Abstract
Many cell types are known to undergo a series of morphological changes during the progression of apoptosis, leading to their disassembly into smaller membrane-bound vesicles known as apoptotic bodies (ApoBDs). In particular, the formation of circular bulges called membrane blebs on the surface of apoptotic cells is a key morphological step required for a number of cell types to generate ApoBDs. Although apoptotic membrane blebbing is thought to be regulated by kinases including ROCK1, PAK2 and LIMK1, it is unclear whether these kinases exhibit overlapping roles in the disassembly of apoptotic cells. Utilising both pharmacological and CRISPR/Cas9 gene editing based approaches, we identified ROCK1 but not PAK2 or LIMK1 as a key non-redundant positive regulator of apoptotic membrane blebbing as well as ApoBD formation. Functionally, we have established an experimental system to either inhibit or enhance ApoBD formation and demonstrated the importance of apoptotic cell disassembly in the efficient uptake of apoptotic materials by various phagocytes. Unexpectedly, we also noted that ROCK1 could play a role in regulating the onset of secondary necrosis. Together, these data shed light on both the mechanism and function of cell disassembly during apoptosis.
Subject terms: Biochemistry, Cell biology
Introduction
Apoptosis is a form of programmed cell death required for development and maintenance of tissue homeostasis. Remarkably, even though billions of cells undergo apoptosis on a daily basis, apoptotic cells are rarely observed due to the cell clearance mechanisms in place to remove apoptotic cells efficiently by phagocytes [1–3]. Defects in apoptotic cell clearance has been linked to various autoimmune and chronic inflammatory diseases [1, 4] including systemic lupus erythematosus [5, 6], rheumatoid arthritis [7], colitis [8] and atherosclerosis [9]. The pathogenicity associated with uncleared apoptotic cells is mainly due to their progression into secondary necrosis when the plasma membrane becomes permeabilised, leading to the release of proinflammatory and immunogenic intracellular contents [1, 3, 5, 6].
During apoptosis, many cell types will undergo morphological changes, resulting in the fragmentation of the cell into smaller membrane-bound vesicles termed apoptotic bodies (ApoBDs) that are typically 1–5 μm in diameter [4, 10]. This process is known as apoptotic cell disassembly and is regulated by three distinct morphological steps, namely apoptotic membrane blebbing which involves the generation of circular bulges from the plasma membrane, followed by the formation of string-like membrane protrusions termed apoptopodia, and finally the appearance of discrete ApoBDs [4, 10–12]. The formation of ApoBDs has been proposed to regulate a number of processes including prompt clearance of apoptotic cells, whereby previous studies showed that the inhibition of ApoBD formation by pharmacological approaches can limit the efficiency of apoptotic cell engulfment [13, 14]. It should be noted that the contents of ApoBDs may also contribute to the development of autoimmune disorders [15, 16]. For example, ApoBDs generated from mouse NIH3T3 fibroblasts was found to contain high levels of histone H3 protein, a potent damage-associated molecular pattern (DAMP) [17] and an autoantigen [18].
The formation of apoptotic membrane blebs is the best characterised step of the apoptotic cell disassembly process and has been proposed to be regulated by a number of kinases [19–23]. Among these the role of Rho-associated kinase 1 (ROCK1) in promoting apoptotic membrane blebbing has been demonstrated initially based on the inhibition of ROCK1 kinase activity with Y-27632 treatment in apoptotic NIH3T3 fibroblasts [19] and human Jurkat T cells [20], and with the overexpression of the active ROCK1 kinase domain in healthy Jurkat T cells [20]. Furthermore, ROCK1 was found to be activated during apoptosis by caspase 3-mediated cleavage of the auto-inhibitory domain, leading to myosin light chain (MLC) phosphorylation and subsequent actinomyosin contraction necessary for membrane blebbing [19, 20]. Besides ROCK1, p21 activated kinase 2 (PAK2) has been shown to be activated by caspase-mediated cleavage in apoptotic cells [21, 22] and expression of dominant negative PAK mutants in Jurkat T cells caused abnormal apoptotic morphology with only rounded cells and defective ApoBD formation [22]. In contrast to this study [22], microinjection of active PAK2 fragment into serum-starved NIH3T3 fibroblasts failed to promote membrane blebbing [19], suggesting that active PAK2 fragment is not sufficient to drive membrane blebbing. Lastly, Lim domain kinase 1 (LIMK1) has also been reported to regulate apoptotic membrane blebbing following caspase 3-mediated cleavage [23]. Expression of the N-terminal active fragment of LIMK1 induced membrane blebbing in viable Jurkat T cells and human HeLa epithelial cells, and downregulation of LIMK1 using siRNA reduced apoptotic membrane blebbing in Jurkat T cells induced to undergo apoptosis by FasL [23]. Although these studies suggested that a number of kinases could be involved in regulating apoptotic membrane blebbing as well as subsequent ApoBD formation, it is not clear whether they could exhibit redundant role in these apoptotic processes. In this study, we employ both pharmacological and CRISPR/Cas9 gene editing based approaches to address the importance of ROCK1, PAK2 and LIMK1 in apoptotic membrane blebbing and the generation of ApoBDs. Targeting ROCK1 but not PAK2 or LIMK1 pharmacologically and genetically inhibited the formation of dynamic membrane blebbing and ApoBDs during the progression of apoptosis. Surprisingly, loss of LIMK1 promoted the formation of ApoBDs. Furthermore, we investigated the functional importance of apoptotic cell disassembly in the uptake of apoptotic materials by phagocytes. We found that cell lines that exhibit either a defect or enhanced ApoBD formation were less or more readily engulfed, respectively, by phagocytes. Unexpectedly, we also observed that ROCK1 is important in regulating the onset of secondary necrosis and leakage of intracellular contents.
Results
Pharmacological inhibition of ROCK1 but not PAK2 or LIMK1 inhibits apoptotic membrane blebbing
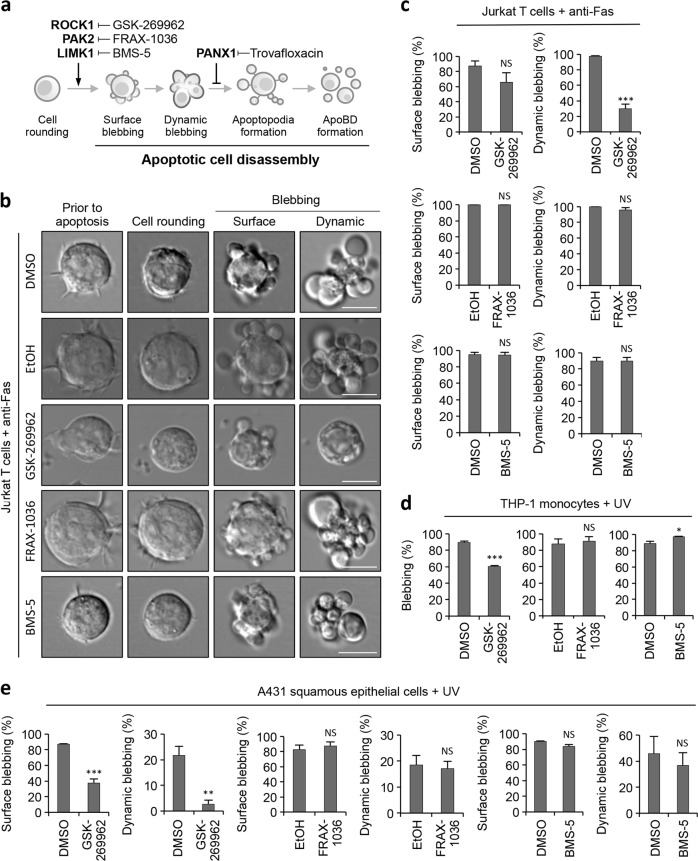
To examine the role of ROCK1, PAK2 and LIMK1 in apoptotic membrane blebbing initially, we used pharmacological compounds to target these kinases (Fig. 1a). Human Jurkat T cells were used as a model as we have previously characterised the apoptotic cell disassembly process of this cell line in detail [12, 24]. Jurkat T cells were treated with GSK-269962, a potent ROCK1 inhibitor [25], during anti-Fas induced apoptosis and the formation of apoptotic membrane blebs monitored by time-lapse differential interference contrast (DIC) microscopy (Fig. 1b). Although GSK-269962 treatment had minimal effect on surface blebbing, a significant reduction (~60%) in dynamic blebbing [12] was observed (Fig. 1b, c). In contrast, neither treatment of Jurkat T cells with PAK2 inhibitor FRAX-1036 [26] nor LIMK1 inhibitor BMS-5 [27] during the progression of apoptosis had an effect on surface and dynamic blebbing (Fig. 1b, c). It should be noted that both PAK2 and LIMK1 have been described to regulate cell cycle progression [28–31], and the activity of FRAX-1036 and BMS-5 on Jurkat T cells was first confirmed by cell cycle analysis (Supplementary Fig. 1a and b), as well as by immunoblot analysis of PAK2 autophosphorylation and phosphorylation of cofilin in FRAX-1036 and BMS-5 treated cells, respectively (Supplementary Fig. 1c and d). Next, we examined the role of ROCK1, PAK2 and LIMK1 in regulating apoptotic membrane blebbing of human THP-1 monocytic cells and A431 squamous epithelial cells. It should be noted that both THP-1 and A431 cells have been shown previously to undergo membrane blebbing during apoptotic cell disassembly [10, 32]. Similar to Jurkat T cells, GSK-269962 but not FRAX-1036 or BMS-5 was effective in inhibiting membrane blebbing of apoptotic THP-1 and A431 cells (Fig. 1d, e). Collectively, these data suggest that ROCK1 but not PAK2 or LIMK1 is the key regulator of apoptotic membrane blebbing in these suspension (Jurkat T and THP-1) and adherent (A431) cells.
Fig. 1.
Inhibition of ROCK1 but not PAK2 or LIMK1 pharmacologically impairs apoptotic membrane blebbing. a Schematic showing morphological steps and molecular regulators of apoptotic cell disassembly. Pharmacological compounds targeting regulators of apoptotic cell disassembly are also depicted. b Time-lapse DIC microscopy images monitoring surface and dynamic blebbing of human Jurkat T cells treated with anti-Fas to induce apoptosis and in the presence of vehicle controls, GSK-269962 (1 µM, ROCK1 inhibitor), FRAX-1036 (2.5 µM, PAK2 inhibitor) or BMS-5 (10 µM, LIMK1 inhibitor). c Quantitation of live microscopy data from (a) to determine the percentage of apoptotic Jurkat T cells that underwent surface and dynamic blebbing (n = 3). Human THP-1 monocytes (d) and human A431 squamous epithelial cells (e) were induced to undergo apoptosis by UV irradiation and in the presence of vehicle controls, GSK-269962 (1 µM), FRAX-1036 (2.5 µM) or BMS-5 (10 µM). Percentage of apoptotic cells that underwent blebbing was quantified based on live microscopy analysis (n = 3). Error bars represent s.e.m. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, NS = P > 0.05, unpaired Student’s two-tailed t-test
Apoptotic membrane blebbing and cell disassembly is regulated by ROCK1
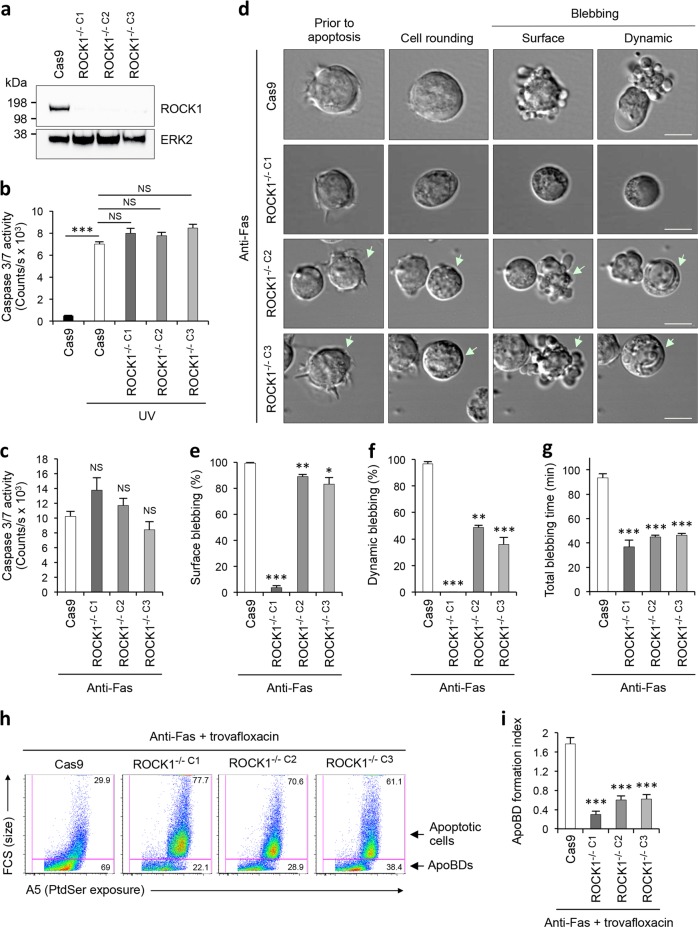
To gain further insights into the role of ROCK1 in mediating apoptotic membrane blebbing as well as ApoBD formation, we used an inducible CRISPR/Cas9 gene editing system [33] to disrupt ROCK1 expression in Jurkat T cells. ROCK1 gene was targeted at exon one and three clonal ROCK1−/− cell lines (ROCK1−/− C1, ROCK1−/− C2, ROCK1−/− C3) were confirmed by immunoblotting (Fig. 2a). All three ROCK1−/− cell lines showed similar sensitivity to both intrinsic and extrinsic apoptotic stimuli (UV [34] and anti-Fas [35] treatments, respectively), as measured by caspase 3/7 activity (Fig. 2b, c). Next, anti-Fas treated ROCK1−/− cell lines were examined by time-lapse DIC microscopy to monitor their ability to undergo apoptotic membrane blebbing in comparison to Cas9 expressing control (Cas9) cells. ROCK1−/− cell lines exhibited a reduction in both surface and dynamic blebbing during the progression of apoptosis (Fig. 2d–f), where ROCK1−/− C1 cells showed a greater reduction in apoptotic cells undergoing surface blebbing compared to ROCK1−/− C2 and ROCK1−/− C3 cells (Fig. 2e). We further confirmed the blebbing defect in a nonclonal ROCK1−/− Jurkat T cells (isgROCK1 cells) (Supplementary Fig. 2a–c). This defect in apoptotic membrane blebbing was also accompanied by an overall shortened blebbing time in all ROCK1−/− cell lines (Fig. 2g). Thus, these results suggest that ROCK1 is necessary for apoptotic membrane blebbing.
Fig. 2.
Loss of ROCK1 expression impairs apoptotic membrane blebbing and ApoBD formation. a Loss of ROCK1 protein expression with CRISPR/Cas9-mediated ROCK1 gene disruption in three Jurkat T cell clonal populations (ROCK1−/− C1, ROCK1−/− C2 and ROCK1−/− C3) was validated by immunoblot analysis. Caspase 3/7 activity in ROCK1−/− cells induced to undergo apoptosis by UV irradiation (b) or anti-Fas treatment (c) for 4 h (n = 3). d Time-lapse microscopy images monitoring the morphologies of ROCK1−/− cells undergoing anti-Fas induced apoptosis. Arrow, cell of interest. Quantitation of apoptotic ROCK1−/− cells that underwent surface blebbing (e) and dynamic blebbing (f), and total blebbing time (g) (n = 3). h Representative flow cytometry plots of apoptotic cells and ApoBDs generated by ROCK1−/− cells treated with anti-Fas and trovafloxacin (40 µM, PANX1 inhibitor, to promote apoptopodia-dependent ApoBD formation). i Formation of ApoBDs from apoptotic ROCK1−/− cells treated with anti-Fas and trovafloxacin, as measured by flow cytometry (n = 3). For quantitative analysis of blebbing time, all cells showing surface or dynamic blebbing was included. The duration between the first detection of blebbing to termination of blebbing was measured. ApoBD formation index determined by the number of ApoBDs divided by the number of A5+ apoptotic cells. a–i Cas9, Jurkat T cells expressing Cas9 only was used as a control. Error bars represent s.e.m. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, NS = P > 0.05, unpaired Student’s two-tailed t-test
Since membrane blebbing is a key step of the apoptotic cell disassembly process [11, 24], we next examined whether loss of ROCK1 could impair the formation of ApoBDs. Utilising a previously described model system [10, 11], cells were induced to undergo apoptosis in the presence of the pannexin 1 (PANX1) inhibitor trovafloxacin to promote apoptopodia-dependant ApoBD formation. Under these conditions, we observed a significant reduction in the formation of ApoBDs by all three ROCK1−/− cell lines (Fig. 2h, i) and in isgROCK1 cells (Supplementary Fig. 2d), indicating that ROCK1-mediated apoptotic membrane blebbing is necessary for apoptopodia-dependent apoptotic cell disassembly.
Apoptotic membrane blebbing is not regulated by PAK2 or LIMK1
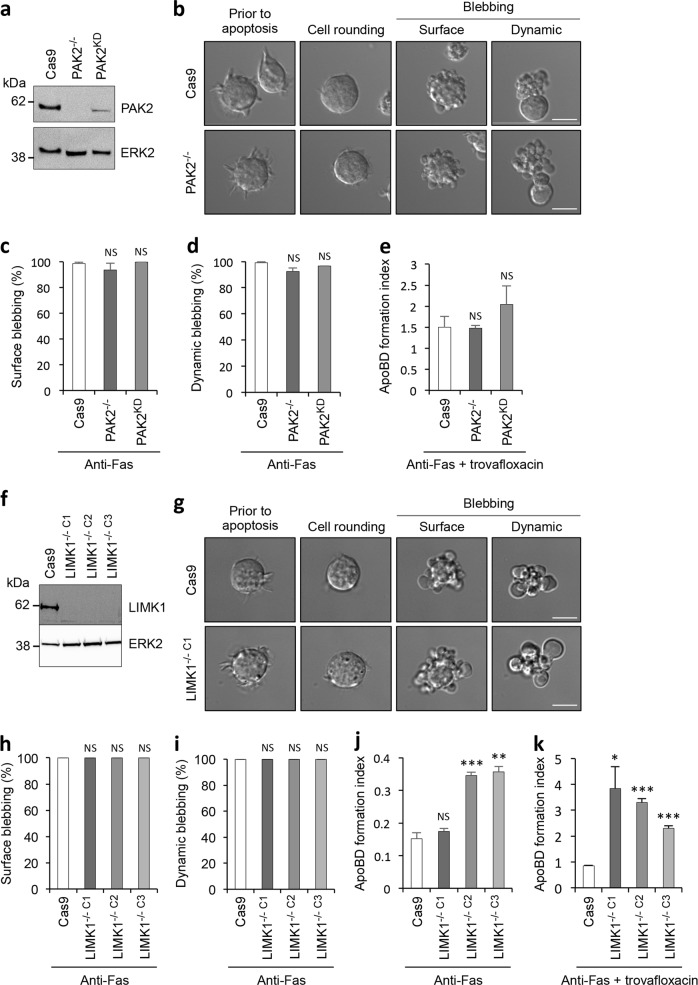
To examine the specific contribution of PAK2 in regulating apoptotic morphology, we generated Jurkat T cell lines that lack PAK2 expression (PAK2−/−) and reduced PAK2 expression (PAK2KD) (Fig. 3a). Apoptotic PAK2−/− and PAK2KD cells showed a comparable level of surface and dynamic blebbing as apoptotic Cas9 control cells (Fig. 3b–d), indicating that PAK2 is not necessary for apoptotic membrane blebbing in Jurkat T cells. Furthermore, loss and reduced PAK2 expression did not affect the ability of Jurkat T cells to generate ApoBD via an apoptopodia-dependent mechanism (Fig. 3e). It should be noted that apoptotic THP-1 monocytes lacking PAK2 expression also exhibited no defects in ApoBD formation (Supplementary Fig. 3a and b), indicating PAK2 is not required for the disassembly of apoptotic cells.
Fig. 3.
Apoptotic membrane blebbing and ApoBD formation is not reduced with loss of PAK2 or LIMK1. a Loss and reduction of PAK2 protein expression with CRISPR/Cas9-mediated PAK2 gene disruption in Jurkat T cells (PAK2−/− and PAK2KD, respectively) was analysed by immunoblot. b Time-lapse microscopy images monitoring apoptotic morphology of Cas9 and PAK2−/− cells treated with anti-Fas to induce apoptosis. Quantitation of apoptotic Cas9, PAK2−/− and PAK2KD cells that underwent surface blebbing (c), dynamic blebbing (d), and apoptotic cell disassembly in the presence of trovafloxacin (e) (n = 3). f Loss of LIMK1 protein expression with CRISPR/Cas9-mediated LIMK1 gene disruption in three Jurkat T cell clonal populations (LIMK1−/− C1, LIMK1−/− C2 and LIMK1−/− C3) was validated by immunoblot analysis. g Time-lapse microscopy images monitoring apoptotic morphology of Cas9 and LIMK1−/− cells treated with anti-Fas to induce apoptosis. Quantitation of apoptotic Cas9 and LIMK1−/− cells that underwent surface blebbing (h), dynamic blebbing (i), and apoptotic cell disassembly in the absence of trovafloxacin (j) or the presence of trovafloxacin (k) (n = 3). Error bars represent s.e.m. Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001, NS = P > 0.05, unpaired Student’s two-tailed t-test
Next, the importance of LIMK1 in regulating the apoptotic cell disassembly process was also examined. Jurkat T cells that lack LIMK1 expression (LIMK1−/− C1, LIMK1−/− C2, LIMK1−/− C3) were generated and confirmed by immunoblotting (Fig. 3f). Consistent with pharmacological inhibition of LIMK1 activity with BMS-5 (Fig. 1b, c), LIMK1−/− cell lines had no defects in surface or dynamic blebbing during the progression of apoptosis (Fig. 3g–i), indicating that LIMK1 is not necessary for apoptotic membrane blebbing in Jurkat T cells. Surprisingly, apoptotic LIMK1−/− cell lines showed an enhancement in ApoBD formation as compared to Cas9 control cells in the absence or presence of PANX1 inhibition (Fig. 3j, k). Taken together, although LIMK1 does not play a key role in apoptotic membrane blebbing, LIMK1 could negatively regulate the formation of ApoBDs in Jurkat T cells.
Apoptotic cell disassembly aids efficient engulfment of apoptotic materials
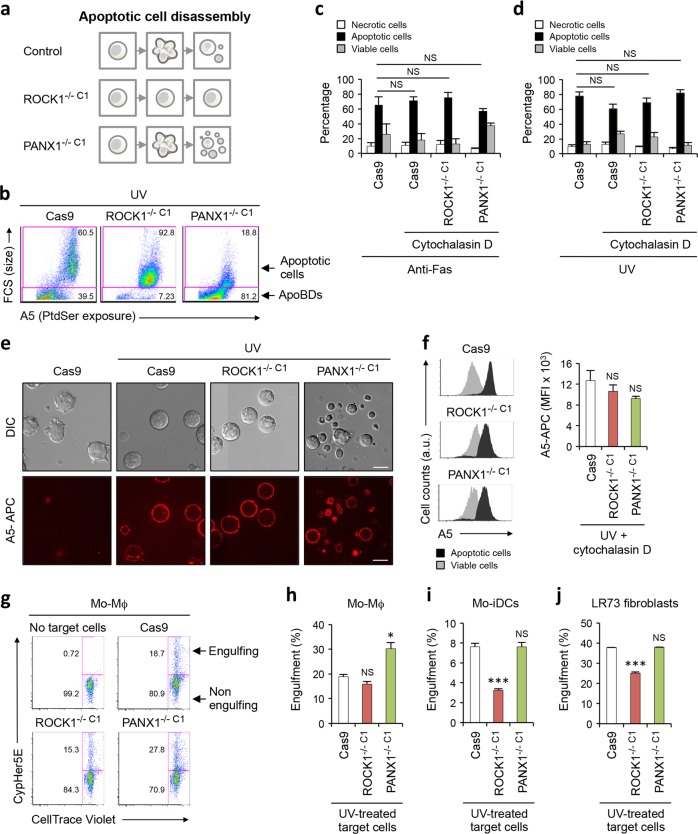
The fragmentation of apoptotic cells into ApoBDs can be observed both in vitro and in vivo [10, 11, 36]. Yet, the functional significance of cell disassembly during apoptosis is not well understood, in particular whether changes in ApoBD formation could alter the ability of phagocytes to engulf apoptotic materials. To this end, we have generated Jurkat T cell lines that exhibit a deficiency or enhanced cell disassembly during apoptosis by targeting the key positive and negative regulators, ROCK1 (Figs. 1 and 2) and PANX1 [11] respectively (Fig. 4a, b, Supplementary Fig. 4a–c). It is important to note that Cas9 control, ROCK1−/− C1 and PANX1−/− C1 cell lines all showed similar sensitivity to both intrinsic and extrinsic apoptotic stimuli (Fig. 4c, d), and expose comparable levels of the key ‘eat-me’ signal phosphatidylserine (PtdSer, important for phagocyte recognition and uptake) on the surface of apoptotic cells and ApoBDs (Fig. 4e, f). When engulfment assays were performed using human monocytic-derived macrophages (Mo-Mϕ) as the phagocytes, and UV-treated Cas9, ROCK1−/− C1 or PANX1−/− C1 cells as target cells, Mo-Mϕ showed a comparable efficiency in engulfing apoptotic ROCK1−/− C1 and Cas9 samples, whereas enhanced ability to engulf apoptotic PANX1−/− C1 sample (Fig. 4g, h). Similar engulfment assays were also performed using other types of phagocytes including human monocytic-derived immature dendritic cells (Mo-iDCs) and hamster LR73 ovary fibroblasts (Fig. 4i, j). Both Mo-iDCs and LR73 cells showed a reduced ability to engulf apoptotic ROCK1−/− C1 sample, while these phagocytes engulfed apoptotic PANX1−/− C1 and Cas9 samples to a similar level (Fig. 4i, j). Additionally, LR73 cells also showed a reduced ability to engulf apoptotic isgROCK1 cells (Supplementary Fig. 5a). In contrast to ROCK1 deficient cells, apoptotic PAK2−/− and LIMK1−/− C1 & C3 Jurkat T cells, which exhibit no impairment in ApoBD formation (Fig. 3e and j), apoptosis induction (Supplementary Fig. 5b and e) and PtdSer exposure (Supplementary Fig. 5c and f), were readily engulfed by LR73 cells (Supplementary Fig. 5d and g). Collectively, these data indicate that apoptotic cell disassembly could facilitate efficient engulfment of apoptotic materials.
Fig. 4.
Role of apoptotic cell disassembly in efferocytosis. a Schematic showing the disassembly of apoptotic Cas9, ROCK1−/− C1 and PANX1−/− C1 Jurkat T cells. b Cas9, ROCK1−/− C1 and PANX1−/− C1 cells were induced to undergo apoptosis by UV irradiation and the formation of apoptotic cells and ApoBDs measured by flow cytometry. The levels of viable, apoptotic and necrotic cells in Cas9, ROCK1−/− C1 and PANX1−/− C1 cells treated with anti-Fas treatment (c) or UV irradiation (d) for 4 h to induce apoptosis was determined by flow cytometry (n = 3). Actin polymerisation inhibitor cytochalasin D (10 µM) was used to stop the disassembly of apoptotic cells in all samples to ensure fair comparison between cell lines. e Live DIC and confocal images monitoring the exposure of PtdSer by UV-irradiated Cas9, ROCK1−/− C1 and PANX1−/− C1 cells via A5-APC staining. f Flow cytometry analysis of the exposure of PtdSer by apoptotic Cas9, ROCK1−/− C1 and PANX1−/− C1 cells based on A5-APC staining. g Flow cytometry analysis of the uptake of UV-treated CypHer5E-stained Cas9, ROCK1−/− C1 and PANX1−/− C1 cells by CellTrace Violet-stained human Mo-Mϕ. Quantitation of the ability of CellTrace Violet-stained human Mo-Mϕ (h), human Mo-iDCs (i) and hamster LR73 fibroblasts (j) to engulf UV-treated CypHer5E-stained Cas9, ROCK1−/− C1 and PANX1−/− C1 cells, as measured by flow cytometry (n = 3). Error bars represent s.e.m. Data are representative of at least three independent experiments. *P < 0.05, ***P < 0.001, NS = P > 0.05, unpaired Student’s two-tailed t-test
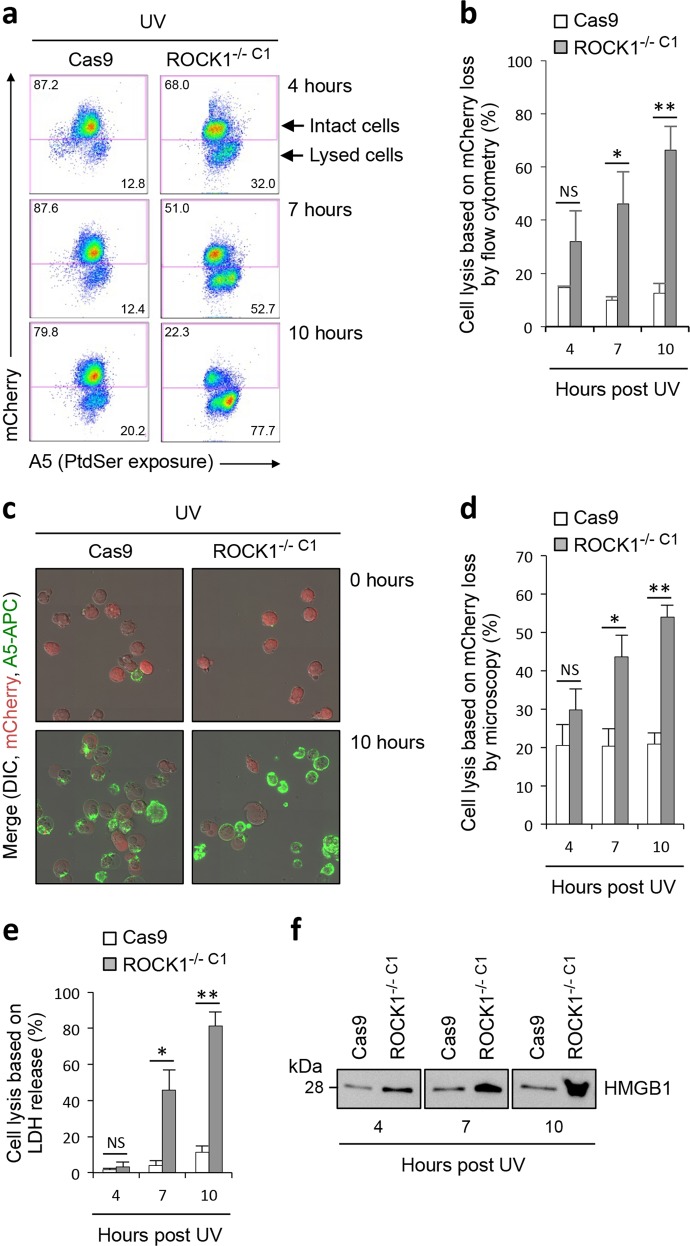
ROCK1 is involved in regulating the onset of secondary necrosis
Cytoskeletal rearrangement such as microtubule bundling and actin polymerisation during apoptosis have been proposed to maintain the integrity of the plasma membrane [37, 38]. Given the well-established role of ROCK1 in regulating actin reorganisation [39], we asked if the loss of ROCK1 expression could affect the membrane integrity of apoptotic cells. To address this, we monitored the loss of mCherry (~29 kDa, expressed in both Cas9 and ROCK1−/− C1 cells) signal from apoptotic cells as a measure of plasma membrane permeabilisation by flow cytometry (Fig. 5a). Interestingly, at 7 and 10 but not 4 h post induction of apoptosis, a significant higher amount of ROCK1−/− C1 cells were permeabilised as compared to Cas9 control cells (Fig. 5a, b). Using a similar experimental approach, membrane permeabilisation was also confirmed by live time-lapse confocal microscopy analysis, with more ROCK1−/− C1 cells permeabilised at 7 and 10 h post apoptosis induction (Fig. 5c, d). We next monitored the release of the cytosolic protein lactic dehydrogenase (LDH, ~140 kDa) into the culture supernatant to assess cell permeabilisation. Consistent with flow cytometry and live microscopy based analysis, higher levels of LDH was also detected in the ROCK1−/− C1 cell culture supernatant at 7 and 10 but not 4 h post apoptosis induction (Fig. 5e), further demonstrating the loss of ROCK1 promotes earlier onset of secondary necrosis. Lastly, we assessed the release of high-mobility group box 1 (HMGB1), a nuclear protein associated with DAMP signalling [40, 41], from apoptotic cells into the culture supernatants. Higher levels of HMGB1 was observed in the culture supernatant of ROCK1−/− C1 cells as compared to Cas9 cells at 4, 7 and 10 h post induction of apoptosis (Fig. 5f, ponceau staining in Supplementary Fig. 6a). Recent studies have described the ability of gasdermin E (GSDME) following caspase 3-mediated cleavage, to oligomerize and form pores targeting the plasma membrane to mediate secondary necrosis [42, 43]. Thus, we first monitored the processing of GSDME in ROCK1−/− C1 over 4 h post apoptosis induction. ROCK1−/− C1 cells showed comparable level of caspase-cleaved GSDME to Cas9 control cells (Supplementary Fig. 6b), indicating it is unlikely that the early onset of secondary necrosis in ROCK1−/− C1 cells was due to accelerated processing and activation of GSDME. Furthermore, given the previously described role of cytoskeletal rearrangement in maintaining membrane integrity of apoptotic cells [37, 38], we also examined the microtubule network during the progression of apoptosis by time-lapse microscopy. Notably, disassembly of microtubule network was observed for both ROCK1−/− C1 and Cas9 control cells undergoing apoptosis (Supplementary Fig. 6c). Collectively, these data indicate that the loss of ROCK1 could lead to earlier apoptotic membrane permeabilisation and leakage of intracellular contents, possibly via a mechanism independent to the function of GSDME and apoptotic microtubule network.
Fig. 5.
Loss of ROCK1 expression promotes early onset of secondary necrosis. a Flow cytometry analysis of intact (mCherryHigh) and membrane permeabilised (mCherryLow) Cas9 and ROCK1−/− C1 cells 4, 7 and 10 h post UV irradiation. Levels of cell lysis over time were quantified based on the loss of mCherry in cells by flow cytometry (b) and live microscopy (c and d), as well as the release of LDH (e) and HMGB1 (f) into the culture supernatant. b, d and e Error bars represent s.e.m. (n = 3). Data are representative of at least three independent experiments. *P < 0.05, **P < 0.01, NS = P > 0.05, unpaired Student’s two-tailed t-test
Discussion
While morphologic changes like membrane blebbing are hallmarks of apoptotic cell death [44, 45] and play a key role in facilitating the disassembly of dying cells [10, 11], the molecular regulators and the function of this process are not fully defined [4]. Understanding the mechanistic basis of apoptotic cell disassembly is important especially as the formation of ApoBDs have been implicated in various disease settings including inflammation [8], infection [46], atherosclerosis [47] and autoimmunity [5, 6, 48]. In this study, using both pharmacologic and genetic approaches, we have shown that ROCK1 but not PAK2 or LIMK1 is a key positive regulator of apoptotic membrane blebbing and ApoBD formation in selected number of cell types. Moreover, we have established experimental systems to show that apoptotic cell disassembly could aid efficient engulfment of apoptotic materials.
The formation of apoptotic membrane blebs have been proposed to be regulated by caspase-activated kinases, namely ROCK1, PAK2 and LIMK1, through inhibitor, overexpression or siRNA-mediated knockdown studies [19, 20, 22, 23]. In this study using time-lapse imaging, newer generations of kinase inhibitors and gene editing approaches, ROCK1 but not PAK2 or LIMK1 was found to be a key positive regulator of apoptotic membrane blebbing in cell types including T cells, monocytes and epithelial cells. ApoBD formation by Jurkat T cells was also largely dependent on ROCK1 but not PAK2. In contrast to Rudel et al. (1997) and in line with [19], PAK2 is redundant for apoptotic membrane blebbing and cell disassembly. It is interesting to note that through bioinformatics analysis using the caspase-site prediction program Cascleave [49], we did not identify any putative caspase cleavage site in PAK2 that was previously shown to regulate its activation and function in apoptosis [21, 22] (Supplementary Fig. 3c), further questioning the role of PAK2 in apoptotic membrane blebbing and ApoBD formation. Although the data presented in this study also suggest LIMK1 is redundant for apoptotic membrane blebbing, the loss of LIMK1 expression in Jurkat T cells unexpectedly resulted in an enhancement in ApoBD formation. Previous studies have shown that LIMK1 is rendered active during apoptosis [23] and can function to block actin filament depolymerisation and severing by deactivating cofilin [50]. Thus, LIMK1 may negatively regulate ApoBD formation through maintaining actin filaments, possibly at later stages of apoptosis when apoptopodia are disrupted to release distinct ApoBDs. It should be noted that MLC kinase (MLCK) has also been implicated in regulating apoptotic membrane blebbing through the phosphorylation of MLC [51] however, since ROCK1 was shown previously to be necessary and sufficient for MLC phosphorylation during apoptosis [20], the role of MLCK was not investigated further in this study.
Apoptotic cells can aid their clearance via the release of ‘find-me’ signals to facilitate the recruitment of phagocytes towards sites of cell death, and exposure of ‘eat-me’ signals on the cell surface to mediate recognition and uptake by phagocytes [1, 3]. Whether the formation of apoptotic membrane blebs can also aid cell clearance has been investigated in a number of previous studies, whereby inhibition of ROCK1 activity in apoptotic Jurkat T cells with Y-27632 did not affect their engulfment by mouse peritoneal macrophages [52], while rat PC12 and monkey COS-7 cells treated with Y-27632 during the progression of apoptosis impaired the ability of PMA-differentiated THP-1 macrophages to engulf these apoptotic targets [14, 52, 53]. Moreover, it should be noted that ROCK1 is a negative regulator of apoptotic cell uptake for bone marrow-derived macrophages and LR73 fibroblasts, and inhibition of ROCK1 activity in these phagocytic cells has been shown previously to enhance the engulfment of apoptotic thymocytes [54, 55]. Thus, the contrasting observations in previous studies targeting the role of apoptotic membrane blebbing in cell clearance may be attributed to factors including the type of apoptotic targets and phagocytes used, as well as the use of pharmacological inhibitors in co-culture experiments. In this study, we utilised genetically modified cell lines that exhibit an enhancement or impairment in ApoBD formation and demonstrated that apoptotic cell disassembly is another process during the progression of apoptosis that could aid the clearance of apoptotic cells. It is interesting to note that although an enhancement in apoptotic cell disassembly promoted Mo-Mϕ-mediated engulfment of apoptotic materials, loss of ROCK1-mediated apoptotic disassembly did not impair apoptotic cell uptake by Mo-Mϕ, supporting the concept that macrophages are able to engulf their targets in their entirety [56]. In contrast to Mo-Mϕ, Mo-iDC and LR73 fibroblasts were less efficient in engulfing apoptotic cells that did not undergo apoptotic cell disassembly, highlighting that different types of phagocytes may have differential preference for engulfing whole or fragmented apoptotic cells. Notably, previous studies have shown that professional phagocytes such as the brain microglia are able to rapidly engulf cerebellar granule neurons in their entirety while non-professional phagocytes such as primary lens epithelial cells and baby hamster kidney cells are less efficient at engulfing whole apoptotic neurons [56].
Maintaining plasma membrane integrity in apoptotic cells prior to removal by phagocytes is crucial as membrane permeabilisation can lead to the exposure of intracellular contents, which could drive pathogenicity in various autoimmune and inflammatory conditions [1, 5, 6, 9]. Apoptotic microtubule networks and more recently DFNA5/GSDME have been proposed to regulate the permeability of the apoptotic cell plasma membrane [38, 42, 43]. Interestingly, loss of ROCK1 in apoptotic Jurkat T cells also resulted in an early onset of plasma membrane permeabilisation at 7 and 10 h post UV-induced apoptosis, suggesting that ROCK1 is also involved in regulating the integrity of the plasma membrane. It should be noted that apoptotic membrane blebbing would have ceased at approximately 4 h post apoptosis induction in Jurkat T cells [11], indicating that the activity of ROCK1 during early stages of apoptosis may have a lasting impact on the apoptotic cell post membrane blebbing. Furthermore, it is interesting to noted that previous studies using NIH3T3 fibroblasts showed that inhibition of ROCK1 by Y-27632 reduces plasma membrane permeabilisation at 4 but not 24 h post TNF-induced apoptosis [17]. Whether the discrepancies between these studies are due to differences in the type of cells being used, timing in monitoring membrane permeabilisation or apoptotic stimuli requires further investigation. Nevertheless, it is clear that ROCK1 may also play a role in controlling membrane integrity of apoptotic cells and the exact mechanism underpinning this process remains to be determined.
Overall, this study demonstrates that ROCK1 is a non-redundant regulator of both apoptotic membrane blebbing and ApoBD formation, and further establishes the importance of ApoBD formation in cell clearance.
Materials and methods
Reagents
Reagents were obtained as follows: trovafloxacin (Sigma-Aldrich, MO), cytochalasin D (Sigma-Aldrich), anti-Fas (clone CH11, Millipore, MA), A5-PE and A5-APC (BD Biosciences, CA), TO-PRO-3 (Life Technologies, NY), carboxyfluorescein diacetate succinimidyl ester (CFSE, Life Technologies), CypHer5E (GE Lifesciences, IL), CellTrace Violet (Life Technologies), RNase I/TI (Thermo-Fisher Scientific, MA), propidium iodide (Sigma-Aldrich), GSK-269962 (Tocris bioscience, UK), FRAX-1036 (Selleckchem, TX), and BMS-5 (Enzo, NY).
Cell culture
Human Jurkat T cells and THP-1 monocytes were obtained from ATCC and cultured in complete RPMI media. Complete RPMI constituted of RPMI 1640 medium (Life Technologies), 10% (vol/vol) foetal bovine serum (FBS, Bovogen, New Zealand), penicillin (50 U/ml) and streptomycin (50 mg/ml) (Life Technologies), and 0.2% (vol/vol) MycoZap (Lonza, Switzerland). Hamster LR73 ovary fibroblasts (gift from Prof. Kodi Ravichandran) were cultured in alpha-MEM (Life Technologies) supplemented with 10% FBS, penicillin (50 U/ml) and streptomycin (50 mg/ml). Human A431 epidermoid cells (Lonza) were cultured in MEM (Lonza) supplemented with 10% FBS, penicillin (50 U/ml) and streptomycin (50 mg/ml), and non-essential amino acid and L-glutamine (Thermofisher Scientific, MA).
Isolation of human peripheral blood mononuclear cells (PBMCs) and generation of human monocytes-derived macrophages (Mo-Mϕ) and immature dendritic cells (Mo-iDCs)
Buffy coat was obtained with donor consent from the Australian Red Cross Blood Service (Melbourne, Australia) and appropriate ethics approval was granted by the La Trobe University Human Ethics Committee. PBMCs were isolated from buffy coat using Ficoll density gradient separation as previously reported [57]. To isolate monocytes, PBMCs were plated and incubated at 37 °C and 5% CO2 for 2 h, and washed three times with 1 × PBS (Life Technologies) to remove non-adherent cells. To generate Mo-Mϕs, monocytes were treated with MCSF (1000 U/ml, Miltenyi Biotec, Germany) in 5% RPMI for 7 days. Monocytes were differentiated into Mo-iDCs with GMCSF (1000 U/ml, Miltenyi Biotec) and IL-4 (400 U/ml, Miltenyi Biotec) in complete RPMI for 7 days.
CRIPSR/Cas9 gene editing
A doxycycline-inducible sgRNA vector CRISPR/Cas9 system was used to generate gene disruptions in Jurkat T and THP-1 cells as previously described [33]. Jurkat T and THP-1 cells stably expressing Cas9 endonuclease and mCherry were generated by lentiviral transduction using pFUCas9mCherry plasmid. Targeting gRNAs were designed using mit.edu.au/CRIPSR software [58]. The following oligonucleotide pairs corresponding to guide RNA sequences ROCK1 exon 1: 5′TCCCCAGCAGCAACATGTCGACTG-3′, 5′AAACCAGTCGACATGTTGCTGCTG-3′, PAK2 exon 1: 5′TCCCAGCAGCACCATCTTTAGCAC-3′, 5′AAACGTGCTAAAGATGGTGCTGCT-3′, LIMK1 exon 3: 5′TCCCAAGGACTACTGGGCCCGCTA-3′, 5′AAACTAGCGGGCCCAGTAGTCCTT-3′, LIMK1 exon 4: 5′TCCC TGGGACCTTTATCGGTGACGGGG-3′, 5′AAACCCCCGTCACCGATAAAGGTCCCA-3′, PANX1 exon 1: 5′TCCCGATGGTCACGTGCATTGCGG-3′, 5′AAACCCGCAATGCACGTGACCATC-3′ were annealed and ligated in pFgh1tUTG plasmid digested with BsmB1 restriction enzyme. For LIMK1, two gRNAs targeting exon 2 and exon 4 were used to disrupt all isoforms. Cas9 mCherry cells were spin infected with lentiviral supernatant containing appropriate gRNA pFgh1tUTG constructs. To generate clonal knockout cell lines, cells were treated with doxycycline (1 µg/ml) in complete RPMI for 72 h to induce gRNA synthesis. Cells expressing both mCherry (indicative of Cas9 expression) and GFP (indicative of cells containing the gRNA insert) were single-cell sorted into 96 well plates containing complete RPMI using FACS Aria II (BD Biosciences). To generate nonclonal ROCK1 knockout cells (isgROCK1 cells), Jurkat T cells expressing both mCherry (indicative of Cas9 expression) and GFP (indicative of cells containing the gRNA insert) were bulk sorted and treated with doxycycline (1 µg/ml) in complete RPMI for 7 days. Notably, the expression of ROCK2 was unaffected in isgROCK1 cells treated with doxycycline (1 µg/ml) and in clonal ROCK1−/− cells lines as examined by immunoblot (described below) analysis (Supplementary Fig. 2e, f).
Immunoblotting
Cells were lysed at 4 °C in lysis buffer (20 mM HEPES pH 7.4, 1%, IGEPAL® CA-630, 10% glycerol, 1% Triton X-100, 150 mM NaCl, 50 mM NaF, protease inhibitor cocktail tablet (Roche, CH), phosphatase inhibitor cocktail tablet (Roche)), analysed by SDS-PAGE and immunoblotted using the following antibodies: anti-ROCK1 (1:1000, clone H-85, Santa Cruz, TX), anti-PAK2 (1:500, clone 3B5 Abcam, Cambridge, UK), anti-LIMK1 (1:1000, Cell Signalling Technologies, MA), anti-HMGB1 (1:1000, Cell Signalling Technologies), anti-PANX1 (1:1000, clone D9M1C, Cell Signalling Technologies), anti-phospho-Cofilin (Ser3) (1:1000, clone 77G2, Cell Signalling Technologies), anti-Cofilin (1:1000, clone D3F9, Cell Signalling Technologies), anti-phospho-PAK2 (1:1000, Cell Signalling Technologies), anti-ROCK2 (1:1000, clone H-85, Santa Cruz, TX) or anti-ERK2 (1:1000, clone D-2, Santa Cruz) in 3% BSA in PBST (0.1% Tween). Blots were incubated in secondary HRP-conjugated sheep anti-mouse (1:5000, Millennium Science, AU) or donkey anti-rabbit (1:5000, Millennium Science) antibodies in 5% milk in PBST (0.1% Tween). HRP signal was developed using ECL (GE Lifesciences) and monitored using the Syngene G:Box gel documentation and analysis system (Syngene, MD). To re-probe membranes, primary and secondary antibodies were removed using stripping buffer (10% SDS, 0.5 M Tris HCl pH 6.8 0.2 v/v ß-mercaptoethanol) at 50 °C for 30 min. Membranes were re-blocked with 5% milk in PBST (0.1% Tween) prior to immunoblotting.
Cell cycle arrest assay
Cells were treated with FRAX-1036 (2.5 µM), or BMS-5 (10 µM) in complete RPMI for 24 h. Cell cycle analysis was conducted using propidium iodide DNA staining as previously described [59]. Briefly, treated cells were fixed in 70% ethanol. Fixed cells were treated with RNase I/TI (62.5 μg/μl) in 1 × PBS for 20 min. Cell samples were further stained with propidium iodide (1 µg/ml) and analysed using BD FACSCanto II Flow cytometer (BD Biosciences). Data was analysed using FlowJo software (version 9.8.5, FlowJo, OR).
Quantitative PCR
Cells were pelleted and lysed in TRIzol (Invitrogen, Thermo Fisher Scientific) and processed according to manufactures instructions. cDNA was generated from reconstituted RNA (1 ug total) using the iScript Select cDNA Synthesis Kit (Bio-Rad). For PAK1, PAK4 and LIMK2 amplification, reverse transcription PCR (RT-PCR) was performed using the Power SYBR Green System (Applied Biosystems). The following primer sequences were used to amplify cDNA, PAK1, forward: 5′ATACTGGATGGCACCAGAGG′ PAK1, reverse: 5′TTGGTGGCAATGAGGTACAA3′, PAK4, forward: 5′CTACGGGCCAGAGGTAGACA3′, PAK4, reverse: 5′TTGTGCAGGTTCTTCAGTCG3′, LIMK1, forward: 5′ATGCACATCAGTCCCAACAA3′, LIMK1, reverse: 5′TCAGTGTCCCCTCCAGATTC3′ ubiquitin C, forward: TGAAGAGAATCCACAAGGAATTGA, and ubiquitin C, reverse: CAACAGGACCTGCTGAACACTG. The house keeping gene ubiquitin C was used as a control for gene expression. Data was analysed using the ΔΔCT method. Quantitative PCR analysis indicated no significant difference in expression of PAK1 and PAK4 in the PAK2−/− cells as compared to Cas9 control (Supplementary Fig. 7a and b). LIMK2 expression was reduced in LIMK1−/− C1–C3 (Supplementary Fig. 7c), which may be due to clonal differences. Notably, only LIMK1 has been previously shown to be caspase-cleaved and activated during apoptosis [23], thus changes in LIMK2 expression is unlikely to affect the apoptotic cell disassembly process.
Induction of apoptosis
Cells in 1% BSA/RPMI or complete RPMI were induced to undergo apoptosis with anti-Fas treatment (0.5 µg/ml, Merck, Germany) and incubated at 37 °C in humidified atmosphere with 5% CO2 for 4 h or as indicated. In certain experiments, apoptosis was induced by UV irradiation (150 mJ/cm2) using Stratagene UV Stratalinker 1800 (Agilent Technologies, CA).
Time-lapse confocal microscopy
Mammalian cells were seeded in a 4 or 8 chambered NuncTM Lab-TekTM II chambered coverglass (Nunc, Denmark) prior to induction of apoptosis and drug treatment. For THP-1 cells, coverslips were pre-treated with Poly-l-lysine (Sigma Aldrich). A431 cells were seeded 16 h prior to induction of apoptosis and drug treatment. To detect PtdSer exposure, cells were stained with A5-APC in 1% BSA/RPMI containing 1 × A5 binding buffer (BD Biosciences). Time-lapse DIC and confocal imaging was performed at 37 °C in humidified atmosphere with 5% CO2 using Spinning Disc Confocal microscope (Zeiss, Germany) with ×63 oil immersion objective. Images were obtained at intervals of 2 min or as indicated. Image processing and analysis was carried out using Zeiss imaging software (Zeiss).
Quantification of cell viability, ApoBD formation and cell lysis by flow cytometry
Samples were stained with A5-PE and TO-PRO-3, or A5-APC only in 1 × A5 binding buffer (BD Biosciences) at room temperature (RT) in dark for 5 min [24]. Samples were analysed using BD FACSCanto II Flow cytometer (BD Biosciences). Data was analysed using FlowJo software (version 9.8.5, FlowJo, OR) as previously described [24].
Engulfment assay
Phagocytes including Mo-Mϕ, Mo-iDC and LR73 cells were stained with CellTrace Violet (Life Technologies) and staining solution was replaced with complete RPMI. Target cells (Cas9 only, ROCK1−/− and PANX1−/− Jurkat T cells) were stained with CypHer5E (GE Lifesciences) [60] and induced to undergo apoptosis by UV irradiation (150 mJ/cm2) in complete RPMI. Phagocytes and target cells were co-cultured in 1:1 ratio for 5 h at 37 °C, 5% CO2 in humidified atmosphere. Mo-Mϕ and Mo-iDCs were harvested using cell dissociation reagent (Sigma Aldrich), and LR73 were detached with typsin-EDTA (Life Technologies). Cells were placed on ice till analysis by BD FACSCanto II Flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (version 9.8.5).
LDH release assay
LDH release assay was performed as previously described [61] using a LDH Cytotoxic Assay Kit II (Abcam). The release of cytosolic enzyme LDH was detected in culture supernatants to determine cell lysis. Briefly, Cas9 and ROCK1−/− Jurkat T cells were induced to undergo apoptosis by UV irradiation in a 96 well plate and incubated in complete RPMI at 37 °C, 5% CO2 in humidified atmosphere. Cell suspension was centrifuged at 300 × g for 10 min to remove cells and the resultant supernatant centrifuged at 3000 × g for 20 min to remove cell debris [62]. Culture supernatant was added to LDH reaction mix for 30 min at RT. The absorbance of product was measured at 450 nm using SpecraMax M5e Plate reader (Molecular Devices, CA) and data was analysed using SoftMaxPro 5.2 software (Molecular Devices).
Caspase-Glo 3/7 assay
Caspase activity in samples was determined using the Caspase-Glo 3/7 Assay (Promega, Madison, WI) according to manufacturer’s instruction. Briefly, cells were added to an opaque 96 well plate (30,000 cells in 50 μl) and induced to undergo apoptosis by anti-Fas treatment or UV irradiation, and incubated for 4 h at 37 °C, 5% CO2 in humidified atmosphere. Equal volume of Caspase-Glo3/7 reagent was added and incubated at RT in dark for 30 min. Caspase 3/7 activity, as measured by the luminescence, was determined using SpecraMax M5e Plate reader (Molecular Devices) and data was analysed using SoftMaxPro 5.2 software (Molecular Devices).
Protein precipitation
Protein precipitation was performed using trichloroacetic acid (TCA) [63]. Cas9 and ROCK1−/− Jurkat T cells (3 × 106 cells in 1 ml) were induced to undergo apoptosis by UV irradiation in RPMI supplemented with Insulin-Transferrin-Selenium (Gibco), and incubated at 37 °C, 5% CO2 in humidified atmosphere. Cell suspension was centrifuged at 300 × g for 10 min to remove cells and the resultant supernatant centrifuged at 3000 × g for 20 min to remove cell debris [62]. TCA (12% vol/vol) was added to the supernatant and incubated on ice for 30 min. Precipitated protein was pelleted at 18,000 × g at 4 °C for 30 min and washed with ice cold acetone. Protein precipitates were redissolved in NuPAGE LDS sample buffer (Life Technologies) prior to analysis by SDS-PAGE and immunoblotting.
Statistics
Data are presented as means + s.e.m. Statistical significance was determined by unpaired student’s two-tailed T test. P values < 0.05 were considered significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary information
Acknowledgements
We would like to thank the LIMS BioImaging Facility for assistance with microscopy. We would also like to thank Alyce Mayfosh for assistance with quantitative PCR. This work was supported by grants from the National Health & Medical Research Council of Australia (GNT1125033 and GNT1140187) and Australian Research Council (DP170103790) to I.K.H.P.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by S. Kumar
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41418-019-0342-5) contains supplementary material, which is available to authorized users.
References
- 1.Poon IKH, Lucas CD, Rossi AG, Ravichandran KS. Apoptotic cell clearance: basic biology and therapeutic potential. Nat Rev Immunol. 2014;14:166–80. doi: 10.1038/nri3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hochreiter-Hufford A, Ravichandran KS. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harb Perspect Biol. 2013;5:a008748. doi: 10.1101/cshperspect.a008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagata S. Apoptosis and clearance of apoptotic cells. Annu Rev Immunol. 2018;36:489–517. doi: 10.1146/annurev-immunol-042617-053010. [DOI] [PubMed] [Google Scholar]
- 4.Atkin-Smith GK, Poon IKH. Disassembly of the dying: mechanisms and functions. Trends Cell Biol. 2017;27:151–62. doi: 10.1016/j.tcb.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 5.Shao WH, Cohen PL. Disturbances of apoptotic cell clearance in systemic lupus erythematosus. Arthritis Res Ther. 2011;13:202. doi: 10.1186/ar3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann I, Kolowos W, Voll RE, Manger B, Gaipl U, Neuhuber WL, et al. Impaired uptake of apoptotic cells into tingible body macrophages in germinal centers of patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:191–201. doi: 10.1002/1529-0131(200201)46:1<191::AID-ART10027>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 7.Gaipl US, Franz S, Voll RE, Sheriff A, Kalden JR, Herrmann M. Defects in the disposal of dying cells lead to autoimmunity. Curr Rheuma Rep. 2004;6:401–7. doi: 10.1007/s11926-004-0016-1. [DOI] [PubMed] [Google Scholar]
- 8.Lee CS, Penberthy KK, Wheeler KM, Juncadella IJ, Vandenabeele P, Lysiak JJ, et al. Boosting apoptotic cell clearance by colonic epithelial cells attenuates inflammation in vivo. Immunity. 2016;44:807–20. doi: 10.1016/j.immuni.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ait-Oufella H, Pouresmail V, Simon T, Blanc-Brude O, Kinugawa K, Merval R, et al. Defective mer receptor tyrosine kinase signaling in bone marrow cells promotes apoptotic cell accumulation and accelerates atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:1429–31. doi: 10.1161/ATVBAHA.108.169078. [DOI] [PubMed] [Google Scholar]
- 10.Atkin-Smith GK, Tixeira R, Paone S, Mathivanan S, Collins C, Liem M, et al. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat Commun. 2015;6:7439. doi: 10.1038/ncomms8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poon IKH, Chiu YH, Armstrong AJ, Kinchen JM, Juncadella IJ, Bayliss DA, et al. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature. 2014;507:329–34. doi: 10.1038/nature13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tixeira R, Caruso S, Paone S, Baxter AA, Atkin-Smith GK, Hulett MD, et al. Defining the morphologic features and products of cell disassembly during apoptosis. Apoptosis. 2017;22:475–7. doi: 10.1007/s10495-017-1345-7. [DOI] [PubMed] [Google Scholar]
- 13.Witasp E, Uthaisang W, Elenström-Magnusson C, Hanayama R, Tanaka M, Nagata S, et al. Bridge over troubled water: milk fat globule epidermal growth factor 8 promotes human monocyte-derived macrophage clearance of non-blebbing phosphatidylserine-positive target cells. Cell Death Differ. 2007;14:1063. doi: 10.1038/sj.cdd.4402096. [DOI] [PubMed] [Google Scholar]
- 14.Orlando KA, Stone NL, Pittman RN. Rho kinase regulates fragmentation and phagocytosis of apoptotic cells. Exp Cell Res. 2006;312:5–15. doi: 10.1016/j.yexcr.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Fransen JH, Hilbrands LB, Ruben J, Stoffels M, Adema GJ, van der Vlag J, et al. Mouse dendritic cells matured by ingestion of apoptotic blebs induce T cells to produce interleukin-17. Arthritis Rheum. 2009;60:2304–13. doi: 10.1002/art.24719. [DOI] [PubMed] [Google Scholar]
- 16.Fransen JH, Hilbrands LB, Jacobs CW, Adema GJ, Berden JH, Van Der Vlag J. Both early and late apoptotic blebs are taken up by DC and induce IL-6 production. Autoimmunity. 2009;42:325–7. doi: 10.1080/08916930902828049. [DOI] [PubMed] [Google Scholar]
- 17.Wickman GR, Julian L, Mardilovich K, Schumacher S, Munro J, Rath N, et al. Blebs produced by actin-myosin contraction during apoptosis release damage-associated molecular pattern proteins before secondary necrosis occurs. Cell Death Differ. 2013;20:1293–305. doi: 10.1038/cdd.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fournel S, Neichel S, Dali H, Farci S, Maillère B, Briand JP, et al. CD4+T cells from (New Zealand Black x New Zealand White)F1 lupus mice and normal mice immunized against apoptotic nucleosomes recognize similar Th cell epitopes in the C terminus of histone H3. J Immunol. 2003;171:636–44. doi: 10.4049/jimmunol.171.2.636. [DOI] [PubMed] [Google Scholar]
- 19.Coleman ML, Sahai EA, Yeo M, Bosch M, Dewar A, Olson MF. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat Cell Biol. 2001;3:339–45. doi: 10.1038/35070009. [DOI] [PubMed] [Google Scholar]
- 20.Sebbagh M, Renvoizé C, Hamelin J, Riché N, Bertoglio J, Bréard J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat Cell Biol. 2001;3:346. doi: 10.1038/35070019. [DOI] [PubMed] [Google Scholar]
- 21.Lee N, MacDonald H, Reinhard C, Halenbeck R, Roulston A, Shi T, et al. Activation of hPAK65 by caspase cleavage induces some of the morphological and biochemical changes of apoptosis. Proc Natl Acad Sci USA. 1997;94:13642–7. doi: 10.1073/pnas.94.25.13642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudel T, Bokoch GM. Membrane and morphological changes in apoptotic cells regulated by caspase-mediated activation of PAK2. Science. 1997;276:1571–4. doi: 10.1126/science.276.5318.1571. [DOI] [PubMed] [Google Scholar]
- 23.Tomiyoshi G, Horita Y, Nishita M, Ohashi K, Mizuno K. Caspase-mediated cleavage and activation of LIM-kinase 1 and its role in apoptotic membrane blebbing. Genes Cells. 2004;9:591–600. doi: 10.1111/j.1356-9597.2004.00745.x. [DOI] [PubMed] [Google Scholar]
- 24.Jiang L, Tixeira R, Caruso S, Atkin-Smith GK, Baxter AA, Paone S, et al. Monitoring the progression of cell death and the disassembly of dying cells by flow cytometry. Nat Protoc. 2016;11:655–63. doi: 10.1038/nprot.2016.028. [DOI] [PubMed] [Google Scholar]
- 25.Doe C, Bentley R, Behm DJ, Lafferty R, Stavenger R, Jung D, et al. Novel Rho kinase inhibitors with anti-inflammatory and vasodilatory activities. J Pharm Exp Ther. 2007;320:89–98. doi: 10.1124/jpet.106.110635. [DOI] [PubMed] [Google Scholar]
- 26.Koval AB, Wuest WM. An optimized synthesis of the potent and selective Pak1 inhibitor FRAX-1036. Tetrahedron Lett. 2016;57:449–51. [Google Scholar]
- 27.Ross-Macdonald P, de Silva H, Guo Q, Xiao H, Hung CY, Penhallow B, et al. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol Cancer Ther. 2008;7:3490–8. doi: 10.1158/1535-7163.MCT-08-0826. [DOI] [PubMed] [Google Scholar]
- 28.May M, Schelle I, Brakebusch C, Rottner K, Genth H. Rac1-dependent recruitment of PAK2 to G2 phase centrosomes and their roles in the regulation of mitotic entry. Cell Cycle. 2014;13:2211–21. doi: 10.4161/cc.29279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar R, Vadlamudi RK. Emerging functions of p21-activated kinases in human cancer cells. J Cell Physiol. 2002;193:133–44. doi: 10.1002/jcp.10167. [DOI] [PubMed] [Google Scholar]
- 30.Davila M, Jhala D, Ghosh D, Grizzle WE, Chakrabarti R. Expression of LIM kinase 1 is associated with reversible G1/S phase arrest, chromosomal instability and prostate cancer. Mol Cancer. 2007;6:40. doi: 10.1186/1476-4598-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sumi T, Hashigasako A, Matsumoto K, Nakamura T. Different activity regulation and subcellular localization of LIMK1 and LIMK2 during cell cycle transition. Exp Cell Res. 2006;312:1021–30. doi: 10.1016/j.yexcr.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 32.Moss DK, Betin VM, Malesinski SD, Lane JD. A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation. J Cell Sci. 2006;119:2362–74. doi: 10.1242/jcs.02959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kueh AJ, Herold MJ. Using CRISPR/Cas9 technology for manipulating cell death regulators. Methods Mol Biol. 2016;1419:253–64. doi: 10.1007/978-1-4939-3581-9_18. [DOI] [PubMed] [Google Scholar]
- 34.Heck DE, Vetrano AM, Mariano TM, Laskin JD. UVB light stimulates production of reactive oxygen species: unexpected role for catalase. J Biol Chem. 2003;278:22432–6. doi: 10.1074/jbc.C300048200. [DOI] [PubMed] [Google Scholar]
- 35.Ogasawara J, Watanabe-Fukunaga R, Adachi M, Matsuzawa A, Kasugai T, Kitamura Y, et al. Lethal effect of the anti-Fas antibody in mice. Nature. 1993;364:806–9. doi: 10.1038/364806a0. [DOI] [PubMed] [Google Scholar]
- 36.Mayer Christian T., Gazumyan Anna, Kara Ervin E., Gitlin Alexander D., Golijanin Jovana, Viant Charlotte, Pai Joy, Oliveira Thiago Y., Wang Qiao, Escolano Amelia, Medina-Ramirez Max, Sanders Rogier W., Nussenzweig Michel C. The microanatomic segregation of selection by apoptosis in the germinal center. Science. 2017;358(6360):eaao2602. doi: 10.1126/science.aao2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levee MG, Dabrowska MI, Lelli JL, Hinshaw DB. Actin polymerization and depolymerization during apoptosis in HL-60 cells. Am J Physiol. 1996;271(6 Pt 1):C1981–92. doi: 10.1152/ajpcell.1996.271.6.C1981. [DOI] [PubMed] [Google Scholar]
- 38.Sánchez-Alcázar JA, Rodríguez-Hernández Aacute, Cordero MD, Fernández-Ayala DJM, Brea-Calvo G, Garcia K, et al. The apoptotic microtubule network preserves plasma membrane integrity during the execution phase of apoptosis. Apoptosis. 2007;12:1195–208. doi: 10.1007/s10495-006-0044-6. [DOI] [PubMed] [Google Scholar]
- 39.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, et al. Rho‐associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J. 1996;15:2208–16. [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi ME, Manfredi AA. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol Rev. 2007;220:35–46. doi: 10.1111/j.1600-065X.2007.00574.x. [DOI] [PubMed] [Google Scholar]
- 41.Chen G, Ward MF, Sama AE, Wang H. Extracellular HMGB1 as a proinflammatory cytokine. J Inter Cytokine Res. 2004;24:329–33. doi: 10.1089/107999004323142187. [DOI] [PubMed] [Google Scholar]
- 42.Rogers C, Fernandes-Alnemri T, Mayes L, Alnemri D, Cingolani G, Alnemri ES. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat Commun. 2017;8:14128. doi: 10.1038/ncomms14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y, Gao W, Shi X, Ding J, Liu W, He H, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 44.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–57. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kroemer G, Galluzzi L, Vandenabeele P, Abrams J, Alnemri ES, Baehrecke EH, et al. Classification of cell death: recommendations of the Nomenclature Committee on Cell Death 2009. Cell Death Differ. 2009;16:3–11. doi: 10.1038/cdd.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arslan SY, Son KN, Lipton HL. During infection, Theiler’s virions are cleaved by caspases and disassembled into pentamers. J Virol. 2016;90:3573–83. doi: 10.1128/JVI.03035-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aprahamian T, Rifkin I, Bonegio R, Hugel B, Freyssinet JM, Sato K, et al. Impaired clearance of apoptotic cells promotes synergy between atherogenesis and autoimmune disease. J Exp Med. 2004;199:1121–31. doi: 10.1084/jem.20031557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korb LC, Ahearn JM. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. J Immunol. 1997;158:4525–8. [PubMed] [Google Scholar]
- 49.Song J, Tan H, Shen H, Mahmood K, Boyd SE, Webb GI, et al. Cascleave: towards more accurate prediction of caspase substrate cleavage sites. Bioinformatics. 2010;26:752–60. doi: 10.1093/bioinformatics/btq043. [DOI] [PubMed] [Google Scholar]
- 50.Arber S, Barbayannis FA, Hanser H, Schneider C, Stanyon CA, Bernard O, et al. Regulation of actin dynamics through phosphorylation of cofilin by LIM-kinase. Nature. 1998;393:805–9. doi: 10.1038/31729. [DOI] [PubMed] [Google Scholar]
- 51.Mills JC, Stone NL, Erhardt J, Pittman RN. Apoptotic membrane blebbing is regulated by myosin light chain phosphorylation. J Cell Biol. 1998;140:627–36. doi: 10.1083/jcb.140.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiratsuchi A, Mori T, Nakanishi Y. Independence of Plasma Membrane Blebbing from Other Biochemical and Biological Characteristics of Apoptotic Cells 1. J Biochem. 2002;132:381–6. doi: 10.1093/oxfordjournals.jbchem.a003233. [DOI] [PubMed] [Google Scholar]
- 53.Orlando KA, Pittman RN. Rho kinase regulates phagocytosis, surface expression of GlcNAc, and Golgi fragmentation of apoptotic PC12 cells. Exp Cell Res. 2006;312:3298–311. doi: 10.1016/j.yexcr.2006.06.033. [DOI] [PubMed] [Google Scholar]
- 54.Nakaya M, Tanaka M, Okabe Y, Hanayama R, Nagata S. Opposite effects of rho family GTPases on engulfment of apoptotic cells by macrophages. J Biol Chem. 2006;281:8836–42. doi: 10.1074/jbc.M510972200. [DOI] [PubMed] [Google Scholar]
- 55.Tosello-Trampont AC, Nakada-Tsukui K, Ravichandran KS. Engulfment of apoptotic cells is negatively regulated by Rho-mediated signaling. J Biol Chem. 2003;278:49911–9. doi: 10.1074/jbc.M306079200. [DOI] [PubMed] [Google Scholar]
- 56.Parnaik R, Raff MC, Scholes J. Differences between the clearance of apoptotic cells by professional and non-professional phagocytes. Curr Biol. 2000;10:857–60. doi: 10.1016/s0960-9822(00)00598-4. [DOI] [PubMed] [Google Scholar]
- 57.Phan TK, Lay FT, Hulett MD. Importance of phosphoinositide binding by human β-defensin 3 for Akt-dependent cytokine induction. Immunol Cell Biol. 2018;96:54–67. doi: 10.1111/imcb.1017. [DOI] [PubMed] [Google Scholar]
- 58.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diamond RA, Demaggio S. In Living Color: Protocols in Flow Cytometry and Cell Sorting. Berlin: Springer Berlin Heidelberg; 2000. p. 802.
- 60.Beletskii A, Cooper M, Sriraman P, Chiriac C, Zhao L, Abbot S, et al. High-throughput phagocytosis assay utilizing a pH-sensitive fluorescent dye. Biotechniques. 2005;39:894–7. doi: 10.2144/000112001. [DOI] [PubMed] [Google Scholar]
- 61.Poon IK, Baxter AA, Lay FT, Mills GD, Adda CG, Payne JA, et al. Phosphoinositide-mediated oligomerization of a defensin induces cell lysis. Elife. 2014;3:e01808. doi: 10.7554/eLife.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Atkin-Smith GK, Paone S, Zanker DJ, Duan M, Phan TK, Chen W, et al. Isolation of cell type-specific apoptotic bodies by fluorescence-activated cell sorting. Sci Rep. 2017;7:39846. doi: 10.1038/srep39846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosenberg IM. Protein analysis and purification: benchtop techniques. Basel: Birkhäuser; 2005. p. 520.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.