Abstract
Deregulation of E3 ubiquitin ligases is intimately implicated in breast cancer pathogenesis and progression, but the underlying mechanisms still remain elusive. Here we report that RING finger protein 144A (RNF144A), a poorly characterized member of the RING-in-between-RING family of E3 ubiquitin ligases, functions as a tumor suppressor in breast cancer. RNF144A was downregulated in a subset of primary breast tumors and restoration of RNF144A suppressed breast cancer cell proliferation, colony formation, migration, invasion in vitro, tumor growth, and lung metastasis in vivo. In contrast, knockdown of RNF144A promoted malignant phenotypes of breast cancer cells. Quantitative proteomics and biochemical analysis revealed that RNF144A interacted with and targeted heat-shock protein family A member 2 (HSPA2), a putative oncoprotein that is frequently upregulated in human cancer and promotes tumor growth and progression, for ubiquitination and degradation. Notably, the ligase activity-defective mutants of RNF144A impaired its ability to induce ubiquitination and degradation of HSPA2, and to suppress breast cancer cell proliferation, migration, and invasion as compared with its wild-type counterpart. Moreover, RNF144A-mediated suppression of breast cancer cell proliferation, migration, and invasion was rescued by ectopic HSPA2 expression. Clinically, low RNF144A and high HSPA2 expression in breast cancer patients was correlated with aggressive clinicopathological characteristics and decreased overall and disease-free survival. Collectively, these findings reveal a previously unappreciated role for RNF144A in suppression of breast cancer growth and metastasis, and identify RNF144A as the first, to our knowledge, E3 ubiquitin ligase for HSPA2 in human cancer.
Subject terms: Oncogenes, Ubiquitin ligases, Tumour-suppressor proteins
Introduction
Breast cancer is the most commonly diagnosed cancer and the leading cause of cancer death among females worldwide [1]. Accumulating evidence shows that deregulation of the ubiquitin–proteasome system (UPS), a key mechanism for regulating intracellular protein degradation and turnover, is implicated in the pathogenesis and progression of breast cancer [2]. The specificity of protein degradation by UPS is determined by E3 ubiquitin ligases, which catalyze the transfer of ubiquitin from an E2 enzyme to a target protein [3]. The human genome encodes ~600 putative E3 ligases, which are classified into two main families based on their structure and catalytic mechanism, including homologous to the E6-AP C terminus (HECT) and really interesting new gene (RING) finger [3–5]. More recently, another mechanistically distinct family of E3 ligases has been identified, termed RING-in-between-RING (RBR) E3 ligases, which are characterized by the presence of three consecutive structural domains (known as the RBR feature), including a typical N-terminal RING finger domain (RING1) and a structurally unique C-terminal RING2 domain, separated by an in-between RING domain [6]. In contrast to the HECT- and RING finger-type E3 ligases [7], the RBR E3 ligases employ a RING/HECT hybrid mechanism to conjugate ubiquitin to target proteins [6].
The RBR ubiquitin ligases comprise 14 members in the human genome and are highly conserved in all eukaryotes [8]. To date, the biological functions and physiological substrates for the vast majority of the RBR family ligases remain undefined. A case in point is the RING finger protein 144A (RNF144A), a poorly characterized member of the RBR E3 ligase family [9, 10]. Two recent studies showed that RNF144A possesses intrinsic ubiquitin ligase activities and promotes apoptosis in response to DNA damage through inducing ubiquitination and degradation of cytosolic DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) [11, 12]. In line with these observations, we recently demonstrated that RNF144A is epigenetically silenced in breast cancer cells by promoter hypermethylation [13], and that RNF144A targets poly(ADP-ribose) polymerase 1 (PARP1), a critical DNA repair protein, for proteasomal degradation and regulates the sensitivity of breast cancer cells to PARP inhibitor [14]. However, the functional and mechanistic role of RNF144A in breast cancer development and progression remains unexplored.
Heat-shock protein family A member 2 (HSPA2), also known as heat-shock 70 kDa protein 2 (HSP70-2), is a member of the evolutionarily conserved heat-shock protein superfamily [15]. HSPA2 was originally recognized as a testis-specific chaperone protein with a fundamental role in spermatogenesis [16]. Subsequent studies demonstrated that HSPA2 is upregulated in various types of human cancers and promotes cancer cell growth, angiogenesis, migration, invasion, and metastasis through distinct mechanisms [17–26]. Consequently, elevated levels of HSPA2 in human cancers are associated with disease progression and poor prognosis [18, 24–26]. A recent work showed that hypoxia-inducible factor 1α (HIF1α) directly binds to the HSPA2 promoter and transcriptionally upregulates its expression in response to hypoxia [27]. HSPA2 is also subjected to proteasome-dependent degradation in human and mouse germ cells [28–30]. However, the mechanism for regulating HSPA2 stability in human cancers is unclear.
In this study, we report that RNF144A is downregulated in a subset of primary breast tumors and suppresses breast cancer growth and metastasis through, at least in part, targeting HSPA2 for ubiquitination and degradation in an ubiquitin ligase activity-dependent manner. Moreover, the expression levels of RNF144A and HSPA2 are inversely correlated with the prognosis and clinicopathologic characteristics of breast cancer patients. These findings uncover a previously unknown role for the RNF144A-HSPA2 axis in breast cancer growth and metastasis.
Materials and methods
Cell culture and reagents
The well-characterized human breast cancer (MCF-7, T47D, BT474, SK-BR-3, Hs578T, HCC1937, MDA-MB-231, MDA-MB-436, MDA-MB-468), colon carcinoma HCT116, lung carcinoma A549, hepatocellular carcinoma HepG2, and human embryonic kidney 293T (HEK293T) cell lines were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). MDA-MB-231-derived LM2-4173 and LM2-4175 cells were kindly provided by Guohong Hu (University of Chinese Academy of Sciences, Shanghai, China). Both cell lines have increased metastatic activity to the lungs compared with parental MDA-MB-231 cells [31]. All of cell lines were authenticated by short tandem repeat profiling. Cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) or RPMI1640 medium containing 10% fetal bovine serum (FBS; ExCell Biol, Shanghai, China) and 1% penicillin/streptomycin. Culture media and supplements were obtained from BasalMedia (Shanghai, China). MG-132 and cycloheximide (CHX) were purchased from Selleck (Houston, USA) and Cell Signaling Technology (Danvers, USA), respectively. Other chemicals and regents were purchased from Sigma-Aldrich (St. Louis, USA) unless otherwise noted.
Tissue samples
A total of 60 pairs of primary breast tumor tissues and adjacent normal breast tissues and 166 primary breast cancer specimens were obtained from breast cancer patients who underwent surgery at Shanghai Cancer Center, Fudan University. No patients had received chemotherapy or radiotherapy before surgery. Adjacent normal breast tissues were histologically confirmed as cancer free. Characterization of clinicpathological features of these breast cancer patients was described in Supplementary Tables S1-3. Samples were collected with written informed consent from all patients under Institutional Review Board-approved protocols. All procedures were conducted in accordance with the Declaration of Helsinki and International Ethical Guidelines for Biomedical Research Involving Human Subjects.
DNA constructs, transfection, and viral transduction
Myc-DDK-tagged-RNF144A cDNA, RNF144A short hairpin RNA (shRNA) lentiviral constructs (shRNF144A #1), and corresponding control constructs were purchased from Origene (Rockville, USA). Flag-His-tagged human HSPA2 cDNA was obtained from Vigene Biosciences (Rockville, USA). HSPA2 shRNA (shHSPA2) and RNF144A shRNA (shRNF144A #2) lentiviral constructs were obtained from GenePharma (Shanghai, China). To generate HA-HSPA2 construct, HSPA2 cDNAs were amplified by PCR and then subcloned into the lentiviral vector pCDH-CMV-MCS-EF1-Puro (System Biosciences, Mountain View, USA). Expression vectors encoding Flag-RNF144A (wild-type, C20A/C23A, and C198A), GST-RNF144A (full-length and deletion constructs), and V5-ubiquitin have been described previously [14]. All constructs were verified by DNA sequencing (HuaGene Biotech, Shanghai, China). The detailed information of DNA constructs and the primers used for molecular cloning is provided in Supplementary Tables S4 and S5. Transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, USA) or Neofect DNA transfection reagents (TengyiBio, Shanghai, China). Viral production and infection were done as described previously [32, 33]. To generate stable cell lines, cells were selected with 2 μg/ml puromycin (Cayman, Ann Arbor, USA) for 1–2 weeks.
Cell proliferation, apoptosis, cell cycle analysis, and colony-formation assays
Cell proliferation assays were performed using Cell Counting Kit-8 (CCK-8) (Dojindo, Shanghai, China) according to the manufacturer’s instructions. For detection of apoptosis, cells were collected and fixed in 75% ethanol overnight. After phosphate-buffered saline (PBS) wash, cells were stained with Annexin V-PE/7-AAD Apoptosis Detection Kit (Yeasen, Shanghai, China) and analyzed on a BD FACSCanto II flow cytometer (BD Bioscience, San Jose, USA). Apoptotic cells (Annexin V-PE-positive and 7-AAD-negative for early apoptosis and both Annexin V-PE and 7-AAD positive for late apoptosis) were quantified. Cell cycle progression was analyzed by fluorescence-activated cell sorting (FACS) as described previously [32]. For colony-formation assays, cells were grown onto six-well plates for 14 days with media replacement every 3 days. Cells were stained with 1% crystal violet staining solution and the number of colonies formed in each well was counted under a microscope.
Wound-healing and Transwell migration and invasion assays
Wound-healing assays were performed as described previously [33–35]. Briefly, cells were seeded in six-well plates. When cells were grown to confluency, the wound was created by 200 μl tips, the floated cells were removed through PBS washing, and the culture medium were replaced by DMEM containing 0.1% FBS. Images were taken at the indicated time points and the wound-closure ratios were calculated. Transwell migration and invasion assays were carried out as described previously [32, 33]. Cells were resuspended in DMEM medium containing 0.1% FBS and then loaded onto the upper well of transwell chambers in the absence (migration) or presence (invasion) of precoated Matrigel (Corning Biocoat, Tewksbury, USA). The lower side of the separating filter was filled with complete medium containing 10% FBS as a source of chemoattractant. Cells were stained with 1% crystal violet staining solution at the indicated times. All assays were conducted in triplicate and repeated at least three times.
In vivo tumor growth and lung metastasis
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Fudan University and animal care was in accordance with institutional guidelines. For subcutaneous inoculation, cells (1 × 107 and 2 × 106 cells for RNF144A-overexpressing and RNF144A-depleted MDA-MB-231 cells, respectively) in 200 μl PBS were injected directly into the mammary fat pad of 6-week-old female BALB/c athymic nude mice (State Key Laboratory of Oncogenes and Related Genes, Shanghai, China). As RNF144A is emerging as a tumor suppressor gene in breast cancer cells, knockdown of RNF144A was supposed to enhance xenograft tumor growth. If the same number of cells (1 × 107 cells) was used for both RNF144A-overexpressing and RNF144A-depleted experiments, the size of tumors formed by MDA-MB-231–shRNF144A cells would exceed the maximum size (15 mm in any dimension) permitted by the IACUC of Fudan University Shanghai Cancer Center. Therefore, we reduced cell number for RNF144A-knockdown xenograft experiments. The tumors were measured twice a week after appearance of tumors and the tumor volume was calculated by the formula of (length × width2)/2. Mice were killed by cervical dislocation after 8 weeks of the inoculation. The tumors were excised and weighed. For experimental lung metastasis assays, 2 × 106 cells were resuspended in 0.1 ml of PBS and were injected into tail veins of nude mice. The number of lung metastatic foci was quantified after 6 weeks of injection. The lungs were then excised and fixed in formalin overnight. Paraffin-embedded sections were subjected to hematoxylin-eosin staining to examine the presence of micrometastases.
Quantitative real-time PCR
Total RNA was isolated from cell lines and tissue samples using TRIzol reagent (Invitrogen), and cDNA was synthesized using PrimeScript RT Master Mix (Takara, Dalian, China) according to the manufacturer’s protocol. The resultant cDNA was subjected to quantitative real-time PCR (qPCR) using SYBR Premix Ex Taq (Tli RNaseH Plus) (Takara) on an Eppendorf Mastercycler ep realplex4 instrument. The values for specific genes were normalized to actin housekeeping controls and data were present as mean ± SE. qPCR primers were synthesized in HuaGene Biotechnology (Shanghai, China) and sequences are described in Supplementary Table S6.
Quantitative proteomics
Isobaric tags for relative and absolute quantitation (iTRAQ) coupled with liquid chromatography-tandem mass spectrometry (LC-MS/MS) was employed to identify the differentially expressed proteins as described previously [36]. Briefly, proteins were digested and labeled using the 8-plex iTRAQ reagents (AB SCIEX, Shanghai, China) according to the manufacturer’s instructions. The labeled peptides were fractionated with high pH reversed-phase LC on a UPLC system (Waters, Milford, USA), followed by analysis using Nano-LC-MS/MS system [36]. Protein identification and quantification analysis were performed with Proteome Discovery (v.1.3, Thermo Fisher Scientific) and ProteinPilot (version 4.5, AB SCIEX). All data were searched against the Swiss-Prot human database. Proteins with a fold change >1.5 or <0.7 with a Student’s t-test p-value < 0.05 were selected as differently expressed proteins.
Antibodies, immunoblotting, immunoprecipitation, and immunofluorescence
The information for primary antibodies used in this study is summarized in Supplementary Table S7. Immunoblotting, immunoprecipitation (IP), and indirect immunofluorescence (IF) staining were performed as described previously [32, 33]. Briefly, protein extracts were prepared using RIPA buffer containing 1 × protease inhibitor cocktail and 1 × phosphatase inhibitor cocktails (Bimake, Houston, USA). Quantification of protein concentrations was performed using the BCA Protein Assay Kit (Yeasen). Cellular extracts were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), transferred to polyvinylidene difluoride membranes (Millipore), and incubated with the indicated antibodies. Antibody signals were detected using the electrochemiluminescence reagents (Yeasen). For IP analysis, total 1–2 mg of protein extracts was incubated with the indicated antibodies overnight at 4 °C, followed by addition of 30 µl of protein A/G magnetic beads (Bimake) and incubation for another 2 h. Immunoprecipitates were collected by centrifugation at 4000 r.p.m. for 5 min, washed with lysis buffer for three to five times, and then dissolved in a sample buffer for SDS-PAGE. For indirect IF staining, cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% Triton X-100, and blocked in 10% normal goat serum in PBS. Cells were incubated with primary antibodies, washed three times in PBS, and then incubated with secondary antibody conjugated with 555-Alexa (red) or 488-Alexa (green) (Cell Signaling Technology), respectively. The blue DNA dye 4’, 6-diamidino-2-phenylindole (DAPI; Abcam, Shanghai, China) was used as nuclear staining. Microscopic analyses were performed using a LEICA SP5 confocal microscope (Leica Microsystems, Buffalo Grove, USA).
Immunohistochemistry
Immunohistochemistry (IHC) staining was carried out using EnVision Detection Systems Peroxidase/DAB (DAKO, Santa Clara, USA) following the manufacturer’s recommendations.
The primary antibody against human RNF144A (Lifespan, #LS-C162648) or human HSPA2 (Proteintech, #12797-1-AP) was diluted at dilution of 1:100 and then incubated at 4 °C overnight in a humidified container. Interpretation of the IHC results was performed by two independent pathologists who were blinded to the clinicopathological information. Slides were evaluated using light microscopy and a standard semiquantitative immunoreactivity score as described previously [37]. By recording the percentage of positive staining (0 = negative, 1 ≤ 10%, 2 = 10–50%, 3 ≥ 50%) and staining intensity (0 = no, 1 = weak, 2 = moderate, 3 = strong) for each sample, immunoreactivity score (IRS) (0–9) was calculated by multiplying positive staining percentage with staining intensity. Low and high expression were defined as IRS of < 6 and ≥ 6, respectively [37].
GST pull-down assay
GST and GST-RNF144A were expressed in Escherichia coli strain BL21 (DE3) (Tiangen Biotech, Beijing, China) and purified using the Glutathione Sepharose 4B batch method (GE Healthcare, Piscataway, USA) as described previously [14]. Equal amounts of GST and GST-RNF144A proteins were incubated with an equal aliquot of cellular lysates from MCF-7 cells in 500 μl of GST-binding buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8, 1% Nonidet P-40, 1 × phosphatase, and protease inhibitor cocktails) at 4 °C overnight. After washing three to five times with the binding buffer, proteins were eluted in 2 × SDS loading buffer and subjected to SDS-PAGE. The interaction was detected by immunoblotting using the indicated antibodies.
In vivo ubiquitination and cycloheximade analyses
In vivo ubiquitination assays was performed as described previously [33]. Briefly, cells were transfected with expression plasmids encoding HA-HSPA2, V5-ubiquitin, and Flag-RNF144A alone or in combination. After 48 h of transfection, cells were treated with 10 µM MG-132 for 6 h and then total cellular lysates were subjected to IP and immunoblotting analyses with the indicated antibodies. For CHX-chase assays [32, 33], cells were treated with 100 µg/ml of CHX and collected at the indicated times for immunoblotting.
Statistical analysis
All data are presented as the mean ± SE from at least three independent experiments, unless otherwise indicated. The unpaired two-tailed Student’s t-test was used to compare data between two groups. The correlation between RNF144A/HSPA2 expression and clinicopathologic factors of breast cancer patients was analyzed by the χ2-test or Fisher’s exact test. Survival curves were obtained using the Kaplan–Meier method and statistics analysis was performed using the log-rank test. Cox proportional hazards regression model was performed for univariate and multivariate survival analyses. Correlation coefficients were calculated using the Pearson’s test. P-values of <0.05 were considered statistically significant.
Results
RNF144A is downregulated in primary breast tumors
To examine the relevance of RNF144A to breast cancer pathogenesis and progression, we first analyzed the expression levels of RNF144A in 30 pairs of primary breast tumors and matched normal breast tissues (Supplementary Table S1) by qPCR. Results showed that the mRNA levels of RNF144A were downregulated in 20 out of 30 (66.7%) breast tumor samples as compared with normal breast tissues (Supplementary Fig. S1A). To substantiate our results, we next analyzed RNF144A mRNA levels in Oncomine and The Cancer Genome Atlas (TCGA) databases. In the Curtis et al. [38] dataset, RNF144A mRNA levels were downregulated by 2.131-fold in mammary carcinomas relative to normal breast tissues (Supplementary Fig. S1B). TCGA database analysis showed that RNF144A mRNA levels were reduced in 72 out of 113 (63.7%) breast tumors compared with normal breast counterparts (Supplementary Fig. S1C). A meta-analysis of recently published ten gene expression profiling studies also showed a widespread downregulation of RNF144A in breast cancer tissues (Supplementary Fig. S1D). To verify whether reduced RNF144A expression in primary breast tumors was reflected at the protein level, we analyzed the expression status of RNF144A protein in another 30 pairs of human breast tumors and matched normal breast tissues (Supplementary Table S2) by IHC staining. Results showed that RNF144A expression levels were downregulated in 23 out of 30 (76.6%) breast cancer tissues when compared with their normal breast tissues (Supplementary Fig. S1E and S1F). These results suggest that RNF144A is downregulated in a subset of primary breast tumors.
RNF144A suppresses breast cancer cell proliferation and colony formation in vitro and xenograft tumor growth in vivo
Breast cancer is a highly heterogeneous disease, which is clinically divided into three major molecular subtypes according to the expression status of estrogen receptor α (ERα), progesterone receptor (PR), and human epidermal growth receptor 2 (HER2), including luminal, HER2-positive, and triple-negative breast cancer (TNBC) [33, 39]. TNBC is the most aggressive type of breast cancer with high recurrence and metastasis rates [40]. To delineate the functional role of RNF144A in breast cancer, we first analyzed the protein levels of RNF144A in nine representative breast cancer cell lines by immunoblotting. As shown in Fig. 1a, luminal (MCF-7 and T47D) and HER2-positive (BT474 and SK-BR-3) breast cancer cell lines expressed relatively higher levels of RNF144A than TNBC cell lines did. We next stably expressed RNF144A in MDA-MB-231 and Hs578T cells by lentiviral infections (Fig. 1b) or depleted RNF144A in MCF-7 and MDA-MB-231 cells using shRNF144A (Fig. 1c and Supplementary Fig. S2A). CCK-8 assays showed that overexpression of RNF144A in MDA-MB-231 and Hs578T cells suppressed cell proliferation (Fig. 1d). Although RNF144A has been shown to regulate apoptosis following DNA damage [12], FACS analysis using Annexin V-PE/7-AAD Apoptosis Detection Kit revealed that there was no significant difference in the percentage of apoptotic cells between empty vector- and RNF144A-expressing cells under basal conditions (Supplementary Fig. S2B and S2C). Cell cycle analysis and colony-formation assays showed that overexpression of RNF144A in MDA-MB-231 and Hs578T cells arrested cell cycle in the G1/S phase (Supplementary Fig. S2D) and decreased the number of formed colonies (Fig. 1e, f), respectively. Conversely, knockdown of RNF144A in MCF-7 and MDA-MB-231 cells enhanced cell proliferation (Fig. 1g), promoted G1/S phase transition (Supplementary Fig. S2E), and enhanced the ability of cells to form colonies (Fig. 1h, i) as compared with shNC-infected cells.
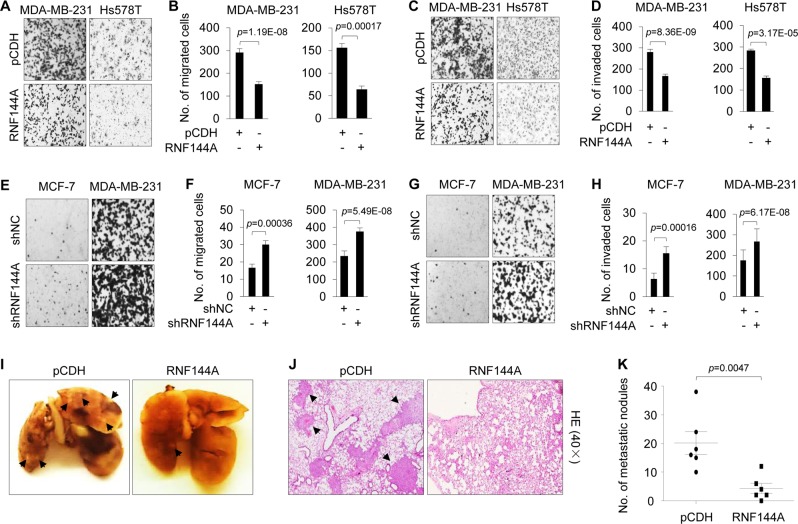
Fig. 1.
RNF144A suppresses breast cancer cell proliferation and colony formation in vitro and tumor growth in vivo. a Expression of RNF144A, ER, PR, and HER2 in nine representative breast cancer cell lines analyzed by immunoblotting. Vinculin was used as a loading control. b MDA-MB-231 and Hs578T cells stably expressing pCDH and Flag-RNF44A were subjected to immunoblotting analysis with the indicated antibodies. c Immunoblotting analysis of RNF144A expression in MCF-7 and MDA-MB-231 cells stably expressing shNC and shRNF144A. d MDA-MB-231 and Hs578T cells stably expressing pCDH and Flag-RNF144A (1 × 104 cells per well) were subjected to cell proliferation assays using CCK-8. e, f Colony-formation assays of MDA-MB-231 and Hs578T cells (1000 cells per well) expressing pCDH and Flag-RNF44A. Images were obtained using an EPSON V370 scanner. Representative images of the colonies (e) and quantitative results (f) are shown. g Cell proliferation assays of MDA-MB-231 (2 × 103 cells per well) and MCF-7 (1 × 104 cells per well) stably expressing shNC and shRNF144A using CCK-8. h, i Colony-formation assays of MCF-7 (1000 cells per well) and MDA-MB-231 (500 cells per well) cells stably expressing shNC and shRNF144A. Images were obtained using an EPSON V370 scanner. Representative images of the colonies (h) and quantitative results (i) are shown. In d, f, g, and i, the quantified results are presented as means ± SE (n = 3). j, k Stable MDA-MB-231 cells expressing pCDH and Flag-RNF144A (1 × 107 cells) were subcutaneously injected into mammary fat pads of 6-week-old female BALB/c nude mice (n = 8). Images of the xenografted tumors (j) and tumor weight (k) are shown. l, m MDA-MB-231 cells stably expressing shNC and shRNF144A (2 × 106 cells) were subcutaneously injected into mammary fat pads of 6-week-old female BALB/c nude mice (n = 7). Images of the xenografted tumors (l) and tumor weight (m) are shown
To investigate whether RNF144A could regulate tumorigenic capacity of breast cancer cells in vivo, MDA-MB-231 cells stably expressing pCDH and Flag-RNF144A were subcutaneously injected into mammary fat pads of 6-week-old female BALB/c nude mice. In support of in vitro findings, tumors from RNF144A-overexpressing MDA-MB-231 cells grew much slower than their control cells (Fig. 1j, k). Conversely, knockdown of RNF144A in MDA-MB-231 cells increased tumor growth in xenograft mouse models (Fig. 1l, m). These results suggest that RNF144A suppresses breast cancer cell proliferation and colony formation in vitro and xenograft tumor growth in vivo.
RNF144A suppresses breast cancer cell migration and invasion in vitro and lung metastasis in vivo
As an important property of breast cancer cells is their invasive and metastatic behavior, we next examined whether RNF144A could modulate the invasive and metastatic phenotype of breast cancer cells. Transwell migration assays revealed that overexpression of RNF144A in MDA-MB-231 and Hs578T cells resulted in a significant reduction in the number of migrated cells through insert membrane compared with empty vector (Fig. 2a, b). Consistently, wound-healing assays showed that RNF144A-overexpressing MDA-MB-231 cells had lower closure of the wound area compared with their control cells (Supplementary Fig. S3A). Moreover, RNF144A-expressing MDA-MB-231 and Hs578T cells showed a reduced invasion through Matrigel-coated invasion chambers as compared with empty vector-expressing controls (Fig. 2c, d). In contrast, shRNF144A-infected MCF-7 and MDA-MB-231 cells exhibited an enhanced migratory (Fig. 2e, f and Supplementary Fig. S3B) and invasive (Fig. 2g, h) phenotype as compared with shNC-infected cells. These results indicate that RNF144A suppresses breast cancer cell migration and invasion in vitro, two key events in tumor metastasis.
Fig. 2.
RNF144A suppresses migratory and invasive potential of breast cancer cells in vitro and lung metastatic capacity in vivo. a–d MDA-MB-231 and Hs578T cells stably expressing pCDH and Flag-RNF144A were subjected to Transwell cell migration assays (a, b; 1 × 105 cells per well) and Matrigel invasion assays (c, d; 2 × 105 cells per well). After the indicated times (24 h and 48 h for MDA-MB-231 and Hs578T cells, respectively), migrated and invaded cells were fixed and stained with 1% crystal violet. Cells were counted under an inverted microscope at ×200 magnification. Representative images of migration and invasion are shown in a and c, respectively. Quantitative results of migration and invasion are shown in b and d, respectively. The quantified results are presented as means ± SE (n = 3). e–h MCF-7 and MDA-MB-231 cells stably expressing shNC and shRNF144A (1 × 105 cells per well) were subjected to Transwell migration assays (e, f) and Matrigel invasion assays (g, h). After 24 h, migrated and invaded cells were fixed and stained with 1% crystal violet. Cells were counted under an inverted microscope at ×200 magnification. Representative images of migration and invasion are shown in e and g, respectively. Quantitative results of migration and invasion are shown in f and h, respectively. The quantified results are presented as means ± SE (n = 3). i–k MDA-MB-231 cells stably expressing pCDH and RNF144A (2 × 106 cells) were injected into nude mice (n = 6) through the tail vein and lungs were collected after 8 weeks of injection. Representative images of metastatic lung tumors (i) and of HE-stained sections of lung tissues (j), and quantitative results of metastatic lung nodules (k) are shown
To examine whether RNF144A affects the metastatic potential of breast cancer cells in vivo, MDA-MB-231 cells stably expressing pCDH and Flag-RNF144A were injected into nude mice through tail veins and the number of lung metastatic foci was determined after 8 weeks of injection [41]. As shown in Fig. 2i–k, more lung metastatic colonies were observed in mice bearing MDA-MB-231-pCDH tumors than that in mice bearing MDA-MB-231-RNF144A tumors. Collectively, these results establish that RNF144A suppresses breast cancer cell migration and invasion in vitro and the formation of lung metastases in xenograft mouse models.
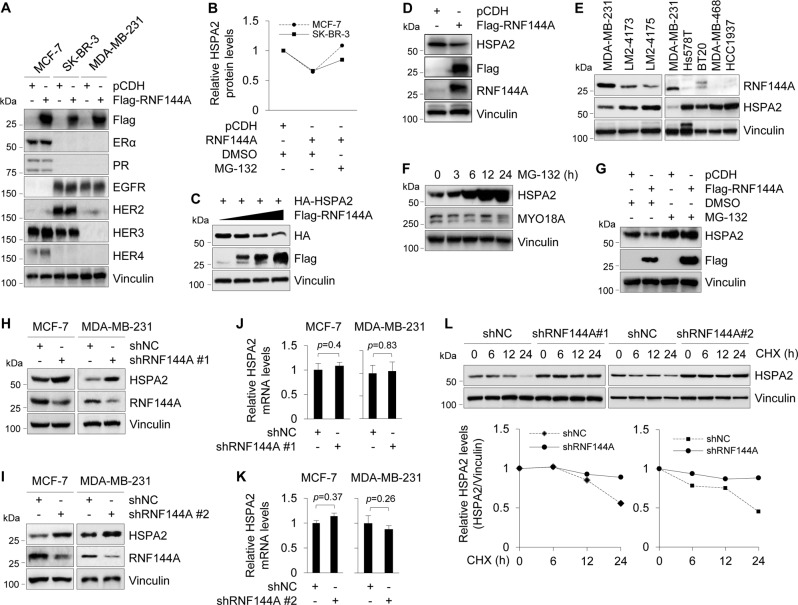
RNF144A targets oncoprotein HSPA2 for proteasome-dependent degradation
To reveal the molecular mechanisms by which RNF144A suppresses breast cancer growth and metastasis, we first examined whether RNF144A could affect the expression levels of ER, PR, and epidermal growth factor receptor family members, which are putative drivers of breast cancer pathogenesis and progression [42, 43]. Results showed that overexpression of RNF144A in MCF-7, SK-BR-3, and MDA-MB-231 cells had no remarkable effects on the expression levels of these proteins (Fig. 3a). Given that RNF144A contains a conserved RBR domain architecture (Supplementary Fig. S4A) and possesses intrinsic E3 ubiquitin ligase activities [11, 12, 14], we next sought to identify the bona fide substrates of RNF144A using iTRAQ-based quantitative proteomics [36]. To do this, RNF144A-overepxressing MCF-7 and SK-BR-3 cells and its corresponding control cells were treated with or without 10 μM of proteasome inhibitor MG-132 for 6 h and then subjected to iTRAQ labeling and LC-MS/MS analysis (Supplementary Fig S4B). Using this approach, we identified HSPA2 as a common potential substrate for RNF144A in both MCF-7 and SK-BR-3 cell lines (Fig. 3b). HSPA2 attracted our attention, because it has been documented that HSPA2 is overexpressed in human breast cancer tissues and is essential for breast cancer cell growth, migration, and invasion [19, 22, 44].
Fig. 3.
RNF144A targets oncoprotein HSPA2 for proteasome-dependent degradation. a Cells stably expressing pCDH and Flag-RNF144A were subjected to immunoblotting analysis with the indicated antibodies. Vinculin was used as loading control. b MCF-7 and SK-BR-3 cells stably expressing pCDH and RNF144A were treated with or without 10 μM MG-132 for 6 h as indicated in Supplementary Fig. S4B and then subjected to iTRAQ-based quantitative proteomics. Quantitative results of HSPA2 protein levels are representative of the mean value of two independent experiments. c HEK293T cells were transfected with HA-HSPA2 expression vector alone or in combination with increasing doses of Flag-RNF144A plasmid DNA. After 48 h of transfection, lysates were subjected to immunoblotting with the indicated antibodies. d MDA-MB-231 cells stably expressing pCDH and Flag-RNF144A were subjected to immunoblotting analysis with the indicated antibodies. e Immunoblotting analysis of RNF144A and HSPA2 in the indicated breast cancer cell lines. f MDA-MB-231 cells were treated with 10 μM of MG-132 for the indicated time points and subjected to immunoblotting analysis with the indicated antibodies. g MDA-MB-231 cells stably expressing pCDH and Flag-RNF144A were treated with DMSO or 10 μM of MG-132 for 6 h and then subjected to immunoblotting analysis with the indicated antibodies. h, i Lysates from stable MCF-7 and MDA-MB-231 cells expressing shNC and shRNF144A were subjected to immunoblotting analysis with the indicated antibodies. j, k qPCR analysis of HSPA2 mRNA levels in MCF-7 and MDA-MB-231 cells stably expressing shNC and shRNF144A. l MCF-7 cells stably expressing shNC and shRNF144A were treated with 100 μg/ml of CHX for the indicated times and then subjected to immunoblotting with the indicated antibodies. Quantitative results of HSPA2 protein levels (HSPA2/vinculin) are representative of the mean value of two independent experiments and are shown in the lower panel
To demonstrate whether RNF144A regulates HSPA2 expression levels, HEK293T cells were transfected with HA-HSPA2 expression vector alone or in combination with increasing doses of Flag-RNF144A. As shown in Fig. 3c, ectopic expression of Flag-RNF144A led to a reduction in the protein levels of HA-HSPA2 in a dose-dependent manner. In agreement with these results, stable expression of RNF144A in MDA-MB-231 cells downregulated endogenous HSPA2 levels in vitro (Fig. 3d) and in xenograft tumors in mice (Supplementary Fig. S4C). The specificity of HSPA2 antibody was verified by immunoblotting in HSPA2-deficient and HSPA2-overexpressing cell lines [45] (Supplementary Fig. S4D). Moreover, an inverse correlation of protein expression levels between RNF144A and HSPA2 was observed in isogenic MDA-MB-231 sublines and in other breast cancer cell lines (Fig. 3e).
Recent work shows that HSPA2 is regulated by the ubiquitin–proteasome pathway in human and mouse germ cells [28–30]. In agreement with these results, we found that HSPA2 protein levels were increased in MDA-MB-231 cells following MG-132 treatment in a time-dependent manner (Fig. 3f). RNF144A-mediated HSPA2 downregulation in MDA-MB-231 cells was restored following MG-132 treatment (Fig. 3g, compare lane 4 with 2), indicating that RNF144A induces HSPA2 degradation through the proteasome-dependent pathway. In support of these results, knockdown of RNF144A in MCF-7 and MDA-MB-231 cells resulted in an accumulation of endogenous HSPA2 protein (Fig. 3h, i). In contrast, there was no significant change in HSPA2 mRNA levels following RNF144A knockdown in both cell lines (Fig. 3j, k). Moerover, CHX chase assays demonstrated that knockdown of RNF144A in MCF-7 cells extended the half-life of HSPA2 protein (Fig. 3l). Collectively, these findings demonstrated that RNF144A targets HSPA2 for proteasome-mediated degradation.
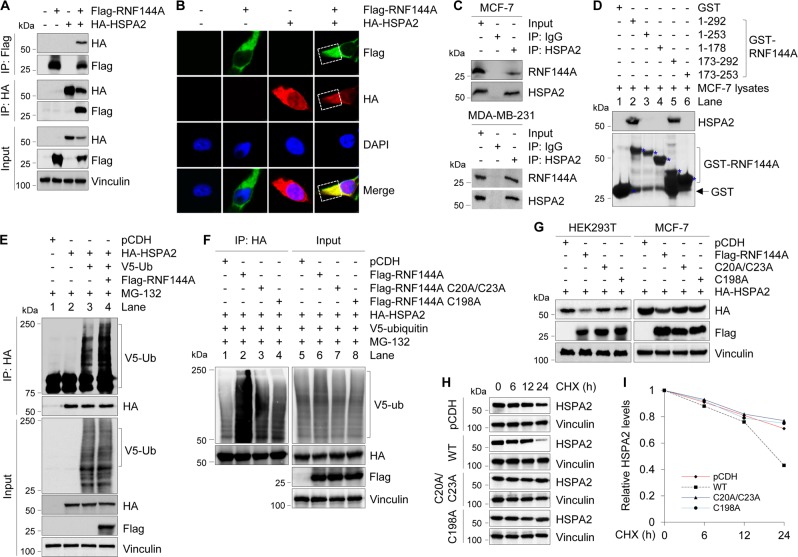
RNF144A interacts with HSPA2 and promotes its ubiquitination
To gain mechanistic insights into how RNF144A regulates HSPA2 degradation, we next examined whether RNF144A interacts with HSPA2. To this end, HEK293T cells were transfected with Flag-RNF144A, HA-HSPA2 expression vector alone or in combination, and immunoprecipiated with either anti-Flag or anti-HA antibody. Immunoblotting analysis showed that Flag-RNF144A and HA-HSPA2 were co-immunoprecipiated when co-expressed (Fig. 4a, lane 4), indicating that those two proteins can interact with each other in vivo. Immunofluorescent staining showed that Flag-RNF144A and HA-HSPA2 colocalized mainly in the cytoplasm in co-transfected HEK293T cells (Fig. 4b). Moreover, there was an interaction between HSPA2 and RNF144A at the endogenous level in MCF-7 and MDA- MB-231 cells (Fig. 4c). GST pull-down assays showed that endogenous HSPA2 interacted with GST-RNF144A but not GST (Fig. 4d, compare lane 2 with 1) and this interaction was mediated by the C-terminal region (amino acids 253–292) of RNF144A (Fig. 4d).
Fig. 4.
RNF144A interacts with HSPA2 and promotes its polyubiquitination. a HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, total cellular lysates were subjected to IP and immunoblotting analysis with the indicated antibodies. b HEK293T cells were transfected with the indicated expression vectors and indirect IF staining was carried out after 48 h of transfection with the indicated antibodies. DAPI was used as a nuclear staining. c MCF-7 and MDA-MB-231 cells were treated with 10 μM MG-132 for 6 h to enhance basal levels of endogenous RNF144A and HSPA2. Then, total cellular lysates were subjected to IP analysis with control IgG or an anti-HSPA2 antibody, followed by immunoblotting with the indicated antibodies. d Lysates from MCF-7 cells were incubated with purified GST or GST-RNF144A protein and then subjected to immunoblotting analysis with the indicated antibodies. e, f HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were incubated with 10 μM of MG-132 for 6 h and then subjected to sequential IP and immunoblotting analysis with the indicated antibodies. g HEK293T and MCF-7 cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were subjected to immunoblotting analysis with the indicated antibodies. h, i HEK293T cells were transfected with the indicated expression vectors. After 48 h of transfection, cells were treated with 100 μg/ml of CHX for the indicated times and then subjected to immunoblotting with the indicated antibodies (h). Quantitative results of HSPA2 protein levels (HSPA2/vinculin) are representative of the mean value of two independent experiments (i)
As RNF144A is a RBR-type E3 ubiquitin ligase [11, 12, 14], we next tested whether RNF144A induces ubiquitination of HSPA2. As shown in Fig. 4e, HA-HSPA2 was ubiquitinated in the presence of V5-ubiquitin (compare lane 3 with 2). Moreover, co-expression of Flag-RNF144A further increased HSPA2 ubiquitination (compare lane 4 with 3). Next, we generated two E3 ligase-defective mutants by substitution of consensus cysteine (C) to alanine (A) at the residues 20/23 (C20A/C23A) and 198 (C198A) within the RING1 and RING2 domains, respectively [12, 14], and tested their effects on HSPA2 ubiquitination and degradation. Results showed that E3 ligase-defective mutants impaired the ability of RNF144A to ubiquitinate HSPA2 (Fig. 4f, compare lanes 3 and 4 with 2) and to downregulate HSPA2 expression (Fig. 4g) compared with its wild-type counterpart. Consistently, wild-type RNF144A, but not by its E3 ligase-defective mutants, significantly reduced the half-life of HSPA2 (Fig. 4h, i). These results suggest that RNF144A targets HSPA2 for ubiquitination and degradation in a ubiquitin ligase activity-dependent manner.
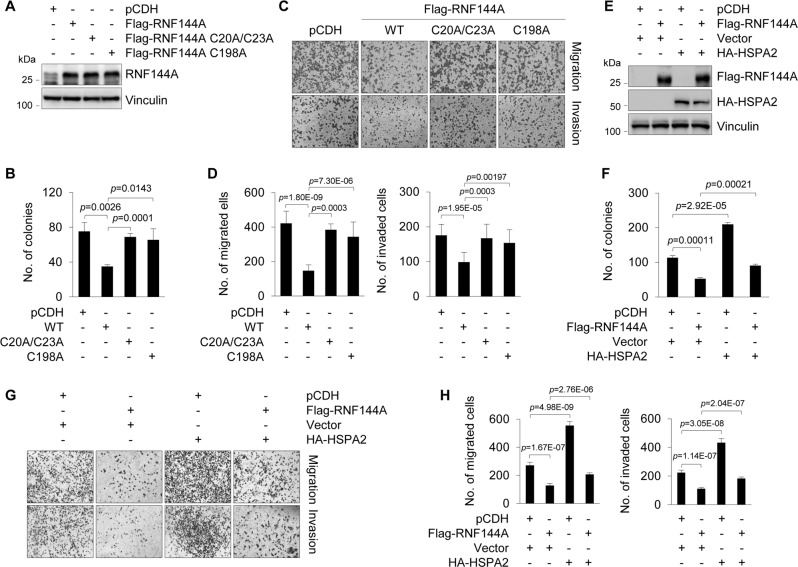
RNF144A suppresses breast cancer cell proliferation, migration, and invasion partially through HSPA2
Following these observations, we next addressed whether HSPA2 is indeed involved in tumor suppressor functions of RNF144A in breast cancer cells. First, we generated MDA-MB-231 cell lines stably expressing wild-type Flag-RNF144A or its E3 ligase-defective mutants (C20A/C23A and C198A) by lentiviral infections (Fig. 5a). Functional assays showed that E3 ligase-defective mutants of RNF144A impaired the ability of RNF144A to suppress colony formation (Fig. 5b and Supplementary Fig. S5A), migration, and invasion (Fig. 5c, d) of MDA-MB-231 cells. Second, we infected MDA-MB-231 cells with lentiviral vectors encoding Flag-RNF144A, HA-HSPA2 alone or in combination (Fig. 5e). As expected, ectopic expression of HSPA2 could partially rescue RNF144A-mediated suppression of breast cancer cell migration and invasion (Fig. 5g, h), and colony formation (Fig. 5f and Supplementary Fig. S5B). These results suggest that RNF144A suppresses breast cancer progression through, at least in part, antagonizing oncogenic activities of HSPA2.
Fig. 5.
RNF144A suppresses breast cancer cell proliferation, migration, and invasion partially through HSPA2. a Establishment of stable MDA-MB-231 cell lines expressing empty vector, wild-type, and E3 ligase-defective mutants of Flag-RNF144A by lentiviral infection. Expression of RNF114A in resultant cell lines was validated by immunoblotting. b Established MDA-MB-231 stable cells (a) were subjected to colony-formation assays as described in Fig. 1. Quantitative results are shown (representative images of the colonies are shown in Supplementary Fig. S5A). c, d Established MDA-MB-231 stable cells (a) were subjected to Transwell cell migration and Matrigel invasion assays as described in Fig. 2. Representative images of migration and invasion, and quantitative results of migration and invasion are shown in c and d, respectively. e MDA-MB-231 cells were infected with the indicated lentiviral expression vectors and their expression status was verified by immunoblotting with indicated antibodies. f Established MDA-MB-231 cells (e) were subjected to colony-formation assays. Quantitative results are shown (representative images of the colonies are shown in Supplementary Fig. S5B). g, h Established MDA-MB-231 cells (e) were subjected to Transwell migration and Matrigel invasion assays. Representative images are shown in g and the corresponding quantitative results are shown in h. The quantified results are presented as means ± SE (n = 3)
RNF144A and HSPA2 are inversely associated with prognosis and clinicopathological characters of breast cancer patients
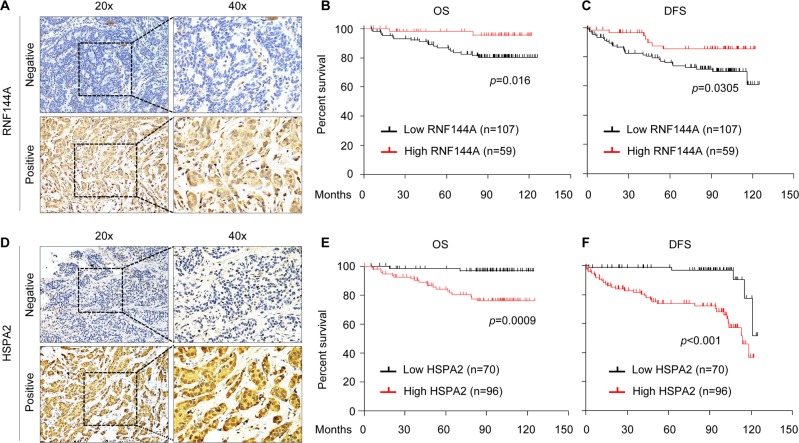
To examine the clinical significance of the RNF144A-HSPA2 axis in breast cancer, we first evaluated RNF144A expression status in a tissue microarray containing 166 surgical specimens from patients diagnosed with invasive breast cancer (Supplementary Table S3) by IHC staining (Fig. 6a). According to the RNF144A expression scores >6 or <6 [37], patients were divided into RNF144A-high and RNF144A-low subgroups. In this context, 35.5% (59/166) of patients had high RNF144A expression, whereas 64.5% (107/166) of patients had low RNF144A expression. Kaplan–Meier survival analyses revealed that patients with low RNF144A expression had poorer overall survival (OS, Fig. 6b) and disease-free survival (DFS; Fig. 6c). Moreover, the expression levels of RNF144A were negatively associated with larger tumor size, the presence of lymph node metastasis, and higher clinical stage, but not with age, menopausal status, and histologic grade of patients with breast cancer (Supplementary Table S8). Univariate and multivariate Cox regression analyses showed that RNF144A was an independent predictor of poor OS and DFS (Supplementary Tables S9 and S10, respectively).
Fig. 6.
HSPA2 and RNF144A inversely associate with prognosis and clinicopathological features of breast cancer patients. a Representative images showing positive and negative RNF144A immunostaining in breast cancer tissues. b, c Kaplan–Meier curves for OS (b) and DFS (c) of breast cancer patients (n = 166) with low or high expression of RNF144A. d Representative images showing positive and negative HSPA2 immunostaining in breast cancer tissues. e, f Kaplan–Meier curves of OS (e) and DFS (f) according to HSPA2 expression in 166 human breast tumors
Accumulating evidence shows that elevated levels of HSPA2 in human cancer are associated with disease progression and poor prognosis [18, 24–26]. To confirm these findings, we next analyzed HSPA2 expression by IHC staining in the same tissue microarray used for RNF144A staining (Fig. 6d). Of the 166 patients studied, 96 (57.8%) had high HSPA2 expression, whereas 70 (42.1%) showed low HSPA2 expression. Kaplan–Meier survival analyses revealed that patients with high HSPA2 expression had poorer OS (Fig. 6e) and DFS (Fig. 6f) than those with low HSPA2 expression. Moreover, HSPA2 expression levels were significantly correlated with the presence of lymph node metastasis and clinical stage (Supplementary Table S11), indicating that high HSPA2 expression is associated with more aggressive breast cancer. However, there was no significant association between HSPA2 expression levels and patient age, menopausal status, and histology grade. Moreover, univariate and multivariate Cox regression analyses revealed that elevated expression of HSPA2 was an independent predictor of OS (Supplementary Table S12) and RFS (Supplementary Table S13). Together, these results suggest that the HSPA2 expression is associated with poor prognosis of breast cancer patients.
Finally, we examined the correlation between RNF144A and HSPA2 in those 166 breast tumor samples, and observed a negative correlation between RNF144A and HSPA2, as 65.4% (70/107) of RNF44A-low samples had HSPA2-high expression, whereas 55.9% (33/59) RNF44A-high samples had HSPA2-low expression (Supplementary Table S14). In summary, our results suggest that RNF144A is a bona fide E3 ubiquitin ligase for HSPA2 and suppresses breast cancer growth and metastasis through, at least in part, ubiquitination and degradation of HSPA2 (Fig. 7).
Fig. 7.

The proposed working model. Low expression of RNF144A in breast cancer cells results in enhanced HSPA2 oncoprotein stability, thus contributing to breast cancer progression. Conversely, high expression of RNF144A suppresses breast cancer progression through targeting HSPA2 for ubiquitination and subsequent degradation
Discussion
In this study, we report several key findings concerning the RNF144A-HSPA2 axis in breast cancer progression and prognosis. First, it establishes RNF144A as a novel breast cancer suppressor, which is downregulated in primary breast tumors, suppresses breast cancer growth and progression, and correlates with favorable prognosis of breast cancer patients. One of our earlier studies identified RNF144A as a transcriptional repression target of metastasis-associated protein 1 (MTA1) gene, a key molecule driving tumor invasion and metastasis, through microarray-based gene expression profiling analyses of wild-type and MTA1-knockout mouse embryonic fibroblasts [46]. In addition, Ho et al. [12] showed that RNF144A promotes DNA damage-induced apoptosis, a key biological process tightly linked to cancer development. More recently, we demonstrated that RNF144A is epigenetically silenced in breast cancer cells by promoter hypermethylation [13]. In agreement with these results, we provide multiple lines of evidence that RNF144A functions as a novel tumor suppressor in breast cancer. qPCR, IHC staining, and public database analyses showed that RNF144A is downregulated in primary breast tumors (Supplementary Fig. S1). Loss- and gain-of-function experiments further revealed that RNF144A suppresses breast cancer cell migration, invasion, and colony formation in vitro, and tumor growth and lung colonization in vivo (Figs. 2 and 3). More importantly, low expression of RNF144A is associated with poor prognosis and aggressive clinicophathological features of breast cancer patients (Fig. 6a–c and Supplementary Table S8). In support of our findings, Parkin, the best studied RBR family ligase, has been shown to be downregulated in breast cancer tissues and to inhibit breast tumor progression through targeting HIF1α for ubiquitination and degradation [37].
Second, it provides mechanistic insights into tumor suppressor functions of RNF144A in breast cancer through targeting oncoprotein HSPA2 for ubiquitination and degradation in a ubiquitin ligase activity-dependent manner. Accumulating evidence shows that HSPA2 is upregulated in multiple types of human cancers and its expression levels are associated with disease progression and poor prognosis [17–26]. Although it has been shown that HLA-B-associated transcript 3, also known as BCL2-associated athanogene 6, negatively regulates the polyubiquitination and stability of HSPA2 in human and mouse germ cells [28–30], how the stability of HSPA2 is regulated in human cancers remains unexplored. In this study, we identified HSPA2 as a novel ubiquitination substrate of RNF144A by quantitative proteomic and biochemical analyses (Figs. 3 and 4). To our knowledge, RNF144A is the first identified E3 ubiquitin ligase for HSPA2 ubiquitination and degradation in human cancers. Moreover, HSPA2, in addition to DNA-PKcs [12] and PARP1 [14], is the third identified ubiquitination substrate for RNF144A ligase. Functional experiments further demonstrated that HSPA2 is indeed involved in RNF144A-mediated breast cancer proliferation, migration, and invasion (Fig. 5). Consistent with its oncogenic activities [17–26], clinical data showed that breast cancer patients with elevated HSPA2 expression had poor clinical outcome and malignant clinicopathologic parameters (Fig. 6d–f and Supplementary Table S11). These findings establish that RNF144A suppresses breast cancer progression through, at least in part, blocking HSPA2-mediated oncogenic signaling.
In conclusion, findings presented here suggest that RNF144A is an E3 ubiquitin ligase for HSPA2 and suppresses the aggressive phenotype of breast cancer through controlling HSPA2-mediated oncogenic activities (Fig. 7). These results may offer novel therapeutic opportunities in breast cancer through restoration of RNF144A expression by DNA methylation inhibitors or suppression of oncoprotein HSPA2-mediated oncogenic signaling by specific small-molecule inhibitors.
Supplementary information
Acknowledgements
We sincerely acknowledge the staff members of the Pathology Core Facility (Shanghai Cancer Center), the Proteomic Center (Institutes of Biomedical Sciences), the Animal Resource Center (State Key Laboratory of Oncogene and Related Gene), and members in the Li laboratory for their excellent technical assistance. The work in the Li laboratory is supported, in whole or in part, by the National Natural Science Foundation of China (No. 81372847, 81572584, and 81772805), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (No. 2013-06), the Science and Technology Innovation Action Plan of Shanghai Municipal Science and Technology Commission (No. 16JC1405400), and start-up fund for new investigators from Fudan University.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by V. D'Angiolella
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yin-Long Yang, Ye Zhang, Dou-Dou Li, Fang-Lin Zhang
Contributor Information
Da-Qiang Li, Email: daqiangli1974@fudan.edu.cn.
Zhi-Min Shao, Email: zhimingshao@yahoo.com.
Supplementary information
The online version of this article (10.1038/s41418-019-0400-z) contains supplementary material, which is available to authorized users.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen C, Seth AK, Aplin AE. Genetic and expression aberrations of E3 ubiquitin ligases in human breast cancer. Mol Cancer Res. 2006;4:695–707. doi: 10.1158/1541-7786.MCR-06-0182. [DOI] [PubMed] [Google Scholar]
- 3.Buetow L, Huang DT. Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat Rev Mol Cell Biol. 2016;17:626–42. doi: 10.1038/nrm.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lipkowitz S, Weissman AM. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat Rev Cancer. 2011;11:629–43. doi: 10.1038/nrc3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 6.Walden H, Rittinger K. RBR ligase-mediated ubiquitin transfer: a tale with many twists and turns. Nat Struct Mol Biol. 2018;25:440–5. doi: 10.1038/s41594-018-0063-3. [DOI] [PubMed] [Google Scholar]
- 7.Mészáros Bálint, Kumar Manjeet, Gibson Toby J., Uyar Bora, Dosztányi Zsuzsanna. Degrons in cancer. Science Signaling. 2017;10(470):eaak9982. doi: 10.1126/scisignal.aak9982. [DOI] [PubMed] [Google Scholar]
- 8.Marin I, Lucas JI, Gradilla AC, Ferrus A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol Genom. 2004;17:253–63. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- 9.Spratt DE, Walden H, Shaw GS. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J. 2014;458:421–37. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8:209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho SR, Lee YJ, Lin WC. Regulation of RNF144A E3 ubiquitin ligase activity by self-association through its transmembrane domain. J Biol Chem. 2015;290:23026–38. doi: 10.1074/jbc.M115.645499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho SR, Mahanic CS, Lee YJ, Lin WC. RNF144A, an E3 ubiquitin ligase for DNA-PKcs, promotes apoptosis during DNA damage. Proc Natl Acad Sci USA. 2014;111:E2646–2655. doi: 10.1073/pnas.1323107111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Yang YL, Zhang FL, Liao XH, Shao ZM, Li DQ. Epigenetic silencing of RNF144A expression in breast cancer cells through promoter hypermethylation and MBD4. Cancer Med. 2018;7:1317–25. doi: 10.1002/cam4.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Liao XH, Xie HY, Shao ZM, Li DQ. RBR-type E3 ubiquitin ligase RNF144A targets PARP1 for ubiquitin-dependent degradation and regulates PARP inhibitor sensitivity in breast cancer cells. Oncotarget. 2017;8:94505–18. doi: 10.18632/oncotarget.21784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scieglinska D, Krawczyk Z. Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans. Cell Stress Chaperon. 2015;20:221–35. doi: 10.1007/s12192-014-0548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eddy EM. Role of heat shock protein HSP70-2 in spermatogenesis. Rev Reprod. 1999;4:23–30. doi: 10.1530/ror.0.0040023. [DOI] [PubMed] [Google Scholar]
- 17.Garg M, Kanojia D, Seth A, Kumar R, Gupta A, Surolia A, et al. Heat-shock protein 70-2 (HSP70-2) expression in bladder urothelial carcinoma is associated with tumour progression and promotes migration and invasion. Eur J Cancer. 2010;46:207–15. doi: 10.1016/j.ejca.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Garg M, Kanojia D, Saini S, Suri S, Gupta A, Surolia A, et al. Germ cell-specific heat shock protein 70-2 is expressed in cervical carcinoma and is involved in the growth, migration, and invasion of cervical cells. Cancer. 2010;116:3785–96. doi: 10.1002/cncr.25218. [DOI] [PubMed] [Google Scholar]
- 19.Jagadish N, Agarwal S, Gupta N, Fatima R, Devi S, Kumar V, et al. Heat shock protein 70-2 (HSP70-2) overexpression in breast cancer. J Exp Clin Cancer Res. 2016;35:150. doi: 10.1186/s13046-016-0425-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagadish N, Parashar D, Gupta N, Agarwal S, Suri V, Kumar R, et al. Heat shock protein 70-2 (HSP70-2) is a novel therapeutic target for colorectal cancer and is associated with tumor growth. BMC Cancer. 2016;16:561. doi: 10.1186/s12885-016-2592-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta N, Jagadish N, Surolia A, Suri A. Heat shock protein 70-2 (HSP70-2) a novel cancer testis antigen that promotes growth of ovarian cancer. Am J Cancer Res. 2017;7:1252–69. [PMC free article] [PubMed] [Google Scholar]
- 22.Rohde M, Daugaard M, Jensen MH, Helin K, Nylandsted J, Jaattela M. Members of the heat-shock protein 70 family promote cancer cell growth by distinct mechanisms. Genes Dev. 2005;19:570–82. doi: 10.1101/gad.305405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh S, Suri A. Targeting the testis-specific heat-shock protein 70-2 (HSP70-2) reduces cellular growth, migration, and invasion in renal cell carcinoma cells. Tumour Biol. 2014;35:12695–706. doi: 10.1007/s13277-014-2594-5. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Chen W, Duan CJ, Zhang CF. Overexpression of HSPA2 is correlated with poor prognosis in esophageal squamous cell carcinoma. World J Surg Oncol. 2013;11:141. doi: 10.1186/1477-7819-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Y, Zhao H, Li XS, Kang HR, Ma JX, Yao FF, et al. Expression of HSPA2 in human hepatocellular carcinoma and its clinical significance. Tumour Biol. 2014;35:11283–7. doi: 10.1007/s13277-014-2430-y. [DOI] [PubMed] [Google Scholar]
- 26.Zhang H, Gao H, Liu C, Kong Y, Wang C. Expression and clinical significance of HSPA2 in pancreatic ductal adenocarcinoma. Diagn Pathol. 2015;10:13. doi: 10.1186/s13000-015-0253-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang WJ, Xia LM, Zhu F, Huang B, Zhou C, Zhu HF, et al. Transcriptional upregulation of HSP70-2 by HIF-1 in cancer cells in response to hypoxia. Int J Cancer. 2009;124:298–305. doi: 10.1002/ijc.23906. [DOI] [PubMed] [Google Scholar]
- 28.Sasaki T, Marcon E, McQuire T, Arai Y, Moens PB, Okada H. Bat3 deficiency accelerates the degradation of Hsp70-2/HspA2 during spermatogenesis. J Cell Biol. 2008;182:449–58. doi: 10.1083/jcb.200802113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bromfield E, Aitken RJ, Nixon B. Novel characterization of the HSPA2-stabilizing protein BAG6 in human spermatozoa. Mol Hum Reprod. 2015;21:755–69. doi: 10.1093/molehr/gav041. [DOI] [PubMed] [Google Scholar]
- 30.Bromfield EG, Aitken RJ, McLaughlin EA, Nixon B. Proteolytic degradation of heat shock protein A2 occurs in response to oxidative stress in male germ cells of the mouse. Mol Hum Reprod. 2017;23:91–105. doi: 10.1093/molehr/gaw074. [DOI] [PubMed] [Google Scholar]
- 31.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–24. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang FL, Cao JL, Xie HY, Sun R, Yang LF, Shao ZM, et al. Cancer-associated MORC2-mutant M276I regulates an hnRNPM-mediated CD44 splicing switch to promote invasion and metastasis in triple-negative breast cancer. Cancer Res. 2018;78:5780–92. doi: 10.1158/0008-5472.CAN-17-1394. [DOI] [PubMed] [Google Scholar]
- 33.Sun R, Xie HY, Qian JX, Huang YN, Yang F, Zhang FL, et al. FBXO22 possesses both protumorigenic and antimetastatic roles in breast cancer progression. Cancer Res. 2018;78:5274–86. doi: 10.1158/0008-5472.CAN-17-3647. [DOI] [PubMed] [Google Scholar]
- 34.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2:329–33. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 35.Liao XH, Zhang Y, Dong WJ, Shao ZM, Li DQ. Chromatin remodeling protein MORC2 promotes breast cancer invasion and metastasis through a PRD domain-mediated interaction with CTNND1. Oncotarget. 2017;8:97941–54. doi: 10.18632/oncotarget.18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X, Zheng W, Wang W, Shen H, Liu L, Lou W, et al. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br J Cancer. 2017;117:1846–54. doi: 10.1038/bjc.2017.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang S, et al. Parkin targets HIF-1alpha for ubiquitination and degradation to inhibit breast tumor progression. Nat Commun. 2017;8:1823. doi: 10.1038/s41467-017-01947-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 40.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. 2016;13:674–90. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W, Slingerland JM. Links between oestrogen receptor activation and proteolysis: relevance to hormone-regulated cancer therapy. Nat Rev Cancer. 2014;14:26–38. doi: 10.1038/nrc3622. [DOI] [PubMed] [Google Scholar]
- 43.Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L. Treatment of HER2-positive breast cancer: current status and future perspectives. Nat Rev Clin Oncol. 2011;9:16–32. doi: 10.1038/nrclinonc.2011.177. [DOI] [PubMed] [Google Scholar]
- 44.Scieglinska D, Piglowski W, Chekan M, Mazurek A, Krawczyk Z. Differential expression of HSPA1 and HSPA2 proteins in human tissues; tissue microarray-based immunohistochemical study. Histochem Cell Biol. 2011;135:337–50. doi: 10.1007/s00418-011-0791-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scieglinska D, Piglowski W, Mazurek A, Malusecka E, Zebracka J, Filipczak P, et al. The HspA2 protein localizes in nucleoli and centrosomes of heat shocked cancer cells. J Cell Biochem. 2008;104:2193–206. doi: 10.1002/jcb.21778. [DOI] [PubMed] [Google Scholar]
- 46.Ghanta KS, Li DQ, Eswaran J, Kumar R. Gene profiling of MTA1 identifies novel gene targets and functions. PLoS ONE. 2011;6:e17135. doi: 10.1371/journal.pone.0017135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.