Abstract
Mitochondria are highly dynamic organelles and respond to stress by changing their fission-fusion cycle, undergoing mitophagy, or releasing apoptotic proteins to initiate cell death. The molecular mechanisms that sense different stresses and coordinate distinct effectors still await full characterization. Here, we show that PGAM5, which exists in an equilibrium between dimeric and multimeric states, dephosphorylates BCL-xL to inhibit apoptosis or FUNDC1 to activate mitofission and mitophagy in response to distinct stresses. In vinblastine-treated cells, PGAM5 dephosphorylates BCL-xL at Ser62 to restore BCL-xL sequestration of BAX and BAK and thereby resistance to apoptosis. Selenite-induced oxidative stress increases the multimerization of PGAM5, resulting in its dissociation from BCL-xL, which causes increased BCL-xL phosphorylation and apoptosis. Once freed, the more multimeric and active PGAM5 dephosphorylates FUNDC1 to initiate mitofission and mitophagy. The reciprocal interaction of PGAM5 with FUNDC1 and BCL-xL, controlled by PGAM5 multimerization, serves as a molecular switch between mitofission/mitophagy and apoptosis.
Subject terms: Cell biology, Autophagy
Introduction
Mitochondria are bioenergetic and biosynthetic organelles which play a central role in cell signaling and homeostasis [1], Mitochondria are also a major source and target of reactive oxygen species [2, 3]. In response to bioenergetic demand and oxidative stress, mitochondria change their morphology through fission and fusion. The damaged or superfluous mitochondria are selectively removed by mitophagy. If the damage is severe and the damaged mitochondria cannot be effectively removed, the mitochondrial outer membrane will be permeabilized and the released mitochondrial proteins will trigger apoptosis. Mitochondrial outer membrane permeabilization (MOMP) is regulated by BCL-2-family proteins including the anti-apoptotic BCL-2 and BCL-xL and the pro-apoptotic BAX and BAK as well as the BH3-only proteins [4]. BCL-2/BCL-xL inhibit MOMP by sequestering either the BH3-only proteins to prevent BAX/BAK activation or the activated BAX/BAK to prevent oligomeric pore formation in the MOM.
Mitochondria are highly dynamic organelles that undergo cycles of fission and fusion to maintain mitochondrial networks and inheritance. The mitochondria segregated by fission are either fused back to the network, or engulfed by mitophagy if they were damaged. Two distinct mitophagy pathways that have been described are the PINK1/Parkin and the mitophagy receptors-mediated pathways [5]. In particular, the mitophagy receptors NIX, FUNDC1 and PHB2 interact with the autophagosome protein LC3 to mediate mitophagy [6–8].
As a mitochondrial Ser/Thr phosphatase, PGAM5 participates in multiple processes that regulate mitochondrial homeostasis. It dephosphorylates Drp1 to promote mitochondrial fission and necrosis in response to TNF-α [9]. It binds AIF to trigger mitophagic cell death [10]. Its deficiency suppresses the PINK1-mediated mitochondrial degeneration in Drosophila [11] and causes a Parkinson’s-like phenotype and resistance to metabolic stress in mammals [12, 13]. We have shown that PGAM5 dephosphorylates FUNDC1 to promote mitophagy in response to mitochondrial stresses [14]. PGAM5 also could interact with anti-apoptotic protein BCL-xL [15]. Interestingly, the function of BCL-xL is regulated by phosphorylation of many Ser/Thr residues, including Ser49 by PLK3 [16], Thr47 and Thr115 by JNK [17], Ser73 by CDK2 [18] and Ser14 by MST1 [19]. Here, we show that different stresses regulate the multimeric state of PGAM5, which in turn regulates its interaction with BCL-xL and FUNDC1, and their phosphorylation and function. We thus reveal a molecular switch mediated by the reciprocal PGAM5 interaction with and modification of BCL-xL and FUNDC1 that controls mitochondrial morphology, mitophagy and apoptosis.
Results
PGAM5 dephosphorylates BCL-xL at Ser62
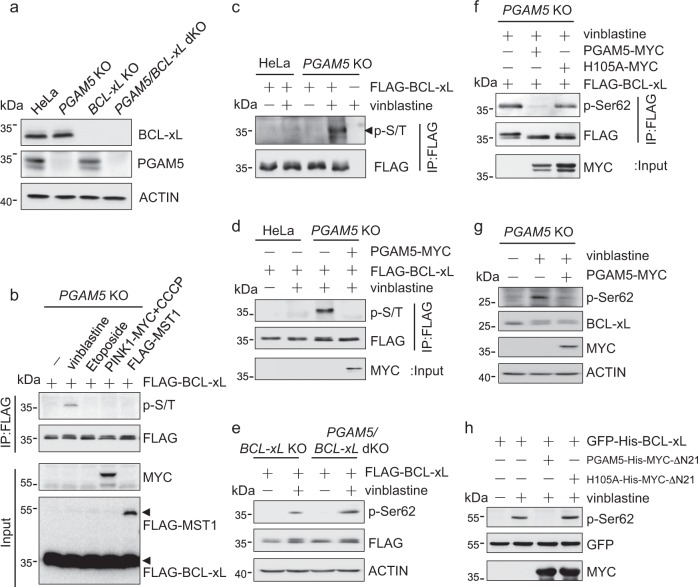
PGAM5 dephosphorylates BCL-xL peptides [20] and interacts with BCL-xL protein [15]. To determine whether PGAM5 is able to dephosphorylate BCL-xL protein in cells, we generated BCL-xL knockout, PGAM5 knockout and BCL-xL/PGAM5 double knockout HeLa cell lines (Fig. 1a). FLAG-BCL-xL expressed in PGAM5 knockout cells was phosphorylated after treatment with vinblastine, a chemotherapy drug inducing mitotic arrest and BCL-xL phosphorylation (Fig. 1b, c) [21, 22]. Without vinblastine BCL-xL phosphorylation was undetectable even when the cells co-expressed the kinases PINK1 or MST1 that could phosphorylate BCL-xL [16, 19, 23]. Interestingly, co-expressing PGAM5-MYC diminished the vinblastine-induced BCL-xL phosphorylation (Fig. 1d), suggesting that PGAM5 dephosphorylates BCL-xL.
Fig. 1.
The vinblastine-induced phosphorylation of BCL-xL at Ser62 is reversed by the mitochondrial phosphatase PGAM5. a Western blots of lysates from wild-type, PGAM5 knockout, BCL-xL knockout or PGAM5/BCL-xL double knockout HeLa cells using the indicated antibodies. b PGAM5 knockout HeLa cells expressing FLAG-BCL-xL with or without co-expression of PINK1-MYC or FLAG-MST1 were treated with vinblastine (500 nM), Etoposide (50 uM), CCCP (50 μM) or the vehicle for 24 h. The cells were lysed for immunoprecipitation (IP) with anti-FLAG antibody and western blotting with antibodies recognizing the FLAG or MYC epitope or phospho-Ser/Thr (p-S/T). c Wild-type or PGAM5 knockout HeLa cells expressing FLAG-BCL-xL were treated with vinblastine or the vehicle for 24 h, lysed and immunoprecipitated with anti-FLAG antibody. Total and phospho-BCL-xL were detected by western blotting with anti-FLAG or anti-p-S/T antibody, respectively. d Wild-type or PGAM5 knockout HeLa cells expressing PGAM5-MYC and/or FLAG-BCL-xL were treated with vinblastine or the vehicle. The FLAG-BCL-xL in the cell lysate was enriched by IP, and the level of phospho-BCL-xL was determined by western blotting with the p-S/T antibody. e BCL-xL single or PGAM5/BCL-xL double knockout HeLa cells expressing FLAG-BCL-xL were treated with vinblastine or the vehicle for 24 h, lysed and western blotted with the indicated antibodies including one that recognizes the phospho-Serine62 of BCL-xL (p-Ser62). f PGAM5 knockout HeLa cells expressing PGAM5-MYC or the phosphatase-dead H105A mutant, and/or FLAG-BCL-xL were treated with vinblastine, lysed, immunoprecipitated with anti-FLAG antibody, and western blotted with the p-Ser62 antibody to detect the level of BCL-xL phosphorylation at Ser62. g PGAM5 knockout HeLa cells with or without PGAM5-MYC expression were treated with vinblastine and lysed. The endogenous phospho-Ser62-BCL-xL was detected by western blotting using the p-Ser62 antibody. Note that the endogenous BCL-xL in the vinblastine-treated cells was detected as double bands by western blotting using the anti-BCL-xL antibody. Although the nature of the double bands is unknown, it is unlikely due to the Ser62 phosphorylation, which was detected by the anti-phospho-Ser62 antibody as an extra-band above the double bands. h PGAM5/BCL-xL double knockout HeLa cells expressing GFP-His-BCL-xL were treated with vinblastine and lysed. The immunoprecipitated GFP-His-BCL-xL was incubated with purified PGAM5-His-MYC-ΔN21 protein or the phosphatase-dead H105A mutant. The phospho-Ser62-BCL-xL was detected by western blotting using the p-Ser62 antibody
Earlier studies suggest that vinblastine induces BCL-xL phosphorylation at Ser62 via JNK or CDK1 [21, 22]. Using an antibody specific to the phospho-Ser62 of BCL-xL (Fig. S1a), we found that the phosphorylation of Ser62 of FLAG-BCL-xL was increased in PGAM5 knockout HeLa cells (Figs. 1e and S1b) or PGAM5 knockdown HepG2 cells (Fig. S1c). Moreover, wild-type PGAM5, but not the phosphatase-dead mutant (H105A), reversed the vinblastine-induced phosphorylation of FLAG-BCL-xL at Ser62 (Figs. 1f and S1d). Wild-type PGAM5 also blocked the phosphorylation of endogenous BCL-xL at Ser62 in vinblastine-treated cells, suggesting that PGAM5 targets the phospho-Ser62 of BCL-xL (Figs. 1g and S1e). To further verify that the Ser62-phosphorylated BCL-xL is a substrate of PGAM5, we isolated GFP-His-BCL-xL from PGAM5/BCL-xL knockout cells after vinblastine treatment and incubated it with purified PGAM5-His-MYC-ΔN21 protein or the phosphatase-dead mutant. The recombinant phosphatase completely dephosphorylated the BCL-xL at Ser62, whereas the phosphatase-dead mutant did not (Fig. 1h). In contrast to BCL-xL, ectopically expressed PGAM5 neither interacted with BCL-2 (Fig. S1f, g) nor reduced the vinblastine-induced BCL-2 phosphorylation (Fig. S1h). Our results demonstrate that vinblastine induces BCL-xL phosphorylation at Ser62, which is directly and specifically reversed by the phosphatase PGAM5.
Dephosphorylation of BCL-xL by PGAM5 enhances its interaction with pro-apoptotic BAX/BAK proteins and anti-apoptotic activity
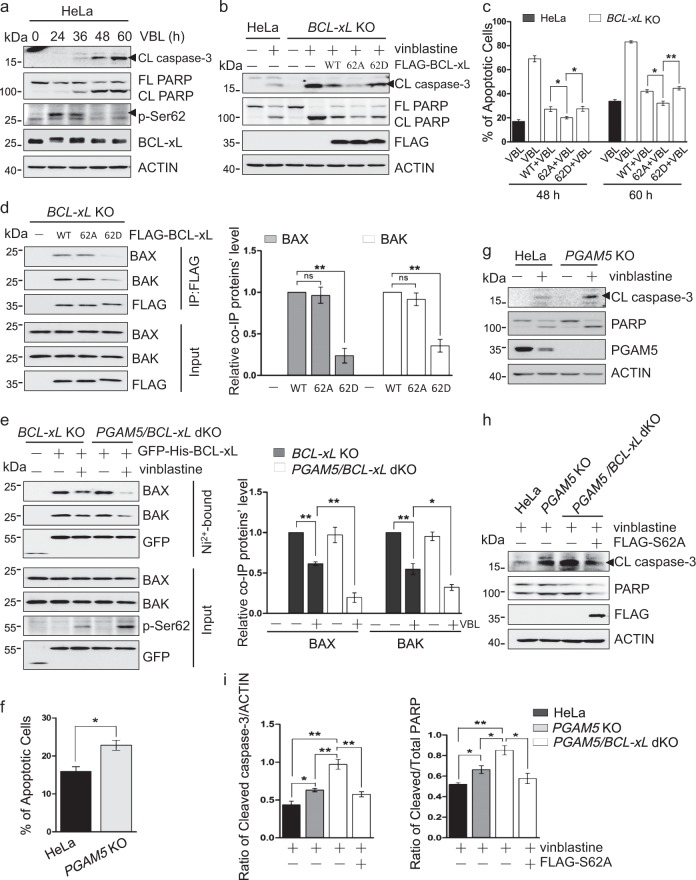
To determine the impact of Ser62 phosphorylation on BCL-xL anti-apoptotic activity, we performed a temporal analysis of HeLa cells following treatment with vinblastine. Vinblastine induced BCL-xL Ser62 phosphorylation from 24 to 36 hours, whereas caspase-3 and PARP cleavage began to increase at 36 hours (Figs. 2a and S2a). This suggests that BCL-xL phosphorylation occurs prior to caspase activation. As expected, a pan-caspase inhibitor Z-VAD-FKM inhibited vinblastine-induced apoptosis (Fig. S2b, c). To address the functional significance of the BCL-xL phosphorylation, we expressed FLAG-tagged BCL-xL, phosphorylation mimic mutant (S62D) or nonphosphorylatable mutant (S62A) in BCL-xL knockout cells. The caspase-3 or PARP cleavage data show that BCL-xL knockout cells are more sensitive to apoptotic drug vinblastine than wild-type cells (Figs. 2b and S2d). Expression of nonphosphorylatable BCL-xL inhibited the apoptosis, whereas expression of wild-type or phosphorylation mimic BCL-xL had lesser effects (Figs. 2b and S2d), which were further confirmed by the Annexin V and PI staining at two different time points (Fig. 2c). A recent report showed that the phosphorylation of BCL-xL at Ser62 lowers its binding affinity for pro-apoptotic BCL-2-family proteins [24]. As expected, less pro-apoptotic BAX/BAK bound to the phosphorylation mimic mutant compared to the wild-type protein or the nonphosphorylatable mutant (Fig. 2d). In PGAM5 knockout cells, vinblastine induced stronger BCL-xL phosphorylation at Ser62, and consequently the interaction between BCL-xL and BAX/BAK was significantly reduced compared to the wild-type cells (Fig. 2e). The PGAM5 knockout cells were more sensitive to vinblastine as more BAX was localized to mitochondria and more cytochrome c was released to the cytosol, resulting in more apoptosis (Figs. S2e, f and 2f, g). Importantly, BCL-xL deficiency made the cells more sensitive to vinblastine, whereas ectopic expression of nonphosphorylatable mutant BCL-xL significantly reduced the level of vinblastine-induced apoptosis (Fig. 2h, i).
Fig. 2.
Dephosphorylation of BCL-xL by PGAM5 increases its anti-apoptotic activity. a HeLa cells were treated with vinblastine (VBL) for the indicated times, lysed and analyzed by western blotting with the indicated antibodies. CL caspase-3, cleaved caspase-3; FL PARP, full-length PARP; CL PARP, cleaved PARP. b HeLa or BCL-xL knockout HeLa cells with or without FLAG-BCL-xL, FLAG-BCL-xL-S62A, or FLAG-BCL-xL-S62D expression were treated with vinblastine for 48 h, lysed and analyzed by western blotting with the indicated antibodies. (c) HeLa or BCL-xL knockout HeLa cells expressing FLAG-BCL-xL or its mutants were treated with vinblastine for indicated times. After treatment, the cells were collected and stained with FITC-annexin V and propidium iodide (PI), then analyzed by flow cytometry. The percentages of apoptotic cells (annexin V+/PI− and annexin V+/PI+) were calculated. Data are the mean ± SEM of three independent experiments. **p < 0.01; *p < 0.05. d BCL-xL knockout HeLa cells were transfected with FLAG-tagged wild-type BCL-xL (WT) or the S62A or S62D mutant for 24 h, lysed and immunoprecipitated with anti-FLAG antibody. The co-immunoprecipitated endogenous BAX and BAK proteins were detected by western blotting. Right: densitometric quantification of co-immunoprecipitated BAX or BAK. Normalized to the bands from the wild type FLAG-BCL-xL expressing samples. Data are the mean ± SEM of three experiments. **p < 0.01. e BCL-xL or PGAM5/BCL-xL knockout HeLa cells were transfected with GFP-His-BCL-xL for 24 h, treated with vinblastine for 24 h, and lysed. The GFP-His-BCL-xL protein was isolated using Ni2+-chelating beads, eluted, and analyzed by western blotting with the indicated antibodies to detect the GFP-His-BCL-xL and the co-isolated BAX and BAK. Right: densitometric quantification of co-immunoprecipitated BAX or BAK. Normalized to the bands from the GFP-His-BCL-xL expressing untreated BCL-xL knockout HeLa cells samples. Data are the mean ± SEM of three experiments. *p < 0.05; **p < 0.01. f Wild type or PGAM5 knockout HeLa cells were treated with 500 nM vinblastine for 48 h, then stained with annexin V and propidium iodide (PI), and analyzed by flow cytometry. Data are the mean ± SEM of three experiments. *p < 0.05. g HeLa or PGAM5 knockout HeLa cells were treated without or with 500 nM vinblastine for 48 h, lysed and analyzed by western blotting with the indicated antibodies. h HeLa, PGAM5 knockout or PGAM5/BCL-xL double knockout cells, transfected with FLAG-BCL-xL-S62A if indicated, were treated with vinblastine for 48 h, lysed and analyzed by western blotting with the indicated antibodies. i The ratio of cleaved caspase-3 to ACTIN and the ratio of cleaved PARP to total PARP in (h) were calculated, respectively. Data are the mean ± SEM of three experiments. *p < 0.05; **p < 0.01
One implication of our findings is that although PGAM5 can inhibit vinblastine-induced apoptosis by reactivating BCL-xL using its phosphatase activity, it may promote cell death under other stress conditions through other proteins [25, 26]. We monitored PARP cleavage in cells treated with another apoptotic stimulus, H2O2, or staurosporine. The results revealed that PGAM5 knockout protected cells from H2O2 or staurosporine, but not vinblastine (Fig. S2g, h), which suggests that PGAM5 is important for protecting cells stressed by BCL-xL phosphorylation, but not other stressors. Together, our data show that the dephosphorylation of BCL-xL by PGAM5 augments the anti-apoptosis activity of BCL-xL by increasing its binding to BAX/BAK.
The interaction with PGAM5 is critical for BCL-xL dephosphorylation
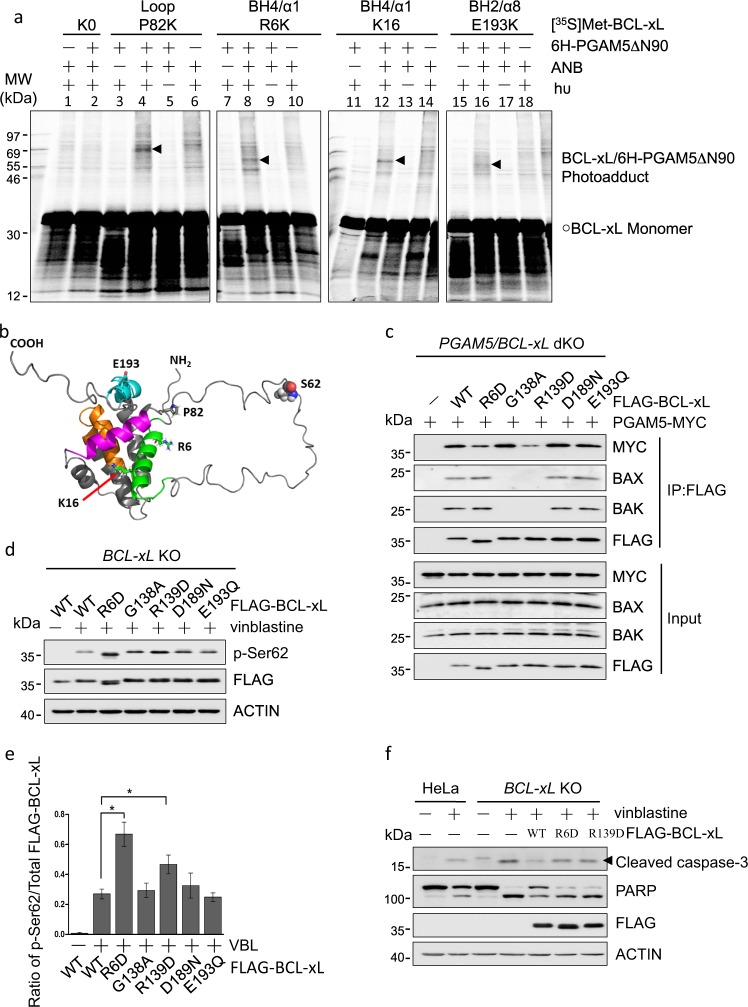
To determine whether BCL-xL dephosphorylation depends on the physical interaction with PGAM5 and the PGAM5-binding site in BCL-xL, we used a photocrosslinking approach to determine which residues in BCL-xL are near PGAM5 when the proteins interact. We synthesized radioactive BCL-xL proteins, each with a photoreactive lysine (εANB-Lys) at a specific position. We incubated each ANB-labeled BCL-xL protein with a His6-tagged truncated PGAM5 protein (6H-PGAM5ΔN90) [27], and photolyzed the sample to activate the photoprobe, which can then form a covalent linkage with any PGAM5 atoms that are in close proximity. The resulting radioactive BCL-xL-PGAM5 photoadduct was separated from the unreacted BCL-xL protein using SDS-PAGE and visualized by phosphor-imaging. When the εANB-Lys was used to replace P82 in the disordered loop between α-helices 1 and 2 (α1 and α2) where the phosphorylation site (S62) is located, a BCL-xL-PGAM5-specific photoadduct with the molecular weight close to the heterodimer was detected (Fig. 3a, lane 4). This photoadduct was not formed when either the ANB probe, the light (hυ) or the 6H-PGAM5ΔN90 was omitted (lanes 3, 5, or 6), or when a BCL-xL mutant not containing any lysine including the photoreactive lysine was used (lane 2). The BCL-xL-PGAM5-specific photoadduct was also formed when the R6 or K16 in the BH4 domain/α1 or the E193 in the BH2 domain/α8 was replaced by εANB-Lys (Fig. 3a, lanes 8, 12, and 16) but not in the control reactions that lack the ANB probe, the light or the 6H-PGAM5ΔN90. These data suggest that the PGAM5 binding site in BCL-xL is not limited to the disordered loop where the substrate phospho-S62 is located but extends to the folded regions including the BH domains (Fig. 3b). Moreover, PGAM5 contains a putative BH3 domain that may interact with the canonical BH3-binding groove in BCL-xL [28], and the BH3 domain in BCL-xL is critical to the interaction [29]. Therefore, we propose a model for BCL-xL/PGAM5 binding (Figs. 3b and S3a). Thus, the phospho-S62 in the disordered loop of BCL-xL binds to the active site in the PGAM5 phosphatase; the canonical BH3-binding groove formed by the BH1 to 3 domains binds to the BH3 domain of PGAM5; and the BH4 domain binds to an unknown region of PGAM5.
Fig. 3.
The interaction of PGAM5 with BCL-xL is critical for dephosphorylation of BCL-xL. a Photocrosslinking reaction of [35S]Met-labeled BCL-xL proteins, each with the indicated single Lys in the indicated domain replaced by an εANB-Lys, to His6-tagged PGAM5 protein lacking the N-terminal 90 residues (6H-PGAM5ΔN90). The control reactions omitted the ANB probe, the light (hυ) that activates the ANB probe, or the PGAM5 protein. Cross-linked products were analyzed by SDS-PAGE, alongside with another control reaction containing a Lys-null BCL-xL protein (K0). Radioactive proteins on the gel were detected by phosphor-imaging. The circle indicates the [35S]Met-labeled BCL-xL monomer and arrowheads indicate the BCL-xL/6H-PGAM5ΔN90 photoadducts. The molecular weights (MW) of radioactive protein standards are indicated in kDa on the left side. b The BCL-xL structure, generated from PDB entry 1LXL [39] using the PyMOL program. The NH2 and COOH termini are labeled. The BH1, 2, 3 and 4 domains are colored in orange, cyan, magenta and green, respectively. The residues that were replaced by εANB-Lys in the photocrosslinking experiment are shown in stick form. The residue S62, which is located in the disordered loop and can be dephosphorylated by PGAM5, is shown in sphere form. c PGAM5/BCL-xL double knockout HeLa cell lines were co-transfected with PGAM5-MYC and FLAG-BCL-xL (WT) or the arginine-6 to aspartic acid (R6D), glycine-138 to alanine (G138A), arginine-139 to aspartic acid (R139D), aspartic acid-189 to asparagine (D189N), or glutamic acid-193 to glutamine (E193Q) mutant as indicated for 24 h. The cells were lysed and immunoprecipitated with anti-FLAG antibody. Co-immunoprecipitated PGAM5-MYC and endogenous BAX and BAK were detected by immunoblotting. d BCL-xL knockout HeLa cells were transfected with FLAG-BCL-xL or the indicated mutants for 24 h, treated with 500 nM vinblastine for 24 h, lysed and analyzed by immunoblotting with the indicated antibodies. e The ratio of p-Ser62 to total WT or mutant FLAG-BCL-xL were calculated from the respective band intensities in the blots in (d). Data are the mean ± SEM of three experiments. *p < 0.05. f HeLa or BCL-xL knockout HeLa cells expressing FLAG-BCL-xL (WT, R6D or R139D) were treated with vinblastine for 48 h, lysed and analyzed by immunoblotting with the indicated antibodies
To test our model, we mutated residues within the potential PGAM5-binding sites in BCL-xL: R6D in BH4; G138A and R139D in BH1; and D189N and E193Q in BH2 domains. Immunoprecipitation analysis of cells expressing each mutant revealed that R6D and R139D mutations weakened the BCL-xL interaction with PGAM5, while other mutations did not (Figs. 3c and S3b). In vinblastine-treated cells, more S62-phosphorylation was detected for R6D and R139D mutants than wild-type BCL-xL or other mutants (Fig. 3d, e), suggesting that BCL-xL interaction with PGAM5 is important for the dephosphorylation by PGAM5. Consistent with our previous report [30], G138A or R139D mutation in the BH3-binding groove abolished the BCL-xL binding to BAX/BAK in untreated cells, whereas R6D mutation did not (Fig. 3c). Vinblastine treatment caused more caspase activation in cells expressing R6D than those expressing wild-type BCL-xL (Figs. 3f and S3c). This is consistent with lower levels of BAX/BAK binding to R6D mutant, which was caused by higher levels of S62-phosphorylation of the mutant than the wild-type BCL-xL (Fig. S3d, e). Furthermore, similar levels of caspase activation occurred in vinblastine-treated R6D or R139D-expressing cells (Fig. 3f). Therefore, the interaction of PGAM5 with BCL-xL is critical for BCL-xL dephosphorylation and anti-apoptotic function.
Selenite increases PGAM5 multimerization to decrease its interaction with and dephosphorylation of BCL-xL
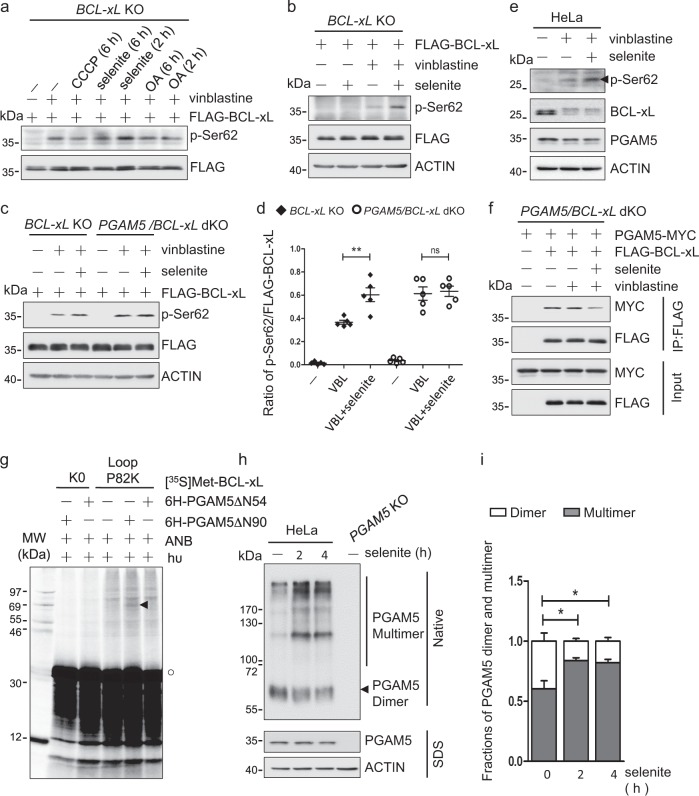
Although mitotic arrest increases BCL-xL phosphorylation, thus reducing its anti-apoptotic activity, PGAM5 binds BCL-xL and dephosphorylates it to counter this stress and protect cells. However, an additional stress may tip the balance to augment cell death. Indeed, selenite, a reagent that targets mitochondria and triggers ROS production, enhanced the vinblastine-induced phosphorylation of BCL-xL (Figs. 4a and S4a), although selenite alone did not induce BCL-xL phosphorylation (Fig. 4b). Moreover, selenite failed to enhance the phosphorylation of BCL-xL in the absence of PGAM5, suggesting that selenite inhibited the dephosphorylation of BCL-xL by PGAM5 (Fig. 4c, d). In accordance with the increased phosphorylation of BCL-xL, less BAX/BAK was bound with BCL-xL in selenite-treated cells (Fig. S4b, c). Short-term treatment with selenite affected neither the protein level of BCL-xL and PGAM5 in cells nor the PGAM5 phosphatase activity in vitro (Figs. 4e and S4d). However, co-IP analysis revealed that the interaction between BCL-xL and PGAM5 was significantly attenuated by selenite treatment (Figs. 4f and S4e).
Fig. 4.
Selenite induces PGAM5 multimerization to inhibit PGAM5 interaction with and dephosphorylation of BCL-xL. a BCL-xL knockout HeLa cells expressing FLAG-BCL-xL were treated with 500 nM vinblastine for 24 h and 10 μM CCCP, 5 μM selenite or 50 nM oligomycin and 500 nM antimycin (OA) for the indicated times, lysed and analyzed by immunoblotting with the indicated antibodies. b BCL-xL knockout HeLa cells expressing FLAG-BCL-xL were treated with 500 nM vinblastine for 24 h and/or 5 μM selenite for 2 h, lysed and analyzed by the indicated antibodies. c BCL-xL and PGAM5/BCL-xL knockout HeLa cells expressing FLAG-BCL-xL were treated without or with 5 μM selenite for 2 h and 500 nM vinblastine for 24 h, lysed and analyzed by the indicated antibodies. d The ratio of p-Ser62 to total FLAG-BCL-xL were calculated from the respective band intensities in the respective blots in (c) measured by ImageJ. Data from five experiments and the mean ± SEM are shown. **p < 0.01. e HeLa cells were treated with or without selenite for 2 h and vinblastine for 24 h, lysed and analyzed by immunoblotting with the indicated antibodies. f PGAM5/BCL-xL double knockout HeLa cells expressing FLAG-BCL-xL and/or PGAM5-MYC were treated with vinblastine (500 nM, 24 h) and selenite (5 μM, 2 h) as indicated, lysed and immunoprecipitated with anti-FLAG antibody. The co-precipitated PGAM5-MYC was detected by immunoblotting with anti-MYC antibody. g Photocrosslinking reaction of a [35S]Met-BCL-xL protein, with the single Lys (P82K) in the loop domain replaced by an εANB-Lys, to a 6H-PGAM5 protein lacking the N-terminal 54 or 90 residues (ΔN54 or ΔN90). The products were analyzed by SDS-PAGE, alongside control reactions containing a Lys-null BCL-xL protein (K0) or omitting the PGAM5 protein. Radioactive proteins on the gel were detected by phosphor-imaging. The circle indicates the [35S]Met-labeled BCL-xL monomer and the arrowhead indicates the BCL-xL/6H-PGAM5ΔN90 photoadduct. Molecular weights (MW) of protein standards are shown in kDa on the left side. h HeLa and PGAM5 knockout HeLa cells were treated with or without selenite for the indicated times, lysed, analyzed by native PAGE or denaturing (SDS) PAGE and immunoblotted with the indicated antibodies. i The fractions of PGAM5 dimer and multimer were determined from the intensities of the respective bands in the native gel immunoblot in (h) measured by ImageJ. Data are the mean ± SEM of three experiments. *p < 0.05
Interestingly, the interaction of BCL-xL and PGAM5 still decreased in the selenite-treated cells even without vinblastine (Fig. S4f). This implies that mitochondrial oxidative stress may alter the BCL-xL/PGAM5 interaction independent of BCL-xL phosphorylation. Structural analyses show that PGAM5 can transform from a dimer to a dodecamer [20, 27, 31]. Intriguingly, BCL-xL underwent photocrosslinking to PGAM5ΔN90, which forms homodimers, but not to PGAM5ΔN54, which forms higher-order multimers (Figs. 4g and S4g). PGAM5ΔN54, but not ΔN90, contains a highly conserved motif, WDPNWD, located in the N-terminal region of PGAM5 and required for assembly of the dodecameric complex in vitro [20, 27]. Using a WDPNWD-deletion mutant to verify whether this motif is important for PGAM5 oligomerization in cells, we found that wild-type PGAM5 formed dimers and several higher-order multimers, whereas the mutant is detected only as monomers and dimers (Fig. S4h). Furthermore, selenite enhanced the homotypic interaction of PGAM5, as more PGAM5 multimers and fewer dimers were detected in the selenite-treated cells compared to the control cells (Fig. 4h, i). Together, these results suggest a model in which selenite increases PGAM5 multimerization, reducing its association with and dephosphorylation of BCL-xL.
PGAM5 dissociates from BCL-xL to promote mitochondrial fission and mitophagy through the FUNDC1 pathway
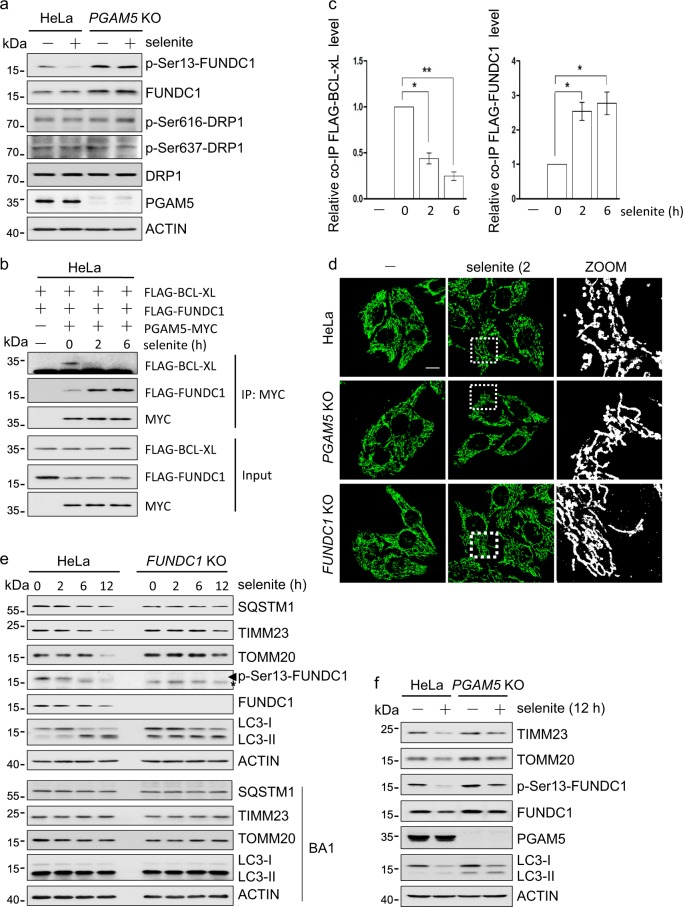
We reported that PGAM5 could dephosphorylate FUNDC1 to promote mitochondrial fission and mitophagy, and this was inhibited by BCL-xL in the absence of stress [14, 29, 32]. Here, we observed that selenite significantly enhanced the dephosphorylation of FUNDC1 at Ser13, which was blocked by PGAM5 knockout (Figs. 5a and S5a). In comparison, neither selenite nor PGAM5 knockout affected the phosphorylation of DRP1 at Ser637 and Ser616 (Figs. 5a and S5a). In the presence of selenite, the PGAM5-BCL-xL interaction decreased, whereas the PGAM5-FUNDC1 interaction increased (Fig. 5b, c). As expected, dephosphorylation of FUNDC1 caused mitochondrial localization of DRP1 as well as mitochondrial fission (mitofission) shortly after selenite addition, and both were blocked by FUNDC1 knockout (Figs. 5d and S5d, b, c). Similarly, PGAM5 knockout prevented mitofission following selenite treatment (Figs. 5d and S5d), suggesting that the selenite-induced mitofission is associated with the dephosphorylation of FUNDC1 by PGAM5.
Fig. 5.
PGAM5 dephosphorylates FUNDC1 to regulate mitochondrial fission and mitophagy in response to selenite. a HeLa and PGAM5 knockout HeLa cells were treated with selenite or vehicle for 2 h, lysed and analyzed by immunoblotting with the indicated antibodies. b HeLa cells were transfected with FLAG-BCL-xL, FLAG-FUNDC1 and PGAM5-MYC, treated with selenite for the indicated times, lysed and immunoprecipitated with anti-MYC antibody. The co-precipitated FLAG-BCL-xL and FLAG-FUNDC1 were detected by immunoblotting with anti-FLAG antibody. c Densitometric quantification of co-immunoprecipitated FLAG-BCL-xL or FLAG-FUNDC1 in (b). Normalized to the band from the untreated samples. Data are the mean ± SEM of three experiments. *p < 0.05; **p < 0.01. d HeLa and FUNDC1 or PGAM5 knockout HeLa cells were treated with selenite for 2 h, fixed and immunostained with a fluorescent dye-conjugated antibody against the mitochondrial protein cytochrome c (green). Confocal fluorescence images of the cells are shown (scale bar, 10 μm.) e HeLa and FUNDC1 knockout HeLa cells were treated with 5 μM selenite in the absence or presence of bafilomycin A1 (BA1) for the indicated times, lysed and analyzed by immunoblotting with the indicated antibodies. Asterisk indicates a nonspecific band detected by the anti-Ser13-FUNDC1 antibody. f HeLa and PGAM5 knockout HeLa cells were treated with 5 μM selenite or vehicle for 12 h, lysed and analyzed by immunoblotting with the indicated antibodies
Since FUNDC1 is a mitophagy receptor [7], we examined the role of FUNDC1 in selenite-induced mitophagy. The mitochondrial proteins TIMM23 and TOMM20 and mitophagy adaptor SQSTM1 were degraded when cells were treated with selenite for 12 hours. Their degradation, a biochemical hallmark of mitophagy, was reduced by FUNDC1 knockout (Figs. 5e and S5e) or by bafilomycin A1, a lysosomal ATPase inhibitor (Fig. 5e). Similarly, PGAM5 knockout prevents selenite-induced FUNDC1 dephosphorylation and mitophagy (Figs. 5f and S5f). These results indicate that in response to selenite, the PGAM5 dimer dissociates from BCL-xL, multimerizes, and enhances mitofission and mitophagy through dephosphorylation of FUNDC1.
Reciprocal interaction of PGAM5 with BCL-xL and FUNDC1 dictates mitochondrial and cellular fate
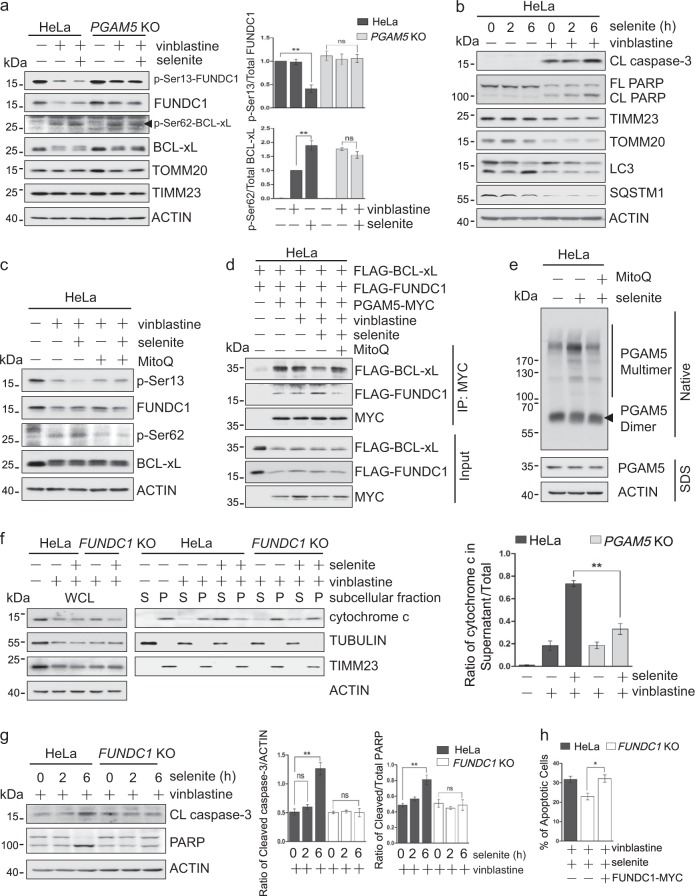
We have shown that PGAM5 dephosphorylates BCL-xL at Ser62 to enhance its interaction with pro-apoptotic BAX/BAK, thereby preventing apoptosis. On the other hand, PGAM5 dephosphorylates FUNDC1 to promote mitophagy, an early response to mitochondrial stress that can also prevent apoptosis. A fundamental question is whether the reciprocal interaction of PGAM5 with BCL-xL and FUNDC1 serves as a molecular switch to determine mitochondrial and cellular fate in response to mitochondrial stress. We took further advantage of vinblastine and selenite to manipulate the protein phosphorylation status. FUNDC1, which is phosphorylated by CK2 in untreated cells at Ser13 [14], was dephosphorylated with selenite and/or vinblastine treatment (Fig. 6a). In contrast, the dephosphorylation of BCL-xL, which is phosphorylated at Ser62 in vinblastine-treated cells, was inhibited by selenite (Fig. 6a). Moreover, apoptotic markers were increased in cells treated with a combination of selenite and vinblastine (Figs. 6b and S6a). Previous reports suggest that selenite perturbs the redox system and generates ROS, leading to mitofission, mitophagy and apoptosis [32, 33]. To test whether mitochondrial ROS (mito-ROS) is important for the selenite-modulated dephosphorylation of BCL-xL or FUNDC1 by PGAM5, we used MitoSox staining and found that selenite increased mito-ROS, which was reversed by mito-ROS scavenging agent MitoQ (Fig. S6b). MitoQ blocked FUNDC1 dephosphorylation while enhancing BCL-xL dephosphorylation (Figs. 6c and S6c), likely by modulating the PGAM5 interaction with BCL-xL and FUNDC1. Indeed, MitoQ strongly reversed the effects of selenite, including the decreased BCL-xL/PGAM5 interaction, the increased FUNDC1/PGAM5 interaction, and the PGAM5 multimerization (Figs. 6d, e and S6d). As expected, selenite inhibited BCL-xL dephosphorylation, resulting in less BAX/BAK bound to BCL-xL; this effect was also rescued by MitoQ (Fig. S6e). Consequently, MitoQ inhibited the selenite-induced apoptosis (Fig. S6f). These results together suggest that selenite-induced mitochondrial oxidative stress inhibits dephosphorylation of BCL-xL by PGAM5 to promote apoptosis.
Fig. 6.
Mitochondrial oxidative stress regulates dephosphorylation of BCL-xL and FUNDC1 by PGAM5 to promote apoptosis during mitotic arrest. a HeLa and PGAM5 knockout HeLa cells were treated with selenite (5 uM, 2 h) and/or vinblastine (500 nM, 24 h), lysed and analyzed by immunoblotting with the indicated antibodies. Right: the ratio of p-Ser13 to total FUNDC1 or p-Ser62 to total BCL-xL were calculated from the respective band intensities in the respective blots measured by ImageJ, normalized to the values from untreated Hela cells or vinblastine-treated HeLa cells, respectively. Data are the mean ± SEM of three experiments. **p < 0.01. b HeLa cells were treated with 500 nM vinblastine for 24 h in combination with 5 μM selenite for the indicated times. After removal of selenite, the cells were cultured for an additional 24 h, then lysed and analyzed by immunoblotting with the indicated antibodies. c HeLa cells were treated with vinblastine with or without MitoQ before addition of selenite as indicated, lysed and analyzed by immunoblotting with the indicated antibodies. d HeLa cells expressing FLAG-BCL-xL, FLAG-FUNDC1 and/or PGAM5-MYC were treated with vinblastine with or without MitoQ before addition of selenite as indicated, lysed and immunoprecipitated with anti-MYC antibody. The co-precipitated FLAG-BCL-xL and FLAG-FUNDC1 were detected by immunoblotting with anti-FLAG antibody. e HeLa cells were treated with selenite for 2 h, with MitoQ pretreatment if indicated, lysed, analyzed by native or denaturing SDS PAGE, then immunoblotted with the indicated antibodies. f HeLa and FUNDC1 knockout HeLa cells were treated with 500 nM vinblastine for 36 h and 5 μM selenite for 2 h, lysed and fractionated. The cytochrome c in the heavy membrane pellet (P) and supernatant (S) were detected by immunoblotting along with the cytoplasmic protein tubulin and the mitochondrial protein TIMM23. Right: the ratio of cytochrome c in supernatant to total cytochrome c were calculated. Data are the mean ± SEM of three experiments. **p < 0.01. g HeLa and FUNDC1 knockout HeLa cells were treated with 500 nM vinblastine for 48 h and 5 μM selenite for the indicated times, lysed and analyzed by immunoblotting with the indicated antibodies. Right; the ratio of cleaved caspase-3 to ACTIN and cleaved to total PARP were calculated from the respective band intensities measured by ImageJ. Data are the mean ± SEM of three experiments. **p < 0.01. h HeLa or FUNDC1 knockout HeLa cells, transfected with FUNDC1-MYC if indicated, were treated with vinblastine for 48 h and selenite 6 h. After treatment, the cells were collected and stained with FITC-annexin V and propidium iodide (PI), then analyzed by flow cytometry. The percentages of apoptotic cells (annexin V+/PI− and annexin V+/PI+) were calculated. Data are the mean ± SEM of three experiments. *p < 0.05
Interestingly, FUNDC1 knockout attenuated the vinblastine- and selenite-induced cytochrome c release and apoptosis (Fig. 6f, g). Moreover, FUNDC1 knockout cells expressing FUNDC1-MYC were obviously more susceptible to apoptosis induced by vinblastine and selenite (Fig. 6h). This seems contradictory to the anti-apoptotic role of FUNDC1-dependent mitophagy. Since previous studies show that inhibition of mitofission partially prevents the release of cytochrome c [34], we reasoned that FUNDC1-dependent mitofission might participate in the selenite-mediated aggravation of apoptosis in vinblastine-treated cells. Treating cells with selenite or vinblastine or their combination, we found that vinblastine failed to induce FUNDC1 dephosphorylation, but selenite increased FUNDC1 dephosphorylation, the FUNDC1/DRP1 interaction, and mitofission in the presence of vinblastine (Fig. S6g–i). In summary, selenite causes mitochondrial oxidative stress which exacerbates vinblastine-induced apoptosis by promoting FUNDC1 dephosphorylation by PGAM5. This in turn induces mitochondrial fission while also inhibiting the dephosphorylation of BCL-xL by PGAM5, thus increasing BAX/BAK-mediated MOMP.
Discussion
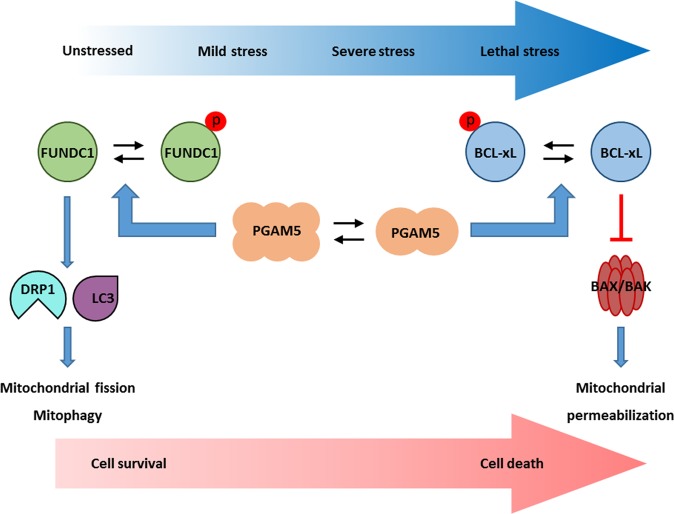
It is known that different stresses trigger different mitochondrial and cellular responses, but the mechanisms that sense the stresses and coordinate effectors to execute graded responses are poorly understood. Our study sheds light on these mechanisms. We show that PGAM5, which exists in an equilibrium between dimeric and oligomeric states that can sense stress, dephosphorylates either BCL-xL to “switch off” apoptosis or FUNDC1 to “switch on” mitofission and mitophagy for survival (Fig. 7). Our data suggest that in unstressed cells, BCL-xL is nonphosphorylated and strongly interacts with pro-apoptotic molecules such as BAX/BAK to promote cell survival [24]. BCL-xL also interacts with PGAM5 to inhibit FUNDC1 dephosphorylation and subsequent mitophagy [29]. However, the stress from the mitotic arrest induced by vinblastine likely activates the kinases CDK1 and/or JNK [21, 22], which phosphorylate BCL-xL to reduce its interaction with BAX/BAK, thereby promoting apoptosis. To mitigate apoptosis, the phosphatase PGAM5 reverses the BCL-xL phosphorylation. Phospho-BCL-xL is an atypical substrate of PGAM5 as it interacts with PGAM5 through sites other than the phospho-site, and can therefore sequester the phosphatase even after dephosphorylation. Moreover, the PGAM5 dimer bound by BCL-xL has lower catalytic activity than the multimer, which does not bind BCL-xL [20]. This further increases the retention time of BCL-xL on PGAM5. Therefore, BCL-xL may function as a suicide inhibitor of PGAM5, preventing it from acting on another substrate (phospho-FUNDC1) in mitotically arrested cells.
Fig. 7.
Reciprocal interaction of PGAM5 with BCL-xL and FUNDC1 functions as a molecular switch to sense different stresses and generate appropriate responses to dictate mitochondrial and cellular fate. In unstressed cells, BCL-xL is not phosphorylated and binds the mitochondrial phosphatase PGAM5, whereas FUNDC1 is inhibited by phosphorylation. Severe stress such as the mitotic arrest induced by vinblastine is sensed by certain kinases that phosphorylate BCL-xL to reduce its interaction with BAX and BAK, and hence its anti-apoptotic function. PGAM5 in the dimeric state binds and dephosphorylates BCL-xL to reactivate its anti-apoptotic function. Mild stress such as mitochondrial ROS induced by selenite switches PGAM5 to the multimeric state, which cannot bind and reactivate BCL-xL to prevent mitochondrial damage. However, the multimeric PGAM5 binds and dephosphorylates FUNDC1 to activate mitochondrial fission and subsequent mitophagy to eliminate the damaged mitochondria, thus promoting cell survival. Lethal stress such as a combination of mitochondrial ROS and mitotic arrest promotes cell death by inactivating BCL-xL and blocking FUNDC1-dependent mitophagy but not mitochondrial fission
Our data indicate that PGAM5 may serve as a sensor for oxidative stress caused by selenite to determine mitochondrial and cellular fates. Selenite-induced oxidative stress alters the multimeric status of mitochondrial PGAM5, thus freeing PGAM5 from BCL-xL (Figs. 4h, 5b and 7). This change is strongly inhibited by MitoQ, a potent scavenger that diminishes mito-ROS (Fig. 6d, e). It is possible that mito-ROS directly target PGAM5, likely at the cysteine residues [35, 36], to induce oligomerization. Indeed, our unpublished data show that mutations of cysteines 12 and 229 reduce the oligomerization in response to selenite treatment. Thus, the cell responds to these distinct stresses by altering the multimerization state of PGAM5, which in turn alters the interaction of PGAM5 with BCL-xL and FUNDC1 and hence their phosphorylation state. This balance modulates mitochondrial integrity, morphology and recycling, and ultimately makes a life-or-death decision for the cell.
When the cell experiences both mitotic and oxidative stresses, the balance is tipped toward death. The underlying molecular mechanism we revealed here is that BCL-xL loses the ability to bind to PGAM5 and thus loses the chance to be reactivated by dephosphorylation to inhibit BAX/BAK-dependent apoptosis in the mitotically arrested and oxidized cell. Furthermore, FUNDC1, which is activated by PGAM5-mediated dephosphorylation cannot complete mitophagy to remove the damaged mitochondria because vinblastine prevents polymerization of microtubules, which inhibits fusion of autophagosomes with endosomes [37]. However, FUNDC1 can promote mitofission through recruitment of DRP1, which results in mitochondrial fragmentation that in turn facilitates mitochondrial permeabilization and apoptosis. Further dissecting the structural and molecular details of these important stress-dependent processes will provide better understanding of the life-or-death balance of the cell.
Martials and methods
Cell culture and transfection
HeLa cells were cultured in DMEM supplemented with 10% fetal bovine serum (Hyclone) and 1% penicillin-streptomycin at 37 °C under 5% CO2. Plasmids were transfected into cells with polyethylenimine according to the lab manufacturer’s protocols.
CRISPR/Cas9-mediated gene-knockout
The CRISPR/Cas9 system was used to generate FUNDC1, PGAM5, and BCL-xL single and PGAM5/BCL-xL double knockout HeLa cell lines. The oligonucleotide pairs used were as follows: BCL-xL (5′ -CACCGCAGGCGACGAGTTTGAACTG-3′ and 5′ -AAACCAGTTCAAACTCGTCGCCTGC-3′); PGAM5 (5′ -CACCGCGCACGTTGATCAGAGACAG-3′ and 5′ -AAAC GCAGGCGACGAGTTTGAACTGGTTTC-3′); FUNDC1 (5′ - CACCGCTGGCGGTATCATGGCGACC-3′ and 5′ - AAACGGTCGCCATGATACCGCCAGC-3′). Single clones were picked and expanded after selection with 2.5 µg/ml. Knockout of the respective genes was confirmed by sequencing the edited genomic regions after amplification by PCR. The loss of the respective proteins was verified by immunoblotting with appropriate antibodies.
Flow cytometry
Cells were stained with 100 nM MitoSox (ThermoFisher) for 15 min at 37 °C, washed 2 times with warm PBS, and then collected for analysis with flow cytometer (BD Calibur). Cells were collected and washed 2 times with warm PBS, then stained with FITC-Annexin V for 15 min at room temperature. Samples were stained with PI before analysis with a flow cytometer.
Immunoblotting
Cells treated with the indicated agents were harvested and lysed in lysis buffer (150 mM NaCl, 20 mM Tris, pH 7.4, 1 mM EGTA, 1 mM EDTA, 1% SDS, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4, and 10 μM PMSF). Cell lysates containing equivalent amounts of protein (20 µg) were subjected to SDS-PAGE, and transferred to nitrocellulose membranes, which were blocked by 5% milk and probed with the indicated primary antibodies, followed by the appropriate HRP-conjugated secondary antibodies (KPL, 1912I). Immunoreactive bands were detected by a chemiluminescence kit (Engreen Biosystem, 29100) and visualized by a chemiluminescence imager (JUNYI). The following antibodies were used: Phospho-(Ser/Thr) (1:1000, Abcam, ab117253); BCL-xL (1:1000, Cell Signaling Technology, 2762); phospho-BCL-xL (pSer62) (1:1000, Sigma, SAB4504352); PGAM5 (1:1000, Abcam, ab131552); ACTIN (1:3000, Sigma, A5441); Tubulin (1:3000, MBL, PM054); TIMM23 (1:1000, BD Biosciences, 611222); TOMM20 (1:1000, BD Biosciences, 612278); COX IV (1:1000, Proteintech, 11242-1-AP); Cytochrome c (1:1000, BD Biosciences, 556433); MFN1 (1:1000, Abgent, ASC11815); Drp1(1:1000, BD Biosciences, 611113); phospho-DRP1 (pSer637) (1:1000, Cell Signaling Technology,4867S); BECN1 (1:1000, BD Biosciences, 612113); LC3 (1:1000, MBL, PM036); SQSTM1 (1:2000, MBL,PM045); BAX (1:1000, Santa Cruz, SC-493); BAK (1:1000, Santa Cruz, SC-S17309); Cleaved Caspase-3 (1:1000, Cell Signaling Technology, 9664S); PARP (1:1000, Cell Signaling Technology, 9542); MYC (1:2000, Santa Cruz, B1115); FLAG (1:2000, Sigma, F1804); GFP (1:2000, Santa Cruz Biotechnology, A2115); and FUNDC1 (1:1000, AVIVA, ARP53280_P050). The anti-FUNDC1 (p-Ser13) polyclonal antibody (1:1,000) was affinity purified after immunizing rabbits with purified FUNDC1 phosphopeptides
Immunoprecipitation
Cells treated with the indicted agents were lysed with 0.5 ml of lysis buffer (20 mM Tris, pH 7.4, 137 mM NaCl, 2 mM EDTA, 10% glycerol, 1% NP-40; Shanghai Sangon Biotech, A510110) plus protease inhibitor cocktail (Roche Applied Science, 11206893001) for 50 min on ice. After centrifugation at 12,000 g for 10 min, the lysates were incubated with Ni2+-chelating agarose beads (GE Healthcare) for 2 hours or specific antibody and then protein A/G-agarose beads (Abmart, A10001) overnight at 4 °C. The beads were washed 5 times with lysis buffer, and the immune complexes were eluted with reducing SDS sample buffer for 5 min at 95 °C and analyzed by SDS-PAGE and immunoblotting.
Immunofluorescence microscopy
After treatment, cells were fixed with freshly prepared 4% paraformaldehyde (Dingguo Changsheng Biotechnology) at 37 °C for 20 min, then permeabilized with 0.2% Triton X-100 (Shanghai Sangon Biotech, TB0198) at 4 °C. After blocking in goat serum for 1 h, cells were incubated with primary antibodies for 2 h at room temperature, washed with PBS, and stained with FITC- or CY5-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. Cell images were captured with a TCS SP5 Leica confocal microscope.
Native PAGE
Cells were lysed in RIPA lysis buffer (50 mM Tris-HCL, pH 7.4, 150 mM NaCl, 0.5% NP40 and PMSF). After centrifugation at 12,000 g for 10 min, supernatant proteins were quantified and diluted with 2× native PAGE sample buffer (125 mM Tris-HCl, pH 6.8; 30% glycerol; and 0.1% bromophenol blue). 15 µg protein was used for native-PAGE using gels made without SDS. Gels were run with 25 mM Tris, pH 8.4, 192 mM glycine and 1% deoxycholate (DOC) in a chamber. After electrophoresis, proteins were transferred to nitrocellulose membranes for immunoblotting.
Phosphatase assay
We were unable to purify recombinant full-length PGAM5 protein that contains a N-terminal transmembrane sequence because it formed aggregates when expressed in Escherichia coli BL21 cells. We thus expressed a PGAM5 protein lacking the transmembrane sequence (His-PGAM5-ΔN21) and the phosphatase-dead mutant (His-PGAM5 (H105A)-ΔN21) in Escherichia coli BL21 cells. Using Plasmids for His-PGAM5-ΔN21 and His-PGAM5 (H105A)-ΔN21 were from Dr. Xiaodong Wang’s lab, and the proteins were expressed and purified from Escherichia coli BL21 cells.
Ni2+ beads were used as described above to enrich GFP-His-BCL-xL (pSer62). After three washes with phosphatase buffer (50 mM imidazole, pH 7.2, 0.2 mM EGTA, 0.02% 2-mecaptoethanol, and 0.1 mg/ml BSA), the beads were incubated with the recombinant His-tagged PGAM5 protein or the phosphatase-dead mutant (H105A) (200 ng) at 30 °C for 1 hour, and subjected to immunoblotting using anti-phospho-BCL-xL (pSer62) antibody.
Photocrosslinking
The photocrosslinking method for studying interactions between BCL-2 family proteins has been described in detail. For this study [38], [35S]Met-labeled BCL-xL proteins, each with a single Nε-(5-azido-2-nitrobenzoyl)-lysine (εANB-Lys) incorporated at a specific location, were synthesized from RNA encoding the corresponding single-Lys BCL-xL mutants using an in vitro translation system [38]. 10 μl of each resulting BCL-xL protein was incubated at 37 °C for 1.5 h with 3 μM purified His6-tagged PGAM5ΔN90 or ΔN54 protein lacking the N-terminal 90 or 54 residues in a 20-μl reaction containing buffer A. The samples were photolyzed to induce crosslinking via the ANB probe. The resulting photoadducts of the [35S]Met-labeled BCL-xL protein and the His6-tagged PGAM5 protein were enriched on Ni2+-nitrilotriacetic acid agarose, eluted and analyzed with reducing SDS-PAGE and phosphor-imaging. The following control reactions were carried out in parallel: the lysine-null (K0) BCL-xL control lacked the εANB-Lys residue; the minus ANB control had BCL-xL with an unmodified Lys residue instead of the εANB-Lys; the minus light (hυ) control had an ANB probe which was neutralized by DTT before photolysis; and the minus 6H-PGAM5ΔN90 or ΔΝ54 control lacked the His6-tagged protein. The His6-tagged PGAM5ΔN90 and ΔN54 proteins were expressed and purified using plasmids from Dr. Apirat Chaikuad as described [27].
Subcellular fractionation
The cells were collected and resuspended in hypotonic buffer (210 mM sucrose, 70 mM mannitol, 1 mM EDTA, 1 mM EGTA, 1.5 mM MgCl2, 10 mM HEPES, pH 7.2, 10 mM KCl). After gentle homogenization with a Dounce homogenizer, the cell lysates were subjected to differential and gradient centrifugation (800 × g,10 min and 8000 × g, 10 min). The resulting membrane fractions were lysed by 1% SDS, and then analyzed together with the soluble fraction by immunoblotting.
Statistical analysis
Data were analyzed from three independent experiments and are shown as the mean ± SEM. Statistical analyses were performed using Student’s two-tailed t-test, with p values < 0.05 being considered significant. Significance levels relative to the controls are indicated by *, ** or *** for p < 0.05; 0.01 or 0.001, respectively, in the figures. All analyses were performed with GraphPad Prism software.
Supplementary information
Acknowledgements
This research was supported by Grants 301520103904, 91754114, 31271529, 31671441 and 3192100015 from the Natural Science Foundation of China to YZ and QC, Grants 2016YFA0500201 and 2016YFA0100503 from the Ministry of Science and Technology of China to QC, Grant QYZDJ-SSW-SMC004 from the CAS Key Project of Frontier Science, 111 Project from the Ministry of Education and the State Administration of Foreign Experts Affairs of the People’s Republic of China (B08011), Grant R01GM062964 from the United States National Institutes of Health and Grant HR16-026 from the Oklahoma Center for the Advancement of Science and Technology to JL, and by an Institutional Development Award from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103640.
Author contributions
KM conceived and designed the experiments with YZ, JL and QC; KM performed most of the experiments and data analysis with assistance from RC, HC, MC, TZ, BC, CM and QL. ZZ performed the photocrosslinking experiment. KM, QC, JL and YZ wrote the manuscript. All authors provided intellectual input to the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jialing Lin, Email: jialing-lin@ouhsc.edu.
Yushan Zhu, Email: zhuys@nankai.edu.cn.
Quan Chen, Email: chenq@nankai.edu.cn, Email: chenq@ioz.ac.cn.
Supplementary information
The online version of this article (10.1038/s41418-019-0396-4) contains supplementary material, which is available to authorized users.
References
- 1.Galluzzi L, Kepp O, Trojel-Hansen C, Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ Res. 2012;111:1198–207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- 2.Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic Biol Med. 2010;48:749–62. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalkavan H, Green DR. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018;25:46–55. doi: 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–22. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 6.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012;14:177–85. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 8.Wei Y, Chiang WC, Sumpter R, Jr., Mishra P, Levine B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell. 2017;168:224–38 e210. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–43. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 10.Lenhausen AM, Wilkinson AS, Lewis EM, Dailey KM, Scott AJ, Khan S, et al. Apoptosis inducing factor binding protein PGAM5 triggers mitophagic cell death that is inhibited by the ubiquitin ligase activity of X-linked inhibitor of apoptosis. Biochemistry. 2016;55:3285–302. doi: 10.1021/acs.biochem.6b00306. [DOI] [PubMed] [Google Scholar]
- 11.Imai Y, Kanao T, Sawada T, Kobayashi Y, Moriwaki Y, Ishida Y, et al. The loss of PGAM5 suppresses the mitochondrial degeneration caused by inactivation of PINK1 in Drosophila. PLoS Genet. 2010;6:e1001229. doi: 10.1371/journal.pgen.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekine S, Yao A, Hattori K, Sugawara S, Naguro I, Koike M, et al. The ablation of mitochondrial protein phosphatase Pgam5 confers resistance against metabolic stress. EBioMedicine. 2016;5:82–92. doi: 10.1016/j.ebiom.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu W, Karuppagounder SS, Springer DA, Allen MD, Zheng L, Chao B, et al. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson’s-like movement disorder. Nat Commun. 2014;5:4930. doi: 10.1038/ncomms5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen G, Han Z, Feng D, Chen Y, Chen L, Wu H, et al. A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Mol cell. 2014;54:362–77. doi: 10.1016/j.molcel.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 15.Lo SC, Hannink M. PGAM5, a Bcl-XL-interacting protein, is a novel substrate for the redox-regulated Keap1-dependent ubiquitin ligase complex. J Biol Chem. 2006;281:37893–903. doi: 10.1074/jbc.M606539200. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Beauchemin M, Bertrand R. Bcl-xL phosphorylation at Ser49 by polo kinase 3 during cell cycle progression and checkpoints. Cell Signal. 2011;23:2030–8. doi: 10.1016/j.cellsig.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kharbanda S, Saxena S, Yoshida K, Pandey P, Kaneki M, Wang Q, et al. Translocation of SAPK/JNK to mitochondria and interaction with Bcl-x(L) in response to DNA damage. J Biol Chem. 2000;275:322–7. doi: 10.1074/jbc.275.1.322. [DOI] [PubMed] [Google Scholar]
- 18.Megyesi J, Tarcsafalvi A, Seng N, Hodeify R, Price PM. Cdk2 phosphorylation of Bcl-xL after stress converts it to a pro-apoptotic protein mimicking Bax/Bak. Cell Death Discov. 2016;2:pii: 15066. doi: 10.1038/cddiscovery.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Del Re DP, Matsuda T, Zhai P, Maejima Y, Jain MR, Liu T, et al. Mst1 promotes cardiac myocyte apoptosis through phosphorylation and inhibition of Bcl-xL. Mol Cell. 2014;54:639–50. doi: 10.1016/j.molcel.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkins JM, McConnell C, Tipton PA, Hannink M. A conserved motif mediates both multimer formation and allosteric activation of phosphoglycerate mutase 5. J Biol Chem. 2014;289:25137–48. doi: 10.1074/jbc.M114.565549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terrano DT, Upreti M, Chambers TC. Cyclin-dependent kinase 1-mediated Bcl-xL/Bcl-2 phosphorylation acts as a functional link coupling mitotic arrest and apoptosis. Mol Cell Biol. 2010;30:640–56. doi: 10.1128/MCB.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan M, Goodwin M, Vu T, Brantley-Finley C, Gaarde WA, Chambers TC. Vinblastine-induced phosphorylation of Bcl-2 and Bcl-XL is mediated by JNK and occurs in parallel with inactivation of the Raf-1/MEK/ERK cascade. J Biol Chem. 2000;275:29980–5. doi: 10.1074/jbc.M003776200. [DOI] [PubMed] [Google Scholar]
- 23.Arena G, Gelmetti V, Torosantucci L, Vignone D, Lamorte G, De Rosa P, et al. PINK1 protects against cell death induced by mitochondrial depolarization, by phosphorylating Bcl-xL and impairing its pro-apoptotic cleavage. Cell Death Differ. 2013;20:920–30. doi: 10.1038/cdd.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Follis AV, Llambi F, Kalkavan H, Yao Y, Phillips AH, Park CG, et al. Regulation of apoptosis by an intrinsically disordered region of Bcl-xL. Nat Chem Biol. 2018;14:458–65. doi: 10.1038/s41589-018-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu W, Jing L, Wang Q, Lin CC, Chen X, Diao J, et al. Bax-PGAM5L-Drp1 complex is required for intrinsic apoptosis execution. Oncotarget. 2015;6:30017–34. doi: 10.18632/oncotarget.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hawk MA, Gorsuch CL, Fagan P, Lee C, Kim SE, Hamann JC, et al. RIPK1-mediated induction of mitophagy compromises the viability of extracellular-matrix-detached cells. Nat Cell Biol. 2018;20:272–84. doi: 10.1038/s41556-018-0034-2. [DOI] [PubMed] [Google Scholar]
- 27.Chaikuad A, Filippakopoulos P, Marcsisin SR, Picaud S, Schroder M, Sekine S, et al. Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly. Structure. 2017;25:1089–99 e1083. doi: 10.1016/j.str.2017.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammond PW, Alpin J, Rise CE, Wright M, Kreider BL. In vitro selection and characterization of Bcl-X(L)-binding proteins from a mix of tissue-specific mRNA display libraries. J Biol Chem. 2001;276:20898–906. doi: 10.1074/jbc.M011641200. [DOI] [PubMed] [Google Scholar]
- 29.Wu H, Xue D, Chen G, Han Z, Huang L, Zhu C, et al. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014;10:1712–25. doi: 10.4161/auto.29568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding J, Mooers BH, Zhang Z, Kale J, Falcone D, McNichol J, et al. After embedding in membranes antiapoptotic Bcl-XL protein binds both Bcl-2 homology region 3 and helix 1 of proapoptotic Bax protein to inhibit apoptotic mitochondrial permeabilization. J Biol Chem. 2014;289:11873–96. doi: 10.1074/jbc.M114.552562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz K, Thaker TM, Agnew C, Miller-Vedam L, Trenker R, Herrera C, et al. Functional role of PGAM5 multimeric assemblies and their polymerization into filaments. Nat Commun. 2019;10:531. doi: 10.1038/s41467-019-08393-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen M, Chen Z, Wang Y, Tan Z, Zhu C, Li Y, et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. 2016;12:689–702. doi: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang F, Nie C, Yang Y, Yue W, Ren Y, Shang Y, et al. Selenite induces redox-dependent Bax activation and apoptosis in colorectal cancer cells. Free Radic Biol Med. 2009;46:1186–96. doi: 10.1016/j.freeradbiomed.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 34.Estaquier J, Arnoult D. Inhibiting Drp1-mediated mitochondrial fission selectively prevents the release of cytochrome c during apoptosis. Cell Death Differ. 2007;14:1086–94. doi: 10.1038/sj.cdd.4402107. [DOI] [PubMed] [Google Scholar]
- 35.Nie C, Tian C, Zhao L, Petit PX, Mehrpour M, Chen Q. Cysteine 62 of Bax is critical for its conformational activation and its proapoptotic activity in response to H2O2-induced apoptosis. J Biol Chem. 2008;283:15359–69. doi: 10.1074/jbc.M800847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vermeij WP, Backendorf C. Reactive oxygen species (ROS) protection via cysteine oxidation in the epidermal cornified cell envelope. Methods Mol Biol. 2014;1195:157–69. doi: 10.1007/7651_2013_51. [DOI] [PubMed] [Google Scholar]
- 37.Kochl R, Hu XW, Chan EY, Tooze SA. Microtubules facilitate autophagosome formation and fusion of autophagosomes with endosomes. Traffic. 2006;7:129–45. doi: 10.1111/j.1600-0854.2005.00368.x. [DOI] [PubMed] [Google Scholar]
- 38.Lin J, Johnson AE, Zhang Z. Photocrosslinking Approach to Investigate Protein Interactions in the BCL-2 Family. Methods Mol Biol. 2019;1877:131–49. doi: 10.1007/978-1-4939-8861-7_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–41. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.