Abstract
Extracellular biosynthesis of silver nanoparticles (AgNPs) was explored using Thiosphaera pantotropha since this strain exhibits both nitrate- and nitrite-reductase enzyme activity (NaR and NiR, respectively). Optimal AgNP synthesis was achieved using 2 mM AgNO3, culture supernatant of nutrient broth grown T. pantotropha, and incubation at 37 °C and 180 rpm. Under these conditions, the localized surface plasmon resonance peak of silver at 404 nm matched well with the average size of the spherical AgNPs based on FEG-TEM micrographs, i.e., 14.6 nm (range: 5–51 nm). The zeta potential of −33.6 mV indicated good stability of the biosynthesized nanoparticles. The XRD spectra demonstrated the simultaneous presence of face-centered cubic crystal structure of AgNPs and AgCl NPs. Ag+ ions were possibly reduced by the NaR and NiR enzymes released into the culture media. The FTIR spectra confirmed the stabilization of the AgNPs by biomolecules present in the culture supernatant of T. pantotropha. The synthesized Ag/AgCl NPs exhibited good antibacterial efficacy against both Gram-negative (Escherichia coli and Pseudomonas aeruginosa) and Gram-positive bacteria (Bacillus subtilis and Staphylococcus aureus). The minimum inhibitory concentration (MIC) was 2.5 µg/ml for all the bacteria except B. subtilis (MIC of 10 µg/ml). The minimum bactericidal concentration (MBC) was 2.5, 10, 20, and 5 µg/ml for E. coli, P. aeruginosa, B. subtilis, and S. aureus, respectively. At MBC and higher AgNP concentration, both plating and CLSM imaging confirmed the absence of viable bacteria in treated water. The biogenic AgNPs depicted IC50 of 34.8 µg/ml for MCF-7 cells.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02218-0) contains supplementary material, which is available to authorized users.
Keywords: Antibacterial activity, Confocal laser scanning microscopy, In vitro cytotoxic effect, Localized surface plasmon resonance, Biogenic silver nanoparticles
Introduction
There is increasing interest in developing nanomaterial synthesis protocols due to their potential for application in various fields, such as biosensors, catalysis, medicine, and water treatment (Li et al. 2013; Bharti et al. 2015; Singh and Bahadur 2015). Among metallic nanomaterials, silver nanoparticles (AgNPs) have been widely explored in diverse applications, particularly as antimicrobial agents. The unique properties of AgNPs are ascribed to their high surface area to volume ratio, which imparts high reactivity (Agnihotri et al. 2013). The effectiveness of nanomaterials is dependent on the shape and size of the nanomaterials (Agnihotri et al. 2014). Hence, various synthesis routes have been explored for the preparation of nanomaterials with desired characteristics. Due to various limitations associated with physical and chemical approaches, the synthesis of AgNPs using biomimetic approaches holds promise (Iravani et al. 2014). Among various biological agents, bacteria have attracted considerable attention through both intracellular and extracellular synthesis routes due to ease of culturing bacteria, short generation time, and mild experimental conditions (Jeevan et al. 2012).
The present study focused on the synthesis of AgNPs using a non-pathogenic bacteria, Thiosphaera pantotropha. This bacteria has the unique ability to utilize nitrate, nitrite, or nitrogen oxide as terminal electron acceptor and is capable of heterotrophic nitrification and aerobic denitrification (Gupta 1997). Unlike most denitrifying bacteria, T. pantotropha possesses two types of nitrate reductase (NaR) enzymes, i.e., a membrane-bound NaR enzyme (only under anaerobic condition) and a periplasmic NaR enzyme (under aerobic condition) and nitrite reductase (NiR) enzyme, which further reduces nitrite to N2 (Gupta 1997). Due to these properties, T. pantotropha was explored for extracellular biosynthesis of AgNPs. This strain has not been previously employed for synthesis of nanoparticles. Various researchers demonstrated extracellular biosynthesis of AgNPs by adding AgNO3 to the culture supernatant (Thamilselvi and Radha 2013; Gopinath et al. 2017; Ghareib et al. 2016; Shanthi et al. 2016) and reported nitrite accumulation in the mixture due to nitrate-reductase activity (Thamilselvi and Radha 2013). Purified NaR enzyme isolated from a fungus, along with redox mediator and stabilizer was also found to promote AgNP synthesis in vitro (Jain et al. 2011). Only one study using Streptomyces sp. LK3 culture supernatant demonstrated AgNP synthesis accompanied by conversion of nitrate to N2 (Karthik et al. 2014).
The present study focusses on extracellular biosynthesis of AgNPs using T. pantotropha and determination of biocidal activity of the synthesized AgNPs against various bacteria and MCF-7, a human breast cancer cell line. The effect of metal precursor (AgNO3) concentration, temperature, and nutrient media on AgNP synthesis was explored using various characterization techniques. Subsequently, the nitrate- and nitrite-reductase activity in the culture supernatant was also analyzed to demonstrate the mechanism of AgNP biosynthesis. Finally, these nanoparticles were tested for their antibacterial effectiveness against two representative Gram-negative and Gram-positive bacteria, in terms of, zone of inhibition (ZOI), minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). The bacterial species selected for antimicrobial studies were Escherichia coli, Pseudomonas aeruginosa, Bacillus subtilis and Staphylococcus aureus based on their common occurrence in contaminated water, contaminated food, hospital acquired infections and predominant occurrence in drinking water biofilms formed in the water distribution system. Complete killing of bacterial strains was further confirmed by testing the presence/absence of viable but non-culturable (VBNC) state bacteria by confocal laser scanning microscopy (CLSM). Cytotoxic potential of the synthesized AgNPs was determined using the 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl- tetrazolium bromide (MTT) assay using the MCF-7 cell line.
Materials and methods
Chemicals and reagents
Silver nitrate (AgNO3, > 99% pure) was purchased from Merck (India). Nutrient broth (NB) media and nutrient agar (NA) were procured from HiMedia, Mumbai, India. Gupta and Kshirsagar media (GK media, details given in Electronic supplementary information (ESI), section S1) components were procured from Merck (India). Brucine sulfate and sulfanilic acid reagent, N-(1-naphthyl)ethylenediamine (NEED), and sulfanilamide were purchased from Merck (India). Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS) and penicillin–streptomycin, and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-tetrazolium bromide (MTT) were procured from HiMedia. Dimethyl sulfoxide (DMSO) was procured from Merck (India).
Source of microorganism and their growth and maintenance
T. pantotropha (ATCC 35512) was procured from American Type Culture Collection (ATCC, USA). Two strains of Gram-positive bacteria (B. subtilis, MTCC 441 and S. aureus, NCIM 5022) and Gram-negative bacteria (P. aeruginosa, GeneBank accession No. KF751345 and E. coli, MTCC 443) were used to evaluate the antimicrobial activity of AgNPs. E. coli and B. subtilis were purchased from Institute of Microbial Technology (Chandigarh, India) and S. aureus was procured from National Centre for Industrial Microorganisms (NCIM, Pune, India). P. aeruginosa (RS1) was isolated from oily sludge, as discussed elsewhere (Jasmine and Mukherji 2014). Both NB (1.3%) and GK media were used for growth and maintenance of T. pantotropha (Phatak et al. 2016), while only NB (1.3%) was used for maintenance of all the other strains. The GK media is a defined media containing 1.64 g/l sodium acetate as the carbon source with nitrogen provided simultaneously in the form of nitrite, nitrate, and ammonium salts (details provided in ESI). For preparing the inoculum for initiating a batch culture, each pure culture was grown up to end of log phase (24 h) and was harvested by centrifugation at 10,000 rpm for 10 min. The cells were washed thrice and resuspended in phosphate buffer (pH 7.2) to achieve unit absorbance at 600 nm (~ 1 × 109 CFU/ml bacterial concentration). A batch culture was started by adding 0.1 ml of the inoculum (adjusted to unit absorbance) to 100 ml media (NB/GK). The cultures were incubated for 24 h in an orbital shaker cum incubator set at 180 rpm and 37 °C. For biogenic AgNP synthesis, the T. pantotropha culture after 24 h incubation was centrifuged, and the culture supernatant was used for AgNP synthesis. The protein concentration and enzyme activity in the supernatant were also determined, as discussed later. For testing antibacterial activity, each of the four strains selected were harvested, washed, resuspended in phosphate buffer to achieve unit absorbance at 600 nm and appropriately diluted to achieve the desired initial concentration.
The MCF-7 cell line was procured from National Centre for Cell Science (NCCS), Pune, India. The MCF-7 cell lines were grown in 75 cm2 tissue culture flasks as a monolayer in DMEM, supplemented with 10% FBS, and 1% penicillin–streptomycin and were incubated at 37 °C in a 5% CO2 incubator. All the cultures were subcultured periodically to ensure viability of the cultures.
Method optimization for biosynthesis of silver nanoparticles
The bacterial strain, T. pantotropha was grown in NB by incubating the culture at 37 °C for 24 h, and the culture supernatant was collected and filtered using a 0.45 µm membrane filter. The filtered culture supernatant was used for biosynthesis of AgNPs in the presence of varying AgNO3 concentration, maintaining a ratio of culture supernatant to AgNO3 solution as 1:1 (v/v) while stirring the mixture. Growth media with AgNO3 was kept as the positive control, while only AgNO3 solution served as the negative control. The mixtures were incubated in an orbital shaker operated at 180 rpm to facilitate nanoparticle synthesis. Optimization of AgNP biosynthesis was done by varying the parameters, including AgNO3 concentration (1, 2, and 5 mM), temperature (37 °C, 60 °C, and 90 °C) and growth medium (NB and GK media). The AgNPs were centrifuged at 12,000 rpm for 30 min and were redispersed in DI water, characterized and stored at 4 °C for future use.
Characterization of silver nanoparticles
The absorbance spectra of AgNPs were recorded over the wavelength range, 200–800 nm using a UV–visible spectrophotometer (Shimadzu UV 1800, Japan). The size and shape of AgNPs were characterized using FEG-TEM (JEOL, JEM-2100F, Japan) and FEG-SEM (JEOL, JSM-7600F, Japan). The crystallinity of nanoparticles was determined by recording the XRD spectra with CuKα radiation (λ = 1.5418 Å) in an X-ray diffractometer (XRD, PANalytical X’pert PRO, Netherlands), operated over the 2θ range, 10°–80°. The XRD spectra were analyzed using the X’pert Highscore software. The stability of the nanoparticles was tested by measuring the zeta potential using zeta potential analyzer (Zeta pals, Brookhaven, USA). The functional groups involved in the synthesis of AgNPs were confirmed by FTIR spectroscopy (3000 Hyperion Microscope with Vertex 80 FTIR System, Bruker, Germany) in the wavenumber range 500–4500 cm−1. The yield of AgNPs was determined by acid digestion of the AgNP suspension followed by determination of silver concentration using ICP-AES (ARCOS, SPECTRO Analytical Instruments, Germany).
Mechanism of biosynthesis of silver nanoparticles
Nitrate- and nitrite-reductase enzyme activity
The NaR and NiR enzyme activity in the T. pantotropha culture supernatant were quantified to illustrate the mechanism of AgNP biosynthesis. Initially the protein content was estimated in 24 h grown T. pantotropha culture supernatant using Bradford method (Bradford 1976). Subsequently, NaR enzyme assay was performed using the culture supernatant, as described by Saifuddin et al. (2009). The assay reagents used for estimating the enzyme activity included 60 mg/l nitrate and 5% propanol in 0.1 M phosphate buffer (pH 7.5). NaR and NiR enzyme activity was determined by mixing the assay reagents and the culture supernatant in 1:1 (v/v) ratio. For quantification of NaR and NiR enzyme activity in the culture supernatant, separate enzyme assays were performed using a similar protocol with 14 mg/l (~ 225.8 µM) nitrate and 10 mg/l (~ 217 µM) nitrite concentration. The activity of these enzymes was demonstrated by measuring the change in nitrate and nitrite concentration in the reaction mixture over a period of 3 h. Nitrate concentration was determined using the brucine sulfanilic acid method (United States Environment Protection Agency 1971). The quantification of nitrite was done using the Griess reaction-based two-step diazotization reaction using sulfanilamide and NEED solution (Clesceri et al. 1999). The control reaction consisted of culture supernatant boiled in a water bath for 10 min to deactivate the enzymes. The reduced form of nicotinamide adenine dinucleotide (NADH) in the culture supernatant was also quantified to establish the mechanism of NADH-dependent nitrate reduction. NADH concentration was measured by measuring the absorbance at 340 nm using UV–Vis spectrophotometer (Held 2007). The concentration of chloride in the nutrient media was measured using argentometric titration (Clesceri et al. 1999).
Biocidal potential of biosynthesized AgNPs
Zone Inhibition test
Antibacterial effect of AgNPs was tested using zone inhibition test employing AgNP- (20 µg) loaded discs (0.2 µm pore size, 0.8 cm diameter, Pall corporation), as reported by Ruparelia et al. (2008). Briefly, the bacterial suspension (105 CFU/ml) was uniformly spread on nutrient agar plates before placing the discs. The AgNP-loaded discs and control disc (without AgNPs) were placed on each plate and were incubated at 37 °C for 24 h. The zone of inhibition (ZOI) was measured using a ruler and reported as mean ± standard deviation (SD) based on three replicate measurements for each bacterial strain.
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) of AgNPs
MIC is the lowest concentration of a test material that inhibits growth of microorganisms and MBC is the lowest concentration that kills 99.9% of the population (Ruparelia et al. 2008). The method reported by Ruparelia et al. (2008) was employed for determining the MIC and MBC of AgNPs. Briefly, growth studies were conducted for each representative bacterial strain in batch mode in the presence of varying concentration of AgNPs, i.e., 1–60 µg/ml and initial bacterial concentration (No) of 103 CFU/ml. A positive control containing NB medium with AgNPs only and negative control containing only bacteria inoculated in NB were also maintained. For MIC determination, bacterial growth was measured as an increase in absorbance at 600 nm over 24 h. MBC was determined by observing the presence or absence of bacterial growth in agar plates. Each plate was inoculated with a 100 µl aliquot withdrawn from each test flask with AgNP concentration higher than MIC.
Testing for presence of viable but non-culturable (VBNC) bacteria in the treated samples
Confocal laser scanning microscopy (CLSM) was used to explore the possible presence of bacteria in the viable but non-culturable (VBNC) state in the treated samples, which showed the absence of culturable bacteria. Bacterial viability was tested using a live/dead staining kit (BacLight kit, Invitrogen, Molecular Probes) containing nucleic acid stains SYTO 9 and propidium iodide (PI). SYTO 9 is a green fluorescent nucleic acid stain with excitation/emission maxima at 480/500 nm, which labels all cells. In comparison, PI is a red-fluorescent nucleic acid stain with excitation/emission maxima at 490/635 nm which stains damaged cells. For imaging, the samples were treated with 20 μl of the stain mixture (SYTO 9/PI) and incubated at 4 °C in the dark for 15 min. Subsequently, the samples were placed on the coverslip and examined under the confocal microscope (U-LH100HG, Olympus Fluoview, Japan) at 100 × using FITC and PI filter for CLSM imaging.
Cytotoxic potential of AgNPs against breast cancer cell line (MCF-7)
The in vitro cytotoxic potential of AgNPs was tested against MCF-7 cell lines using MTT assay (Jeyaraj et al. 2013). Briefly, the cytotoxic effect was tested on the MCF-7 cell line (104 cells/ml) at varying concentration of AgNPs. MCF-7 cells were grown in 96-well plates containing DMEM with 10% FBS and 1% penicillin–streptomycin. The plates were incubated in a CO2 (5%) incubator for 24 h. Subsequently, the cells were washed and exposed to varying concentrations of AgNPs (0–90 µg/ml) and were incubated in a CO2 incubator for 24 h. Further, the AgNP-treated cells were tested for cell viability using the MTT assay. In this assay, MTT reagent (10 µl, 5 mg/ml) was added to each well, and the plate was incubated at 37 °C for 4 h. DMSO (100 µl) was added to the formazan crystal formed, and absorbance of each well was recorded at 570 nm. The concentration of AgNPs showing 50% cell viability with respect to the control (IC50) was determined by plotting % cell viability (Eq. 1) versus concentration of AgNPs (Jeyaraj et al. 2013):
| 1 |
Results and discussion
Extracellular biosynthesis of silver nanoparticles
Synthesis of AgNPs was indicated by a color change of the reaction mixture to dark brown within 24 h. Stable appearance of dark brown color in the solution confirmed the synthesis of AgNPs. No change in solution color was observed in the negative control and positive control, which revealed that the reduction of Ag+ ion was solely due to components secreted by the bacterial cells in nutrient media. A localized surface plasmon resonance (LSPR) peak in the range of 400–450 nm in UV–Vis spectra revealed the formation of AgNPs. Biological components responsible for the reduction of metal ions for the biosynthesis of nanoparticles may include proteins, enzymes, soluble redox mediators, and secondary metabolites (Durán et al. 2005). These biomolecules are also reported to play a vital role in the capping of the nanoparticles, thereby providing stability to the dispersion (Durán et al. 2005; Saifuddin et al. 2009). Peptides are frequently reported to remain associated with the surface of nanoparticles (Sharma et al. 2019).
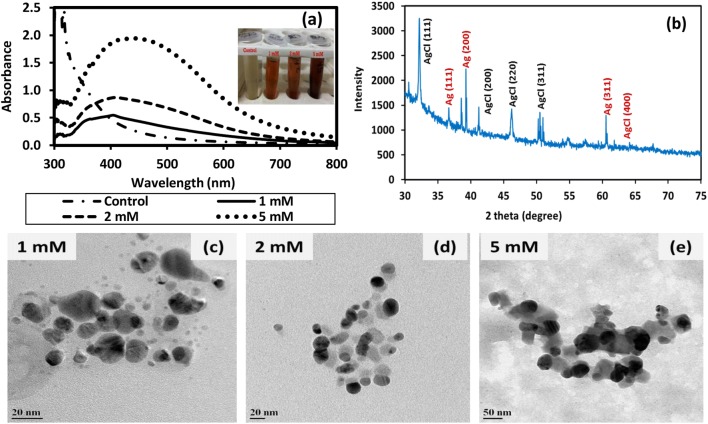
Changing the various parameters affected the characteristics of AgNPs formed. An increase in the concentration of AgNO3 revealed increased synthesis of AgNPs by exhibiting higher absorbance, as confirmed from the absorbance spectra (Fig. 1a). No peak was observed in the nutrient broth control in the wavelength range 390–450 nm where the LSPR peak of AgNPs is expected (Fig. 1a). AgNPs synthesized using varying AgNO3 concentrations (i.e., 1, 2, and 5 mM in the final mixture) exhibited localized surface plasmon resonance (LSPR) peaks at 400, 404 and 440 nm, respectively. The bathochromic-shift in LSPR peak indicated an increase in size with increasing concentration of AgNO3 up to 5 mM, which was also observed by other researchers (Bar et al. 2009). Microscopic imaging of AgNPs was recorded using FEG-TEM (Fig. 1c–e) and FEG-SEM (Fig. S1). The size (mean ± SD and size range) of AgNPs based on FEG-TEM image analysis were 9.6 ± 4.9 nm (2–26 nm), 14.6 ± 4.6 nm (5–51 nm) and 32.4 ± 10.5 nm (16–72 nm) and the corresponding zeta potential values (Fig. S1) were −18.2, −33.6 and −54 mV for AgNPs synthesized using 1, 2 and 5 mM AgNO3, respectively. The yield of AgNPs determined using ICP-AES was found to be 72%, 82% and 84% for AgNO3 concentration of 1, 2 and 5 mM, respectively.
Fig. 1.
Characterization of silver and silver chloride nanoparticles (Ag/AgCl NPs) synthesized using culture supernatant of nutrient broth (NB)-grown T. pantotropha and AgNO3 at 37 °C and pH 7 a UV–Vis spectra of NB control and Ag/AgCl NPs synthesized at varying AgNO3 concentration, b XRD spectra of Ag/AgCl NPs synthesized using 2 mM AgNO3, and FEG-TEM images of Ag/AgCl NPs synthesized using c 1 mM AgNO3, d 2 mM AgNO3 and e 5 mM AgNO3. The inset in a depicts the visual appearance of the NB control and Ag/AgCl NP suspensions synthesized at varying AgNO3 concentration
The full width at half maxima and position of the LSPR peak not only depends on the size and shape of the nanoparticles but also depends on the chemical properties of the nanocrystalline surface (Basavaraja et al. 2008). Hence, nanoparticles of the same size synthesized by different methods may exhibit some variation in their LSPR peak position. However, typically the peak for AgNPs lies in the wavelength range 390–450 nm. Agnihotri et al. (2014) reported the characteristic wavelength for LSPR peak of spherical AgNPs at 393, 394, 398, 401, 406, 411, 420, 429, 449, and 462 nm for AgNPs with the average sizes 5, 7, 10, 15, 20, 30, 50, 63, 85, and 100 nm, respectively. Thus, the LSPR peak at 404 nm observed for the AgNPs synthesized by adding 2 mM AgNO3 in the culture supernatant derived from T. pantotropha grown on nutrient broth matches well with the average AgNP size of 14.6 nm. Using Cunninghamella phaeospora culture supernatant, Ghareib et al. (2016) also reported an LSPR peak at 415 nm for AgNPs of average size 14 nm (size range 6–32 nm). In another study by Maiti et al. (2014), AgNPs with size range 10–40 nm synthesized using Lycopersicon esculentum extracts exhibited the LSPR peak at 410 nm. In the present study, the AgNPs synthesized using varying AgNO3 concentrations, i.e., 1, 2, and 5 mM exhibited LSPR peak at 400, 404, and 440 nm, respectively. The LSPR peak position and average size based on TEM (9.6 nm, 14.6 nm, and 32.4 nm, respectively) were as expected based on the literature. Minor variation in LSPR peak position may be due to the complex composition of the media used for biosynthesis of AgNPs. Compared to chemically synthesized AgNPs (Agnihotri et al. 2014), the size range of biologically synthesized AgNPs was found to be much wider. This may be attributed to the complexity of the media used for synthesis (nutrient broth along with biomolecules released by T. pantotropha). The use of biomolecules purified from the culture supernatant may result in the synthesis of nanoparticles with a narrower size distribution. However, the interaction between nutrient media components together with the biomolecules released may have facilitated the synthesis. This synergistic effect may be lost when purified biomolecules are used for synthesis. Moreover, the complexity of the synthesis process would also increase.
To determine crystallinity, AgNPs synthesized using 2 mM AgNO3 were characterized using XRD, where the characteristic peaks of AgNPs appeared at 38.12°, 44.43°, 64.53°, 77.55° (Fig. 1b), corresponding to the planes (111), (200), (220) and (311) of face-centered cubic (fcc) crystal structure of silver (JCPDS reference code 00-004-483). Additional peaks were also found at 2θ values of 27.90°, 32.28°, 46.29°, 54.73°, and 66.07°, which corresponded to (111), (200), (220), (311) planes of AgCl NPs demonstrating fcc crystal structure (Durán et al. 2016). The presence of chloride peak in the EDS spectra also confirmed the formation of AgCl NPs (Fig. S1), which may be attributed to high chloride concentration in the NB media (chloride concentration in NB = 1500 ± 0.47 mg/l).
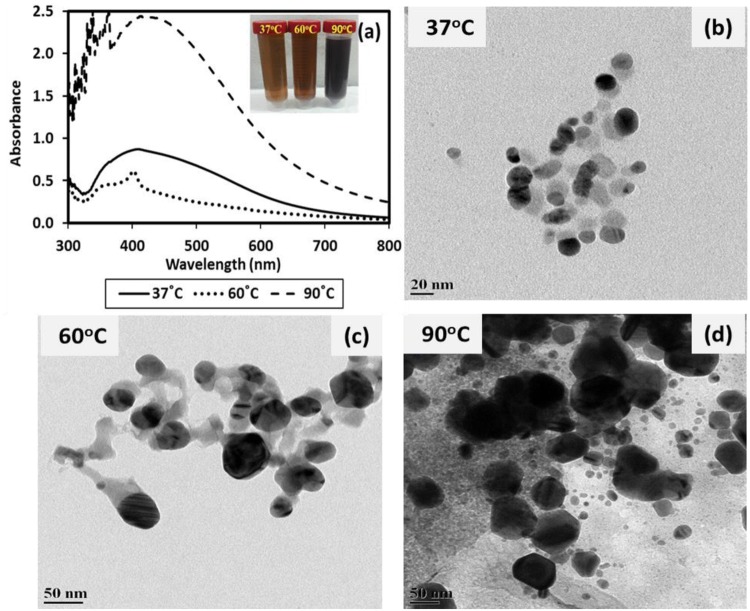
To study the effect of temperature, AgNPs were also synthesized with 2 mM AgNO3 concentration at various temperatures (37 °C, 60 °C, and 90 °C). The UV–Vis spectra exhibited the increasing trend in absorbance of AgNPs and LSPR peaks at 404, 405 and 415 nm for AgNPs synthesized at 37 °C, 60 °C, and 90 °C, respectively (Fig. 2a) exhibiting a bathochromic shift and indicating the larger size of AgNPs (Fig. 2a). This trend was also confirmed in FEG-TEM (Fig. 2b–d) and FEG-SEM (Fig. S2) images, which exhibited size (mean ± SD and size range) of 14.6 ± 4.0 nm (6–42 nm), 28.2 ± 9.09 nm (11–64 nm) and 33.3 ± 13.4 nm (12–92 nm) for AgNPs synthesized at 37 °C, 60 °C, and 90 °C, respectively. However, a comparable yield of 82.2%, 78.2%, and 79.6% was obtained for AgNPs synthesized at 37 °C, 60 °C, and 90 °C, respectively. Hence, the increasing absorbance values for AgNPs at higher temperature (Fig. 2a) may be ascribed to more intense color due to the charring of biomolecules at elevated temperatures (Fig. 2a inset). Since enzymes are sensitive to high temperatures, enzyme-mediated biosynthesis is unlikely to promote AgNP synthesis at elevated temperatures. However, extracellular polymeric substances (EPS) secreted in the growth medium may have acted as a reducing agent and contributed to AgNP synthesis, as also reported by other researchers (Saravanan et al. 2017). The zeta potential values were −33.6, −55 and −50 mV for nanoparticles synthesized at 37, 60 and 90 °C, respectively. Multiple peaks observed during zeta potential determination (Fig. S2) may be attributed to the presence of complex biomolecules in the culture supernatant (Skoglund et al. 2017).
Fig. 2.
Characterization of silver and silver chloride nanoparticles (Ag/AgCl NPs) synthesized using culture supernatant of nutrient broth (NB)-grown T. pantotropha and 2 mM AgNO3 at neutral pH and various incubation temperature a UV–Vis spectra of Ag/AgCl NPs synthesized at 37 °C, 60 °C and 90 °C, and FEG-TEM images of Ag/AgCl NPs synthesized at b 37 °C, c 60 °C and d 90 °C. The inset in a depicts the visual appearance of the AgNP suspensions synthesized at varying temperatures
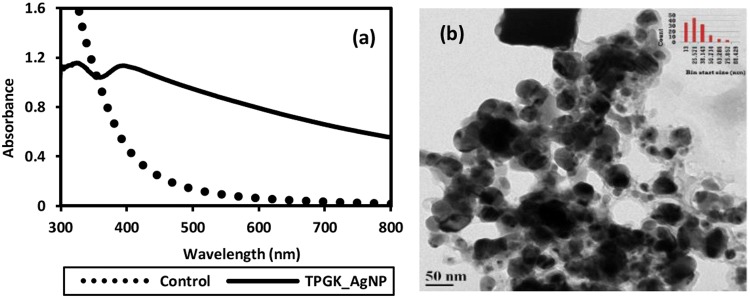
In addition to AgNP synthesis in NB as discussed earlier (Fig. 1), GK media culture supernatant was also used for biosynthesis of AgNPs. Nanoparticles synthesized using GK media culture supernatant are shown in Fig. 3. A broad LSPR peak was found at ~ 405 nm in the UV–Vis spectra (Fig. 3a), which revealed a non-homogenous size distribution of AgNPs. FEG-TEM micrographs (Fig. 3b) also depicted wide variability in the size of AgNPs with an average size of 37 ± 17.8 nm (range: 11–81 nm). FEG-SEM micrographs (Fig. S3 inset) also revealed non-homogeneous size distribution. EDS spectra (Fig. S3) exhibited a peak for Ag as well as for Cl, thus demonstrating the synthesis of Ag/AgCl NPs. The chloride content in GK media was quantified as 301 ± 0.94 mg/l. Hence, the synthesis of AgCl NPs may have occurred as a result of the reaction between Ag+ and Cl− present in GK media along with other biological components released into the growth medium. The zeta potential was −35 mV. The yield of nanoparticles (74.8%) was marginally lower than that for nanoparticles synthesized using NB grown culture supernatant.
Fig. 3.
a UV–Vis spectra of silver and silver chloride nanoparticles (Ag/AgCl NPs) and b FEG-TEM image of Ag/AgCl NPs synthesized using 2 mM AgNO3 at 37 °C and neutral pH using the culture supernatant of GK media-grown T. pantotropha
Mechanism of biosynthesis of silver nanoparticles
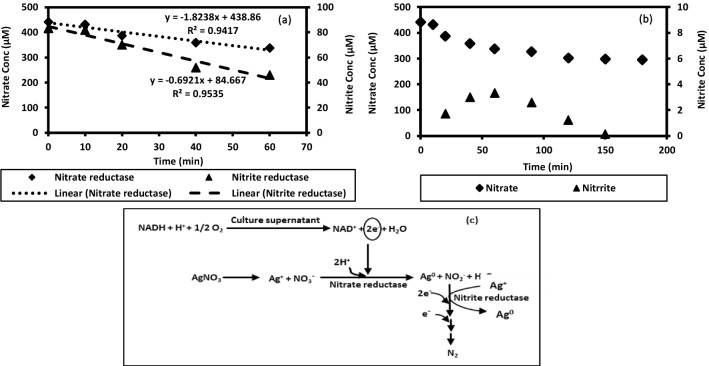
Although the exact underlying mechanism is still not well defined, the mechanism of biosynthesis of nanoparticles may be attributed to the reduction of metal ions by protein/enzymes/other biomolecules secreted by the cells (Durán et al. 2005). The protein content and enzyme activity were analyzed in 24 h old culture supernatant. The protein concentration in NB and NB grown culture supernatant was quantified as 43 ± 1.2 µg/ml and 170.5 ± 3.6 µg/ml, respectively, whereas, the protein concentration in GK media culture supernatant was quantified as 25.3 ± 2.4 µg/ml. No protein was present in the GK media. These results confirmed that extracellular proteins and enzymes released into the culture medium by T. pantotropha contributed to nanoparticle synthesis. Nanoparticle synthesis mediated by NaR enzyme is widely reported (Durán et al. 2005; Kumar et al. 2007; Saifuddin et al. 2009). NaR enzyme is a NADH-dependent molybdoenzyme that reduces nitrate to nitrite by accepting two electrons released by oxidation of NADH (Kumar et al. 2007). The released electrons may also reduce Ag+ ions to AgNPs. The NaR and NiR enzyme activity in the culture supernatant of T. pantotropha was confirmed through enzyme assays (Fig. 4a). The concentration profile of nitrate and nitrite due to NaR and NiR activity over time is shown in Fig. 4a, b. NaR activity was determined by monitoring the nitrate disappearance rate as 1164.8 nM/min. In this study, the nitrite production rate was not quantified as a measure of NaR activity since nitrite generated may get simultaneously utilized by the NiR enzyme. Nitrate reduction in T. pantotropha is also known to proceed via NO as an intermediate (Bell et al. 1990). The total enzyme activity (TEA, U/ml) for NaR enzyme was determined as 61.1 U/ml, and specific enzyme activity was determined as 358.4 U/mg of total protein. One unit (U) is defined as the amount of enzyme that catalyzes the reaction of 1 nmol of substrate (nitrate or nitrite) per min. NiR enzyme activity in the culture supernatant was also confirmed using nitrite as the substrate where the decrease in nitrite concentration was quantified over a 1 h period (Fig. 4a). NiR enzyme activity in the culture supernatant was determined as 692 nM/min. The total enzyme activity (TEA, U/ml) for NiR enzyme was determined as 31.05 U/ml, and specific enzyme activity was determined as 182.1 U/mg of total protein. Based on these results, a proposed schematic describing the mechanism of AgNP biosynthesis is shown in Fig. 4c. No prior study has shown the NaR and NiR enzyme activities in the culture supernatant of T. pantotropha as demonstrated in this work.
Fig. 4.
a Concentration of nitrate and nitrite during nitrate reductase and nitrite-reductase enzyme assay using the culture supernatant of nutrient broth (NB)-grown T. pantotropha, b concentration profile of nitrate and nitrite over time in a reaction mixture containing 30 mg/l nitrate and culture supernatant of NB-grown T. pantotropha and c proposed mechanism for the biosynthesis of silver nanoparticles using T. pantotropha culture supernatant
The coenzyme NADH linked with NaR and NiR enzyme facilitates nitrate and nitrite reduction by providing electrons. A similar mechanism may be involved in the reduction of Ag+ ions to Ag0 (AgNPs). The NADH concentration was also quantified as 6.41 ± 0.28 μM (mean ± SE) in the culture supernatant. Thus, it can be concluded that the NADH-dependent NaR and NiR enzymes may have played a role in the reduction of silver ions for synthesis of nanoparticles. The observations of this study are consistent with the results reported by Saifuddin et al. (2009), where NaR enzyme activity was reported as 152 nmol/h/ml in Bacillus subtilis (ATCC 6633) culture supernatant used for nanoparticle synthesis (Saifuddin et al. 2009).
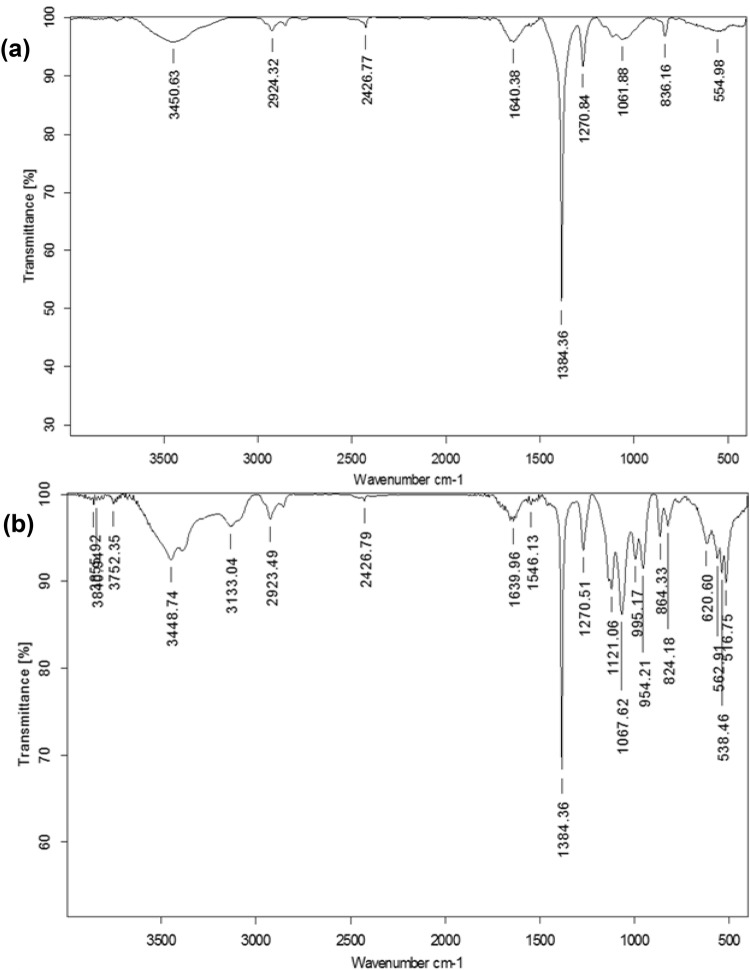
In addition to the reduction of Ag+ ions, presence of biomolecules, and extracellular polymeric substances (EPS) secreted by bacteria in the NB and GK media culture supernatant may also have contributed to stability by capping the nanoparticles. The stability may be attributed to the presence of aromatic amino acids of proteins, which arise due to electron excitations in tryptophan and tyrosine residues/peptides/their residues (Durán et al. 2005; Thamilselvi and Radha 2013). The FTIR spectra (Fig. 5a, b) revealed bands at 3450 and 3448 cm−1, which may be attributed to stretching vibration of primary amine, thus, signifying the presence of amide bonds/amino groups (Thamilselvi and Radha 2013). The bands at 3133 cm−1 and 2923 cm−1 correspond to primary and secondary amines, respectively (Jeevan et al. 2012). The bands observed at 1384 and 1067 cm−1 correspond to –C–N stretching vibrations of aromatic and aliphatic amines, respectively, while the band at 1640 cm−1 reveals the presence of –C=O and –N–H stretching vibrations in the amide linkages, thus, indicating the presence of proteins/peptides on the surface (Jain et al. 2011; Jeevan et al. 2012; Mohanta and Behera 2014). The bands at 824 cm−1 correspond to the stretching vibration of C–Cl group. Additional bands in Fig. 5b were possibly observed due to the complex nature of biomolecules and other undefined components present in NB. The contribution of biomolecules to the stability of nanoparticles was also revealed by a significant drop in total organic carbon (TOC), i.e., 59% and 62% reduction in the NB and GK media culture supernatant during AgNP synthesis, respectively.
Fig. 5.
FTIR spectra of silver and silver chloride nanoparticles (Ag/AgCl NPs) biosynthesized at 37 °C and pH 7 using 2 mM AgNO3 and the culture supernatant of T. pantotropha grown in a GK media and b nutrient broth
Antimicrobial effectiveness of AgNPs
E. coli, P. aeruginosa, B. subtilis and S. aureus, were used for testing the antibacterial activity of biogenic AgNPs. Coliforms (including E. coli) are indicator organisms representing fecal contamination in drinking water (Ashbolt 2015; Bitton 2005). It is a common practice to use non-pathogenic surrogates, such as coliforms and E. coli for estimating the effectiveness of disinfection (Bitton 2005). However, some strains of E. coli are also known to be pathogenic. Various microorganisms occurring in the environment are potential opportunistic pathogens that may cause diseases in immune-compromised people. Pseudomonas aeruginosa is among the six bacterial species recently designated as an emerging threat due to their association with hospital-acquired infections (Ramachandran et al. 2014). Due to its oligotrophic nature, P. aeruginosa may regrow in treated water, and its presence is commonly reported in drinking water biofilms. Contamination of water in swimming pools and spas with P. aeruginosa is known to cause folliculitis (Ashbolt 2015). Bacillus spp. are commonly detected in most drinking-water supplies. Although most Bacillus spp. are harmless, a few are pathogenic to humans and animals (Gorchev and Ozolins 2011). S. aureus is also of potential concern due to its transmission via animals to water. Hence, it is also considered a useful reference pathogen (Ashbolt 2015). Various other studies have employed these bacteria for testing the effectiveness of chemically synthesized silver nanoparticles (Agnihotri et al. 2013; Ruparelia et al. 2008), such that choice of these microorganisms also allowed comparison of the effectiveness of these biogenic AgNPs with chemically synthesized AgNPs.
Zone of inhibition (ZOI) test
The Ag/AgCl NP- (20 μg) loaded discs exhibited potent antimicrobial activity, as evident from the clear ZOI. However, no ZOI was observed for the control disc. The average ZOI was determined as 16.4 ± 1.8, 14.9 ± 0.7, 12.7 ± 0.8 and 12.4 ± 0.7 mm for E. coli, P. aeruginosa, B. subtilis, and S. aureus, respectively, indicating E. coli as the most sensitive strain.
Minimum inhibitory concentration test and minimum bactericidal concentration test
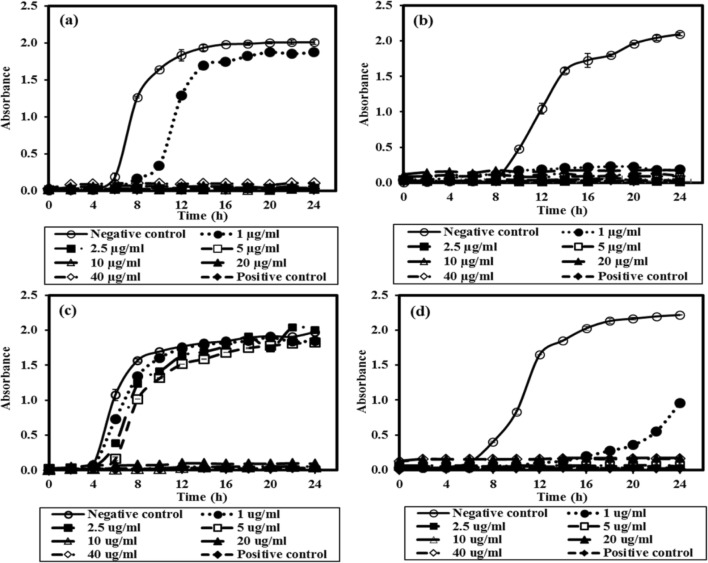
The Ag/AgCl NPs exhibited strong bacteriostatic and bactericidal potential against the tested bacterial strains. At low concentration of Ag/AgCl NPs, an increase in absorbance at 600 nm could be observed visibly due to bacterial growth. In contrast, no increase in absorbance was observed at 2.5 µg/ml or higher concentration of Ag/AgCl NPs (14 nm) for all the selected bacterial strains, except B. subtilis. Complete growth inhibition was observed at 10 µg/ml for B. subtilis (Fig. 6), demonstrating B. subtilis as the most resistant strain among the tested bacteria. At Ag/AgCl NP concentration below the MIC, an increase in nanoparticle concentration caused an increase in the lag phase for all the cultures. Suresh et al. (2010) reported MIC values of 2.0, 0.5, and 3.0 µg/mL for E. coli, B. subtilis, and S. oneidensis, respectively, with biogenic AgNPs (size: 4 nm) synthesized using culture supernatant of Shewanella oneidensis (Suresh et al. 2010). In contrast, the MIC values using chemically synthesized AgNPs of comparable size (~ 15 nm) were much higher for all the strains, i.e., 30, 50 and 100 µg/ml for E. coli MTCC 443, B. subtilis and S. aureus, respectively for No of 105–106 CFU/ml (Agnihotri et al. 2014). In a similar study, Ruparelia et al. (2008) reported MIC values of AgNPs (~ 3 nm) as 40, 40 and 120 µg/ml for E. coli MTCC 443, B. subtilis and S. aureus for No of 103–104 CFU/ml, respectively (Ruparelia et al. 2008).
Fig. 6.
Growth profile of various bacterial strains at varying concentration of silver and silver chloride nanoparticles synthesized using culture supernatant of nutrient broth-grown T. pantotropha and 2 mM AgNO3 at 37 °C and pH 7 aE. coli, bP. aeruginosa, cB. subtilis and dS. aureus
Plating was done for the various bacterial strains treated with AgNP concentration equal to or greater than the MIC. After incubation of the plates, the MBC values were found to be 2.5, 10 µg/ml, 20 and 5 µg/ml for E. coli, P. aeruginosa, B. subtilis and S. aureus, respectively. In contrast, MBC values using 3 nm AgNPs were much higher for all the strains, i.e., 60, 60 and 160 µg/ml for E. coli MTCC 443, B. subtilis and S. aureus for No of 103–104 CFU/ml, respectively (Ruparelia et al. 2008), although AgNPs of smaller size are expected to enhance antimicrobial activity. In another study, MBC values of chemically synthesized AgNPs of comparable size (~ 15 nm) were much higher, i.e., 50, 60 and 110 µg/ml for E. coli MTCC 443, B. subtilis and S. aureus, for No of 105–106 CFU/ml, respectively (Agnihotri et al. 2014). The results of the present study confirmed that the synthesized Ag/AgCl NPs are significantly more effective as compared to chemically synthesized AgNPs. This higher antimicrobial activity may be attributed to the rapid interaction of bacterial cells due to the presence of biomolecules on the surface of Ag/AgCl NPs. These biomolecules may have facilitated effective penetration into the cell, causing a series of biochemical reactions, finally leading to cell death.
Mechanism of inactivation of bacteria
The interaction between bacteria and the AgNPs could have triggered a change in cell permeability, thereby leading to a series of changes in the bacterial cells. AgNPs may also disrupt protein and enzyme synthesis by binding to the disulfide linkage present at the active site of the proteins and enzymes. These may have led to the breakage of the cell wall as a result of contact with AgNPs and pit formation in the plasma membrane (Bharti et al. 2015). AgNPs are also known to interfere with DNA replication by interacting with the phosphate group of the DNA molecules, thereby inducing cell death (Agnihotri et al. 2013). Complete inactivation of bacterial cells was confirmed using the Live/Dead BacLight staining assay (Fig. S4) (Das et al. 2012). In CLSM imaging, the Ag/AgCl NP-treated cells for concentration corresponding to MBC exhibited only red fluorescence (Fig. S4 a, c, e, g), thereby demonstrating the presence of dead bacterial cells (Fig. S4 b, d, f, h). SYTO 9 can penetrate and stain nucleic acid in all cells and exhibit green fluorescence. In contrast, PI only penetrates cells with damaged membranes and also masks the fluorescence due to SYTO 9.
In vitro cytotoxic potential of AgNPs
The Ag/AgCl NPs also exhibited a cytotoxic effect on MCF-7 cell line. The IC50 value of AgNPs was 34.8 µg/ml, and complete killing of the cells was observed at 90 µg/ml (Fig. S5). The IC50 value on MCF-7 cell line was only slightly lower than for chemically synthesized AgNPs which exhibited IC50 of 40 µg/ml, as reported by Bharti et al. (2019) The cytotoxicity of these biogenic silver AgNPs was in the range reported by other researchers. Based on in vitro studies using MCF-7 cell line and biogenic AgNPs, Vivek et al. (2012) reported IC50 of 50 µg/ml while Jeyaraj et al. (2013) reported IC50 value of 20 µg/ml (22 nm AgNPs synthesized using Sesbania grandiflora (L) leaf extract).
Conclusion
Silver nanoparticles were successfully synthesized with T. pantotropha culture supernatant using AgNO3 concentration of 2 mM at 37 °C. The presence of a single peak at 404 nm in UV–Vis spectra revealed synthesis of spherical AgNPs with 2 mM AgNO3 (yield 82%) with an average size of 14.6 nm. The Ag/AgCl NPs demonstrated good antimicrobial activity showing MIC values of 2.5 µg/ml or lower for all strains tested except for B. subtilis, which exhibited MIC value of 10 µg/ml. The MBC values were 2.5, 5, 10, and 20 µg/ml for E. coli, S. aureus, P. aeruginosa and B. subtilis, respectively. CLSM imaging further confirmed the complete inactivation of all the bacterial strains and the absence of VBNC state bacteria. The cytotoxic effect of these biogenic AgNPs measured through in vitro studies was in the range reported by other researchers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The authors would like to acknowledge SAIF, IIT Bombay for providing FEG-TEM, FEG-SEM and FTIR facilities; MEMS Department, IIT Bombay for providing the XRD Central facility and zeta potential analyzer used for characterization of nanoparticles; and BSBE, IIT Bombay for providing the CLSM Central facility for imaging of bacterial cells.
Author contributions
SB performed all the experiments, analyzed the results and prepared the manuscript. Prof. SM supervised the work and revised the manuscript. Prof. SM co-supervised the research work. All authors discussed the experiments and the results and contributed to the final manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
References
- Agnihotri S, Mukherji S, Mukherji S. Immobilized silver nanoparticles enhance contact killing and show highest efficacy: elucidation of the mechanism of bactericidal action of silver. Nanoscale. 2013;5:7328–7340. doi: 10.1039/C3NR00024A. [DOI] [PubMed] [Google Scholar]
- Agnihotri S, Mukherji S, Mukherji S. Size-controlled silver nanoparticles synthesized over the range 5–100 nm using the same protocol and their antibacterial efficacy. RSC Adv. 2014;4:3974–3983. doi: 10.1039/c3ra44507k. [DOI] [Google Scholar]
- Ashbolt NJ. Microbial contamination of drinking water and human health from community water systems. Curr Environ Health Rep. 2015;2:95–106. doi: 10.1007/s40572-014-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar H, Bhui DK, Sahoo GP, et al. Green synthesis of silver nanoparticles using latex of Jatropha curcas. Colloids Surf A Physicochem Eng Asp. 2009;339:134–139. doi: 10.1016/j.colsurfa.2009.02.008. [DOI] [Google Scholar]
- Basavaraja S, Balaji SD, Lagashetty A, et al. Extracellular biosynthesis of silver nanoparticles using the fungus Fusarium semitectum. Mater Res Bull. 2008;43:1164–1170. doi: 10.1016/j.materresbull.2007.06.020. [DOI] [Google Scholar]
- Bell LC, Richardson DJ, Ferguson SJ. Periplasmic and membrane-bound respiratory nitrate reductase in Thiosphaera pantotropha. FEBS Lett. 1990;265:85–87. doi: 10.1016/0014-5793(90)80889-Q. [DOI] [PubMed] [Google Scholar]
- Bharti S, Agnihotri S, Mukherji S, Mukherji S. Effectiveness of immobilized silver nanoparticles in inactivation of pathogenic bacteria. J Environ Res Dev. 2015;9:849–856. [Google Scholar]
- Bharti S, Mukherji S, Mukherji S. Water disinfection using fixed bed reactors packed with silver nanoparticle immobilized glass capillary tubes. Sci Total Environ. 2019;689:991–1000. doi: 10.1016/j.scitotenv.2019.06.482. [DOI] [PubMed] [Google Scholar]
- Bitton G. Wastewater Microbiology. 3. New York: Wiley-Liss Inc.; 2005. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clesceri LS, Greenbaerg AE, Eaton AD. Standard methods for the examination of water and wastewater. 20. Washington, D.C: American Public Health Association American Water Works Association, Water Environment Federation; 1999. [Google Scholar]
- Das SK, Khan MMR, Guha AK, et al. Silver-nano biohybride material: synthesis, characterization and application in water purification. Bioresour Technol. 2012;124:495–499. doi: 10.1016/j.biortech.2012.08.071. [DOI] [PubMed] [Google Scholar]
- Durán N, Marcato PD, Alves OL, et al. Mechanistic aspects of biosynthesis of silver nanoparticles by several Fusarium oxysporum strains. J Nanobiotechnology. 2005;3:8. doi: 10.1186/1477-3155-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durán N, Nakazato G, Seabra AB. Antimicrobial activity of biogenic silver nanoparticles, and silver chloride nanoparticles: an overview and comments. Appl Microbiol Biotechnol. 2016;100:6555–6570. doi: 10.1007/s00253-016-7657-7. [DOI] [PubMed] [Google Scholar]
- Ghareib M, Tahon MA, Saif MM, Abdallah WES. Rapid extracellular biosynthesis of silver nanoparticles by Cunninghamella phaeospora culture supernatant. Iran J Pharm Res. 2016;15:915–924. [PMC free article] [PubMed] [Google Scholar]
- Gopinath V, Priyadarshini S, Loke MF, et al. Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity. Arab J Chem. 2017;10:1101–1117. doi: 10.1016/j.arabjc.2015.11.011. [DOI] [Google Scholar]
- Gorchev HG, Ozolins G (2011) WHO guidelines for drinking-water quality. In: WHO chronicle, 4th edn. pp 104–108 [PubMed]
- Gupta AB. Thiosphaera pantotropha: a sulphur bacterium capable of simultaneous heterotrophic nitrification and aerobic denitrification. Enzyme Microb Technol. 1997;21:589–595. doi: 10.1016/S0141-0229(97)00070-7. [DOI] [Google Scholar]
- Held P (2007) Determination of NADH concentrations with the synergy TM 2 Multi-detection microplate reader using fluorescence or absorbance. BioTek Instruments, 1–6. https://www.biotek.com/resources/docs/NADH_App_Note.pdf
- Iravani S, Korbekandi H, Mirmohammadi SV, et al. Synthesis of silver nanoparticles: chemical, physical and biological methods. Res Pharm Sci. 2014;9:385–406. [PMC free article] [PubMed] [Google Scholar]
- Jain N, Bhargava A, Majumdar S, et al. Extracellular biosynthesis and characterization of silver nanoparticles using Aspergillus flavus NJP08: a mechanism perspective. Nanoscale. 2011;3:635–641. doi: 10.1039/c0nr00656d. [DOI] [PubMed] [Google Scholar]
- Jasmine J, Mukherji S. Evaluation of bioaugmentation and biostimulation effects on the treatment of refinery oily sludge using 2(n) full factorial design. Environ Sci Process Impacts. 2014;16:1889–1896. doi: 10.1039/C4EM00116H. [DOI] [PubMed] [Google Scholar]
- Jeevan P, Ramya K, Rena E. Extracellular biosynthesis of silver nanoparticles by culture supernatant of Pseudomonas aeruginosa. Indian J Biotechnol. 2012;11:72–76. [Google Scholar]
- Jeyaraj M, Sathishkumar G, Sivanandhan G, et al. Biogenic silver nanoparticles for cancer treatment: an experimental report. Colloids Surf B Biointerfaces. 2013;106:86–92. doi: 10.1016/j.colsurfb.2013.01.027. [DOI] [PubMed] [Google Scholar]
- Karthik L, Kumar G, Kirthi AV, et al. Streptomyces sp. LK3 mediated synthesis of silver nanoparticles and its biomedical application. Bioprocess Biosyst Eng. 2014;37:261–267. doi: 10.1007/s00449-013-0994-3. [DOI] [PubMed] [Google Scholar]
- Kumar SA, Abyaneh MK, Gosavi SW, et al. Nitrate reductase-mediated synthesis of silver nanoparticles from AgNO3. Biotechnol Lett. 2007;29:439–445. doi: 10.1007/s10529-006-9256-7. [DOI] [PubMed] [Google Scholar]
- Li J, Kuang D, Feng Y, et al. Green synthesis of silver nanoparticles-graphene oxide nanocomposite and its application in electrochemical sensing of tryptophan. Biosens Bioelectron. 2013;42:198–206. doi: 10.1016/j.bios.2012.10.029. [DOI] [PubMed] [Google Scholar]
- Maiti S, Krishnan D, Barman G, et al. Antimicrobial activities of silver nanoparticles synthesized from Lycopersicon esculentum extract. J Anal Sci Technol. 2014;5:1–7. doi: 10.1186/s40543-014-0040-3. [DOI] [Google Scholar]
- Mohanta YK, Behera SK. Biosynthesis, characterization and antimicrobial activity of silver nanoparticles by Streptomyces sp. SS2. Bioprocess Biosyst Eng. 2014;37:2263–2269. doi: 10.1007/s00449-014-1205-6. [DOI] [PubMed] [Google Scholar]
- Phatak PS, Trivedi S, Garg A, et al. Start-up of sequencing batch reactor with Thiosphaera pantotropha for treatment of high-strength nitrogenous wastewater and sludge characterization. Environ Sci Pollut Res. 2016;23:20065–20080. doi: 10.1007/s11356-016-6412-z. [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Chalasani AG, Lal R, Roy U. A broad-spectrum antimicrobial activity of Bacillus subtilis RLID 12.1. Sci World J. 2014 doi: 10.1155/2014/968487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruparelia JP, Chatterjee AK, Duttagupta SP, Mukherji S. Strain specificity in antimicrobial activity of silver and copper nanoparticles. Acta Biomater. 2008;4:707–716. doi: 10.1016/j.actbio.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Saifuddin N, Wong CW, Yasumira AAN. Rapid biosynthesis of silver nanoparticles using culture supernatant of bacteria with microwave irradiation. E-J Chem. 2009;6:61–70. doi: 10.1155/2009/734264. [DOI] [Google Scholar]
- Saravanan C, Rajesh R, Kaviarasan T, et al. Synthesis of silver nanoparticles using bacterial exopolysaccharide and its application for degradation of azo-dyes. Biotechnol Rep. 2017;15:33–40. doi: 10.1016/j.btre.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanthi S, David Jayaseelan B, Velusamy P, et al. Biosynthesis of silver nanoparticles using a probiotic Bacillus licheniformis Dahb1 and their antibiofilm activity and toxicity effects in Ceriodaphnia cornuta. Microb Pathog. 2016;93:70–77. doi: 10.1016/j.micpath.2016.01.014. [DOI] [PubMed] [Google Scholar]
- Sharma D, Kanchi S, Bisetty K. Biogenic synthesis of nanoparticles: a review. Arab J Chem. 2019;12:3576–3600. doi: 10.1016/j.arabjc.2015.11.002. [DOI] [Google Scholar]
- Singh S, Bahadur D. Catalytic and antibacterial activity of Ag decorated magnetic core shell nanosphere. Colloids Surf B Biointerfaces. 2015;133:58–65. doi: 10.1016/j.colsurfb.2015.05.033. [DOI] [PubMed] [Google Scholar]
- Skoglund S, Hedberg J, Yunda E, et al. Difficulties and flaws in performing accurate determinations of zeta potentials of metal nanoparticles in complex solutions—four case studies. PLoS ONE. 2017;12:1–19. doi: 10.1371/journal.pone.0181735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suresh AK, Pelletier DA, Wang W, et al. Silver nanocrystallites: biofabrication using Shewanella oneidensis, and an evaluation of their comparative toxicity on Gram-negative and Gram-positive bacteria. Environ Sci Technol. 2010;44:5210–5215. doi: 10.1021/es903684r. [DOI] [PubMed] [Google Scholar]
- Thamilselvi V, Radha KV. Synthesis of silver nanoparticles from Pseudomonas putida NCIM 2650 in silver nitrate supplemented growth medium and optimization using response surface methodology. Dig J Nanomater Biostruct. 2013;8:1101–1111. [Google Scholar]
- United States Environment Protection Agency (1971) Method 352.1: Nitrogen, Nitrate (Colorimetric, Brucine) by Spectrophotometer
- Vivek R, Thangam R, Muthuchelian K, et al. Green biosynthesis of silver nanoparticles from Annona squamosa leaf extract and its in vitro cytotoxic effect on MCF-7 cells. Process Biochem. 2012;47:2405–2410. doi: 10.1016/j.procbio.2012.09.025. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.