Abstract
Aging-related cognitive ability impairments are one of the main threats to public health, and impaired hippocampal neurogenesis is a major cause of cognitive decline during aging. However, the regulation of adult neurogenesis in the hippocampus requires further study. Here, we investigated the role of microRNA-153 (miR-153), a highly conserved microRNA in mice and humans, in adult neurogenesis. During the passaging of neural stem cells (NSCs) in vitro, endogenous miR-153 expression was downregulated, with a decrease in neuronal differentiation ability. In addition, miR-153 overexpression increased the neurogenesis of NSCs. Further studies showed that miR-153 regulated neurogenesis by precisely targeting the Notch signaling pathway through inhibition of Jagged1 and Hey2 translation. In vivo analysis demonstrated that miR-153 expression was decreased in the hippocampi of aged mice with impaired cognitive ability, and that miR-153 overexpression in the hippocampus promoted neurogenesis and markedly increased the cognitive abilities of the aged mice. Overall, our findings revealed that miR-153 affected neurogenesis by regulating the Notch signaling pathway and elucidated the function of miR-153 in aging-related, hippocampus-dependent cognitive ability impairments, and neurodegenerative diseases.
Subject terms: Epigenetics, Cognitive ageing
Introduction
With aging, the brain gradually loses its ability to respond to external stimuli, and decreased brain plasticity is accompanied by individual cognitive dysfunctions [1, 2]. The hippocampus is closely associated with cognitive function and is considered a key area affected by aging [3–6]. Within the hippocampus, adult neurogenesis mainly occurs in the subgranular zone (SGZ) of the dentate gyrus (DG), which contains adult neural stem cells (NSCs) [7–9]. Adult NSCs maintain the ability to differentiate into neurons, and aging is associated with a decreased number of newborn neurons in the DG, as well as an increased number of astrocytes [10–12]. Moreover, the decrease in neurogenesis is closely associated with a decline in hippocampus-dependent cognition [2, 13, 14], suggesting that aging-related neurogenesis impairments may contribute to hippocampal functional loss. Therefore, understanding the molecular mechanisms underlying NSC neurogenesis in the adult hippocampus is of crucial importance for ameliorating hippocampus-dependent cognitive impairment.
NSCs specifically express a series of microRNAs (miRNAs) that are involved in the maintenance of self-renewal and differentiation. Let-7b targets Hmga2, TLX, and cyclin D1 to precipitate the cell cycle exit of NSCs and promote their differentiation [15–20]. Furthermore, several miRNAs are considered to be closely associated with brain aging [21], such as miR-219, which increases the myelin plasticity of aging mouse brains and delays aging-induced cognitive impairment [22]. Although these studies have shown that miRNAs can regulate NSC differentiation and are closely associated with the cognitive impairment caused by aging, whether miRNAs can directly regulate neurogenesis in the adult hippocampus to affect age-related cognitive decline remains unclear.
MiR-153 regulates neurobehavioral development in zebrafish and inhibits gliogenesis during mouse embryonic development [23, 24]; however, few studies have focused on the function of miR-153 in adult neurogenesis. Here, we found that miR-153 promotes NSC neurogenesis in vitro and in vivo and that miR-153 functions by specifically modifying the Notch signaling pathway, which is considered a master controller of NSC differentiation by promoting gliogenesis and inhibiting neurogenesis [25–27], though targeting Jag1 and Hey2. Moreover, miR-153 mediated the aging-related, hippocampus-dependent impairment of learning and memory abilities, while exogenous miR-153 overexpression in the aged mouse hippocampus enhanced neurogenesis and rescued cognitive decline.
Results
miR-153 is abundantly expressed in neural tissues and downregulated with neurogenesis decrease
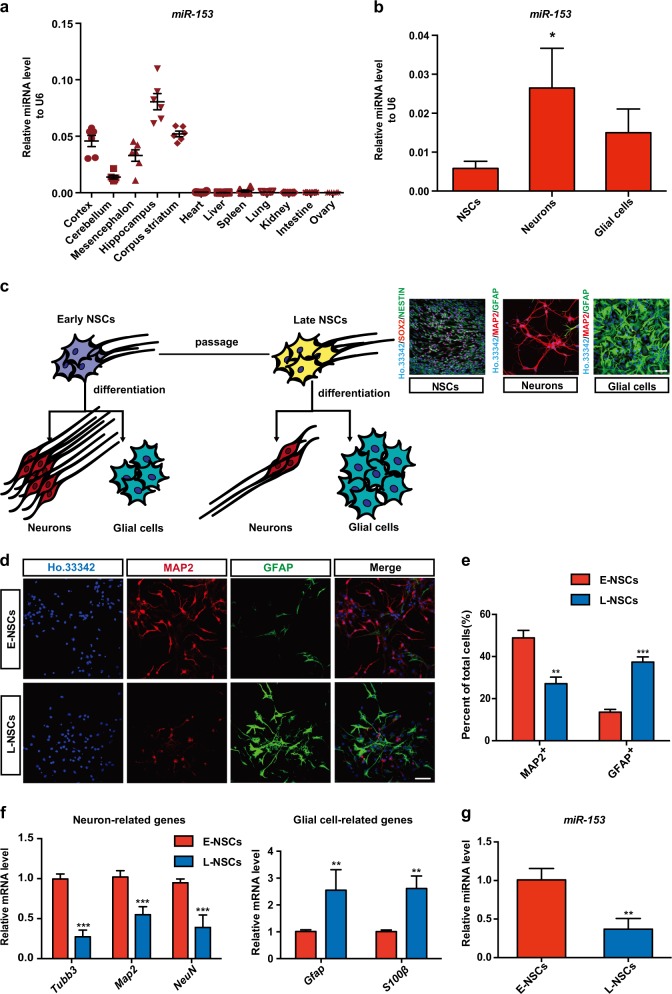
To understand the function of miR-153, we measured miR-153 expression in different mouse tissues and found abundant expression in neural tissues, especially in the hippocampus (Fig. 1a). Therefore, we next isolated NSCs for culture and differentiation in vitro and found significantly high miR-153 expression in neurons (Fig. 1b), indicating a possible linkage between miR-153 and neuronal differentiation. Previous studies have shown that the neurogenesis capacity decreases as NSCs are cultured in vitro [28]. In our study, we also found this phenomenon and divided NSCs into early NSCs (E-NSCs) and late NSCs (L-NSCs) according to their neuronal differentiation ability (Fig. 1c). Two days after differentiation, L-NSCs differentiated less into neurons and more often into glial cells, as indicated by immunofluorescence staining (Fig. 1d, e). Quantitative RT-PCR (qRT-PCR) showed downregulation of the neuron-related genes Tubb3, Map2, and NeuN and upregulation of the glial cell-related genes Gfap and S100β in the L-NSC group (Fig. 1f). Interestingly, we found significantly lower miR-153 expression in L-NSCs compared with that in E-NSCs (Fig. 1g). Therefore, we speculated that miR-153 plays an important role in neuronal differentiation.
Fig. 1.
miR-153 is abundantly expressed in neural tissues and downregulated with neurogenesis decreased. a Quantitative RT-PCR detection of miR-153 in different mouse tissues, n = 6. b Quantitative RT-PCR detection of miR-153 in neural stem cells (NSCs), neurons, and glial cells; immunostaining images of SOX2 (red), NESTIN (green), and Ho.33342 (blue) in NSCs and of the neuron marker MAP2 (red), the glial cell marker GFAP (green), and Ho.33342 (blue) in neurons and glial cells are shown. Scale bar: 50 μm. c Model of neural differentiation: early NSCs (E-NSCs) exhibited a high level of differentiation into neurons, while late NSCs (L-NSCs) differentiated into fewer neurons. d Representative images showing staining for MAP2 (red), GFAP (green), and Ho.33342 (blue) to evaluate neuron differentiation. Scale bar: 50 μm. e Percentage of MAP2-positive and GFAP-positive cells in neural differentiation. f Quantitative RT-PCR detection of neuron-related genes (Tubb3, Map2, NeuN) and glial cell-related genes (Gfap, S100β) in the neural differentiation of E-NSCs and L-NSCs. g Quantitative RT-PCR detection of miR-153 in E-NSCs and L-NSCs. Data information: the data shown are the mean ± SEM, n = 3; ANOVA; ***p < 0.001, **p < 0.01, *p < 0.05. Gapdh or U6 was used as an internal control for normalization in quantitative RT-PCR
miR-153 promotes neurogenesis and inhibits gliogenesis
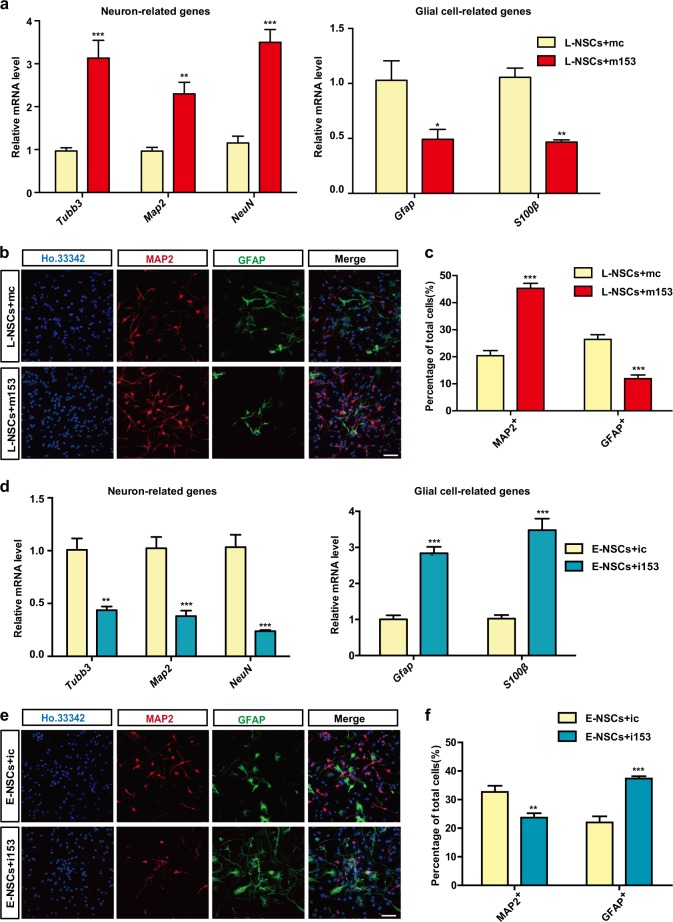
To determine the function of miR-153 in neuronal differentiation, we overexpressed miR-153 in L-NSCs through transfection with miR-153 mimics (m153). Flow-cytometry analysis showed that more than 94% of cells were cy5-positive after transfection with fluorescently labeled miRNA mimics or inhibitors, indicating that the mimics and inhibitors could be efficiently transfected into NSCs (Fig. S1a–e). To determine the function of miR-153 in neuronal differentiation, we overexpressed miR-153 in L-NSCs through transfection with m153. Two days after differentiation, in the m153 group, the mRNA levels of neuron-related genes were significantly increased, while those of glial cell-related genes were decreased (Fig. 2a; Fig. S1f), and a higher percentage of MAP2-positive neurons and a lower percentage of GFAP-positive glial cells were observed (Fig. 2b, c). In contrast, when miR-153 was inhibited in E-NSCs with a miR-153 inhibitor (i153), the mRNA levels of neuron-related genes were downregulated, and those of glial cell-related genes were upregulated (Fig. 2d). The percentage of MAP2-positive neurons was decreased, while the percentage of GFAP-positive glial cells was increased in the i153 group (Fig. 2e, f). To further investigate the function of miR-153 in neurogenesis, we also used a lentivirus, lenti-sponge-153-wpre (lenti-sp-153-wpre), to inhibit miR-153 function by sponging miR-153 in NSCs [29, 30]. The sponge-153 was effectively expressed in NSCs as the wpre level was significantly increased (Fig. S1g). The NSCs infected with lenti-sp-153-wpre were subjected to qRT-PCR and immunofluorescence analyses, and the results were similar to those obtained for the i153 group (Fig. S1h–j). These data suggest that miR-153 critically regulates neurogenesis and gliogenesis.
Fig. 2.
miR-153 promotes neurogenesis and inhibits gliogenesis. a Quantitative RT-PCR detection of neuron-related genes (Tubb3, Map2, NeuN) and astrocyte-related genes (Gfap, S100β) in the neural differentiation of L-NSCs + mc or L-NSCs + m153 (L-NSCs transfected with control or miR-153 mimics). b Immunofluorescence analysis of neural differentiation with L-NSCs + mc or L-NSCs + m153 as evaluated by MAP2 (red), GFAP (green), and Ho.33342 (blue). Scale bar: 50 μm. c Percentages of MAP2-positive and GFAP-positive cells in differentiation. d Quantitative RT-PCR detection of neuron-related genes and glial cell-related genes in the neural differentiation of E-NSCs + ic or E-NSCs + i153 (E-NSCs transfected with ctrl or miR-153 inhibitor). e Immunofluorescence analysis of neural differentiation with E-NSCs + ic or E-NSCs + i153 as evaluated by MAP2 (red), GFAP (green), and Ho.33342 (blue). Scale bar: 50 μm. f Percentages of MAP2-positive and GFAP-positive cells in neural differentiation. Data information: the data shown are the mean ± SEM, n = 3; ANOVA; ***p < 0.001, **p < 0.01, *p < 0.05. Gapdh was used as an internal control for normalization in quantitative RT-PCR. L-NSCs late NSCs, E-NSCs early NSCs, mc control mimics, m153 miR-153 mimics, ic control inhibitor, i153 miR-153 inhibitor
Neurogenesis is regulated by miR-153 through the Notch signaling pathway
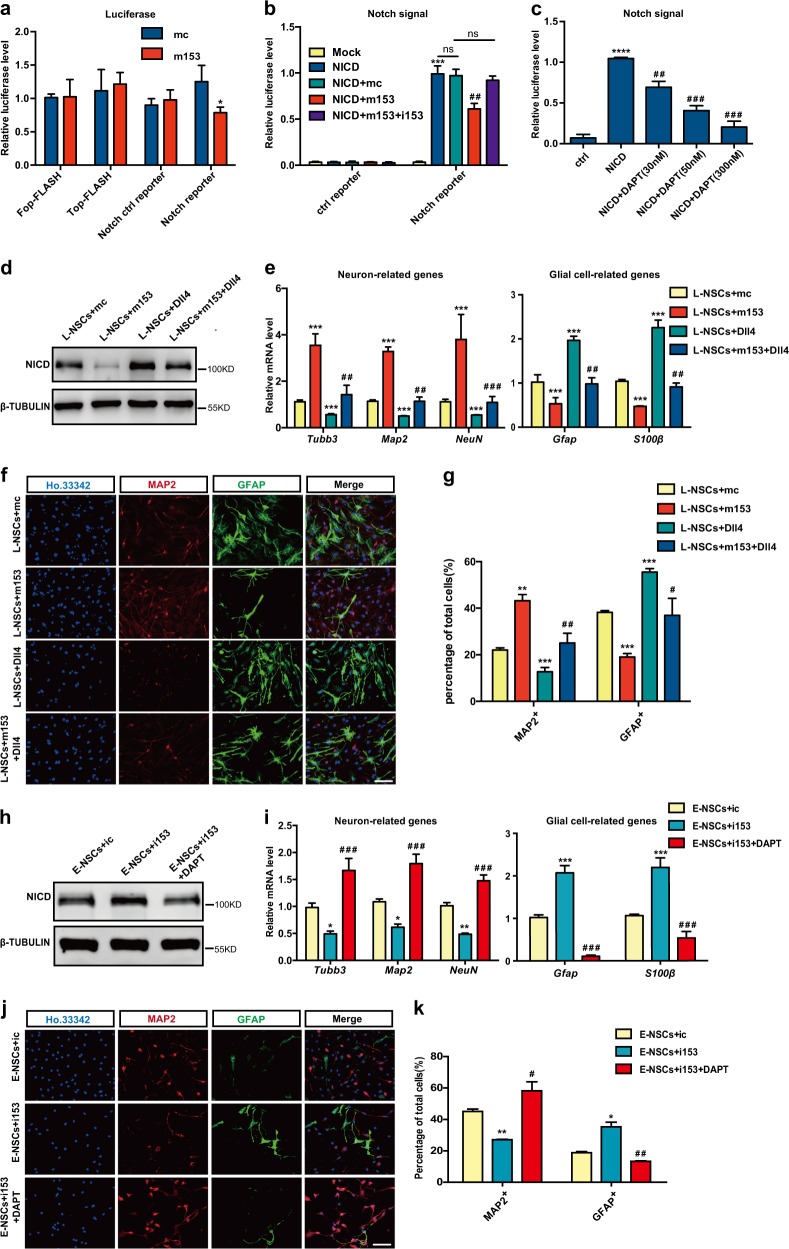
The Wnt and Notch signaling pathways play important roles in the neural differentiation of NSCs [27, 31]. To explore whether miR-153 regulates the Wnt and Notch signaling pathways, luciferase reporter activity in 293FT cells was analyzed with the TOP/FOP-FLASH reporter (Wnt reporter) [32] and Notch reporter. No difference in the activation of the TOP-Flash reporter was observed between the m153 and control groups, but the Notch reporter-dependent luciferase activity was decreased in the m153 group (Fig. 3a), suggesting that miR-153 regulates the Notch signaling pathway, but not the Wnt signaling pathway. To confirm the function of miR-153 in the Notch signaling pathway, 293FT cells were transfected with m153 and the Notch reporter, and the Notch intracellular domain (NICD) was used to activate the Notch signaling pathway. miR-153 could observably inhibit the luciferase activity of the Notch reporter, and this effect could be offset by the miR-153 inhibitor (Fig. 3b). DAPT, a specific inhibitor of Notch signaling [33], served as a positive control for the Notch reporter assay (Fig. 3c). The above data indicate that miR-153 could downregulate the activity of the Notch signaling pathway.
Fig. 3.
Neurogenesis is regulated by miR-153 through the Notch signaling pathway. a Relative luciferase activities of the Wnt reporter TOP-FLASH and the Notch reporter in the mc and m153 groups; FOP-FLASH was used as the negative ctrl of the Wnt reporter. b Relative luciferase activities of the Notch reporter in the mc and m153 groups; NICD is the active form of the Notch reporter. The data shown are the mean ± SEM, n = 3; ANOVA; ***p < 0.001, ##p < 0.01, “*” means compared with the mock group, “#” means compared with the NICD group. c Experiment to positively detect Notch signaling pathway activity. The data shown are the mean ± SEM, n = 3; ANOVA; ****p < 0.0001, ###p < 0.001, ##p < 0.01, “*” means compared with the mc group, “#” means compared with the NICD group. d Western blot analysis of NICD expression in the L-NSCs + mc, L-NSCs + m153, L-NSCs + Dll4 (treated with Dll4, 5 ng/µl), and L-NSCs + m153 + Dll4 groups. e Quantitative RT-PCR detection of neuron-related genes (Tubb3, Map2, NeuN) and glial cell-related genes (Gfap, S100β) involved in neural differentiation in the L-NSCs + mc, L-NSCs + m153, L-NSCs + Dll4, and L-NSCs + m153 + Dll4 groups. f Immunofluorescence analysis of the neural differentiation in the L-NSCs + mc, L-NSCs + m153, L-NSCs + Dll4, and L-NSCs + m153 + Dll4 groups as evaluated with MAP2 (red), GFAP (green) and Ho.33342 (blue). Scale bar: 50 μm. g Percentages of MAP2+ and GFAP+ cells in neural differentiation. h Western blot analysis of NICD expression in the E-NSCs + ic, E-NSCs + i153, E-NSCs + DAPT (treated with DAPT, 50 nM) and E-NSCs + i153 + DAPT groups. i Quantitative RT-PCR detection of neuron-related genes and glial cell-related genes in the neural differentiation of the E-NSCs + ic, E-NSCs + i153, E-NSCs + DAPT, and E-NSCs + i153 + DAPT groups. j Immunofluorescence analysis of the neural differentiation of E-NSCs + ic, E-NSCs + i153, E-NSCs + DAPT, and E-NSCs + i153 + DAPT as evaluated with MAP2 (red), GFAP (green), and Ho.33342 (blue). Scale bar: 50 μm. k Percentages of MAP2-positive and GFAP-positive cells in neural differentiation. Data information: the data shown are the mean ± SEM, n = 3; ANOVA; ***/###p < 0.001, **/##p < 0.01, */#p < 0.05; “*” means compared with the mc or ic group, “#” means compared with the m153 or i153 group in e, g, i, k. Gapdh was used as an internal control for the normalization in quantitative RT-PCR. L-NSCs late NSCs, E-NSCs early NSCs, mc control mimics, m153 miR-153 mimics, ic control inhibitor, i153 miR-153 inhibitor
To confirm the effect of miR-153 on the Notch signaling pathway in terms of neural differentiation, we used Dll4, a recombinant Notch ligand protein that can activate Notch signaling [34], to rescue the function of miR-153 in NSCs. Western blot assays showed that the protein level of NICD was upregulated in the Dll4 group (NSC + Dll4) and downregulated in the m153 group, and Dll4 could offset the effect of miR-153 on NICD expression (Fig. 3d; Fig. S2). In the Dll4 group, the mRNA levels of neuron-related genes and the percentage of MAP2-positive neurons were decreased, while the mRNA levels of glial cell-related genes and the percentage of GFAP-positive glial cells were increased. Moreover, Dll4 could offset the changes by m153 in the mRNA levels of neuron- and glial cell-related genes and the percentages of MAP2-positive neurons and GFAP-positive glial cells (Fig. 3e–g). In addition, when miR-153 was inhibited in NSCs, the NICD protein level was increased, and DAPT treatment rescued NICD expression (Fig. 3h). DAPT treatment was sufficient for rescuing the neuronal differentiation impaired by miR-153 inhibition, as shown by the upregulation of neuron-related genes, downregulation of glial cell-related genes, increased percentage of MAP2-positive neurons and decreased percentage of GFAP-positive glial cells (Fig. 3i–k). Taken together, these results suggest that miR-153 promotes NSC neurogenesis and inhibits gliogenesis by downregulating the Notch signaling pathway.
Dual regulatory effects of miR-153 on Jag1 and Hey2 in neurogenesis
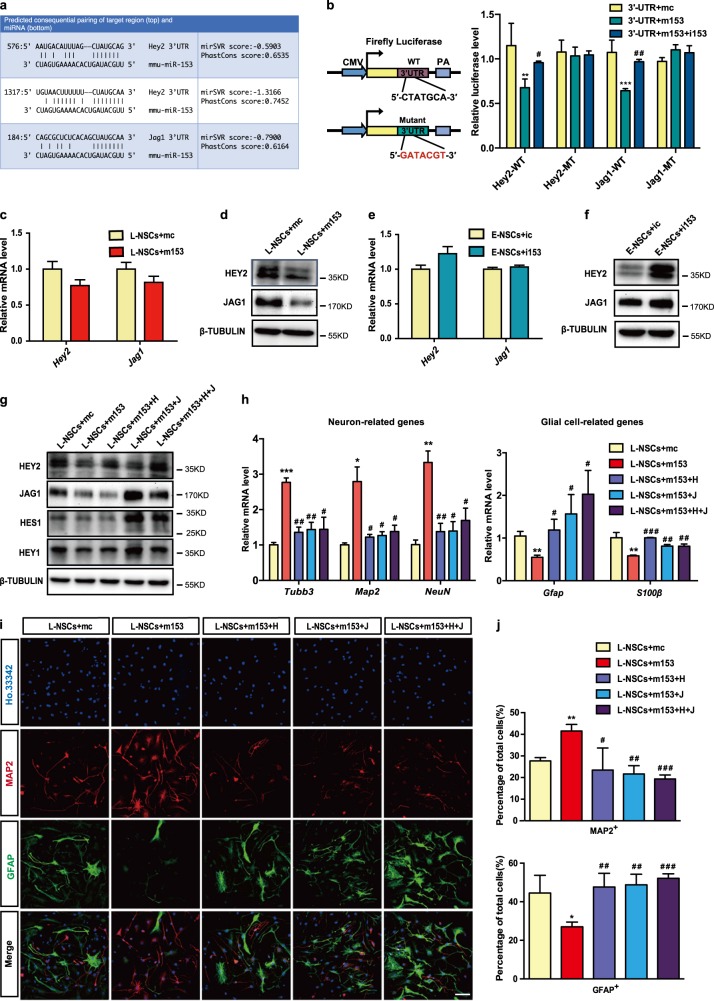
Because miR-153 regulated the activity of the Notch signaling pathway, we used TargetScan to search for potential miR-153 target genes that are key molecules in the Notch signaling pathway and found that miR-153 might bind to the 3′UTRs of Hey2 and Jag1 (Fig. 4a). We also found that the seed sequence of miR-153 is highly conserved among different species, and that the miR-153-binding sites in the 3′UTRs of Hey2 and Jag1 are also conserved (Fig. S3a). Jag1 is a ligand of the Notch signaling pathway [35], and Hey2 is a downstream target gene of the Notch signaling pathway [36]. To confirm the binding of miR-153 to Jag1 and Hey2, we performed luciferase assays in 293FT cells. We generated wild-type (WT) and mutant 3′UTR (MT) reporters of Hey2 or Jag1 and found that miR-153 inhibited the luciferase activity of the Hey2 and Jag1 WT reporters, but not that of the MT reporters, and the effect of miR-153 could be offset by miR-153 inhibitor (Fig. 4b). Furthermore, the protein levels, but not the mRNA levels, of both Hey2 and Jag1 were significantly decreased by overexpressing miR-153 in NSCs (Fig. 4c, d). Moreover, inhibition of miR-153 in NSCs upregulated the protein levels of Hey2 and Jag1 (Fig. 4e, f). These results support the idea that Hey2 and Jag1 are targets of miR-153. To elucidate the roles of Hey2 and Jag1 in neural differentiation, we knocked down Hey2 and Jag1 in NSCs. qRT-PCR analysis showed that Hey2 or Jag1 knockdown increased the expression of neuron-related genes and decreased that of glial cell-related genes (Fig. S3b–e). Knockdown of Jag1 in NSCs resulted in decreased protein levels of Hes1 and Hey1, downstream genes of the Notch signaling pathway (Fig. S3d). These data suggest that the effect of knocking down Hey2 or Jag1 on neural differentiation was similar to that of overexpressing miR-153.
Fig. 4.
Dual regulatory effects of miR-153 on Jag1 and Hey2 in neurogenesis. a Summary of the miR-153 target sites in the 3′UTRs of Jag1 and Hey2. In the double strands of the sequence, the upper strand is miR-153, and the lower strand is the binding site of the 3′UTR mRNA. b MiR-153 specifically represses its targets in the luciferase assay. WT-UTR indicates the WT 3’UTR. MT-UTR indicates the 3′UTR containing the mutant binding site of miR-153. c Quantitative RT-PCR detection of Jag1 and Hey2 in the L-NSCs + mc and L-NSCs + m153 groups. d Western blot analysis of the protein levels of JAG1 and HEY2 in the L-NSCs + mc and L-NSCs + m153 groups normalized to β-TUBULIN. e Quantitative RT-PCR detection of Jag1 and Hey2 in the E-NSCs + ic and E-NSCs + i153 groups. f Western blot analysis of the protein levels of JAG1 and HEY2 in the E-NSCs + ic and E-NSCs + i153 groups normalized to β-TUBULIN. g Western blot analysis of the protein levels of HEY2, JAG1, HEY1, and HES1 in rescue experiments normalized to β-TUBULIN. h Quantitative RT-PCR detection of neuron-related genes and glial cell-related genes in the neural differentiation of the L-NSCs + mc, L-NSCs + m153, L-NSCs + m153 + H, L-NSCs + m153 + J and L-NSCs + m153 + H + J groups. i Immunofluorescence analysis of neural differentiation by means of MAP2 (red), GFAP (green), and Ho.33342 (blue). Scale bar: 50 μm. j Percentages of MAP2-positive and GFAP-positive cells in neural differentiation. Data information: the data shown are the mean ± SEM, n = 3; ANOVA; ***/###p < 0.001, **/##p < 0.01, */#p < 0.05; “*” means compared with the mc group, and “#” means compared with the m153 group. Gapdh was used as an internal control for the normalization in quantitative RT-PCR. L-NSCs late NSCs, E-NSCs early NSCs, mc control mimics, m153 miR-153 mimics, ic control inhibitor, i153 miR-153 inhibitor, H Hey2, J Jag1
To further confirm that miR-153 regulates neural differentiation by targeting Hey2 and Jag1, we next used Hey2 and Jag1 to rescue the effects of miR-153 in NSCs (Fig. S3f; Fig. 4g). Western blot assays showed that HES1 and HEY1 expression was downregulated with miR-153 overexpression, and Jag1 overexpression restored HES1 and HEY1 expression, but not that of HEY2 (Fig. 4g). The overexpression of Jag1 and Hey2 alone or together could rescue the mRNA levels of neuron-related genes and glial cell-related genes (Fig. 4h), as well as the percentages of MAP2-positive neurons and GFAP-positive glial cells (Fig. 4i, j). These results demonstrate that miR-153 simultaneously targets Jag1 and Hey2 for dual regulation of the Notch signaling pathway, thereby promoting NSC neurogenesis.
MiR-153 expression is decreased in the hippocampi of aged mice with impaired cognitive ability
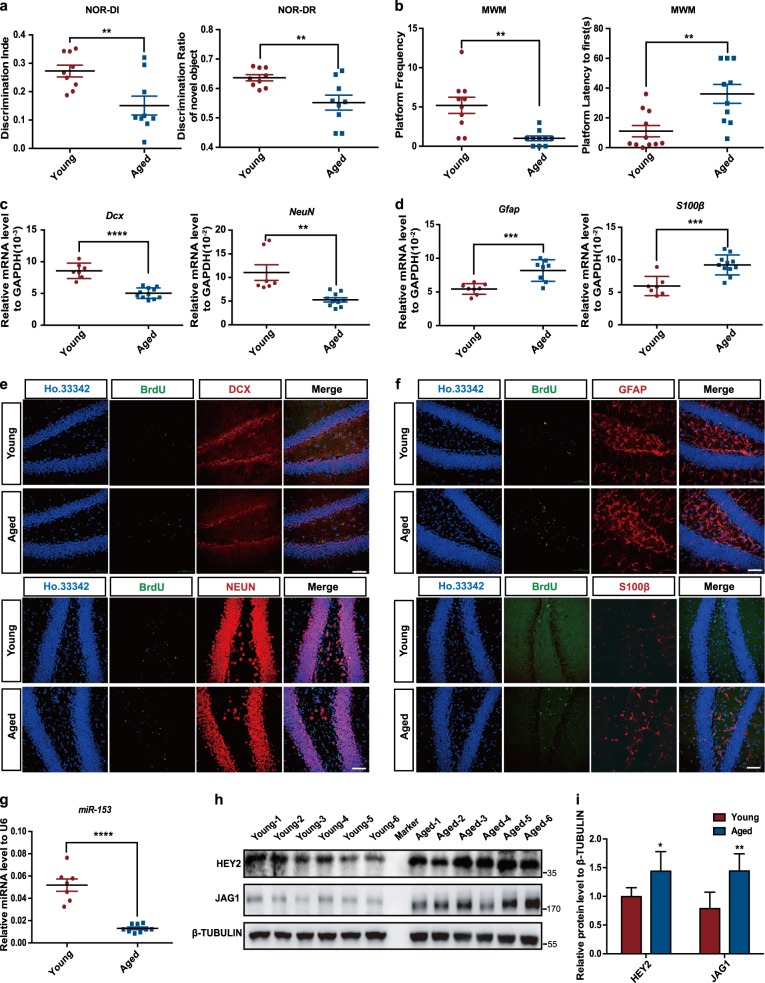
Previous studies have shown that aging leads to decreased neurogenesis and cognitive decline in adult mammals [37, 38]. Novel object recognition experiments showed that the discrimination index and recognition of novel objects by aged (16–18 months old) mice were significantly lower than those of young (8–10 weeks old) mice (Fig. 5a), and there was no difference in the preferences for the two objects between the two groups of mice (Fig. S4a). In the water maze experiment, the young mice crossed the platform more times and took less time to arrive at the platform during testing, while all mice showed similar swim speeds (Fig. 5b; Fig. S4b, c). Then, we examined the neuron- and glial cell-related genes in the hippocampi of young and aged mice. Consistent with previous reports [12, 39], in aged mice, the mRNA levels of a newborn neuronal marker (Doublecortin, Dcx) and a mature neuronal marker (NeuN) were downregulated (Fig. 5c), while those of glial cell-related genes (Gfap, S100β) were upregulated (Fig. 5d) in aged mice. To further examine neurogenesis in the hippocampus, we injected BrdU (50 mg/kg) intraperitoneally into the mice for 7 days. After 3 weeks, immunofluorescence staining results showed that in aged mice, the percentages of DCX+/BrdU+ newborn neurons and NEUN+/BrdU+ neurons were significantly decreased (Fig. 5e; Fig. S4d), while the percentages of GFAP+/BrdU+ and S100β+ /BrdU+ glial cells were significantly increased (Fig. 5f; Fig. S4e).
Fig. 5.
miR-153 expression is decreased in the hippocampi of aged mice with impaired cognitive ability. a Discrimination index and discrimination ratio of the novel object recognition task; young, n = 9; aged, n = 9; **p < 0.01. b Platform crossings and the amounts of time required for the mice to first reach the platform area during the water maze test; young, n = 10; aged, n = 10. c, d Quantitative RT-PCR detection of the newborn neuronal marker Dcx (young, n = 7; aged, n = 11), the mature neuronal marker NeuN (young, n = 7; aged, n = 10), and the glial cell markers Gfap (young, n = 8; aged, n = 8) and S100β (young, n = 7; aged, n = 11) in the hippocampi of young and aged mice. e Immunofluorescence analysis of neurons in the hippocampi of young and aged mice as evaluated by BrdU (green), DCX (red), NEUN (red), and Ho.33342 (blue). Scale bar: 50 μm. f Immunofluorescence analysis of astrocytes in the hippocampi of young and aged mice as evaluated by BrdU (green), GFAP (red), S100β (red), and Ho.33342 (blue). Scale bar: 50 μm. g Quantitative RT-PCR detection of miR-153 in the hippocampi of young and aged mice; young, n = 7; aged, n = 9. h Western blot analysis of the protein levels of JAG1 and HEY2 in the hippocampi of young and aged mice normalized to β-TUBULIN. i Statistical analysis of the grayscale levels of JAG1 and HEY2 by ImageJ; young, n = 6; aged, n = 6. Data information: the data shown are the mean ± SEM; Student’s t test; ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05. young mice 8–10 weeks old, aged mice 16–18 months old, NOR novel object recognition, MWM Morris water maze
Furthermore, we observed a decrease in miR-153 expression in the hippocampal tissues of aged mice (Fig. 5g). Consistently, western blot analysis showed that HEY2 and JAG1 were clearly upregulated in the aged mice (Fig. 5h, i). Collectively, these observations suggest that decreased miR-153 expression in the hippocampi of aged mice leads to decreased hippocampal neurogenesis and hippocampus-dependent cognitive dysfunction.
Inhibiting miR-153 decreases the hippocampal neurogenesis and impairs the cognitive function of young mice
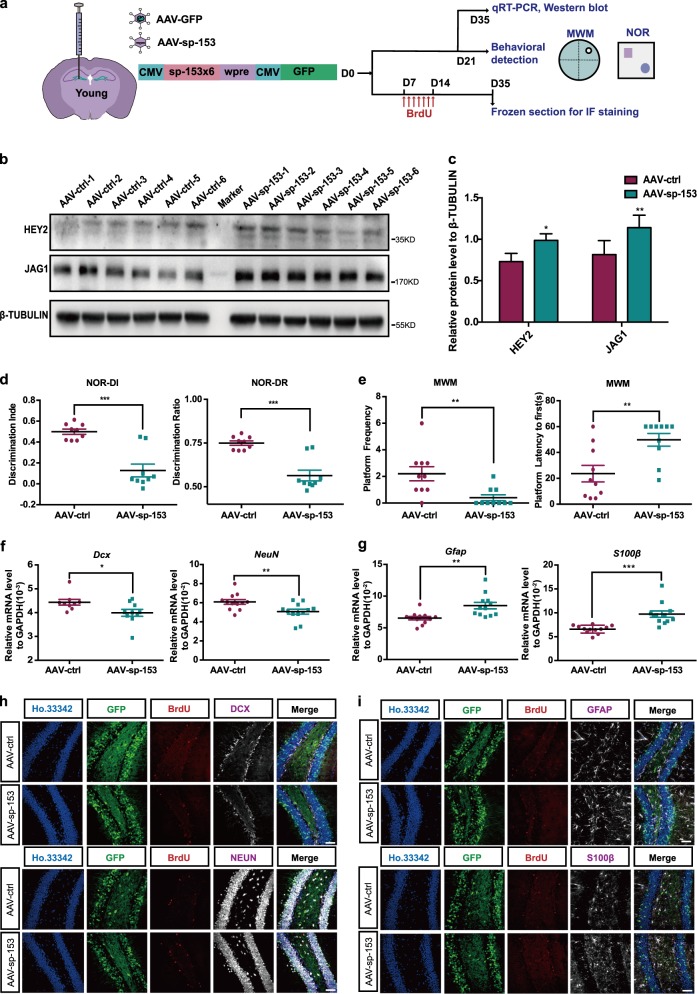
Because miR-153 exhibited a higher expression in the hippocampi of young mice than that in aged mice, we explored whether miR-153 is necessary for neurogenesis and the cognitive functions of young mice. We injected an adeno-associated virus, AAV-sponge-153 (AAV-sp-153), to inhibit miR-153 in the hippocampi of young mice, and AAV-GFP was used as a control (AAV-ctrl) (Fig. 6a). More than 70% of the cells were GFP-positive (Fig. S5a), and both HEY2 and JAG1 were upregulated in the mouse hippocampus after miR-153 inhibition (Fig. 6b, c). Regarding cognitive function, the novel object test showed that the discrimination indices and recognition of novel objects were significantly deteriorated in mice of the AAV-sp-153 group (Fig. 6d; Fig. S5b). The water maze experiment showed that the number of platform crossings made by mice in the AAV-sp-153 group was significantly decreased, and the time required for mice in the AAV-sp-153 group to first reach the platform area was increased (Fig. 6e; Fig. S5c). These experiments collectively show that miR-153 inhibition in the hippocampi of young mice impaired their hippocampus-dependent cognitive ability.
Fig. 6.
Inhibition of miR-153 decreases hippocampal neurogenesis and impairs the cognitive functions of young mice. a Schematic of virus injection into the hippocampi of young mice and diagram of the detection process. MWM Morris water maze, NOR novel object recognition. b Western blot analysis of the protein levels of JAG1 and HEY2 in the hippocampi of AAV-ctrl and AAV-sp-153 young mice normalized to β-TUBULIN. c Statistical analysis of the grayscale levels of JAG1 and HEY2 by ImageJ; AAV-ctrl, n = 6; AAV-sp-153, n = 6. d Discrimination index and discrimination ratio of novel objects; AAV-ctrl, n = 10; AAV-sp-153, n = 9. e Platform crossings and amounts of time required for the mice to first reach the platform area during the water maze test; AAV-ctrl, n = 10; AAV-sp-153, n = 10. f, g Quantitative RT-PCR detection of the newborn neuronal marker Dcx (AAV-ctrl, n = 9; AAV-sp-153, n = 10), the mature neuronal marker NeuN (AAV-ctrl, n = 12; AAV-sp-153, n = 12), and the glial cell markers Gfap (AAV-ctrl, n = 11; AAV-sp-153, n = 11) and S100β (AAV-ctrl, n = 12; AAV-sp-153, n = 12) in the hippocampi of AAV-ctrl and AAV-sp-153 young mice. h Immunofluorescence analysis of neurons in the hippocampi of AAV-ctrl and AAV-sp-153 young mice as evaluated by GFP (green), BrdU (red), DCX (white), NEUN (white), and Ho.33342 (blue). Scale bar: 50μm. i Immunofluorescence analysis of astrocytes in the hippocampi of AAV-ctrl and AAV-sp-153 mice as evaluated by GFP (green), BrdU (red), GFAP (white), S100β (white), and Ho.33342 (blue). Scale bar: 50 μm. Data information: the data shown are the mean ± SEM; Student’s t test; ***p < 0.001, **p < 0.01, *p < 0.05. AAV-sp-153 AAV-sponge-153, young mice 8–10 weeks old
Subsequently, we examined changes in neurogenesis in young mice injected with AAV-sp-153. In AAV-sp-153 mice, the mRNA levels of Dcx and NeuN were downregulated (Fig. 6f), while those of Gfap and S100β were upregulated (Fig. 6g). Furthermore, the percentages of DCX + /BrdU + /GFP + newborn neurons and NEUN + /BrdU + /GFP + neurons were decreased (Fig. 6h; Fig. S5d), and those of GFAP + /BrdU + /GFP + and S100β + /BrdU + /GFP + glial cells were significantly increased (Fig. 6i; Fig. S5e). In addition, we also injected a retrovirus, Retro-sponge-153 (Retro-sp-153), inhibiting miR-153, which infected only proliferating cells [40], into the hippocampi of young mice (Fig. S6a). More than 13% of cells were GFP-positive 5 weeks after injection (Fig. S6b). The mice injected with Retro-sp-153 showed phenotypes similar to those of AAV-sp-153 mice in terms of neurogenesis and cognitive ability (Fig. S6c–i). The above results indicate that inhibition of miR-153 decreases adult hippocampal neurogenesis and impairs the cognitive abilities of young mice.
miR-153 increases adult neurogenesis in the hippocampus and improves the cognitive abilities of aged mice
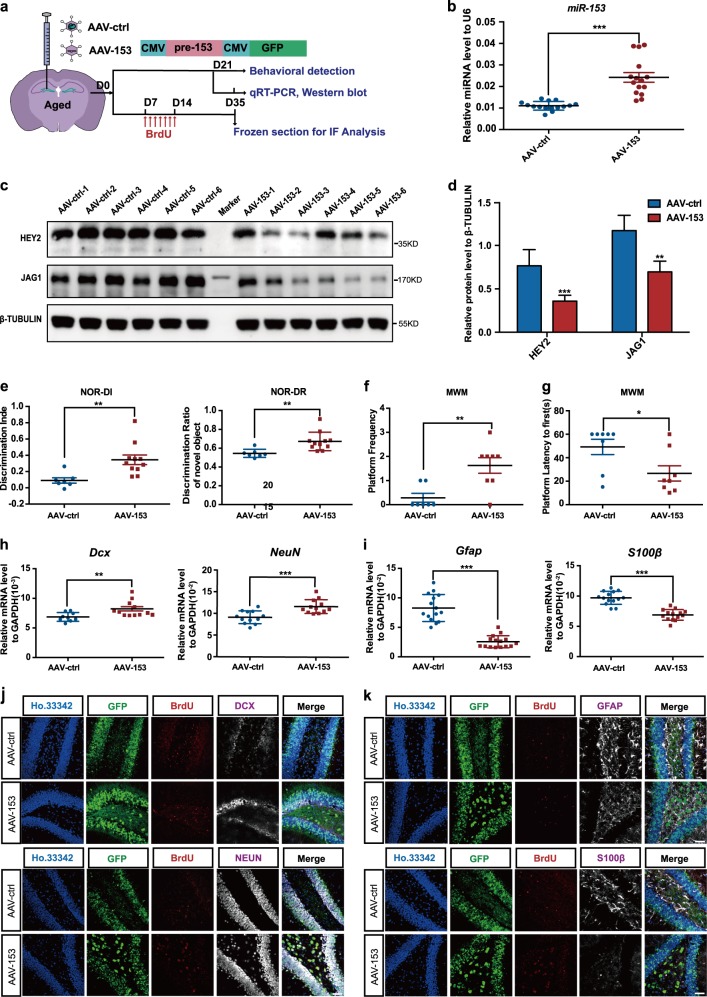
To investigate the potential benefit of increasing miR-153 expression on aging-related cognitive decline, an adeno-associated virus overexpressing miR-153 (AAV-153) was injected into the hippocampi of aged mice (Fig. 7a); more than 75% of the cells were GFP positive (Fig. S7a). qRT-PCR confirmed miR-153 overexpression in the mouse hippocampus (Fig. 7b). Western blot assays showed that HEY2 and JAG1 were both downregulated by miR-153 overexpression (Fig. 7c, d). Regarding cognitive function, the novel object test showed that the discrimination indices and recognition of novel objects were significantly improved in the AAV-153 group (Fig. 7e; Fig. S7b). The water maze experiment showed that the number of platform crossings made by mice in the AAV-153 group was significantly increased, and the time required for mice in the AAV-153 group to first reach the platform area was shortened (Fig. 7f, g; Fig. S7c, d). These experiments collectively show that miR-153 overexpression in the hippocampi of aged mice could improve the hippocampus-dependent cognitive ability of aged mice.
Fig. 7.
miR-153 increases adult neurogenesis in the hippocampus and enhances the cognitive abilities of aged mice. a Schematic of virus injection into the hippocampi of aged mice and diagram of the detection process. b Quantitative RT-PCR detection of miR-153 in the hippocampi of AAV-ctrl and AAV-153 aged mice; AAV-ctrl, n = 15; AAV-153, n = 15. c Western blot analysis of the protein levels of JAG1 and HEY2 in the hippocampi of AAV-ctrl and AAV-153 aged mice normalized to β-TUBULIN. d Statistical analysis of the grayscale levels of JAG1 and HEY2 by ImageJ; AAV-ctrl, n = 6; AAV-153, n = 6. e Discrimination index and discrimination ratio of novel objects; AAV-ctrl, n = 7; AAV-153, n = 11. f, g Platform crossings and amounts of time required for the mice to first reach the platform area during the water maze test; AAV-ctrl, n = 7; AAV-153, n = 8. h, i Quantitative RT-PCR detection of the newborn neuronal marker Dcx (AAV-ctrl, n = 9; AAV-153, n = 12), the mature neuronal marker NeuN (AAV-ctrl, n = 12; AAV-153, n = 12), and the glial cell markers Gfap (AAV-ctrl, n = 15; AAV-153, n = 15) and S100β (AAV-ctrl, n = 14; AAV-153, n = 14) in the hippocampi of AAV-ctrl and AAV-153 aged mice. j Immunofluorescence analysis of neurons in the hippocampi of AAV-ctrl and AAV-153 aged mice as evaluated by GFP (green), BrdU (red), DCX (white), NEUN (white), and Ho.33342 (blue). Scale bar: 50 μm. k Immunofluorescence analysis of astrocytes in the hippocampi of AAV-ctrl and AAV-153 mice as evaluated by GFP (green), BrdU (red), GFAP (white), S100β (white), and Ho.33342 (blue). Scale bar: 50 μm. Data information: the data shown are the mean ± SEM; Student’s t test; ***p < 0.001, **p < 0.01, *p < 0.05. aged mice 16–18 months old, NOR novel object recognition, MWM Morris water maze
Subsequently, we investigated whether the application of exogenous miR-153 counteracted aging-related declines in neurogenesis. In the AAV-153 mice, the mRNA levels of Dcx and NeuN were upregulated (Fig. 7h), while those of Gfap and S100β were downregulated (Fig. 7i). Furthermore, the percentages of DCX + /BrdU + /GFP + newborn neurons and NEUN + /BrdU + /GFP + neurons were increased (Fig. 7j; Fig. S7e), while those of GFAP + /BrdU + /GFP + and S100β + /BrdU + /GFP + glial cells were decreased (Fig. 7k; Fig. S7f). We also detected NSC markers in the hippocampus, and immunofluorescence staining and qRT-PCR showed no significant difference between AAV-ctrl and AAV-153 mice (Fig. S7g–i). These results indicated that miR-153 overexpression did not deplete the NSC pool.
Furthermore, we injected a retrovirus overexpressing miR-153 (Retro-153) into the hippocampi of aged mice (Fig. S8a), and more than 10% of the cells were GFP positive (Fig. S8b). qRT-PCR indicated that miR-153 was overexpressed in the mouse hippocampus (Fig. S8c). In addition, the mice injected with Retro-153 showed phenotypes similar to those of AAV-153 mice in regards to their cognitive abilities and neurogenesis (Fig. S8d–j). These results indicate that miR-153 could promote adult hippocampal neurogenesis and improve the cognitive abilities of aged mice by inhibiting two important Notch pathway molecules, Jag1 and Hey2.
To investigate whether miR-153 converts glial cells into neurons, we overexpressed miR-153 in astrocytes (Fig. S9a). Immunofluorescence staining indicated that no astrocytes transdifferentiated into neurons after miR-153 overexpression (Fig. S9b, c). In addition, to further determine whether miR-153 overexpression in astrocytes affects the neural differentiation of NSCs or whether miR-153 overexpression in NSCs affects neuronal transdifferentiation of astrocytes, we co-cultured NSCs with astrocytes [41]. miR-153 overexpression in astrocytes did not improve the rate of NSC neuronal differentiation (Fig. S9d–g), and miR-153 overexpression in NSCs did not promote the transdifferentiation of astrocytes into neurons (Fig. S9h–j). The above results indicate that miR-153 enhances the learning and memory abilities of aged mice by promoting NSC neurogenesis in the hippocampus.
Discussion
Neurological diseases are often caused by impairment of hippocampal neurogenesis [37, 42]. Recent studies have shown that endogenous adult NSCs could restore brain capabilities by undergoing neurogenesis [14]; hence, understanding the mechanisms of NSC differentiation into neuronal lineages is necessary to improve neurogenesis in neurological disease states. Many studies have found critical roles of miRNAs in neuronal differentiation [15, 21]. However, the mechanism of adult hippocampal neurogenesis is still unclear. In our study, during the culture of NSCs in vitro, their neurogenesis decreased, while gliogenesis increased. At the same time, miR-153 expression was also downregulated. Moreover, miR-153 overexpression promoted neuronal differentiation and inhibited the glial differentiation of NSCs, suggesting that miR-153 vivifies NSC neurogenesis.
In further studies, we found that miR-153 regulates neurogenesis through the Notch signaling pathway. Excessive activation of the Notch signaling pathway leads to decreased neurogenesis and increased gliogenesis [33, 43]. Previous studies have shown that miRNAs can target the Notch signaling pathway to regulate neural differentiation [44]. Our study found that miR-153 simultaneously targets two key molecules at different stages of the Notch signaling pathway to precisely regulate the pathway. miR-153 was able to decrease the protein level of Jag1 and regulate the activity of the Notch signaling pathway, thereby inhibiting the expression of certain downstream transcription factors, such as Hes1 and Hey1 [45]. MiR-153 was also able to target Hey2, a downstream transcription factor of the Notch signaling pathway. Astrocytes can reportedly negatively regulate nerve regeneration by regulating the Notch signaling pathway through Jag1 [43], while Hey2 can help maintain neural progenitors at the embryonic development stage and inhibit neuronal differentiation by promoting differentiation into glial cells at the postnatal stage [36]. In our study, miR-153 inhibited the protein levels of JAG1 and HEY2 but not their mRNA levels, indicating that miR-153 functions by inhibiting protein translation and not by degrading the mRNAs of Jag1 and Hey2. Usually, if the binding between an miRNA and its target gene is complete, the miRNA will destabilize the target gene’s mRNA, whereas if the binding is incomplete, the miRNA will inhibit the protein translation of the target gene [46, 47]. The mode of action between miR-153 and Jag1 or Hey2 may be due to incomplete binding between miR-153 and Jag1 or Hey2, which is consistent with a previous study. Collectively, our results clearly demonstrated that miR-153 targets not only Jag1 but also Hey2, thus exerting combined effects that promote neurogenesis in vitro and in vivo.
In adults, aging leads to decreased hippocampal NSC neurogenesis, which impairs hippocampus-dependent cognition, as well as learning and memory abilities [2]. Studies have found that miRNAs are involved in aging and neurogenesis [48]. However, the mechanisms by which miRNAs regulate aging-related adult hippocampal neurogenesis as well as decreased cognitive, learning, and memory abilities require further study. Previous studies have shown that miR-153 can regulate the differentiation of glial cells during mouse embryonic development by acting on Nfia (nuclear factor-1A) and Nfib (nuclear factor-1B) [23]. However, no studies are currently available on the function of miR-153 in adult neurogenesis. Here, we found that miR-153 plays an important role in adult hippocampal neurogenesis. Upon reduced neurogenesis caused by adult brain aging, miR-153 expression is dramatically decreased. Overexpression of miR-153 in the hippocampus significantly increased the neurogenesis, and learning and memory abilities of aged mice. The above studies have shown that miR-153 tends to promote the neurogenesis of adult NSCs and can rescue the cognitive impairment of aged mice. Considering the convenience of using miRNAs as therapeutic agents, miR-153 may provide the basis for a class of drugs used to treat aging-related cognitive decline and learning and memory impairment.
Patients with Alzheimer’s disease (AD) exhibit progressive cognitive impairment due to critical neuron loss [49, 50]. Studies have reported that the Notch pathway influences the AD disease process [51], and amyloid precursor protein (APP) can induce the glial differentiation of neural progenitor cells (NPCs) through Notch signaling [52]. Our studies demonstrated that miR-153 improves neurogenesis, inhibits the gliogenesis of NSCs and relieves cognitive dysfunction in mice by inhibiting the Notch signaling pathway. These results suggest that miR-153 has great potential to increase neurogenesis in patients with AD and thus improve their learning, memory and cognitive abilities.
Materials and methods
Cell culture and differentiation
NSCs were isolated from the forebrain of C57 mice at E13.5 as described previously [53]. NSCs were cultured in the DMEM/F12 (Gibco) supplemented with 2% B27 (Invitrogen), 1% GlutaMAX (Invitrogen), 1% NEAA (Invitrogen), 20 ng/ml fibroblast growth factor (bFGF, Sino_Biological), and 20 ng/ml epidermal growth factor (EGF, Sino_Biological), with passaging every 4 days. For differentiation, embryonic NSCs were dissociated into single cells using Accutase (Gibco) and plated on polyornithine (Gibco)- and laminin (Sigma, 10 ng/ml)-coated plates in the NSC medium, without bFGF or EGF. The cells were allowed to differentiate for 48 h and then immunostained for neuronal and glial markers. To directly differentiate NSCs into pure neurons, neurospheres were dissociated, plated on polyornithine- and laminin-coated plates in the neurobasal medium (Gibco) supplemented with 2% B27 (Invitrogen) and 1% GlutaMAX (Invitrogen) and treated with 2.5 ng/ml cytosine arabinoside (Sigma, c1768) after 2 days. The neuron purity was verified with anti-MAP2 staining (> 98% of cells were Map2-positive after 6 days of culture). To directly differentiate NSCs into pure astrocytes, neurospheres were dissociated and plated on poly-D-lysine-coated (100 µg/ml, Sigma) plates in DMEM/F12 medium supplemented with 2% B27 and 10% FBS (Gibco). The glial cell purity was verified with anti-GFAP staining (> 95% of cells were GFAP-positive after 4 days of culture).
microRNA mimic
miRNA mimics and inhibitors are synthesized small, double-stranded RNA molecules that are used to mimic or inhibit endogenous mature miRNA molecules when transfected into cells. The control/miR-153-cy5 mimics and control/miR-153-cy5 inhibitor were labeled with cy5 fluorescein. The miR-153 mimics, control mimics, miR-153 inhibitor, control inhibitor, control/miR-153-cy5 mimics, and control/miR-153-cy5 inhibitor were obtained from RiboBio.
Transfection
For miR-153 treatment-coupled differentiation, NSCs (6 × 105) were first transfected with 5 µl of an miR-153 mimic (20 µM) or 10 µl of inhibitor (20 µM) with FuGENE HD reagent. The medium was changed to the neural differentiation medium after 8 h of transfection, and the NSCs were kept in this medium for 48 h. For astrocytes, astrocytes (6 × 104) were plated in poly-D-lysine-coated 24-well plates and transfected with 5 µl of control or miR-153 mimics (20 µM) with FuGENE HD reagent; the medium was changed to neural differentiation medium after 8 h of transfection, and detection was performed after 3 days.
FACS analysis
FACS analysis was performed as described previously [54]. Fluorescently labeled mimic (control/miR-153-cy5 mimics) or inhibitor (control/miR-153-cy5 inhibitor) was used to determine the transfection efficiency in vitro by flow cytometry. To determine the percentage of cy5-positive NSCs after transfection, day 4 NSCs were dissociated into single cells with trypsin-EDTA (Gibco, 0.05%) and neutralized with serum. Then, the cells were resuspended in PBS and subjected to FACS analysis, which was performed on a FACSCalibur instrument (BD Biosciences) at a 625-nm excitation wavelength with standard emission filters. A fluorescence control was established with NSCs that were not transfected with the cy5 mimics or inhibitor. The data were analyzed by FlowJo 7.6 software, and more than 94% of the cells were transfected with the mimics or inhibitors.
Co-culture experiments
For the NSC-astrocyte co-culture experiments, astrocytes were isolated from the cortices of C57 mice at 1–2 days after birth and cultured in the DMEM/F12 medium supplemented with 2% B27, 10% FBS, bFGF (10 ng/ml), and EGF (10 ng/ml) as described previously [55]. NSCs were plated on polyornithine- and laminin-coated glass coverslips in 24-well plates and allowed to attach at 37 °C in a 5% CO2 incubator for 2 days. Then, transwell inserts (0.4 μm, Corning, 3470-Clear) containing astrocytes transfected with miR-153 or control mimics were transferred into 24-well plates containing NSCs. The medium was then changed to allow for differentiation; after 48 h of incubation, the neuron morphology was analyzed. For the astrocyte-NSC co-culture experiments, astrocytes were plated on poly-D-lysine-coated glass coverslips and cultured at 37 °C in a 5% CO2 incubator for 2 days. After washing, transwell inserts containing NSCs transfected with miR-153 or control mimics were transferred into 24-well plates containing astrocytes. After 48 h of incubation, the neuron morphology was analyzed.
Western blot analysis
Western blot was performed as described previously [56]. The cells were collected, washed with PBS (Gibco, China), and then lysed with sodium dodecyl sulfate (SDS, Amersco). For mouse hippocampal protein extraction, tissues were incubated on ice for 30 min with RIPA lysis buffer (Beyotime), and the protein concentrations were measured using the Pierce BCA Protein Assay Kit (Thermo). The protein samples were separated on SDS-polyacrylamide gels and transferred onto PVDF membranes. The membranes were incubated with primary antibodies overnight at 4 °C and with secondary antibodies for 2 h. The antibodies used for western blotting are listed in Supplementary Table 1.
qRT-PCR analysis
qRT-PCR was performed as described previously [57]. The total RNA was extracted from cells or the mouse hippocampus using TRIzol (TaKaRa) according to the manufacturer’s instructions. The quality of the total RNAs was determined based on the OD260/OD280 ratio with the NanoDrop spectrophotometer (MALCOM), and the OD260/OD280 ratio for each sample was controlled between 1.9 and 2.1. The total RNA (500 ng) was reverse transcribed into cDNA using the PrimeScript RT reagent kit (TaKaRa). qRT-PCR analysis was performed using SYBR Green qRT-PCR Master Mix (Bio-Rad) on the Agilent Stratagene Mx3000P machine. The primers used for qRT-PCR are listed in Supplementary Table 2. The efficacy of each primer and the scope of the Ct value for each detected gene are listed in Supplementary Table 2. miRNA levels were measured using the Bulge-Loop TM miRNA qRT-PCR Primer Set (RiboBio) according to the manufacturer’s instructions. Relative expression was calculated using the 2−ΔΔct method by normalizing with Gapdh or U6 expression and presented as the fold change relative to the control.
Immunofluorescence staining
For cell immunofluorescence staining, cells cultured on coverslips were washed with PBS, fixed with 4% paraformaldehyde (Amersco) for 20 min, and then permeabilized with 0.2% Triton X-100 (Amersco) for 8 min. The cells were then washed with PBS three times and blocked in 10% FBS for 1 h. For immunofluorescence staining of brain sections, the mouse brains were prepared as frozen sections using a freezing microtome (Leica CM3050 S) and stored in freezing medium. The sites of stereotaxic injections are listed in the Supplementary Material. Brain slices were removed from the freezing medium, washed three times with PBS, permeabilized and blocked in a solution containing 10% FBS and 0.1% Triton X-100 for 1 h. For BrdU immunofluorescence staining, BrdU (Sigma, B5002) was intraperitoneally injected into mice (50 mg/kg) seven times on 7 sequential days, and brain slices were obtained after 3 weeks. The brain slices were incubated with 2 N HCl (Sinopharm) for 45 min before permeabilization and blocking. Both cell and brain tissue immunofluorescence staining involved overnight incubation with primary antibodies at 4 °C. Next, the cells and brain slices were stained with fluorescent secondary antibodies and Hoechst 33342 dye for 2 h. Images were acquired using a fluorescence microscope (Nikon). The antibodies used for western blot analysis are listed in Supplementary Table 1.
Dual luciferase assay
For the dual luciferase assay of Hey2 and Jag1, 293FT cells (3 × 104) were plated in 24-well plates, and cells in each well were transfected with luciferase reporters (100 ng), Renilla (10 ng), and 1 µl of mimics (20 µM); the luciferase assay was performed after 48 h. Luciferase reporters were generated by cloning the 3′UTR of WT or mutant target genes into pGL3-based vectors. The WT miR-153 target site 5′-CTATGCA-3′ was mutated to 5′-GATACGT-3′. Mutant vectors were obtained using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent Technologies). The luciferase activity was measured as described previously [58]. For the dual luciferase assay of the signaling pathway, Wnt signaling pathway activity was detected by the TOP-FLASH and FOP-FLASH vectors [32]. The activity of the Notch signaling pathway was detected using a Notch pathway reporter kit (Bioscience), and DAPT (Selleck, S2215) was used to inhibit the Notch signaling pathway.
Animal studies
The mice used in this study were obtained from Shanghai SLAC laboratory Animal Co., Ltd. (SLAC, http://www.slac.com.cn) and maintained in a pathogen-free environment throughout the experiments. All procedures involving animals were approved by the Laboratory Animal Care Committee of Tongji University in accordance with the Guide for the Care and Use of Laboratory Animals (NIH) [54]. In this study, all efforts were made to minimize the number of animals used and their suffering.
Vector construction and virus
For the lenti-sponge-153-wpre vector, six repeats of the sponge-153 sequence and wpre were cloned into the lentivirus background vector (sponge-153 sequence is listed in Supplementary Table 3). For the generation of shHey2 and shJag1, 21-base pair Hey2- and Jag1-specific regions (shRNA sequences are listed in Supplementary Table 3) for RNA interference were designed and cloned into the pLKO.1-TRC cloning vector. In the AAV-153, Retro-153, AAV-sponge-153, and Retro-sponge-153 constructs, pre-153 and six repeats of the sponge-153 sequence were driven by the CMV promoter, and the vector contained a GFP sequence also driven by the CMV promoter. In total, 75% of the cells in the hippocampal DG were positive after injection with AAV-153. Lenti-and retroviral packaging was performed as described previously [59, 60]. The lentivirus and retrovirus were concentrated by ultracentrifugation at 3000g for 30 min at 4 °C, postprecipitated with virus precipitation solution (ExCell Bio, EMB810A-1, EMB100A-1) for 24 h and resuspended in 30 μl of PBS. The adeno-associated virus AAV-9 contained AAV-GFP, AAV-153, and AAV-sp-153, which were provided by Hanbio Company.
Virus and stereotaxic injections
For stereotaxic injections, 8–10 weeks old (young) or 16–18 months old (aged) C57 mice were used, and the site of stereotaxic injections can be found in the Supplementary Methods. The behavioral evaluations were performed 3 weeks after injection.
Mouse behavior evaluation
Novel object recognition and water maze tasks were used to measure spatial memory retention. The experimental details of the novel object recognition and water maze tasks can be found in the Supplementary Methods.
Statistical analysis
The data are expressed as the mean ± error (SEM), and significance was set at p < 0.05 (see each figure for details). ANOVA and Student’s t test statistical analyses were performed unless otherwise specified using GraphPad software, and the data in the figures reflect at least three independent experiments performed on different days. To quantify the immunofluorescence staining of cells expressing type-specific markers, at least 15 sections from at least three independent experiments were collected, and at least 1000 cells were collected. Quantification of GFP-positive cells and phenotypic quantification of GFP-positive cells in the granule layer were performed using a Nikon-A1R microscope with NIS-Elements software in at least three sections containing the DG from at least three different animals. The exact n values are listed in the figure legends.
Softwares
GraphPad Prism6 was used for data calculations and statistics. Targetscan (http://www.targetscan.org/vert_72/) was used to search for potential miR-153 target genes. ImageJ (https://imagej.en.softonic.com) was used for western blot grayscale analysis, and FlowJo 7.6 was used to analyze the FACS data.
Supplementary information
Acknowledgements
This work was supported by the Ministry of Science and Technology (grant 2016YFA0101300), the National Natural Science Foundation of China (grants 81530042, 31721003, 31871495, 31571529, 31571519, 31701110, and 31671533), and Shanghai Municipal Medical and Health Discipline Construction Projects (grant 2017ZZ02015).
Author contributions
The author contributions can be found in the Supplementary Material. All authors read and approved the final paper.
Data availability
The additional data or reagents are available from the corresponding author upon reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by N. Bazan
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiajie Xi, Phone: +0086-21-65988876, Email: jiajiexi_bio@163.com.
Jiuhong Kang, Phone: +0086-21-65988876, Email: jhkang@tongji.edu.cn.
Supplementary information
The online version of this article (10.1038/s41418-019-0388-4) contains supplementary material, which is available to authorized users.
References
- 1.Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, et al. Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat Med. 2014;20:659–63. doi: 10.1038/nm.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Villeda SA, Luo J, Mosher KI, Zou B, Britschgi M, Bieri G, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngwenya LB, Heyworth NC, Shwe Y, Moore TL, Rosene DL. Age-related changes in dentate gyrus cell numbers, neurogenesis, and associations with cognitive impairments in the rhesus monkey. Front Syst Neurosci. 2015;9:102. doi: 10.3389/fnsys.2015.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennis CV, Suh LS, Rodriguez ML, Kril JJ, Sutherland GT. Human adult neurogenesis across the ages: an immunohistochemical study. Neuropathol Appl Neurobiol. 2016;42:621–38. doi: 10.1111/nan.12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spalding KL, Bergmann O, Alkass K, Bernard S, Salehpour M, Huttner HB, et al. Dynamics of hippocampal neurogenesis in adult humans. Cell. 2013;153:1219–27. doi: 10.1016/j.cell.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kempermann G, Kuhn HG, Gage FH. Experience-induced neurogenesis in the senescent dentate gyrus. J Neurosci. 1998;18:3206–12. doi: 10.1523/JNEUROSCI.18-09-03206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eriksson PS, Perfilieva E, Bjork-Eriksson T, Alborn AM, Nordborg C, Peterson DA, et al. Neurogenesis in the adult human hippocampus. Nat Med. 1998;4:1313–7. doi: 10.1038/3305. [DOI] [PubMed] [Google Scholar]
- 8.Kempermann G, Kuhn HG, Gage FH. More hippocampal neurons in adult mice living in an enriched environment. Nature. 1997;386:493–5. doi: 10.1038/386493a0. [DOI] [PubMed] [Google Scholar]
- 9.Roy NS, Wang S, Jiang L, Kang J, Benraiss A, Harrison-Restelli C, et al. In vitro neurogenesis by progenitor cells isolated from the adult human hippocampus. Nat Med. 2000;6:271–7. doi: 10.1038/73119. [DOI] [PubMed] [Google Scholar]
- 10.Encinas JM, Michurina TV, Peunova N, Park JH, Tordo J, Peterson DA, et al. Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell. 2011;8:566–79. doi: 10.1016/j.stem.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.BM J. Neuronal loss, neurofibrillary tangles and granulovacuolar degeneration in the hippocampus with aging and dementia. A qualitative study. Acta Neuropathol. 1977;37:111–8. doi: 10.1007/BF00692056. [DOI] [PubMed] [Google Scholar]
- 12.Mouton PR, Long JM, Lei DL, Howard V, Jucker M, Calhoun ME, et al. Age and gender effects on microglia and astrocyte numbers in brains of mice. Brain Res. 2002;956:30–35. doi: 10.1016/s0006-8993(02)03475-3. [DOI] [PubMed] [Google Scholar]
- 13.Drapeau E, Mayo W, Aurousseau C, Le Moal M, Piazza PV, Abrous DN. Spatial memory performances of aged rats in the water maze predict levels of hippocampal neurogenesis. Proc Natl Acad Sci USA. 2003;100:14385–90. doi: 10.1073/pnas.2334169100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burghardt NS, Park EH, Hen R, Fenton AA. Adult-born hippocampal neurons promote cognitive flexibility in mice. Hippocampus. 2012;22:1795–808. doi: 10.1002/hipo.22013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szulwach KE, Li XK, Smrt RD, Li YJ, Luo YP, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–U181. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishino J, Kim I, Chada K, Morrison SJ. Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf Expression. Cell. 2008;135:227–39. doi: 10.1016/j.cell.2008.09.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao C, Sun G, Li S, Lang MF, Yang S, Li W, et al. MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc Natl Acad Sci USA. 2010;107:1876–81. doi: 10.1073/pnas.0908750107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata M, Kurokawa D, Nakao H, Ohmura T, Aizawa S. MicroRNA-9 modulates Cajal-Retzius cell differentiation by suppressing Foxg1 expression in mouse medial pallium. J Neurosci. 2008;28:10415–21. doi: 10.1523/JNEUROSCI.3219-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY) 2010;2:415–31. doi: 10.18632/aging.100176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington’s disease. J Neurosci. 2008;28:14341–6. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Kim MS, Jia B, Yan J, Zuniga-Hertz JP, Han C, et al. Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature. 2017;548:52–57. doi: 10.1038/nature23282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santa-Maria I, Alaniz ME, Renwick N, Cela C, Fulga TA, Van Vactor D, et al. Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J Clin Invest. 2015;125:681–6. doi: 10.1172/JCI78421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuyama J, Bunt J, Richards LJ, Iwanari H, Mochizuki Y, Hamakubo T, et al. MicroRNA-153. Stem Cell Rep. 2015;5:365–77. doi: 10.1016/j.stemcr.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. Transcriptome-wide analysis of small RNA expression in early zebrafish development. RNA. 2012;18:915–29. doi: 10.1261/rna.029090.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu TS, Costello MA, Talsma CE, Flack CG, Crowley JG, Hamm LL, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer Res. 2011;71:6061–72. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baik SH, Fane M, Park JH, Cheng YL, Fann DYW, Yun UJ, et al. Pin1. Ann Neurol. 2015;77:504–16. doi: 10.1002/ana.24347. [DOI] [PubMed] [Google Scholar]
- 27.Grandbarbe L, Bouissac J, Rand M, de Angelis MH, Artavanis-Tsakonas S, Mohier E. Delta-Notch signaling controls the generation of neurons/glia from neural stem cells in a stepwise process. Development. 2003;130:1391–402. doi: 10.1242/dev.00374. [DOI] [PubMed] [Google Scholar]
- 28.Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, et al. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- 29.Ebert MS, Sharp PA. MicroRNA sponges: progress and possibilities. Rna-a Publication of the Rna. Society. 2010;16:2043–50. doi: 10.1261/rna.2414110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–6. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zechner D, Fujita Y, Hulsken J, Muller T, Walther I, Taketo MM, et al. beta-catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev Biol. 2003;258:406–18. doi: 10.1016/s0012-1606(03)00123-4. [DOI] [PubMed] [Google Scholar]
- 32.Liu XF, Li XY, Zheng PS, Yang WT. DAX1 promotes cervical cancer cell growth and tumorigenicity through activation of Wnt/beta-catenin pathway via GSK3beta. Cell Death Dis. 2018;9:339. doi: 10.1038/s41419-018-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chi Z, Zhang J, Tokunaga A, Harraz MM, Byrne ST, Dolinko A, et al. Botch promotes neurogenesis by antagonizing Notch. Dev Cell. 2012;22:707–20. doi: 10.1016/j.devcel.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salewski RP, Buttigieg J, Mitchell RA, van der Kooy D, Nagy A, Fehlings MG. The generation of definitive neural stem cells from PiggyBac transposon-induced pluripotent stem cells can be enhanced by induction of the NOTCH. Stem Cells Dev. 2013;22:383–96. doi: 10.1089/scd.2012.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Jagged - a mammalian ligand that activates Notch1. Cell. 1995;80:909–17. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 36.Sakamoto M, Hirata H, Ohtsuka T, Bessho Y, Kageyama R. The basic helix-loop-helix genes Hesr1/Hey1 and Hesr2/Hey2 regulate maintenance of neural precursor cells in the brain. J Biol Chem. 2003;278:44808–15. doi: 10.1074/jbc.M300448200. [DOI] [PubMed] [Google Scholar]
- 37.van Praag H, Schinder AF, Christie BR, Toni N, Palmer TD, Gage FH. Functional neurogenesis in the adult hippocampus. Nature. 2002;415:1030–4. doi: 10.1038/4151030a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akers KG, Martinez-Canabal A, Restivo L, Yiu AP, De Cristofaro A, Hsiang HL, et al. Hippocampal neurogenesis regulates forgetting during adulthood and infancy. Science. 2014;344:598–602. doi: 10.1126/science.1248903. [DOI] [PubMed] [Google Scholar]
- 39.Li G, Zhang X, Cheng H, Shang X, Xie H, Zhang X, et al. Acupuncture improves cognitive deficits and increases neuron density of the hippocampus in middle-aged SAMP8 mice. Acupunct Med. 2012;30:339–45. doi: 10.1136/acupmed-2012-010180. [DOI] [PubMed] [Google Scholar]
- 40.Grande A, Sumiyoshi K, Lopez-Juarez A, Howard J, Sakthivel B, Aronow B, et al. Environmental impact on direct neuronal reprogramming in vivo in the adult brain. Nat Commun. 2013;4:2373. doi: 10.1038/ncomms3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh HJ, Shin Y, Chung S, Hwang DW, Lee DS. Convective exosome-tracing microfluidics for analysis of cell-non-autonomous neurogenesis. Biomaterials. 2017;112:82–94. doi: 10.1016/j.biomaterials.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 42.Wang R, Dineley KT, Sweatt JD, Zheng H. Presenilin 1 familial Alzheimer’s disease mutation leads to defective associative learning and impaired adult neurogenesis. Neuroscience. 2004;126:305–12. doi: 10.1016/j.neuroscience.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 43.Magnusson JP, Goritz C, Tatarishvili J, Dias DO, Smith EM, Lindvall O, et al. A latent neurogenic program in astrocytes regulated by Notch signaling in the mouse. Science. 2014;346:237–41. doi: 10.1126/science.346.6206.237. [DOI] [PubMed] [Google Scholar]
- 44.Rani N, Nowakowski TJ, Zhou H, Godshalk SE, Lisi V, Kriegstein AR, et al. A primate lncRNA mediates notch signaling during neuronal development by sequestering miRNA. Neuron. 2016;90:1174–88. doi: 10.1016/j.neuron.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonev B, Stanley P, Papalopulu N. MicroRNA-9 modulates Hes1 ultradian oscillations by forming a double-negative feedback loop. Cell Rep. 2012;2:10–8. doi: 10.1016/j.celrep.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iacomino G, Siani A. Role of microRNAs in obesity and obesity-related diseases. Genes Nutr. 2017;12:23. doi: 10.1186/s12263-017-0577-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Somel M, Guo S, Fu N, Yan Z, Hu HY, Xu Y, et al. MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 2010;20:1207–18. doi: 10.1101/gr.106849.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boekhoorn K, Joels M, Lucassen PJ. Increased proliferation reflects glial and vascular-associated changes, but not neurogenesis in the presenile Alzheimer hippocampus. Neurobiol Dis. 2006;24:1–14. doi: 10.1016/j.nbd.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 50.Mueller SG, Weiner MW. Selective effect of age, Apoe4, and Alzheimer’s disease on hippocampal subfields. Hippocampus. 2009;19:558–64. doi: 10.1002/hipo.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li SH, Gao P, Wang LT, Yan YH, Xia Y, Song J, et al. Osthole stimulated neural stem cells differentiation into neurons in an alzheimer’s disease cell model via upregulation of MicroRNA-9 and rescued the functional impairment of hippocampal neurons in APP/PS1 transgenic mice. Front Neurosci. 2017;11:340. doi: 10.3389/fnins.2017.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugaya K. Mechanism of glial differentiation of neural progenitor cells by amyloid precursor protein. Neurodegener Dis. 2008;5:170–2. doi: 10.1159/000113693. [DOI] [PubMed] [Google Scholar]
- 53.Ahlenius H, Kokaia Z. Isolation and generation of neurosphere cultures from embryonic and adult mouse brain. Methods Mol Biol. 2010;633:241–52. doi: 10.1007/978-1-59745-019-5_18. [DOI] [PubMed] [Google Scholar]
- 54.Li G, Jiapaer Z, Weng R, Hui Y, Jia W, Xi J, et al. Dysregulation of the SIRT1/OCT6 axis contributes to environmental stress-induced neural induction defects. Stem Cell Rep. 2017;8:1270–86. doi: 10.1016/j.stemcr.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schildge S, Bohrer C, Beck K, Schachtrup C. Isolation and culture of mouse cortical astrocytes. J Vis Exp. 2013;71:e50079. [DOI] [PMC free article] [PubMed]
- 56.Guo X, Xu Y, Wang Z, Wu Y, Chen J, Wang G, et al. A Linc1405/Eomes complex promotes cardiac mesoderm specification and cardiogenesis. Cell Stem Cell. 2018;22:893–908 e896. doi: 10.1016/j.stem.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 57.Xi J, Wu Y, Li G, Ma L, Feng K, Guo X, et al. Mir-29b mediates the neural tube versus neural crest fate decision during embryonic stem cell neural differentiation. Stem Cell Rep. 2017;9:571–86. doi: 10.1016/j.stemcr.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res. 2018;46:3906–20. doi: 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guo XD, Liu QD, Wang GY, Zhu SC, Gao LF, Hong WJ, et al. microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res. 2013;23:142–56. doi: 10.1038/cr.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen W, Liu N, Zhang H, Zhang H, Qiao J, Jia W, et al. Sirt6 promotes DNA end joining in iPSCs derived from old mice. Cell Rep. 2017;18:2880–92. doi: 10.1016/j.celrep.2017.02.082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The additional data or reagents are available from the corresponding author upon reasonable request.