Abstract
The cullin-RING ubiquitin E3 ligase (CRL) family consists of ~250 complexes that catalyze ubiquitylation of proteins to achieve cellular regulation. All CRLs are inhibited by the COP9 signalosome complex (CSN) through both enzymatic (deneddylation) and nonenzymatic (steric) mechanisms. The relative contribution of these two mechanisms is unclear. Here, we decouple the mechanisms using CSNAP, the recently discovered ninth subunit of the CSN. We find that CSNAP reduces the affinity of CSN toward CRL complexes. Removing CSNAP does not affect deneddylation, but leads to global effects on the CRL, causing altered reproductive capacity, suppressed DNA damage response, and delayed cell cycle progression. Thus, although CSNAP is only 2% of the CSN mass, it plays a critical role in the steric regulation of CRLs by the CSN.
Subject terms: Biochemistry, Proteolysis
Introduction
Protein degradation is one of the essential mechanisms that enables reshaping of the proteome landscape in response to various stimuli [1]. The specificity of this process is largely mediated by E3 ligases that ubiquitinate target proteins [2, 3]. One of the largest E3 ubiquitin ligase families, responsible for ubiquitination of 20% of the proteins degraded by the 26S proteasome, is comprised of cullin-RING ligases (CRLs) [4]. This family encompasses ~250 distinct complexes that are built in a modular fashion around a central cullin scaffold, which is associated with a specific substrate receptor, adaptor protein, and a RING protein that recruits the E2 enzyme (reviewed in [2, 5]). Seven different cullins have been identified in humans, each interacting with a dedicated set of receptors, forming CRL complexes that target a single or a small group of substrate proteins. At any given time, various CRLs are active, and their dynamic assembly and disassembly enables cellular adaptation in response to regulatory inputs.
In spite of the great diversity of CRLs in terms of composition and substrate specificity, all complexes are regulated by the COP9 signalosome complex (CSN) [2]. The CSN regulates CRLs by means of two independent mechanisms, catalytic and non-catalytic. The first involves enzymatic deconjugation of the ubiquitin-like protein Nedd8 from the cullin subunit (deneddylation) [6]. The latter is mediated through physical binding to CRLs, sterically precluding interactions with E2 enzymes and ubiquitination of substrates [7–9]. By inhibiting CRL activity, both mechanisms control the gateway to the exchange cycle that remodels CRL composition [10–12].
The CSN is a highly conserved complex that exists in all eukaryotes [13, 14]. Three types of subunits constitute this complex: two MPN subunits (for Mpr1p and Pad1p N terminal) CSN5 and CSN6 [15], six PCI subunits (for proteasome, COP9, and initiation factor 3); CSN1–CSN4, CSN7, and CSN8 [16]; and an additional small, non-PCI or MPN subunit that we recently discovered and termed CSNAP, for CSN acidic protein [17]. The CSNAP protein, which exists in one-to-one stoichiometry with the other CSN subunits, consists of only 57 amino acids (molecular weight: 6.2 kDa) that link together the two distinct structural elements of the CSN by mutually binding the MPN subunits CSN5 and CSN6, and the PCI subunit CSN3 [17]. Given the small size of CSNAP, a natural question that arises is whether it is actually crucial for CSN function and, if so, what is its functional role? In this study, we address these questions by combining biochemical and cell biology approaches, together with mass spectrometry (MS) analysis.
Using the above approaches, we discovered that manipulating CSNAP enables us to uncouple the steric and catalytic activities of the CSN complex. Although it is only 2% of the CSN mass, we find that removing CSNAP has a global effect on the cell cycle, cell viability, and DNA damage response. This effect is due to a reduction in the Kd of CSN-CRL binding, leaving deneddylation activity unchanged. These findings provide a role for CSNAP, and points to the affinity of CSN-CRL interactions as a critical component for proteostasis.
Results
CSNAP alters the strength of CSN-CRL interaction
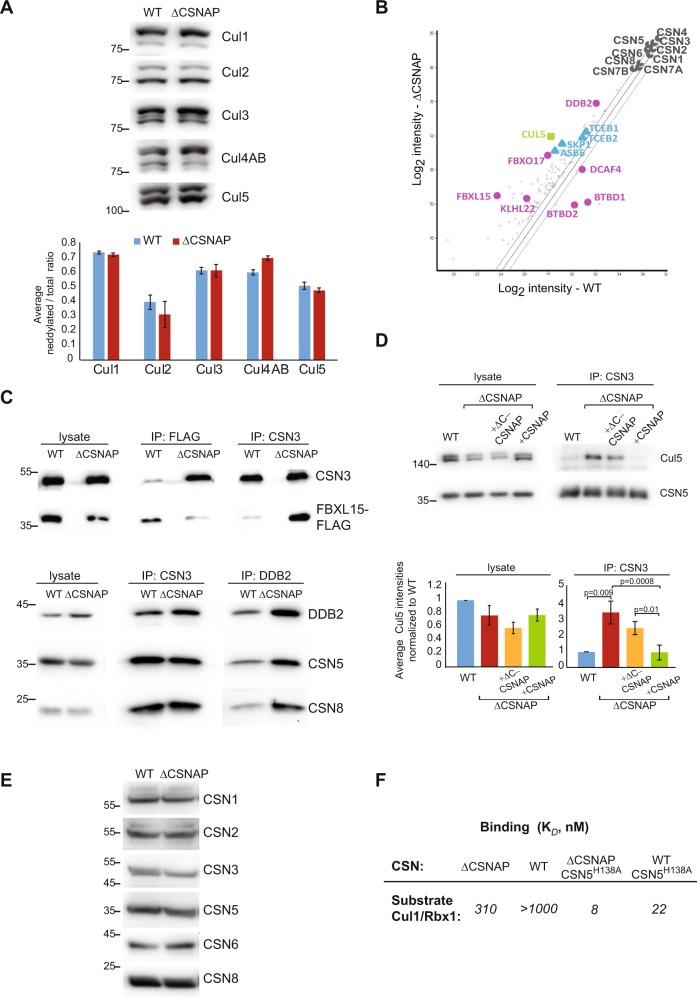
To investigate the impact of CSNAP on both the enzymatic and steric activities of CSN, we initially examined the complex’s deneddylation activity, using HAP-1 cell lines lacking CSNAP (ΔCSNAP cells) [17]. Comparison of the deneddylated/neddylated ratio between WT and ΔCSNAP cells showed that in the absence of CSNAP there are only minor changes of <15% of the cullin’s deneddylated fraction (Fig. 1a). This result is in accordance with our previous finding, showing that WT and ΔCSNAP cells exhibit a similar rate of deneddylation [17] and with studies that compared the rate of deneddylation of endogenous CSN prepared from HEK293 cells, with that of recombinant CSN lacking CSNAP (CSNΔCSNAP) [7, 8]. Thus, it can be concluded that CSNAP does not significantly affect the catalytic capacity of the CSN complex.
Fig. 1.
CSNAP modifies the strength of CSN-CRL interactions. a CSN catalytic activity is not significantly affected by the absence of CSNAP. A representative western blot of WT and ΔCSNAP cell extracts visualized using antibodies against various cullins (top) and a plot demonstrating the average deneddylated fraction. The graph represents the averages of three independent experiments, with standard errors. b CSN and its interacting proteins were pulled down using an antibody against CSN3 from WT and ΔCSNAP cells. Immunoprecipitated proteins were then analyzed by label-free proteomics approach using three biological replicates. Scatter plot comparing log2 intensities of proteins in ΔCSNAP and WT samples show that a number of CRL proteins were found to be over- or underrepresented in the pulldown of the CSN and CSNΔCSNAP complexes. In contrast, the ratio of average intensities for CSN subunits did not exceed the fold change of ΔCSNAP/WT > 1.5, which was considered to be the cutoff for fold change. c Validation of the proteomics data for FBXL15/CSN and DDB2/CSN interactions. Reciprocal immunoprecipitation shows a tighter CSN3 interaction with FBXL15 and DDB2, in the absence of CSNAP. The immunoprecipitated proteins were detected by western blot, using the relevant antibodies. Densitometry analysis of three independent experiments is shown in Fig. S1a. d Validation of the proteomics data for Cul5/CSN interaction. Immunoprecipitation using anti-CSN3 shows stronger interaction in the absence of CSNAP or its C-terminal interacting domain, while overexpression of full-length CSNAP in ΔCSNAP cells rescues the weaker Cul5/CSN interaction, characteristic to WT cells. Due to the lack of appropriate antibody reciprocal immunoprecipitation could not be performed. The densitometry analysis shows the average Cul5 intensities normalized to WT from three independent experiments with standard errors, significance was calculated using two-way ANOVA accounting for treatment and batch (p = 0.000469) followed by Tukey’s post hoc test. e The levels of CSN subunits are comparable in WT and ΔCSNAP cells (30 μg total protein loaded); thus, the differences in the amount of the pulled-down proteins are likely due to different interaction affinities. Representative blot out of three repeats. f Determination of the dissociation constant (Kd) for the CSN and CSNΔCSNAP complexes, and dansyl-labeled Cul1-Nedd8/Rbx1. The absence of CSNAP causes tighter binding to cullin1/Rbx1
To examine whether the steric activity of CSNΔCSNAP is affected by the absence of CSNAP, we applied label-free quantification of protein intensities from pull-down assays, coupled with MS analysis of WT and ΔCSNAP cells. We reasoned that if CSNAP impacts the CSN-CRL interaction, differences in the array of protein binding partners will be revealed. Our results indicated that multiple CRL components are significantly enriched by pull-downs of ΔCSNAP cells, in comparison with immunoprecipitation of WT cells (Fig. 1b and Table S1). These mainly include substrate receptors (DDB2, FBOX17, FBXL15, and KLH22) and adapter proteins (TCEB2, TCEB1, SKP1, and ASB6). In the WT cells, only three proteins, DCAF4, BTBD2, and BTBD1 were enriched; all are CRL substrate receptor proteins. To validate these results, we carried out co-immunoprecipitation experiments. The results obtained for the ΔCSNAP and WT cell lines confirmed that FBXL15 and DDB2 are enriched in the CSNΔCSNAP pull-down experiment, in comparison with the WT complex (Fig. 1c, Fig. S1a). Moreover, the weaker interaction between the CSN and Cul5, typical to WT cells, could be rescued when CSNAP-Cerulean was overexpressed in ΔCSNAP cells, but not by the truncated form, which lacks the C-terminal region that is crucial for the protein’s integration into the CSN [17] (ΔC-CSNAP-Cerulean) (Fig. 1d, Fig. S1b). Notably, these results did not arise from changes in the expression levels of CSN subunits, as all CSN subunits (except for CSNAP, depleted from the cells) displayed insignificant differences when the two cell lines were compared (Fig. 1b), a finding that was further validated by western blot analysis (Fig. 1e, Fig. S1c). Taken together, the data suggest that CSNAP plays a role in tuning CSN-CRL interactions in cells.
To further assess the contribution of CSNAP to the CSN/CRL interaction, we utilized a quantitative in vitro binding assay to determine the affinity between Cul1-Rbx1, and recombinant CSNΔCSNAP or CSN complexes [10] (Fig. S2a, S2b). In this assay, the environmentally-sensitive dye dansyl was conjugated to the C-terminus of Cul1, and an increase in fluorescence upon CSN binding was detected [10]. Both WT CSN5 and the well characterized CSN5-H138A mutant [7, 8, 10] (CSN5H138A) were used, as the latter binds Cul1-Rbx1 ~30-fold more tightly, enabling us to reach saturation. The results indicate that CSN complexes display decreased affinity to Cul1-Rbx1, in comparison with CSNΔCSNAP (Fig. 1f, Fig. S2c). Taken together, the results imply that CSNAP reduces the affinity of the CSN towards certain CRL complexes.
CSNAP is required for proper cell cycle progression and viability
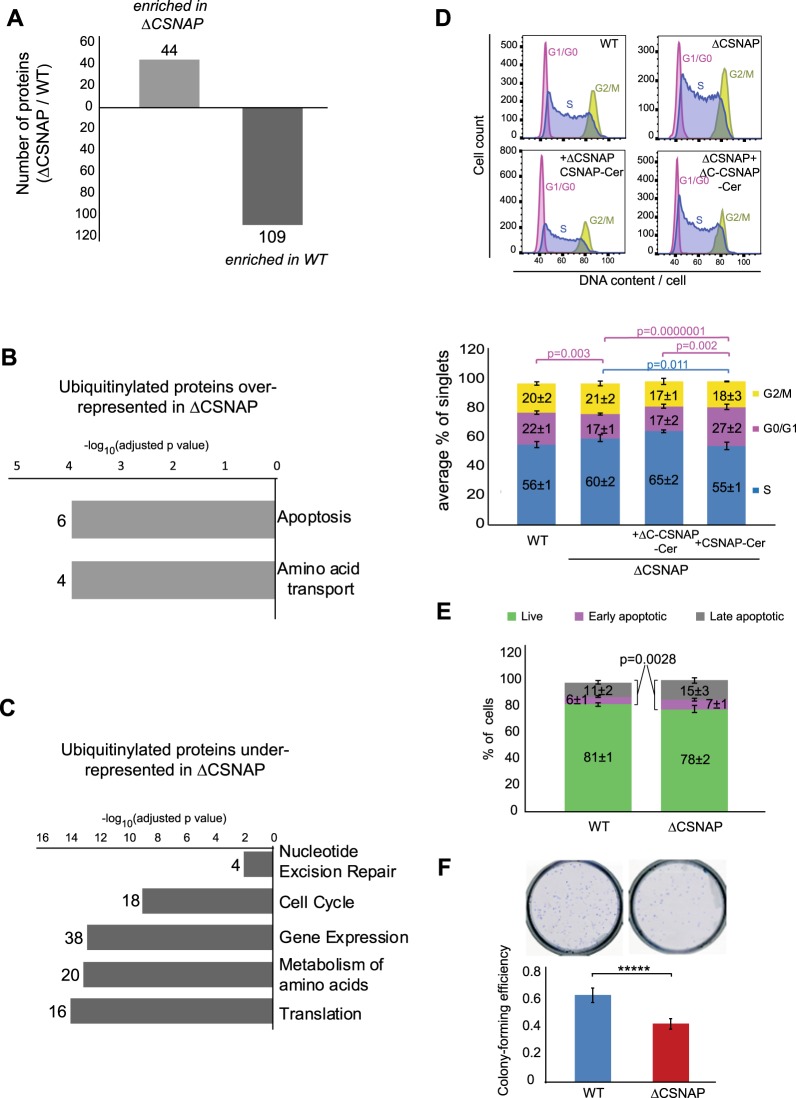
The apparent difference in Kd for CSNΔCSNAP binding to SCF, compared with CSN, led us to question whether such a change in affinity can influence the repertoire of active CRLs and, as a consequence, the array of ubiquitinated proteins. We therefore performed a large-scale analysis of protein ubiquitination, relying on the enrichment of ubiquitinated tryptic peptides [18]. The relative differences in the ubiquitination of WT and ΔCSNAP cells was quantified, using the SILAC (stable isotope labeling by amino acids in cell culture) approach [19]. The results indicated that differences exist in the extent of ubiquitination in ΔCSNAP and WT cell lines (Fig. 2a and Table S2A and B). In total, 109 ubiquitinated proteins were found to be enriched only in WT and not in ΔCSNAP cells, while 44 ubiquitinated proteins, were abundant in cells lacking CSNAP. Forty-nine out of the one hundred and fifty three identified proteins, whose ubiquitination levels were different between the two cell types, are known substrates of the different CRL complexes (Table S3) [20–23]. These results suggest that reducing the affinity between CSN and CRL modifies CRL assembly and, consequently, the repertoire of ubiquitinated proteins. Whether this is dependent on direct or indirect effects requires further elucidation.
Fig. 2.
The absence of CSNAP impairs cell cycle progression and viability. a Protein ubiquitinylation was monitored in stable heavy and light isotope labelled ΔCSNAP and WT cells. The bar plot shows the number of differentially ubiquitinated (up or down) proteins that appeared in at least three out of four independent experiments. In total 1563 proteins with single or multiple ubiquitinylation sites were identified. b, c Pathway analysis of the differentially ubiqiutinylated proteins in ΔCSNAP and WT cells (using Reactome 2016) indicate the enrichment of several sets of proteins. The numbers on the left of each bar refer to the number of proteins found to be enriched and the –log10(adjusted p value) y-axis reflects the probability of overlap with annotated gene sets, (Table S5A, B shows the ratio of overlapping genes/number of genes in each annotated group). d Validation of the influence of the absence of CSNAP on cell cycle. Histograms of BrdU and propidium iodide stained asynchronous cells show that the lack of CSNAP results in a S-G2 shifted phenotype. The percentage of S phase population can be rescued by the expression of CSNAP-Cerulean, but not when its C-terminal CSN interacting domain is absent. The figure shows a representative experiment (50,000 cells collected each), and the bar chart demonstrates the distribution of the cell populations showing four (ΔCSNAP + ΔC-CSNAP-Cer and ΔCSNAP + CSNAP-Cer), or ten replicates (WT and ΔCSNAP). Significance levels were calculated using one-way ANOVA (S phase p = 0.0102, G0/G1 phase p = 2.8e−07), followed by Tukey's post hoc test. e ΔCSNAP cells harbor larger populations of dead cells (early and late apoptotic) compared with WT cells. The bar charts show populations as average percentage of single cells ± standard errors, measured by flow cytometry using annexin V-FITC and propidium iodide staining. Significance was calculated using a paired Student’s t-test, using 14 replicates. (Early + late apoptotic: p = 0.0028). f Cells lacking CSNAP have lower colony forming potential than WT cells. The graph represents the results from 14 biological replicates, significance was calculated using paired Student’s t-test (p = 0.00039). Images above show a representative experiment
Functional annotations revealed that among the ubiquitinated proteins identified as being enriched in WT, or ΔCSNAP cells, 17% and 14% are clustered in the cell cycle and apoptosis pathways, respectively (Fig. 2b, c, Tables S4, S5A and S5B). We confirmed this data by assessing the cell cycle distribution of both ΔCSNAP and WT cells, using flow cytometry analysis. The results indicated that compared with WT cells, ΔCSNAP cells remained relatively stalled in the S phase in expense to G0/G1 (Fig. 2d), a phenotype that can be prevented by exogenous expression of CSNAP-Cerulean, but not by the CSN-binding deficient form of the protein (ΔC-CSNAP-Cerulean). Analysis of apoptotic cell populations indicated that ΔCSNAP cells also harbor a higher percentage of early and late apoptotic cells (Fig. 2e). In addition, colony formation assays [24] showed that the viability of cells lacking CSNAP is significantly reduced, in comparison with that of WT cells (Fig. 2f). Taken together, our results suggest that the absence of CSNAP influences CSN-CRL interactions in a manner that affects, protein ubiquitination and, therefore likely, cell cycle coordination.
Cellular protein levels are influenced by CSNAP
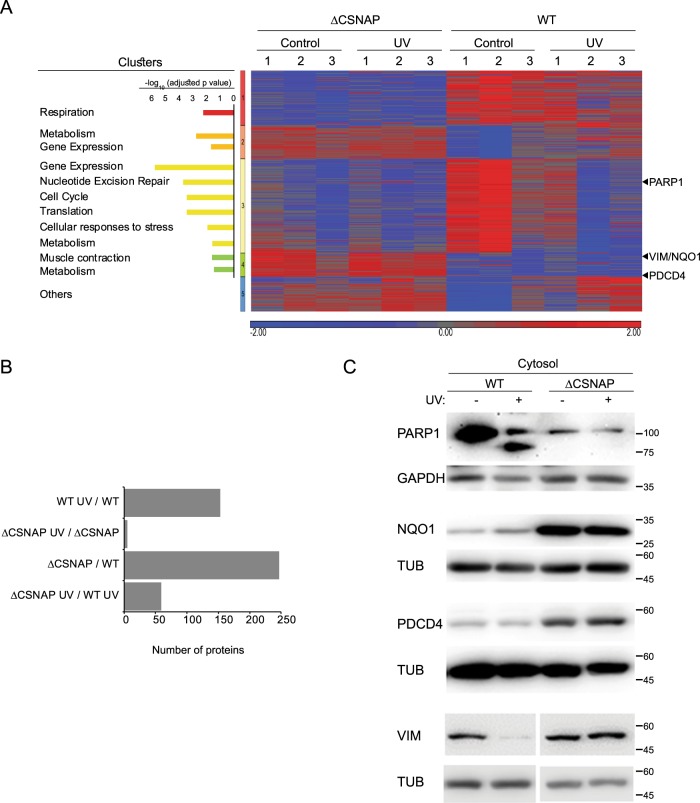
Considering the dependence of the ubiquitinated proteome on the presence of CSNAP, we wished to examine whether the impact of this subunit would also be detected in a global proteome analysis. To this end, we performed label-free quantification [25] of the proteomes of WT and ΔCSNAP cells. Given that the CSN complex and protein ubiquitination are vital to the DNA damage response [26–29], we performed the analysis both prior to and following exposure of the cells to UV irradiation. Data were analyzed by two-way ANOVA, taking into consideration both the UV treatment, and the type of cell being treated (WT or ΔCSNAP). Proteins that were considered significantly differentially expressed were clustered into five groups according to their cellular functions (Fig. 3a and Tables S6A–D and S7). Remarkably, we noticed that cellular pathways that were enriched in this experiment are in accordance with those identified in the SILAC-based ubiquitinylation analysis (Fig. 2b, c). Among these proteins we could identify known substrates of various CRL complexes (Table S8), which are known to be involved in ubiquitination, apoptosis, cell cycle regulation, and DNA damage response. This observation may explain the detected phenotypic effects.
Fig. 3.
The absence of CSNAP affects proteome remodeling following UV damage. a Proteomes of untreated or UV exposed WT and ΔCSNAP cells 4 h post damage were analyzed using label free proteomics approach. Proteomics data of three biological replicates, after logarithmic transformation and flooring, were analyzed by two-way ANOVA using the two factors: strain and UV treatment, as well as their interaction. Proteins with a p-value below 0.05 and an absolute fold change above 1.5 were considered as being differentially expressed. Heatmap of differentially expressed proteins grouped to five clusters. Pathway analysis of the clusters indicate up- and downregulation of several cellular functions in ΔCSNAP cells. The bar chart on the left expresses the significance levels of the enrichment analysis of the proteins using the protein coding part of the human genome. b Comparison of the differentially expressed proteins in the proteome in untreated and UV-exposed WT and ΔCSNAP cells. The bar plot shows each of the four pair-wise comparisons, highlighting that in ΔCSNAP cells the DNA damage response is compromised. c Expression levels of four representative proteins: PARP1, NQO1, PDCD4, and vimentin, analyzed by western blots of WT and ΔCSNAP cell lysates. UV-treated samples were tested 2 h post damage
Examination of the five clusters indicated that even under normal conditions, there are clear differences in protein levels among WT and ΔCSNAP cells. A particularly striking observation was that the proteome response associated with UV irradiation was nearly abolished in ΔCSNAP cells (Fig. 3b). To validate the proteomics results we performed western blot analysis. Specifically, we monitored the levels of four proteins that displayed differential levels between WT and ΔCSNAP cells: the quinone reductase enzyme, NQO1, the tumor suppressor PDCD4, and the filament protein vimentin, which appear in Cluster 4, and PARP1, a member of the PARP family that appears in Cluster 3 (Fig. 3a; see arrows on the right). The results confirmed that unlike WT cells, higher levels (due to expression or lack of degradation) of NQO1, PDCD4, and vimentin are obtained in ΔCSNAP cells (Figs. 3c and S3a). Likewise, the blots validated that the abundance of PARP1 in WT cells is high under normal conditions, and is decreased following UV-induced DNA damage; while in ΔCSNAP cells, regardless of UV irradiation, low levels of PARP1 are maintained (Figs. 3c and S3a). We also noticed that in WT cells, a cleavage product of PARP1 was detected 2 h post damage, a phenomenon that was not observed in ΔCSNAP cells at that time point; this result will be further discussed in the next section. In summary, our findings suggest that the lack of CSNAP following UV-treatment elicited a strong and specific influence on downstream effectors of the DNA damage repair process.
CSNAP is required for DNA repair
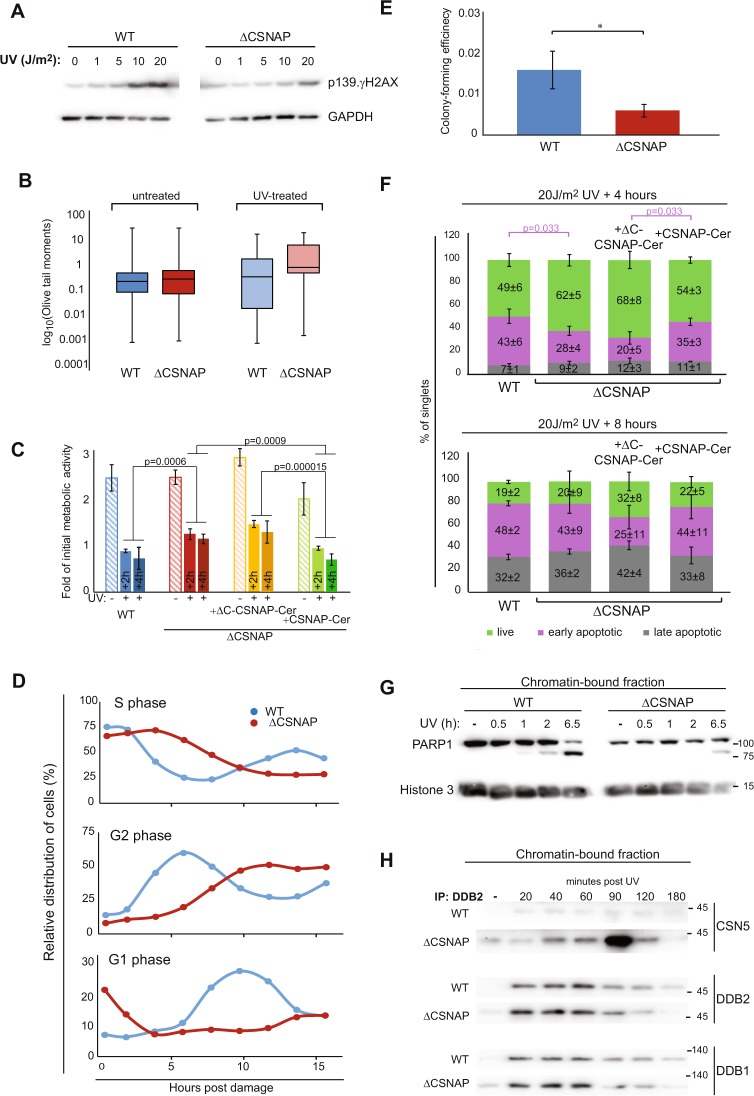
Building on our results reflecting the compromised protein remodeling capability following DNA damage in ΔCSNAP cells, and the identification of multiple DNA damage related proteins in both the SILAC-based ubiqitinylation and total proteome analysis (Table S9), we wished to explore the DNA repair response in these cells. Initially, we tested the UV-dose response of WT and ΔCSNAP cells, comparing the extent of Ser139 phosphorylation on γH2AX, which serves as an indicator for UV-induced signal transduction [30]. In WT cells, increasing levels of γH2AX phosphorylation on Ser139 were observed from 10 J/m2, but in cells lacking CSNAP partial response was detected after exposure of cells to 20 J/m2 (Fig. 4a), suggesting attenuated DNA damage repair. Next, we measured the DNA repair capacity following UV irradiation using the comet assay [31]. The results indicated that ΔCSNAP cells display a longer tail moment, which is associated with the accumulation of both single- and double-strand DNA breaks (Fig. 4b). Following DNA damage, cells would reduce their rates of proliferation, in order to enable DNA damage repair [32]. We therefore measured cell proliferation before and after treatment with UV irradiation, and found that, as expected, a pronounced proliferation arrest was detected in WT cells, however, in cells lacking CSNAP, nor in ΔCSNAP cells exogenously expressing ΔC-CSNAP-Cerulean, which is not incorporated into the CSN complex the reduction in activity was less significant (Fig. 4c). Nevertheless, cell rescue was achieved in ΔCSNAP cells by overexpressing the full-length CSNAP protein.
Fig. 4.
Recovery following exposure to UV irradiation is affected by the absence of CSNAP. a DNA damage response is attenuated in ΔCSNAP cells. Analysis of dose-dependent induction of γH2AX Ser139 phosphorylation. Western blots were performed 1.5 h post damage. Representative results out of five experiments indicating reduced γH2AX Ser139 phosphorylation in ΔCSNAP cells. b Unlike WT cells, damaged DNA accumulates in ΔCSNAP cells following UV exposure. The genotoxic effect of UV was measured using alkaline comet assay. DNA damage, expressed as Olive tail moments were calculated and presented as a box-whisker plot. Within each biological replicate, the proportion of Olive tail moments larger than 3 was calculated. The proportions in ΔCSNAP and WT samples were compared using a two-way ANOVA, accounting for group and batch (UV treated samples p = 0.00142). Data are shown before and after UV treatment (representative plot of 6 replicates, each). c Comparison of metabolic activity of WT and ΔCSNAP cells before and after UV irradiation. The plot shows metabolic activities of WT and ΔCSNAP cells, before and after UV induced DNA damage, 26 and 28 h (2 and 4 h post UV, respectively) after seeding, calculated as a fold of initial activity for each cell line. Metabolic activity of both WT and ΔCSNAP cells is reduced in response to DNA damage but a difference in the decrease in activity is detected, a pronounced degree of metabolic reduction is observed for WT cells or ΔCSNAP cells expressing full length CSNAP-Cerulean. The graph represents the averages of three independent experiments, with standard errors. Significance was calculated using a two-way ANOVA test accounting for treatment, time, cell type, and batch (effect of treatment: p < 2e−16, effect of time: p = 0.07905, effect of cell type: p = 1.27e−08), another ANOVA was run only on the treated samples, followed by a Tukey's post hoc test for cell type. d UV-exposed ΔCSNAP cells stay longer in S and G2 phases. Comparison of the relative distribution of cell populations in different phases of the cell cycle, as calculated from flow cytometry histograms of double thymidine-synchronized cells following exposure to UV irradiation (5 J/m2). UV-induced DNA damage causes longer cell cycle phases in cells lacking CSNAP. The graph shows a representative experiment out of three. e Cells lacking CSNAP exhibit a compromised recovery after exposure to high-dose UV. WT and ΔCSNAP cells were exposed to UV irradiation, prior to incubation in culturing conditions for 8 days. Colonies were stained and counted. ΔCSNAP cells exhibit ~2.7-fold less colony-forming potential following UV damage, in comparison with WT cells. The graph represents average results from seven biological replicates with standard errors. Significance was calculated using paired Student’s t-test (p = 0.035). f The early apoptotic response is delayed in ΔCSNAP cells, following UV damage. The bar charts represent the average percentage of live, early, and late apoptotic cells detected by three independent flow cytometry experiments for each time points ± standard errors. A significant difference is seen in the percentage of early apoptotic cell populations between WT and ΔCSNAP cells after 4 h of UV exposure, calculated using two-way ANOVA, accounting for treatment and batch (effect of treatment in early apoptosis p = 0.00506) followed by a Tukey’s post hoc test. Overexpression of CSNAP-Cerulean in cells lacking CSNAP rescues the late-onset apoptosis, but not when its C-terminal interacting domain is missing. Eight hours after UV irradiation the distribution of apoptotic cells is similar in both WT and ΔCSNAP cells. g PARP1 cleavage is delayed in cells lacking CSNAP. Chromatin-bound fractions were monitored by western blot for caspase-mediated PARP1 cleavage, a marker for commitment to apoptosis. h CSNΔCSNAP exhibits increased affinity toward DDB2, in comparison with the CSN complex. WT and ΔCSNAP cells were exposed to UV irradiation, and DDB2 was immunoprecipitated from the chromatin-bound fraction at different time points post UV damage. Western blot analyses show tighter CSN-CRL binding when CSNAP is absent. Representative blot out of four repeats
Considering that widespread DNA damage induces cell cycle arrest [32], we evaluated the cell cycle distribution of WT and ΔCSNAP cells exposed to mild (5 J/m2) UV irradiation following a double thymidine block, which induces a G1/S-phase arrest. After their release from cell cycle synchronization, untreated ΔCSNAP cells proceeded to the S phase significantly more slowly than WT cells, and reached the G2 phase with a delay of ∼4 h (Fig. S3b). However, following the induction of DNA damage, CSNAP-depleted cells, unlike the WT cells that displayed a slight delay in progression, remained stalled in the S and G2 phases (Fig. 4d). This scenario could be due to impaired checkpoint control, rather than exclusively due to a faulty DNA repair mechanism. We therefore validated that the activation of the UV-induced kinase, Chk1, is not dependent on CSNAP (Fig. S3c). Similarly, comparison of the colony-forming potential of WT and ΔCSNAP cells following UV irradiation, indicated a significant, 2.7-fold reduction in the number of colonies of cells lacking CSNAP (Fig. 4e), which is significantly higher than the ~1.5 reduction fold observed under normal condition (Fig. 2f). This finding suggests that the accumulation of damaged DNA compromises cell cycle progression and reproductive ability in ΔCSNAP cells.
Next, we determined whether the absence of CSNAP affects DNA damage-induced cellular apoptosis. To this end, we measured the populations of live, early apoptotic, and late apoptotic cells in UV-exposed WT and ΔCSNAP cultures 4 and 8 h post damage, using flow cytometry. We found that the population of early apoptotic cells 4 h following UV exposure is significantly enlarged in WT cells (Fig. 4f), a phenomenon that does not occur at that time point in ΔCSNAP cells. Whereas, 8 h post damage the apoptotic populations of ΔCSNAP cells become comparable with that of WT cells, suggesting a delayed onset of apoptotic response in the absence of CSNAP.
Cleavage of PARP1 by caspases is considered to be a hallmark of apoptosis [33–35], and in agreement with the above results, a cleavage product of the protein was detected only in WT but not in ΔCSNAP cells (Fig. 3c). To further examine this phenomenon, we monitored the appearance of the 89 kDa cleavage product of PARP1 following UV irradiation, in a time-dependent manner. We found that in WT cells, the presence of the 89 kDa fragment could already be detected 1 h following DNA damage (Fig. 4g, Fig. S3d). The levels of the cleavage product increased over time, concomitantly with the reduction of the full-length PARP1 (113 kDa) protein. In ΔCSNAP cells, however, the relative abundance of PARP1 was lower than in WT cells, even prior to UV irradiation, and the formation of the 89 kDa cleavage product was only detected after 6.5 h. Therefore, delayed PARP1 cleavage in ΔCSNAP cells may explain the slow activation of the early apoptotic response.
Previous studies have shown that CSN is physically recruited to DNA damage sites on the chromatin, and on its path partners with the Cul4A–Rbx1–DDB1–DDB2 complex (CRL4ADDB2) [26, 29, 36]. The CSN/ CRL4ADDB2 association is rapidly relieved at the DNA lesion site, to induce activation of the CRL4ADDB2 complex. Thus, we examined the associations of both CSNΔCSNAP and the WT complex with CRL4ADDB2 components, following the induction of DNA damage. Time-course analysis of DDB2 pull-downs from chromatin-bound fractions following UV irradiation indicated that, as expected, both DDB2 and DDB1 are rapidly recruited to chromatin (Fig. 4h). In WT cells, we could detect CSN release from the DDB2 complex following UV irradiation. However, in ΔCSNAP cells, although release was observed after 20 min it was less significant and rapid restoration of CSN/DDB2 interaction was detected, compromising the activation of the DNA damage response through CRL4ADDB2. This observation is consistent with our in vitro binding data, which showed a stronger binding between Cul1/Rbx1 and CSNΔCSNAP versus CSN (Fig. 1f) and between CSN3 and DDB2 (Fig. 1b, c). Overall, these results support the view that the affinity of CSN for CRL complexes is enhanced in the absence of CSNAP.
Discussion
Here, we investigated the functional contribution of CSNAP, the smallest and last to be discovered CSN subunit to the steric and catalytic functions of the CSN. We find that CSNAP attenuates CSN binding interactions with CRL (Fig. 5). Efficient dissociation from CRL assemblies is essential for reconfiguration of new CRL compositions in order to respond to changing regulatory inputs. Therefore, a hypothesis emerging from this study is that the increased affinity of CSNΔCSNAP for CRLs will affect the dynamic plasticity of CRL configuration. Indeed, we find that the absence of CSNAP alters cell cycle progression and reduces cellular viability. In addition, the attenuated DNA damage repair following UV irradiation of CSNΔCSNAP indicates a reduced capacity of ΔCSNAP cells to adapt to cellular stimuli. Together these results show that CSNAP contributes to the steric regulation of CRL by CSN, with global cellular effects.
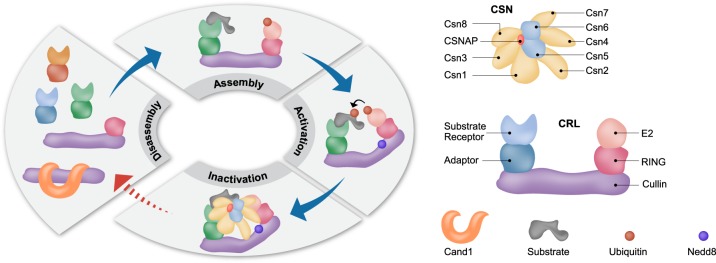
Fig. 5.
CSNAP influences the strength of the CSN-CRL interaction. Diagram representing the CRL cycle. CRLs form dynamic complexes with various adaptors and substrate receptors. The conjunction of Nedd8 to a conserved lysine residue in the cullin subunit, induces a conformational change that activates the CRL complex, promoting ubiquitin transfer to the substrate. The CSN complex inactivates CRL assemblies by two independent mechanisms, catalytic and non-catalytic. The first involves catalytic removal of the Nedd8 conjugate, while the second is mediated through physical binding to CRLs, sterically precluding interactions with E2 enzymes and ubiquitination substrates. Subsequently, after CSN dissociation, CRLs can be disassembled and assembled into new configurations, or bind Cand1. This cycle enables CRL adaptation according to cellular need, enabling specific substrates to be ubiquitinated. Our results indicate that CSNAP reduces the affinity of CSN for CRL, thus enabling efficient disassembly and remodeling of CRL complexes. In the absence of CSNAP, the disassembly and assembly steps of the cycle are compromised, as designated by the red dashed lines, affecting the reconfiguration of CRL assemblies, and their ability to respond to cellular stimuli
Our data indicate that the Kd for CSN binding to Cul1 is at least three fold higher than for the complex lacking CSNAP (Fig. 1f). Given that the Kd is in the micromolar range, and that the cellular cullin and CSN concentrations are ~2.2 and 0.45 μM, respectively [10], the change in Kd would be expected to impact the free CSN and CRL pools. Unneddylated cullins bind Cand1 or Cand2, the F-box protein exchange factors that mediates CRL recycling [12, 37, 38] (Fig. 5). A portion of unneddylated cullins, however, were shown to remain unbound to Cand1 [38–40] and some SCF substrates are efficiently degraded independently of these exchange factors [12, 41]. Hence, free unneddylated cullins may be directly available for configuration of new CRL modules.
In line with this assumption, it was demonstrated recently that the presence of unneddylated Cul1 is important for maintaining the substrate receptor pool and promoting rapid assembly and activation of Cul1-Skp1-F-box complexes [39]. Moreover, prolonging CSN-CRL interaction using irreversible neddylation inhibits CRL activity [42]. Likewise, strengthening the CRL-CSN interaction using the metabolite inositol hexakisphosphate promotes CRL inactivation [42]. Taken together with the present results, it is reasonable to conclude that modulation of CSN-CRL binding is an important mode of CRL regulation.
The recent crystal structure of free CSNΔCSNAP indicated that PCI and MPN subunits form largely distinct substructures [43]. The six PCI subunits comprise the base of the CSN, with their C-terminal ends forming an elaborate bundle above which the heterodimer CSN5/CSN6 sits. Previously, we discovered that CSNAP tethers together these two distinct structural elements, by mutually binding CSN5/CSN6, and the PCI subunit CSN3. Both CSN5 and CSN3 directly interact with the CRL assembly, CSN3 with the substrate receptor [7, 44], and CSN5 with the Nedd8 cullin modification [6]. Although, it is possible that through these interactions, CSNAP modulates CSN-CRL interactions, our results show that even in the absence of a substrate receptor CSNAP impacts CSN/Cul1–Rbx1. Thus, unraveling the precise structural contribution of CSNAP to CSN-CRL binding affinity awaits high-resolution structural analyses; however, it is reasonable to speculate that CSNAP shifts the CSN conformational equilibrium toward low affinity states.
Given that CRLs are involved in regulating numerous cellular processes, including cell-division cycle and cellular proliferation, the correlation between aberrant CRL function and cancer is not surprising, making this system an attractive target for therapeutic intervention (reviewed in refs. [45–47]). For example Cul4/CRBN has been implicated as the target of the anti-myeloma agent lenalidomide [48], and the neddylation inhibitor MLN4924 is an anti-cancer drug currently in clinical trials [4]. As a direct regulator of CRLs, the CSN constitutes another objective for drug development, with a major focus on inhibiting CSN5, the catalytic subunit [49–54]. Our latest findings lead us to propose CSNAP as a new therapeutic avenue. Preventing CSNAP integration within the CSN complex would be expected to impair cell cycle progression and the adaptive response to oncogenic stress conditions.
Materials and methods
Cell cultures, transfections, and UV-C exposure
HAP1 WT and ΔCSNAP CRISPR cell lines were purchased from Haplogene GmbH, Austria, and cultured in a humidified CO2 incubator at 37 °C in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 10% fetal calf serum, penicillin–streptomycin, and Mycozap (Lonza). As previously reported [55], the HAP1 haploid-enriched cells diploidized over several weeks of passage as shown in Table S10. HAP1 cells were transfected with phyg-CSNAP-Cerulean, phyg-ΔN-CSNAP-Cerulean, or phyg-FBXL15-FLAG, using the JetPrime reagent (Polyplus). Cerulean-expressing cell lines were isolated and sorted for low-medium expression levels by fluorescence-activated cell sorting (FACSAria Fusion; BD Biosciences), and expanded in complete IMDM. For UV treatments, plates were washed twice with PBS, and after removal of the liquid, were illuminated with UV-C light (1–20 J/m2).
Immunoprecipitation and FLAG-pull down
For immunoprecipitation experiments, HAP1 cells were lysed in 50 mM Tris pH 7.4, 150 mM NaCl, 0.5% NP40, phosphatase inhibitors (5 mM Na-o-vanadate, 4 mM Na-pyrophosphate, and β-glycerophosphate), and protease inhibitors (1 mM PMSF, 1 mM benzamidine, and 1.4 μg/ml pepstatin A). In all, 0.25–1 mg total protein was incubated with 10 μl anti-CSN3 (Abcam ab79398), anti-DDB2 (Santa Cruz sc-81246) or 35 μl anti-FLAG resin (Sigma A2220) overnight. For immunoprecipitation of the CSN3 or DDB2 antibody 35 μl protein G sepharose slurry was added for 1 h. Bound proteins were washed and eluted with 2× Laemmli sample buffer. Chromatin-bound proteins were purified as previously described [29], using 50 μg/ml digitonin instead of NP40 in the hypotonic lysis buffer. 250 μg of chromatin bound fraction was suspended in 250 μl TBS and rotated overnight at 4 °C with 5 μl of anti-DDB2 (Santa Cruz sc-81246). Then 30 μl of TBS equilibrated Protein G Sepharose resin (GE) was added for 1 h, and after three washes of 300 μl, TBS-bound proteins were eluted in 35 μl 2× Laemmli sample buffer.
Fluorescence assays
Purification of recombinant CSNΔCSNAP complexes was performed as described in ref. [7]. The production of CSN and CSN5H138A involved the generation of a pFBDM vector containing CSN1/His6-CSN5/CSN2/StrepII2x-CSN3/CSNAP or CSN1/His6-CSN5H138A/CSN2/StrepII2x-CSN3/CSNAP, respectively. Baculoviruses produced from each of these vectors were used to coinfect HighFive insect cells with a baculovirus expressing CSN4/CSN7b/CSN6/CSN8 to produce the full complexes. The fluorescent assays to determine the affinity of the CSN complexes for Cul1-dansyl/Rbx1 variants and their deneddylation activity were performed as described in ref. [10].
Western blots
Proteins were separated on 12 or 9% SDS-PAGE, and transferred to PVDF membranes. Primary antibodies used for detection: anti-CSN1 (Enzo PW8285), anti-CSN2 (Abcam ab10462), anti-CSN3 (ab79398), anti-CSN5 (ab495 and ab118841), anti-CSN6 (PW 8295), anti-CSN8 (BML-PW8290), anti-PDCD4 (ab80590), anti-cullin1 (ab75817), anti-cullin2 (ab166917), anti-cullin3 (ab75851), anti-cullin4AB (ab76470), anti-cullin5 (ab184177), anti-DDB1 (Bethyl A300-462A), anti-DDB2 (ab181136), anti-PARP1 (sc-8007), anti-FLAG (Sigma F3165), anti-pChk1 (Cell signaling 2341), anti-GAPDH (Millipore MAB374), anti-vimentin (ab92547), anti-NQO1 (ab28947), anti-tubulin (ab184613), anti-gH2AX pSer139 (Cell Signaling #2577), and anti-histone 3 (ab24834). All western blot analyses were repeated at least three times.
Comet assay
Alkaline single-cell electrophoresis was performed according to the protocol from Trevigen. HAP1 cells were treated with 20 J/m2 UV-C light, and 6 h post-exposure cells were trypsinized, counted, and suspended in ice-cold PBS (-Ca/-Mg) to a density of 2 × 105 cells/ml. Fifty microliters of cell suspension were mixed with 450 μl LM-agarose (Trevigen), and 50 μl of the mix was pipetted onto the comet slide, and incubated in the dark at 4 °C for 30 min. Slides were immersed in a lysis solution (Trevigen) for 1 h at 4 °C, and then equilibrated to an alkaline electrophoresis solution (300 mM NaOH and 2 mM EDTA, pH > 13) for 20 min at room temperature. Slides were run at 1 V/cm (~300 mA constant) in ice-cold alkaline electrophoresis solution for 30 min and then neutralized for 5 min in 400 mM Tris pH 7.5, rinsed in distilled water, immersed in 70% ethanol for 5 min, and dried at room temperature. DNA was stained with SYBR Gold (Invitrogen); the slides were then dried completely, prior to imaging. Images were acquired using an inverted Nikon microscope (Eclipse Ti, Nikon, Japan) using a 20× objective, and with a cooled electron-multiplying charge-coupled device camera (iXon Ultra, Andor, Ireland). Comet parameters were analyzed using the CASP comet software. At least 74 cells were analyzed per sample, in three biological samples, in duplicates. Within each biological replicate, the proportions of Olive tail moments larger than 3 were calculated in both ΔCSNAP and WT samples, and were compared using a two-way ANOVA, accounting for group and batch.
Colony-forming assay
Untreated or 20 J/m2 UV-C exposed WT and ΔCSNAP cells were trypsinized, counted, and plated in 10 cm tissue culture dishes. For untreated cells, 100 cells, and for UV-illuminated cells, 5000 cells were plated per dish. All experiments were done in triplicates. Dishes were incubated in normal growth conditions for 8 days. The plates were then washed twice with PBS, dried, and stained with 0.15% Crystal violet in methanol for 3 min, rinsed with tap water, and air-dried before scanning. Colony counts were measured using OpenCFU software.
Cell cycle analysis
Cells were synchronized to G1/S phase using double thymidine block as previously described [29]. UV treated cells were exposed to 5 J/m2 UV-C at release, and fixed with ethanol at different time points. Cell cycle phases were assessed by flow cytometry (LSRII, BD Biosciences) following propidium iodide staining. Asynchronous cells were analyzed using propidium iodide and BrdU double staining. In total, 105 cells were denatured after fixation using 2 N HCl, 0.5% Triton X-100 in PBS for 30 min, neutralized in 0.1 M Na2B4O7, pH 8.5, and incubated with 5 μl of anti-BrdU-FITC (eBioscience 11-5071-41) in 1% BSA, 0.5% Triton X-100 in PBS for 1 h. Cells were washed in 1% BSA in PBS, resuspended in PBS containing 50 μg/ml propidium iodide and 50 μg/ml RNase A, and analyzed in FACSAria Fusion flow cytometer (BD Biosciences).
Measurement of viable and dead cell populations
Determination of percentage of live, early apoptotic, and late apoptotic cells in WT and ΔCSNAP cultures was performed using Annexin V-FITC Apoptosis Detection Kit (MEBCYTO Apoptosis kit, MBL#4700) by flow cytometry. Single cells (20,000 per sample) were analyzed for AnnexinV-FITC and propidium iodide fluorescence. Early apoptotic cells (annexin V positive, PI negative), late apoptotic/necrotic/dead cells (annexin V positive, PI positive), and live/viable cells (annexin V negative, PI negative) were gated and quantified.
Resazurin assay
WT, ΔCSNAP, ΔCSNAP-Cerulean, and ΔCSNAP-ΔC-Cerulean expressing cells were trypsinized, counted, and seeded in four replicates at a cell density of 5000 cells/well in 24 well plates. Cells in one plate were seeded directly to 30 μg/ml resazurin containing growth medium, and fluorescence intensity (540/600 nm) was measured after 2 h, for initial proliferation value. The other plates were either UV-exposed at 20 J/m2 24 h after seeding or left untreated. Immediately, or 2 h post UV, the growth medium was changed to 30 μg/ml resazurin containing medium, incubated for 2 h, and fluorescence was measured. Proliferation was calculated at each time point normalizing to the initial proliferation value.
SILAC
HAP1 cells were grown in SILAC IMDM (Invitrogen) with 10% dialyzed fetal calf serum (Biological Industries, 04-011-1 A) supplemented with 25 μg/ml light L-lysine and L-arginine (Sigma) or 25 μg/ml heavy L-lysine (L-Lys8-CNLM-291-H-1, Cambridge Isotopes) and L-arginine (L-Arg10-CNLM-539-H-1, Cambridge Isotopes) each, and labeling was swapped between WT and ΔCSNAP cells. Cells were incubated with 5 μM MG132 for 4 h before harvesting. Samples were prepared, as previously described [56]. Briefly, the samples were lysed using 8 M urea, mixed at a 1:1 protein:protein ratio, and digested with trypsin, followed by a desalting step. The resulting peptides were fractionated offline using high pH-reversed phase chromatography, followed by enrichment for K-ε-GlyGly using the Cell Signaling PTMScan® Ubiquitin Remnant Motif (K-ε-GG) Kit #5562 (antibody-based). Each fraction was then analyzed, using online nanoflow liquid chromatography (nanoAcquity) coupled to high-resolution, high-mass accuracy mass spectrometry (Fusion Lumos). Raw data was processed with MaxQuant v1.5.5.1. The data was searched with the Andromeda search engine against the human proteome database appended with common lab protein contaminants, and allowing for GG modifications of lysines. The ratio of H/L (heavy to light) ratio was calculated, and results were log-transformed. The datasets of four (two label swap) experiments were combined in the way, that the experiment with the largest number of proteins identified was merged with proteins that had data for the same proteins and modification sites from the three other experiments. Genes corresponding to proteins that showed fold change above 1.5 or below 0.66 in each of the four experiments were filtered, and were selected only if appeared at least three out of the four experiments (from nonunique proteins only the first in the gene list was included). The resulting protein/gene list was analyzed using EnrichR/Reactome 2016 or Webgestalt/Reactome 2016 for overrepresentation, and the results were filtered using a cutoff of the adjusted p value or FDR < 0.05. The ratio of H/L (heavy to light) ratio was calculated, and results were log-transformed.
Label-free quantitation
WT and ΔCSNAP cells were lysed in 50 mM Tris, pH 7.4, 150 mM NaCl, and 0.5% NP40, supplemented with phosphatase and protease inhibitors as described above. One milligram of total protein was used for immunoprecipitation, using anti-CSN3 antibody (ab79698) as described above in three biological replicates. Proteins were eluted by 75 μl of 0.1 M glycine-HCl, pH 2.5. The beads were washed in 25 mM Tris pH7.4, 150 mM NaCl (TBS), and subjected to on-bead tryptic digestion as follows: 8 M urea in 0.1 M Tris pH 7.9, was added onto TBS washed beads, and incubated for 15 min at room temperature. Proteins were reduced by incubation with dithiothreitol (5 mM, Sigma) for 60 min at room temperature, and alkylated with 10 mM iodoacetamide (Sigma) in the dark for 30 min at room temperature. Urea was diluted to 2 M with 50 mM ammonium bicarbonate. Trypsin (250 ng, Promega, Madison, WI, USA) was added and incubated overnight at 37 °C, followed by addition of 100 ng trypsin for 4 h at 37 °C [11–18]. Digestions were stopped by addition of trifluoroacetic acid (1% final concentration). Following digestion, peptides were desalted using Oasis HLB μElution format (Waters, Milford, MA, USA), vacuum-dried, and stored at −80 °C until further analysis.
ULC/MS grade solvents were used for all chromatographic steps. Each sample was loaded using splitless nano-Ultra Performance Liquid Chromatography (UPLC) (10 kpsi nanoAcquity, Waters). The mobile phase was: (A) H2O + 0.1% formic acid and (B) acetonitrile + 0.1% formic acid. Sample desalting was performed online, using a reversed-phase symmetry C18 trapping column (180 µm internal diameter, 20 mm length, 5 µm particle size, Waters). The peptides were then separated using a T3 HSS nanocolumn (75 µm internal diameter, 250 mm length, 1.8 µm particle size, Waters) at 0.35 µl/min. Peptides were eluted from the column into the mass spectrometer, using the following gradient: 4 to 30% B for 55 min, 30 to 90% B for 5 min, maintained at 90% for 5 min, and then back to initial conditions. The nano-UPLC was coupled online through a nanoESI emitter (10 μm tip, New Objective, Woburn, MA, USA) to a quadrupole orbitrap mass spectrometer (Q Exactive Plus, Thermo Scientific), using a FlexIon nanospray apparatus (Proxeon).
Data was acquired in data-dependent acquisition (DDA) mode, using a Top20 method. MS1 resolution was set to 70,000 (at 400 m/z), mass range of 300–1650 m/z, AGC of 3e6, and maximum injection time was set to 20 ms. MS2 resolution was set to 17,500, quadrupole isolation 1.7 m/z, AGC of 1e6, dynamic exclusion of 30 s, and maximum injection time of 60 ms. Raw data was imported into Expressionist® software version 9.1.3 (Genedata), and processed as described here. The software was used for retention time alignment and peak detection of precursor peptides. A master peak list was generated from all MS/MS events, and sent for database searching using Mascot v2.5.1 (Matrix Sciences). Data was searched against the human sequences UniprotKB (http://www.uniprot.org/), appended with the CSNAP sequence and common laboratory contaminant proteins. Fixed modification was set to carbamidomethylation of cysteines, and variable modifications were set to oxidation of methionines and deamidation of N or Q. Search results were then filtered using the PeptideProphet algorithm, to achieve a maximum false discovery rate of 1% at the protein level. Peptide identifications were imported back to Expressionist to annotate identified peaks. Quantification of proteins from the peptide data was performed, using an in-house script. Data was normalized, based on the total ion current. Protein abundance was obtained by summing the three most intense, unique peptides per protein. A Student’s t-test, after logarithmic transformation, was used to identify significant differences (>1.5-fold) across the biological replica. Fold changes were calculated based on the ratio of arithmetic means of the case versus control samples.
Total proteome analysis and bioinformatics
WT and ΔCSNAP cells were lysed in SDT buffer (4% SDS, 100 mM Tris/HCl pH 7.6, and 0.1 M dithiothreitol) and subjected to tryptic digestion, using a FASP™ Protein Digestion Kit (Expedeon). The resulting peptides were desalted and analyzed on the LC-MS instrument (Q-Exactive Plus) in DDA mode. The raw data was processed in Expressionist by Genedata, using Mascot as the search engine against the uniprot human proteome database, and common protein contaminants. Identifications were filtered to a maximum of 1% FDR on both the peptide and protein levels. Protein inference was performed by an in-house script. Overall, about 4000 proteins were identified and quantified. Proteomics data, after logarithmic transformation and flooring, were analyzed by two-way ANOVA using two factors, strain and UV treatment, as well as their interaction. Proteins with a p value of <0.05 and an absolute fold change > 1.5 were considered to be differentially expressed. The proteins were filtered to keep those that had an absolute fold change of at least 1.5 and a p value of <0.05 in at least one of the following pairwise comparisons: 1. WT UV/WT untreated; 2. ΔCSNAP UV/ΔCSNAP untreated; 3. ΔCSNAP untreated/ WT untreated; 4. ΔCSNAP UV/WT UV. The log intensities of the 347 proteins that passed these criteria (according to ANOVA analysis with all samples, after flooring was used). Intensities were clustered using the k-means algorithm, with Pearson dissimilarity as the distance measure to five clusters. Log2 intensities were standardized, so that each protein displayed zero mean and unit standard deviation. The proteins in each cluster could be obtained by filtering the Excel file. Enrichment analysis of the filtered protein list was performed using Webgestalt overrepresentation analysis using Reactome 2016 pathway as functional database against the protein coding database as reference set, and results were filtered for adjusted p value < 0.05.
All raw data, peak lists, and identifications were deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.
Native mass spectrometry
Nano-electrospray ionization MS experiments were performed on a modified Q Exactive Plus Orbitrap EMR [57]. Prior to MS analysis, 25 µl of the samples (~16 μM) were buffer-exchanged into 0.5 M ammonium acetate (pH 7), using Bio-Rad Biospin columns. Two microliters of the buffer-exchanged samples were mixed with 1 µl MeOH 40%, to reach a final concentration of 13%. Proteins were loaded into gold coated nano-ESI capillaries, prepared in house from borosilicate glass tubes [58]. For the reconstitution of the CSN complex, after buffer exchange, 2 µl of CSNΔCSNAP, were incubated for 3 h on ice with 2 µl of a synthetic CSNAP peptide, dissolved to 50 µM in 250 mM ammonium acetate. The conditions within the MS were adjusted to optimize signals of the intact CSN and preserve non-covalent interactions. The instrument was operated in positive mode at capillary voltage of 1.7 kV. Argon was used as the collision gas in the higher energy collision-induced dissociation cell. Resolution was set to 8750. Forevacuum was set to 1.5 mbar and the trapping gas was set to 3, corresponding to pressures of 8.8 × 10−5 and 1.7 × 10−10 mbar in the HV and UHV regions, respectively. Flatapole bias was set to transmission at 1.5 V. Bent flatapole and axial gradient were set to DC 2.2 V and 37.2 V, respectively. HCD cell bias was set to 150 V. Spectra are shown with no smoothing and without background subtraction.
Supplementary information
Acknowledgements
We thank Dieter A. Wolf for comments on the paper, and Dr. Ron Rotkopf for the statistical analysis. MS is grateful for the financial support of the US National Institutes of Health, grant no. GM121834 and for an Israel Science Foundation (ISF) grant 300/17. MS is the incumbent of the Aharon and Ephraim Katzir Memorial Professorial Chair. GF is the Incumbent of the David and Stacey Cynamon Research fellow Chair in Genetics and Personalized Medicine
Author contribution
MGF-L, GB-N, RIE, MP and MS designed the experiments and analyzed the data. MGF-L and IF performed the cell biology and biochemistry experiments. MGF-L and TMS performed the flow cytometry, and MGF-L and RN performed the microscopy experiments. GB-N performed the mass spectrometry and RIE the Kd measurement experiments. YL and MK performed the SILAC and label-free proteomics analysis. GF performed the bioinformatics analysis of the proteomics data. MGF-L, GB-N and MS wrote the paper.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by D. Guardavaccaro
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41418-019-0392-8) contains supplementary material, which is available to authorized users.
References
- 1.Hershko A, Ciechanover A, Varshavsky A. Basic medical research award. The ubiquitin system. Nat Med. 2000;6:1073–81. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 3.Enchev RI, Schulman BA, Peter M. Protein neddylation: beyond cullin-RING ligases. Nat Rev Mol Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–6. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 5.Skaar JR, Pagan JK, Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–81. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 7.Enchev RI, Scott DC, da Fonseca PC, Schreiber A, Monda JK, Schulman BA, et al. Structural basis for a reciprocal regulation between SCF and CSN. Cell Rep. 2012;2:616–27. doi: 10.1016/j.celrep.2012.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emberley ED, Mosadeghi R, Deshaies RJ. Deconjugation of Nedd8 from Cul1 is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J Biol Chem. 2012;287:29679–89. doi: 10.1074/jbc.M112.352484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fischer ES, Scrima A, Bohm K, Matsumoto S, Lingaraju GM, Faty M, et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–39. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 10.Mosadeghi R, Reichermeier KM, Winkler M, Schreiber A, Reitsma JM, Zhang Y, et al. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. eLife. 2016;5;e12102. [DOI] [PMC free article] [PubMed]
- 11.Reitsma JM, Liu X, Reichermeier KM, Moradian A, Sweredoski MJ, Hess S, et al. Composition and regulation of the cellular repertoire of SCF ubiquitin ligases. Cell. 2017;171:1326–39 e14. doi: 10.1016/j.cell.2017.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X, Reitsma JM, Mamrosh JL, Zhang Y, Straube R, Deshaies RJ. Cand1-mediated adaptive exchange mechanism enables variation in F-Box protein expression. Mol Cell. 2018;69:773–86 e6. doi: 10.1016/j.molcel.2018.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei N, Serino G, Deng XW. The COP9 signalosome: more than a protease. Trends Biochem Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 14.Wei N, Deng XW. The COP9 signalosome. Annu Rev Cell Dev Biol. 2003;19:261–86. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 15.Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–23. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 16.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–5. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 17.Rozen S, Fuzesi-Levi MG, Ben-Nissan G, Mizrachi L, Gabashvili A, Levin Y, et al. CSNAP Is a stoichiometric subunit of the COP9 signalosome. Cell Rep. 2015;13:585–98. doi: 10.1016/j.celrep.2015.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nat Protoc. 2013;8:1950–60. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 20.Emanuele MJ, Elia AE, Xu Q, Thoma CR, Izhar L, Leng Y, et al. Global identification of modular cullin-RING ligase substrates. Cell. 2011;147:459–74. doi: 10.1016/j.cell.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–9. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 22.Zheng N, Zhou Q, Wang Z, Wei W. Recent advances in SCF ubiquitin ligase complex: clinical implications. Biochim Biophys Acta. 2016;1866:12–22. doi: 10.1016/j.bbcan.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koren I, Timms RT, Kula T, Xu Q, Li MZ, Elledge SJ. The eukaryotic proteome is shaped by E3 ubiquitin ligases targeting C-terminal degrons. Cell. 2018;173:1622–35 e14. doi: 10.1016/j.cell.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 25.Shalit T, Elinger D, Savidor A, Gabashvili A, Levin Y. MS1-based label-free proteomics using a quadrupole orbitrap mass spectrometer. J Proteome Res. 2015;14:1979–86. doi: 10.1021/pr501045t. [DOI] [PubMed] [Google Scholar]
- 26.Meir M, Galanty Y, Kashani L, Blank M, Khosravi R, Fernandez-Avila MJ, et al. The COP9 signalosome is vital for timely repair of DNA double-strand breaks. Nucleic Acids Res. 2015;43:4517–30. doi: 10.1093/nar/gkv270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dubois EL, Gerber S, Kisselev A, Harel-Bellan A, Groisman R. UV-dependent phosphorylation of COP9/signalosome in UV-induced apoptosis. Oncol Rep. 2016;35:3101–5. doi: 10.3892/or.2016.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannss R, Dubiel W. COP9 signalosome function in the DDR. FEBS Lett. 2011;585:2845–52. doi: 10.1016/j.febslet.2011.04.027. [DOI] [PubMed] [Google Scholar]
- 29.Fuzesi-Levi MG, Ben-Nissan G, Bianchi E, Zhou H, Deery MJ, Lilley KS, et al. Dynamic regulation of the COP9 signalosome in response to DNA damage. Mol Cell Biol. 2014;34:1066–76. doi: 10.1128/MCB.01598-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci USA. 2006;103:9891–6. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–9. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 32.Gentile M, Latonen L, Laiho M. Cell cycle arrest and apoptosis provoked by UV radiation-induced DNA damage are transcriptionally highly divergent responses. Nucleic Acids Res. 2003;31:4779–90. doi: 10.1093/nar/gkg675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufmann SH, Desnoyers S, Ottaviano Y, Davidson NE, Poirier GG. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–85. [PubMed] [Google Scholar]
- 34.Soldani C, Scovassi AI. Poly(ADP-ribose) polymerase-1 cleavage during apoptosis: an update. Apoptosis. 2002;7:321–8. doi: 10.1023/a:1016119328968. [DOI] [PubMed] [Google Scholar]
- 35.Chaitanya GV, Steven AJ, Babu PP. PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal. 2010;8:31. doi: 10.1186/1478-811X-8-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113:357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 37.Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, et al. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell. 2013;153:206–15. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt MW, McQuary PR, Wee S, Hofmann K, Wolf DA. F-box-directed CRL complex assembly and regulation by the CSN and CAND1. Mol Cell. 2009;35:586–97. doi: 10.1016/j.molcel.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Zhou Y, Tang R, Wang X, Hu Q, Wang Y, et al. Increasing the unneddylated Cullin1 portion rescues the CSN phenotypes by stabilizing adaptor modules to drive SCF assembly. Mol Cell Biol. 2017;37:e00109–17. doi: 10.1128/MCB.00109-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–65. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott DC, Schulman BA. SCF E3 ligase substrates switch from CAN-D to can-ubiquitylate. Mol Cell. 2018;69:721–3. doi: 10.1016/j.molcel.2018.02.019. [DOI] [PubMed] [Google Scholar]
- 42.Scherer PC, Ding Y, Liu Z, Xu J, Mao H, Barrow JC, et al. Inositol hexakisphosphate (IP6) generated by IP5K mediates cullin-COP9 signalosome interactions and CRL function. Proc Natl Acad Sci USA. 2016;113:3503–8. doi: 10.1073/pnas.1525580113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lingaraju GM, Bunker RD, Cavadini S, Hess D, Hassiepen U, Renatus M, et al. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–5. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 44.Cavadini S, Fischer ES, Bunker RD, Potenza A, Lingaraju GM, Goldie KN, et al. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature. 2016;531:598–603. doi: 10.1038/nature17416. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y, Sun Y. Cullin-RING ligases as attractive anti-cancer targets. Curr Pharm Des. 2013;19:3215–25. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Z, Liu P, Inuzuka H, Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–47. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kitagawa K, Kitagawa M. The SCF-type E3 ubiquitin ligases as cancer targets. Curr Cancer Drug Targets. 2016;16:119–29. doi: 10.2174/1568009616666151112122231. [DOI] [PubMed] [Google Scholar]
- 48.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science. 2014;343:305–9. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schlierf A, Altmann E, Quancard J, Jefferson AB, Assenberg R, Renatus M, et al. Targeted inhibition of the COP9 signalosome for treatment of cancer. Nat Commun. 2016;7:13166. doi: 10.1038/ncomms13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YH, Judge AD, Seo D, Kitade M, Gomez-Quiroz LE, Ishikawa T, et al. Molecular targeting of CSN5 in human hepatocellular carcinoma: a mechanism of therapeutic response. Oncogene. 2011;30:4175–84. doi: 10.1038/onc.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pulvino M, Chen L, Oleksyn D, Li J, Compitello G, Rossi R, et al. Inhibition of COP9-signalosome (CSN) deneddylating activity and tumor growth of diffuse large B-cell lymphomas by doxycycline. Oncotarget. 2015;6:14796–813. doi: 10.18632/oncotarget.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Altmann E, Erbel P, Renatus M, Schaefer M, Schlierf A, Druet A, et al. Azaindoles as zinc-binding small-molecule inhibitors of the JAMM Protease CSN5. Angew Chem Int Ed Engl. 2017;56:1294–7. doi: 10.1002/anie.201608672. [DOI] [PubMed] [Google Scholar]
- 53.Cope GA, Deshaies RJ. Targeted silencing of Jab1/Csn5 in human cells downregulates SCF activity through reduction of F-box protein levels. BMC Biochem. 2006;7:1. doi: 10.1186/1471-2091-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lauinger L, Li J, Shostak A, Cemel IA, Ha N, Zhang Y, et al. Thiolutin is a zinc chelator that inhibits the Rpn11 and other JAMM metalloproteases. Nat Chem Biol. 2017;13:709–14. doi: 10.1038/nchembio.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Essletzbichler P, Konopka T, Santoro F, Chen D, Gapp BV, Kralovics R, et al. Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 2014;24:2059–65. doi: 10.1101/gr.177220.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, et al. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics. 2013;12:825–31. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ben-Nissan G, Belov ME, Morgenstern D, Levin Y, Dym O, Arkind G, et al. Triple-stage mass spectrometry unravels the heterogeneity of an endogenous protein complex. Anal Chem. 2017;89:4708–15. doi: 10.1021/acs.analchem.7b00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirshenbaum N, Michaelevski I, Sharon M Analyzing large protein complexes by structural mass spectrometry. J Vis Exp. 2010;e1954. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.