Abstract
Ferroptosis is a specialized iron-dependent cell death that is associated with lethal lipid peroxidation. Modulation of ferroptosis may have therapeutic potential since it has been implicated in various human diseases as well as potential antitumor activities. However, much remains unknown about the underlying mechanisms and genetic determinants of ferroptosis. Given the critical role of kinases in most biological processes and the availability of various kinase inhibitors, we sought to systemically identify kinases essential for ferroptosis. We performed a forward genetic-based kinome screen against ferroptosis in MDA-MB-231 cells triggered by cystine deprivation. This screen identified 34 essential kinases involved in TNFα and NF-kB signaling. Unexpectedly, the DNA damage response serine/threonine kinase ATM (mutated in Ataxia-Telangiectasia) was found to be essential for ferroptosis. The pharmacological or genetic inhibition of ATM consistently rescued multiple cancer cells from ferroptosis triggered by cystine deprivation or erastin. Instead of the canonical DNA damage pathways, ATM inhibition rescued ferroptosis by increasing the expression of iron regulators involved in iron storage (ferritin heavy and light chain, FTH1 and FTL) and export (ferroportin, FPN1). The coordinated changes of these iron regulators during ATM inhibition resulted in a lowering of labile iron and prevented the iron-dependent ferroptosis. Furthermore, we found that ATM inhibition enhanced the nuclear translocation of metal-regulatory transcription factor 1 (MTF1), responsible for regulating expression of Ferritin/FPN1 and ferroptosis protection. Genetic depletion of MTF-1 abolished the regulation of iron-regulatory elements by ATM and resensitized the cells to ferroptosis. Together, we have identified an unexpected ATM-MTF1-Ferritin/FPN1 regulatory axis as novel determinants of ferroptosis through regulating labile iron levels.
Subject terms: Cancer genomics, Cancer metabolism
Introduction
Ferroptosis is a novel form of regulated cell death with human disease relevance
During the past decade, ferroptosis has been recognized as a form of iron-dependent regulated death with distinct morphologic features and molecular mechanisms [1, 2]. Ferroptosis was first found to be triggered by erastin that selectively induced cell death in tumor cells carrying mutated forms of RAS [3]. Later, erastin was found to be a potent inhibitor of xCT, a cystine-glutamate antiporter encoded by the SLC7A11 gene, which encodes the substrate specific subunit of system xc− that is required for the import of extracellular cystine into human cells. The amino acid cysteine usually exists as the dimeric and oxidized form of cystine (CS-SC). Once imported, cystine is reduced to cysteine and used as the limiting component for the synthesis of the tripeptide glutathione (GSH), which is used by glutathione peroxidase 4 (GPX4) to repair lipid peroxidation and prevent ferroptosis. Inhibition of cysteine-dependent GSH synthesis via erastin, other xCT inhibitors, or cystine deprivation leads to inactivation of GPX4, accumulation of lipid-based reactive oxygen species (ROS), and eventually cell death via ferroptosis [4, 5].
While ferroptosis was initially discovered as an erastin-induced death, further investigations have implicated ferroptosis in various pathophysiological processes and diseases. For example, inactivation of GPX4 in mice lead to acute renal failture [6]. The tumor suppression capacity of p53 and BAP1 has been attributed to their ability to trigger ferroptosis [7, 8]. In addition, ferroptosis inhibitors have been found to prevent cell death in Huntington’s diseases [9], acute kidney injury caused by rhabdomyolysis [9], or ischemia-reperfusion injury [10]. Therefore, modulating ferroptosis may have therapeutic potentials in various ferroptosis-associated diseases. In addition, since tumor cells with certain oncogenic mutations are very sensitive to ferroptosis, triggering ferroptosis may also have significant therapeutic potential for ferroptosis-sensitive tumor cells. However, much remains unknown about the underlying mechanisms to optimize the therapeutic potential of modulating ferroptosis.
Genetic determinants of ferroptosis
Several recent excellent reviews have discussed the genetic determinants of the ferroptosis in various biological processes, including cystine and lipid metabolisms as well as iron biology [1, 2]. Many of these genetic determinants were identified using unbiased forward genetic screens under ferroptosis-inducing conditions. For example, a genome-wide siRNA screen of erastin-treated HT-1080 fibrosarcoma identified cysteine-tRNA synthetase, CARS, to be essential for ferroptosis [11]. Further mechanistic studies revealed that inhibition of CARS significantly enhances the transcriptional upregulation of serine metabolisms as well as the transsulfation pathways, in which methionine could be used to produce cysteine. Thus, CARS depletion helps to sustain the cysteine and GSH under erastin and confer the ferroptosis resistance phenotype. Another study using genome-wide CRISPR-based genetic screen identified acyl-CoA synthetase long-chain family member 4 as an essential component for ferroptosis execution through the lipid reprogramming [12]. Even with these studies, much still remains unknown about the processes and determinants of ferroptosis.
The role of kinome in ferroptosis remains poorly understood
Phosphorylation is one of the most important regulatory protein modifications, which is involved in a wide spectrum of cellular processes. The protein kinase regulates phosphorylation via catalyzing the transfer of γ-phosphate from ATP to the tyrosine, threonine, or serine residues of their substrate proteins. Dysregulated phosphorylation events due to oncogenic mutation of various kinases may also lead to tumorigenesis in tumors arising from various tissues [13]. With the impressive clinical benefits of the inhibitors that target these oncogenic kinases, many kinase inhibitors are currently used in clinics or under various stages of development. Thus, identifying kinases essential for ferroptosis may have translational potential for clinical studies. However, other than a few independent studies [1, 14], little is known about the role of kinases in the ferroptosis. To systemically investigate the role of protein kinases in the ferroptosis, we performed a kinome genetic screen to uncover the kinases essential for ferroptosis triggered by cystine deprivation. From our screen, we unexpectedly identified the noncanonical function of ataxia-telangiectasia mutated (ATM) as an essential kinase for ferroptosis through the regulation of the iron metabolism. These findings suggest the possibility to employ ATM inhibitors to reduce the cell death and pathogenesis of ferroptosis-associated human diseases.
Results
Kinome screen of the cystine-deprived ferroptosis
While previous studies of ferroptosis focused on effectors involved in cysteine metabolism [11] or lipid peroxidation [2, 9, 12], several studies have indicated that kinases might play an essential role in this process [1, 14]. To systematically identify kinases essential for ferroptosis, we performed forward genetic screens using a siRNA kinome library (Horizon/Dharmacon siGENOME SMARTpool) targeting 715 human genes annotated as “kinases”. The MDA-MB-231 cell was chosen due to its strong cystine addiction and sensitivity to erastin-induced death [15]. MDA-MB-231 were first transfected with each siRNA in the kinome library for two days, then placed under either regular (200 µM cystine) or cystine-deprived (1 µM cystine) media known to induce ferroptosis. Two days after the media changes, the viability of each well under regular or cystine-deprived media was assessed by CellTiter-Glo®, which measured ATP content as an indicator for cell viability. Two criteria were initially applied to identify kinase hits essential for ferroptosis. First, we calculated the “rescue ratio” as the extent of viability observed after treatment with each siRNAs to that of the nontargeting siRNA control under cystine-deprived conditions. The “rescue ratio” represents the observed degree of improved viability rescued by each siRNA under cystine-deprived condition. The rescue ratio of each siRNA was plotted (Y-axis) against its corresponding effects on the general viability observed under full, regular medium (X-axis, Fig. 1a). Second, we eliminated all siRNAs that led to less than 50% viability under full medium condition. Using ANOVA test (with threshold p < 0.0001), we identified our top 34 hits that rescued to a significant degree the ferroptosis phenotype observed after cystine depletion (Fig. 1b). Of note, we compared the mRNA expression profiles of those top 34 candidates in MDA-MB-231, MCF7 and NCI-ADR-RES cell lines [16, 17], and found most of these hit kinases were expressed in these cell lines at comparable levels (Fig. S1). Our analysis revealed that several kinases are involved in the TNFα pathway. For example, spleen associated tyrosine kinase [18] is essential for the TNFα-induced signaling [19] while TGFβ activated kinase 1, MAP3K7 is activated by TNFα to trigger cell death [19, 20]. Other hit is in the NF-kB pathway, such as interleukin 1 receptor associated kinase 1, which is recruited by myeloid differentiation primary response 88 to regulate NF-kB nuclear translocation [21]. These kinase hits suggest the potential involvement of TNFα and NF-kB signaling in the ferroptosis process.
Fig. 1.
Kinome screening identified 34 kinase essential for cystine-deprived death. a Scatter plot displayed the effects of siRNA-mediated silencing of individual kinase on the viability of MDA-MB-231 under cystine-deprived (Y-axis) and full media (X-axis) upon the siRNA-mediated silencing of individual kinase in MADA-MB-231 breast cancer cells. Y-axis and X-axis are the relative cell viabilities after knockdown each kinase normalized against nontargeting control siRNA in cystine deprived and normal medium, respectively. Each purple dot represents individual kinase-targeting pooled siRNA; each red dot represents significant “hits” from the screen based on the log-transformed normalized viability and calculated by ANOVA (threshold: p < 0.0001). b List of 34 “hit” essential kinases is shown with their normalized viability (against nontarget siRNA) in the control full media or cystine-deprived media (ANOVA p < 0.0001). The kinases were ranked by the normalized viability under cystine deprivation. c STRING analysis of all the hit kinases based on the protein–protein interaction networks, highlighting the essential role of ATM and ATR as highly connecting nodes among the kinases essential for the cystine-deprived death
To examine whether the gene products corresponding to these hits might interact with each other as part of interacting networks, we used STRING analysis [22] (Fig. 1c). Unexpectedly, we found that ATM and ataxia telangiectasia and Rad3-related protein (ATR) were the most connected “nodes” with largest number of interactions with other hits. Both ATM and ATR are kinases critical for the DNA damage responses [23], and one of their downstream targets, p53, has been implicated in regulating ferroptosis [8, 24–26], prompting us to further validate and investigate the role of ATM and ATR in the ferroptosis.
Genetic or pharmacological inhibition of ATM confers resistant to cystine deprivation and erastin
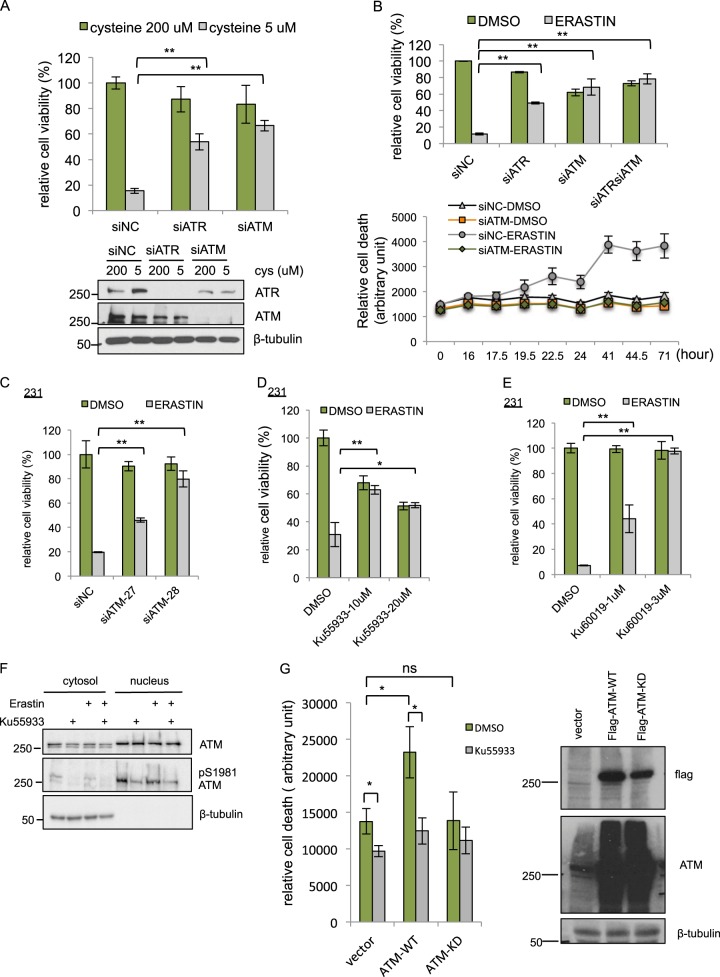
First, we validated the essential role of ATR or ATM in ferroptosis. We found that silencing of either ATM or ATR by pooled siRNAs rescued MDA-MB-231 from cell death triggered by cystine-deprivation (Fig. 2a) and erastin (Fig. 2b). When ATM and ATR were silenced simultaneously, ATM-depletion provided the predominant protective effect under erastin (Fig. 2b; siATR versus siATRsiATM, p-value < 0.01; siATM versus siATRsiATM, p-value = 0.22). Therefore, we decided to focus on the functional role and mechanisms of ferroptosis protection associated with ATM inhibition. In addition, ATM-depletion provided long-term (up to 71 h) protective effect against erastin (Fig. 2b, lower panel). ATM-depletion also protected cells from ferroptosis triggered by different concentrations of various GPX4 inhibitors including RSL3 [27] (Fig. S2a, b), or FIN56 [28] (Fig. S2c, d), suggesting that the ATM may act downstream of xCT and GPX4. To rule out any potential off-targets [29] associated with the original ATM targeting siRNA pool used in our experiments, we confirmed that ferroptosis could be rescued by the two individual siRNAs against ATM in both MDA-MB-231 and RCC4 (renal cell carcinoma) (Fig. 2c and S2e).
Fig. 2.
Validation of ATM as a critical mediator of ferroptosis. a Depletion of ATM and ATR confers resistance to cystine-deprived death. MDA-MB-231 cells were transfected with siNC, siATM, or siATR for 48 h, then incubated with normal full (200 μM cystine) or cystine-deprived (5 μM cystine) medium for 68 h. Cell viability was determined by CellTiter-Glo. Lower panel, the knockdown efficiency of ATM and ATR were determined by Western blots. b Depletion of ATM, ATR, or both ATM/ATR rescued MDA-MB-231 cells from erastin-induced death. MDA-MB-231 cells were transfected with siNC, siATM, siATR, or combined siATM/ATR for 72 h are then incubated with DMSO or erastin (10 μM) for 16 h (upper panel) or indicated time points up to 71 h (lower panel). Cell viability or death was determined by CellTiter-Glo (upper panel) or CellTox Green (lower panel), respectively. c Depletion of ATM by two individual siRNAs (siATM-27 and siATM-28) recued MDA-MB-231 cells from erastin-induced death. d, e ATM inhibitor Ku-55933 (d) and Ku-60019 (e) rescued cells from erastin-induced death. MDA-MB-231 cells were treated with Ku-55933 for 72 h, and then treated with DMSO or erastin for 24 h before viability determination by CellTiter-Glo. (The data are shown from one representative experiment with three biological replicates, and the data were reproduced from at least two independent experiments; Student’s t-test; *p < 0.05; **p < 0.01) f ATM inhibitor Ku-55933 reduced the S1981 phosphorylation of nuclear ATM. The amount of ATM protein or ATM S1981 phosphorylation in the nuclear or cytosol were analyzed by Western blots in the presence of erastin or Ku-55933. g Overexpression of wild-type (WT) but kinase dead (KD) ATM sensitized cells to ferroptosis. Vector, flag-WT-ATM, or flag-KD-ATM was transiently expressed into 293T cells. 24 h after transfection, cells were seeded into 96 well and pretreated with Ku-55933 (10 μM) for additional 24 h. The cell death was monitored by CellTox-Green 18 h after erastin (10 μM) treatment. The data was normalized to time 0 (immediately after addition of erastin). The data presented are mean ± S.D. from four biological replicates (Student’s t-test; *p < 0.05). In parallel, transfected cells lysates were collected for the protein expression of flag-WT-ATM or flag-KD-ATM by Western blots with indicated antibodies (right panel)
After validating that genetic silencing of ATM protected cells from erastin, RSL3, FIN56, or cystine-deprivation induced ferroptosis, we tested whether chemical inhibitors of ATM had similar effects. Three specific and structurally distinct ATM inhibitors, Ku-55933 [30], Ku-60019 [31], and AZD1390 [32] all significantly protected from erastin-induced ferroptosis phenotype in MDA-MB-231cells (Fig. 2d, e, and S2f) and RCC4 cells (Fig. S2g). Of note, Ku-55933 and Ku-60019 seem to exert stronger ferroptosis protection than AZD1390, raising the concerns about nonspecific redox properties of these two ATM inhibitors. However, such concern is not supported by the reported ability of Ku-55933 to accumulate intracellular ROS and depletes GSH [33, 34]. We also noted that, in MDA-MB-231, ATM proteins and S1981 phosphorylated ATM were highly enriched in the nucleus (Fig. 2f) under basal conditions [35, 36]. Importantly, the nuclear and S1981 phosphorylated ATM were not affected by erastin, suggesting that the ATM activation was found in MDA-MB231 at baseline before erastin treatment. However, Ku-55933 successfully reduced ATM phosphorylation at S1981 (Fig. 2f). The presence of nuclear and phosphorylated ATM in MDA-MB-231 suggested that ATM might have already been activated in the absence of ionization radiation or erastin, consistent with a previous study showing spontaneous DNA damage and ATM activation in MDA-MB-231 [37]. During DNA damage, CHK2 (Checkpoint Kinase 2) and tumor suppressor p53 are often considered the canonical signaling substrates of ATM [23]. However, depletion of CHK2 by siRNA did not rescue cells from ferroptosis (Fig. S2h), suggesting that ATM may regulate ferroptosis in a CHK2-independent way. In addition, ATM inhibition rescued cells from ferroptosis in both p53 wild type RCC4 (Fig. S2e, g) and HT-1080 (Fig. S2i, j) or p53 mutant MDA-MB-231 (Fig. 2). To further determine the role of ATM kinase activity in ferroptosis, we overexpressed wild type or kinase dead ATM [38] into 293T cells due to easy transfection and measured their sensitivity to erastin. We found overexpression of wild type, but not kinase dead mutant, ATM sensitized cells to erastin while blocking ATM activity by Ku-55933 reduced cell death (Fig. 2g). Collectively, these data indicate that ATM-depletion or inhibition of its enzymatic activity robustly conferred ferroptosis resistance, in a CHK2- or p53-independent manner.
ATM inhibition regulates the mRNA levels of iron regulators
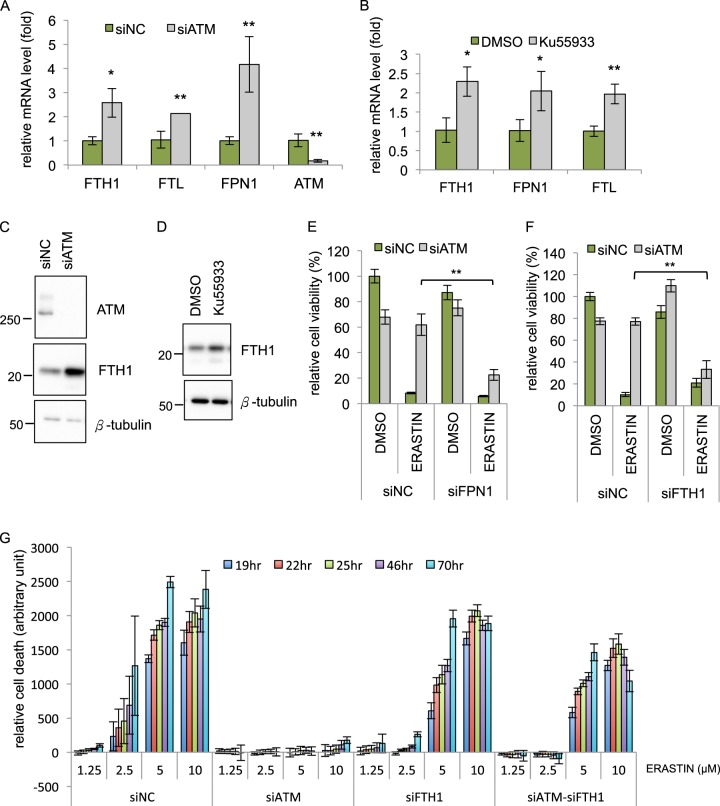
To investigate how ATM inhibition confers ferroptosis protection, we analyzed a published transcriptome data in response to ATM inhibition conducted by Miller’s group [39]. Interestingly, we noted the ATM knockdown led to the upregulation of the SLC40A1, encoding ferroportin (or FPN1), the sole iron exporter. In addition, McDonald et al. also reported altered ferritin and ferroportin expression in the Atm null mouse liver [40]. Since iron metabolism is critical for ferroptosis [4], these data prompted us to examine whether ATM inhibition might affect the expression of various iron regulators. Quantitative RT-PCR revealed that ATM knockdown (Fig. 3a) or ATM inhibition (Fig. 3b) both increased the mRNA expression of FPN1 (Ferroportin), FTH1 (Ferritin heavy chain 1), and FTL (Ferritin light chain). In addition, we observed an increase in FTH1, FTL, SLC7A11/xCT, and GPX4 protein upon ATM knockdown (Figs. 3c, S3a) or increase in FTH1 protein upon treatment with the ATM inhibitor Ku-55933 (Fig. 3d). Of note, induction of SLC7A11/xCT in ATM knockdown cells was associated with a lower reduced GSH (Fig. S3b). Erastin treatments significantly reduced the cellular GSH levels in both siNC and siATM cells, suggesting that SLC7A11/xCT induction in ATM knockdown cells may not be sufficient to abolish the erastin-induced GSH depletion to confer ferroptosis protection (Fig. S3b).
Fig. 3.
The induction of iron regulatory genes by ATM inhibition is essential for protection against ferroptosis a Depletion of ATM upregulates the mRNA expression of FTH1, FTL, and FPN1. MDA-MB-231 cells were transfected with siNC or siATM for 48 h and the mRNA levels of indicated genes were determined by qRT-PCR. b ATM inhibitor Ku-55933 upregulated the mRNA expression of FTH1, FTL, and FPN1. MDA-MB-231 cells were treated with Ku-55933 for 48 h, and the mRNA levels of indicated genes were determined by qRT-PCR. c, d Inhibition of ATM by siRNA (c) or Ku-55933 (d) increased the level of FTH1 protein. Cell lysates from siATM (69 h) or Ku-55933 (48 h) were analyzed by Western blots with indicated antibodies. e, f FPN1 and FTH1 are essential for ferroptosis protection by ATM inhibition. Cells were transfected with indicated siRNA for 78 h, and incubated with DMSO or erastin (10 μM) for 18 h before determining the viability by CellTiter-Glo. (The data are shown from one representative experiment with three biological replicates, and the data were reproduced from at least two independent experiments; Student’s t-test; *p < 0.05; **p < 0.01) g FTH1 is essential for ferroptosis protection by ATM inhibition for longer time course. MDA-MB-231 cells were transfected with indicated siRNA for 72 h, and incubated with DMSO or erastin for 70 h. The cell death was monitored from time 0 to 70 h by CellTox-Green. All the data were normalized to DMSO control in each group (The data are shown from three biological replicates.)
These data clearly indicate that ATM inhibition triggers a coordinated change in the mRNA levels of multiple iron regulators to increase export (increased FPN1), as well as iron sequestration (increased FTH1 and FTL). Consequently, these changes may affect the sensitivity of cells to undergo ferroptosis [4]. To test this possibility, we abolished FTH1 or FPN1 induction by siRNA-mediated silencing upon ATM inhibition. We found that the knockdown of either FPN1 (Fig. 3e) or FTH1 (Fig. 3f, g and S3c for longer time course) significantly reduced the ferroptosis protection offered by ATM knockdown. Collectively, these data show that FTH1 and FPN1 induction is critical to the ferroptosis protection phenotypes upon ATM knockdown.
ATM inhibition reduces cellular levels of labile iron
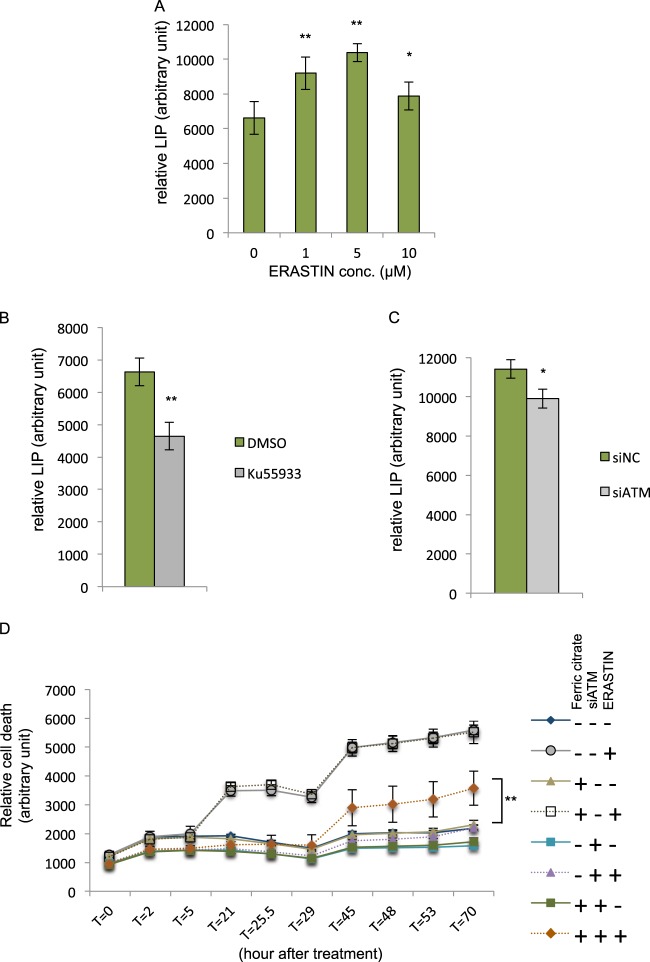
ATM inhibition induced the expression of iron exporter (FPN1) and ferritins (FTH1, FTL) involved in sequestering iron. These changes are expected to affect the levels of labile iron. To test this possibility, we used the fluorescent metal sensor (calcein green) and iron chelator to determine the levels of labile iron pools in MDA-MB-231 cells exposed to varying doses of erastin [41, 42]. Consistent with previous studies, we found that erastin (1, 5, or 10 μM) increased the labile iron pool (Fig. 4a) [42]. Furthermore, chemical inhibition of ATM by Ku-55933 significantly reduced the level of labile iron (Fig. 4b), as expected from the induction of FPN1 and ferritins. Similarly, ATM knockdown by siRNA also reduced the level of labile iron (Fig. 4c). Given the importance of labile iron in ferroptosis, it is likely that a reduction in the labile iron pool contributes to the ferroptosis protection associated with ATM inhibition. Indeed, supplementing ferric citrate in the medium resensitized the ATM knockdown cells to ferroptosis under erastin (Fig. 4d). Taken together, we found ATM inhibition protects against ferroptosis by reducing the labile iron pool.
Fig. 4.
ATM inhibition reduces the intracellular labile iron pool. a Erastin treatment increases intracellular labile iron pool (LIP). MDA-MB-231 cells were treated with indicated concentrations of erastin for 6 h, and incubated with Calcien-AM. The relative LIP was calculated based on the differences in the fluorescence intensity before and after DFO chelation. (The data was acquired from six biological replicates performed in parallel; Student’s t-test; *p < 0.05, **p < 0.01) b ATM inhibition by Ku-55933 decreases intracellular labile iron pool. MDA-MB-231 cells were treated with Ku-55933 (5 μM) for 24 h, and incubated with Calcien-AM to measure the relative LIP. (The data was acquired from eight biological replicates performed in parallel; Student’s t-test; **p < 0.01) c Depletion of ATM by siRNA decreases intracellular labile iron pool. MDA-MB-231 cells were transfected with indicated siRNA for 72 h, and incubated with Calcien-AM to measure the relative LIP. (The data was acquired from four biological replicates performed in parallel; Student’s t-test; *p < 0.05) d Iron supplementation sensitizes ATM-depleted cells against erastin. MDA-MB-231 cells were transfected with siNC or siATM for 72 h, and then incubated with DMSO, erastin (10 μM), with or without ferric citrate (50 μM) for indicated time point. The amount of cell death was evaluated by CellTox-Green at each time point. The data presented are mean ± S.D. from four biological replicates, and the data were reproduced from two independent experiments. P-values were calculated by Student’s t-test, **p < 0.01
MTF1 regulates the iron regulatory transcriptional program of ATM inhibition
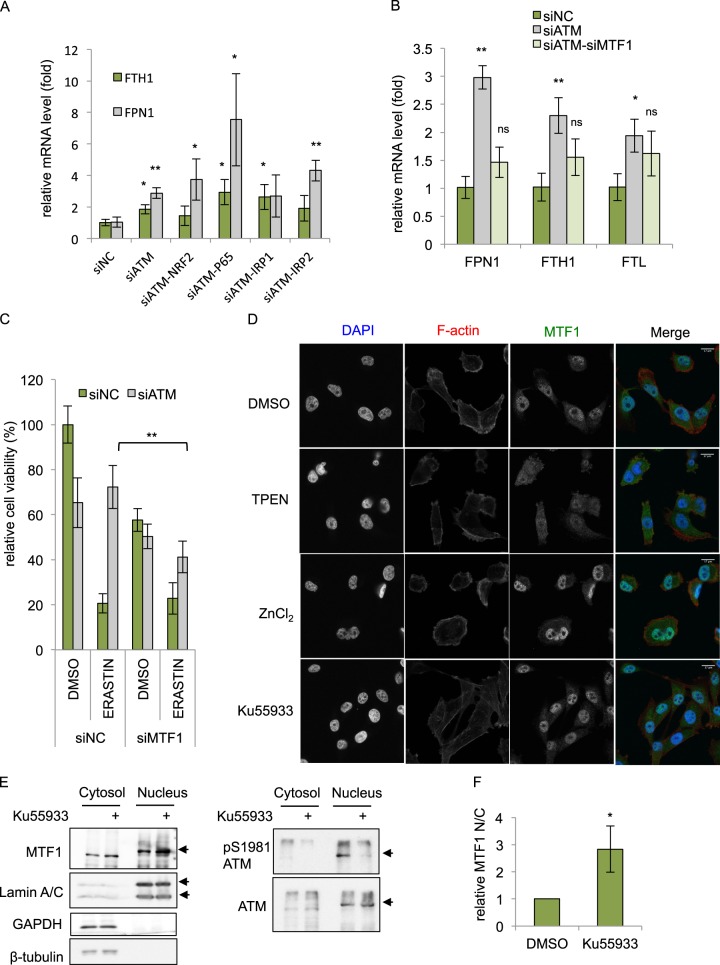
Next, we wished to identify the regulator(s) responsible for the FPN1 and FTH1 induction during ATM inhibition. Based on a literature search, we considered several candidates including iron-responsive element-binding proteins (IRP1 or IRP2) [43], nuclear factor (erythroid-derived 2)-like-2 (NRF2) [44–46], myeloid zinc-finger 1 (MZF1) [46], NF-κB p65 [47, 48], and metal-regulatory transcription factor-1 (MTF1) [49, 50]. To determine the potential role of each candidate protein in the induction of iron regulators upon ATM inhibition, we transfected siRNAs targeting each candidate regulator, together with siATM, and analyzed by qRT-PCR to determine how they affected the FPN1/FTH1 induction. We found that silencing of NRF2, p65, IRP1, and IRP2 only partially or did not significantly affect the FTH1 and FPN1 induction caused by ATM inhibition (Fig. 5a). In contrast, silencing of MTF1 abolished the induction of FPN1, FTH1, and FTL during ATM inhibition (Fig. 5b). Importantly, MTF1 silencing also abolished the ferroptosis protection phenotypes of ATM silencing (Fig. 5c). In addition, we noticed that the protective effect of ATM silencing could also be mitigated by NRF2 silencing (Fig. S4a), consistent with known role of NRF2 in regulating ferroptosis [44]. In contrast, the silencing of MZF1, IRP1, or IRP2 did not abolish the ferroptosis protection by ATM silencing (Fig. S4b–d). In sum, our data validated the function of NRF2 in regulating ferroptosis and also implicated a novel role of MTF1 in the ferroptosis protection phenotypes upon ATM inhibition.
Fig. 5.
MTF1 is responsible for induction of iron regulators and ferroptosis resistance of ATM inhibition. a, b MTF1 but not NRF2, p65, or IRP1/2 is responsible for the induction of FTH1, FTL, and FPN1 mRNA associated with ATM silencing. MDA-MB-231 cells were transfected with indicated siRNAs for 48 h, and the mRNA expression of indicated genes were analyzed by qRT-PCR. The data was acquired from three biological replicates. P-values were calculated by Student’s t-test, *p < 0.05, **p < 0.01, ns stands for non-significant. The level of each mRNA was normalized to β-actin mRNA. The P-value was calculated by comparing the expression of each group to the siNC control. c Depletion of MTF1 sensitized ATM-knockdown cells to ferroptosis. MDA-MB-231 cells transfected with indicated siRNAs for 72 h, treated with DMSO or erastin for 30 h and the viability was determined by CellTiter-Glo. The data was acquired from six biological replicates. P-values were calculated by Student’s t-test, *p < 0.05, **p < 0.01. d The subcellular localization of MTF1 upon TPEN, ZnCl2 or Ku-55933 treatment. MDA-MB-231 cells treated with DMSO, TPEN (5 μM), ZnCl2 (100 μM), or Ku-55933 (5 μM) were fixed and stained with DAPI (nuclear), phalloidin (F-actin), and MTF1 antibody. Representative images are shown to visualize the subcellular locations of MTF1 upon different treatments. e, f Ku-55933 increased nuclear translocation. The cytosol and nuclear fractions of MDA-MB-231 were collected after treatment of Ku-55933 (5 μM) for 24 h. The successful cytosolic and nuclear fractions were assessed GADPH and Lamin A/C, respectively. The relative amount of MTF1 in each fraction was quantified by Western blots (e) and ImageJ (f). The data presented are mean ± S.D. from three biological replicates. P-values were calculated by Student’s t-test, *p < 0.05, **p < 0.01
MTF1 is a metal-regulatory transcription factor that responds to and regulates the cellular levels of various metals, especially zinc [50]. A previous study has shown that MTF1 triggers the expression of the iron exporter FPN1 [49], consistent with our data on its role in the induction of FPN1/FTH1/FTL during ATM inhibition (Fig. 5b). Since MTF1 activity is usually regulated by nuclear translocation [50], we investigated whether the subcellular locations of MTF1 was affected by ATM inhibition. Confocal immunofluorescence assay showed that, at baseline (DMSO), MTF1 was found in both nuclei and cytosol of MDA-MB-231 (Fig. 5d). Zinc chelator (TPEN) restricted MTF1 protein in the cytosol with limited amount in the nucleus as observed with obscure nuclear and cytosol boundary. Conversely, zinc chloride triggered almost complete nuclear translocation (Fig. 5d) as we observed several dense nuclear MTF1 speckles. Importantly, Ku-55933 dramatically enhanced nuclear localization of MTF1, consistent with the activation of the iron regulatory genes by MTF1 during ATM inhibition. The nuclear translocation of MTF1 by ATM inhibition (Ku-55933) was further validated using Western blots of the nuclear and cytosolic fractions. Ku-55933 significantly reduced the ATM phosphorylation (Fig. 5e) and increased the nuclear fractions of MTF1 protein, but not lamin A/C (nuclear loading control) (Fig. 5e) (see also Fig. S5a for uncropped blot and validation of the specificity of MTF1 antibody by siRNA in Fig. S5b), as quantified in Fig. 5f. Together, these data indicate that ATM inhibition induced nuclear translocation of the MTF1 transcription factor. This nuclear translocation of MTF1 is expected to result in an increase in the expression of iron-regulatory genes, which leads to a decrease in labile iron levels and helps prevent ferroptosis.
Discussion
Summary of the findings
Using a siRNA library targeting the human kinome, we have identified 34 kinases that are essential for the full execution of ferroptosis in response to cystine deprivation. In addition to kinases involved in the TNF-α and NF-kB pathway, we uncovered an unexpected role for ATM in regulating ferroptosis. Genetic knockdown and chemical inhibition of ATM both suppress ferroptotic cell death and reduce intracellular labile iron via enhancing iron sequestration (increased FTH1, FTL) and export (increased FPN1). These coordinated changes rely largely on MTF1 transcriptional activity and its nuclear translocation upon ATM inhibition. Together, we uncover a novel regulatory mechanism by which ATM regulates the labile iron levels and determines the sensitivity to ferroptosis (Fig. 6). These findings reveal a novel function of ATM in regulating cellular iron metabolisms and provide important insights into the regulation of ferroptosis. In addition, these findings also have significant therapeutic implications.
Fig. 6.
Working model of ATM regulates ferroptosis. Cystine deprivation or erastin treatment reduces intracellular cystine, cysteine, and GSH (reduced glutathione) level while increases ROS (reactive oxygen species) and oxidized glutathione (GSSG). These changes make the cells susceptible to iron-mediated cell death, ferroptosis. ATM-inactivation increases the nuclear translocation of MTF1 protein, which induces the expression of ferritin (FTH1 and FTL) and ferroportin (FPN1) to sustain low-labile iron condition and confer resistance to ferroptosis
ATM inhibitors may be used to treat ferroptosis related diseases
It has been appreciated that ferroptosis is involved in many human diseases [2], such as acute renal injury [9, 10]. Therefore, blocking ferroptosis may have significant therapeutic benefits. Given the essential role of ATM in ferroptosis, our findings provide a mechanism-based rationale for testing the therapeutic potential of various ATM inhibitors in ferroptosis-related diseases. Many ATM inhibitors have been developed as radiosensitizers [51] to enhance the efficacy of radiation therapies. Of note, AZD1390, a brain-penetrant ATM inhibitor, significantly induced tumor regressions and increased animal survival when treated in combination with ionizing radiation (IR) [32], also rescued MDA-MB-231 cells against ferroptosis (Fig. S2f). As these ATM inhibitors are evaluated for the safety and efficacy for treatment various tumors, our results suggest these ATM inhibitors, selected to be “safe” in the setting of these trials, may be also repurposed to block ferroptosis and improve the outcomes of these ferroptosis-associated diseases. However, it is also important to point out that since ATM inhibitors affect ferroptosis indirectly through regulating labile iron levels, the ferroptosis protection effects of ATM inhibitors may be affected by many other factors impacting iron metabolisms, thus may limiting the efficacy of such ferroptosis inhibitors.
The connection between DNA damage response and ferroptosis
While the involvement of ATM in the ferroptosis is unexpected, it is important to note that ATM phosphorylates and stabilizes p53 during DNA damage [52]. As the gatekeeper of genomic integrity, p53 has been shown to trigger ferroptosis as a part of its tumor suppressor activities [8, 26]. However, other studies have shown that p53 can suppress ferroptosis through the regulation of dipeptidyl peptidase 4 [24] or p21 [25]. Therefore, the regulation of ferroptosis by p53 may be context dependent. In our story, p53 may not be the key determinant of ferroptosis protection through ATM inhibition since we observed these phenotypes in both p53-mutated triple negative breast cancer cells [53] or p53 wild-type HT-1080 and RCC4 cells (Fig. S2e, g, i, and j). Another canonical ATM target CHK2 was also not relevant for the ferroptosis protection (Fig. S2h). Therefore, ATM probably affects ferroptosis through a mechanism distinct from the canonical DNA damage pathways. Our data indicates that ATM inactivation regulates the iron metabolism and reduces labile iron levels through a novel mechanism by affecting MTF1 activities. MTF1 is a metal responsive transcription factor that typically senses and responds to intracellular metal levels [50, 54]. While most of studies focus on the regulation between MTF1 and zinc in mammals, several reports have suggested the potential connections with iron. For example, the expression of ferroportin or hepcidin is regulated by MTF1 [49, 55]. In Drosophila, induction of ferritin was stimulated by treatment of cadmium, copper, or zinc in a MTF1 dependent manner [56]. Overexpression of MTF1 in flies also prolongs their lifespans under iron exposing condition [57]. Taken together, we uncovered a novel crosstalk of between ATM, MTF1, and iron metabolism that regulate the sensitivities of cancer cells to ferroptosis. In fact, we did not find putative ATM phosphorylation motif within MTF1, suggesting that MTF1 might not be a direct substrate of ATM. However, upon the inhibition of ATM, accumulation of ROS and oxidative stress [58–63] may activate c-Jun N-terminal kinase (JNK) [64, 65], and thus leading to activation of MTF1 [66]. Alternatively, delayed dephosphorylation of JNK upon ATM inhibition or inhibition of some protein tyrosine phosphatases during ferroptosis-inducing conditions might also contribute to activation of MTF1 [66–68]. Future study focusing on ATM signaling and phosphoproteomics may be an interesting topic to determine whether JNK or other direct ATM targets are responsible for the activation and translocation of MTF1 upon ATM inhibition. In addition, how does MTF1 selectively regulates zinc, iron, or other metal level in the context of ferroptosis may also be a relevant topic for future investigation.
Iron level as a mechanistic link between oncogenic states and ferroptosis sensitivity
Several studies have shown that ionization radiations increased labile iron level [69, 70]. Importantly, chelation of labile iron protect cells from ionizing radiation [71], indicating the critical role of increased labile iron in the therapeutic benefits. These data are consistent with our data showing that ATM activities can regulate labile iron levels. Reciprocally, our results also suggest that IR-mediated induction of labile iron may sensitize the cells to various ferroptosis-inducing agents. It will be important to determine whether the ATM-MTF1-FNP1/FTH1/FTL regulatory axis is responsible for the IR-induced changes in labile iron. In sum, there is a strong rationale to combine ferroptosis and ionization radiation to enhance the efficacy of IR and reduce resistance for various tumors.
Materials and methods
Cell lines, siRNAs, chemicals, constructs, and antibodies
MDA-MB-231, RCC4, 293T, and HT-1080 cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplied with fetal bovine serum (FBS) and penicillin/streptomycin under 37 °C and 5%CO2. siRNAs targeting ATR-pool (M-003202-05-0005), ATM-pool (M-003201-04-0005), ATM-27 (D-003201-27-0005), ATM-28 (D-003201-28-0005), FTH1 (M-019634-02-0005), FPN1 (M-007576-01-0005), CHK2 (M-003256-06-0005), NRF2 (J-003755-09), RELA (M-003533-02-0005), MTF1 (M-020078-01-0005), MZF1 (M-006578-02-0005), IRP1(M-010037-00-0005), and IRP2 (M-022281-01-0005) were obtained from Horizon/Dharmacon. Erastin (Caymen chemical, CAS 571203-78-6), ZnCl2 (Sigma, #39059), N,N,N′,N′-Tetrakis (2-pyridylmethyl) ethylenediamine (TPEN, Sigma, #P4413), ferric citrate (Sigma, F3388), Ku-55933 (Selleckchem, #S1092), Ku-60019 (Selleckchem, #S1570), RSL3 (Caymen, #19288), FIN56 (Sigma, #SML1740), AZD1390 (Selleckchem, #S8680), ATM-WT (Addgene, #31985), ATM-KD (Addgene, #31986), anti-ATM (Cell Signaling, #2873), anti-ATR (Cell Signaling, #2790), anti-phospho serine 1981 ATM (ABcam, ab81292), anti-FTH1 (Cell Signaling, #4393), anti-FTL (Santa Cruz, #SC-74513), anti-MTF1 (NBP1-86380), anti-β-tubulin (Cell Signaling, #2128), anti-lamin A/C (Cell Signaling, #4777), anti-xCT/SLC7A11 (Cell Signaling, #12691), and anti-GPX4 (ABcam, #ab125066).
Western blot and nuclear/cytosol fraction
Cells were lysed by RIPA buffer and quantified by BCA (Bicinchoninic acid) assay. The proteins of interest were analyzed by Western blot using chemiluminescence (ThermoFisher Scientific SuperSignal Pico) and ChemiDoc Imager (Bio-Rad). For nuclear and cytosol fractionation, the REAP protocol [72] was used.
Determination of cell viability, cell death, and glutathione level
The cells transfected with siRNA (48–72 h) or incubated with chemical inhibitor were further treated with erastin for additional 16–24 h, and the cell viability was determined by the CellTiter-Glo assay (Promega) according to manufacturer’s instruction. For the cell death assay, after siRNA transfection for 72 h, the cells were incubated with erastin (10 µM) and CellTox Green dye (Promega) or supplementation with ferric citrate (Sigma, F3388) as indicated. The amount of cell death was evaluated by fluorescence intensity and plate reader (FLUOstar Optima, BMG LABTECH) at the indicated time points. To measure intracellular reduced GSH, cells were first transfected with siNC or siATM for 48 h and then treated with erastin (2.5 μM) for additional 18 h. The amount of GSH was then detected by GSH/GSSG-Glo assay (Promega, #V6611). The relative amount of reduced GSH was calculated from total and oxidized glutathione (GSSG) and normalized to cell viability assay performed.
Kinome screen
The kinome siRNA library was purchased from Horizon/Dharmacon siGENOME SMARTpool siRNA library (G-003505-E2, Lot 08166) targeting 715 human “kinase” genes in 96-well plate format. Regular (200 µM cystine) or cystine-deprived (1 µM cystine) media were prepared as described [73, 74] using DMEM, high glucose, no glutamine, no methionine, no cystine media (ThermoFisher Scientific, #21013024) with 10% dialyzed FBS (Sigma, F0392), l-Methionine (Sigma, M5308, final concentration 30 mg/L), l-glutamine (ThermoFisher, #25030-081, final concentration 4 mM), and either 200 µM or 1 µM l-Cystine (Sigma, C6727). In brief, 4000 MDA-MB-231 cells were reverse transfected with a pool of 4 siRNA (0.25 nmol) targeting one gene in each well with 0.2 µL of lipofectamine RNAiMAX transfection reagent (ThermoFisher Scientific, #13778150) for 48 h. The cells were then washed twice by PBS with calcium and magnesium (ThermoFisher Scientific, #14040133) to remove remaining cystine in the well and then incubated with either regular (200 µM cystine) or cystine-deprived (1 µM cystine) media. After 48 h of incubation in cystine manipulated medium, the cellular viability of the each well (with different siRNA) was assessed by CellTiter-Glo (Promega, G7571) reagent and luminescent plate reader (FLUOstar Optima, BMG LABTECH). There are at least two biological replicates for each gene with either regular or cystine-deprived treatment. The transfection efficiency for each plate was accessed by the remaining viability of cells transfected with TOX transfection control (Horizon/Dharmacon, D-001500-01), which the cell death is proportional to the transfection efficiency. The cell viability for each kinase siRNA knockdown were normalized against nontargeting siRNA control (Qiagen, AllStars Negative control siRNA, SI03650318) on each plate to eliminate batch effect of each plate. The protein–protein interaction map was generated by STRING version 9.1 [75].
RNA extraction and quantitative RT-PCR
RNA samples collected from MDA-MB-231 cells were extracted by RNeasy kit (Qiagen). cDNA was then synthesized by SuperScript II reverse transcriptase (Thermo Fisher Scientific). The primers used for real time PCR (StepOnePlus, Applied Biosystems) are listed as follows:
FTH1 (F: 5′-AAAGCCATCAAAGAATTGGG; R: 5′-GGGTGTGCTTGTCAAAGAGA), FTL (F: 5′-GGGTCTGTCTCTTGCTTCAAC; R: 5′-GGTTGGCAAGAAGGAGCTAA), FPN1 (F: 5′-ATGGGTTCTCACTTCCTGCT; R: 5′-GCTTCTGTCTTCTCCTGCAA), ACTB (F: 5′-CACTCTTCCAGCCTTCCTTC; R: 5′-GGATGTCCACGTCACACTTC), and ATM (F: 5′-TGGCTTACTTGGAGCCATAA; R: 5′-ATACTGGTGGTCAGTGCCAA).
Immunofluorescence assay
The subcellular localization (cytosol vs. nuclear) of MTF-1 was visualized by MTF-1 antibody staining (NBP1-86380) after MDA-MB-231 cells were treated with DMSO, TPEN (5 μM), ZnCl2 (100 μM), or Ku-55933 (5 μM) for 6 h. The cells were fixed in 4% formaldehyde for 10 min, followed with 1XPBS (phosphate buffered saline) wash, blocking at room temperature for 1 h (5% bovine serum albumin in 1XPBS/0.3% Triton X-100), and incubation with MTF-1 antibody (dilution factor 1:200 in blocking solution) at 4 °C for overnight. The nuclear fraction was marked by DAPI staining, while the cytosol fraction was marked by F-actin (phalloidin) staining. The images were acquired by Zeiss 780 inverted confocal microscopy at the Duke light microscopy core facility.
Measurement of labile iron level
The level of labile iron was measured as previously described [41, 42]. Briefly, MDA-MB-231 cells treated with erastin (1, 5, or 10 μM, 6 h), Ku-55933 (5 μM, 24 h), siRNA (siNC or siATM for 72 h) were incubated with calcein-AM (acetoxymethyl ester, C3099-Thermo) (1 μM) for 15 min. The initial (F0) and chelated fluorescence intensity (FDFO, after incubation with the iron chelator (100 μM, desferoxamine mesylate, D9533-Sigma)) were recorded by plate reader (FLUOstar Optima, BMG LABTECH) to obtain the relative amount of labile iron pool (ΔF = FDFO − F0).
Statistical methods
ANOVA with Tukey honestly significant difference post hoc test were used to analyze kinome screen data. Student’s t-test was used throughout the study unless specifically mentioned to acquire P-values (two-sided). P-value < 0.05 (*) was considered significant. All of the data are presented as mean ± standard deviation (S.D.).
Supplementary information
Acknowledgements
We appreciate the suggestions from members of Chi lab at Duke University and discussion with Dr. Stacey-Lynn Paiva at Yale University. This work was supported in part by DOD grants (W81XWH-17-1-0143, W81XWH-15-1-0486 to JTC), Duke Cancer Institute (DCI) Discovery fund and DCI pilot fund.
Author contributions
PHC, JW, CKD, CCL, SP, NB, YX, and WHY performed the experiments. PHC, CKD, and JTC designed and interpreted the experiments and wrote and revised the manuscript. All authors reviewed and approved the finished manuscript. JTC supervised all aspects of the work. BMP provided reagents and manuscript discussion.
Compliance with ethical standards
Conflict of interest
The authors have no conflicts of interests to declare.
Footnotes
Edited by S. Fulda
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41418-019-0393-7) contains supplementary material, which is available to authorized users.
References
- 1.Xie Y, Hou W, Song X, Yu Y, Huang J, Sun X, et al. Ferroptosis: process and function. Cell Death Differ. 2016;23:369–79. doi: 10.1038/cdd.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockwell BR, Friedmann Angeli JP, Bayir H, Bush AI, Conrad M, Dixon SJ, et al. Ferroptosis: a regulated cell death nexus linking metabolism, redox biology, and disease. Cell. 2017;171:273–85. doi: 10.1016/j.cell.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature. 2007;447:864–8. doi: 10.1038/nature05859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang WS, SriRamaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–31. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–91. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol. 2018;20:1181–92. doi: 10.1038/s41556-018-0178-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520:57–62. doi: 10.1038/nature14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Skouta R, Dixon SJ, Wang J, Dunn DE, Orman M, Shimada K, et al. Ferrostatins inhibit oxidative lipid damage and cell death in diverse disease models. J Am Chem Soc. 2014;136:4551–6. doi: 10.1021/ja411006a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linkermann A, Skouta R, Himmerkus N, Mulay SR, Dewitz C, De Zen F, et al. Synchronized renal tubular cell death involves ferroptosis. Proc Natl Acad Sci USA. 2014;111:16836–41. doi: 10.1073/pnas.1415518111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ. 2016;23:270–8. doi: 10.1038/cdd.2015.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang Y, Tiziani S, Park G, Kaul M, Paternostro G. Cellular protection using Flt3 and PI3Kα inhibitors demonstrates multiple mechanisms of oxidative glutamate toxicity. Nat Commun. 2014;5:3672. doi: 10.1038/ncomms4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, Ding CK, Wu J, Sjol J, Wardell S, Spasojevic I, et al. Cystine addiction of triple-negative breast cancer associated with EMT augmented death signaling. Oncogene. 2017;36:4379. doi: 10.1038/onc.2017.192. [DOI] [PubMed] [Google Scholar]
- 16.Kohn KW, Zeeberg BM, Reinhold WC, Pommier Y. Gene expression correlations in human cancer cell lines define molecular interaction networks for epithelial phenotype. PLoS ONE. 2014;9:e99269. doi: 10.1371/journal.pone.0099269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfister TD, Reinhold WC, Agama K, Gupta S, Khin SA, Kinders RJ, et al. Topoisomerase I levels in the NCI-60 cancer cell line panel determined by validated ELISA and microarray analysis and correlation with indenoisoquinoline sensitivity. Mol Cancer Ther. 2009;8:1878–84. doi: 10.1158/1535-7163.MCT-09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahabeleshwar Ganapati H., Kundu Gopal C. Syk, a Protein-tyrosine Kinase, Suppresses the Cell Motility and Nuclear Factor κB-mediated Secretion of Urokinase Type Plasminogen Activator by Inhibiting the Phosphatidylinositol 3′-Kinase Activity in Breast Cancer Cells. Journal of Biological Chemistry. 2002;278(8):6209–6221. doi: 10.1074/jbc.M208905200. [DOI] [PubMed] [Google Scholar]
- 19.Takada Y, Aggarwal BB. TNF activates Syk protein tyrosine kinase leading to TNF-induced MAPK activation, NF-kappaB activation, and apoptosis. J Immunol. 2004;173:1066–77. doi: 10.4049/jimmunol.173.2.1066. [DOI] [PubMed] [Google Scholar]
- 20.Morioka S, Broglie P, Omori E, Ikeda Y, Takaesu G, Matsumoto K, et al. TAK1 kinase switches cell fate from apoptosis to necrosis following TNF stimulation. J Cell Biol. 2014;204:607–23. doi: 10.1083/jcb.201305070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luftig M, Prinarakis E, Yasui T, Tsichritzis T, Cahir-McFarland E, Inoue J, et al. Epstein–Barr virus latent membrane protein 1 activation of NF-kappaB through IRAK1 and TRAF6. Proc Natl Acad Sci USA. 2003;100:15595–15600. doi: 10.1073/pnas.2136756100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRINGv10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 24.Xie Y, Zhu S, Song X, Sun X, Fan Y, Liu J, et al. The tumor suppressor p53 limits ferroptosis by blocking DPP4 activity. Cell Rep. 2017;20:1692–704. doi: 10.1016/j.celrep.2017.07.055. [DOI] [PubMed] [Google Scholar]
- 25.Tarangelo A, Magtanong L, Bieging-Rolett KT, Li Y, Ye J, Attardi LD, et al. p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 2018;22:569–75. doi: 10.1016/j.celrep.2017.12.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy ME. Ironing out how p53 regulates ferroptosis. Proc Natl Acad Sci USA. 2016;113:12350–2. doi: 10.1073/pnas.1615159113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol. 2008;15:234–45. doi: 10.1016/j.chembiol.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimada K, Skouta R, Kaplan A, Yang WS, Hayano M, Dixon SJ, et al. Global survey of cell death mechanisms reveals metabolic regulation of ferroptosis. Nat Chem Biol. 2016;12:497–503. doi: 10.1038/nchembio.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers JW, Chi JT, Schaner ME, Brown PO, Ferrell JE. Minimizing off-target effects by using diced siRNAs for RNA interference. J RNAi Gene Silenc. 2006;2:181–94. [PMC free article] [PubMed] [Google Scholar]
- 30.Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, et al. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 31.Golding SE, Rosenberg E, Valerie N, Hussaini I, Frigerio M, Cockcroft XF, et al. Improved ATM kinase inhibitor KU-60019 radiosensitizes glioma cells, compromises insulin, AKT and ERK prosurvival signaling, and inhibits migration and invasion. Mol Cancer Ther. 2009;8:2894–902. doi: 10.1158/1535-7163.MCT-09-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durant ST, Zheng L, Wang Y, Chen K, Zhang L, Zhang T, et al. The brain-penetrant clinical ATM inhibitor AZD1390 radiosensitizes and improves survival of preclinical brain tumor models. Sci Adv. 2018;4:eaat1719. doi: 10.1126/sciadv.aat1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sullivan KD, Palaniappan VV, Espinosa JM. ATM regulates cell fate choice upon p53 activation by modulating mitochondrial turnover and ROS levels. Cell Cycle. 2015;14:56–63. doi: 10.4161/15384101.2014.973330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin CS, Wang YC, Huang JL, Hung CC, Chen JY. Autophagy and reactive oxygen species modulate cytotoxicity induced by suppression of ATM kinase activity in head and neck cancer cells. Oral Oncol. 2012;48:1152–8. doi: 10.1016/j.oraloncology.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 35.So S, Davis AJ, Chen DJ. Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J Cell Biol. 2009;187:977–90. doi: 10.1083/jcb.200906064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 37.Liu X, Li F, Huang Q, Zhang Z, Zhou L, Deng Y, et al. Self-inflicted DNA double-strand breaks sustain tumorigenicity and stemness of cancer cells. Cell Res. 2017;27:764–83. doi: 10.1038/cr.2017.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, et al. Activation of the ATM kinase by ionizing radiation and phosphorylation ofp53. Science. 1998;281:1677–9. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 39.Chen WT, Ebelt ND, Stracker TH, Xhemalce B, Van Den Berg CL, Miller KM. ATM regulation of IL-8 links oxidative stress to cancer cell migration and invasion. Elife 2015;4:e07270. [DOI] [PMC free article] [PubMed]
- 40.McDonald CJ, Ostini L, Wallace DF, John AN, Watters DJ, Subramaniam VN. Iron loading and oxidative stress in the Atm−/− mouse liver. Am J Physiol Gastrointest Liver Physiol. 2011;300:G554–560. doi: 10.1152/ajpgi.00486.2010. [DOI] [PubMed] [Google Scholar]
- 41.Cabantchik ZI. In search for symbolic qualities of iron: the metal of life. Front Pharmacol. 2016;7:220. doi: 10.3389/fphar.2016.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao M, Monian P, Pan Q, Zhang W, Xiang J, Jiang X. Ferroptosis is an autophagic cell death process. Cell Res. 2016;26:1021–32. doi: 10.1038/cr.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisenstein RS. Iron regulatory proteins and the molecular control of mammalian iron metabolism. Annu Rev Nutr. 2000;20:627–62. doi: 10.1146/annurev.nutr.20.1.627. [DOI] [PubMed] [Google Scholar]
- 44.Chen PH, Smith TJ, Wu J, Siesser PF, Bisnett BJ, Khan F, et al. Glycosylation of KEAP1 links nutrient sensing to redox stress signaling. EMBO J. 2017;36:2233–50. doi: 10.15252/embj.201696113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kerins Michael John, Ooi Aikseng. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxidants & Redox Signaling. 2018;29(17):1756–1773. doi: 10.1089/ars.2017.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y, Zhang S, Wang X, Guo W, Wang L, Zhang D, et al. Disordered signaling governing ferroportin transcription favors breast cancer growth. Cell Signal. 2015;27:168–76. doi: 10.1016/j.cellsig.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Siegert I, Schodel J, Nairz M, Schatz V, Dettmer K, Dick C, et al. Ferritin-mediated iron sequestration stabilizes hypoxia-inducible factor-1alpha upon LPS activation in the presence of ample oxygen. Cell Rep. 2015;13:2048–55. doi: 10.1016/j.celrep.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Pham CG, Bubici C, Zazzeroni F, Papa S, Jones J, Alvarez K, et al. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–42. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 49.Troadec MB, Ward DM, Lo E, Kaplan J, De Domenico I. Induction of FPN1 transcription by MTF-1 reveals a role for ferroportin in transition metal efflux. Blood. 2010;116:4657–64. doi: 10.1182/blood-2010-04-278614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutherford JC, Bird AJ. Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot Cell. 2004;3:1–13. doi: 10.1128/EC.3.1.1-13.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8:193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 52.Cheng Q, Chen J. Mechanism of p53 stabilization by ATM after DNA damage. Cell Cycle. 2010;9:472–8. doi: 10.4161/cc.9.3.10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 54.Gunther V, Lindert U, Schaffner W. The taste of heavy metals: gene regulation by MTF-1. Biochim Biophys Acta. 2012;1823:1416–25. doi: 10.1016/j.bbamcr.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 55.Balesaria S, Ramesh B, McArdle H, Bayele HK, Srai SK. Divalent metal-dependent regulation of hepcidin expression by MTF-1. FEBS Lett. 2010;584:719–25. doi: 10.1016/j.febslet.2009.12.023. [DOI] [PubMed] [Google Scholar]
- 56.Yepiskoposyan H, Egli D, Fergestad T, Selvaraj A, Treiber C, Multhaup G, et al. Transcriptome response to heavy metal stress in Drosophila reveals a new zinc transporter that confers resistance to zinc. Nucleic Acids Res. 2006;34:4866–77. doi: 10.1093/nar/gkl606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahadorani S, Mukai S, Egli D, Hilliker AJ. Overexpression of metal-responsive transcription factor (MTF-1) in Drosophila melanogaster ameliorates life-span reductions associated with oxidative stress and metal toxicity. Neurobiol Aging. 2010;31:1215–26. doi: 10.1016/j.neurobiolaging.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 58.Barzilai A, Rotman G, Shiloh Y. ATM deficiency and oxidative stress: a new dimension of defective response to DNA damage. DNA Repair. 2002;1:3–25. doi: 10.1016/s1568-7864(01)00007-6. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Lee JH, Paull TT, Gehrke S, D’Alessandro A, Dou Q, et al. Mitochondrial redox sensing by the kinase ATM maintains cellular antioxidant capacity. Sci Signal. 2018;11:538. doi: 10.1126/scisignal.aaq0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Z, Kozlov S, Lavin MF, Person MD, Paull TT. ATM activation by oxidative stress. Science. 2010;330:517–21. doi: 10.1126/science.1192912. [DOI] [PubMed] [Google Scholar]
- 61.Krüger A, Ralser M. ATM is a redox sensor linking genome stability and carbon metabolism. Sci Signal. 2011;4:pe17–pe17. doi: 10.1126/scisignal.2001959. [DOI] [PubMed] [Google Scholar]
- 62.Okuno Y, Nakamura-Ishizu A, Otsu K, Suda T, Kubota Y. Pathological neoangiogenesis depends on oxidative stress regulation by ATM. Nat Med. 2012;18:1208–16. doi: 10.1038/nm.2846. [DOI] [PubMed] [Google Scholar]
- 63.Gregory MA, D’Alessandro A, Alvarez-Calderon F, Kim J, Nemkov T, Adane B, et al. ATM/G6PD-driven redox metabolism promotes FLT3 inhibitor resistance in acute myeloid leukemia. Proc Natl Acad Sci USA. 2016;113:E6669–E6678. doi: 10.1073/pnas.1603876113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider JG, Finck BN, Ren J, Standley KN, Takagi M, Maclean KH, et al. ATM-dependent suppression of stress signaling reduces vascular disease in metabolic syndrome. Cell Metab. 2006;4:377–89. doi: 10.1016/j.cmet.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 65.Shi Y, Nikulenkov F, Zawacka-Pankau J, Li H, Gabdoulline R, Xu J, et al. ROS-dependent activation of JNK converts p53 into an efficient inhibitor of oncogenes leading to robust apoptosis. Cell Death Differ. 2014;21:612–23. doi: 10.1038/cdd.2013.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.LaRochelle O, Gagne V, Charron J, Soh JW, Seguin C. Phosphorylation is involved in the activation of metal-regulatory transcription factor 1 in response to metal ions. J Biol Chem. 2001;276:41879–88. doi: 10.1074/jbc.M108313200. [DOI] [PubMed] [Google Scholar]
- 67.Conrad M, Sandin Å, Förster H, Seiler A, Frijhoff J, Dagnell M, et al. 12/15-lipoxygenase–derived lipid peroxides control receptor tyrosine kinase signaling through oxidation of protein tyrosine phosphatases. Proc Natl Acad Sci USA. 2010;107:15774–9. doi: 10.1073/pnas.1007909107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bar-Shira A, Rashi-Elkeles S, Zlochover L, Moyal L, Smorodinsky NI, Seger R, et al. ATM-dependent activation of the gene encoding MAP kinase phosphatase 5 by radiomimetic DNA damage. Oncogene. 2002;21:849–55. doi: 10.1038/sj.onc.1205127. [DOI] [PubMed] [Google Scholar]
- 69.Wolszczak M, Gajda J. Iron release from ferritin induced by light and ionizing radiation. Res Chem Intermed. 2010;36:549–63. [Google Scholar]
- 70.Moser JC, Rawal M, Wagner BA, Du J, Cullen JJ, Buettner GR. Pharmacological ascorbate and ionizing radiation (IR) increase labile iron in pancreatic cancer. Redox Biol. 2013;2:22–27. doi: 10.1016/j.redox.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berndt C, Kurz T, Selenius M, Fernandes AP, Edgren MR, Brunk UT. Chelation of lysosomal iron protects against ionizing radiation. Biochem J. 2010;432:295–301. doi: 10.1042/BJ20100996. [DOI] [PubMed] [Google Scholar]
- 72.Suzuki K, Bose P, Leong-Quong RY, Fujita DJ, Riabowol K. REAP: a two minute cell fractionation method. BMC Res Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang X, Wu J, Ding CK, Lu M, Keenan MM, Lin CC, et al. Cystine deprivation triggers programmed necrosis in VHL-deficient renal cell carcinomas. Cancer Res. 2016;76:1892–903. doi: 10.1158/0008-5472.CAN-15-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tang X, Keenan MM, Wu J, Lin CA, Dubois L, Thompson JW, et al. Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet. 2015;11:e1005158. doi: 10.1371/journal.pgen.1005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRINGv9.1: protein–protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.