Abstract
Autophagy, a main intracellular catabolic process, is induced in response to a variety of cellular stresses to promptly degrade harmful agents and to coordinate the activity of prosurvival and prodeath processes in order to determine the fate of the injured cells. While the main components of the autophagy machinery are well characterized, the molecular mechanisms that confer selectivity to this process both in terms of stress detection and cargo engulfment have only been partly elucidated. Here, we discuss the emerging role played by the E3 ubiquitin ligases of the TRIM family in regulating autophagy in physiological and pathological conditions, such as inflammation, infection, tumorigenesis, and muscle atrophy. TRIM proteins employ different strategies to regulate the activity of the core autophagy machinery, acting either as scaffold proteins or via ubiquitin-mediated mechanisms. Moreover, they confer high selectivity to the autophagy-mediated degradation as described for the innate immune response, where TRIM proteins mediate both the engulfment of pathogens within autophagosomes and modulate the immune response by controlling the stability of signaling regulators. Importantly, the elucidation of the molecular mechanisms underlying the regulation of autophagy by TRIMs is providing important insights into how selective types of autophagy are altered under pathological conditions, as recently shown in cancer and muscular dystrophy.
Subject terms: Macroautophagy, Ubiquitin ligases
Facts
TRIMs participate to the autophagic response to stress stimuli by regulating both autophagy induction and cargos recognition.
TRIMs modulate autophagy levels in ubiquitin-dependent and independent manners by interacting with either the autophagy core machinery or upstream signaling pathways.
TRIMs act as selective substrate receptors for the intracellular degradation of pathogens, immune signaling proteins, protein aggregates, and muscle proteins.
Open questions
What are the molecular mechanisms that turn on and off the autophagic activity of TRIMs during stress responses?
Is TRIM-regulated selective autophagy based on TRIM-interacting motifs shared by different types of autophagic cargos?
How are the autophagic and proteasomal functions of TRIMs coordinated during stress responses?
Is the TRIM-regulated selective autophagy a therapeutic target for human diseases?
Introduction
Autophagy is the main intracellular catabolic process that is induced in response to a variety of physiological and pathological stresses [1]. Autophagy allows the prompt degradation of damaged cell structures and invading pathogens within lysosomes [2, 3]. Moreover, several pathways activated under stressful conditions are directly controlled by autophagy, which, by regulating signaling protein stability, is able to fine-tuning the intensity and duration of defence response [1].
In the last 20 years, the identification of the main components of the autophagy core machinery, named autophagy-related proteins (ATGs), has contributed to uncover many aspects of how autophagosomes are formed [1, 4]. More recently, considerable efforts have been devoted to elucidate how selectivity of the process is achieved both in terms of activation by different types of stress/damage agents and sequestration of target materials within autophagosomes. Many pieces of evidence point to the E3 ubiquitin ligases as key players that signal to the autophagy machinery under stress conditions [5]. Here, we review how autophagy is regulated by TRIpartite Motif (TRIM) proteins, a large family of E3 ubiquitin ligases that play pivotal role in conferring selectivity to this process.
The TRIM protein family
The TRIM proteins are a large family of E3 ubiquitin ligases (>80 members) characterized by the presence of three domains: RING finger, B-Box-type zinc finger (B1 box and B2 box), and Coiled-Coil region (Fig. 1) [6]. The RING finger contains the catalytic site, the B-boxes act as protein–protein interaction domains and together with the Coiled-Coil mediate protein homo- or hetero-oligomerization of TRIMs [7]. Depending on the interaction with the E2 conjugating enzymes, TRIMs are able to catalyze the formation of different ubiquitin chains that may have either degradative or regulative roles [8]. Specific TRIMs have also been shown to act as E3 ligase for ubiquitin-like proteins, such as SUMO [9]. Moreover, eight human TRIMs are RING-less (underlined in Fig. 1), suggesting that they may either indirectly contribute to ubiquitination as scaffold proteins in association with other TRIMs or play other functions not related to ubiquitination.
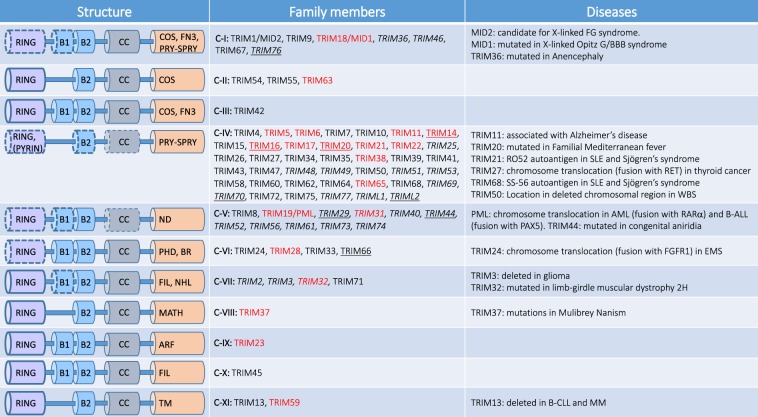
Fig. 1. Classification of TRIM proteins.
TRIM proteins are characterized by the presence of a tripartite domain consisting of a RING finger, two B-boxes (B1 and B2) and a coiled-coil domain. TRIM proteins are divided into 11 subclasses (from C-I to C-XI) on the basis on their C-terminal domain. C-terminal domains expressed by TRIM subgroups are: ARF ADP-ribosylation factor-like, BR bromodomain, COS C-terminal subgroup one signature, FIL filamin-type immunoglobulin, NHL NHL domain, FN3 fibronectin type 3, PRY-SPRY PRY-SPRY domain, MATH meprin and tumor-necrosis factor receptor-associated factor homology, PHD plant homeodomain, TM transmembrane. Some members of TRIM subclasses lack the RING domain (underlined), B-boxes or Coiled-Coil (CC) domains (italics), ND: TRIM lacking a unique C-terminal domain. As indicated in the main text, members of the C-VI constitute the TIF1 family of chromatin binding proteins. TRIM proteins that play a role in autophagy are indicated in red. FG: X‐linked multiple congenital anomalies syndrome. SLE systemic lupus erythematosus, WBS Williams–Beuren syndrome, AML acute myeloid leukemia, B-ALL B-cell precursor acute lymphoblastic leukemia, EMS 8p11 myeloproliferative syndrome, B-CLL B-cell chronic lymphocytic leukemia, MM multiple myeloma.
TRIM proteins are classified into 11 subfamilies, based on their C-terminal domains, which may mediate both substrates recognition and regulative protein–protein interactions [10, 11] (Fig. 1). The PRY-SPRY, a domain with an immunoglobulin-like fold, is present in approximately two-thirds of the TRIM proteins. The NHL (named after the identification in NCL-1, HT2A, and LIN-41 proteins) consists of five or six repeats that fold into a barrel-like β-propeller structure and mediate both protein and RNA interactions. The tandem plant homeodomain and bromodomain characterize the transcriptional intermediary factor 1 (TIF1) subgroup of TRIM family members involved in chromatin binding. Other domains that are present alone or in different combinations [10, 11] are less represented, such as: Meprin and TRAF-homology domain, ADP-ribosylation factor (ARF) family domain, C-terminal subgroup one signature, filamin-type immunoglobulin, fibronectin type 3 repeat, and transmembrane (TM) region.
TRIM proteins are involved in a wide range of molecular functions, ranging from transcriptional regulation to signal transduction pathways, in the context of several cellular processes, such as growth, differentiation, development, oncogenesis, and immunity [12]. The majority of TRIMs show multiple alternatively spliced mRNAs, and are nonubiquitously expressed, suggesting that they are involved in cell type specific functions [13].
An interesting feature of this family of proteins is the rapid increase in number that has occurred recently in evolution, probably by gene duplications [10, 14, 15]. This expansion has occurred in jawed fishes that have more than hundred TRIM genes with respect to lower organisms, such as fruit flies, with only seven TRIMs. Since this expansion occurred when the adaptive immune system emerged, it has been proposed that “new” TRIM proteins may have evolved in parallel with the increasing complexity of the immune system [16–18]. This hypothesis is now corroborated by a large set of experimental data that support the major role played by many TRIMs in the immune response, where they control several inflammatory pathways [13]. It is in the context of the immune response that the strict link between TRIM proteins and autophagy has originally emerged, as described in details below.
The autophagy core machinery
Macroautophagy, hereafter defined as autophagy, allows degradation of intracellular components by enwrapping them in double-membrane vesicles, called autophagosomes, and delivering them to lysosome for degradation [3]. Five main steps are required to allow the autophagosome-mediated degradation: initiation/nucleation, elongation, cargo interaction, closure and fusion with the endolysosomal compartment (Fig. 2) [19, 20]. These events are under the control of distinct autophagy proteins [4, 21–23].
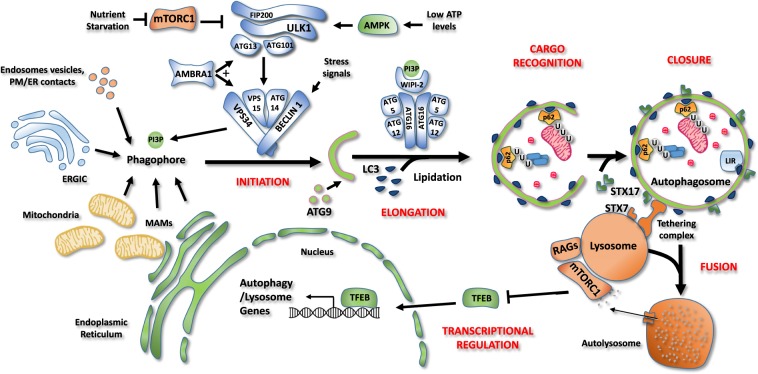
Fig. 2. Schematic representation of the autophagy core machinery.
Description of the different steps of the autophagy process. Initiation. Autophagosome formation is under the control of the ULK1 and BECLIN 1 complexes. Under nutrient-rich conditions, mTORC1 associates with and inhibits ULK1 complex (ATG101, ATG13, FIP200, and ULK1/2). During nutrient starvation, mTORC1 is inhibited, allowing ULK1 complex to induce autophagy. When ATP levels are low, the AMP-activated protein kinase (AMPK) acts as a positive regulator of the ULK1 complex. Once active, ULK1 phosphorylates BECLIN 1 and stimulates the lipid kinase activity of the BECLIN 1 complex (BECLIN 1, ATG14, VPS34, VPS15). This complex is responsible for the production of phosphatidylinositol 3-phosphate (PI3P) by the Class III PI-3-kinase, VPS34. PI3P allows the nucleation of an isolation membrane, also called phagophore, which may originate from endoplasmatic reticulum (ER), plasma membrane, Golgi complex, endosomes vesicles or endoplasmatic reticulum-mitochondrial contact sites (MAMs). Elongation, cargo recognition and closure. Autophagosome expansion and closure requires the fusion of the isolation membrane with ATG9-positive vesicles and the insertion of ATG8 family members (LC3, GABARAP, GATE-16) on autophagosome membranes upon lipidation. WIPI-2 binds PI3P on forming vesicles as well as ATG16, thus allowing the recruitment of both ATG12-5-16 complex, which is necessary for LC3 lipidation. LC3 allows cargo recruitment by interacting with the adaptor proteins, such as the ubiquitin binding protein p62 and proteins containing the LC3-interacting region (LIR) motif. Then, phagophore closes to form a double-membrane vesicle, named autophagosome. Fusion. Finally, autophagosomes are recruited to lysosomes by a tethering complex and fuse through SNARE proteins SINTAXIN 7 (STX79 and SINTAXIN 17 (STX17), forming a single membrane vesicle, named autolysome, where sequestered materials are degraded. Transcriptional regulation. Autophagy process is tightly regulated also at transcriptional level by several transcription factors. Under amino acid sufficiency, mTORC1 inhibits the transcription factor EB (TFEB) causing its retention in the cytosol and blocking its transcriptional activity. Upon nutrient starvation, mTORC1 is inactivated, leading to TFEB nuclear translocation and transcription of genes involved in lysosomal biogenesis and autophagosome formation.
The BECLIN 1 complex, composed by class III phosphatidylinositol (PI)-3-kinase VPS34 and the cofactors BECLIN-1, VPS15, and ATG14, allows the formation of PI-3-phosphate (PI3P), a signal for the assembly of the autophagosome precursor membrane (isolation membrane, also called phagophore) [23–27]. Autophagosomes are mainly formed at specialized endoplasmic reticulum (ER) structures, such as ER-Golgi intermediate compartment, ER-mitochondria, and ER-plasma membrane contact sites [23–27].
The ULK complex, composed by the protein kinases ULK1/2 and the cofactors ATG13, FIP200, ATG101, is required for the activation of the BECLIN 1 complex through the phosphorylation of its components (BECLIN-1, VPS34, and ATG14) as well as of AMBRA1, a cofactor that promotes BECLIN-1 complex stability/activity [28–32].
The ULK1 and the BECLIN 1 complexes are regulated by a large variety of stress signaling pathways, which modulate their activity mainly via posttranslational modifications, as described for the nutrient and energy sensor kinases mTOR and AMPK, and various E3 ubiquitin ligases, such as TRAF6, NEDD4L, CULLIN 3, and CULLIN 4 [33–36].
Generation of phagophores and their subsequent expansion and closure requires the recruitment of ATG9-positive vesicles and membrane insertion of ATG8 family members (LC3, GABARAP, and GATE-16) [4, 37, 38]. LC3 lipidation is mediated by a ubiquitination-like cascade consisting of the LC3 protease ATG4, the E1 activating enzyme ATG3, the E2 conjugating enzyme ATG7, and the E3 ligase ATG5/ATG12/ATG16 complex [4, 37]. The LC3 lipidation machinery is recruited to the isolation membrane by the PI3P interacting protein WIPI-2 which directly associates to ATG16 [39]. Besides regulating autophagosome formation, LC3 interacts with cargos receptors that provide selectivity to autophagy degradation (see below) [40, 41].
The fusion of autophagosomes with the endolysosomal compartment requires tethering factors, PI3P, Rab GTPases, and SNARE proteins [42]. Tethering proteins, including EPG5, HOPS, and PLEKHM1, simultaneously bind to ATG8s on autophagosomes and RAB7 on lysosomes [43]. They promote the assembly of trans-SNARE complexes formed by STX17-SNAP29-VAMP7/8 on autophagosomes and STX7-SNAP29-YKT6 on lysosome, which mediate membrane fusion [42–45].
Although autophagy induction is mainly regulated by cytosolic pathways, transcriptional and epigenetic events also play a role in sustaining the autophagic response to stress stimuli [19, 46]. Various transcription regulators have been shown to regulate the expression of autophagy genes, including TFEB, TFE3, ZKSCAN3, FOXO3, NF-κB, E2F1, p53, PPARa, FXR, SIRT1, and CARM1 [19, 46]. In particular, how the activity of TFEB is regulated during the autophagy response has been well-characterized. TFEB is phosphorylated by growth-factors/nutrients regulated kinases, such as mTOR, ERK, AKT, and PKCβ, and sequestered in the cytosol [47]. Nutrient starvation or cytosolic calcium increase triggers TFEB dephosphorylation and nuclear translocation to induce the expression of a variety of autophagy/lysosome genes [47].
Selective autophagy
Although it has long been considered as a nonspecific degradation process, it is now evident that autophagy is highly capable to discriminate the components that have to be sequestered within autophagosomes [48, 49]. These types of selective autophagy pathways have been named according to the targeted cellular material: protein aggregates (aggrephagy) [50], pathogens (xenophagy) [51], damaged or unnecessary organelles, such as mitochondria (mitophagy) [52], ER (ER-phagy or reticulophagy) [53], nuclear envelope (nucleophagy) [54], lipid droplets (lipophagy) [55], lysosomes (lysophagy) [56], and other cellular structures, such as ribosomes (ribophagy), the midbody ring, ferritin (ferritinophagy), zymogen granules (zymophagy), stress granules (granulophagy), proteasome (proteaphagy), and centriolar satellites [5, 57, 58]. The targeted cellular components are recognized by so-called autophagy receptors, which share the property to interact with autophagosomes via LC3-interacting regions (LIR) [40].
There are two main mechanisms by which autophagy receptors work: (1) they are selectively expressed on the target structures, as in mitochondria (BNIP3, NIX, FUNDC1, AMBRA1, PROHIBITIN) [59] and ER (FAM134B, SEC62, RTN3L, CCPG1, ATL3, TEX264) [60]; (2) they bind components of the target structure upon their posttranslational modifications [5]. In this case, ubiquitination is so far the best characterized modification that mediates cargo recognition [57, 61]. Ubiquitin is recognized by a group of proteins that share the presence of both an LIR domain and a ubiquitin binding domain (p62, NBR1, OPTN, NDP52, TAX1BP1) [61]. These receptors have been described to mediate the degradation of ubiquitinated structures, including organelles, such as mitochondria (OPTN, NDP52, TAX1BP1) and peroxisomes (NBR1), protein structures, such as protein aggregates (p62, NBR1, OPTN), the residual mitotic structure called midbody (p62, NBR1), and bacteria (p62, OPTN, NDP52) [57].
As described in the following paragraphs, novel important aspects of how selective autophagy works have been clarified thanks to the identification of TRIM proteins as regulators of both autophagy induction and substrate recognition under various stress conditions.
Regulation of autophagy by TRIM proteins
It is noteworthy that TRIM proteins have been directly linked to several steps of the autophagic process, ranging from upstream signaling pathways to autophagosome formation and transcriptional regulation of autophagy genes (Table 1).
Table 1.
TRIM proteins involved in autophagy regulation.
| Protein | Role in autophagy | Ref. |
|---|---|---|
| TRIM5α | Autophagy Induction and selective degradation of HIV capsid proteins | [72, 82] |
| Selective inhibition of autophagic degradation of TAK1 in cytoplasmic bodies | [80] | |
| TRIM6 | Bulk autophagy induction | [72] |
| TRIM11 | Autophagic degradation of misfolded proteins | [119] |
| Autophagic degradation of AIM2 during viral infection | [94] | |
| TRIM14 | Inhibition of autophagic degradation of cGAS during viral infection | [105] |
| TRIM16 | Autophagy induction and selective degradation of protein aggregates and damaged endolysomal compartments | [75] |
| Autophagy-mediated IL-1β secretion | [98] | |
| TRIM17 | Inhibition of bulk autophagy by promoting BECLIN-1/MCL-1 interaction | [79] |
| Autophagic degradation of midbody remnant | [79] | |
| TRIM18 | Autophagy inhibition by preventing TFEB activation through proteasomal degradation of PP2A | [85] |
| TRIM19 | Autophagy inhibition by limiting AMPK activity in tumors and viral infection | [65] |
| TRIM20 | Autophagy induction and selective degradation of inflammasome by IFN-γ | [74] |
| TRIM21 | Autophagy Induction and selective degradation of IRF3 upon IFN-γ treatment | [74] |
| Ubiquitin-mediated autophagic degradation of IKKβ during HTLV-1 infection | [113] | |
| Autophagic degradation of opsonized Salmonella enterica | [117] | |
| TRIM2 | Bulk autophagy induction | [72] |
| Induction of BECLIN 1 gene expression during M. tuberculosis infection | [84] | |
| TRIM23 | Stimulation of p62 activity by TBK1-mediated phosphorylation during viral infection | [109] |
| TRIM28 | Autophagy inhibition by proteasomal degradation of AMPK in cancer cells | [62] |
| Autophagy induction through VPS34 SUMOylation upon HDAC inhibitor treatment | [83] | |
| Mitophagy induction by inhibiting miRNAs targeting autophagy genes | [88] | |
| TRIM31 | Induction of unconventional autophagy in intestinal epithelia upon lipopolysaccharide stimulation | [89] |
| TRIM32 | Autophagy induction by promoting ULK1 activity through unanchored ubiquitination during muscle atrophy | [76] |
| Autophagic degradation of TRIF protein to downregulate Toll-like receptor signaling | [112] | |
| Stimulation of the cargo binding properties of p62 monoubiquitination | [121] | |
| TRIM37 | Bulk autophagy Inhibition by promoting mTOR/RAG-B activity | [64] |
| TRIM38 | SUMOylation-mediated inhibition of STING degradation by chaperone-mediated autophagy during viral infection | [106] |
| TRIM49 | Bulk autophagy induction | [72] |
| TRIM50 | Autophagy induction through nondegradative ubiquitination of BECLIN 1 | [73] |
| TRIM59 | Autophagy inhibition by promoting the proteasomal degradation of TRAF6 | [81] |
| Inhibition of BECLIN 1 expression by repressing NF-κB pathway | [81] | |
| TRIM63 | Autophagic degradation of nicotinic acetylcholine receptor during muscle atrophy | [120] |
| TRIM65 | Inhibition of miRNAs targeting ATG7 by proteasomal degradation of the RISC complex protein TNRC6A | [87] |
TRIM28, TRIM37, and TRIM19 controls autophagy by targeting upstream signaling pathways conveying on ULK1 and BECLIN 1 complexes, such as those regulated by AMPK and mTOR [62–65] (Fig. 3).
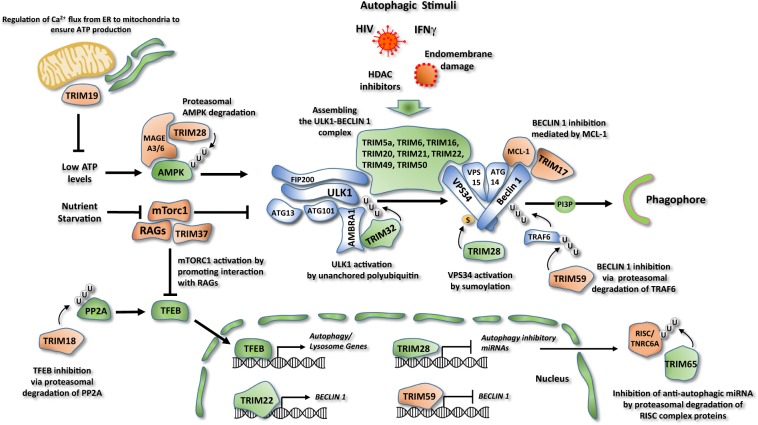
Fig. 3. Regulation of the autophagy core machinery by TRIM proteins.
Regulation of AMPK and mTOR activity. TRIM28 stimulates AMPK proteasomal degradation, through its degradative ubiquitination. The binding and ubiquitination of AMPK requires the interaction of TRIM28 with the protein MAGE-A3/6. TRIM37 is a negative regulator of autophagy that activates mTOR pathway by promoting the interaction between mTORC1 and RAG proteins. TRIM19 negatively regulates autophagy by controlling Ca2+ flux from ER to mitochondria to ensure ATP production. Regulation of BECLIN 1 and ULK1 complex. TRIM5α, 6, 16, 20, 21, 22, 49, and 50 associate with both autophagic proteins ULK1 and BECLIN 1 assembling these factors in a complex and promoting their functions. Most of these TRIMs act in response to selective stimuli like HIV infection (TRIM5α), Endomembrane damage (TRIM16), interferon γ (IFN-γ) stimulation (TRIM20, TRIM21), and HDAC inhibition (TRIM50). A group of TRIM proteins regulates autophagy initiation preferably acting only on ULK1 or BECLIN 1. TRIM32 interacts with the autophagy cofactor AMBRA1 and promotes ULK1 kinase function by unanchored polyubiquitin. TRIM17 inhibits BECLIN 1 complex by promoting the interaction of BECLIN 1 with its negative regulator MCL-1. TRIM59 ubiquitinates and promotes the proteasomal degradation of the E3 ubiquitin ligase TRAF6, inhibiting BECLIN 1 activity. TRIM28 positively regulates autophagy promoting VPS34 activation by sumoylation. Regulation of autophagy at transcriptional level. TRIM59 negatively regulates the expression of BECLIN 1, while TRIM22 stimulates autophagy by promoting BECLIN 1 expression. TRIM28 inhibits the expression of a series of autophagy inhibitory miRNAs. TRIM18 ubiquitinates and targets PP2A for proteasomal degradation, causing inhibition of TFEB activity. TRIM65 ubiquitinates and induces the degradation of TNRC6A, a component of the RNA-induced silencing complex (RISC), inhibiting the expression of a series of antiautophagic miRNAs. Proautophagic factors are colored in green, antiautophagic factors in red.
TRIM28 was shown to regulate autophagy by controlling AMPK stability in cancer cells [62]. TRIM28 also known as Krüppel-associated box-associated protein 1 (KAP1) or TIF1-β shares many structural features with three other TRIM proteins, TRIM24 (TIF1α), TRIM33 (TIF1γ), and TRIM66 (TIF1δ), and together constitute the TIF1 family, which is involved in transcriptional repression and DNA repair [66].
TRIM28 expression is upregulated in a large set of cancers, including cervical, gastric, ovarian, glioma, breast, and hepatocellular carcinoma, which often correlates with worse overall patient survival, suggesting that TRIM28 supports cancer progression [66]. In addition to its roles in transcription and DNA damage, the tumor promoting function of TRIM28 has been ascribed to its ability to inhibit the tumor suppressive activity of autophagy in the early stages of tumorigenesis [62]. This is due to the ability of TRIM28 to target the proautophagic regulator AMPK to proteasome degradation [62]. Interestingly, the binding and ubiquitination of AMPK require the interaction of TRIM28 to MAGE-A3/6, proteins exclusively present in the testis, which are however frequently expressed in tumors, including colon, lung, and breast cancers [62]. The relevance of this regulation is further highlighted by the finding that a high level of MAGE-A/TRIM28 prevents the response of melanoma patients to anti-CTL4 based immunotherapy, because it impairs the autophagic activity required for the anticancer immune response [63]. This inhibition is not only associated to the degradation of AMPK but also of HMGB1 (high-mobility group box 1), an autophagy regulator of immunogenic cell death required for dendritic cell-mediated priming of the adaptive immune response [63].
TRIM37 has been characterized as a negative regulator of autophagy by modulating the activity of the mTOR pathway. TRIM37 gene mutations result in Mulibrey (muscle-liver-brain-eye) nanism, which is a rare, autosomal-recessive growth disorder of prenatal onset with characteristic dysmorphic features, pericardial constriction, and hepatomegaly [67]. At molecular level, TRIM37 deficiency leads to (1) dysregulated centriole duplication during cell cycle, (2) defects in TRAF2 activity in the NF-κB pathway, (3) degradation of the peroxisome import protein PEX5, (4) impaired mTOR activity and consequent high basal autophagy [67]. In the latter case, TRIM37 was shown to interact with mTOR and the amino acid sensor RAG-B and to be required for their association in nutrient-rich conditions [64]. In TRIM37 deficient cells, mTOR inhibition results in higher autophagy levels and the activation of the proautophagic transcription factor TFEB [64]. As to whether deregulated autophagy contributes to Mulibrey nanism represents an important question to be addressed.
TRIM19 regulates autophagy by controlling intracellular Ca2+ levels [65]. TRIM19, best known as promyelocytic leukemia (PML) gene, is a tumor suppressor that was initially identified inactivated in acute PML by chromosome translocation giving rise to the PML–RARα fusion protein, and later on found mutated in a wider range of tumors [68]. PML mainly resides in the nucleus where it forms nuclear structures, called PML-nuclear bodies, involved in proliferation, apoptosis, genome stability, and antiviral response [68].
PML-deficient tumors show high resistance to chemotherapy and this feature has been correlated to an upregulation of basal autophagy levels. Indeed, in normal conditions PML negatively regulates autophagy by localizing at the ER-mitochondria contact sites in a p53-dependent manner [65]. In keeping with its localization, a reduced IP3R-mediated Ca2+ transfer from the ER to mitochondria has been observed in PML-deficient cancer cells [65]. This causes an impaired activity of the tricarboxylic acid cycle dehydrogenases and subsequent decrease in mitochondrial respiration and ATP production, which results in AMPK activation and autophagy induction. Consistently, restoration of the Ca2+ signal by overexpression of the mitochondrial Ca2+ uniporter restores basal levels of autophagy in cancer cells [65].
The autophagy inhibitory role of PML was also confirmed in other cell systems [69–71]. PML is responsible for reducing autophagy levels in mesenchymal stem cells, a decrease that is required to induce the differentiation of these cells in adipocytes [71]. Moreover, in HeLa and mouse embryonic fibroblasts infected with enterovirus 71 (EV71), PML-deficient cells show enhanced viral replication associated with an increased formation of autophagosomes, which are hijacked by EV71 to accomplish its replication [69]. Interestingly, the EV71 protease 3Cpro favors viral replication by promoting PML cleavage and consequent disruption of PML bodies [69]. In addition to the negative role of PML in autophagy, it is worth to mention that, in acute PML cells, the PML–RARα fusion can become an autophagy target, whose degradation is triggered following tumor cell treatment with all-trans retinoic acid and arsenic trioxide [70].
A large set of TRIM proteins directly regulate the autophagy machinery, as initially elucidated by the seminal work of Mandell et al. [72]. TRIM5α, 6, 16, 20, 21, 22, 49, and 50 are able to associate with both ULK1 and BECLIN 1 and act as platforms to assemble these factors in a complex to promote their activation [72–75] (Fig. 3). So far, evidence that the autophagy complex formation triggered by these TRIMs is mediated by their ubiquitination activity has been only provided for TRIM16 and TRIM50, which target ULK1 and BECLIN 1 for K63-linked polyubiquitination, respectively [73, 75]. Moreover, the presence of TRIMs that lack part of the catalytic domain suggest that the direct ubiquitination of substrates may not always be required for this proautophagic function [72]. Notably, most TRIMs have been characterized to mediate autophagy activation in response to selective stimuli, such as TRIM5α upon HIV infection, TRIM16 upon endomembrane damage, TRIM20 and 21 upon Interferon (IFN)-γ stimulation, TRIM50 upon spermidine treatment [72–75].
Not all the TRIMs that stimulate autophagy act as scaffolds to assemble ULK1 and BECLIN 1 in a single complex. We have recently demonstrated that the E3 ubiquitin ligase TRIM32 is an exclusive ULK1 activator [76]. TRIM32 gene is mutated in limb-girdle muscular dystrophy 2H (LGMD2H), a late-onset autosomal-recessive myopathy [77]. We found that the E3 ubiquitin ligase activity of TRIM32 is required to regulate autophagy in muscles upon atrophy induction and, in this way, protects muscle cells from excessive ROS production and induction of the proatrophic genes [76]. We also observed that the interaction of TRIM32 with ULK1, which requires the presence of the autophagy cofactor AMBRA1, stimulates ULK1 kinase activity through the formation of unanchored K63-linked polyubiquitin chains that associate to the carboxy-terminal domain of ULK1 [76]. Notably, mutations in TRIM32 responsible for LGMD2H affect its ability to bind ULK1 and to induce autophagy in muscle cells, resulting in a dysregulated activation of the atrophic process [76, 78].
A subset of TRIM proteins that negatively regulate autophagy initiation by preferably acting on the BECLIN 1 complex has been also identified [79–81].
TRIM17 inhibits BECLIN 1-dependent autophagy by promoting the interaction of BECLIN-1 with its negative regulator MCL-1 [79]. Intriguingly, TRIM17 was also found to act as a positive regulator of the selective type of autophagy targeting the midbody [79]. These different functions of TRIM17 suggest that TRIM proteins may be able to direct the autophagy machinery to selective cargos and, at the same time, to inhibit its activity on others. This opposite activity of TRIM proteins has been recently corroborated by studies on TRIM5α, where this mechanism has been named “deselective autophagy” [80]. On one hand, TRIM5α promotes autophagy during HIV infection (see below for details) [72, 82]. On the other hand, TRIM5α was found to induce the formation of cytoplasmic bodies, where the autophagy receptor p62 and the associated protein kinase TAK1 localized in stress conditions. Notably, these p62 bodies are protected from autophagic degradation, so as to allow TAK1 to activate NF-κB-mediated signaling pathways [80].
At variance with its inhibitory role on AMPK, TRIM28 was reported to stimulate prosurvival autophagy by regulating VPS34 activity when cells are treated with anticancer agents, such as histone deacetylase (HDAC) inhibitors [83]. In details, HDAC inhibition leads to HSP70 acetylation, a posttranslational modification that promotes the recruitment of TRIM28 to the BECLIN 1 complex. Here, TRIM28 acts as an E3 ligase for the ubiquitin-like protein SUMO to VPS34. SUMOylation occurs in the catalytic domain of VPS34 and is required for maintaining VPS34 in an open conformation, thus stimulating the lipid kinase activity of VPS34 [83].
TRIM59 inhibits BECLIN 1 activity by triggering the proteasomal degradation of TRAF6, an E3 ubiquitin ligase able to stimulate BECLIN 1 activity through K63-linked polyubiquitination [81]. Moreover, TRIM59 has been reported to negatively regulate the expression of BECLIN 1 through the inhibition of NF-κB pathway [81].
The ability of TRIM proteins to regulate autophagy at transcriptional level is not limited to TRIM59. TRIM22 was shown to stimulate autophagy in response to Mycobacterium tuberculosis infection by promoting BECLIN 1 expression in NF-κB dependent manner [84].
TRIM18 is involved in the regulation of the expression of autophagy genes by controlling TFEB activity [85]. TRIM18, best known as MID1, is a gene whose mutations are associated with the X-linked form of Opitz Syndrome, a developmental disorder characterized by midline defects and intellectual disability [86]. MID1 targets PP2A to proteasomal degradation and, in this way, it inhibits TFEB dephosphorylation and nuclear translocation [85]. This inhibition is released by the fasting-induced hormone FGF21, which mobilizes calcium from the ER via the inositol trisphosphate receptor, and allows the transcription repressor complex DREAM to shuttle to the nucleus [85]. DREAM then inhibits the expression of the MID1 gene, leading to stabilization of PP2A and the activation of TFEB [85].
TRIM proteins also regulate autophagy by regulating miRNA expression [87]. For example, TRIM65 induces the ubiquitination and degradation of TNRC6A, a component of the RNA-induced silencing complex [87]. Decreased TNRC6A levels inhibits the expression of a series of miRNAs including miR-138-5p, which was shown to target the expression of ATG7 [87]. The relevance of the TRIM65/TNRC6A/miR-138-5p pathway in autophagy regulation was confirmed in non-small cell lung cancer cells, where TRIM65-regulated autophagy contributes to the resistance to cisplatin treatment [87]. Moreover, TRIM28 was shown to sustain mitophagy at transcriptional level [88]. In fact, TRIM28 represses the expression of a series of miRNAs that target autophagy genes, such as Nix/Bnip3l, Ulk1, Gabarapl2, Atg12, Beclin 1, and Bcl-2 [88]. As a consequence, the hemato-specific knockout of mouse Trim28 displayed fatal hyporegenerative anemia, characterized by erythrocyte deficiency and accumulation of erythroblasts in the bone marrow, which failed to activate mitophagy to degrade mitochondria during differentiation [88].
In addition to regulate the activity of the autophagy core machinery, recent evidences show that TRIMs may also trigger an unconventional form of autophagy, in which autophagosome formation is independent of ATG5/ATG7 [89]. In particular, TRIM31 was found to promote unconventional autophagy in response to lipopolysaccharide stimulation by interacting with phosphatidylethanolamine in a palmitoylation-dependent manner [89]. Importantly, TRIM31 expression is required to block Shigella infection in intestinal epithelial cells and is downregulated in patients with Crohn’s disease, suggesting that its dysregulation may contribute to the uncontrolled bacterial replication in inflammatory bowel diseases [89].
Regulation of autophagic cargo recognition by trim proteins
Studies on TRIM proteins have profoundly improved our understanding on how autophagy contributes to the cellular response to a variety of stress cues by either controlling the stability of specific signaling proteins or by selectively removing damaged cellular components. As described in the following paragraphs, most of the TRIMs that work as substrate receptors are involved in the regulation of the immune responses, i.e., inflammasome, cytosolic DNA sensing, and other NF-κB-regulated pathways (Table 1).
Regulation of inflammasome by TRIM proteins
Inflammasomes are protein complexes formed by (1) sensor/scaffold proteins (e.g., NLRP1, NLRP3, AIM2, NLRC4, and Pyrin/TRIM20), (2) the adaptor ASC (also known as PYCARD), (3) the procaspase 1. Inflammasome activation is triggered by a wide range of pathogen-associated or danger-associated molecular patterns (PAMPs or DAMPs) which are recognized by different sensor proteins [90]. Once assembled, caspase 1 is activated and is responsible for the maturation of interleukin (IL)-1β and IL-18 through proteolytic cleavage. Caspase 1 also targets gasdermin D, which is responsible for the activation of pyroptosis, a specific cell death modality [91, 92].
Autophagy is known to act predominantly as a negative regulator of inflammasome-mediated responses [93]. This feature was initially ascribed to the ability of autophagy to promptly remove inflammasome stimuli, such damaged mitochondria or invading pathogens. More recently, it has become evident that autophagy can also directly target inflammasome components for lysosomal degradation, whose recognition is mainly mediated by TRIMs (Fig. 4) [74, 94, 95]. In this manner, autophagy may limit inflammasome activity and avoid excessive or prolonged inflammation [96]. For example, during viral infection, TRIM11 binds to AIM2, an inflammasome sensor activated by double-stranded DNA, and promotes its selective engulfment within autophagosomes [94]. Interestingly, AIM2 is not ubiquitinated by TRIM11, but is the auto-polyubiquitinated TRIM11 in complex with AIM2 that is recognized by the autophagic cargo receptor p62 and captured by the nascent autophagosome [94]. This regulation is specific for AIM2, because NLRP3 is not affected by TRIM11 [94].
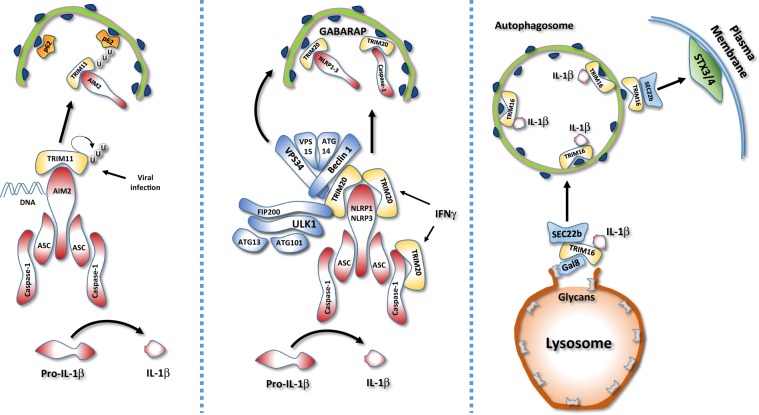
Fig. 4. Regulation of inflammasome by TRIM-mediated autophagy.
TRIM proteins regulate inflammasome stability and IL-1β secretion. Left: During viral infection, TRIM11 binds to the inflammasome DNA sensor AIM2 and promotes its selective autophagic degradation. TRIM11 auto-polyubiquitination allows the AIM2/TRIM11 complex to be recognized by the selective autophagy cargo receptor, p62 and sequestered into the autophagosome. Middle: In response to interferon γ stimulation, TRIM20 negatively regulates inflammasome activity by directly targeting NLRP1, NLRP3, and procaspase 1 for autophagic degradation. Right: In response to lysosomal damage, Galectin 8 (Gal8) recruits TRIM16 that in turns binds to IL-1β and mediates its engulfment within the authophagosome. In addition, TRIM16 interacts with the SNARE protein Sec22b on the forming autophagosome, allowing the fusion with the plasma membrane and IL-1β secretion, by interacting with syntaxin 3/4 (STX34).
An important role in the TRIM-mediated crosstalk between autophagy and inflammasome is played by TRIM20, a peculiar TRIM in which the RING domain is substituted by the PYRIN domain, a region evolutionarily related to the death domain [97]. Importantly, mutations in the TRIM20 gene, also known as PYRIN and MEFV, are associated with the autoinflammatory disease familial Mediterranean fever [97]. TRIM20 has been shown to play both positive and negative roles in the regulation of inflammasome depending on the stimuli [74, 95]. TRIM20 acts as an inflammasome sensor to stimulate IL-1β production in response to actin cytoskeleton modifications [95]. On the other hand, TRIM20 mediates the inhibition of inflammasome activity when cells are treated with IFN-γ by targeting NLRP1, NLRP3, and procaspase 1 for autophagic degradation [74]. This function is mediated by the ability of TRIM20 to interact with both upstream autophagy regulators, such as ULK1, BECLIN 1, and ATG16, and the ATG8 proteins GABARAP and GABARAPL1 [74].
IL-1β secretion is another important step at which TRIM-dependent autophagy controls inflammasome activity [98, 99]. Once cleaved by caspase 1, a mature IL-1β is secreted by at least two different mechanisms: (1) Gasdermin mediated pore formation [100] (2) Secretory autophagy [99]. Recently, important aspects of the molecular mechanism underlying the autophagy-dependent release of IL-1β in response to lysosomal damage have been elucidated [98]. Following lysosomal membranes damage, glycan groups are exposed and rapidly recognized by galectin 8, which, in turn, recruits TRIM16 [98]. Here, TRIM16 also binds IL-1β and mediates its engulfment within LC3-II positive vesicles. Moreover, TRIM16 allows the recruitment of the SNARE protein SEC22B on the forming autophagosomes, which is required to tether them to the plasma membrane, by interacting with SYNTAXIN 3 and SYNTAXIN 4, instead of SYNTAXIN 17 that is present on lysosomes. Of note, this unconventional secretion pathway is not specific for IL-1β, but also involved in the extracellular release of other cytosolic proteins, such as FERRITIN [98].
Regulation of the STING signaling pathway by TRIM proteins
TRIM proteins were reported to regulate both positively and negatively the activity of various components of the cGAS/STING pathway by modulating either protein stability in a proteasome-dependent manner or protein activity by regulative ubiquitination [101]. More recently, they have also emerged as key players of the crosstalk between autophagy and cytosolic DNA response (Fig. 5).
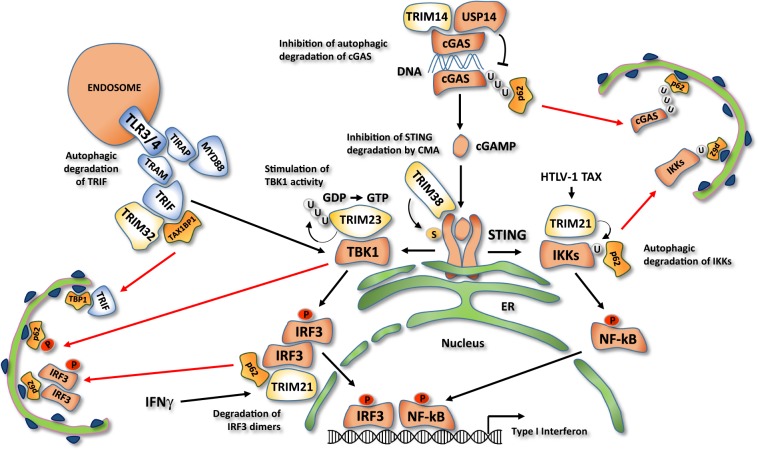
Fig. 5. Regulation of STING signaling pathway by TRIM-mediated autophagy.
cGAS senses cytosolic DNA and generates the second messenger cGAMP, which binds to STING, promoting its oligomerization. STING is therefore able to interact with protein kinases TBK1 and IKK and stimulate the expression of type I IFN and inflammatory cytokines. TRIM14 recruits the deubiquitinase USP14, which cleaves polyubiquitin from the cytosolic DNA sensor cGAS and promotes its p62-mediated autophagic degradation. TRIM38 is required for the sumoylation of the downstream effector STING, preventing its degradation by the chaperone-mediated autophagy. Auto-ubiquitination of TRIM23 stimulates the STING activated protein kinase TBK1, promoting its ability to phosphorylate the selective autophagy cargo receptor p62. In response to interferon γ stimulation (IFNγ), TRIM21 inhibits STING signaling pathway by promoting the autophagic degradation of IRF3 dimers. In HTLV-1-infected cells, the viral protein TAX promotes monoubiquitination of IKKs by TRIM21 which allows their interaction with the autophagy receptor p62 and the sequestration into the autophagosome. TRIM32 inhibits Toll-like receptors 3/4 activity. TRIM32 mediates the autophagic degradation of TRIF, a critical adaptor protein of Toll-like receptors 3/4, by interacting with the selective autophagy receptor TAX1BP1.
The cGAS/STING signaling pathway is essential to detect DNA released into the cytosol during pathogen replication or organelle damage [102]. cGAS senses cytosolic DNA and generates the second messenger cGAMP, which binds to STING, an ER-resident TM protein, and promotes its conformational change and oligomerization [103]. STING is thus able to interact with the protein kinases TBK1 and IKKs and stimulate the phosphorylation of transcription factors IRF3 and NF-κB, thus inducing the expression of type I IFN and inflammatory cytokines [104].
TRIM14 and TRIM38 contribute to enhance the activity of the STING pathway by preventing the autophagic degradation of cGAS and STING, respectively [105, 106]. In particular, TRIM14 recruits the deubiquitinase USP14 on cGAS to decrease its K48-linked ubiquitination, which mediates p62-mediated autophagic degradation [105]. Instead, TRIM38 is required for STING sumoylation, which impedes STING degradation by the chaperone-mediated autophagy, a selective form of autophagy that does not require autophagosome formation but targets proteins directly inside the lysosome through LAMP2A [106]. TRIM38 also mediates cGAS sumoylation, but, in this case, this modification prevents cGAS proteasomal degradation. The relevance of TRIM14 and TRIM38 in the activation of the cGAS/STING pathways has been confirmed in the context of HSV1 infection, where both TRIMs are required for the efficient activation of type I IFN response [105, 106].
TRIM23 regulates the activity of TBK1, a protein kinase involved in the STING signaling pathway and other important immune responses [107]. TBK1 also plays a direct role in autophagy by phosphorylating p62/SQSTM1 family members, which is required for their binding of both upstream autophagy regulators and ubiquitinated cargos [108]. TRIM23 is a protein with both E3 ubiquitin ligase and ARF GTPase activities, which are both required to stimulate TBK1- and p62-mediated selective autophagy [109]. In particular, K27-linked auto-polyubiquitination of the ARF domain is essential for the GTP hydrolysis activity of TRIM23 and the activation of TBK1, by facilitating dimerization and its ability to phosphorylate p62 [109]. As a proof of the importance of this regulation, the Us11 Protein of Herpes Simplex Virus 1 is able to inhibit the antiviral autophagy response by disassembling the TRIM23–TBK1 complex [110].
An opposite role on cGAS/STING and other innate immune pathways is played by TRIM21 [74]. TRIM21 has long been recognized as a suppressor of type I IFN and identified as an autoantigen associated with Sjögren syndrome and systemic lupus erythematosus [111]. Recently, TRIM21 was found to bind to the ATG8-like proteins GABARAP and p62, and, in this way, to promote the autophagy degradation of IRF3 dimers, thus highlighting a novel mechanism by which autophagy may restrain type I IFN response [74].
Regulation of other NF-κB signaling pathways by TRIM proteins
In addition to the STING pathway, TRIM proteins are involved in the autophagy regulation of other NF-κB-regulated pathways to avoid an excessive or prolonged inflammatory responses [112, 113]. Inflammatory cytokines and Toll-like receptors activate NF-κB by through the protein kinases IKKs and the regulatory nonenzymatic scaffold protein NEMO [114]. IKKs phosphorylates NF-κB to promote its dimerization and nuclear translocation, and the NF-κB inhibitor IκB to trigger its ubiquitin-mediated proteasomal degradation [115].
During HTLV-1 infection, TRIM-regulated autophagy is responsible for preventing a persistent activation of NF-κB induced by the HTLV-1 protein TAX through the degradation of IKKs (Fig. 5) [113]. In particular, TRIM21 was found to interact with the active form of IKKβ and to mediate its monoubiquitination. Ubiquitinated IKKβ is then sequestered within autophagosomes and delivered to lysosome for degradation [113].
Upstream regulators of IKK are also targeted by proautophagic TRIMs (Fig. 5), as shown for TRIM32, which interacts with TRIF, a critical adaptor protein of Toll-like receptors 3/4, and mediates its autophagic degradation in an E3 activity-independent manner [112]. In particular, TRIM32 acts a scaffold protein to recruit TRIF within autophagosomes by interacting with TAX1BP1, a receptor for selective autophagy [112].
Regulation of selective degradation of pathogens by TRIM proteins
An important autophagic contribution of TRIM proteins to the innate immune response is definitely their ability to directly bind pathogens’ structures and deliver them to the autophagosomes for degradation, a process defined as xenophagy [101].
A first evidence of a direct role of TRIM in xenophagy came from studies of the role of TRIM5α in HIV infection [72, 82]. Independently of its E3 ubiquitin ligase activity, TRIM5a acts as a bridge between the HIV-1 p24 capsid, to which binds through its SPRY domain, and members of the ATG8 family, to trigger viral degradation [72].
The antiviral function of TRIM5α has been long considered to be reserved to non-human primates and only in part dependent on its ability to trigger xenophagy [116]. However, an important work from Ribeiro et al. has confirmed that the proautophagic activity of TRIM5α is relevant in the immune response to HIV-1 infection in human Langerhans cells, a specific subset of dendritic cells [82]. In these cells, TRIM5α is required to recruit the autophagic machinery to cytosolic Birbeck granules, where HIV-1 is sequestered by the lectin receptor Langerin, and to ensure the autophagic degradation of the invading HIV-1. This mechanism is specific for Langerhans cells, because, in dendritic cells expressing the lectin receptor DC-SIGN, HIV-1 infection leads to the dissociation of TRIM5α from DC-SIGN, thus inhibiting its autophagy-mediated degradation [82].
Interestingly, TRIM-mediated recognition of pathogens could also occur indirectly by interacting with cellular structures associated to the invading agent [75]. For example, TRIM16 restricts M. tuberculosis infection in macrophages by localizing to the bacteria-containing phagosomes [75]. TRIM16 does not interact with bacterial components but associates with vesicle membranes that are damaged by the M. tuberculosis secretion systems leading to the exposure of glycan residues to the cytosol. Glycan residues are then recognized by galectin 3, which in turn recruits TRIM16 and allow M. tuberculosis ubiquitination, a prerequisite for autophagosomal engulfment [75].
A different strategy to indirectly recognize pathogens is adopted by TRIM21 through a process called antibody-dependent intracellular neutralization [117]. Taking advantage of the ability to bind the Fc region of immunoglobulins, TRIM21 associates with opsonized Salmonella enterica when enters in epithelial cells, allowing autophagosome engulfment and autolysosomal degradation [117].
Other autophagic cargos recruited by TRIMs
In addition to the important autophagic roles played in the immune response, TRIMs act as autophagy receptors in response to other critical cellular stresses. For example, TRIM16 has been shown to localize to stress-induced protein aggregates where it induces autophagy, by interacting with ULK1 and ATG16L1, and acts as autophagy receptor by recruiting p62 and LC3B (aggrephagy) [118]. In addition, TRIM16 participates to p62-mediated NRF2 stabilization to induce the expression of a set of genes that convert misfolded proteins into less toxic protein aggregates [118]. In turn, NRF2 is able to stimulate TRIM16 expression in a positive feedback mechanism. In this way, TRIM16 maintains proteostasis and protects cells from oxidative and proteotoxic stress-induced cell death [118].
The role of TRIM proteins in mitigating proteotoxic stress has been highlighted in the context of tumorigenesis, where it was demonstrated that NRF2 induces TRIM11 expression, which is required for survival of mammary epithelial cancer cells by promoting both proteasome and autophagy-mediated degradation of misfolded proteins [119].
TRIM proteins are also involved in the recognition of autophagy cargos in tissue-specific contexts. An important indication has arisen from the study of the role of TRIM63, best known as MuRF1, upon induction of muscle atrophy by denervation [120]. In this damage condition, the levels of the nicotinic acetylcholine receptor (CHRN) on the postsynaptic membrane of the nerve–muscle synapse is decreased by MuRF1-mediated autophagy upon endosomal internalization [120]. In MuRF1 knockout mice, CHRN-positive vesicles failed to be engulfed by autophagosomes upon denervation, thus impairing their lysosomal degradation and allowing them to be recycled on sarcolemma [120].
TRIM proteins can also indirectly contribute to cargo degradation by regulating the activity of the autophagy receptor p62. An interesting example is represented by TRIM32, which mono-ubiquitinates p62 to promote its recruitment to autophagosomes [121]. This modification is also required to stimulate the degradation of TRIM32 itself, thus establishing a feedback loop to terminate the autophagy response. Importantly, a TRIM32 mutant causing the LGMD2H disease is unable to mono-ubiquitylate p62 and undergo autophagic degradation, suggesting that impairment of p62 activation by TRIM32 may contribute to the development of muscular dystrophies [121].
Concluding remarks
The identification of TRIM family proteins as important players of autophagy has greatly contributed to understand the key homeostatic role played by this process in a variety of stress responses, uncovering new specific intracellular targets which are delivered to lysosomes for degradation. Selective autophagy requires a large set of substrate receptors that not only bind substrates in response to cell damages and stress signals, but also recruit the autophagy machinery in loco to form new autophagosomes. The large family of TRIM ubiquitin ligases satisfy both requirements. They selectively bind to substrates that have to be degraded, including pathogens, damaged or unneeded cellular structures, components of stress signaling pathways, and concomitantly to ATG8 family members that decorate nascent autophagosomes. Moreover, most of proautophagic TRIMs interact with ULK1 and BECLIN 1 complexes favouring their activation by either acting as a recruitment platform for these upstream autophagy regulators or through their regulative ubiquitination. However, not all the TRIM proteins act as positive regulators, but some of them were shown to inhibit either the autophagic core machinery or upstream signaling pathways. In this regard, the finding that TRIM5α and TRIM17 may favor or prevent selective autophagy depending on the specific targets opens the possibility that TRIM proteins may play more complex roles in autophagy regulation.
Many aspects of the role of TRIMs in selective autophagy remain to be elucidated. Few autophagic targets of TRIMs have been identified so far, which still limits our understanding of the molecular mechanisms underlying selective cargo recognition in response to stress stimuli/damage and their ability to activate the proautophagic function of TRIMs. Proteomic characterization of autophagic cargos regulated by TRIMs is therefore an emerging requisite to obtain a more comprehensive view of TRIM-regulated autophagy. In addition, most of the autophagic TRIMs are E3 ubiquitin ligases that also play a role in proteasome-mediated degradation. Elucidating as to whether these different activities of TRIMs are coordinated will contribute to better clarify the crosstalk between these catabolic processes during stress responses.
To date, most of the available information on the role of TRIM in autophagy has been obtained in context of innate immunity, where specific TRIMs contribute to modulate intensity and duration of inflammatory responses.
A contribution of TRIM proteins to different types of selective autophagy is expected to emerge in the near future in other cell contexts frequently exposed to physiological and pathological stress conditions, with neurological and cardiovascular systems representing the most likely candidates.
Acknowledgements
This work was supported in part by grants from AIRC (IG2015 n. 17404 to GMF and IG2018 n. 21880 to MP), the Italian Ministry of University and Research (FIRB Accordi di Programma 2011 to MP and PRIN 2015 20152CB22L to GMF), the Italian Ministry of Health (Ricerca Corrente to MP and GMF, and Ricerca Finalizzata GR-2011-02350886 to AR), Fondazione Fibrosi Cistica (Progetto FFC#8/2018 to MP), Regione Lazio (Progetto “Gruppi di Ricerca” 2018 to MP), Transautophagy COST Action CA15138 to MA and GMF, and Fondazione Umberto Veronesi to MA.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by F. Pentimalli
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mauro Piacentini, Email: mauro.piacentini@uniroma2.it.
Gian Maria Fimia, Email: gianmaria.fimia@inmi.it.
References
- 1.Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349–64. doi: 10.1038/s41580-018-0003-4. [DOI] [PubMed] [Google Scholar]
- 3.Morishita H, Mizushima N. Diverse cellular roles of autophagy. Annu Rev Cell Dev Biol. 2019;35:453–75. doi: 10.1146/annurev-cellbio-100818-125300. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 5.Khaminets A, Behl C, Dikic I. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 2016;26:6–16. doi: 10.1016/j.tcb.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 6.Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42:297–311. doi: 10.1016/j.tibs.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 7.Esposito D, Koliopoulos MG, Rittinger K. Structural determinants of TRIM protein function. Biochem Soc Trans. 2017;45:183–91. doi: 10.1042/BST20160325. [DOI] [PubMed] [Google Scholar]
- 8.Napolitano LM, Jaffray EG, Hay RT, Meroni G. Functional interactions between ubiquitin E2 enzymes and TRIM proteins. Biochem J. 2011;434:309–19. doi: 10.1042/BJ20101487. [DOI] [PubMed] [Google Scholar]
- 9.Napolitano LM, Meroni G. TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- 10.Ozato K, Shin DM, Chang TH, Morse HC., 3rd TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8:849–60. doi: 10.1038/nri2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–51. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tocchini C, Ciosk R. TRIM-NHL proteins in development and disease. Semin Cell Dev Biol. 2015;47-48:52–9. doi: 10.1016/j.semcdb.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 13.Versteeg GA, Rajsbaum R, Sanchez-Aparicio MT, Maestre AM, Valdiviezo J, Shi M, et al. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity. 2013;38:384–98. doi: 10.1016/j.immuni.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 2011;7:e1002388. doi: 10.1371/journal.pgen.1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vunjak M, Versteeg GA. TRIM proteins. Curr Biol. 2019;29:R42–4. doi: 10.1016/j.cub.2018.11.026. [DOI] [PubMed] [Google Scholar]
- 16.Boudinot P, van der Aa LM, Jouneau L, Du Pasquier L, Pontarotti P, Briolat V, et al. Origin and evolution of TRIM proteins: new insights from the complete TRIM repertoire of zebrafish and pufferfish. PLoS One. 2011;6:e22022. doi: 10.1371/journal.pone.0022022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Aa LM, Levraud JP, Yahmi M, Lauret E, Briolat V, Herbomel P, et al. A large new subset of TRIM genes highly diversified by duplication and positive selection in teleost fish. BMC Biol. 2009;7:7. doi: 10.1186/1741-7007-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meroni G. Genomics and evolution of the TRIM gene family. Adv Exp Med Biol. 2012;770:1–9. doi: 10.1007/978-1-4614-5398-7_1. [DOI] [PubMed] [Google Scholar]
- 19.Di Malta C, Cinque L, Settembre C. Transcriptional regulation of autophagy: mechanisms and diseases. Front Cell Dev Biol. 2019;7:114. doi: 10.3389/fcell.2019.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 21.Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. Molecular definitions of autophagy and related processes. EMBO J. 2017;36:1811–36. doi: 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonioli M, Di Rienzo M, Piacentini M, Fimia GM. Emerging mechanisms in initiating and terminating autophagy. Trends Biochem Sci. 2017;42:28–41. doi: 10.1016/j.tibs.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 23.Ktistakis NT, Tooze SA. Digesting the expanding mechanisms of autophagy. Trends Cell Biol. 2016;26:624–35. doi: 10.1016/j.tcb.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 24.Levine B, Liu R, Dong X, Zhong Q. Beclin orthologs: integrative hubs of cell signaling, membrane trafficking, and physiology. Trends Cell Biol. 2015;25:533–44. doi: 10.1016/j.tcb.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–93. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 26.Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nascimbeni AC, Giordano F, Dupont N, Grasso D, Vaccaro MI, Codogno P, et al. ER-plasma membrane contact sites contribute to autophagosome biogenesis by regulation of local PI3P synthesis. EMBO J. 2017;36:2018–33. doi: 10.15252/embj.201797006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan DF, Chun MG, Vamos M, Zou H, Rong J, Miller CJ, et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59:285–97. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol. 2010;191:155–68. doi: 10.1083/jcb.201002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–50. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, et al. The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14. Autophagy. 2016;12:547–64. doi: 10.1080/15548627.2016.1140293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wold MS, Lim J, Lachance V, Deng Z, Yue Z. ULK1-mediated phosphorylation of ATG14 promotes autophagy and is impaired in Huntington’s disease models. Mol Neurodegener. 2016;11:76. doi: 10.1186/s13024-016-0141-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonioli M, Albiero F, Nazio F, Vescovo T, Perdomo AB, Corazzari M, et al. AMBRA1 interplay with cullin E3 ubiquitin ligases regulates autophagy dynamics. Dev Cell. 2014;31:734–46. doi: 10.1016/j.devcel.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 34.Hill SM, Wrobel L, Rubinsztein DC. Post-translational modifications of Beclin 1 provide multiple strategies for autophagy regulation. Cell Death Differ. 2019;26:617–29. doi: 10.1038/s41418-018-0254-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, et al. Cul3-KLHL20 ubiquitin ligase governs the turnover of ULK1 and VPS34 complexes to control autophagy termination. Mol Cell. 2016;61:84–97. doi: 10.1016/j.molcel.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 36.Nazio F, Carinci M, Valacca C, Bielli P, Strappazzon F, Antonioli M, et al. Fine-tuning of ULK1 mRNA and protein levels is required for autophagy oscillation. J Cell Biol. 2016;215:841–56. doi: 10.1083/jcb.201605089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol Cell. 2014;55:238–52. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J Mol Biol. 2019. [Epub ahead of print]. [DOI] [PubMed]
- 41.Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol Cell. 2019;76:268–85. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 42.Zhao YG, Zhang H. Autophagosome maturation: an epic journey from the ER to lysosomes. J Cell Biol. 2019;218:757–70. doi: 10.1083/jcb.201810099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McEwan DG, Richter B, Claudi B, Wigge C, Wild P, Farhan H, et al. PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 2015;17:58–71. doi: 10.1016/j.chom.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 44.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Matsui T, Jiang P, Nakano S, Sakamaki Y, Yamamoto H, Mizushima N. Autophagosomal YKT6 is required for fusion with lysosomes independently of syntaxin 17. J Cell Biol. 2018;217:2633–45. doi: 10.1083/jcb.201712058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fullgrabe J, Ghislat G, Cho DH, Rubinsztein DC. Transcriptional regulation of mammalian autophagy at a glance. J Cell Sci. 2016;129:3059–66. doi: 10.1242/jcs.188920. [DOI] [PubMed] [Google Scholar]
- 47.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–96. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin M, Liu X, Klionsky DJ. SnapShot: selective autophagy. Cell. 2013;152:368. doi: 10.1016/j.cell.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fimia GM, Kroemer G, Piacentini M. Molecular mechanisms of selective autophagy. Cell Death Differ. 2013;20:1–2. doi: 10.1038/cdd.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway O, Akpinar HA, Rogov V, Kirkin V. Selective autophagy receptors in neuronal health and disease. J Mol Biol. 2019. [Epub ahead of print]. [DOI] [PubMed]
- 51.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pickles S, Vigie P, Youle RJ. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28:R170–85. doi: 10.1016/j.cub.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hubner CA, Dikic I. ER-phagy and human diseases. Cell Death Differ. 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 54.Papandreou ME, Tavernarakis N. Nucleophagy: from homeostasis to disease. Cell Death Differ. 2019;26:630–9. doi: 10.1038/s41418-018-0266-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zechner R, Madeo F, Kratky D. Cytosolic lipolysis and lipophagy: two sides of the same coin. Nat Rev Mol Cell Biol. 2017;18:671–84. doi: 10.1038/nrm.2017.76. [DOI] [PubMed] [Google Scholar]
- 56.Hung YH, Chen LM, Yang JY, Yang WY. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 57.Gatica D, Lahiri V, Klionsky DJ. Cargo recognition and degradation by selective autophagy. Nat Cell Biol. 2018;20:233–42. doi: 10.1038/s41556-018-0037-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holdgaard SG, Cianfanelli V, Pupo E, Lambrughi M, Lubas M, Nielsen JC, et al. Selective autophagy maintains centrosome integrity and accurate mitosis by turnover of centriolar satellites. Nat Commun. 2019;10:4176. doi: 10.1038/s41467-019-12094-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villa E, Marchetti S, Ricci JE. No parkin zone: mitophagy without parkin. Trends Cell Biol. 2018;28:882–95. doi: 10.1016/j.tcb.2018.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Wilkinson S. Emerging principles of selective ER autophagy. J Mol Biol. 2019. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 61.Grumati P, Dikic I. Ubiquitin signaling and autophagy. J Biol Chem. 2018;293:5404–13. doi: 10.1074/jbc.TM117.000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pineda CT, Ramanathan S, Fon Tacer K, Weon JL, Potts MB, Ou YH, et al. Degradation of AMPK by a cancer-specific ubiquitin ligase. Cell. 2015;160:715–28. doi: 10.1016/j.cell.2015.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shukla SA, Bachireddy P, Schilling B, Galonska C, Zhan Q, Bango C, et al. Cancer-germline antigen expression discriminates clinical outcome to CTLA-4 blockade. Cell. 2018;173:624. doi: 10.1016/j.cell.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang W, Xia Z, Farre JC, Subramani S. TRIM37 deficiency induces autophagy through deregulating the MTORC1-TFEB axis. Autophagy. 2018;14:1574–85. doi: 10.1080/15548627.2018.1463120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Missiroli S, Bonora M, Patergnani S, Poletti F, Perrone M, Gafa R, et al. PML at mitochondria-associated membranes is critical for the repression of autophagy and cancer development. Cell Rep. 2016;16:2415–27. doi: 10.1016/j.celrep.2016.07.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Czerwinska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci. 2017;24:63-017–0374-4. doi: 10.1186/s12929-017-0374-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brigant Benjamin, Metzinger-Le Meuth Valérie, Rochette Jacques, Metzinger Laurent. TRIMming down to TRIM37: Relevance to Inflammation, Cardiovascular Disorders, and Cancer in MULIBREY Nanism. International Journal of Molecular Sciences. 2018;20(1):67. doi: 10.3390/ijms20010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lallemand-Breitenbach V, de The H. PML nuclear bodies: from architecture to function. Curr Opin Cell Biol. 2018;52:154–61. doi: 10.1016/j.ceb.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 69.Chen D, Feng C, Tian X, Zheng N, Wu Z. Promyelocytic leukemia restricts enterovirus 71 replication by inhibiting autophagy. Front Immunol. 2018;9:1268. doi: 10.3389/fimmu.2018.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Isakson P, Bjoras M, Boe SO, Simonsen A. Autophagy contributes to therapy-induced degradation of the PML/RARA oncoprotein. Blood. 2010;116:2324–31. doi: 10.1182/blood-2010-01-261040. [DOI] [PubMed] [Google Scholar]
- 71.Morganti C, Missiroli S, Lebiedzinska-Arciszewska M, Ferroni L, Morganti L, Perrone M, et al. Regulation of PKCbeta levels and autophagy by PML is essential for high-glucose-dependent mesenchymal stem cell adipogenesis. Int J Obes. 2019;43:963–73. doi: 10.1038/s41366-018-0167-1. [DOI] [PubMed] [Google Scholar]
- 72.Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fusco C, Mandriani B, Di Rienzo M, Micale L, Malerba N, Cocciadiferro D, et al. TRIM50 regulates Beclin 1 proautophagic activity. Biochim Biophys Acta Mol Cell Res. 2018;1865:908–19. doi: 10.1016/j.bbamcr.2018.03.011. [DOI] [PubMed] [Google Scholar]
- 74.Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, et al. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. J Cell Biol. 2015;210:973–89. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chauhan S, Kumar S, Jain A, Ponpuak M, Mudd MH, Kimura T, et al. TRIMs and Galectins globally cooperate and TRIM16 and Galectin-3 co-direct autophagy in endomembrane damage homeostasis. Dev Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Rienzo M, Antonioli M, Fusco C, Liu Y, Mari M, Orhon I, et al. Autophagy induction in atrophic muscle cells requires ULK1 activation by TRIM32 through unanchored K63-linked polyubiquitin chains. Sci Adv. 2019;5:eaau8857. doi: 10.1126/sciadv.aau8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shieh PB, Kudryashova E, Spencer MJ. Limb-girdle muscular dystrophy 2H and the role of TRIM32. Handb Clin Neurol. 2011;101:125–33. doi: 10.1016/B978-0-08-045031-5.00009-8. [DOI] [PubMed] [Google Scholar]
- 78.Di Rienzo M, Piacentini M, Fimia GM. A TRIM32-AMBRA1-ULK1 complex initiates the autophagy response in atrophic muscle cells. Autophagy. 2019;15:1674–6. doi: 10.1080/15548627.2019.1635385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mandell MA, Jain A, Kumar S, Castleman MJ, Anwar T, Eskelinen EL, et al. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. J Cell Sci. 2016;129:3562–73. doi: 10.1242/jcs.190017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kehl SR, Soos BA, Saha B, Choi SW, Herren AW, Johansen T, et al. TAK1 converts Sequestosome 1/p62 from an autophagy receptor to a signaling platform. EMBO Rep. 2019;20:e46238. doi: 10.15252/embr.201846238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Han T, Guo M, Gan M, Yu B, Tian X, Wang JB. TRIM59 regulates autophagy through modulating both the transcription and the ubiquitination of BECN1. Autophagy. 2018;14:2035–48. doi: 10.1080/15548627.2018.1491493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ribeiro CM, Sarrami-Forooshani R, Setiawan LC, Zijlstra-Willems EM, van Hamme JL, Tigchelaar W, et al. Receptor usage dictates HIV-1 restriction by human TRIM5alpha in dendritic cell subsets. Nature. 2016;540:448–52. doi: 10.1038/nature20567. [DOI] [PubMed] [Google Scholar]
- 83.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, et al. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci USA. 2013;110:6841–6. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lou J, Wang Y, Zheng X, Qiu W. TRIM22 regulates macrophage autophagy and enhances Mycobacterium tuberculosis clearance by targeting the nuclear factor-multiplicity kappaB/beclin 1 pathway. J Cell Biochem. 2018;119:8971–80. doi: 10.1002/jcb.27153. [DOI] [PubMed] [Google Scholar]
- 85.Chen L, Wang K, Long A, Jia L, Zhang Y, Deng H, et al. Fasting-induced hormonal regulation of lysosomal function. Cell Res. 2017;27:748–63. doi: 10.1038/cr.2017.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zanchetta ME, Meroni G. Emerging roles of the TRIM E3 ubiquitin ligases MID1 and MID2 in cytokinesis. Front Physiol. 2019;10:274. doi: 10.3389/fphys.2019.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pan X, Chen Y, Shen Y, Tantai J. Knockdown of TRIM65 inhibits autophagy and cisplatin resistance in A549/DDP cells by regulating miR-138-5p/ATG7. Cell Death Dis. 2019;10:429. doi: 10.1038/s41419-019-1660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barde I, Rauwel B, Marin-Florez RM, Corsinotti A, Laurenti E, Verp S, et al. A KRAB/KAP1-miRNA cascade regulates erythropoiesis through stage-specific control of mitophagy. Science. 2013;340:350–3. doi: 10.1126/science.1232398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ra EA, Lee TA, Won Kim S, Park A, Choi HJ, Jang I, et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat Commun. 2016;7:11726. doi: 10.1038/ncomms11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–20. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 91.Qiu S, Liu J, Xing F. ‘Hints’ in the killer protein gasdermin D: unveiling the secrets of gasdermins driving cell death. Cell Death Differ. 2017;24:588–96. doi: 10.1038/cdd.2017.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Frank D, Vince JE. Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 2019;26:99–114. doi: 10.1038/s41418-018-0212-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. Autophagy and Inflammation. Annu Rev Immunol. 2018;36:73–101. doi: 10.1146/annurev-immunol-042617-053253. [DOI] [PubMed] [Google Scholar]
- 94.Liu T, Tang Q, Liu K, Xie W, Liu X, Wang H, et al. TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-dependent selective autophagy. Cell Rep. 2016;16:1988–2002. doi: 10.1016/j.celrep.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 95.Xu H, Yang J, Gao W, Li L, Li P, Zhang L, et al. Innate immune sensing of bacterial modifications of Rho GTPases by the pyrin inflammasome. Nature. 2014;513:237–41. doi: 10.1038/nature13449. [DOI] [PubMed] [Google Scholar]
- 96.Deretic V, Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14:243–51. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. The pyrin Inflammasome in health and disease. Front Immunol. 2019;10:1745. doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kimura T, Jia J, Kumar S, Choi SW, Gu Y, Mudd M, et al. Dedicated SNAREs and specialized TRIM cargo receptors mediate secretory autophagy. EMBO J. 2017;36:42–60. doi: 10.15252/embj.201695081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Claude-Taupin A, Bissa B, Jia J, Gu Y, Deretic V. Role of autophagy in IL-1beta export and release from cells. Semin Cell Dev Biol. 2018;83:36–41. doi: 10.1016/j.semcdb.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Place DE, Kanneganti TD. Cell death-mediated cytokine release and its therapeutic implications. J Exp Med. 2019;216:1474–86. doi: 10.1084/jem.20181892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Gent M, Sparrer KMJ, Gack MU. TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol. 2018;5:385–405. doi: 10.1146/annurev-virology-092917-043323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Motwani M, Pesiridis S, Fitzgerald KA. DNA sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet. 2019;20:657–74. doi: 10.1038/s41576-019-0151-1. [DOI] [PubMed] [Google Scholar]
- 103.Kato K, Omura H, Ishitani R, Nureki O. Cyclic GMP-AMP as an endogenous second messenger in innate immune signaling by cytosolic DNA. Annu Rev Biochem. 2017;86:541–66. doi: 10.1146/annurev-biochem-061516-044813. [DOI] [PubMed] [Google Scholar]
- 104.Ablasser Andrea, Chen Zhijian J. cGAS in action: Expanding roles in immunity and inflammation. Science. 2019;363(6431):eaat8657. doi: 10.1126/science.aat8657. [DOI] [PubMed] [Google Scholar]
- 105.Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, et al. TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell. 2016;64:105–19. doi: 10.1016/j.molcel.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 106.Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, et al. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity. 2016;45:555–69. doi: 10.1016/j.immuni.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 107.Louis C, Burns C, Wicks I. TANK-binding kinase 1-dependent responses in health and autoimmunity. Front Immunol. 2018;9:434. doi: 10.3389/fimmu.2018.00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Herhaus L, Dikic I. Expanding the ubiquitin code through post-translational modification. EMBO Rep. 2015;16:1071–83. doi: 10.15252/embr.201540891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sparrer KMJ, Gableske S, Zurenski MA, Parker ZM, Full F, Baumgart GJ, et al. TRIM23 mediates virus-induced autophagy via activation of TBK1. Nat Microbiol. 2017;2:1543–57. doi: 10.1038/s41564-017-0017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu X, Matrenec R, Gack MU, He B. Disassembly of the TRIM23-TBK1 complex by the Us11 protein of herpes simplex virus 1 impairs autophagy. J Virol. 2019;93. 10.1128/JVI.00497-19. [DOI] [PMC free article] [PubMed]
- 111.Rhodes DA, Isenberg DA. TRIM21 and the function of antibodies inside cells. Trends Immunol. 2017;38:916–26. doi: 10.1016/j.it.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 112.Yang Q, Liu TT, Lin H, Zhang M, Wei J, Luo WW, et al. TRIM32-TAX1BP1-dependent selective autophagic degradation of TRIF negatively regulates TLR3/4-mediated innate immune responses. PLoS Pathog. 2017;13:e1006600. doi: 10.1371/journal.ppat.1006600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Niida M, Tanaka M, Kamitani T. Downregulation of active IKK beta by Ro52-mediated autophagy. Mol Immunol. 2010;47:2378–87. doi: 10.1016/j.molimm.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sun SC. The non-canonical NF-kappaB pathway in immunity and inflammation. Nat Rev Immunol. 2017;17:545–58. doi: 10.1038/nri.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Napetschnig J, Wu H. Molecular basis of NF-kappaB signaling. Annu Rev Biophys. 2013;42:443–68. doi: 10.1146/annurev-biophys-083012-130338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ganser-Pornillos BK, Pornillos O. Restriction of HIV-1 and other retroviruses by TRIM5. Nat Rev Microbiol. 2019;17:546–56. doi: 10.1038/s41579-019-0225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rakebrandt N, Lentes S, Neumann H, James LC, Neumann-Staubitz P. Antibody- and TRIM21-dependent intracellular restriction of Salmonella enterica. Pathog Dis. 2014;72:131–7. doi: 10.1111/2049-632X.12192. [DOI] [PubMed] [Google Scholar]
- 118.Jena KK, Kolapalli SP, Mehto S, Nath P, Das B, Sahoo PK, et al. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018;37. 10.15252/embj.201798358. [DOI] [PMC free article] [PubMed]
- 119.Chen L, Brewer MD, Guo L, Wang R, Jiang P, Yang X. Enhanced degradation of misfolded proteins promotes tumorigenesis. Cell Rep. 2017;18:3143–54. doi: 10.1016/j.celrep.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Khan MM, Strack S, Wild F, Hanashima A, Gasch A, Brohm K, et al. Role of autophagy, SQSTM1, SH3GLB1, and TRIM63 in the turnover of nicotinic acetylcholine receptors. Autophagy. 2014;10:123–36. doi: 10.4161/auto.26841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Overå Katrine Stange, Garcia-Garcia Juncal, Bhujabal Zambarlal, Jain Ashish, Øvervatn Aud, Larsen Kenneth Bowitz, Deretic Vojo, Johansen Terje, Lamark Trond, Sjøttem Eva. TRIM32, but not its muscular dystrophy-associated mutant, positively regulates and is targeted to autophagic degradation by p62/SQSTM1. Journal of Cell Science. 2019;132(23):jcs236596. doi: 10.1242/jcs.236596. [DOI] [PMC free article] [PubMed] [Google Scholar]