Abstract
Cellular senescence is implicated in aging or age-related diseases. Sonic hedgehog (Shh) signaling, an inducer of embryonic development, has recently been demonstrated to inhibit cellular senescence. However, the detailed mechanisms to activate Shh signaling to prevent senescence is not well understood. Here, we demonstrate that Protein arginine methyltransferase 7 (PRMT7) promotes Shh signaling via GLI2 methylation which is critical for suppression of cellular senescence. PRMT7-deficient mouse embryonic fibroblasts (MEFs) exhibited a premature cellular senescence with accompanied increase in the cell cycle inhibitors p16 and p21. PRMT7 depletion results in reduced Shh signaling activity in MEFs while PRMT7 overexpression enhances GLI2-reporter activities that are sensitive to methylation inhibition. PRMT7 interacts with and methylates GLI2 on arginine residues 225 and 227 nearby a binding region of SUFU, a negative regulator of GLI2. This methylation interferes with GLI2-SUFU binding, leading to facilitation of GLI2 nuclear accumulation and Shh signaling. Taken together, these data suggest that PRMT7 induces GLI2 methylation, reducing its binding to SUFU and increasing Shh signaling, ultimately leading to prevention of cellular senescence.
Subject terms: Cell biology, Molecular biology
Introduction
Cellular senescence involves an irreversible cell cycle arrest accompanying profound alterations in gene expression including tumor suppressor/cell cycle inhibitor activation and secretory protein production, as well as chromatin abnormality [1–4]. Cellular senescence is implicated in regeneration defects associated with aging or age-related diseases [5, 6]. In contrast, it is considered as a safeguard against cancer [5, 7]. Therefore, much attention is paid to elucidate regulatory pathways underlying cellular senescence [8–11]. A recent emerging paradigm proposes that defective sonic hedgehog (Shh) signaling associated with the suppression of cell cycle inhibitor p16INK4a (p16 hereafter) leads to replicative senescence [12].
Shh signaling is composed of 4 major components; the 12-transmembrane domain receptor protein Patched 1 (PTCH1), the 7-transmembrane domain protein Smoothened (SMO) as the activating membrane component, Glioma-associated oncogene (GLI) transcription factors, and the suppressor of fused protein (SUFU), a negative regulator of GLI [13]. In vertebrate, Shh signaling is distinctively dependent on primary cilia and the ciliary localization of signaling components such as SMO and SUFU/GLI2 are regulated by ligand-triggered signaling [14, 15]. Upon Shh binding to PTCH1, activated SMO inhibits SUFU association with GLI2 leading to GLI2 nuclear translocation from cytoplasm and target gene expression [15]. SUFU inhibits Shh signaling in cytoplasm by sequestering GLI2 [16, 17]. In addition, the activity and stability of GLI2 is tightly controlled by posttranslational modifications such as phosphorylation, ubiqitination, sumoylation and acetylation [18–22].
Protein arginine methyltransferases (PRMTs) are responsible for arginine methylation of histone or non-histone proteins to regulate diverse biological processes, including cell proliferation, differentiation and senescence [23–27]. A recent study showed that PRMT1 methylates GLI1 on R597 enhancing its transcriptional activity in pancreatic ductal adenocarcinoma [28]. However, whether GLI2 and GLI3 are regulated by arginine methylation is currently unknown. Among 9 PRMTs, multiple PRMTs such as PRMT1, PRMT4, PRMT5, PRMT6 and PRMT7 have been shown to be implicated in regulation of cellular senescence through suppression of cell cycle inhibitors [29–33]. Specifically, PRMT7 is implicated in maintenance of muscle stem cell function. PRMT7-deficient muscle stem cells exhibit impaired cell growth and premature cellular senescence through decreased expression of a DNA methyltransferase DNMT3b, leading to p21 derepression [30]. However, the role of PRMT7 in senescence of fully differentiated cells such as fibroblasts and neurons remain unexplored. Here, we demonstrate a critical role of PRMT7 in Shh signaling activation and prevention of cellular senescence in mouse embryonic fibroblasts (MEFs). PRMT7-deficient MEFs exhibited premature cellular senescence accompanying increased expression of the cell cycle inhibitors, p16 and p21. PRMT7 depletion reduced GLI2 activity, while PRMT7 overexpression enhanced it. PRMT7 deficiency impaired the nuclear accumulation of GLI2. PRMT7 interacted with GLI2 and methylated at arginine 225 and 227, inhibiting GLI2-SUFU interaction and thus facilitating GLI2-mediated Shh signaling.
Results
PRMT7 deficiency results in premature senescence of MEFs
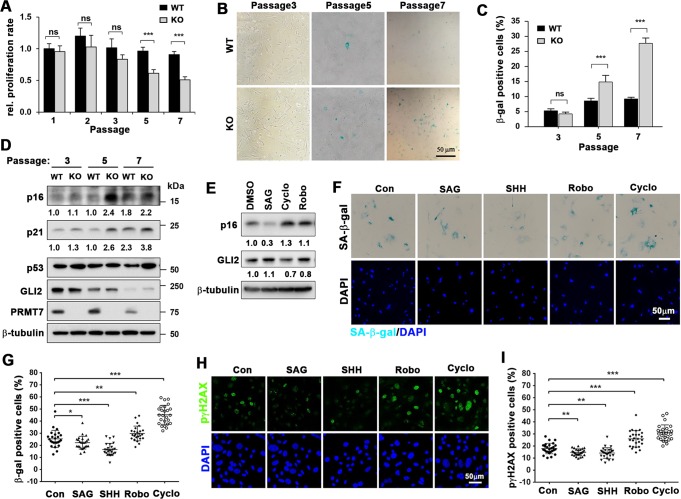
In consideration of a previous study on cellular senescence of PRMT7-deficient muscle stem cells [30], we examined whether PRMT7 is also involved in cellular senescence in MEFs. First, the growth potential of PRMT7+/+ (WT) and PRMT7−/− (KO) MEFs was assessed through serial passaging. For each passage, 5 × 104 cells were seeded and cultured for 2 days, followed by cell counting. KO MEFs exhibited progressively decreased proliferation starting at passage 5 (p5), while WT MEFs maintained the proliferative capacity (Fig. 1a). Next, p3 to p7 MEFs were stained for senescence marker β-galactosidase (SA-β-gal). p5 and p7 KO MEFs had more SA-β-gal-positive cells, compared with WT (Fig. 1b, c). The expression of p16 and p21 was substantially increased in p5 and p7 KO MEFs and p53 was mildly elevated (Fig. 1d). PRMT7 was dramatically decreased in p7 WT MEFs coinciding with high p16 and p21 levels. Interestingly, GLI2 was progressively reduced and almost diminished in p7 MEFs. In a previous study, PRMT7-deficient muscle stem cells undergo cellular senescence upon activation due to decreased Dnmt3b expression [30]. However, Dnmt3b was increased in p3 KO MEFs and thereafter it was not altered, relative to WT cells (Supplementary Fig. 1), suggesting that PRMT7 regulates MEF senescence through other mechanism than Dnmt3b. We then examined the effect of Shh signaling agonist and inhibitors on p16 expression and cellular senescence (Fig. 1e–i). Thus, p5 MEFs were treated with DMSO, a Shh agonist SAG, or two Shh antagonists, Cyclopamine (Cyclo) and Robotnikinin (Robo) for 48 h. The SAG treatment reduced p16 protein levels, while antagonists further increased it (Fig. 1e). Furthermore, the staining for SA-β-gal and a phosphorylated form of γH2AX (p-γH2AX), a marker for DNA damage, revealed that SAG or Shh treatment significantly reduced SA-β-gal-postive cells or p-γH2AX-positive cells, while Shh antagonists elevated cellular senescence (Fig. 1f–i). Taken together, these data suggest that PRMT7-deficient MEFs show p16 and p21 upregulation, and undergo premature cellular senescence which are inversely related with Shh signaling activity.
Fig. 1.
PRMT7 deficiency leads to premature cellular senescence of MEFs. a Relative proliferative capacity of WT and KO MEFs cells during cell passaging. The cell number of the first passage WT cells was set to 1.0. The determinants represent the means ± SEM. n = 3. ns = not significant, ***P < 0.001. The experiment was performed at least three times with similar results. b Staining for senescence-associated β-galactosidase activity (SA-β-gal) with MEFs at cell passage number 3, 5, and 7 (p3, p5, and p7). Size bar = 50 μM. c SA-β-gal positive cells were counted and plotted as percentile. Cells in at least 6 random areas were analyzed and performed with 3 different MEF batches. Data are presented as mean ± SEM. ANOVA test for comparison with the control cells. ***P < 0.001. d Immunoblot analysis for senescence-associated proteins such as p53, p21, and p16, and β-tubulin used as a loading control. The relative levels of p16 and p21 are listed below and the relative signal intensity to the loading control of WT cells at p3 was set to 1.0. e Immunoblot analysis for p16 and GLI2 in WT MEFs at p5 treated with SAG or two Shh antagonists, cyclopamine (Cyclo) or robotnikinin (Robo) for 48 h. The relative signal intensity to the loading control from vehicle treated cells was set to 1.0. f Staining for SA-β-gal of MEFs at p7 treated with vehicle, SAG, Shh, Robo, or Cyclo for 48 h. Size bar = 50 μM. g SA-β-gal positive cells were counted and plotted as percentile. Data are presented as mean ± SEM. ANOVA test for comparison with the control cells. n = 25 fields for each condition. *P < 0.05, **P < 0.01, ***P < 0.001. h Immunostaining for a DNA damage marker p-γH2AX of MEFs at p7 treated with vehicle, SAG, Shh, Robo, or Cyclo for 48 h. Size bar = 50 μM. i Quantification of p-γH2AX-positive cells. Data are presented as mean ± SEM. ANOVA test for comparison with the control cells. n = 26 fields for each condition. **P < 0.01, ***P < 0.001
PRMT7 promotes Shh signaling activity through GLI2
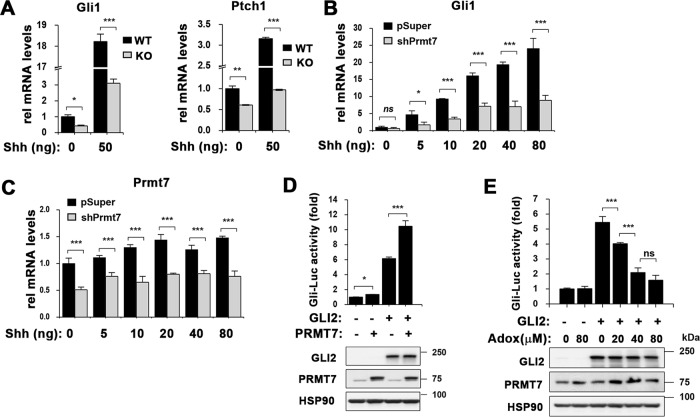
To assess the role of PRMT7 in Shh signaling, WT and KO MEFs were serum-starved and treated with Shh (50 ng/ml) for 12 h, followed by quantitative RT-PCR (qRT-PCR) for GLI2 targets, Gli1 and Ptch1 (Fig. 2a). KO MEFs exhibited significantly reduced Gli1 and Ptch1 expression in control-condition without Shh treatment, relative to WT MEFs. In response to Shh treatment, WT MEFs robustly increased Gli1 and Ptch1 expression while this response was blunted in KO MEFs. To further confirm, control or PRMT7-depleted 10T1/2 cells were serum starved and treated with increasing Shh concentration for 12 h, followed by qRT-PCR (Fig. 2b). PRMT7 depletion significantly reduced Shh-induced increase in Gli1 levels, relative to controls. PRMT7 expression was decreased about 50% by shPRMT7, compared with the control pSuper (Fig. 2c). PRMT7 reexpression restored Gli1 expression in PRMT7-depleted cells, supporting for the specificity of shPRMT7 (Supplementary Fig. 2). These data suggest that PRMT7 is required for Shh signaling activation.
Fig. 2.
PRMT7 deficiency blunts Shh signaling activity. a qRT-PCR analysis for Shh signaling components, Gli1 and Ptch1. PRMT7+/+ (WT) and PRMT7−/− (KO) MEFs were treated with 50 ng/ml recombinant Shh for 12 h. Data are presented as means ± SD. n = 3, ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001. b qRT-PCR analysis for Gli1 mRNA expression in control shRNA-expressing or PRMT7 shRNA-expressing 10T1/2 cells treated with increasing amounts of Shh for 12 h. ns = not significant, *P < 0.05, ***P < 0.001 for the comparison to control by ANOVA test. n = 3. c qRT-PCR analysis for PRMT7 mRNA expression with the total RNAs shown in panel b. Data are presented as means ± SD. n = 3, ***P < 0.001. d GLI2-responsive luciferase activities in 10T1/2 cells transfected with indicated expression vectors (upper panel). The protein expressions of GLI2 and PRMT7 were determined by western blot (lower panel). Data are presented means ± SD. n = 3, *P < 0.05, ***P < 0.001. e The effect of arginine methylation inhibition on GLI-reporter activities. Control-expressing or GLI2-expressing 10T1/2 cells were treated with control vehicle or a PRMT inhibitor, Adox with increasing concentration for 24 h (upper panel). The protein expressions of GLI2 and PRMT7 were determined by western blot (lower panel). Data are presented means ± SD. n = 3, ns = not significant, ***P < 0.001
Next, we asked whether PRMT7 regulates GLI2 activity by using a GLI-luciferase reporter containing the Gli1 promoter region (Gli-Luc). 10T1/2 cells were co-transfected with Gli-Luc plus either control, GLI2 or PRMT7 vectors for 48 h, followed by luciferase assay. PRMT7 expression elevated slightly but significantly Gli-reporter activities without GLI2 transfection. GLI2 transfection greatly enhanced Gli-reporter activities which were further enhanced by PRMT7 coexpression (Fig. 2d). To analyze the role of arginine methylation in GLI2 activities, 10T1/2 cells were transfected with vectors for Gli-Luc and control or GLI2 and 36 h later, cell were treated with DMSO or Adox, a methylation inhibitor for 24 h. Notably, PRMT inhibition elicited a dose-dependent inhibition of GLI2 activities (Fig. 2e). The expression of GLI2 protein and PRMT7 was not greatly altered by Adox treatment. These data suggest that PRMT7 regulates GLI2 transcription activities likely through arginine methylation.
PRMT7 is important for the localization of GLI2 in primary cilium in response to Shh signaling
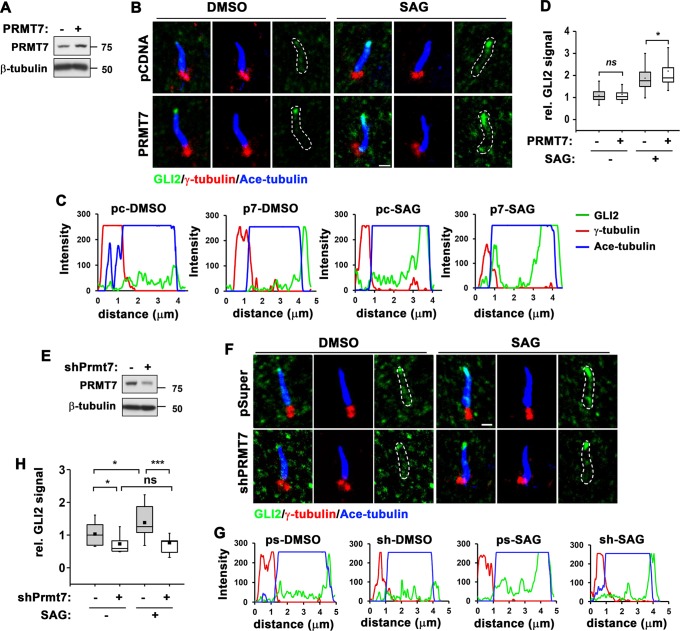
Control or PRMT7-overexpressing 10T1/2 cells were serum starved and treated with DMSO or SAG for 12 h, followed by immunostaining for GLI2, Ace-tubulin and γ-tubulin, a marker for the basal bodies (Fig. 3a–d). GLI2 proteins were recruited to primary cilium in response to SAG treatment and PRMT7 overexpression enhanced GLI2 ciliary localization in response to SAG, as demonstrated in the relative GLI2 signal in cilium. In a converse experiment, control or PRMT7-depleted 10T1/2 cells were serum starved and treated with DMSO or SAG for 12 h, followed by immunostaining for GLI2, Ace-tubulin and γ-tubulin. PRMT7-depleted cells exhibited less GLI2 levels in primary cilium and reduced recruitment upon SAG application (Fig. 3e–h). These data suggest that PRMT7 is important for GLI2 ciliary localization.
Fig. 3.
PRMT7 inhibition reduces ciliary localization of GLI2 in response to SAG. a Immunoblot for PRMT7 expression in 10T1/2 cells transfected with control or PRMT7 expression vector. b Immunostaining for GLI2, acetylated-tubulin (a marker for primary cilium, Ace-tubulin) and γ-tubulin (a marker for the basal body of the primary cilium) in control or PRMT7 overexpressing cells treated with vehicle or SAG for 12 h. Scale bar: 1 μm. c The signal intensity of GLI2 from the base to the tip of primary cilium shown in panel b was quantified. d The relative signal intensity of GLI2 in primary cilium shown in panel b was indicated. The value from the vehicle-treated control cells was set to 1.0. Data are presented as means ± SD. n ≥ 15. ns; not significant. *P < 0.05. e Immunoblot for PRMT7 in 10T1/2 cells expressing control or PRMT7 shRNA. f Immunostaining for GLI2, Ace-tubulin and γ-tubulin in control or PRMT7-depleted cells treated with vehicle or SAG for 16 h. Scale bar: 1 μm. g The signal intensity of GLI2 from the base to the tip of primary cilium shown in panel f was quantified. h The relative signal intensity of GLI2 in primary cilium shown in panel f was indicated. The value from the vehicle-treated control cells was set to 1.0. Data are presented as means ± SD. n ≥ 15. ns = not significant. *P < 0.05. ***P < 0.001
PRMT7 deficiency attenuates Gli2 nuclear accumulation in response to SAG treatment
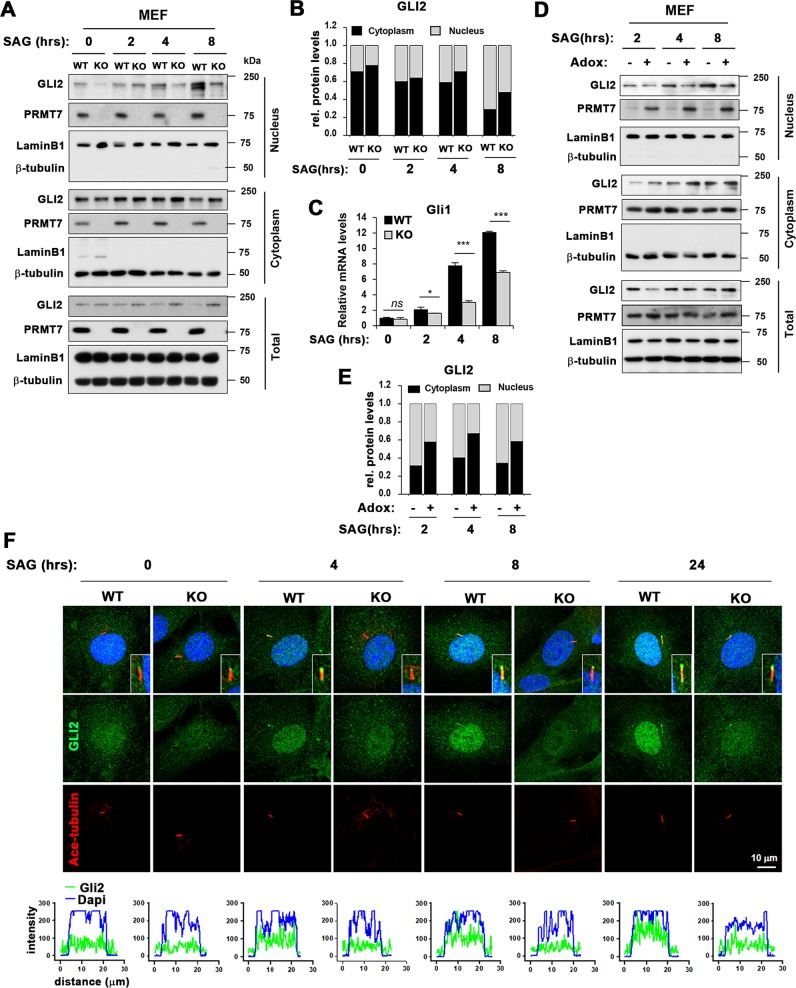
To examine the effect of PRMT7 deficiency in GLI2 activities, WT and KO MEFs were starved for 16 h and treated with vehicle or 100 nM SAG. WT MEFs exhibited a robust nuclear accumulation of GLI2 after 8 h of SAG treatment which was almost abrogated in KO MEFs (Fig. 4a, b). Interestingly, total and cytoplasmic GLI2 levels were elevated in KO cells starting from 4 h of SAG treatment. The quantification of GLI2 levels in each fraction are shown in Fig. 4b. The decreased nuclear localization is consistent with the decreased Gli1 expression in KO MEFs (Fig. 4c). To further define, WT MEFs were pre-treated with Adox (80 μM) for 2 h, prior to the SAG (100 nM) treatment. Similarly, to PRMT7 deficiency, the Adox treatment attenuated GLI2 nuclear accumulation in response to SAG, while more GLI2 were found in the cytoplasmic fraction (Fig. 4d, e). Surprisingly, Adox treatment enhanced the level of nuclear PRMT7, suggestive of a potential role for arginine methylation in PRMT7 protein trafficking. Upon signal activation, GLI2 and SUFU accumulate in primary cilium and the subsequent dissociation of GLI2 from SUFU allows nuclear accumulation of GLI2, leading to downstream target gene expression [15]. To address the role of PRMT7 in GLI2 translocation, WT and KO MEFs were serum starved and treated with SAG for indicated hours and immunostained for GLI2 and Ace-tubulin (Fig. 4f). A robust localization of GLI2 in cilia and nucleus was observed starting at 4 h of SAG treatment in WT cells, while KO cells displayed greatly reduced levels of GLI2 in cilia and nucleus. This is consistent with the data obtained with cellular fractionation assay. These data suggest that PRMT7 is necessary for GLI2 nuclear translocation in response to signal activation.
Fig. 4.
PRMT7 is required for GLI2 nuclear accumulation. a Cellular fractionation analysis of MEFs treated with DMSO or SAG. The highly confluence of WT or KO MEFs were serum-starved for 18 h and treated with DMSO or SAG for indicated period, followed by cellular fractionation assay and immunoblotted for GLI2 and PRMT7. β-tubulin and LaminB1 were used as internal loading control for total and cytoplasm, and nucleus, respectively. b Quantification for GLI2 levels in cytoplasm or nucleus shown in panel a. c Control qRT-PCR for Gli1 expression in the replica sets shown in panel a. Data represent mean ± SD. n = 3, ANOVA test for comparison, *P < 0.05, ***P < 0.001. d Cellular fractionation analysis for GLI2, in response to SAG and/or Adox treatment. The highly confluence of WT MEFs were serum-starved for 18 h. Subsequently, cells were pre-treated with 80 μM Adox for 2 h prior to SAG treatment. e Quantification for GLI2 levels in cytoplasm or nucleus shown in panel d. f Immunostaining for Ace-tubulin and GLI2 with MEFs treated with vehicle DMSO or SAG for indicated time. The lower panel shows the quantification of GLI2 expression in the nucleus labeled with Dapi. The GLI2 localization in primary cilium is shown in the inset
PRMT7 binds to the N-terminal region of GLI2 and regulates its nuclear accumulation
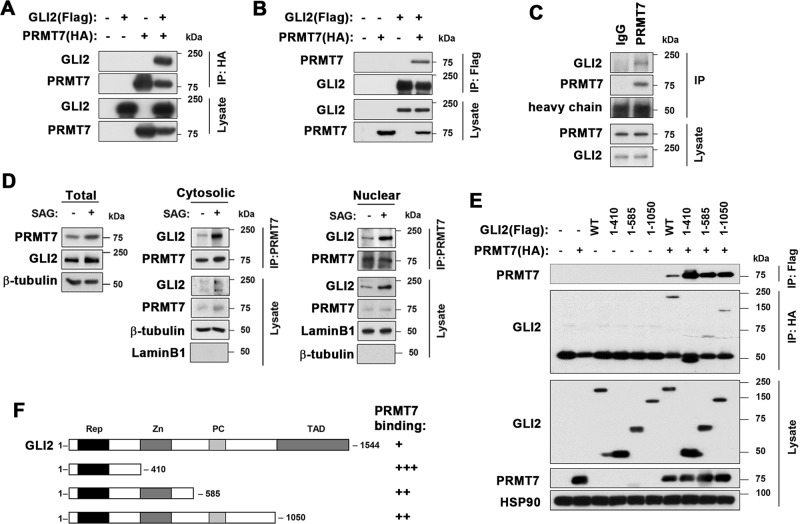
To examine the interaction of GLI2 and PRMT7, 293 T cells were cotransfected with control, Flag-tagged GLI2 and/or HA-tagged PRMT7, followed by immunoprecipitation. HA-PRMT7 and Flag-GLI2 proteins reciprocally co-immunoprecipitated (Fig. 5a, b). Furthermore, 10T1/2 cell lysates were subjected to immunoprecipitation with control IgG or anti-PRMT7 antibodies and GLI2 was coprecipitated with PRMT7 (Fig. 5c). The cellular fractionation of 10T1/2 cells starved and treated with vehicle or SAG for 8 h showed that PRMT7 and GLI2 interacts in cytosol and nucleus and their interaction was greatly increased by SAG treatment (Fig. 5d). To define which domain(s) of GLI2 is important for PRMT7 binding, 293 T cells were cotransfected with vectors for PRMT7 and various regions of GLI2, followed by immunoprecipitation (Fig. 5e). The relative binding of GLI2 fragments from 3 independent experiments summarized in Fig. 5f. The N-terminal domain (1–410) of GLI2 showed the strongest interaction with PRMT7, whereas 1–585 and 1–1050 of GLI2 fragments interacted less efficiently with PRMT7 compared with N-terminal region. Interestingly, the full-length GLI2 interacted less efficiently than fragments, suggesting for a negative role of the C-terminal transactivation domain in PRMT7 interaction.
Fig. 5.
PRMT7 interacts with GLI2 and regulates its nuclear localization. a, b Co-immunoprecipitation for the physical interaction between GLI2 and PRMT7 in 293 T cells transfected with control, Flag-tagged GLI2 or HA-tagged PRMT7. Anti-HA antibody was used for coimmunoprecipitation in panel a, and anti-Flag antibody was subjected for co-immunoprecipitation in panel b. c Coimmunoprecipitation of endogenous PRMT7 with GLI2 in 10T1/2 cells. d Coimmunoprecipitation of GLI2 with PRMT7 in cytosolic and nuclear fractions of control or SAG-treated cells. e Coimmunoprecipitation of PRMT7 with Flag-tagged various deletion constructs of Gli2 indicated with amino acid numbers. f The schematic representation of GLI2 domain structure and the summary of the relative binding affinity with PRMT7 from three experimental sets. Rep repressor, Zn zinc finger, PC processing domain, TAD transcription activation domain
GLI2 methylation on arginine residues 225 and 227 by PRMT7 is important to suppress cellular senescence
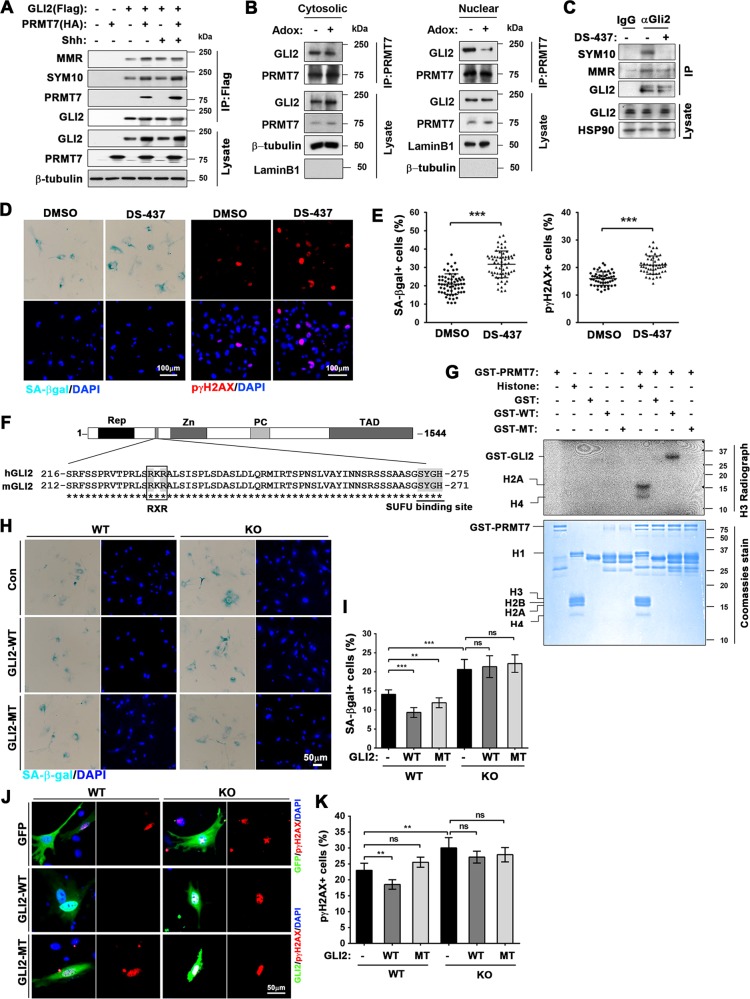
PRMT7 belongs to class III methyltransferase that can catalyze monomethylation and symmetric dimethylation of arginine [34]. To examine whether PRMT7 regulates GLI2 by methylation, 293 T cells were cotransfected with Flag-GLI2 or HA-PRMT7 and treated with Shh for 24 h, followed by immunoprecipitation with anti-Flag antibodies and immunoblotting with mono-methyl (MMR) or symmetrically dimethyl-arginine (SYM10) antibodies (Fig. 6a). The mono-methylation and symmetric di-methylation of GLI2 was observed in control cells, likely mediated by endogenous PRMT7. PRMT7 coexpression elevated both methylation types in GLI2 and the Shh treatment increased PRMT7 interaction with GLI2 and SYM10-positive GLI2. Next, control or Adox-treated 10T1/2 cells were subjected to cellular fractionation and immunoprecipitation (Fig. 6b). Coprecipitated GLI2 levels with PRMT7 was specifically decreased in the nuclear fraction, while cytosolic GLI2 was slightly elevated. Furthermore, p5 WT MEFs were treated with a PRMT5 and PRMT7 inhibitor DS-437 for 48 h, followed by control IgG or anti-GLI2 antibody immunoprecipitation (Fig. 6c). DS-437 treatment almost abolished or reduced the levels of dimethylated-GLI2 or monomethylated-GLI2. In addition, p5 WT MEFs were treated with DMSO or DS-437 for 48 h followed by SA-βgal or p-γH2AX staining (Fig. 6d, e). Similar to KO MEFs, DS-437 treatment enhanced significantly SA-βgal and p-γH2AX-positive cells, compared with control. These data supported that the methyltransferase activity of PRMT7 is important to suppress cellular senescence.
Fig. 6.
PRMT7 methylates GLI2 on arginine residues 225 and 227. a Immunoprecipitation with anti-Flag antibody for Flag-tagged GLI2 and immunoblotting with anti-monomethylated arginine (MMR) and symmetric dimethyl-arginine (SYM10) antibodies. 293 T cells transfected with indicated expression vectors were treated with control or Shh for 24 h. b Cellular fractions from 10T1/2 cells treated with either DMSO or Adox was subjected for immunoprecipitation with anti-PRMT7 antibody, followed by immunoblotting with anti-GLI2 antibody. c Immunoprecipitation with anti-GLI2 antibody and immunoblotting GLI2 with anti-SYM10 or MMR in MEF cell at p5 treated with DMSO or DS-437. d Staining for SA-β-gal and p-γH2AX of p5 MEFs treated with vehicle DMSO or DS-437 for 48 h. Size bar = 100 μM. e SA-β-gal and p-γH2AX-positive cells per filed were counted and plotted as percentile. n = 62 (SA-β-gal/DMSO), 62 (SA-β-gal/DS-437), 50 (p-γH2AX/DMSO), 50 (p-γH2AX/DS-437). Data are presented as mean ± SEM. ANOVA test for comparison with the control cells. ***P < 0.001. f A schematic representation of GLI2 domain structure and the alignment of the region containing SUFU binding site in mouse and human GLI2. Conserved amino acids are indicated by asterisks. g Autoradiography for in vitro methylation using bacterially purified GST-PRMT7 and GST-WT or GST-MT (R225/227 K). Purified histones were used as arginine methylation control. h Staining for SA-β-gal of WT or KO MEFs at p5 transfected with con GFP, GLI2-WT, or GLI2-RK. Size bar = 50 μm. i SA-β-gal-positive cells were counted per filed and plotted as percentile. n = 30 fields for each condition. Data are presented as mean ± SEM. ANOVA test for comparison with the control cells. ***P < 0.001. j Immunostaining for p-γH2AX, GFP or Flag expression in WT or KO MEFs at p5 transfected with GFP, Flag-GLI2-WT or Flag-GLI2-RK. Size bar = 50 μM. k p-γH2AX-positive cells in GFP-, Flag-positive cells were counted per filed and plotted as percentile. n = 47 (WT/con), 68 (WT/GLI2-WT), 78 (WT/GLI2-RK), 45 (KO/con), 50 (KO/GLI2-WT), 66 (KO/GLI2-RK). Data are presented as mean ± SEM. ANOVA test for comparison with the control cells. ***P < 0.001
One of regulatory mechanisms for GLI2 activity is its interaction with SUFU via N-terminal domain containing an SYGH core motif in GLI2 [35], which also interacts with PRMT7. The sequence analysis of the N-terminal domain of mouse and human GLI2 revealed a potential PRMT7 methylation motif, RxR [36] with the key arginine residues at 225 and 227 of mouse GLI2 (RKR) (Fig. 6f). This motif was recently identified as a nuclear localization signal (NLS) in addition to a canonical NLS localized in the region of 562–578 [37]. To determine GLI2 methylation by PRMT7, we generated an arginine to lysine mutation (R/K) which preserves its nuclear localization signal characteristic. GST-tagged GLI2 fragments from 201 to 280 with wildtype sequence (GTS-GLI2 (201–280) WT; GST-WT) or with R225/227 K (GST-GLI2 (201–280) MT; GST-MT) were bacterially purified and followed by in vitro methylation assay with immunoprecipitated HA-tagged PRMT7 or PRMT5 (Supplementary Fig. 3). Both PRMTs could methylate control histones. PRMT7, but not PRMT5 methylated GST-WT which was diminished by R225/227 K mutation. This result was confirmed with bacterially purified GST-tagged PRMT7 (Fig. 6g). These data indicated that PRMT7 interacts with the N-terminal region of GLI2 and methylates it on arginine 225 and 227.
Next, we have transfected p5 WT and KO MEFs with vectors for GFP, Flag-GLI2-WT or Flag-GLI2-R225/227 K (MT), followed by SA-β-gal staining (Fig. 6h, i) and coimmunostaining for p-γH2AX and Flag or GFP (Fig. 6j, k). GLI2-WT expression led to mildly but significantly decreased SA−β-gal or p-γH2AX-positive cells in WT MEFs, compared with control cells, while GLI2-MT expression exhibited scant effects on cellular senescence. Furthermore, the expression of GLI2-WT or GLI2-MT in KO MEFs had no significant effects on cellular senescence. These results support for the importance of PRMT7-mediated GLI2 methylation in suppression of cellular senescence.
PRMT7 suppresses SUFU interaction with GLI2 thereby promoting GLI2 activity
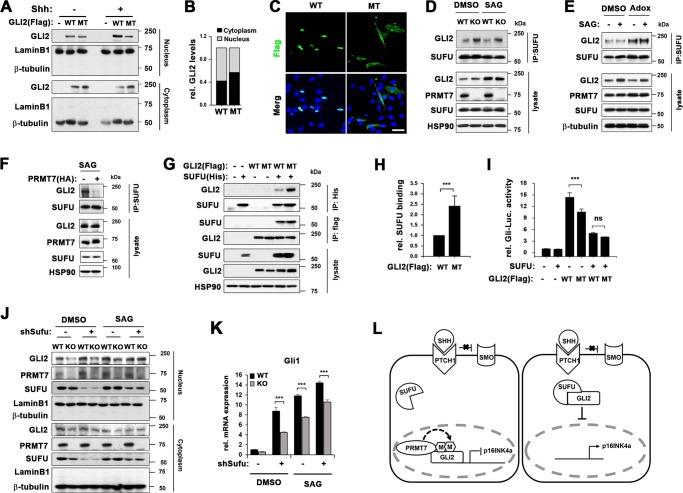
To determine the role of PRMT7-mediated GLI2 methylation in Shh signaling, 10T1/2 cells were transfected with Flag-GLI2 wildtype (WT) or Flag-GLI2 R225/227 K mutant (MT) for 36 h and starved for 12 h followed by Shh treatment for 12 h. Cellular fractionation revealed that more WT proteins were found in nuclear fraction by Shh treatment, while MT failed to accumulate in nucleus in response to Shh (Fig. 7a, b). The immunostaining showed that WT was mostly found in the nucleus, while MT was localized more in cytoplasm (Fig. 7c), suggesting that PRMT7 methylates GLI2 on R225/227 and this methylation is critical for GLI2 nuclear localization in response to Shh. SUFU, a major negative regulator of Shh pathway, interacts with and prevents GLI2 nuclear localization thereby inhibiting its transcriptional activity and in cells lacking SUFU, Shh signaling is constitutively activated [38, 39]. To address the relationship between SUFU and PRMT7, WT and KO MEFs were starved for 16 h and treated with DMSO or SAG for 8 h. As previously reported, GLI2 and SUFU coprecipitated in WT and KO MEFs and the SAG-treatment reduced GLI2 interaction with SUFU in WT MEFs. In contrast, more GLI2 proteins were precipitated with SUFU in both unstimulated and stimulated KO MEFs, compared with WT (Fig. 7d). 10T1/2 cells were starved and pretreated with Adox for 2 h, prior to the SAG treatment for 8 h. The PRMT inhibition dramatically increased the GLI2 levels bound to SUFU without any specific change in total SUFU or GLI2 levels (Fig. 7e). 10T1/2 cells were transfected with control or PRMT7 vector for 36 h and serum-starved for 16 h, followed by SAG treatment for 8 h (Fig. 7f). PRMT7-overexpressing 10T1/2 cells exhibited substantially decreased binding of GLI2 to SUFU in response to SAG. Conversely, PRMT7-depleted 10T1/2 cells had more GLI2 bound to SUFU (Supplementary Fig. 4), which is consistent with the result obtained with KO MEFs. These data suggest that PRMT7 negatively regulates GLI2 binding to SUFU in the Shh pathway.
Fig. 7.
PRMT7 negatively regulates GLI2 binding to SUFU. a Cellular fractionation of wildtype GLI2 (WT) or GLI2 R225/227 K mutant (MT), which were treated with Shh for 24 h after 16 h starvation. b Quantification of cytoplasmic or nucleic WT or MT shown in panel a. c Confocal microscopy for Flag-GLI2 in 10T1/2 cells transfected with WT or MT. Scale bar, 50 μm. d Coimmunoprecipitation with anti-SUFU antibody in WT and KO MEFs, which were treated with DMSO or SAG for 8 h, followed by immunoblotting with anti-GLI2 antibody. e Coimmunoprecipitation with anti-SUFU antibody in 10T1/2 cells, which were pretreated with DMSO or Adox prior to SAG treatment for 8 h, followed by immunoblotting with anti-GLI2 antibody. f Coimmunoprecpitation with anti-SUFU antibody in control-expressing or PRMT7-expressing 10T1/2 cells, which were treated with SAG for 8 h, followed by immunoblotting with anti-GLI2 antibody. g Immunoprecipitation analysis with anti-His antibody or anti-Flag antibody in 293 T cells, which were transfected with Flag-tagged WT or MT, and/or His-tagged SUFU, followed by immunoblotting with anti-Flag antibody or anti-His antibody, respectively. h Quantification for the relative SUFU binding of WT or MT shown in panel h. The results are shown as mean ± SD of triplicate experiments. ***P < 0.001. i Gli-reporter assay in 10T1/2 cells transfected with SUFU, and WT or MT as indicated. Data represent means of triplicates ± SD. n = 3, **P < 0.01, ***P < 0.001. j Cellular fractionation of WT or KO MEFs infected with lentivirus expressing control or SUFU shRNA and 36 h later, treated with DMSO or SAG for 8 h. k qRT-PCR for Gli1 expression in the replica sets shown in panel k. Data represent mean ± SD. n = 3, ANOVA test for comparison, ***P < 0.001. l A working hypothesis of the mechanism by which PRMT7 regulates GLI2 activity. PRMT7 methylates GLI2 at R225/227 and interferes with GLI2 binding to SUFU thereby activating GLI2 mediated inhibition of p16 and cellular senescence
Next, 293 T cells were cotransfected with His-tagged SUFU plus GLI2-WT or GLI2-MT, followed by immunoprecipitation. GLI2-MT proteins interacted more efficiently with SUFU, compared with GLI2-WT (Fig. 7g, h). To determine the functional significance, Gli-reporter assay was performed in 10T1/2 cells. WT enhanced reporter activity almost 15-fold, while GLI2-MT attenuated it (Fig. 7i). SUFU coexpression greatly reduced reporter activities in both GLI2-WT and GLI2-MT-expressing cells. WT and KO MEFs were infected with control or SUFU shRNA-expressing lentivirus serum starved and treated with DMSO or SAG for 8 h, followed by cellular fractionation. SUFU depletion resulted in increased nuclear GLI2 levels in both WT and KO MEFs and the SAG treatment restored nuclear GLI2 levels in KO cells similar to the level of wildtype cells (Fig. 7j). Furthermore, Gli1 mRNA levels were greatly elevated by SUFU depletion in both WT and KO MEFs and SAG treatment further elevated it (Fig. 7k). Reintroduction of SUFU in SUFU-depleted cells partially repressed Gli1 mRNA expression, supporting for the specificity of shSUFU (Supplementary Fig. 5). These data suggest that PRMT7 facilitates GLI2 nuclear translocation through interfering with SUFU interaction. Based on the data, we propose a working hypothesis that PRMT7 methylates GLI2 on R225/227, thereby suppressing its SUFU binding and increasing Shh signaling, ultimately leading to prevention of cellular senescence (Fig. 7l).
Discussion
A critical regulatory mechanism of Shh signaling is the removal of a conserved negative regulator SUFU from GLI proteins. GLI proteins are bound to SUFU with the SYGH core motif in the N-terminal region which is conserved in all GLI proteins [40]. The signaling activation triggers the removal of SUFU from GLI proteins leading to its nuclear translocation to augment target gene transcription [40, 41]. In this study, we demonstrate a positive role of PRMT7 in Shh signaling through methylation of GLI2 on R225/227. GLI2 methylation facilitates its dissociation from SUFU and nuclear localization where it can activate target gene expression. The cilia formation is normal in PRMT7-depleted or deficient fibroblasts, thus the defective Shh signaling activity observed in PRMT7 deficient MEFs is not likely due to the cilia function. We provide several lines of evidence suggesting that PRMT7 promotes Shh signaling activity. (1) PRMT7 depletion causes decreased expression of target genes in response to Shh or SAG. (2) Gli-reporter activity is upregulated by PRMT7 or downregulated by the treatment with a methylation inhibitor. (3) PRMT7 expression and levels are enhanced by Shh signaling, similarly to GLI2. (4) PRMT7 and Gli2 form complexes which is further enhanced by SAG treatment. (5) PRMT7 deficient cells show impaired nuclear localization of GLI2. How Shh signaling regulates PRMT7 levels currently unclear. Since PRMT7 proteins seem to form complexes with SUFU/GLI2 it is conceivable that SUFU might be involved in PRMT7 stability control.
Our data demonstrate that GLI2 is methylated by PRMT7 at R225 and R227 which appears to be critical for its SUFU binding and nuclear localization. These arginine residues are localized at a newly identified NLS thus it is possible that GLI2 R225/227 K mutant could interfere with its function as a NLS. Since the general NLSs contain lysine or arginine residues, arginines were switched to lysines in GLI2 mutant thus it is less likely the defective nuclear localization is due to the arginine to lysine switch. In addition, GLI2 R225/227 K mutant interacted better with SUFU, relative to the wildtype GLI2. This is consistent with the currently accepted models that SUFU interaction with GLI proteins interferes with nuclear translocation of GLI2 [40, 41]. An additional mechanism might be a sequestration of SUFU by PRMT7 so that GLI2 can be freed and translocate into nucleus. In consistent with this hypothesis, SUFU depletion restored the nuclear GLI2 levels in PRMT7 deficient MEF cells. In addition, Gli1 expression was significantly enhanced by SUFU depletion in PRMT7 deficient MEF cells. Our data are in alignment with previously published results showing that SUFU removal activates GLI proteins [40, 41].
The detailed regulatory mechanisms for 3 GLI proteins differ and various non-canonical pathways also involved in regulation of GLI activity. A previous study has shown that GLI1 can be methylated at R597 by PRMT1 to promote its transcriptional activity by enhancing GLI1 binding to its target genes which plays a critical role in pancreatic adenocarcinoma [28]. However, this arginine residue is not conserved in GLI2 and GLI3 proteins. Thus, it is currently unclear whether PRMT1 can modulate GLI2 and GLI3 in a similar fashion. Conversely, R225/227 of GLI2 are not conserved in GLI1 protein, while GLI3 protein contains equivalent arginine residues, suggesting that PRMT7 might also regulate GLI3 activation. The arginine methylation of GLI2 by PRMT7 might crosstalk with other post-translational modification such as its phosphorylation regulating its transcriptional activity. In the Esophageal adenocarcinoma, human GLI1 is phosphorylated at the serine residue 84 by S6 kinase, which enhances GLI1 function through inhibition of SUFU binding [42]. The sequence analysis revealed that the serine residue 84 of human GLI1 is analogous with the serine residue 230 of the mouse GLI2 which is located in the close proximity of R225/R227 suggesting a potential crosstalk between phosphorylation by S6K1 and arginine methylation of GLI2 by PRMT7 to augment the GLI2 transcriptional activation. However, the existence and function of this crosstalk of phosphorylation and methylation needs to be addressed in future. Our current study is consistent with previous reports proposing an important role of Shh signaling in maintaining cellular replicative state [43, 44]. In conclusion, our data suggest that PRMT7 can suppress cellular senescence through regulation of GLI2 activity.
Materials and methods
Cell culture, transfection, and luciferase assay
Isolation of MEFs was carried out as previously described [45]. MEFs and 10T1/2 cells were cultured in DMEM medium containing 10% fetal bovine serum (FBS, Gibco). For serum starvation condition, the cells were switched to DMEM medium containing 0.5% FBS for 12–16 h and treated with the indicated compounds. Transient transfection of 10T1/2 cells were performed by using lipofectamine 2000 (Invitrogen) as described previously [46]. Lentivirus-shRNA were infected into 10T1/2 or MEF cells with polybrene (Sigma). PRMT7 and SUFU knockdown shRNA sequences are listed in Table S1. Luciferase assays were conducted as previously described [47, 48]. All experiments were performed as triplicates and repeated at least three times. To generate the point mutants for the arginine to lysine replacement at R225 and R227 (R225K, R227K) of mouse GLI2, the site directed mutagenesis were performed by using Quick-Change II XL Site-Direct Mutagenesis Kit (Stratagene). Primers used in this study are listed in Supplementary table S1.
Immunocytochemistry, SA-β-galactosidase staining
Immunocytochemistry was performed as previously described [49, 50]. Briefly, the vehicle DMSO or SAG-treated MEFs or 10T1/2 cells were fixed, permeabilized and processed for incubation with antibodies listed in Table S2. SA-β-galactosidase staining were performed by using Senescence β-Galactosidase Staining Kit from Cell Signaling Technology. Briefly, MEFs at the cell passage number 3, 5, and 7 were fixed, followed by incubating with staining solution pH 6.0, containing X-gal, at 37 ℃ overnight. Images were captured under LSM710 confocal microscope system (Carl Zeiss) or Nikon ECLIPS TE-2000U and NIS-Elements F software (Nikon). The colocalization probability was quantified with Pearson’s coefficient using ZEN software.
RNA analysis
Quantitative RT-PCR analysis was analyzed as previously describe [51, 52]. Briefly, total RNAs from 10T1/2 and MEF cells were isolated with Easy-BLUE (iNtRON). cDNA samples were generated from 500 ng of total RNA with the PrimeScript RT kit (TaKaRa). Quantitative RT-PCR was performed using SYBR premix Ex tag (TaKaRa) following Thermal Cycler Dice Rel Time system analysis. All gene expressions were normalized with GAPDH as a loading control. The primer sequences are shown in Supplementary table S1.
Immunoblotting, immunoprecipitation, and cellular fractionation
Immunoblot and coimmunoprecipitation analysis were conducted as previously described [46, 50], Briefly, cells were lysed in extraction buffer (50 mM Tris-HCl, pH7.4, 150 mM NaCl, 1.2 mM MgCl2, 1 mM EGTA, 1% Triton X-100, 10 mM NaF, 1 mM Na3VO4) or RIPA (Thermo) buffer with complete protease inhibitor cocktail (PI, Roche). Protein samples were separated on polyacrylamide gel (SDS-PAGE) followed by immunoblotting. Cellular fractionation was carried out as previously described [53]. Briefly, 5 × 105−1 × 107 cells were suspended in 400 μl buffer A (10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.5 mM PMSF and PI) and incubated on ice for 10 min. Cells were lysed by adding 0.2% Nonidet P (NP)−40 and incubated on ice for 5 min. After centrifugation, the cytosolic fraction was collected and pellets were resuspended in 100 μl buffer B (20 mM HEPES, pH 7.9, 420 mM NaCl, 10 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF and PI). Immunoblots were quantified by using the ImageJ software. Antibodies used in this study are listed in Supplementary table S2.
In vitro methylation assay
In vitro methylation assay was performed as previously described [54]. Briefly, the expression vectors for HA-PRMT7, HA-PRMT7 or HA-PRMT5 were transfected to 293 T cells and immunoprecipitated, followed in vitro methylation assay with GST-GLI2 fragment as substrate. GST-tagged PRMT7 and GST-tagged GLI2 fragment (aa 201–280) were individually generated using with pVFT2S vectors, transformed to Escherichia coli BL21 (DE3) cells and induced by treating 0.5 mM IPTG (isopropyl-β-D-thiogalactopyranoside). GST-tagged recombinant proteins were then purified using glutathione sepharose 4B (GE Healthcare) and dialyzed against phosphate-buffered saline. In vitro methylation assay was performed with purified GST-PRMT7 proteins and histones, GST, GST-Gli2 201–280 WT or RK mutant proteins in the presence of 2 μl [3H]AdoMet in 40 μl reaction buffer (50 mM potassium HEPES, pH7.5, 10 nM NaCl, 1 mM DTT) at 23 ℃ for 20–22 h. Samples were subjected to SDS–PAGE and methylation levels were examined by radiography.
Statistical analysis
Statistical differences between two or multiple groups were analyzed by one-way ANOVA test. All results are representative of three independent experiments. Data are expressed as means ± standard deviation (SD) or ± standard error of mean (SEM), as indicated in the figure legends. Differences were considered statistically significant as *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Acknowledgements
This research was supported by the National Research Foundation of Korea Grant funded by the Korean Government (MSIP) (NRF-2016R1A2B2007179; NRF-2017M3A9D8048710; NRF-2016R1A5A2945889; NRF-2019R1A2C2006233) to J.S.K. and (NRF-2018R1D1A1B07041884) to T.A.V.
Author contributions:
T.A.V., H.J.J., B.K.K., Y.E.L. contributed to the experimental design, research and data analysis. T.A.V., H.C. and J.S.K. wrote the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by E. Baehrecke
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41418-019-0334-5) contains supplementary material, which is available to authorized users.
References
- 1.Di Leonardo A, Linke SP, Clarkin K, Wahl GM. DNA damage triggers a prolonged p53-dependent G1 arrest and long-term induction of Cip1 in normal human fibroblasts. Genes Dev. 1994;8:2540–51. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 2.Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM. Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a) Mol Cell. 2004;14:501–13. doi: 10.1016/s1097-2765(04)00256-4. [DOI] [PubMed] [Google Scholar]
- 3.Ogryzko VV, Hirai TH, Russanova VR, Barbie DA, Howard BH. Human fibroblast commitment to a senescence-like state in response to histone deacetylase inhibitors is cell cycle dependent. Mol Cell Biol. 1996;16:5210–8. doi: 10.1128/mcb.16.9.5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 5.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Childs BG, Durik M, Baker DJ, van Deursen JM. Cellular senescence in aging and age-related disease: from mechanisms to therapy. Nat Med. 2015;21:1424–35. doi: 10.1038/nm.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collado M, Blasco MA, Serrano M. Cellular senescence in cancer and aging. Cell. 2007;130:223–33. doi: 10.1016/j.cell.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Ohtani N, Hara E. Roles and mechanisms of cellular senescence in regulation of tissue homeostasis. Cancer Sci. 2013;104:525–30. doi: 10.1111/cas.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–70. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 10.Salama R, Sadaie M, Hoare M, Narita M. Cellular senescence and its effector programs. Genes Dev. 2014;28:99–114. doi: 10.1101/gad.235184.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shay JW, Pereira-Smith OM, Wright WE. A role for both RB and p53 in the regulation of human cellular senescence. Exp Cell Res. 1991;196:33–9. doi: 10.1016/0014-4827(91)90453-2. [DOI] [PubMed] [Google Scholar]
- 12.Bishop CL, Bergin AM, Fessart D, Borgdorff V, Hatzimasoura E, Garbe JC, et al. Primary cilium-dependent and -independent Hedgehog signaling inhibitsp16(INK4A) Mol Cell. 2010;40:533–47. doi: 10.1016/j.molcel.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 13.Lee RT, Zhao Z, Ingham PW. Hedgehog signalling. Development. 2016;143:367–72. doi: 10.1242/dev.120154. [DOI] [PubMed] [Google Scholar]
- 14.Breunig JJ, Sarkisian MR, Arellano JI, Morozov YM, Ayoub AE, Sojitra S, et al. Primary cilia regulate hippocampal neurogenesis by mediating sonic hedgehog signaling. Proc Natl Acad Sci USA. 2008;105:13127–32. doi: 10.1073/pnas.0804558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 17.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191:415–28. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhatia N, Thiyagarajan S, Elcheva I, Saleem M, Dlugosz A, Mukhtar H, et al. Gli2 is targeted for ubiquitination and degradation by beta-TrCP ubiquitin ligase. J Biol Chem. 2006;281:19320–6. doi: 10.1074/jbc.M513203200. [DOI] [PubMed] [Google Scholar]
- 19.Coni S, Antonucci L, D’Amico D, Di Magno L, Infante P, De Smaele E, et al. Gli2 acetylation at lysine 757 regulates hedgehog-dependent transcriptional output by preventing its promoter occupancy. PLoS One. 2013;8:e65718. doi: 10.1371/journal.pone.0065718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L, Pan Y, Wang B. Small ubiquitin-like Modifier (SUMO) modification inhibits GLI2 protein transcriptional activity in vitro and in vivo. J Biol Chem. 2012;287:20483–9. doi: 10.1074/jbc.M112.359299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pan Y, Bai CB, Joyner AL, Wang B. Sonic hedgehog signaling regulates Gli2 transcriptional activity by suppressing its processing and degradation. Mol Cell Biol. 2006;26:3365–77. doi: 10.1128/MCB.26.9.3365-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan Y, Wang C, Wang B. Phosphorylation of Gli2 by protein kinase A is required for Gli2 processing and degradation and the Sonic Hedgehog-regulated mouse development. Dev Biol. 2009;326:177–89. doi: 10.1016/j.ydbio.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–72. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 25.Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65:8–24. doi: 10.1016/j.molcel.2016.11.003. [DOI] [PubMed] [Google Scholar]
- 26.Jeong HJ, Lee HJ, Vuong TA, Choi KS, Choi D, Koo SH, et al. Prmt7 deficiency causes reduced skeletal muscle oxidative metabolism and age-related obesity. Diabetes. 2016;65:1868–82. doi: 10.2337/db15-1500. [DOI] [PubMed] [Google Scholar]
- 27.Pyun JH, Kim HJ, Jeong MH, Ahn BY, Vuong TA, Lee DI, et al. Cardiac specific PRMT1 ablation causes heart failure through CaMKII dysregulation. Nat Commun. 2018;9:5107. doi: 10.1038/s41467-018-07606-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Hsu JM, Kang Y, Wei Y, Lee PC, Chang SJ, et al. Oncogenic functions of Gli1 in pancreatic adenocarcinoma are supported by its PRMT1-mediated methylation. Cancer Res. 2016;76:7049–58. doi: 10.1158/0008-5472.CAN-16-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banasavadi-Siddegowda YK, Russell L, Frair E, Karkhanis VA, Relation T, Yoo JY, et al. PRMT5-PTEN molecular pathway regulates senescence and self-renewal of primary glioblastoma neurosphere cells. Oncogene. 2017;36:263–74. doi: 10.1038/onc.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blanc RS, Vogel G, Chen T, Crist C, Richard S. PRMT7 preserves satellite cell regenerative capacity. Cell Rep. 2016;14:1528–39. doi: 10.1016/j.celrep.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Gao Y, Zhao Y, Zhang J, Lu Y, Liu X, Geng P, et al. The dual function of PRMT1 in modulating epithelial-mesenchymal transition and cellular senescence in breast cancer cells through regulation of ZEB1. Sci Rep. 2016;6:19874. doi: 10.1038/srep19874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kleinschmidt MA, de Graaf P, van Teeffelen HA, Timmers HT. Cell cycle regulation by the PRMT6 arginine methyltransferase through repression of cyclin-dependent kinase inhibitors. PLoS One. 2012;7:e41446. doi: 10.1371/journal.pone.0041446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang L, Tian H, Chang N, Yi J, Xue L, Jiang B, et al. Loss of CARM1 is linked to reduced HuR function in replicative senescence. BMC Mol Biol. 2013;14:15. doi: 10.1186/1471-2199-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zurita-Lopez CI, Sandberg T, Kelly R, Clarke SG. Human protein arginine methyltransferase 7 (PRMT7) is a type III enzyme forming omega-NG-monomethylated arginine residues. J Biol Chem. 2012;287:7859–70. doi: 10.1074/jbc.M111.336271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dunaeva M, Michelson P, Kogerman P, Toftgard R. Characterization of the physical interaction of Gli proteins with SUFU proteins. J Biol Chem. 2003;278:5116–22. doi: 10.1074/jbc.M209492200. [DOI] [PubMed] [Google Scholar]
- 36.Feng Y, Maity R, Whitelegge JP, Hadjikyriacou A, Li Z, Zurita-Lopez C, et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine- and arginine-rich regions. J Biol Chem. 2013;288:37010–25. doi: 10.1074/jbc.M113.525345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Torrado B, Grana M, Badano JL, Irigoin F. Ciliary entry of the Hedgehog transcriptional activator Gli2 is mediated by then nuclear import machinery but differs from nuclear transport in being Imp-alpha/beta1-independent. PLoS One. 2016;11:e0162033. doi: 10.1371/journal.pone.0162033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia J, Kolterud A, Zeng H, Hoover A, Teglund S, Toftgard R, et al. Suppressor of fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev Biol. 2009;330:452–60. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ding Q, Fukami S, Meng X, Nishizaki Y, Zhang X, Sasaki H, et al. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–22. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 40.Han Y, Shi Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci USA. 2015;112:6383–8. doi: 10.1073/pnas.1421628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng H, Jia J, Liu A. Coordinated translocation of mammalian Gli proteins and suppressor of fused to the primary cilium. PLoS One. 2010;5:e15900. doi: 10.1371/journal.pone.0015900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer Cell. 2012;21:374–87. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subkhankulova T, Zhang X, Leung C, Marino S. Bmi1 directly represses p21Waf1/Cip1 in Shh-induced proliferation of cerebellar granule cell progenitors. Mol Cell Neurosci. 2010;45:151–62. doi: 10.1016/j.mcn.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 44.Tamayo-Orrego L, Wu CL, Bouchard N, Khedher A, Swikert SM, Remke M, et al. Evasion of cell senescence leads to medulloblastoma progression. Cell Rep. 2016;14:2925–37. doi: 10.1016/j.celrep.2016.02.061. [DOI] [PubMed] [Google Scholar]
- 45.Jeong MH, Ho SM, Vuong TA, Jo SB, Liu G, Aaronson SA, et al. Cdo suppresses canonical Wnt signalling via interaction with Lrp6 thereby promoting neuronal differentiation. Nat Commun. 2014;5:5455. doi: 10.1038/ncomms6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vuong TA, Lee SJ, Leem YE, Lee JR, Bae GU, Kang JS. SGTb regulates a surface localization of a guidance receptor BOC to promote neurite outgrowth. Cell Signal. 2019;55:100–8. doi: 10.1016/j.cellsig.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 47.Lee SJ, Leem YE, Go GY, Choi Y, Song YJ, Kim I, et al. Epicatechin elicits MyoD-dependent myoblast differentiation and myogenic conversion of fibroblasts. PLoS One. 2017;12:e0175271. doi: 10.1371/journal.pone.0175271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee SJ, Vuong TA, Go GY, Song YJ, Lee S, Lee SY, et al. An isoflavone compound daidzein elicits myoblast differentiation and myotube growth. J Funct Foods. 2017;38:438–46. [Google Scholar]
- 49.Tran P, Ho SM, Kim BG, Vuong TA, Leem YE, Bae GU, et al. TGF-beta-activated kinase 1 (TAK1) and apoptosis signal-regulating kinase 1 (ASK1) interact with the promyogenic receptor Cdo to promote myogenic differentiation via activation of p38MAPK pathway. J Biol Chem. 2012;287:11602–15. doi: 10.1074/jbc.M112.351601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee SY, Go GY, Vuong TA, Kim JW, Lee S, Jo A, et al. Black ginseng activates Akt signaling, thereby enhancing myoblast differentiation and myotube growth. J Ginseng Res. 2018;42:116–21. doi: 10.1016/j.jgr.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vuong TA, Leem YE, Kim BG, Cho H, Lee SJ, Bae GU, et al. A Sonic hedgehog coreceptor, BOC regulates neuronal differentiation and neurite outgrowth via interaction with ABL and JNK activation. Cell Signal. 2017;30:30–40. doi: 10.1016/j.cellsig.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Kwon YR, Jeong MH, Leem YE, Lee SJ, Kim HJ, Bae GU, et al. The Shh coreceptor Cdo is required for differentiation of midbrain dopaminergic neurons. Stem Cell Res. 2014;13:262–74. doi: 10.1016/j.scr.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 53.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim HJ, Jeong MH, Kim KR, Jung CY, Lee SY, Kim H, et al. Protein arginine methylation facilitates KCNQ channel-PIP2 interaction leading to seizure suppression. Elife. 2016;5:e17159. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.