process of validation it is complicated to obtain a good accuracy maybe due to different phenotypic characterization.
IS THE AIEC PHENOTYPE AN ACQUIRED TRAIT OF ESCHERICHIA COLI STRAINS FROM THE GUT?
By looking at recently published data, it becomes believable that the AIEC phenotype is not permanent, yet one might suspect that one E. coli can acquire the AIEC phenotype under particular conditions and inversely, one AIEC strain without specific triggers might turn to a non-AIEC strain or to some extent modify its virulence level. One observation in line with this hypothesis is the fact that very genetically close E. coli strains (identical pulsed field gel electrophoresis profiles) can be classified as either AIEC or non-AIEC[11,36]. Indicating that these strains have evolved to a pathogenic condition via nearly imperceptible genetic, transcriptomic, or epigenomic changes that may occur in particular cases. Furthermore, Elhenawy et al[51] recently demonstrated that one AIEC strain (NRG857c) evolved during host-to-host transmission in mice models, resulting in a diversified population of isolates with two predominant phenotypes: (1) Hypermotile isolates; and (2) Isolates with improved acetate utilization. The first phenotype was due to the presence of an insertion sequence upstream of the flagellar regulator flhDC, which resulted in hypermotile strains with enhanced IECs invasion. However, the presence of this insertion was reversible in the absence of host selection, suggesting that with the absence of particular conditions, the AIEC virulence may be altered. In the same way, Proença et al[54] observed that under continuous macrophage pressure, one commensal strain evolved to increased intracellular survival due to the incorporation of a transposable element insertion. Thus, their observations reinforce the hypothesis of intra-host E. coli evolution to an adherent invasive phenotype and the importance of conducting experiments simulating disease conditions as much as possible, since the AIEC marker may only be detected under selective pressure conditions.
Taking all of these outcomes into account, one may consider that AIEC strains originated from non-AIEC strains from the gut. For that reason, in the foreseeable future, other approaches beyond genes or SNPs prevalence should be analyzed when looking for AIEC molecular markers. These approaches include transcriptomics, epigenetics, and the study of AIEC under conditions in which they behave differently from other pathotypes, perhaps during interactions with host cells. Nowadays, two studies on transcriptomics[35,48] have been conducted. One study described findings in which the AIEC LF82 strain growing in contact with bile salts caused an increase in the expression of genes involved in ethanolamine utilization in comparison to K-12 and also demonstrated that AIEC strains grew more after incubation with minimum media with bile salts supplemented with ethanolamine than non-AIEC[48]. Therefore, reinforcing the idea that AIEC strains may adapt their metabolism according to gut conditions and that experimental methods need to be carefully considered when drawing conclusions about AIEC molecular traits. Nonetheless, the gene expression analysis of other non-AIEC and AIEC strains apart from K-12 and LF82 in the presence of bile salts has not been provided; thus, it is not possible to say that it is an AIEC-specific trait nor an adaptive method common among AIEC strains. Besides, Zhang et al[35] identified potential coding regions that could be applied as signature transcripts. Nevertheless, it is worth noting that they compared only one AIEC strain (LF82) with one commensal (HS) strain during growth in Luria broth. Thereby, the differences found between the strains could be strain-specific or perceptible due to the phylogenetic distance of the strains rather than to the AIEC phenotype. Given the extent of these studies and although there are some transcripts with a stimulating role in AIEC virulence, a candidate transcript suitable to be considered a universal and specific AIEC probe has not yet been determined. It is against this background that we encourage scientists to compare closely related strains and conduct the protocol in a more accurate environment in order to obtain a more a reliable interpretation of the gut context. For instance, previor to bacterial adhesion and invasion, bacteria need to cross the mucosal layer. As a consequence, an assay examining bacteria capacity to disrupt and translocate through the mucus should also be contemplated.
CONCLUSION
Although the factors that constitute an AIEC strain remain an enigma, the outcomes obtained by several lines of research over the last 15 years provide meaningful information on AIEC genetics. Gene prevalence, amino acid substitutions, and gene expression have been studied for both known and unknown genetic elements. In summary, research studies presented and discussed in this review demonstrate that AIEC is a diverse pathotype considering gene content and point mutations, and gene expression studies insinuated that the AIEC phenotype may be determined by particular differences in gene expression, but these need further verification using other AIEC strains.
The discovery of an AIEC biomarker would significantly ease further epi-demiological studies in order to better determine AIEC prevalence and abundance and, discover environmental and animal reservoirs and transmission pathways in addition to facilitating clinical studies in CD patients. For example, studying the variations in abundance in relation to the state of the disease or in response to treatment might be useful. This type of biomarker would represent a rapid and cost-effective way to identify AIEC carriers, who could be treated with AIEC-directed therapies. So far, the diversity among AIEC strains challenges the correlation of individual virulence factors with pathotype in a way that is predictive. Moreover, AIEC classification as a non-AIEC from the gut that turns to pathogenic in particular conditions is gaining significance, but much remains to be learned about the host-pathogen interactions that govern AIEC infection biology. As a consequence, new approaches need to be performed in order to increase the probability of finding an AIEC molecular signature (these include but are not limited to SNPs in non-coding sequences, transcriptomics, metabolomics, and epigenomics). Nonetheless, all of these studies should be conducted using AIEC strains identified according to a standardized method, and the proposed methods should be tested in diverse strain collections from different geographical regions.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Biochemistry and molecular biology
Country/Territory of origin: Spain
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Peer-review started: December 23, 2019
First decision: February 18, 2020
Article in press: April 1, 2020
P-Reviewer: Barnich N, Zhang L S-Editor: Zhang L L-Editor: A E-Editor: Zhang YL
Contributor Information
Carla Camprubí-Font, Laboratory of Molecular Microbiology, Department of Biology, University of Girona, Girona 17003, Spain.
Margarita Martinez-Medina, Laboratory of Molecular Microbiology, Department of Biology, University of Girona, Girona 17003, Spain. marga.martinez@udg.edu.
References
- 1.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, Darfeuille-Michaud A. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest. 2007;117:1566–1574. doi: 10.1172/JCI30504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hébuterne X, Hofman P, Darfeuille-Michaud A. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut. 2010;59:1355–1362. doi: 10.1136/gut.2010.207456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, Darfeuille-Michaud A. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J Exp Med. 2009;206:2179–2189. doi: 10.1084/jem.20090741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chassaing B, Rolhion N, de Vallée A, Salim SY, Prorok-Hamon M, Neut C, Campbell BJ, Söderholm JD, Hugot JP, Colombel JF, Darfeuille-Michaud A. Crohn disease--associated adherent-invasive E. coli bacteria target mouse and human Peyer's patches via long polar fimbriae. J Clin Invest. 2011;121:966–975. doi: 10.1172/JCI44632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chassaing B, Koren O, Carvalho FA, Ley RE, Gewirtz AT. AIEC pathobiont instigates chronic colitis in susceptible hosts by altering microbiota composition. Gut. 2014;63:1069–1080. doi: 10.1136/gutjnl-2013-304909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drouet M, Vignal C, Singer E, Djouina M, Dubreuil L, Cortot A, Desreumaux P, Neut C. AIEC colonization and pathogenicity: influence of previous antibiotic treatment and preexisting inflammation. Inflamm Bowel Dis. 2012;18:1923–1931. doi: 10.1002/ibd.22908. [DOI] [PubMed] [Google Scholar]
- 8.Bretin A, Lucas C, Larabi A, Dalmasso G, Billard E, Barnich N, Bonnet R, Nguyen HTT. AIEC infection triggers modification of gut microbiota composition in genetically predisposed mice, contributing to intestinal inflammation. Sci Rep. 2018;8:12301. doi: 10.1038/s41598-018-30055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 2007;1:403–418. doi: 10.1038/ismej.2007.52. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, Klapproth JM. Invasive Escherichia coli are a feature of Crohn's disease. Lab Invest. 2007;87:1042–1054. doi: 10.1038/labinvest.3700661. [DOI] [PubMed] [Google Scholar]
- 11.Martinez-Medina M, Aldeguer X, Lopez-Siles M, González-Huix F, López-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis. 2009;15:872–882. doi: 10.1002/ibd.20860. [DOI] [PubMed] [Google Scholar]
- 12.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, Jiang ZD, Dupont HL, Garneau P, Harel J, Rishniw M, Simpson KW. Multidrug resistance is common in Escherichia coli associated with ileal Crohn's disease. Inflamm Bowel Dis. 2013;19:141–150. doi: 10.1002/ibd.22971. [DOI] [PubMed] [Google Scholar]
- 13.Céspedes S, Saitz W, Del Canto F, De la Fuente M, Quera R, Hermoso M, Muñoz R, Ginard D, Khorrami S, Girón J, Assar R, Rosselló-Mora R, Vidal RM. Genetic Diversity and Virulence Determinants of Escherichia coli Strains Isolated from Patients with Crohn's Disease in Spain and Chile. Front Microbiol. 2017;8:639. doi: 10.3389/fmicb.2017.00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raso T, Crivellaro S, Chirillo MG, Pais P, Gaia E, Savoia D. Analysis of Escherichia coli isolated from patients affected by Crohn's disease. Curr Microbiol. 2011;63:131–137. doi: 10.1007/s00284-011-9947-8. [DOI] [PubMed] [Google Scholar]
- 15.Negroni A, Costanzo M, Vitali R, Superti F, Bertuccini L, Tinari A, Minelli F, Di Nardo G, Nuti F, Pierdomenico M, Cucchiara S, Stronati L. Characterization of adherent-invasive Escherichia coli isolated from pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:913–924. doi: 10.1002/ibd.21899. [DOI] [PubMed] [Google Scholar]
- 16.Conte MP, Longhi C, Marazzato M, Conte AL, Aleandri M, Lepanto MS, Zagaglia C, Nicoletti M, Aloi M, Totino V, Palamara AT, Schippa S. Adherent-invasive Escherichia coli (AIEC) in pediatric Crohn's disease patients: phenotypic and genetic pathogenic features. BMC Res Notes. 2014;7:748. doi: 10.1186/1756-0500-7-748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gullberg E, Söderholm JD. Peyer's patches and M cells as potential sites of the inflammatory onset in Crohn's disease. Ann N Y Acad Sci. 2006;1072:218–232. doi: 10.1196/annals.1326.028. [DOI] [PubMed] [Google Scholar]
- 18.Low D, Tran HT, Lee IA, Dreux N, Kamba A, Reinecker HC, Darfeuille-Michaud A, Barnich N, Mizoguchi E. Chitin-binding domains of Escherichia coli ChiA mediate interactions with intestinal epithelial cells in mice with colitis. Gastroenterology. 2013;145:602–12.e9. doi: 10.1053/j.gastro.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lapaquette P, Bringer MA, Darfeuille-Michaud A. Defects in autophagy favour adherent-invasive Escherichia coli persistence within macrophages leading to increased pro-inflammatory response. Cell Microbiol. 2012;14:791–807. doi: 10.1111/j.1462-5822.2012.01768.x. [DOI] [PubMed] [Google Scholar]
- 20.Negroni A, Colantoni E, Vitali R, Palone F, Pierdomenico M, Costanzo M, Cesi V, Cucchiara S, Stronati L. NOD2 induces autophagy to control AIEC bacteria infectiveness in intestinal epithelial cells. Inflamm Res. 2016;65:803–813. doi: 10.1007/s00011-016-0964-8. [DOI] [PubMed] [Google Scholar]
- 21.Bringer MA, Barnich N, Glasser AL, Bardot O, Darfeuille-Michaud A. HtrA stress protein is involved in intramacrophagic replication of adherent and invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect Immun. 2005;73:712–721. doi: 10.1128/IAI.73.2.712-721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meconi S, Vercellone A, Levillain F, Payré B, Al Saati T, Capilla F, Desreumaux P, Darfeuille-Michaud A, Altare F. Adherent-invasive Escherichia coli isolated from Crohn's disease patients induce granulomas in vitro. Cell Microbiol. 2007;9:1252–1261. doi: 10.1111/j.1462-5822.2006.00868.x. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen HT, Dalmasso G, Müller S, Carrière J, Seibold F, Darfeuille-Michaud A. Crohn's disease-associated adherent invasive Escherichia coli modulate levels of microRNAs in intestinal epithelial cells to reduce autophagy. Gastroenterology. 2014;146:508–519. doi: 10.1053/j.gastro.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 24.Wine E, Ossa JC, Gray-Owen SD, Sherman PM. Adherent-invasive Escherichia coli, strain LF82 disrupts apical junctional complexes in polarized epithelia. BMC Microbiol. 2009;9:180. doi: 10.1186/1471-2180-9-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Craven M, Egan CE, Dowd SE, McDonough SP, Dogan B, Denkers EY, Bowman D, Scherl EJ, Simpson KW. Inflammation drives dysbiosis and bacterial invasion in murine models of ileal Crohn's disease. PLoS One. 2012;7:e41594. doi: 10.1371/journal.pone.0041594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Medina M, Mora A, Blanco M, López C, Alonso MP, Bonacorsi S, Nicolas-Chanoine MH, Darfeuille-Michaud A, Garcia-Gil J, Blanco J. Similarity and divergence among adherent-invasive Escherichia coli and extraintestinal pathogenic E. coli strains. J Clin Microbiol. 2009;47:3968–3979. doi: 10.1128/JCM.01484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miquel S, Peyretaillade E, Claret L, de Vallée A, Dossat C, Vacherie B, Zineb el H, Segurens B, Barbe V, Sauvanet P, Neut C, Colombel JF, Medigue C, Mojica FJ, Peyret P, Bonnet R, Darfeuille-Michaud A. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One. 2010:5. doi: 10.1371/journal.pone.0012714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nash JH, Villegas A, Kropinski AM, Aguilar-Valenzuela R, Konczy P, Mascarenhas M, Ziebell K, Torres AG, Karmali MA, Coombes BK. Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics. 2010;11:667. doi: 10.1186/1471-2164-11-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause DO, Little AC, Dowd SE, Bernstein CN. Complete genome sequence of adherent invasive Escherichia coli UM146 isolated from Ileal Crohn's disease biopsy tissue. J Bacteriol. 2011;193:583. doi: 10.1128/JB.01290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clarke DJ, Chaudhuri RR, Martin HM, Campbell BJ, Rhodes JM, Constantinidou C, Pallen MJ, Loman NJ, Cunningham AF, Browning DF, Henderson IR. Complete genome sequence of the Crohn's disease-associated adherent-invasive Escherichia coli strain HM605. J Bacteriol. 2011;193:4540. doi: 10.1128/JB.05374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dogan B, Suzuki H, Herlekar D, Sartor RB, Campbell BJ, Roberts CL, Stewart K, Scherl EJ, Araz Y, Bitar PP, Lefébure T, Chandler B, Schukken YH, Stanhope MJ, Simpson KW. Inflammation-associated adherent-invasive Escherichia coli are enriched in pathways for use of propanediol and iron and M-cell translocation. Inflamm Bowel Dis. 2014;20:1919–1932. doi: 10.1097/MIB.0000000000000183. [DOI] [PubMed] [Google Scholar]
- 32.Desilets M, Deng X, Rao C, Ensminger AW, Krause DO, Sherman PM, Gray-Owen SD. Genome-based Definition of an Inflammatory Bowel Disease-associated Adherent-Invasive Escherichia coli Pathovar. Inflamm Bowel Dis. 2016;22:1–12. doi: 10.1097/MIB.0000000000000574. [DOI] [PubMed] [Google Scholar]
- 33.Deshpande NP, Wilkins MR, Mitchell HM, Kaakoush NO. Novel genetic markers define a subgroup of pathogenic Escherichia coli strains belonging to the B2 phylogenetic group. FEMS Microbiol Lett. 2015:362. doi: 10.1093/femsle/fnv193. [DOI] [PubMed] [Google Scholar]
- 34.O'Brien CL, Bringer MA, Holt KE, Gordon DM, Dubois AL, Barnich N, Darfeuille-Michaud A, Pavli P. Comparative genomics of Crohn's disease-associated adherent-invasive Escherichia coli. Gut. 2017;66:1382–1389. doi: 10.1136/gutjnl-2015-311059. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Rowehl L, Krumsiek JM, Orner EP, Shaikh N, Tarr PI, Sodergren E, Weinstock GM, Boedeker EC, Xiong X, Parkinson J, Frank DN, Li E, Gathungu G. Identification of Candidate Adherent-Invasive E. coli Signature Transcripts by Genomic/Transcriptomic Analysis. PLoS One. 2015;10:e0130902. doi: 10.1371/journal.pone.0130902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camprubí-Font C, Lopez-Siles M, Ferrer-Guixeras M, Niubó-Carulla L, Abellà-Ametller C, Garcia-Gil LJ, Martinez-Medina M. Comparative genomics reveals new single-nucleotide polymorphisms that can assist in identification of adherent-invasive Escherichia coli. Sci Rep. 2018;8:2695. doi: 10.1038/s41598-018-20843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Medina M, Garcia-Gil J, Barnich N, Wieler LH, Ewers C. Adherent-invasive Escherichia coli phenotype displayed by intestinal pathogenic E. coli strains from cats, dogs, and swine. Appl Environ Microbiol. 2011;77:5813–5817. doi: 10.1128/AEM.02614-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camprubí-Font C, Ewers C, Lopez-Siles M, Martinez-Medina M. Genetic and Phenotypic Features to Screen for Putative Adherent-Invasive Escherichia coli. Front Microbiol. 2019;10:108. doi: 10.3389/fmicb.2019.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazeille E, Chassaing B, Buisson A, Dubois A, de Vallée A, Billard E, Neut C, Bommelaer G, Colombel JF, Barnich N, Darfeuille-Michaud A, Bringer MA. GipA Factor Supports Colonization of Peyer's Patches by Crohn's Disease-associated Escherichia Coli. Inflamm Bowel Dis. 2016;22:68–81. doi: 10.1097/MIB.0000000000000609. [DOI] [PubMed] [Google Scholar]
- 40.Dogan B, Belcher-Timme HF, Dogan EI, Jiang ZD, DuPont HL, Snyder N, Yang S, Chandler B, Scherl EJ, Simpson KW. Evaluation of Escherichia coli pathotypes associated with irritable bowel syndrome. FEMS Microbiol Lett. 2018;365 doi: 10.1093/femsle/fny249. [DOI] [PubMed] [Google Scholar]
- 41.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gibold L, Garenaux E, Dalmasso G, Gallucci C, Cia D, Mottet-Auselo B, Faïs T, Darfeuille-Michaud A, Nguyen HT, Barnich N, Bonnet R, Delmas J. The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn's disease-associated Escherichia coli. Cell Microbiol. 2016;18:617–631. doi: 10.1111/cmi.12539. [DOI] [PubMed] [Google Scholar]
- 44.Iebba V, Conte MP, Lepanto MS, Di Nardo G, Santangelo F, Aloi M, Totino V, Checchi MP, Longhi C, Cucchiara S, Schippa S. Microevolution in fimH gene of mucosa-associated Escherichia coli strains isolated from pediatric patients with inflammatory bowel disease. Infect Immun. 2012;80:1408–1417. doi: 10.1128/IAI.06181-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dreux N, Denizot J, Martinez-Medina M, Mellmann A, Billig M, Kisiela D, Chattopadhyay S, Sokurenko E, Neut C, Gower-Rousseau C, Colombel JF, Bonnet R, Darfeuille-Michaud A, Barnich N. Point mutations in FimH adhesin of Crohn's disease-associated adherent-invasive Escherichia coli enhance intestinal inflammatory response. PLoS Pathog. 2013;9:e1003141. doi: 10.1371/journal.ppat.1003141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camprubí-Font C, Ruiz Del Castillo B, Barrabés S, Martínez-Martínez L, Martinez-Medina M. Amino Acid Substitutions and Differential Gene Expression of Outer Membrane Proteins in Adherent-Invasive Escherichia coli. Front Microbiol. 2019;10:1707. doi: 10.3389/fmicb.2019.01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sepehri S, Khafipour E, Bernstein CN, Coombes BK, Pilar AV, Karmali M, Ziebell K, Krause DO. Characterization of Escherichia coli isolated from gut biopsies of newly diagnosed patients with inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:1451–1463. doi: 10.1002/ibd.21509. [DOI] [PubMed] [Google Scholar]
- 48.Delmas J, Gibold L, Faïs T, Batista S, Leremboure M, Sinel C, Vazeille E, Cattoir V, Buisson A, Barnich N, Dalmasso G, Bonnet R. Metabolic adaptation of adherent-invasive Escherichia coli to exposure to bile salts. Sci Rep. 2019;9:2175. doi: 10.1038/s41598-019-38628-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 50.Masseret E, Boudeau J, Colombel JF, Neut C, Desreumaux P, Joly B, Cortot A, Darfeuille-Michaud A. Genetically related Escherichia coli strains associated with Crohn's disease. Gut. 2001;48:320–325. doi: 10.1136/gut.48.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Elhenawy W, Tsai CN, Coombes BK. Host-Specific Adaptive Diversification of Crohn's Disease-Associated Adherent-Invasive Escherichia coli. Cell Host Microbe. 2019;25:301–312.e5. doi: 10.1016/j.chom.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 52.Casadesús J, Low D. Epigenetic gene regulation in the bacterial world. Microbiol Mol Biol Rev. 2006;70:830–856. doi: 10.1128/MMBR.00016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blyn LB, Braaten BA, Low DA. Regulation of pap pilin phase variation by a mechanism involving differential dam methylation states. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Proença JT, Barral DC, Gordo I. Commensal-to-pathogen transition: One-single transposon insertion results in two pathoadaptive traits in Escherichia coli -macrophage interaction. Sci Rep. 2017;7:4504. doi: 10.1038/s41598-017-04081-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simpson KW, Dogan B, Rishniw M, Goldstein RE, Klaessig S, McDonough PL, German AJ, Yates RM, Russell DG, Johnson SE, Berg DE, Harel J, Bruant G, McDonough SP, Schukken YH. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect Immun. 2006;74:4778–4792. doi: 10.1128/IAI.00067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bertuccini L, Costanzo M, Iosi F, Tinari A, Terruzzi F, Stronati L, Aloi M, Cucchiara S, Superti F. Lactoferrin prevents invasion and inflammatory response following E. coli strain LF82 infection in experimental model of Crohn's disease. Dig Liver Dis. 2014;46:496–504. doi: 10.1016/j.dld.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 57.De la Fuente M, Franchi L, Araya D, Díaz-Jiménez D, Olivares M, Álvarez-Lobos M, Golenbock D, González MJ, López-Kostner F, Quera R, Núñez G, Vidal R, Hermoso MA. Escherichia coli isolates from inflammatory bowel diseases patients survive in macrophages and activate NLRP3 inflammasome. Int J Med Microbiol. 2014;304:384–392. doi: 10.1016/j.ijmm.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chalopin T, Brissonnet Y, Sivignon A, Deniaud D, Cremet L, Barnich N, Bouckaert J, Gouin SG. Inhibition profiles of mono- and polyvalent FimH antagonists against 10 different Escherichia coli strains. Org Biomol Chem. 2015;13:11369–11375. doi: 10.1039/c5ob01581b. [DOI] [PubMed] [Google Scholar]
- 59.Assa A, Vong L, Pinnell LJ, Rautava J, Avitzur N, Johnson-Henry KC, Sherman PM. Vitamin D deficiency predisposes to adherent-invasive Escherichia coli-induced barrier dysfunction and experimental colonic injury. Inflamm Bowel Dis. 2015;21:297–306. doi: 10.1097/MIB.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 60.Lapaquette P, Darfeuille-Michaud A. Abnormalities in the handling of intracellular bacteria in Crohn's disease. J Clin Gastroenterol. 2010;44 Suppl 1:S26–S29. doi: 10.1097/MCG.0b013e3181dd4fa5. [DOI] [PubMed] [Google Scholar]
- 61.Chassaing B, Etienne-Mesmin L, Bonnet R, Darfeuille-Michaud A. Bile salts induce long polar fimbriae expression favouring Crohn's disease-associated adherent-invasive Escherichia coli interaction with Peyer's patches. Environ Microbiol. 2013;15:355–371. doi: 10.1111/j.1462-2920.2012.02824.x. [DOI] [PubMed] [Google Scholar]
- 62.Fang X, Monk JM, Nurk S, Akseshina M, Zhu Q, Gemmell C, Gianetto-Hill C, Leung N, Szubin R, Sanders J, Beck PL, Li W, Sandborn WJ, Gray-Owen SD, Knight R, Allen-Vercoe E, Palsson BO, Smarr L. Metagenomics-Based, Strain-Level Analysis of Escherichia coli From a Time-Series of Microbiome Samples From a Crohn's Disease Patient. Front Microbiol. 2018;9:2559. doi: 10.3389/fmicb.2018.02559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cieza RJ, Hu J, Ross BN, Sbrana E, Torres AG. The IbeA invasin of adherent-invasive Escherichia coli mediates interaction with intestinal epithelia and macrophages. Infect Immun. 2015;83:1904–1918. doi: 10.1128/IAI.03003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rolhion N, Carvalho FA, Darfeuille-Michaud A. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Mol Microbiol. 2007;63:1684–1700. doi: 10.1111/j.1365-2958.2007.05638.x. [DOI] [PubMed] [Google Scholar]
- 65.Chassaing B, Darfeuille-Michaud A. The σE pathway is involved in biofilm formation by Crohn's disease-associated adherent-invasive Escherichia coli. J Bacteriol. 2013;195:76–84. doi: 10.1128/JB.01079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kittana H, Gomes-Neto JC, Heck K, Sughroue J, Xian Y, Mantz S, Muñoz RRS, Cody LA, Schmaltz RJ, Anderson CL, Moxley RA, Hostetter JM, Fernando SC, Clarke J, Kachman SD, Cressler CE, Benson AK, Walter J, Ramer-Tait AE. Establishing the phenotypic basis of adherent-invasive Escherichia coli (AIEC) pathogenicity in intestinal inflammation. bioRxiv. 2019 [Google Scholar]
- 67.Barnich N, Bringer MA, Claret L, Darfeuille-Michaud A. Involvement of lipoprotein NlpI in the virulence of adherent invasive Escherichia coli strain LF82 isolated from a patient with Crohn's disease. Infect Immun. 2004;72:2484–2493. doi: 10.1128/IAI.72.5.2484-2493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vazeille E, Bringer MA, Gardarin A, Chambon C, Becker-Pauly C, Pender SL, Jakob C, Müller S, Lottaz D, Darfeuille-Michaud A. Role of meprins to protect ileal mucosa of Crohn's disease patients from colonization by adherent-invasive E. coli. PLoS One. 2011;6:e21199. doi: 10.1371/journal.pone.0021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rolhion N, Darfeuille-Michaud A. Adherent-invasive Escherichia coli in inflammatory bowel disease. Inflamm Bowel Dis. 2007;13:1277–1283. doi: 10.1002/ibd.20176. [DOI] [PubMed] [Google Scholar]
- 70.Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Jezek GE, Islas-Islas M, Torres AG. Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int J Med Microbiol. 2008;298:397–409. doi: 10.1016/j.ijmm.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 71.Boudeau J, Barnich N, Darfeuille-Michaud A. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol Microbiol. 2001;39:1272–1284. doi: 10.1111/j.1365-2958.2001.02315.x. [DOI] [PubMed] [Google Scholar]
- 72.Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun. 1999;67:4499–4509. doi: 10.1128/iai.67.9.4499-4509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lapaquette P, Glasser AL, Huett A, Xavier RJ, Darfeuille-Michaud A. Crohn's disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell Microbiol. 2010;12:99–113. doi: 10.1111/j.1462-5822.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glasser AL, Boudeau J, Barnich N, Perruchot MH, Colombel JF, Darfeuille-Michaud A. Adherent invasive Escherichia coli strains from patients with Crohn's disease survive and replicate within macrophages without inducing host cell death. Infect Immun. 2001;69:5529–5537. doi: 10.1128/IAI.69.9.5529-5537.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]