Abstract
A reciprocal communication between the implantation-competent blastocyst and the receptive uterus is essential to successful implantation and pregnancy success. Progesterone (P4) signaling via nuclear progesterone receptor (PR) is absolutely critical for pregnancy initiation and its success in most eutherian mammals. Here we show that a nuclear protein high-mobility group box-1 (HMGB1) plays a critical role in implantation in mice by preserving P4-PR signaling. Conditional deletion of uterine Hmgb1 by a Pgr-Cre driver shows implantation defects accompanied by decreased stromal cell Hoxa10 expression and cell proliferation, two known signatures of inefficient responsiveness of stromal cells to PR signaling in implantation. These mice evoke inflammatory conditions with sustained macrophage accumulation in the stromal compartment on day 4 of pregnancy with elevated levels of macrophage attractants Csf1 and Ccl2. The results are consistent with the failure of exogenous P4 administration to rescue implantation deficiency in the mutant females. These early defects are propagated throughout the course of pregnancy and ultimately result in substantial subfertility. Collectively, the present study provides evidence that nuclear HMGB1 contributes to successful blastocyst implantation by sustaining P4-PR signaling and restricting macrophage accumulation to attenuate harmful inflammatory responses.
Subject terms: Physiology, Reproductive disorders
Introduction
Embryo implantation is a key to pregnancy success [1, 2]. Defects during implantation either terminate pregnancy or compromise decidualization, placentation, and parturition, thus impacting pregnancy outcomes [1, 3, 4]. In mice, the uterus becomes receptive to blastocyst implantation on day 4 of pregnancy (day 1 = vaginal plug) followed by blastocyst attachment on day 5 [1, 2]. Blood vessels enter the uterus from the mesometrium with orientation of the uterus along a mesometrial–antimesometrial (M-AM) axis. On the evening of day 4, planar cell polarity signaling allows luminal epithelial (LE) evaginations toward the AM pole to form a specialized crypt (implantation chamber) along with preexisting glands, establishing a direct communication between the blastocyst and glands [5]. This process is accompanied by increased endometrial vascular permeability at the site of blastocyst apposition, which can be visualized as blue bands after an injection of a blue dye solution [6].
Progesterone (P4) signaling is absolutely required for the uterus to acquire receptivity for blastocyst implantation and pregnancy maintenance [1]. P4 activates progesterone receptor (PR) with the participation of coactivators and co-chaperones to maintain hormone binding and subsequent transcriptional activation [7, 8]. Following ovulation, serum P4 levels begin to rise from the newly formed corpora lutea from day 3 onward in mice. With increasing P4 levels, stromal cell proliferation becomes evident on day 3 and become more intense on day 4 following preimplantation ovarian secretion of estrogen. With blastocyst attachment on day 5, stromal cells surrounding the implanting blastocyst undergo proliferation and differentiation into decidual cells (decidualization) [1].
High-mobility group box-1 (HMGB1) is a nuclear protein highly conserved spanning simplest multicellular species to humans [9]. HMGB1 binds to DNA through its two DNA-binding domains called HMG boxes with no sequence specificity to maintain chromatin structures and regulate gene transcriptions [9–12]. Although HMGB1 lacks transcriptional activator domains and does not function as a transcriptional factor, it can support the transcriptional activities of nuclear hormone receptors including PR and glucocorticoid receptor (GR) by binding to their response elements on target genes [13–15]. Systemic deletion of HMGB1 results in neonatal death in mice due to reduced GR activation [15], providing evidence for the critical role of HMGB1 and its association with nuclear receptors. However, tissue specific functions of nuclear HMGB1 in adults remain poorly understood.
There is evidence that HMGB1 functions as a damage-associated molecular pattern molecule in cells in vitro under stress conditions, or in mice exposed to LPS [16, 17]. Under these conditions, HMGB1 is translocated from the nucleus into the cytosol or released into extracellular compartments [17–19]. After secretion, HMGB1 exerts inflammatory signals by activating receptor for advanced glycation end-products (RAGE), toll-like receptor 2 (TLR2), or TLR4 [16, 17]. Recent studies using liver specific deletion of HMGB1 show that secreted liver HMGB1 activates RAGE under LPS or acetaminophen stimuli and contributes to sepsis development or tumorigenesis, respectively [20, 21]. On the other hand, there is a report that hepatocyte-specific Hmgb1 deletion show normal cellular functions and gene expression under physiological conditions [22]. These findings indicate that the complexities of HMGB1 functions are context and tissue dependent.
We show here that HMGB1 is highly expressed and retained in stromal cell nuclei in the pregnant uterus and confers PR activation. Females with uterine deletion of Hmgb1 show severe subfertility and give birth to small litters. One cause of this phenotype is inefficient PR signaling with reduced levels of Hoxa10, a P4 responsive gene in the stroma. Interestingly, we also observed higher levels of Ccl2 and Csf1 in Hmgb1-deleted stroma with increased accumulation of macrophages prior to implantation. Normally, macrophage accumulation is suppressed upon elevation of serum P4 levels preceding implantation [23, 24]. These results suggest compromised P4-PR signaling in the absence of HMGB1 and unfold a new function of HMGB1 in the pregnant uterus that previously has not been recognized.
Methods and materials
Mice
PgrCre/+ and Hmgb1f/f mouse lines were generated as previously described [22, 25]. Hmgb1d/d mice were generated by mating floxed females with PgrCre/+ males. All mice used in this study were housed under a constant 12-h/12-h light/dark cycle in the Cincinnati Children’s Animal Care Facility according to NIH and institutional guidelines for the use of laboratory animals. All protocols were approved by the Cincinnati Children’s Animal Care and Use Committee. Mice were provided ad libitum with autoclaved Laboratory Rodent Diet 5010 (Purina) and UV light-sterilized reverse osmosis/deionized constant-circulation water.
Analysis of pregnancy events
Pregnancy events were assessed as previously described [3–5, 26–28]. Three adult females were randomly chosen and housed with a WT fertile male overnight in separate cages; the morning of finding the presence of a vaginal plug was considered successful mating (day 1 of pregnancy). Plug-positive females were then housed separately from males until processed for experiments. Litter size, pregnancy rate, and outcomes were monitored in timed pregnancy. Blue reaction was performed by injecting intravenously a blue dye solution (1% Chicago Blue in Saline, 100 µL/mouse) 4 min prior to mice being sacrificed. The distinct blue bands along the uterus indicated the site of implantation. For confirmation of pregnancy in plug-positive day 4 mice or mice showing no blue bands on day 5, one uterine horn was flushed with saline to check for the presence of blastocysts. If blastocysts were present, the contralateral horn was used for experiments and mice without any blastocysts were discarded. Pseudopregnancy was induced by mating females with vasectomized males. For rescue experiments, pregnant mice were injected subcutaneously with P4 in sesame oil (2 mg/100 µL/dose) on the morning of days 3 and 4. Mice were sacrificed after blue dye injection on day 5 of pregnancy.
Isolation of primary stromal cells
Stromal cells from day 4 pregnant uteri were collected by enzymatic digestion as described previously [3, 29]. Uteri from Hmgb1f/f or Hmgb1d/d mice on day 4 of pseudopregnancy were split open longitudinally and cut into small pieces (2–3 mm long). Tissue pieces were incubated with pancreatin (25 mg/mL, Sigma) and dispase (6 mg/mL, Gibco) for 1 h at 4 °C, followed by 1 h at room temperature and 15 min at 37 °C. LE sheets were removed by pipetting the tissues several times. The remaining tissue fragments were incubated in type IV collagenase (300 U/mL, Washington) to free stromal cells. Stromal cells were suspended in DMEM/F12 (Gibco) containing 10% heat-inactivated FBS (Gibco), 50 units/mL penicillin, 50 μg/mL streptomycin, and 1.25 μg/mL fungizone (Pen Strep; Gibco). Cell suspensions were filtered through a 70-μm nylon mesh to remove glands and clumps of epithelial cells. Cells were seeded into 6-well (for RNA extraction) or 24-well (for luciferase assay) plates and the medium was changed 1 h later to remove unattached immune cells. For RNA extraction, cells were washed in PBS and dissolved in TRIzol reagent (Invitrogen) after another 5 h culture.
Histology
Tissue sections from control and experimental groups were processed on the same slide. Frozen sections (12 μm) were fixed in 4% PFA-PBS for 10 min at room temperature and then stained with hematoxylin and eosin for light microscopy analysis.
Immunofluorescence (IF) and microscopy
IF was performed as previously described [5, 28]. IF using frozen sections (12 µm) was performed using the following first antibodies: HMGB1 (1:2000, 6893S, Cell Signaling Technology), CD45 (1:300, 103102, Biolegend), Ki67 (1:300, RM-9106-S, Thermo Fisher Scientific), Cleaved caspase-3 (1:300, 9661s, Cell Signaling Technology), PR (1:300, 8757; Cell Signaling Technology), ERα (1:300, sc-542; Santa Cruz), FOXA2 (1:300, WRAB-FOXA2, Seven Hills Bioreagents), CK-8 (1:1000, TROMA-1, Hybridoma Bank, Iowa), E-Cadherin (E-Cad) (1:300, 3195s, Cell Signaling Technology), GR (1:300, 12041S, Cell Signaling Technology), CD31 (1:300, 553370, BD Biosciences), F4/80 (1:1000, MCA497R, Bio-Rad), CD11b (1:300, 101201, Biolegend), MHC II (1:500, 14-5321-81, eBioscience), CD206 (1:1000, MCA2235T, AbD Serotec), Fluorescein labeled DBA-lectin (1:500, FL-1031-2, Vectorlabs), Gr-1 (1:500, MCA2387, Bio-Rad), and CSF1R (1:1000, sc-692, Santa Cruz). IF of HMGB1 was performed in PFA-fixed paraffin sections (6 µm) using HMGB1 antibody (1:2000, 6893S, Cell Signaling Technology). For signal detection, secondary antibodies conjugated with Cy-2, Cy-5, Alexa 488, or Alexa 594 (1:300, Jackson ImmunoResearch) were used. Nuclear staining was performed using Hoechst 33342 (5 µg/ml, H1399, Thermo Fisher Scientific). Tissue sections from control and experimental groups were processed onto the same slide. Pictures were taken using Nikon Eclipse 90i upright microscope and processed by Nikon Elements Viewer. F4/80-positive cells in the stroma were counted using ImageJ (NIH) and normalized by IF stained cells over total number of Hoechst stained cells in the stroma or the total area of the stroma. Sections from five mice for day 4 and six mice for day 5 were assayed for this analysis as previously described [30].
In situ hybridization
Frozen sections (12 µm) from Hmgb1f/f or Hmgb1d/d mice were processed onto the same slide. In situ hybridization with 35S-labeled cRNA probes was performed as described [3, 26]. Digoxigenin (DIG)-labeled probes were generated according to the manufacturer’s protocol (Roche). In situ hybridization with DIG-labeled probes was performed as described [27]. The primers used for the DIG-labeled probe of Hmgb1 are listed below: 5′-CGGATGCTTCTGTCAACT-3′ and 5′- ACTTCTCCTTCAGCTTGG-3′. Quantification of in situ hybridization with 35S-labeled cRNA probes was performed using ImageJ (NIH).
Cytoplasm/nuclear protein fractionation
Pregnant uterine tissues were homogenized in Buffer B (5 mM EDTA-PBS). Tissue homogenates were centrifuged at 1000 × g, 4 °C for 2 min, and supernatants were discarded. Remaining pellets were resuspended in Buffer A (10 mM HEPES pH 7.9, 10 mM KCl, 0.1 M EDTA, 1 mM DTT, 0.5 mM PMSF) and kept on ice for 20 min. After adding 1/4 volume of Buffer A with 2.5% NP-40 and vortexed, samples were centrifuged at 15,000 × g, 4 °C for 10 min. Supernatants were collected as cytoplasm fractions and pellets were vortexed in Buffer C (20 mM HEPES pH 7.9, 0.4 M NaCl, 1 mM EDTA, 1 mM DTT, 1 mM PMSF) at 4 °C for 25 min. Supernatants were collected as nuclear fractions after centrifugation at 17,000 × g, 4 °C for 5 min. All samples were kept at −20 °C until use.
Immunoblotting
Western blotting was performed as described [3, 26]. The same blots were used for quantitative analysis of each protein. Bands were visualized using an ECL kit (Bio-Rad). α-Tubulin, β-Actin, and Lamin A/C served as a loading control. The following antibodies were used to detect each protein: HMGB1 (1:1000, 6893S, Cell Signaling Technology), α-Tubulin (1:1000, 2144, Cell Signaling Technology), β-Actin (1:1000, sc1615, Santa Cruz), and Lamin A/C (1:1000, sc20681, Santa Cruz). Quantification of HMGB1 bands was performed by ImageJ (NIH) and normalized by the intensity of α-Tubulin bands.
RT-PCR
RT-PCR was performed as described [3, 26] and PCR was run for 25 cycles using the following primers: 5′-AGATGACAAGCAGCCCTAT-3′ and 5′-CTTTTCAGCCTTGACCAC-3′ for Hmgb1; 5′-GCAGATGTACCGCACTGAGATTC-3′ and 5′-ACCTTTGGGCTTACTCCATTGATA-3′ for Rpl7; Rpl7 served as an internal control. Each PCR product was loaded onto a 2% agarose gel containing ethidium bromide with a volume of 3 µL to detect target bands.
Quantitative RT-PCR
RT-qPCR was performed as described [5], using the following primers: 5′-AGAAGCTGTAGTTTTTGTCACC-3′ and 5′-TGCTTGAGGTGGTTGTGGAA-3′ for Ccl2; 5′-CTCTAGCCGAGGCCATGTGGAG-3′ and 5′-GGCCCCCAACAGTCAGCAAG-3′ for Csf1; 5′-GCAGATGTACCGCACTGAGATTC-3′ and 5′-ACCTTTGGGCTTACTCCATTGATA-3′ for Rpl7; Rpl7 served as an internal control.
Luciferase assay for PR
Luciferase assay was performed as previously described [15], using the primary cultured stromal cells. Briefly, 1 × 105 cells were seeded in 24-well dishes. After 48 h, cells were transfected with 400 ng of PRE2-Tk-Luc and 8 ng of pRL-Tk by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For the rescue experiment, 400 ng of Flag-hHMGB1 (#31609, Addgene) was transfected into Hmgb1-deleted cells. Six hours later, media were changed to 10% FBS-containing DMEM/F12 and incubated for 18 h. Cells were then starved for 4 h, followed by the treatment of P4 (1 µM) for 20 h. Collected cells proceeded to luciferase assay according to the manufacturer’s protocol (Promega).
ChIP-qPCR
Day 4 uteri were homogenized by a Dounce tissue grinder in cold PBS with proteinase and phosphatase inhibitors. Cells were fixed in 1% formaldehyde-PBS, then terminated by adding 2 M glycine. After incubation in lysis buffer 1 (50 mM HEPES pH 7.5, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25 % Triton-X, 1× cOmpleteTM (Roche)) and lysis buffer 2 (10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1× cOmplete TM(Roche)), pellets were resuspended in lysis buffer 3 (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.5% N-laurosylsarcosine, 0.1% sodium deoxycholate, 1× cOmpleteTM (Roche)) and sonicated by repeatedly following a program (30 s On and 30 s Off at 50% amplitude) for 24 min. After the samples were centrifuged, the soluble chromatin was retrieved. Immunoprecipitation was performed by incubation with anti-PR antibody (8757; Cell Signaling Technology) coated on DynabeadsTM Protein G (Thermo Fisher Scientific). After washing, DNA was eluted from the beads and subjected to real-time PCR with specific primers. Identification of the PR-binding site on Hoxa10 gene was performed as previously described [31], using the dataset from GSM857546 [32]. The primers designed and used for qPCR are: 5′-ATCGTAAACTCGAACTTCGC-3′ and 5′-GTGGCTCGCTTGCAGATA-3′.
Migration assay
Before assay, Raw264.7 cells were cultured overnight in 1% FBS-DMEM. Cells were then suspended in serum-depleted DMEM at the concentration of 2.5 × 105 cells/mL. In each bottom well, 700 µL of DMEM were added with or without 100 ng/mL Csf1 (R&D), 100 ng/mL Ccl2 (Biolegend), 1 µM Csf1r inhibitor (GW2580; Calbiochem), and 10 µM Ccr2 antagonist (BMS CCR2 22; Calbiochem). Upper inserts received 100 µL of DMEM with or without 300 ng/mL Csf1, 3 µM Csf1r inhibitor, or 30 µM Ccr2 antagonist and then 200 µL of cell suspensions. After a culture of 4 h for Csf1 and 24 h for Ccl2, migrated cells were fixed in 4% PFA-PBS and stained in 0.1% crystal violet for observation. Three fields/well at ×8 magnification were quantified using ImageJ (NIH).
Measurement of serum P4 levels
Sera were collected on day 4 of pregnancy, and hormone levels were measured by enzyme immunoassay kits (Cayman) as previously described [3–5, 26–28].
Tridimensional (3D) visualization of implantation sites
Whole mount staining, tissue clearing and 3D visualization of day 5 and 6 implantation sites were performed as previously described [5]. Anti-E-Cad antibody (1:100, 3195s, Cell Signaling Technology) and anti-F4/80 antibody (1:1000, MCA497R, Bio-Rad) were used to stain epithelia and macrophages, respectively. Anti-rabbit antibody conjugated with Alexa 594 (1:250, Jackson ImmunoResearch) and anti-rat antibody conjugated with Alexa 488 (1:250, Jackson ImmunoResearch) were used as secondary antibodies. 3D images were acquired by a Nikon multiphoton upright confocal microscope (Nikon A1R). To obtain the 3D structure of the tissue, the surface tool in Imaris (version 9.2.0., Bitplane) was used.
Hmgs, Csf1, and Ccl2 expression status examined by RNA-Seq analysis
Whole uterine tissues or enzymatically digested epithelial/stromal cells on day 4 were homogenized in TRIzol to extract total mRNA. After removing genomic DNA, total RNAs were subjected to RNA-Seq by HiSeq2500. The generated sequencing data have been deposited in the NCBI Gene Expression Omnibus (GEO) repository under the accession number: GSE120549 and GSE116096. The expression levels of genes were presented as reads per kilobase per million by Tophat2 [33] and visualized by R.
Statistics
Statistical analyses were performed using a two-tailed Student’s t test or a multiway analysis of variance (ANOVA) followed by Bonferroni’s post hoc test using Prism 6 (GraphPad Software). A value of P < 0.05 was considered statistically significant. Exact times and conditions of each experiment are described in individual figure legends.
Results
HMGB1 is expressed in the periimplantation uterus in a spatiotemporal manner
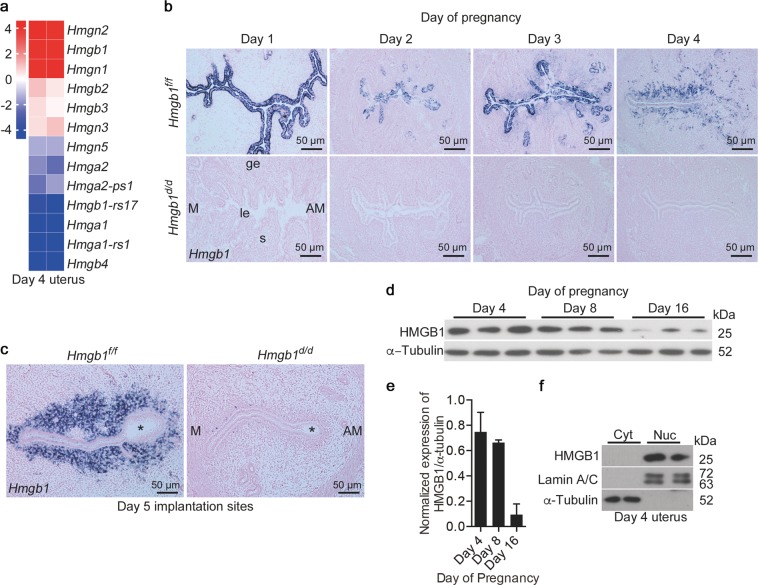
Hmg belongs to a family comprised of several members [34]. Our RNA-seq analysis of day 4 pregnant uteri shows that HMGB1 is one of the most abundant genes among the family members (Fig. 1a). To examine the spatiotemporal expression of Hmgb1 during the periimplantation period, we performed in situ hybridization using DIG (Fig. 1b, c). On day 1 through day 3, Hmgb1 expression is predominantly localized in epithelial cells with some stromal cell localization on day 3. In contrast, Hmgb1 signals are primarily observed in stroma cells on days 4 and 5 of pregnancy (Fig. 1b, c). Western blotting results show that HMGB1 protein levels decrease with pregnancy progression as assessed on days 4, 8, and 16 of pregnancy (Fig. 1d, e).
Fig. 1.
Expression of HMGB1 in early pregnant uteri. a Heatmap shows relative expression levels of Hmg family members in day 4 pregnant uteri. Data are expressed as reads log2 per kilobase per million (RPKM) for RNA-seq analysis, n = 2. b, c In situ hybridization of Hmgb1 expression in Hmgb1f/f and Hmgb1d/d uteri from days 1–5 of pregnancy. Asterisks indicate the location of blastocysts. le luminal epithelium, s stroma, ge glandular epithelium, M mesometrial pole, AM antimesometrial pole. Scale bar: 50 µm. d, e Western blotting of HMGB1 in uterine tissues on days 4, 8, and 16 of pregnancy (d) and quantification of the bands (e). α-Tubulin was used as a loading control. Data are presented as mean ± SEM. f HMGB1 levels in the cytosolic and nuclear fractions from day 4 pregnant uteri. Lamin A/C and α-Tubulin are markers for nuclei and cytosol, respectively. Cyt cytosol, Nuc nuclei. In each group, at least three independent samples from different mice were examined. All uteri were collected at 9–10 a.m. on the indicated day of pregnancy. See also Fig. S1
HMGB1 can be retained in the nucleus, but also in the cytosol in certain tissues under inflammatory conditions [16]. Cytosolic and nuclear extracts from day 4 uteri and immunostaining of sections of days 1–4 uteri show that HMGB1 is localized primarily in the nucleus (Figs. 1f and S1a). In this context, human endometrial cells also show nuclear localization of HMGB1 as depicted in the Protein Atlas [35]. Localization of HMGB1 protein is broader than that of Hmgb1 mRNA expression in mouse uteri (Figs. 1b and S1a), perhaps due to differential stabilities of protein versus mRNA [36].
Mice with uterine deletion of Hmgb1 show defective implantation and severe subfertility
To explore the function of HMGB1 in pregnant uteri, we generated mice with uterine deletion of Hmgb1 (Hmgb1d/d) by crossing Hmgb1 floxed mice (Hmgb1f/f) with PR-Cre transgenic mice (PgrCre/+) [22, 25]. These mice show efficient deletion of Hmgb1 in the pregnant uterus at both mRNA and protein levels (Figs. 1b, c and S1b–d). Some cells in deleted uteri remain HMGB1-positive in the stromal bed on day 4 (Fig. S1d). They are perhaps immune cells as evident by HMGB1 immunostaining in CD45-positive leukocytes in both genotypes (Fig. S1e). The absence of PR in CD45-positive cells provides evidence that the Pgr-Cre driver is unable to delete Hmgb1 in immune cells (Fig. S1e) [31]. The observation that there is no staining of HMGB1 in purified stromal cells from Hmgb1d/d uteri also supports our conjecture (Fig. S1f).
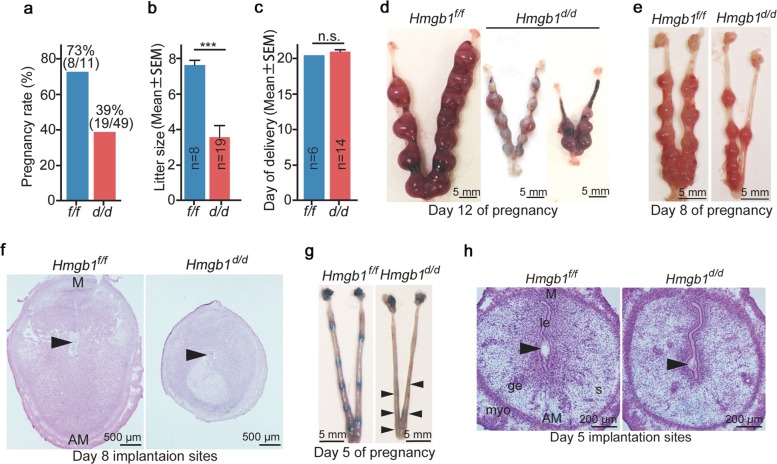
We explored the pregnancy outcome in uterine Hmgb1-deleted (Hmgb1d/d) females. We found that Hmgb1d/d females show severe subfertility: only 39% of plug-positive Hmgb1d/d females produce litters and the litter size is significantly smaller than those in littermate floxed mice (Fig. 2a, b). No differences are noted in birth timing between the two genotypes (Fig. 2c). Further analysis shows numerous embryo resorptions in Hmgb1d/d uteri on day 12 of pregnancy (Fig. 2d). In addition, the implantation sites on day 8 are much smaller in Hmgb1d/d mice (Fig. 2e, f). These observations prompted us to ask if Hmgb1 deletion in uteri compromises the embryo implantation process. To assess this, mice were intravenously injected with blue dye solution on day 5 for visualization of implantation sites [6, 37]. We found that Hmgb1d/d uteri have faint blue bands as compared with littermate floxed uteri (Fig. 2g). Histological analysis shows smaller implantation chambers with shrinkage of some blastocysts (Fig. 2h).
Fig. 2.
Deletion of Hmgb1 in uteri compromises embryo implantation and pregnancy outcome. a Pregnancy rate in Hmgb1f/f and Hmgb1d/d females. The number within brackets indicate females with pups over total number of plug-positive females. b Average number of litter sizes in Hmgb1f/f and Hmgb1d/d females. Hmgb1f/f (n = 8); Hmgb1d/d (n = 19). Data are presented as mean ± SEM, ***P < 0.001 (two-tailed Student’s t test). c Average day of delivery. n = 6 for Hmgb1f/f and n = 14 for Hmgb1d/d. Data are presented as mean ± SEM, n.s. not significant (two-tailed Student’s t test). d Representative images of day 12 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. Scale bar: 5 mm. e Representative images of day 8 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. Scale bar: 5 mm. f Histology of day 8 implantation sites from Hmgb1f/f and Hmgb1d/d females. Arrowheads indicate the location of embryos. M mesometrial pole, AM antimesometrial pole. Scale bar: 500 μm. g Representative images of day 5 implantation sites (blue bands) in Hmgb1f/f and Hmgb1d/d females. Arrowheads indicate blue bands. Scale bar: 5 mm. h Histology of day 5 implantation sites in Hmgb1f/f and Hmgb1d/d females. Arrowheads indicate the location of blastocysts. le luminal epithelium, s stroma, ge glandular epithelium, myo myometrium, M mesometrial pole, AM antimesometrial pole. Scale bar: 200 μm. Each image is a representative from at least three independent experiments. All uteri were collected at 9–10 a.m. on the indicated day of pregnancy
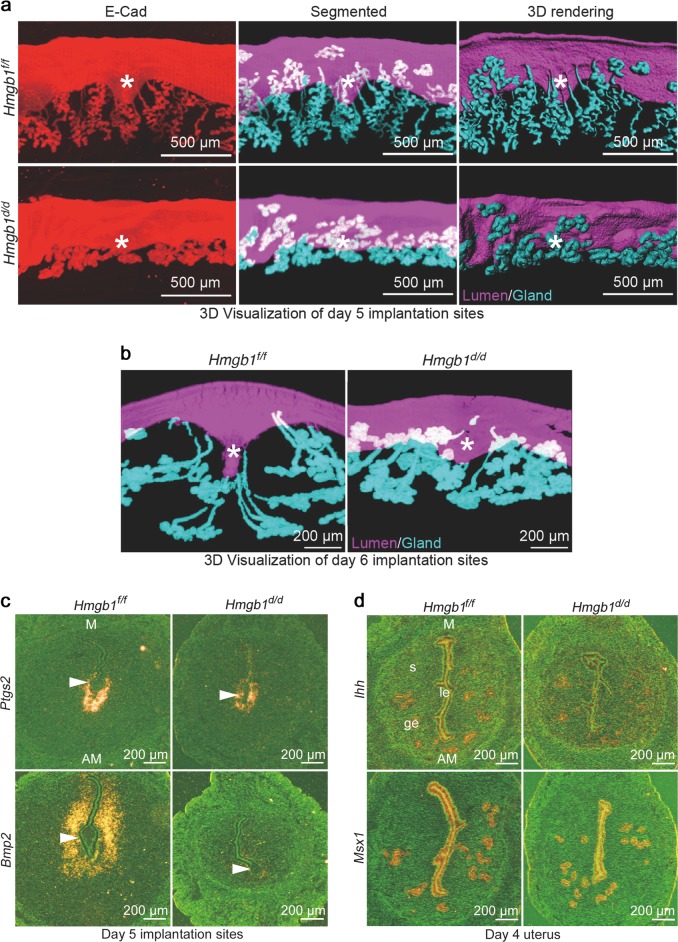
To determine the crypt (implantation chamber) formation and gland-crypt assembly for direct communication of the implanting blastocyst with the glands within the chamber, 3D visualization was employed after fixing and clearing tissues as we recently described [5]. Days 5 and 6 floxed uteri showed well-defined spear-shaped implantation chambers (crypts) with well drawn-out and developed glands [5]. We observed abnormal crypt formation and poorly elongated glands in uteri of Hmgb1d/d mice (Fig. 3a, b). These results suggest that embryo implantation and subsequent development are compromised by Hmgb1 deletion in the uterus. In addition to abnormal crypt formation, the attachment reaction appears defective as indicated by diminished Ptgs2 (encoding cyclooxygenase 2, Cox2) expression along with greatly reduced expression of bone morphogenetic protein (Bmp2), a marker for decidualization [1, 38, 39], in day 5 Hmgb1d/d uteri (Figs. 3c and S2a, b).
Fig. 3.
HMGB1 deletion in uteri causes abnormal decidualization. a Tridimensional (3D) imaging of day 5 implantation sites from Hmgb1f/f and Hmgb1d/d females. Staining of E-Cad (epithelial cell marker), segmented glands and uterine lumen, and 3D rendering of day 5 implantation site. Images were generated by a Nikon A1R Multiphoton Microscope with LWD 10× objective with 3 µm Z-stack. Asterisks indicate the location of embryos. Scale bar: 500 μm. b 3D visualization of day 6 implantation sites from Hmgb1f/f and Hmgb1d/d females. IF of E-Cad was performed to visualize LE and glands. Images were generated as described in a using LWD 16× water objective with 3 µm Z-stack. Asterisks indicate the location of embryos. Scale bar: 200 µm. c In situ hybridization of Ptgs2 and Bmp2 in day 5 implantation sites from Hmgb1f/f and Hmgb1d/d females. Arrowheads indicate the location of blastocysts. M mesometrial pole, AM antimesometrial pole. Scale bar: 200 μm. d In situ hybridization of Msx1 and Ihh in day 4 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. M mesometrial pole, AM antimesometrial pole. Scale bar: 200 μm. Each image is a representative of at least three different animals. All uteri were collected at 9–10 a.m. on the indicated day of pregnancy
These anomalies led us to assess the uterine receptivity in these mice. Two uterine receptivity marker genes Ihh (P4 responsive) and Msx1 (estrogen and P4 unresponsive) that are expressed in the epithelium were examined [1, 3]. We found that their expression levels and patterns are comparable between Hmgb1f/f and Hmgb1d/d mice on day 4 of pregnancy (Fig. 3d). These results led to further quest for the cause of implantation failure in Hmgb1-deleted females.
HMGB1-mediated PR activation is critical for uterine receptivity
The establishment of early pregnancy is accomplished by a dynamic interplay of maternal hormones [1]. Changing levels of ovarian hormones estrogen and P4 direct the establishment of uterine receptivity by guiding cell type specific proliferation and differentiation during days 1–4 of pregnancy [1, 2]. Compared with day 2, the day 3 uterus is marked by the termination of apoptosis in epithelial cells with the onset of stromal cell proliferation under the influence of increasing P4 levels from the newly formed corpora lutea [2, 3, 40]. The observation of defective implantation in Hmgb1-deleted uteri led us to explore whether the hormonal regulation is disturbed in these mice.
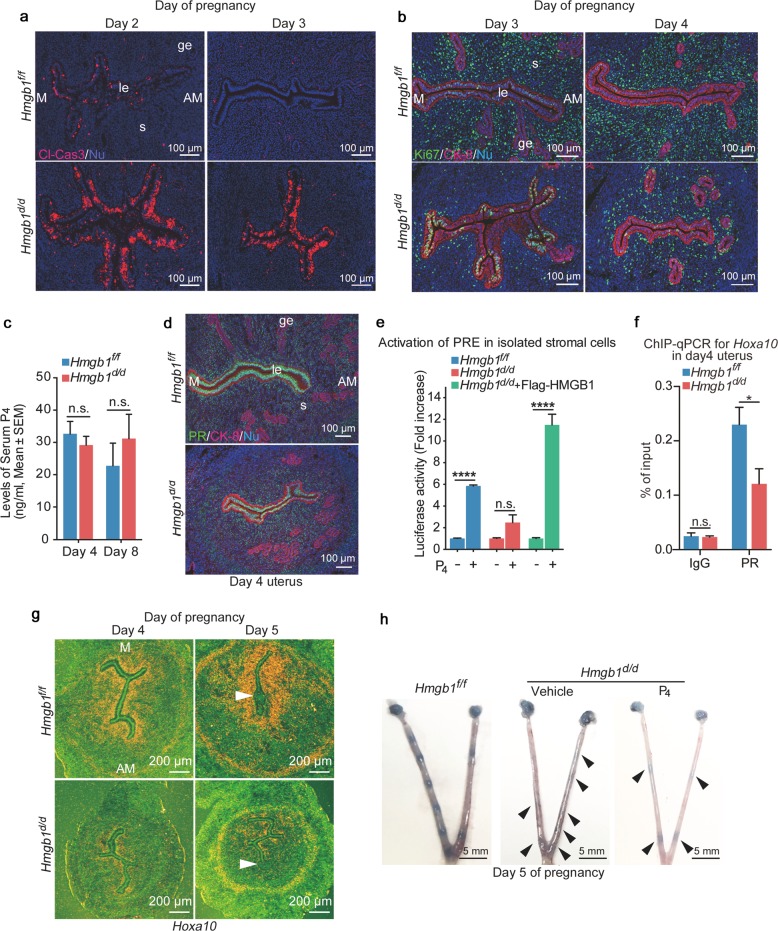
We sought to elucidate if uterine cell transition from day 2 to day 3 is compromised in deleted uteri. In fact, Hmgb1d/d uteri on day 3 show sustained apoptosis in epithelial cells as evident by cleaved caspase-3 signals (Fig. 4a). This aberrant apoptosis is followed by reduced stromal cell proliferation marked by reduced number of Ki67-positive stromal cells on days 3 and 4 in Hmgb1d/d females (Fig. 4b). Collectively, these results show a protracted transition of the uterus from days 2 to 3 in Hmgb1d/d females.
Fig. 4.
Decreased P4 responsiveness in Hmgb1d/d stroma. a IF of cleaved caspase-3 (apoptotic cell marker) in days 2 and 3 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. Cl-Cas3 cleaved caspase-3, le luminal epithelium, s stroma, ge glandular epithelium, M mesometrial pole, AM antimesometrial pole. Scale bar: 100 μm. b IF of Ki67 (cell proliferation marker) and CK-8 (epithelial cell marker) in days 3 and 4 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. le luminal epithelium, s stroma, ge glandular epithelium, M mesometrial pole, AM antimesometrial pole. Scale bar: 100 μm. c Serum P4 levels on days 4 and 8 were analyzed by Progesterone EIA kit. n = 5 for each genotype. Data are presented as mean ± SEM, n.s. not significant (two-way ANOVA followed by Bonferroni’s post hoc test). d IF of PR, and CK-8 (epithelial cell marker) in day 4 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. le luminal epithelium, s stroma, ge glandular epithelium, M mesometrial pole, AM antimesometrial pole. Scale bar: 100 μm. e Decreased PR-PRE activation in Hmgb1d/d stromal cells determined by PRE2-Tk-luciferase assay. 1 µM P4 was used as an agonist for PR. n = 4 for each group. Data are presented as mean ± SEM, ****P < 0.0001 and n.s. not significant (two-way ANOVA followed by Bonferroni’s post hoc test). f ChIP-qPCR targeting on the Hoxa10 locus shows that PR binding to the locus is reduced in Hmgb1d/d uteri on day 4 of pregnancy. Data are presented as mean ± SEM, *P < 0.05 and n.s. not significant (two-way ANOVA followed by Bonferroni’s post hoc test). g In situ hybridization of Hoxa10 in days 4 and 5 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. M mesometrial pole, AM antimesometrial pole. Arrowheads indicate the location of blastocysts. Scale bar: 200 μm. h Day 5 implantation sites (blue bands) in P4-supplemented Hmgb1d/d females. Arrowheads indicate weak blue bands. Scale bar: 5 mm. Each image is a representative from at least three independent experiments. All sera and uteri were collected at 9–10 a.m. on the indicated day of pregnancy. See also Figs. S2–S4
These results led us to examine if the serum levels of P4 or expression of steroid hormone receptors are different between Hmgb1f/f and Hmgb1d/d uteri. We found that serum P4 levels are comparable between the two groups (Fig. 4c) and that there are no apparent changes in PR and estrogen receptor (ERα) expression patterns in day 4 uteri (Figs. 4d and S3a). The expression of FOXA2, a critical transcription factor for uterine gland formation and function [5, 41], is also not affected in Hmgb1d/d uteri (Fig. S3b).
We have previously shown that poor P4 responsiveness in the endometrium accounts for abnormal cell proliferation and pregnancy failures [1, 7]. It was reported that HMGB1 facilitates transcriptional activities of PR by binding to progesterone response elements (PRE) [9, 13, 14]. Therefore, we assessed PR activation in the presence or absence of HMGB1 by PRE-luciferase reporter (PRE2-Tk-Luc) assays using isolated primary stromal cells from floxed and deleted mice. We found that PR activation is substantially reduced in Hmgbd/d stromal cells compared with Hmgb1f/f cells (Fig. 4e). Interestingly, reduced luciferase activity in Hmgb1d/d cells is rescued by transfection of Flag-HMGB1 in isolated stromal cells (Fig. 4e), further demonstrating the importance of HMGB1 in the activation of PRE-mediated transcription. Notably, GR and PR share the binding sequences on their response elements [42]. It is possible that uterine HMGB1 also affects GR activation, although GR is primarily expressed in endothelial or immune cells with no PR expression (Fig. S4a).
We also examined the binding of PR to the gene locus of Hoxa10, a P4-responsive gene that is expressed in the stroma [43]. Hoxa10-deficient females are infertile due to abnormal implantation with reduced stromal cell proliferation and decidualization [44]. We performed ChIP-qPCR for the Hoxa10 locus using a PR antibody and primers designed based on PR-ChIP-seq data [32]. We found poor binding of PR to the Hoxa10 locus in Hmgb1-deleted uteri (Fig. 4f), suggesting that the reduced level of PRE-mediated transcription is caused by defects in PR-PRE binding in the deleted uteri. Consistent with this finding is that Hoxa10 is downregulated in Hmgb1d/d uteri on days 4 and 5 (Figs. 4g and S4b). These results suggest that Hmgb1 deletion compromises stromal functions, conferring effects to interfere with epithelial-stromal interactions required for achieving uterine receptivity and implantation. In the context dependent manner, supplementation of P4 can be effective in improving pregnancy success in the face of reduced serum P4 levels or poor P4 binding to PR [45, 46]. To determine if insufficient PR signaling in Hmgb1d/d mice contributes to implantation failure, exogenous P4 (2 mg/mouse) was injected to Hmgb1d/d females as previously reported by us [46]. We found that this treatment is not effective to rescue defective implantation (Fig. 4h). This corroborates our observation that P4 fails to activate PRE-mediated transcription in Hmgb1-deleted cells. However, this deficit is rescued by replenishment of HMGB1 by transfection in deleted stromal cells (Fig. 4e). These results reinforce that PR-PRE binding is compromised in the uterus missing Hmgb1 and that is why P4 injections cannot rescue implantation defects in these mice.
Hmgb1d/d uteri show increased macrophage accumulation and cytokine levels
Following mating, the mouse uterus shows massive infiltration of leukocytes including macrophages along with increased levels of cytokines as examined on days 1 and 2 of pregnancy. These conditions subside on day 3, the day before embryos enter the uterine cavity with rising serum P4 levels [23, 47, 48]. In the uterus, PR signaling is reported to be associated with macrophage accumulation and P4 inhibits macrophage invasion into the endometrium and this inhibition is compromised in Pgr-deleted mice [24]. As described above, Hmgb1d/d uteri have reduced expression of Hoxa10 (Fig. 4g), a P4-responsive stromal gene. In this context, deletion of Hox genes including Hoxa10 causes increased expression of cytokines and macrophage accumulation in the mouse uterus [49].
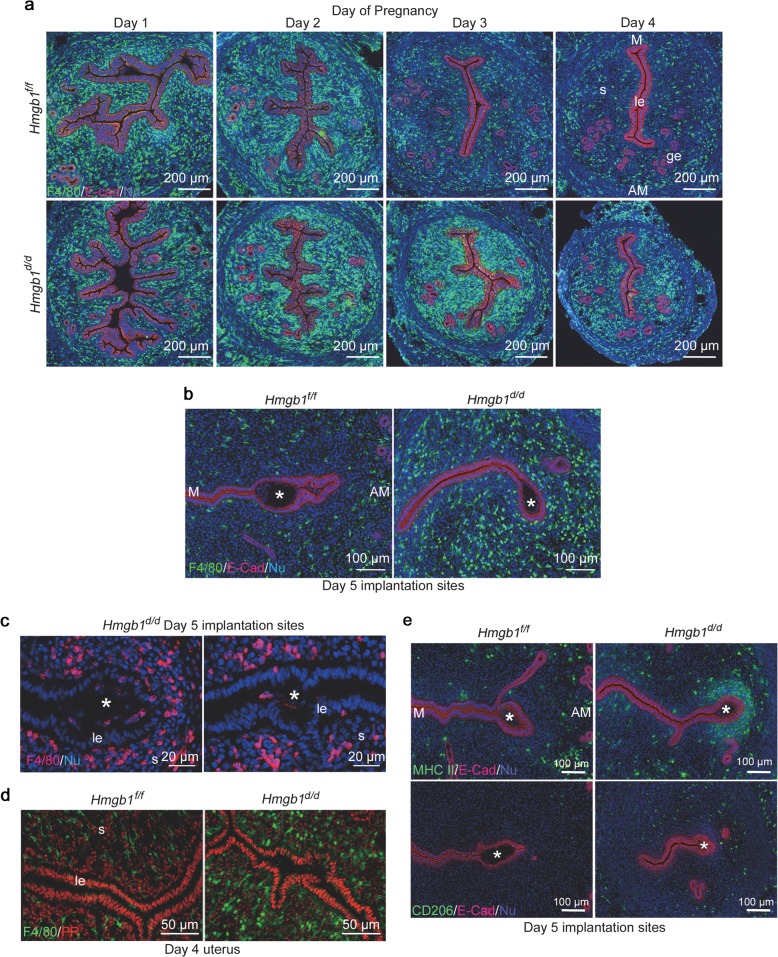
These results led us to ask if macrophage infiltration is affected in Hmgb1d/d uteri. We visualized the macrophage population in the stroma by IF of F4/80, a known macrophage marker. As previously reported [23], a massive accumulation of macrophages into the uterus was observed in Hmgb1f/f and Hmgb1d/d uteri on days 1 and 2. Interestingly, Hmgb1d/d uteri show a large number of macrophages even on day 3 and onward compared with floxed uteri (Figs. 5a, b and S5a, b). The increased macrophage population was observed over a total number of cells as well as per mm2 area in the stromal bed (Fig. S5a, b). Considering the 3D configuration of the uterus, this assessment is an approximation. Nonetheless, 3D visualization of F4/80 and E-Cad co-staining of day 5 uteri further reinforces that more macrophages are enriched throughout the uterus in Hmgb1d/d mice (Movies S1 and S2). We also analyzed the distribution of cells positive for CD11b, which is expressed in myeloid cells including monocytes and macrophages [50–52], and found that these cells accumulate around the implantation site (Fig. S5c). In WT uteri, macrophages accumulate close to the myometrial region, away from the implantation chamber (Figs. 5b and S5c), which is consistent with the previous study [48, 53]. In contrast, we observed abnormal distribution of macrophages in Hmgb1d/d deleted uteri with evidence of some macrophages invading the implantation chamber containing a blastocyst (Figs. 5b, c and S5c). This aberrant macrophage infiltration is not due to Hmgb1 deletion in macrophages, since they are PR-negative (Fig. 5d). Notably, Hmgb1 floxed and deleted mice show comparable number of F4/80-positive cells in the ovary (Fig. S5d). It is also intriguing that NK cells or neutrophils do not show abnormal populations in Hmgb1d/d uteri, suggesting that macrophages specifically accumulate in this milieu (Fig. S5e, f).
Fig. 5.
Deletion of Hmgb1 in uteri results in abnormal macrophage infiltration. a IF of F4/80 (macrophage marker) and E-Cad in days 1–4 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. le luminal epithelium, s stroma, ge glandular epithelium, M mesometrial pole, AM antimesometrial pole. Scale bar: 200 μm. b IF of F4/80 and E-Cad in day 5 pregnant uteri of Hmgb1f/f and Hmgb1d/d females. Asterisks indicate the location of blastocysts. M mesometrial pole, AM antimesometrial pole. Scale bar: 100 μm. c IF of F4/80 at higher magnification of day 5 pregnant uterine sections from Hmgb1d/d females. Invasion of macrophages into the implantation chamber (crypt) was observed. Asterisks indicate the location of blastocysts. le luminal epithelium, s stroma, IS implantation site. Scale bar: 20 µm. d IF of F4/80 and PR in day 4 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. le luminal epithelium, s stroma. Scale bar: 50 µm. e IF of MHC II (M1 macrophage marker; upper panels), CD206 (M2 macrophage marker; lower panels), and E-Cad in day 5 Hmgb1f/f and Hmgb1d/d pregnant uteri. Asterisks indicate the location of blastocysts. M mesometrial pole, AM antimesometrial pole. Scale bar: 100 µm. Each image is a representative from at least three independent experiments. All uteri were collected at 9–10 a.m. on the indicated day of pregnancy. See also Fig. S5, Movies S1 and S2
Macrophages are classified primarily into two subtypes, proinflammatory (M1: MHC II positive) and antiinflammatory (M2: CD206 positive) [54, 55]. In Hmgb1f/f uteri on day 5, both types of macrophages are located at the submyometrial region with few to none around the implanting blastocyst (Fig. 5e). In contrast, Hmgb1d/d uteri show a significant accumulation of MHC II positive cells in the vicinity of the blastocyst, while the distribution of CD206 positive cells is comparable to floxed uteri (Fig. 5e). Taken together, the results show that the Hmgb1-deleted uterine milieu is enriched with macrophage accumulation similar to that of PR or Hox mutants [24, 49].
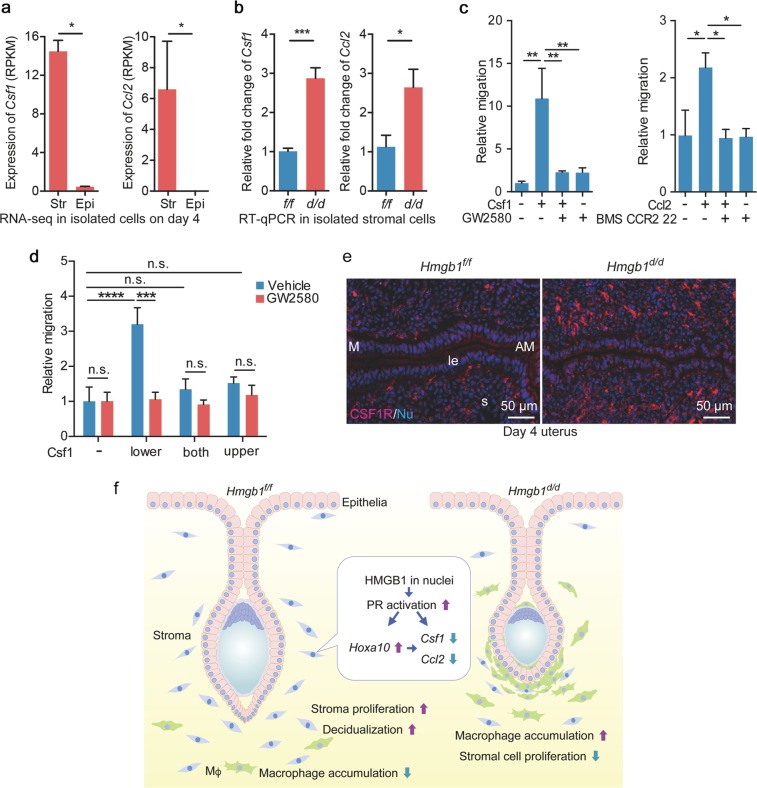
We asked how Hmgb1d/d uteri accumulate and retain macrophages for an extended time in the stroma. Macrophage migration is regulated by chemokines; Ccl2 and Csf1 are considered macrophage attractants [54]. These findings prompted us to examine the levels of chemokines in Hmgb1-deleted uteri. Our RNA-Seq analysis in isolated epithelial and stromal cells from day 4 WT uteri shows that the expression levels of these attractants are higher in stromal cells (Fig. 6a). By analyzing chemokines using qRT-PCR in isolated stromal cells after removing macrophages, we observed that Csf1 and Ccl2 are significantly upregulated in Hmgb1d/d stromal cells (Fig. 6b). We then examined if these chemokines serve as attractants by in vitro migration assays in a macrophage cell line. Indeed, both chemokines in the lower chambers attract macrophages from the upper chambers; CSF1 is more potent than Ccl2 in this assay (Fig. 6c). Interestingly, this effect was absent when Csf1 is added in both the upper and lower chambers or only in the upper chamber where macrophages are seeded on (Fig. 6d), suggesting that high levels of attractants surrounding macrophages suppress macrophage migration toward the outside. These results corroborate a higher level of Csf1 receptor (CSF1R)-positive cells in Hmgb1-deleted stroma (Fig. 6e). These observations suggest that Hmgb1-deleted uteri sustain macrophages in the stroma due to higher levels of attractants.
Fig. 6.
Hmgb1-deleted stromal cells cause increased cytokine levels. a Expression levels (RPKM) of macrophage attractants, Csf1 and Ccl2, were analyzed by RNA-seq analysis in isolated epithelial/stromal cells from day 4 uteri. n = 3 for each group. Str stroma, Epi epithelium. Data are presented as mean ± SEM, *P < 0.05 by two-tailed Student’s t -test. b Quantitative RT-PCR showed increased levels of macrophage attractants (Ccl2 and Csf1). n = 4 for each genotype. Data are presented as mean ± SEM, *P < 0.05 and ***P < 0.001 by two-tailed student’s t test. c Migration assays show that Csf1 and Ccl2 attract macrophages depending on their specific receptors. GW2580 and BMS CCR2 22 were used as Csf1 and Ccl2 receptor antagonists, respectively. These assays were performed in three wells for each group and three fields/wells were quantified. Data are presented as mean ± SEM, *P < 0.05 and **P < 0.01 (one-way ANOVA followed by Bonferroni’s post hoc test). d Migration assays show that Csf1 in the lower chamber attracts macrophages seeded in the upper chamber depending on its specific receptor. The assays were performed in three wells for each group and three fields/wells were quantified. Data are presented as mean ± SEM, *P < 0.05 and **P < 0.01 (two-way ANOVA followed by Bonferroni’s post hoc test). e IF of CSF1R in day 4 pregnant uteri from Hmgb1f/f and Hmgb1d/d females. le luminal epithelium, s: stroma, M mesometrial pole, AM antimesometrial pole. Scale bar: 50 µm. Each image is a representative from at least three independent experiments. f Schematic diagram showing the function of HMGB1 in uteri during pregnancy. All uteri were collected at 9–10 a.m. on day 4 of pregnancy
Discussion
The highlight of this investigation is that nuclear HMGB1 in the uterus is essential to pregnancy success in mice: uterine-specific deletion of Hmgb1 leads to defective implantation due to inefficient PR signaling, creating adverse ripple effects throughout the course of pregnancy (Fig. 6f). These results reveal a previously undocumented role of HMGB1. To our knowledge, this is the first report showing a relationship between HMGB1 and PR in vivo.
HMGB1’s association with steroid hormone receptors, including PR and GR, supports a role for nuclear HMGB1 [13, 14, 56]. HMGB1 has DNA-binding domains and bends DNA structures to facilitate interactions between transcriptional factors and their target sequences [9, 57]. Dean and his associates have shown that HMGB1 increases the DNA-binding affinity of PR and other steroid receptors [13, 14]. Here we show that uterine-specific deletion of Hmgb1 exhibits reduced PR activation. P4-PR signaling is absolutely critical for embryo implantation and maintenance of pregnancy [1]. Systemic Pgr mutant mice show infertility [58], and suppression of PR activation by an antagonist causes pregnancy failure [59]. In addition, deletion of Fkbp52, a co-chaperone of PR, compromises embryo implantation due to reduced binding of P4 to PR [7]. In this study, we provide new insight for PR activation in pregnancy by DNA-binding protein HMGB1. P4 injection has been used to rescue embryo implantation and decidualization attributed by reduced serum P4 levels or poor P4 binding to PR [45, 46]. P4 administration, however, is ineffective to rescue embryo implantation in Hmgb1d/d mice, indicating that HMGB1 deficiency is not replaceable by P4 supplementation in the face of reduced PR-PRE activation. This is compatible with the finding of comparable serum levels of P4 between floxed and deleted mice on day 4.
Although the association of HMGB1 with PR activation is observed in the uterus, how HMGB1 specifically binds to PRE and regulates PR-responsive genes is unknown. There are reports that HMGB1 does not have any binding specificity to DNA sequences [9, 11, 12]. This suggests that HMGB1 cooperates with certain co-regulators to recognize PRE and regulate gene expression. Since HMGB1 also shows association with other transcriptional factors such as Hox proteins and Rel family members [9], it will be intriguing to see the landscape of HMGB1 function in the whole genome, which is yet to be determined.
After mating, a large number of leukocytes, including macrophages, accumulate in the uterus influenced by estrogen and seminal fluid, evoking inflammatory-like responses [60]. Those accumulated immune cells begin to migrate away from the stroma from day 3 onward and gradually assembles in the myometrium [23, 48]. It is thought that this gradual migration from the stroma is critical for successful pregnancy [48, 61] and is associated with rising serum P4 levels from the newly formed corpus lutea [23].
Macrophages are retained in Hmgb1-deleted uteri on day 3 and later. This result places the significance of HMGB1 as a regulator of macrophage accumulation and migration. Csf1 and Ccl2 are considered critical attractants for macrophage migration [54]. Deficient Csf1 levels in Csf1op/op mice were shown to reduce the macrophage population in the uterus, but the phenotype was rescued by Csf1 overexpression [62]. Tissue-specific enhancement of Csf1 expression is accompanied with increased macrophage density [63]. Our results showing increased expression of Csf1 mRNA in deleted uteri suggest that Csf1 is associated with macrophage retention. The regulation of uterine Csf1 expression is unclear. Nonetheless, present evidence indicates that HMGB1 is a regulator of Csf1 induction and macrophage enrichment in the stroma. These results suggest that nuclear HMGB1 plays a critical role in preparation of the uterus for implantation.
Our study presents genetic evidence that HMGB1 is essential for early pregnancy events prior to and during blastocyst implantation. We believe that defective decidualization is a consequence of the derailed implantation process, creating adverse ripple effects [4, 27, 28]. It also remains unclear as to the differential display of HMGB1 protein and mRNA in day 4 uteri. It is possible that Hmgb1 expressed in epithelial cells on days 1–3 is retained on day 4 due to its slower turnover rate, although we do not observe obvious changes in epithelial-receptivity marker genes in Hmgb1d/d day 4 uteri. The function of epithelial HMGB1 can be studied in the future using a Ltf-Cre driver, which deletes genes of interest in the uterine epithelium [64].
Taken together, our study addresses a novel role of HMGB1 in uteri during the periimplantation period. The unappreciated role of HMGB1 in macrophage distribution depicts a new aspect for fertility and pregnancy events. HMGB1 is evolutionally conserved in mammalians including humans with >99% protein identity [10]. In addition, HMGB1 is localized in human endometrial cells [35]. Considering the absolute requirement of P4-PR signaling for the establishment of pregnancy in most of eutherian mammals, our finding showing a role of HMGB1 for PR activation will inspire future studies in humans and other species.
Supplementary information
Acknowledgements
The authors appreciate Katie Gerhardt for editing the manuscript. Pgr-Cre mice were originally obtained from Francesco DeMayo and John P. Lydon (Baylor College of Medicine). Robert F. Schwabe (Columbia University) originally provided the floxed Hmgb1 mouse line. The vector for PRE2-Tk-Luc was a gift from Dean P. Edwards (Baylor College of Medicine). This work was supported in part by NIH grants (HD068524 and DA006668 to SKD) and a March of Dimes Center grant (22-FY17-889). SA was supported by Astellas Foundation for Research on Metabolic Disorder Fellowship for Study Abroad and The Osamu Hayaishi Memorial Foundation Fellowship for Study Abroad. SA is now supported by JSPS Overseas Research Fellowships.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by M. Piacentini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Shizu Aikawa, Wenbo Deng
Supplementary information
The online version of this article (10.1038/s41418-019-0429-z) contains supplementary material, which is available to authorized users.
References
- 1.Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18:1754–67. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dey SK, Lim H, Das SK, Reese J, Paria BC, Daikoku T, et al. Molecular cues to implantation. Endocr Rev. 2004;25:341–73. doi: 10.1210/er.2003-0020. [DOI] [PubMed] [Google Scholar]
- 3.Daikoku T, Cha J, Sun X, Tranguch S, Xie H, Fujita T, et al. Conditional deletion of Msx homeobox genes in the uterus inhibits blastocyst implantation by altering uterine receptivity. Dev Cell. 2011;21:1014–25. doi: 10.1016/j.devcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song H, Lim H, Paria BC, Matsumoto H, Swift LL, Morrow J, et al. Cytosolic phospholipase A2alpha is crucial [correction of A2alpha deficiency is crucial] for ‘on-time’ embryo implantation that directs subsequent development. Development. 2002;129:2879–89. doi: 10.1242/dev.129.12.2879. [DOI] [PubMed] [Google Scholar]
- 5.Yuan J, Deng W, Cha J, Sun X, Borg JP, Dey SK. Tridimensional visualization reveals direct communication between the embryo and glands critical for implantation. Nat Commun. 2018;9:603. doi: 10.1038/s41467-018-03092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Psychoyos A. Endocrine control of egg implantation. In: Greep EGARO, Geiger SR, editors. Handbook of physiology. Washington, D.C.: American Physiology Society; 1973.
- 7.Tranguch S, Cheung-Flynn J, Daikoku T, Prapapanich V, Cox MB, Xie H, et al. Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc Natl Acad Sci USA. 2005;102:14326–31. doi: 10.1073/pnas.0505775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin Q, Kong S, Yan J, Qiu J, He B, Zhou C, et al. Polycomb subunit BMI1 determines uterine progesterone responsiveness essential for normal embryo implantation. J Clin Investig. 2018;128:175–89. doi: 10.1172/JCI92862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bianchi ME, Agresti A. HMG proteins: dynamic players in gene regulation and differentiation. Curr Opin Genet Dev. 2005;15:496–506. doi: 10.1016/j.gde.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Sessa L, Bianchi ME. The evolution of high mobility group box (HMGB) chromatin proteins in multicellular animals. Gene. 2007;387:133–40. doi: 10.1016/j.gene.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Murphy FV, Sweet RM, Churchill ME. The structure of a chromosomal high mobility group protein-DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 1999;18:6610–8. doi: 10.1093/emboj/18.23.6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paull TT, Haykinson MJ, Johnson RC. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 1993;7:1521–34. doi: 10.1101/gad.7.8.1521. [DOI] [PubMed] [Google Scholar]
- 13.Onate SA, Prendergast P, Wagner JP, Nissen M, Reeves R, Pettijohn DE, et al. The DNA-bending protein HMG-1 enhances progesterone receptor binding to its target DNA sequences. Mol Cell Biol. 1994;14:3376–91. doi: 10.1128/mcb.14.5.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boonyaratanakornkit V, Melvin V, Prendergast P, Altmann M, Ronfani L, Bianchi ME, et al. High-mobility group chromatin proteins 1 and 2 functionally interact with steroid hormone receptors to enhance their DNA binding in vitro and transcriptional activity in mammalian cells. Mol Cell Biol. 1998;18:4471–87. doi: 10.1128/mcb.18.8.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calogero S, Grassi F, Aguzzi A, Voigtländer T, Ferrier P, Ferrari S, et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat Genet. 1999;22:276–80. doi: 10.1038/10338. [DOI] [PubMed] [Google Scholar]
- 16.Lotze MT, Tracey KJ. High-mobility group box 1 protein (HMGB1): nuclear weapon in the immune arsenal. Nat Rev Immunol. 2005;5:331–42. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 17.Harris HE, Andersson U, Pisetsky DS. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat Rev Rheumatol. 2012;8:195–202. doi: 10.1038/nrrheum.2011.222. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, et al. HMG-1 as a late mediator of endotoxin lethality in mice. Science. 1999;285:248–51. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 19.Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez C, Huebener P, Pradere JP, Antoine DJ, Friedman RA, Schwabe RF. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J Clin Investig. 2018;128:2436–51. doi: 10.1172/JCI91786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng M, Tang Y, Li W, Wang X, Zhang R, Zhang X, et al. The endotoxin delivery protein HMGB1 mediates caspase-11-dependent lethality in sepsis. Immunity. 2018;49:740–53 e7. doi: 10.1016/j.immuni.2018.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huebener P, Gwak GY, Pradere JP, Quinzii CM, Friedman R, Lin CS, et al. High-mobility group box 1 is dispensable for autophagy, mitochondrial quality control, and organ function in vivo. Cell Metab. 2014;19:539–47. doi: 10.1016/j.cmet.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McMaster MT, Newton RC, Dey SK, Andrews GK. Activation and distribution of inflammatory cells in the mouse uterus during the preimplantation period. J Immunol. 1992;148:1699–705. [PubMed] [Google Scholar]
- 24.Tibbetts TA, Conneely OM, O’Malley BW. Progesterone via its receptor antagonizes the pro-inflammatory activity of estrogen in the mouse uterus. Biol Reprod. 1999;60:1158–65. doi: 10.1095/biolreprod60.5.1158. [DOI] [PubMed] [Google Scholar]
- 25.Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41:58–66. doi: 10.1002/gene.20098. [DOI] [PubMed] [Google Scholar]
- 26.Hirota Y, Daikoku T, Tranguch S, Xie H, Bradshaw HB, Dey SK. Uterine-specific p53 deficiency confers premature uterine senescence and promotes preterm birth in mice. J Clin Investig. 2010;120:803–15. doi: 10.1172/JCI40051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha J, Bartos A, Park C, Sun X, Li Y, Cha SW, et al. Appropriate crypt formation in the uterus for embryo homing and implantation requires Wnt5a-ROR signaling. Cell Rep. 2014;8:382–92. doi: 10.1016/j.celrep.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan J, Cha J, Deng W, Bartos A, Sun X, Ho HH, et al. Planar cell polarity signaling in the uterus directs appropriate positioning of the crypt for embryo implantation. Proc Natl Acad Sci USA. 2016;113:E8079–88. doi: 10.1073/pnas.1614946113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daikoku T, Tranguch S, Friedman DB, Das SK, Smith DF, Dey SK. Proteomic analysis identifies immunophilin FK506 binding protein 4 (FKBP52) as a downstream target of Hoxa10 in the periimplantation mouse uterus. Mol Endocrinol. 2005;19:683–97. doi: 10.1210/me.2004-0332. [DOI] [PubMed] [Google Scholar]
- 30.Ma W, Tan J, Matsumoto H, Robert B, Abrahamson DR, Das SK, et al. Adult tissue angiogenesis: evidence for negative regulation by estrogen in the uterus. Mol Endocrinol. 2001;15:1983–92. doi: 10.1210/mend.15.11.0734. [DOI] [PubMed] [Google Scholar]
- 31.Deng W, Yuan J, Cha J, Sun X, Bartos A, Yagita H, et al. Endothelial cells in the decidual bed are potential therapeutic targets for preterm birth prevention. Cell Rep. 2019;27:1755–68.e4. doi: 10.1016/j.celrep.2019.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rubel CA, Lanz RB, Kommagani R, Franco HL, Lydon JP, DeMayo FJ. Research resource: genome-wide profiling of progesterone receptor binding in the mouse uterus. Mol Endocrinol. 2012;26:1428–42. doi: 10.1210/me.2011-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hock R, Furusawa T, Ueda T, Bustin M. HMG chromosomal proteins in development and disease. Trends Cell Biol. 2007;17:72–9. doi: 10.1016/j.tcb.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–50. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Paria BC, Huet-Hudson YM, Dey SK. Blastocyst’s state of activity determines the “window” of implantation in the receptive mouse uterus. Proc Natl Acad Sci USA. 1993;90:10159–62. doi: 10.1073/pnas.90.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim H, Paria BC, Das SK, Dinchuk JE, Langenbach R, Trzaskos JM, et al. Multiple female reproductive failures in cyclooxygenase 2-deficient mice. Cell. 1997;91:197–208. doi: 10.1016/s0092-8674(00)80402-x. [DOI] [PubMed] [Google Scholar]
- 39.Lee KY, Jeong JW, Wang J, Ma L, Martin JF, Tsai SY, et al. Bmp2 is critical for the murine uterine decidual response. Mol Cell Biol. 2007;27:5468–78. doi: 10.1128/MCB.00342-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huet-Hudson YM, Andrews GK, Dey SK. Cell type-specific localization of c-myc protein in the mouse uterus: modulation by steroid hormones and analysis of the periimplantation period. Endocrinology. 1989;125:1683–90. doi: 10.1210/endo-125-3-1683. [DOI] [PubMed] [Google Scholar]
- 41.Jeong JW, Kwak I, Lee KY, Kim TH, Large MJ, Stewart CL, et al. Foxa2 is essential for mouse endometrial gland development and fertility. Biol Reprod. 2010;83:396–403. doi: 10.1095/biolreprod.109.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strähle U, Klock G, Schütz GA. DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc Natl Acad Sci USA. 1987;84:7871–5. doi: 10.1073/pnas.84.22.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim H, Ma L, Ma WG, Maas RL, Dey SK. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol Endocrinol. 1999;13:1005–17. doi: 10.1210/mend.13.6.0284. [DOI] [PubMed] [Google Scholar]
- 44.Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122:2687–96. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- 45.Sun X, Terakawa J, Clevers H, Barker N, Daikoku T, Dey SK. Ovarian LGR5 is critical for successful pregnancy. FASEB J. 2014;28:2380–9. doi: 10.1096/fj.13-248344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tranguch S, Wang H, Daikoku T, Xie H, Smith DF, Dey SK. FKBP52 deficiency-conferred uterine progesterone resistance is genetic background and pregnancy stage specific. J Clin Invest. 2007;117:1824–34. doi: 10.1172/JCI31622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McMaster MT, Dey SK, Andrews GK. Association of monocytes and neutrophils with early events of blastocyst implantation in mice. J Reprod Fertil. 1993;99:561–9. doi: 10.1530/jrf.0.0990561. [DOI] [PubMed] [Google Scholar]
- 48.Tachi C, Tachi S. Macrophages and implantation. Ann N Y Acad Sci. 1986;476:158–82. doi: 10.1111/j.1749-6632.1986.tb20929.x. [DOI] [PubMed] [Google Scholar]
- 49.Raines AM, Adam M, Magella B, Meyer SE, Grimes HL, Dey SK, et al. Recombineering-based dissection of flanking and paralogous Hox gene functions in mouse reproductive tracts. Development. 2013;140:2942–52. doi: 10.1242/dev.092569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sunderkötter C, Nikolic T, Dillon MJ, Van Rooijen N, Stehling M, Drevets DA, et al. Subpopulations of mouse blood monocytes differ in maturation stage and inflammatory response. J Immunol. 2004;172:4410–7. doi: 10.4049/jimmunol.172.7.4410. [DOI] [PubMed] [Google Scholar]
- 51.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins MK, Tay CS, Erlebacher A. Dendritic cell entrapment within the pregnant uterus inhibits immune surveillance of the maternal/fetal interface in mice. J Clin Invest. 2009;119:2062–73. doi: 10.1172/JCI38714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tagliani E, Shi C, Nancy P, Tay CS, Pamer EG, Erlebacher A. Coordinate regulation of tissue macrophage and dendritic cell population dynamics by CSF-1. J Exp Med. 2011;208:1901–16. doi: 10.1084/jem.20110866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Investig. 2012;122:787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Agresti A, Scaffidi P, Riva A, Caiolfa VR, Bianchi ME. GR and HMGB1 interact only within chromatin and influence each other’s residence time. Mol Cell. 2005;18:109–21. doi: 10.1016/j.molcel.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 57.Laudet V, Stehelin D, Clevers H. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 1993;21:2493–501. doi: 10.1093/nar/21.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–78. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 59.Psychoyos A, Nikas G, Sarantis L, Gravanis A. Hormonal anti-implantation agents: antiprogestins. Hum Reprod. 1995;10(Suppl 2):140–50. doi: 10.1093/humrep/10.suppl_2.140. [DOI] [PubMed] [Google Scholar]
- 60.Robertson SA. Seminal plasma and male factor signalling in the female reproductive tract. Cell Tissue Res. 2005;322:43–52. doi: 10.1007/s00441-005-1127-3. [DOI] [PubMed] [Google Scholar]
- 61.PrabhuDas M, Bonney E, Caron K, Dey S, Erlebacher A, Fazleabas A, et al. Immune mechanisms at the maternal-fetal interface: perspectives and challenges. Nat Immunol. 2015;16:328–34. doi: 10.1038/ni.3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ryan GR, Dai XM, Dominguez MG, Tong W, Chuan F, Chisholm O, et al. Rescue of the colony-stimulating factor 1 (CSF-1)-nullizygous mouse (Csf1(op)/Csf1(op)) phenotype with a CSF-1 transgene and identification of sites of local CSF-1 synthesis. Blood. 2001;98:74–84. doi: 10.1182/blood.v98.1.74. [DOI] [PubMed] [Google Scholar]
- 63.Cohen PE, Nishimura K, Zhu L, Pollard JW. Macrophages: important accessory cells for reproductive function. J Leukoc Biol. 1999;66:765–72. doi: 10.1002/jlb.66.5.765. [DOI] [PubMed] [Google Scholar]
- 64.Daikoku T, Ogawa Y, Terakawa J, Ogawa A, DeFalco T, Dey SK. Lactoferrin-iCre: a new mouse line to study uterine epithelial gene function. Endocrinology. 2014;155:2718–24. doi: 10.1210/en.2014-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.