Abstract
Receptor-mediated mitophagy is a crucial process involved in mitochondria quality control. AMBRA1 is a mitophagy receptor for the selective removal of damaged mitochondria in mammalian cells. A critical unresolved issue is how AMBRA1-mediated mitophagy is controlled in response to cellular stress. Here, we investigated the role of BCL2-family proteins on AMBRA1-dependent mitophagy and showed that MCL1 delays AMBRA1-dependent mitophagy. Indeed, MCL1 overexpression is sufficient to inhibit recruitment to mitochondria of the E3 Ubiquitin ligase HUWE1, a crucial dynamic partner of AMBRA1, upon AMBRA1-mediated mitophagy induction. In addition, we found that during mitophagy induced by AMBRA1, MCL1 levels decreased but were sustained by inhibition of the GSK-3β kinase, which delayed AMBRA1-mediated mitophagy. Also, we showed that MCL1 was phosphorylated by GSK-3β at a conserved GSK-3 phosphorylation site (S159) during AMBRA1-mediated mitophagy and that this event was accompanied by HUWE1-dependent MCL1 degradation. Altogether, our results demonstrate that MCL1 stability is regulated by the kinase GSK-3β and the E3 ubiquitin ligase HUWE1 in regulating AMBRA1-mediated mitophagy. Our work thus defines MCL1 as an upstream stress-sensitive protein, functional in AMBRA1-mediated mitophagy.
Subject terms: Proteins, Autophagy
Introduction
Autophagy is an important eukaryotic process involved in the lysosomal degradation of cytosolic components in both physiological and pathological conditions. During autophagy, the autophagosomes engulf a number of different cargoes and then fuse with lysosomes for subsequent recycling of their content [1]. Several key proteins are involved in the autophagosome formation, such as BECLIN 1 and its positive regulator AMBRA1 [1, 2]. Mitophagy is a selective form of autophagy essential for the elimination of impaired or depolarized mitochondria. It is important for a cell to remove damaged mitochondria, since wanton release of reactive oxygen intermediates leads to inflammasome activation, genotoxic stress, promotion of tumorigenesis, and aging. Accordingly, alteration of the mitophagic pathway may contribute to neurodegenerative or inflammatory diseases, cancer, and decreased lifespan [3]. Damaged mitochondria activate the PINK1/PARKIN pathway, the major mitophagic molecular axis described so far. PINK1, which is constantly degraded, is instead stabilized to mitochondria, resulting in the recruitment of the PARKIN E3 ubiquitin ligase to damaged mitochondria and in the ubiquitination of several outer mitochondria membrane proteins. Mitophagy receptors that bind ubiquitin, such as NDP52 and OPTINEURIN, and the autophagosome-associated protein LC3, then favor the engulfment of damaged mitochondria into autophagosomes [4, 5].
Mitophagy occurs also in cells lacking detectable PARKIN, through alternative mitophagy receptors. For example, BNIP3 (BCL2/adenovirus E1B 19 kDa interacting protein 3) and its homolog NIX/BNIP3L (BCL2/adenovirus E1B 19 kDa interacting protein 3 like) are two mitophagy receptors able to regulate mitochondrial clearance during reticulocyte development. They possess a LC3-interacting region (LIR) that facilitate the binding to LC3B (Microtubule-associated protein 1 A/1B light chain 3B) (for BNIP3) [6] or to the other mATG8-family member Gamma-AminoButyric Acid Receptor-Associated Protein (for NIX/BNIP3L) [7]. FUN14 domain-containing protein 1 is another mitophagy receptor located at the mitochondria, with a LIR domain that mediates its association with LC3B during hypoxia or following mitochondrial membrane depolarization [8]. Prohibitin 2, an inner mitochondria membrane protein, is an another key mitophagy receptor for both PARKIN-mediated mitophagy in mammalian cells and for paternal mitochondrial clearance in C. elegans [9]. Recently, we showed that AMBRA1, also a LIR-containing protein, is a new mitophagic receptor crucial in order to both amplify PARKIN-mediated mitochondrial clearance and regulate PARKIN-independent mitophagy [10, 11]. Similar to BNIP3, NIX/BNIP3L, and FUNDC1, AMBRA1 can be positively regulated by a specific phosphorylation upstream its LIR domain [11].
Since BCL2-family proteins [12–17] can control mitophagy [18, 19] and since we previously demonstrated that AMBRA1 can bind BCL2-family proteins [20, 21], we decided to investigate a putative regulatory role for BCL2-family members on AMBRA1-dependent mitophagy. We thus found that MCL1 is a potent inhibitor of AMBRA1-mediated mitophagy, and discovered that MCL1 destabilization by the Glycogen Synthase kinase-3β (GSK-3β) and the E3 ubiquitin ligase HUWE1 is a key event for this specific mitophagy pathway. In sum, this work unveils a novel mechanism for mitophagy regulation, linking it to the BCL2 death factors.
Results
AMBRA1-dependent mitophagy is inhibited by the anti-apoptotic factor MCL1
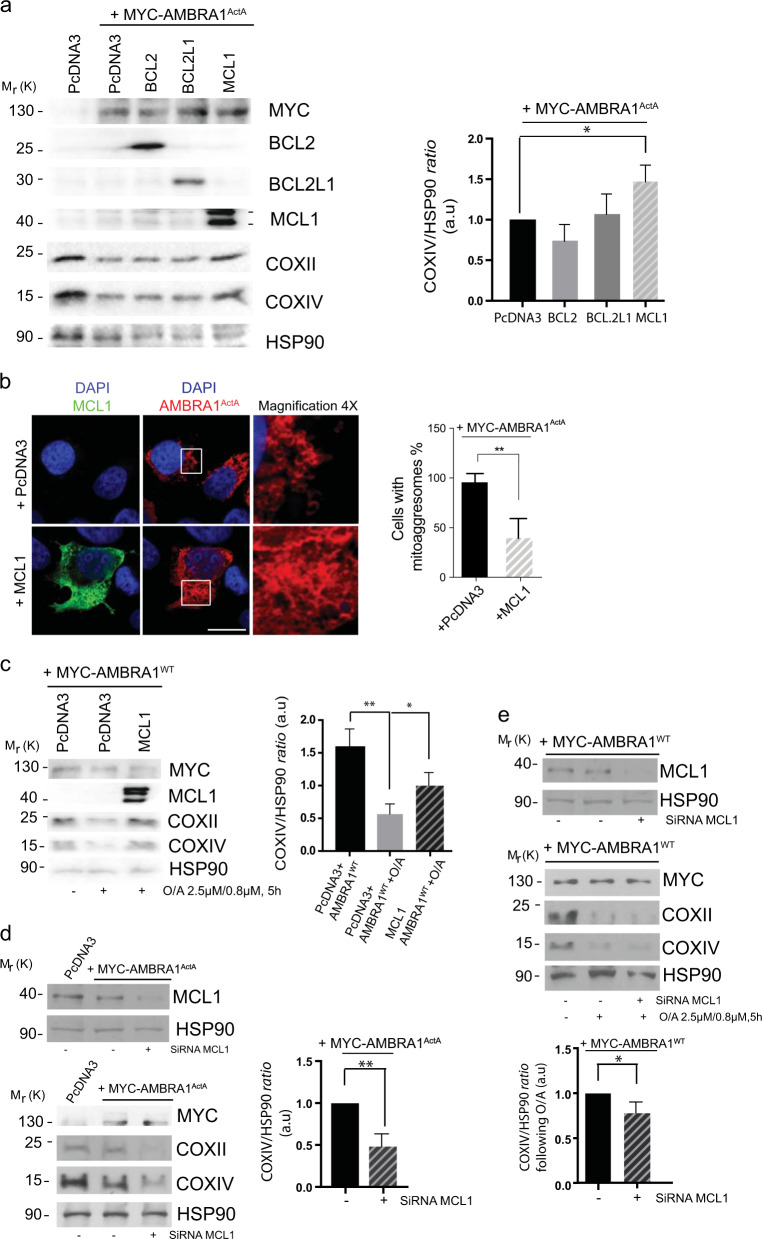
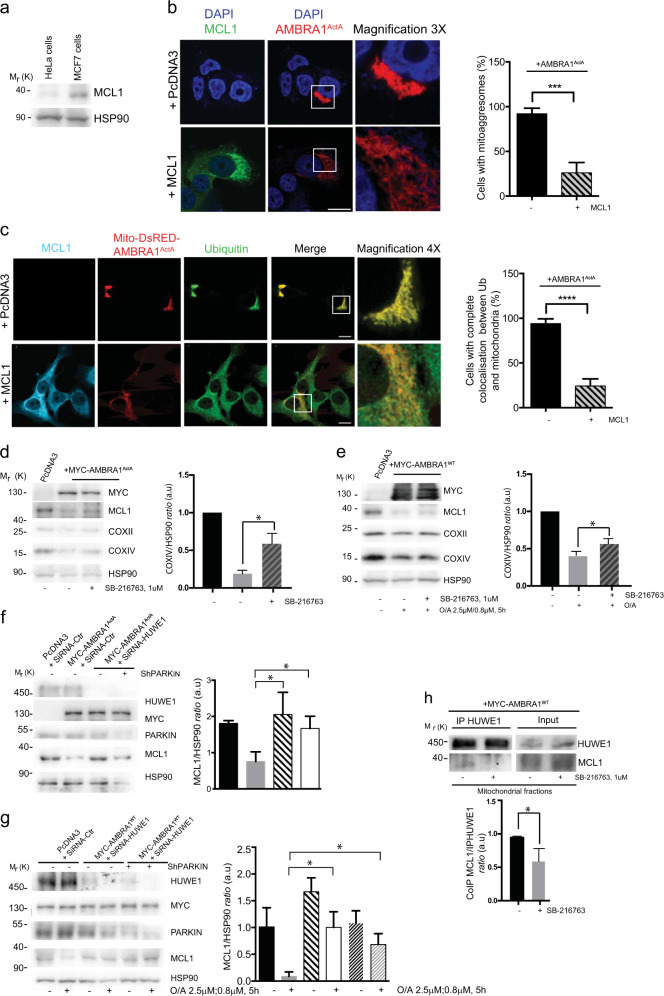
HeLa cells (a well-known PARKIN-free cellular system) rapidly undergo mitophagy when overexpress either AMBRA1ActA, a protein fusion that specifically targets AMBRA1 to the mitochondria, or AMBRA1WT in combination with mitophagy inducers [i.e, Olygocmicin/Actinomycin (O/A), or protonophores such as CCCP or FCCP] [10, 11, 22]. To characterize the role of BCL2-family proteins on AMBRA1-mediated mitophagy, we first checked whether overexpression of these proteins was capable to regulate mitophagy in HeLa cells. Interestingly, we found that MCL1 expression was sufficient to reduce mitophagy induced by AMBRA1ActA. First, by performing a western blot analysis upon the coexpression of AMBRA1ActA and individual BCL2 proteins, we observed a strong decrease in the clearance of both COXII and COXIV, two mitochondrial markers (Fig. 1a and Supplementary Fig. 1a). Of note, no other members of the BCL2-family did significantly inhibit mitophagy in the same conditions. We next confirmed these results by performing a confocal microscopy analysis, by which we checked for AMBRA1ActA-induced mitoaggresome formation, a typical feature of AMBRA1ActA-mediated mitophagy [10]. As illustrated in Fig. 1b, the mitochondrial network is less aggregated around the nucleus when cells are expressing MCL1 in combination with AMBRA1ActA, at variance with cells expressing only AMBRA1ActA. Then, we decided to check whether MCL1 was able to inhibit mitophagy induced by wild-type AMBRA1 (AMBRA1WT) in combination with a mitophagy inducer. As shown in Fig. 1c and Supplementary Fig. 1b, both COXII and COXIV loss is decreased in cells overexpressing AMBRA1WT and MCL1 and treated with O/A for 5 h. Primed by these results, we next decided to interfere MCL1 by using specific siRNAs against human MCL1 in HeLa cells overexpressing AMBRA1ActA or AMBRA1WT cells treated with O/A. Our results indicate that downregulation of MCL1 is sufficient to stimulate AMBRA1-mediated mitophagy, as demonstrated by a more evident reduction of the mitochondrial markers COXII and COXIV, when compared with cells transfected with control siRNA (Fig. 1d, e and Supplementary Fig. 1c, d).
Fig. 1.
MCL1 antagonizes AMBRA1-dependent mitophagy. a HeLa cells were cotransfected with Myc-AMBRA1ActA and PcDNA3, BCL2, BCL2L1 or MCL1. After 24 h of transfection, protein extracts were analyzed using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D.) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent biological replicas. Statistical analysis was performed using Student’s test (*P < 0.05). b HeLa cells were cotransfected with a vector encoding MYC-AMBRA1ActA and PcDNA3 or MCL1. Following 24 h of transfection, cells were fixed and assessed by immunolabeling using anti-MCL1 and MYC antibodies. Nuclei were stained with DAPI 1 µg/µl 20 min. Scale bar: 8 µm. Percentage of AMBRA1ActA-transfected cells with mitoaggresomes has been quantified following pcDNA3 or MCL1 overexpression. Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (**P < 0.01). c HeLa cells were cotransfected with Myc-AMBRA1WT and PcDNA3 or MCL1. After 24 h of transfection, cells were treated with O/A (2.5 μM, 0.8 μM, 5 h), total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D.) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05, **P < 0.01). d HeLa cells were transfected with an empty vector (PcDNA3) or Myc-AMBRA1ActA in combination with SiRNA-CTRL or SiRNA-MCL1. Cells were treated with O/A (2.5 μM, 0.8 μM, 5 h) and total lysates were blotted for the indicated antibodies. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (**P < 0.01). e HeLa cells were transfected with Myc-AMBRA1WT in combination with SiRNA-CTRL or SiRNA-MCL1. Cells were treated with O/A (2.5 μM, 0.8 μM, 5 h) and total lysates were blotted for the indicated antibodies. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05)
In addition, we found that MCL1 overexpression was sufficient to reduce the number of autophagosomes following AMBRA1-mediated mitophagy, as illustrated by a reduction of LC3-II band appearance (Supplementary Fig. 1e, f).
Altogether, these data suggest that the prosurvival factor MCL1 is important for preventing mitophagy governed by the mitophagic receptor AMBRA1.
MCL1 antagonizes mitochondria ubiquitylation following AMBRA1ActA expression by inhibiting mitochondrial translocation of HUWE1
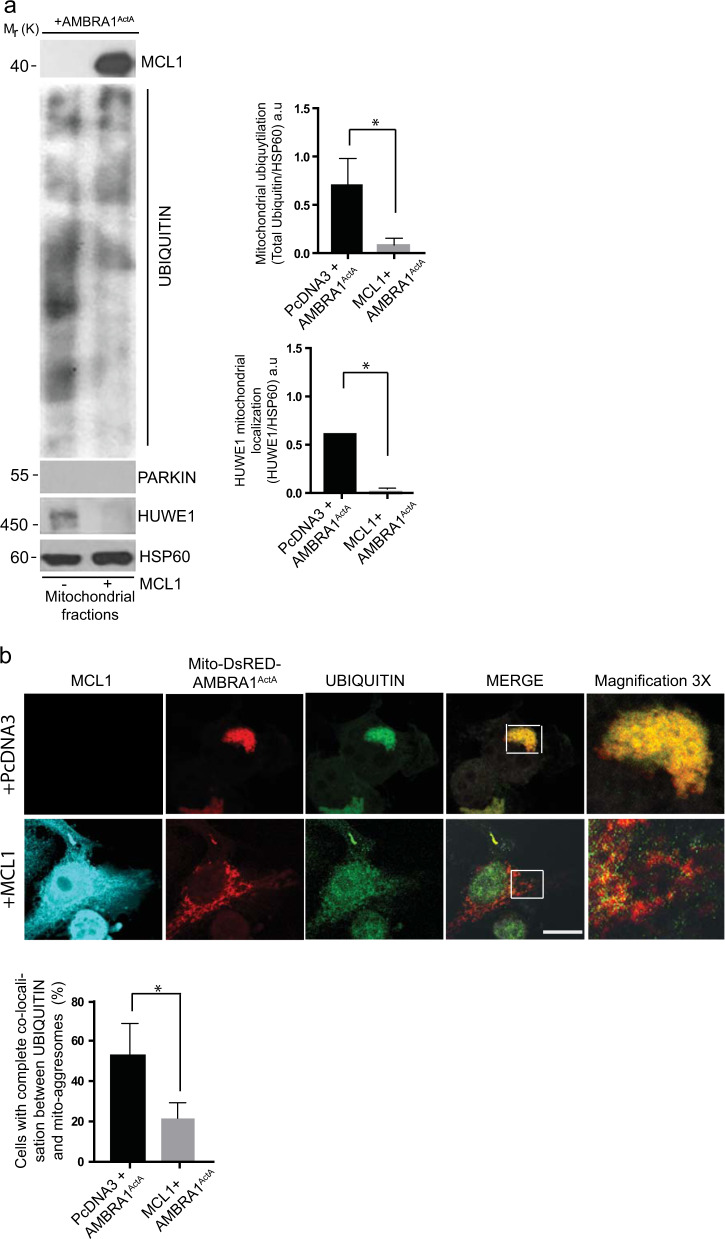
Since we demonstrated that mitochondria ubiquitylation is an early event in AMBRA1-mediated mitophagy [10, 11], in order to better understand the mechanisms by which MCL1 was able to inhibit AMBRA1-mediated mitophagy, we checked for mitochondria ubiquitylation following AMBRA1ActA expression and in the presence or absence of MCL1. By performing a mitochondrial-fractioning assay, we found that mitochondria ubiquitylation was strongly inhibited following MCL1 expression (Fig. 2a), with this suggesting that prosurvival MCL1 inhibits AMBRA1-dependent mitophagy at an early stage in the process, prior to mitochondria tagging by ubiquitin.
Fig. 2.
Upon AMBRA1-mediated mitophagy, MCL1 inhibits HUWE1 translocation to mitochondria and their ubiquitylation. a HeLa cells were cotransfected with Myc-AMBRA1ActA and PcDNA3 or MCL1. After 24 h of transfection, mitochondrial enrichment was performed through differential centrifugations, mitochondrial fractions were analyzed using the indicated antibodies and anti-HSP60 was used as a loading control. The graphs illustrate UBIQUITIN/HSP60 and HUWE1/HSP60 ratio (±S.D.) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05). b HeLa cells were cotransfected with a vector encoding Mito-DsRED-AMBRA1ActA and PcDNA3 or MCL1. Following 24 h of transfection, cells were fixed and assessed by immunolabelling using anti-MCL1 and UBIQUITIN antibodies. Nuclei were stained with DAPI 1 µg/µl 20 min. The merging of the fluorescence signals is illustrated. Scale bar: 8 µm. The graph illustrates the percentage of cells showing a complete colocalization between UBIQUITIN and the mitochondrial network (±S.D). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05)
We recently discovered that AMBRA1-dependent ubiquitylation of mitochondria is carried out by the E3 Ubiquitin ligase HUWE1 [11]; we thus explored whether HUWE1 translocation to mitochondria was affected by MCL1. Indeed, we found that HUWE1 mitochondrial translocation was clearly reduced in the presence of MCL1, at variance with control cells. Of note, the E3 Ubiquitin ligase PARKIN was undetectable in our mitochondrial fractions (Fig. 2a). Next, we strengthened these data by performing a quantification of HUWE1 translocation from cytosol to mitochondria, which confirms that HUWE1 expression clearly decreases in mitochondria when MCL1 is overexpressed (Supplementary Fig. 2).
Moreover, by performing a confocal microscopy analysis, we confirmed that the ubiquitin staining on mitochondria (whose aggregation into mitoaggresomes is almost abolished by MCL1) was also reduced in cells overexpressing MCL1 (Fig. 2b).
In conclusion, we found that prosurvival MCL1 may interfere with the recruitment of HUWE1 to mitochondria, thus inhibiting ubiquitylation of mitochondria, an early event necessary for AMBRA1-mediated mitophagy.
GSK-3β modulates MCL1 protein levels during AMBRA1-mediated mitophagy
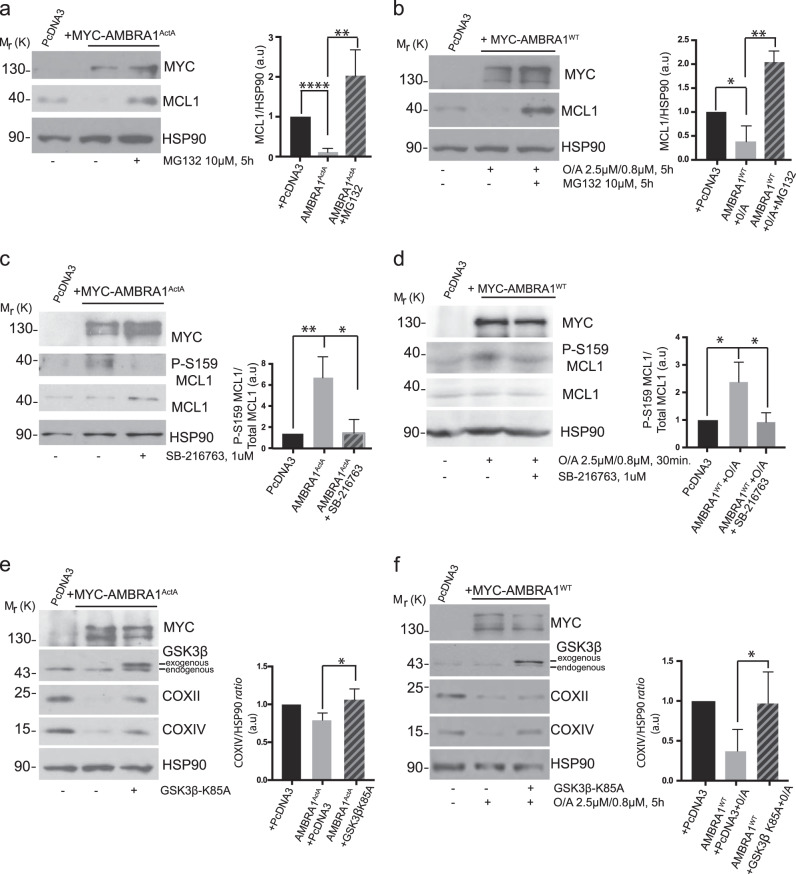
The finding that MCL1 is capable to significantly reduce AMBRA1-mediated mitophagy led us to hypothesize that this prosurvival factor needs to be tightly regulated during that process. We thus checked for its stability during AMBRA1-mediated mitophagy. Indeed, overexpression of AMBRA1ActA is sufficient to induce a clear reduction in the levels of endogenous MCL1 (Fig. 3a). In a similar manner, cells overexpressing AMBRA1WT and then treated with O/A for 5 h, present a decrease in MCL1 protein levels (Fig. 3b). Interestingly, such decrease of MCL1 levels can be blocked by treating both AMBRA1ActA and AMBRA1WT-positive HeLa cells with the proteasome inhibitor MG132 (Fig. 3a, b). These data indicate that MCL1 is degraded by the proteasome upon AMBRA1-mediated mitophagy induction. Since phosphorylation of serine 159 (S159) of MCL1 is correlated to MCL1 degradation [23], we hypothesized that MCL1 is phosphorylated on this site during mitophagy. To check for this modification event, we induced mitophagy in HeLa cells by overexpressing AMBRA1ActA or AMBRA1WT + O/A and then monitored phospho-S159 by means of a specific antibody. As shown in Fig. 3c, d, MCL1 is indeed phosphorylated on S159 in the early phase of the AMBRA1-mediated mitophagy (24 h following AMBRA1ActA expression and 30 min following O/A treatment).
Fig. 3.
MCL1 is phosphorylated on serine 159 by GSK3β during mitophagy and GSK3β inhibition delays AMBRA1-dependent mitophagy. a HeLa cells were transfected with MYC-AMBRA1ActA or PcDNA3 and treated with the proteasome inhibitor MG132 (10 μM, 24 h) or not (DMSO). Total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (****P < 0.0001, **P < 0.01). b HeLa cells were transfected with PcDNA3 or MYC-AMBRA1WT and treated with O/A (2.5 μM, 0.8 μM, 5 h) and proteasome inhibitor (MG132, 10 μM, 5 h) or not (DMSO). Total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an aribitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05, **P < 0.01). c HeLa cells were transfected with MYC-AMBRA1ActA or PcDNA3 and treated with GSK3 inhibitor (SB-216763, 1 μM, 24 h) or not (DMSO). Total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the P-S159MCL1/MCL1 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05). d HeLa cells were transfected with PcDNA3 or MYC-AMBRA1WT and treated with O/A (2.5 μM, 0.8 μM, 5 h) and GSK3 inhibitor (SB-216763, 1 μM, 5 h) or not (DMSO). Total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05, **P < 0.01). e HeLa cells were cotransfected with MYC-AMBRA1ActA and PcDNA3 or GSK3β dead kinase (GSK3β-K85A). After 24 h of transfection, total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05). f HeLa cells were cotransfected with MYC-AMBRA1WT and PcDNA3 or GSK3β dead kinase (GSK3β-K85A) and treated with O/A (2.5 μM, 0.8 μM, 5 h). After 24 h of transfection, total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05)
One protein kinase responsible for MCL1 phosphorylation at S159 is the GSK-3β kinase [23]; we thus speculated that GSK-3β was the kinase responsible for MCL1 phosphorylation and consequently for its degradation, in order to allow mitophagy. We thus checked the effect of GSK3 pharmacological inhibition on MCL1 phospho-S159 during AMBRA1-mediated mitophagy. As expected, S159-phosphorylated MCL1 can be observed during AMBRA1-mediated mitophagy but is reduced when GSK-3β is inhibited by means of SB-216763, a small-molecule inhibitor of GSK-3 (Fig. 3c, d). These results strongly suggest that GSK-3β may be the kinase responsible for MCL1 phosphorylation following mitophagy induction. Primed by this result, we next investigated the contribution of GSK-3β in mitophagy induction upon AMBRA1 expression in HeLa cells. To this end, we interfered genetically the kinase activity of the specific GSK-3β isoform by means of a plasmid encoding GSK-3β K85A, a kinase-dead mutant that inhibits endogenous GSK-3β in a dominant negative manner [24]. As shown in Fig. 3e, f and Supplementary Fig. 3a, b, overexpression of the GSK-3β K85A is sufficient to inhibit both AMBRA1ActA and AMBRA1WT + O/A-mediated mitophagy.
Last, we found that inhibition of GSK-3β was sufficient to reduce the number of autophagosomes following AMBRA1-mediated mitophagy, as shown by a reduction of LC3-II band appearance (Supplementary Fig. 3c, d).
These results strongly imply that GSK-3β is functional to the regulation of AMBRA1-mediated mitophagy and that it acts, at least in part, through MCL1 destablization.
HUWE1 is responsible for MCL1 ubiquitylation and degradation upon AMBRA1-mediated mitophagy
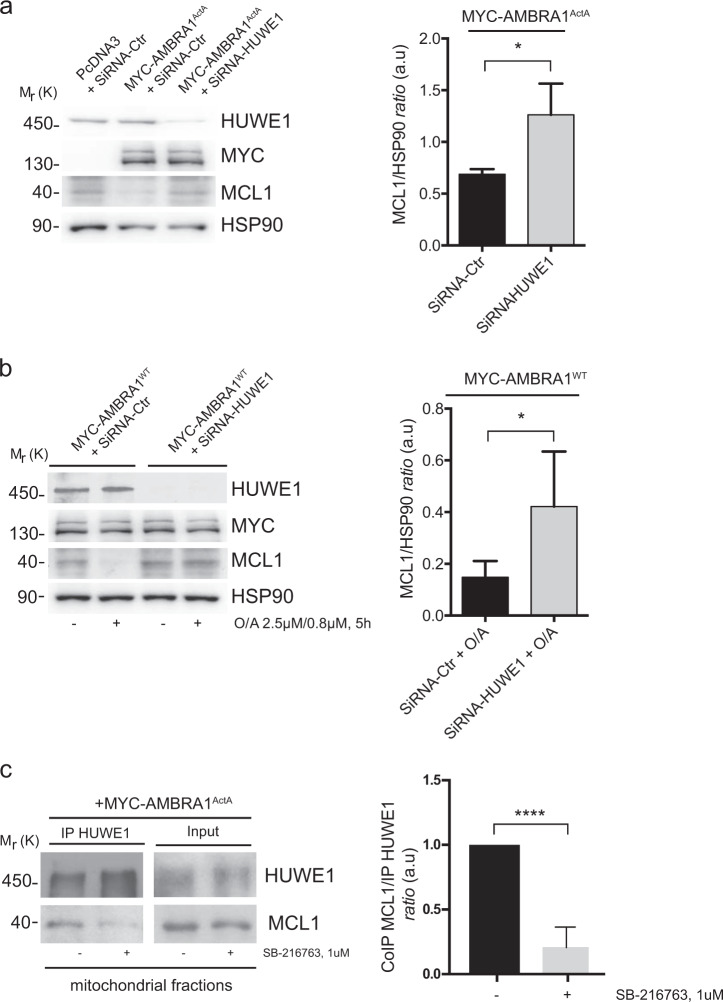
AMBRA1 cooperates with the E3 ubiquitin ligase HUWE1 in order to mediate mitophagy [11]. Interestingly, MCL1 is a well-known substrate of HUWE1 [25]. We could thus candidate HUWE1 as the E3 Ubiquitin ligase responsible for MCL1 degradation during AMBRA-dependent mitophagy. To investigate this possibility, we first downregulated HUWE1 in HeLa cells overexpressing AMBRA1ActA or AMBRA1WT and treated with O/A, and next checked for MCL1 protein levels during mitophagy. As shown in Fig. 4a, b, downregulation of HUWE1 is sufficient to reduce MCL1 degradation upon mitophagy induction. Of note, AMBRA1-mediated mitophagy inhibition, by blocking MCL1 degradation through the use of HUWE1 siRNA, is accompanied by a reduction of the autophagosome number as shown by a reduction of the LC3-II band appearance (Supplementary Fig. 4a, b). HUWE1 should then be able to cooperate with GSK-3β to induce MCL1 degradation following AMBRA1-induced mitophagy. To test this hypothesis, we checked whether HUWE1 was able or not to bind its substrate MCL1 upon AMBRA1ActA-mediated mitophagy dependently or not from GSK-3β inhibitor. Indeed, HUWE1 strongly binds endogenous MCL1 upon AMBRA1ActA-mediated mitophagy and this binding is reduced when GSK-3β is inhibited (Fig. 4c), with this indicating that GSK-3β is a priming kinase important for MCL1 recognition by HUWE1.
Fig. 4.
HUWE1 is responsible for MCL1 degradation during AMBRA1-mediated mitophagy. a HeLa cells transfected with MYC-AMBRA1ActA (18 h) and interfered for HUWE1 (SiRNA-HUWE1) were analyzed by western blot. The quantification results are the mean of three independent experiments (±S.D.). Statistical analysis was performed using Student t-test (**P < 0.01). b HeLa cells transfected with MYC-AMBRA1WT and interfered for HUWE1 (SiRNA-HUWE1) were treated with O/A and analyzed by western blot. The quantification results are the mean of four independent experiments (±S.D.). *P < 0.05. Statistical analysis was performed using Student t-test. c Representative image of HUWE1-MCL1 co-immunoprecipitation (Co-IP), upon mitophagy stimulation (AMBRA1ActA overexpression) in the mitochondrial fractions. The graph illustrates the levels of MCL1 coimmunoprecipitated with HUWE1 in normal condition or following GSK3 inhibition (±S.D.). Statistical analysis was performed using Student t-test (****P < 0.0001)
AMBRA1-mediated mitophagy operates through the HUWE1-GSK3 axis in MCF7 breast cancer cells, in order to degrade MCL1 for a correct mitophagy occurrence
Next, we decided to validate our main findings obtained in HeLa cells in an other cell line, in which high endogenous levels of MCL1 are detected. To this end, we have choosen the MCF7 cells, a breast cancer cell line known to be resistant to some drugs, with this being due, in part, to the high levels of MCL1 they display [26]. In particular, we confirmed that MCL1 was highly expressed in these cells compared with HeLa cells (Fig. 5a). In addition, we showed that MCL1 is able to delay mitoaggresome formation (Fig. 5b), to reduce ubiquitin on mitochondria (Fig. 5c). Moreover, as already demonstarted in HeLa cells, we found that AMBRA1-mediated mitophagy induction is able to induce MCL1 degradation through the HUWE1-GSK3 axis (Fig. 5f–j). These data indicate that AMBRA1-mediated mitophagy induction is sufficient to reduce high levels of endogenous MCL1 in a breast cancer cell line, in order to properly induce mitophagy.
Fig. 5.
MCL1 antagonizes AMBRA1-dependent mitophagy and its HUWE1-dependent degradation, is necessary for a correct mitophagy in MCF7 cells. a Protein extracts of HeLa and MCF7 cells were analyzed using the MCL1 antibody and anti-HSP90 was used as a loading control. b MCF7 cells were cotransfected with a vector encoding MYC-AMBRA1ActA and PcDNA3 or MCL1. Following 24 h of transfection, cells were fixed and assessed by immunolabelling using anti-MCL1 and MYC antibodies. Nuclei were stained with DAPI 1 µg/µl 20 min. Scale bar: 8 µm. Percentage of AMBRA1-ActA transfected cells with mitoaggresomes has been quantified following pcDNA3 or MCL1 overexpression. Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (***P < 0.001). c MCF7 cells were cotransfected with a vector encoding Mito-DsRED-AMBRA1ActA and PcDNA3 or MCL1. Following 24 h of transfection, cells were fixed and assessed by immunolabelling using anti-MCL1 and UBIQUITIN antibodies. Nuclei were stained with DAPI 1 µg/µl 20 min. The merging of the fluorescence signals is illustrated. Scale bar: 8 µm. The graph illustrates the percentage of cells showing a complete colocalization between UBIQUITIN and the mitochondrial network (±S.D.). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (****P < 0,0001). d MCF7 cells were transfected with MYC-AMBRA1ActA or PcDNA3 and treated with GSK3 inhibitor (SB-216763, 1 μM, 24 h) or not (DMSO). Total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05). e MCF7 cells were transfected with PcDNA3 or MYC-AMBRA1WT and treated with O/A (2.5 μM, 0.8 μM, 5 h) and GSK3 inhibitor (SB-216763, 1 μM, 5 h) or not (DMSO). Total extracts were immunoblotted using the indicated antibodies and anti-HSP90 was used as a loading control. The graph illustrates the COXIV/HSP90 ratio (±S.D) as an arbitrary unit (a.u). Each point value represents the mean ± S.D. from three independent experiments. Statistical analysis was performed using Student’s test (*P < 0.05). f MCF7 cells transfected with MYC-AMBRA1ActA (18 h) and interfered for HUWE1 (SiRNA-HUWE1) and PARKIN (Sh-PARKIN), or not, were analyzed by western blot. The quantification results are the mean of three independent experiments (±S.D.). Statistical analysis was performed using Student t-test (*P < 0.05). g MCF7 cells transfected with MYC-AMBRA1WT and interfered for HUWE1 (SiRNA-HUWE1) and PARKIN (Sh-PARKIN), or not, were treated with O/A and analyzed by western blot. The quantification results are the mean of four independent experiments (±S.D.). Statistical analysis was performed using Student's t-test (*P < 0.05). h Representative image of HUWE1-MCL1 co-immunoprecipitation (Co-IP), upon mitophagy stimulation (AMBRA1ActA overexpression) in mitochondrial fractions of MCF7 cells. The graph illustrates the levels of MCL1 coimmunoprecipitated with HUWE1 in normal condition or following GSK3 inhibition (±S.D.). Statistical analysis was performed using Student's t-test (*P < 0.05)
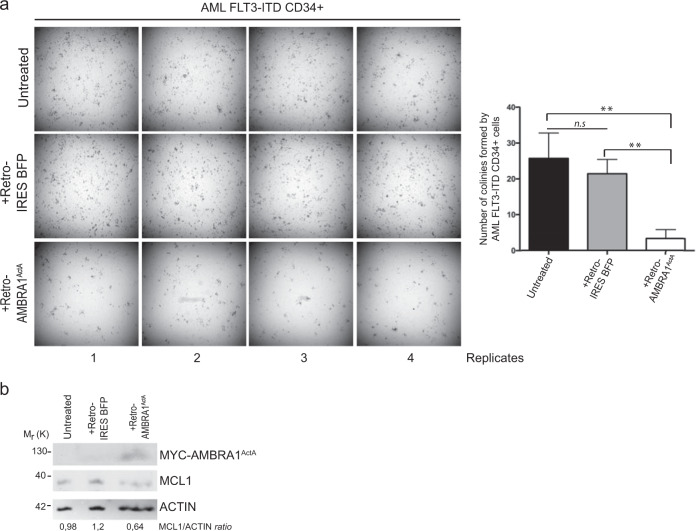
AMBRA1ActA ectopic expression delays the potential of clonogenicity of primary CD34+cells of a CN-AML patient
Interestingly, MCL1 is an essential survival factor for hematopoiesis, and in humans, hematopoietic stem cells express MCL1 at the highest level in response to FMS-like tyrosine kinase-3 (FLT3) signaling. Patients suffering from acute myeloid leukemias (AML) with FMS-like tyrosine kinase-3-internal tandem duplications (FLT3-ITD) have poor outcomes following induction of severals therapies and this is mainly attributed to drug resistance due to the upregulation of MCL1 in primary mononuclear cells (MNC) from AML patients compared with FLT3-wild-type controls [27]. Since therapeutic targeting of MCL1 is a promising strategy for FLT3-ITD-positive AML patients and since we demonstrated that AMBRA1-mediated mitophagy induces MCL1 downregulation, we hypothezized that AMBRA1ActA expression, by mediating MCL1 downregulation, would affect the clonogenic potential of CD34+ AML cells. To this end, we retrovirally infected CD34+ of a paediatric CN-AML patient harboring FLT3-ITD mutation. As shown in Fig. 6, AMBRA1ActA infection in CD34+ cells induced a significant reduction of the in vitro CFUs compare with control cells; this was accompanied by a reduction of MCL1 levels in these primary AMBRA1ActA-postive CD34+ AML cells.
Fig. 6.
Ectopic expression of AMBRA1ActA fusion gene impairs colony formation and MCL1 expression in FLT3-ITD-mutated AML CD34+ cells. a Colony-formation assay (CFU-assay) shows reduced colony-formation capacity after 15 days in culture in AMBRA1ActA (+Retro-AMBRA1ActA) overexpressing CD34+ cells, compared with noninfected (Untreated; t-test P = 0.0068) and empty vector infected (+Retro IRES BFP; t-test P = 0.0028) CD34+ cells. (Four technical replicates are shown). Histograms show the median number of colonies and SDs for the three conditions mentioned above. Difference between untreated and +Retro IRES BFP conditions is not statistically significant (t-test P = 0.41). b Western blot shows expression of ectopic AMBRA1ActA fusion gene in +Retro-AMBRA1ActA infected CD34+ cells along with MCL1 reduction compared with controls cells after 15 days in culture
In sum, targeting MCL1 by overexpressing AMBRA1ActA reduces the clonogenic activity (by >50%) in a primary AML case tested. These findings demonstrate that targeting MCL1 through AMBRA1-mediated mitophagy induction may have the potential to reduce proliferation of CD34+ cells and could be able to suppress human leukemic stem and progenitor cells.
Discussion
Here, we have shown that MCL1, a member of the BCL2-family, inhibits HUWE1 recruitment to mitochondria, a critical event for AMBRA1-mediated mitophagy. We also found that MCL1 stability is regulated during mitophagy by the priming kinase GSK-3β, which phosphorylates MCL1 on S159, this leading to its proteasomal degradation by HUWE1. In sum, we propose that MCL1 is degraded by this axis during mitophagy and that this event is important to remove the MCL1 inhibitory effect on mitophagy (Fig. 7).
Fig. 7.
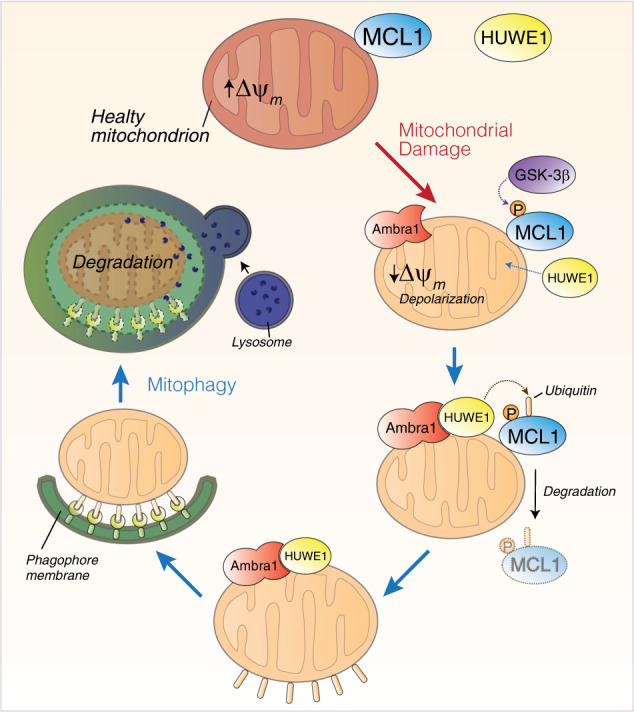
MCL1 antagonizes AMBRA1-dependent mitochondrial quality control. In normal condition, HUWE1 is an E3 ubiquitin ligase mostly found in the cytoplasm. Upon AMBRA1-mediated mitophagy induction, AMBRA1 favors HUWE1 E3 ubiquitin ligase translocation from the cytosol to mitochondria [11]. Here we demonstrate that the prosurvival factor MCL1, when highly expressed, antagonizes the mitochondrial access of the E3 Ubiquitin ligase HUWE1 upon mitophagy induction. In normal condition, MCL1 is thus, most likely, a stress-sensitive protein which is able to preserve the mitochondrial membrane from the E3 ubiquitin ligase HUWE1 access. When mitophagy is induced, by expressing high quantities of AMBRA1 to the mitochondria, MCL1 is phosphorylated by GSK3β on Serine 159 and degraded by the proteasome through HUWE1
Interestingly, MCL1 degradation has been found in serum withdrawal conditions and its deletion exacerbates autophagy, this highlighting a function for MCL1 in autophagy regulation [28]. Our data indicate that MCL1 is also capable to regulate selective autophagy, and in particular mitochondria removal governed by the mitophagy receptor AMBRA1. Hollville et al. demonstrated that MCL1, BCL2L1, and BCL2 are able to inhibit PARKIN-mediated mitophagy by blocking PARKIN translocation to mitochondria [18]. Moreover, Wu et al. demonstrated that BCL2L1 inhibits FUNDC1-mediated mitophagy [19]. Our results are thus in line with the concept that BCL2-family proteins are important regulators of the mitophagic process.
Since both PARKIN and HUWE1 are mainly cytoplasmic E3 ubiquitin ligases, our work together with those of Hollville et al. suggest that a general mechanism for the BCL2-family proteins could exist in order to decide to maintain healthy mitochondria, by inhibiting E3 ubiquitin ligase translocation. One open question is how HUWE1 is recruited to mitochondria. Since HUWE1 is a E3 ubiquitin ligase containing a BH3 domain [25], we can speculate that overexpressed MCL1 is able to compete with HUWE1 for an interaction with another BH3 protein that serves as a “mitochondrial anchor” for HUWE1.
Interestingly, at variance with PARKIN-mediated mitophagy, we found here that MCL1 possesses a unique function, distinct from that of BCL2L1 and BCL2 on AMBRA1-mediated mitophagy.
Further, we validated our main findings also in MCF7 cells, a cancer cell line expressing high levels of endogenous MCL1. Since targeting MCL1 is a current strategy to fight breast cancer, our results indicate that also AMBRA1-mediated mitophagy may be a target to fight such cancer, in which MCL1 is found upregulated.
Moreover, by demonstrating that AMBRA1ActA-infected CD34+ AML cells showed a lower clonogenic potential compared with infected control cells, we highlighted the potential of a novel strategy, based on the stimulation of AMBRA1-mediated mitophagy, for the development of specific MCL1-targeting pathways in order to eliminate residual FLT3-ITD leukemic blasts and stem cells resistant to cytotoxic agents.
Further, deletion of the AMBRA1 gene causes unbalanced cell proliferation accompanied by defects of neuronal development [2]; since MCL1 is strongly involved in neuron survival [29], we can hypothesize that MCL1, by mediating a tight regulation of the balance between mitophagy and apoptosis, may control AMBRA1-mediated mitophagy during brain development. Finally, Hollville et al. [18] found that BH3-only proteins, such as BAD, BIM, PUMA, and NOXA, as well as a BH3-only mimetics, were capable of accelerating PARKIN recruitment in order to impact mitochondria, most likely through neutralization of endogenous BCL2-family proteins. It will be interesting, in the future, to understand whether this happens or not in the context of AMBRA1-mediated mitophagy.
In conclusion, the ability of MCL1 to regulate the mitophagy process driven by AMBRA1 suggests that MCL1 acts as important sensor of mitochondria healthy status, thus impacting on both apoptosis and mitophagy control.
Methods
Antibodies
Rabbit anti-MCL1 (Santa Cruz Biotechnology, sc-819), monoclonal anti-BCL2 (Santa Cruz Biotechnology, sc-7382), mouse monoclonal anti-COXII (Abcam, ab110258); mouse monoclonal anti-COXIV (Abcam, ab33985), mouse monoclonal anti-MYC (9E10; Santa Cruz Biotechnology, sc-40), mouse monoclonal anti-FLAG (Sigma-Aldrich, F3165), rabbit polyclonal anti-FLAG (Sigma-Aldrich, F7425), rabbit polyclonal anti-phospho S159 MCL1 (Cell signaling, #45795), mouse monoclonal anti-GSK3β (E-11) (Santa Cruz Biotechnology, sc-377213), rabbit polyclonal anti-HSP90 (Santa Cruz Biotechnology, sc-7947), rabbit polyclonal anti-HUWE1 (Bethyl, A300-486A), mouse monoclonal anti-UBIQUITIN (Santa Cruz Biotechnology, sc-8017), anti-GAPDH (Santa Cruz Biotechnology sc-47724), and anti-HSP60 (Santa Cruz Biotechnology, sc-13966).
Cloning and plasmids
Construct coding for AMBRA1WT was cloned in pLPCX vector (Clontech, 631511).18. The constructs coding for BCL2L1 and MCL1 were a gift of Muriel Priault (Bordeaux, France). The Myc-AMBRA1ActA plasmid was cloned as described in Strappazzon et al. (2015). The Mito-DsRED construct encodes for human Cox8A mitochondria signal peptide which is fused with wild-type-DsRED in PcDNA3 vector (Invitrogen). The Mito-DsRED-AMBRA1ActA is a bidirectional vector encoding for both Mito-DsRED and FLAG-AMBRA1ActA. Plasmid encoding for HA GSK3-β K85A pcDNA3 was purchased from Addgene.
Interference using SiRNAs
All the RNA-interference was performed using Lipofectamine 2000. We used MCL1 (h) SiRNA from Santa Cruz (sc:43912); SiRNA-HUWE1 from Integrated DNA Technologies (IDT) #150971213.
Cell cultures
HeLa cells were cultured in Dulbecco’s Modified Eagle’s Medium (Lonza, BE12-604F) supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, 10270-106), and 1% penicillin–streptomycin solution (Lonza, 17-602 E) at 37 °C under 5% CO2.
Cell culture transfection
Transient transfections of expression plasmids into HeLa cells were performed using TurboFect according to the supplier’s instructions (Thermo Fisher Scientific, R0532). MCF7 cells were transiently transfected using Lipofectamine 2000 according to the supplier’s instructions (Thermo Fisher Scientific, #11668019).
Cells treatments
In order to induce mitophagy, HeLa cells were treated with the combined treatment O/A [2.5 μM Oligomycin (Calbiochem)/0.8 μM Antimycin A (Sigma)] in fresh growth medium for 5 h. In order to block GSK3 activity, cells were treated with SB-216763 (Sigma-Aldrich S3442) at 1 µM. MG132 (Sigma-Aldrich, M7449) was used to inhibit the proteasome at 10 µM.
Immunocytochemistry
Cells were washed in PBS and fixed with 4% paraformaldehyde in PBS for 15 min. After permeabilization with 0.4% Triton X-100 (Sigma-Aldrich, X100) in PBS for 5 min, cells were blocked in 3% normal goat serum (Sigma-Aldrich, G9023) in PBS and incubated overnight at 4 °C with primary antibodies. We used the antibodies directed against rabbit anti-MCL1, rabbit anti-FLAG-HUWE1 and mouse anti-UBIQUITIN (1:100 overnight). Cells were then washed in blocking buffer and incubated for 1 h with labeled anti-mouse (Alexa Fluor 488; Invitrogen, A11017), anti-rabbit (Alexa Fluor 555; Invitrogen, A21430) or anti-rabbit (Alexa Fluor 647; Invitrogen, A31573) secondary antibodies. Nuclei were stained with 1 μg/ml DAPI and examined under a Zeiss LSM 700 100× oil-immersion objective (CLSM700; Jena, Germany). We used “ZEN 2009 Light edition” software for image analysis. All acquisitions were performed in single z-confocal planes.
Western blot analysis
Cell extracts were centrifuged at 13,000 × g for 10 min at 4 °C. Protein concentrations were determined with the Bio-Rad Protein Assay Kit (Bio-Rad, 5000001). Cell extracts were separated by SDS-PAGE and transferred onto nylon membranes (Immobilon P; Merck-Millipore, IPFL10100). Membranes were incubated with primary antibodies (1:1000, overnight) followed by horseradish peroxidase-conjugated secondary antibody (Bio-Rad, 1706515 and 1721011) and visualized with ECL (Merck-Millipore WBKLS0500).
Co-immunoprecipitation (Co-IP)
After cell lysis, equal amounts of protein were incubated with the primary antibody (Anti-HUWE1) with rotation for 24 h. Then 30 µl of protein A agarose beads (Roche, 11719408001) were added for 60 min. The beads were collected by centrifugation and washed three times with the RIPA buffer. Immuno-complexes were eluted with 30 µl of SDS sample buffer and heated at 95 °C for 10 min.
Cytosol/mitochondria fraction preparation
For cytosol/mitochondria fractionation, cells were lysed by mechanical breaking (~100 potter strokes) into a detergent-free buffer (0.25 M sucrose, 10 mM HEPES pH 7.5, 1 mM EDTA). A first centrifugation (600 × g for 10 min at 4 °C) was employed to pellet nuclei; subsequently, supernatants were recentrifuged at 12,000 × g for 15 min at 4 °C to separate a cytosolic upper phase from a mitochondrial pellet. The latter was lysed in RIPA buffer. All protein extracts were quantified through the Bradford Protein Assay (Bradford solution from Bio-Rad Laboratories) according to the manufacturer’s protocols.
Retrovirus production and infection protocol
CD34+ AML cells were infected using a pMSCV-IRES-Blue FP Retroviral construct (pMSCV-IRES-Blue FP was a gift from Dario Vignali—Addgene plasmid # 52115) carrying a MYC-AMBRA-ActA fusion gene or the Empty vector. Noninfected CD34+ cells were also considered as control. Retroviral particles were generated using pCMV-VSV-G (Envelope; pCMV-VSV-G was a gift from Bob Weinberg—Addgene plasmid # 8454) and pUMVC (Packaging; pUMVC was a gift from Bob Weinberg—Addgene plasmid # 8449) in 293Tcells. MYC-AMBRA-ActA fusion gene was cloned into the multiple cloning site using SnaBI and ApaI restriction enzymes (New England Biolabs). Retrovirus was concentrated using Retro-X™ Concentrator (Takara) and infection was performed after coating the culture plates with RetroNectin (Takara—RetroNectin® GMP grade) following manufacturer protocol. After 24 h in culture, in presence of retroviral particles, cells were washed three times with D-PBS and cultured for colony-forming assay.
Colony formation assay
In vitro colony-forming unit (CFU) assay was performed in order to test clonogenic potential of CD34+ after 15 days of expansion using MethoCult H4434 methylcellulose medium (Stem Cell Technologies, Vancouver, CA). Colonies were counted with CellSens Standard 1.14 (Olympus, Tokyo, JP) with a minimum imposition of 200 nm diameter per colony unit. Unpaired t-test was performed to determine if the means of the datasets were significantly differentent from each other using Bioconductor and GraphPad Prism version 6.00 for MacOSX (GraphPad Software, La Jolla California USA). Frozen bone marrow sample from newly diagnosed AML pediatric patient was collected by Ospedale Pediatrico Bambino Gesù. The patient harbored FLT3-ITD mutation in a CN-AML. Informed consent was obtained from either parents or legal guardians according to the Declaration of Helsinki. Approvals for this study were obtained from the Institutional Review Boards of the institution. MNC were isolated by density gradient centrifugation, diluted in 90% FBS plus 10% dimethyl sulfoxide and stored in liquid nitrogen. CD34+ cells were magnetically separated using MACS CD34+ microbead kit (Miltenyi Biotech, Bergish Gladbach, DE).
Statistical analyses
Data were analyzed and graphs were plotted using GraphPad Prism 6 software. Statistical analyses were performed using the Student two-tailed t-test. Data were shown as means ± SD of three independent experiments. Values of p < 0.05 were considered significant (*p < 0.05; **p < 0.01; ***p < 0.001).
Supplementary information
Acknowledgements
We wish to thank Dr Muriel Priault for kindly providing us the constructs BCL2L1 and MCL1; Prof. M. Campanella for the gift of Short Hairpin PARKIN and Dr V. Stagni and Prof. Barilà for the gift of MCF7 cells. We are grateful to Dr Salvatore Rizza for his help with Fig. 7. We thank M. Acuña Villa for secretarial work and F. Maria Orecchio, S. Verna, T. Maiorino, K. Bruqi for technical assistance.
Funding
This work was also supported by grants: GR2011-02351433 (Italian Ministry of Health), ROCHE (Roche per la ricerca 2017) and 5XMILLE Italian Ministry of Health (2017) to FS; AIRC (MFAG#15523) to AP; MRC, LazioInnova (85-2017-14986) and AIRC (IG#20473) to GM; grants from the Italian Ministry of Education, University and Research (Research Project of National Relevance 2017 ID 2017WC8499) and AIRC (Associazione Italiana Ricerca sul Cancro, 5 × 1000 ID 9962, AIRC IG 2018 ID 21724) to FL. P.L was supported by FUV Grant 2019. Francesco Cecconi’s laboratory is supported by grants from the Bjarne Saxhof Foundation, the Danish Cancer Society (KBVU R72-A4408, R146-A9364), the Novo Nordisk Foundation (7559, 22544), the Lundbeckfonden (R233-2016-3360), the LEO Foundation (LF17024), “Associazione Italiana per la Ricerca sul Cancro” (AIRC IG-15180). FC lab in Copenhagen are part of the Center of Excellence for Autophagy, Recycling and Disease (CARD), funded by the Danmarks Grundforskningsfond (DNRF125).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by H.-U. Simon
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/30/2019
A Correction to this paper has been published: 10.1038/s41418-019-0446-y
Contributor Information
Flavie Strappazzon, Email: f.strappazzon@hsantalucia.it.
Francesco Cecconi, Email: cecconi@cancer.dk.
Supplementary information
The online version of this article (10.1038/s41418-019-0404-8) contains supplementary material, which is available to authorized users.
References
- 1.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;3:344–57. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Nardacci R, Corazzari M, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–5. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Elazar Z. Development. Inheriting maternal mtDNA. Science. 2011;25:1069–70. doi: 10.1126/science.1215480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickrell AM, The roles of PINK1, parkin, and mitochondrial fidelity i n Parkinson's disease. 2015;21:257–73. [DOI] [PMC free article] [PubMed]
- 5.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson ÅB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012;287:19094–104. doi: 10.1074/jbc.M111.322933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarten M, Mohrlüder J, Ma P, Stoldt M, Thielmann Y, Stangler T, et al. Nix directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy. 2009;5:690–8. doi: 10.4161/auto.5.5.8494. [DOI] [PubMed] [Google Scholar]
- 8.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2014;14:177–85. doi: 10.1038/ncb2422. [DOI] [PubMed] [Google Scholar]
- 9.Wei Y, Chiang WC, Sumpter R, Jr, Mishra P, Levine B. Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor. Cell. 2017;168:224–38. doi: 10.1016/j.cell.2016.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strappazzon F, Nazio F, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22:419–32. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Rita A, Peschiaroli A, D Acunzo P, Strobbe D, Hu Z, Gruber J, et al. HUWE1 E3 ligase promotes PINK1/PARKIN-independent mitophagy by regulating AMBRA1 activation via IKKα. Nat Commun. 2018;9:3755. doi: 10.1038/s41467-018-05722-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams JM, Cory S. The BCL-2 arbiters of apoptosis and their growing role as cancer targets. Cell Death Differ. 2018;25:27–36. doi: 10.1038/cdd.2017.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kale J, Osterlund EJ, Andrews DW. BCL-2 family proteins: changing partners in the dance towards death. Cell Death Differ. 2018;25:65–80. doi: 10.1038/cdd.2017.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalkavan H, Green DR. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018;25:46–55. doi: 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukherjee A, Williams DW. More alive than dead: non-apoptotic roles for caspases in neuronal development, plasticity and disease. Cell Death Differ. 2017;24:1411–21. doi: 10.1038/cdd.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opferman JT, Kothari A. Anti-apoptotic BCL-2 family members in development. Cell Death Differ. 2018;25:37–45. doi: 10.1038/cdd.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strasser A, Vaux DL. Viewing BCL2 and cell death control from an evolutionary perspective. Cell Death Differ. 2018;25:13–20. doi: 10.1038/cdd.2017.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hollville E, Carroll RG, Cullen SP, Martin SJ. Bcl-2 family proteins participate in mitochondrial quality control by regulating Parkin/PINK1-dependent mitophagy. Mol Cell. 2014;55:451–66. doi: 10.1016/j.molcel.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Xue D, Chen G, Han Z, Huang L, Zhu C, et al. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014;10:1712–25. doi: 10.4161/auto.29568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, et al. Mitochondrial BCL-2 inhibits AMBRA1-induced autophagy. EMBO J. 2011;30:1195–208. doi: 10.1038/emboj.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strappazzon F, Di Rita A, Cianfanelli V, D’Orazio M, Nazio F, Fimia GM, et al. Prosurvival AMBRA1 turns into a proapoptotic BH3-like protein during mitochondrial apoptosis. Autophagy. 2016;12:963–75. doi: 10.1080/15548627.2016.1164359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Rita A, D’Acunzo P, Simula L, Campello S, Strappazzon F, Cecconi F. AMBRA1-Mediated Mitophagy Counteracts Oxidative Stress and Apoptosis Induced by Neurotoxicity in Human Neuroblastoma SH-SY5Y Cells. Front Cell Neurosci. 2018;12:92. doi: 10.3389/fncel.2018.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–60. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Dominguez I, Itoh K, Sokol SY. Role of glycogen synthase kinase 3b as a negative regulator of dorsoventral axis formation in Xenopus embryos. Proc Natl Acad Sci USA. 1995;92:8498–502. doi: 10.1073/pnas.92.18.8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2015;121:1085–95. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 26.Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M. A survey of the anti-apoptotic bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of bcl-2 antagonists in cancer therapy. Cell Death Dis. 2010;1:e40. doi: 10.1038/cddis.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kasper S, Breitenbuecher F, Heidel F, Hoffarth S, Markova B, Schuler M, et al. Targeting MCL-1 sensitizes FLT3-ITD-positive leukemias to cytotoxic therapies. Blood. Cancer J. 2012;2:e60. doi: 10.1038/bcj.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011;30:395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fogarty LC, Flemmer RT, Geizer BA, Licursi M, Karunanithy A, Opferman JT, et al. Mcl-1 and Bcl-xL are essential for survival of the developing nervous system. Cell Death Differ. 2018;26:1501–15. doi: 10.1038/s41418-018-0225-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.