Avian metapneumoviruses (aMPVs), which have been reported in many countries, cause acute upper respiratory tract disease in chickens and turkeys. Using next-generation sequencing, we report here the complete genome sequence of an aMPV subtype B strain that was isolated from a turkey in Hungary in 1989.
ABSTRACT
Avian metapneumoviruses (aMPVs), which have been reported in many countries, cause acute upper respiratory tract disease in chickens and turkeys. Using next-generation sequencing, we report here the complete genome sequence of an aMPV subtype B strain that was isolated from a turkey in Hungary in 1989.
ANNOUNCEMENT
Avian metapneumoviruses (aMPVs) cause respiratory and reproductive disorders in poultry, most notably turkeys, chickens, and ducks, and are a major economic concern for the industry (1, 2). aMPV is a member of the genus Metapneumovirus within the subfamily Pneumovirinae of the family Paramyxoviridae and contains a negative-sense, nonsegmented RNA genome that is approximately 13.5 to 14 kb long (3, 4). Although only one serotype of aMPV has been described, four subtypes (A, B, C, and D) are recognized based on the levels of genetic and antigenic differences (5, 6). Recently, two distinct aMPVs, isolated from a monk parakeet (7) and a gull (8), were identified and proposed as new subtypes. aMPVs of subtypes A and B are present in Europe and many countries in the world, excluding the United States (9–13). aMPV-C is present mainly in the United States and has been observed to a limited degree in France, South Korea, and China (14–18). To date, aMPV-D has been observed only in France (5, 19). In this study, we report the complete genome of an aMPV-B strain from Hungary.
The aMPV strain Hungary/657/4, which was isolated from a turkey in Hungary in 1989, was obtained from the National Veterinary Services Laboratories of the Animal and Plant Health Inspection Service, U.S. Department of Agriculture (20, 21). The virus was propagated in Vero cells and purified as discussed previously (22). Viral RNA was isolated from the supernatant of virus-infected Vero cells using the QIAamp viral RNA minikit (Qiagen, USA), followed by DNase treatment with the TURBO DNA-free kit (Ambion, USA) to remove host DNA according to the manufacturer’s recommendations. Sequence-independent single-primer amplification (23) was used to produce random amplicons, which were processed using the Nextera XT DNA library preparation kit (Illumina, USA). The distribution size and concentration of the prepared libraries were checked with a Bioanalyzer 2100, using the high-sensitivity DNA kit (Agilent Technologies, Germany), and a Qubit fluorometer, using the double-stranded DNA (dsDNA) high-sensitivity assay kit (Life Technologies, USA), respectively. Next-generation paired-end sequencing (2 × 250 bp) was performed on an Illumina MiSeq instrument using the 500-cycle MiSeq reagent kit v2. The MiSeq run generated 1,209,676 total paired-end reads. A customized workflow on the Galaxy platform (24) was used to perform preprocessing and assembly of the raw sequencing reads, as described previously (25, 26). Briefly, raw read quality was assessed using FastQC v0.63 (27), and residual adapter sequences were trimmed using Cutadapt v1.6 (28). After host and control library reads were removed, overlapping read pairs were joined with PEAR v0.9.6. (29). Digital normalization via median k-mer abundance was performed using the khmer v1.1-1 package (cutoff, 100; k-mer size, 20) (30). De novo assembly was performed utilizing MIRA3 v0.0.1 (31) with default settings. The final genome consensus was called from the raw aMPV reads, which were aligned with the de novo-generated contig by BWA-MEM (32) mapping of trimmed but unnormalized reads to the genome scaffold. This consensus had an average depth of coverage of 49-fold, with a maximum of 456-fold. The complete genome sequence of the isolate designated Hungary/657/4 was 13,508 nucleotides (nt) long (100% genome coverage, based on the VCO3/60616 reference genome [GenBank accession number AB548428]) and had a GC content of 43.2%. Genome annotation was performed using Geneious v11.1.5 and was confirmed by alignment with published aMPV genomes, all with default settings. The genome has the typical genetic structure of all metapneumovirus strains and contains eight open reading frames (3′-N-P-M-F-M2-SH-G-L-5′), which encode nine proteins of 1,176 nt, 840 nt, 765 nt, 1,617 nt, 561/222 nt, 543 nt, 1,245 nt and 6,015 nt. BLAST comparison to the currently available full-length aMPV genome sequences showed the highest (99.92%) nucleotide identity with the pathogenic strain VCO3/60616 (33), belonging to subtype B (Fig. 1). The VCO3/60616 strain is from the seventh passage of the field strain 86004, which was isolated from a turkey affected by turkey rhinotracheitis in France in 1986 (34, 35). Detailed analysis showed that Hungary/657/4 had the same 18 nucleotide substitutions as the pathogenic strain VCO3/60616, distinguishing them from the attenuated vaccine strain VCO3/50, which was established by another 50 passages in Vero cells (33).
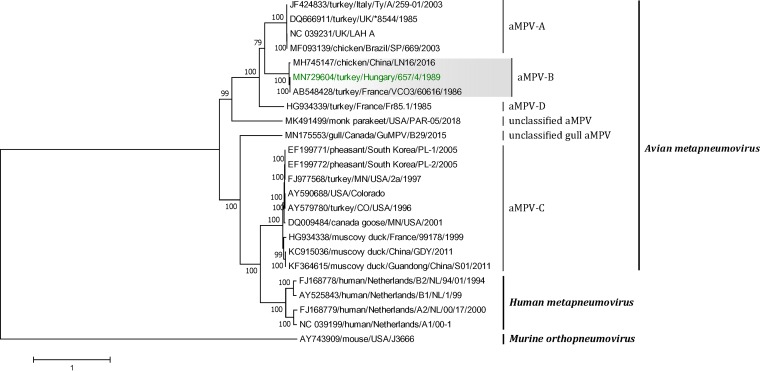
FIG 1.
Phylogenetic analysis of the aMPV-B Hungary/657/4 isolate within the genus Metapneumovirus, based on the complete genome sequences constructed with the maximum likelihood method using the general time reversible model in MEGA v7.0.26. The tree with the highest log likelihood (−113,969.82) is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 24 nucleotide sequences (sequences from Murine orthopneumovirus are included as an outgroup). All positions containing gaps and missing data were eliminated. There was a total of 12,392 positions in the final data set.
Amino acid analysis showed that the fusion protein cleavage site contained 4 basic amino acids (99RKKR↓F102), which are conserved among all wild-type aMPV-B strains (35, 36). Currently, there are only two aMPV-B full-genome sequences available, from strains that were isolated in France in 1986 (GenBank accession number AB548428) (33) and in China in 2016 (GenBank accession number MH745147) (10). This complete genome sequence information is useful for in-depth understanding of the evolution of aMPV, as well as for planning vaccination strategies.
Data availability.
The complete genome sequence of aMPV-B Hungary/657/4 has been deposited in GenBank under accession number MN729604. Raw data were deposited in the SRA under accession number SRR10518066, BioSample number SAMN13354755, and BioProject number PRJNA590745.
ACKNOWLEDGMENTS
This study was supported by USDA CRIS project 6040-32000-072 and APHIS interagency agreement 60-6040-5-009.
The mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Brown PA, Allée C, Courtillon C, Szerman N, Lemaitre E, Toquin D, Mangart JM, Amelot M, Eterradossi N. 2019. Host specificity of avian metapneumoviruses. Avian Pathol 48:311–318. doi: 10.1080/03079457.2019.1584390. [DOI] [PubMed] [Google Scholar]
- 2.Jones RC, Rautenschlein S. 2013. Avian metapneumovirus, p 112–138. In Swayne DE. (ed), Diseases of poultry, 13th ed. Wiley-Blackwell, Ames, IA. [Google Scholar]
- 3.Rima B, Collins P, Easton A, Fouchier R, Kurath G, Lamb RA, Lee B, Maisner A, Rota P, Wang L, ICTV Report Consortium . 2017. ICTV virus taxonomy profile: Pneumoviridae. J Gen Virol 98:2912–2913. doi: 10.1099/jgv.0.000959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, Briese T, Brown PA, Bukreyev A, Balkema-Buschmann A, Buchholz UJ, Chabi-Jesus C, Chandran K, Chiapponi C, Crozier I, de Swart RL, Dietzgen RG, Dolnik O, Drexler JF, Dürrwald R, Dundon WG, Duprex WP, Dye JM, Easton AJ, Fooks AR, Formenty PBH, Fouchier RAM, Freitas-Astúa J, Griffiths A, Hewson R, Horie M, Hyndman TH, Jiāng D, Kitajima EW, Kobinger GP, Kondō H, Kurath G, Kuzmin IV, Lamb RA, Lavazza A, Lee B, Lelli D, Leroy EM, Lǐ J, Maes P, Marzano S-YL, Moreno A, Mühlberger E, Netesov SV, Nowotny N, Nylund A, Økland AL, Palacios G, Pályi B, Pawęska JT, Payne SL, Prosperi A, Ramos-González PL, Rima BK, Rota P, Rubbenstroth D, Shī M, Simmonds P, Smither SJ, Sozzi E, Spann K, Stenglein MD, Stone DM, Takada A, Tesh RB, Tomonaga K, Tordo N, Towner JS, van den Hoogen B, Vasilakis N, Wahl V, Walker PJ, Wang L-F, Whitfield AE, Williams JV, Zerbini FM, Zhāng T, Zhang Y-Z, Kuhn JH. 2019. Taxonomy of the order Mononegavirales: update 2019. Arch Virol 164:1967–1980. doi: 10.1007/s00705-019-04247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bäyon-Auboyer MH, Arnauld C, Toquin D, Eterradossi N. 2000. Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup. J Gen Virol 81:2723–2733. doi: 10.1099/0022-1317-81-11-2723. [DOI] [PubMed] [Google Scholar]
- 6.Seal BS. 2000. Avian pneumoviruses and emergence of a new type in the United States of America. Anim Health Res Rev 1:67–72. doi: 10.1017/s1466252300000062. [DOI] [PubMed] [Google Scholar]
- 7.Retallack H, Clubb S, DeRisi JL. 2019. Genome sequence of a divergent avian metapneumovirus from a monk parakeet (Myiopsitta monachus). Microbiol Resour Announc 8:e00284-19. doi: 10.1128/MRA.00284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canuti M, Kroyer ANK, Ojkic D, Whitney HG, Robertson GJ, Lang AS. 2019. Discovery and characterization of novel RNA viruses in aquatic North American wild birds. Viruses 11:768. doi: 10.3390/v11090768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tucciarone CM, Andreopoulou M, Franzo G, Prentza Z, Chaligiannis I, Cecchinato M. 2017. First identification and molecular characterization of avian metapneumovirus subtype B from chickens in Greece. Avian Dis 61:409–413. doi: 10.1637/11631-032017-CaseR. [DOI] [PubMed] [Google Scholar]
- 10.Yu M, Xing L, Chang F, Bao Y, Wang S, He X, Wang J, Wang S, Liu Y, Farooque M, Pan Q, Wang Y, Gao L, Qi X, Hussain A, Li K, Liu C, Zhang Y, Cui H, Wang X, Gao Y. 2019. Genomic sequence and pathogenicity of the first avian metapneumovirus subtype B isolated from chicken in China. Vet Microbiol 228:32–38. doi: 10.1016/j.vetmic.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 11.Franzo G, Tucciarone CM, Enache M, Bejan V, Ramon G, Koutoulis KC, Cecchinato M. 2017. First report of avian metapneumovirus subtype B field strain in a Romanian broiler flock during an outbreak of respiratory disease. Avian Dis 61:250–254. doi: 10.1637/11557-121216-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 12.Tucciarone CM, Franzo G, Lupini C, Alejo CT, Listorti V, Mescolini G, Brandão PE, Martini M, Catelli E, Cecchinato M. 2018. Avian metapneumovirus circulation in Italian broiler farms. Poult Sci 97:503–509. doi: 10.3382/ps/pex350. [DOI] [PubMed] [Google Scholar]
- 13.Rizotto LS, Scagion GP, Cardoso TC, Simão RM, Caserta LC, Benassi JC, Keid LB, Oliveira TMFS, Soares RM, Arns CW, Van Borm S, Ferreira HL. 2017. Complete genome sequence of an avian metapneumovirus subtype A strain isolated from chicken (Gallus gallus) in Brazil. Genome Announc 5:e00688-17. doi: 10.1128/genomeA.00688-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seal BS, Sellers HS, Meinersmann RJ. 2000. Fusion protein predicted amino acid sequence of the first US avian pneumovirus isolate and lack of heterogeneity among other US isolates. Virus Res 66:139–147. doi: 10.1016/s0168-1702(99)00133-1. [DOI] [PubMed] [Google Scholar]
- 15.Toquin D, Guionie O, Jestin V, Zwingelstein F, Allee C, Eterradossi N. 2006. European and American subgroup C isolates of avian metapneumovirus belong to different genetic lineages. Virus Genes 32:97–103. doi: 10.1007/s11262-005-5850-3. [DOI] [PubMed] [Google Scholar]
- 16.Lee E, Song MS, Shin JY, Lee YM, Kim CJ, Lee YS, Kim H, Choi YK. 2007. Genetic characterization of avian metapneumovirus subtype C isolated from pheasants in a live bird market. Virus Res 128:18–25. doi: 10.1016/j.virusres.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 17.Wei L, Zhu S, Yan X, Wang J, Zhang C, Liu S, She R, Hu F, Quan R, Liu J. 2013. Avian metapneumovirus subgroup C infection in chickens, China. Emerg Infect Dis 19:1092–1094. doi: 10.3201/eid1907.121126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lwamba HC, Alvarez R, Wise MG, Yu Q, Halvorson D, Njenga MK, Seal BS. 2005. Comparison of the full-length genome sequence of avian metapneumovirus subtype C with other paramyxoviruses. Virus Res 107:83–92. doi: 10.1016/j.virusres.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Sun S, Chen F, Cao S, Liu J, Lei W, Li G, Song Y, Lu J, Liu C, Qin J, Li H. 2014. Isolation and characterization of a subtype C avian metapneumovirus circulating in Muscovy ducks in China. Vet Res 45:74. doi: 10.1186/s13567-014-0074-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez R, Njenga MK, Scott M, Seal BS. 2004. Development of a nucleoprotein-based enzyme-linked immunosorbent assay using a synthetic peptide antigen for detection of avian metapneumovirus antibodies in turkey sera. Clin Diagn Lab Immunol 11:245–249. doi: 10.1128/cdli.11.2.245-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collins MS, Gough RE, Alexander DJ. 1993. Antigenic differentiation of avian pneumovirus isolates using polyclonal antisera and mouse monoclonal antibodies. Avian Pathol 22:469–479. doi: 10.1080/03079459308418936. [DOI] [PubMed] [Google Scholar]
- 22.Yu Q, Estevez C, Song M, Kapczynski D, Zsak L. 2010. Generation and biological assessment of recombinant avian metapneumovirus subgroup C (aMPV-C) viruses containing different length of the G gene. Virus Res 147:182–188. doi: 10.1016/j.virusres.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Chrzastek K, Lee DH, Smith D, Sharma P, Suarez DL, Pantin-Jackwood M, Kapczynski DR. 2017. Use of sequence-independent, single-primer-amplification (SISPA) for rapid detection, identification, and characterization of avian RNA viruses. Virology 509:159–166. doi: 10.1016/j.virol.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, Grüning B, Guerler A, Hillman-Jackson J, Von Kuster G, Rasche E, Soranzo N, Turaga N, Taylor J, Nekrutenko A, Goecks J. 2016. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 44:W3–W10. doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimitrov KM, Sharma P, Volkening JD, Goraichuk IV, Wajid A, Rehmani SF, Basharat A, Shittu I, Joannis TM, Miller PJ, Afonso CL. 2017. A robust and cost-effective approach to sequence and analyze complete genomes of small RNA viruses. Virol J 14:72. doi: 10.1186/s12985-017-0741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goraichuk IV, Dimitrov KM, Sharma P, Miller PJ, Swayne DE, Suarez DL, Afonso CL. 2017. Complete genome sequences of four avian paramyxoviruses of serotype 10 isolated from rockhopper penguins on the Falkland Islands. Genome Announc 5:e00472-17. doi: 10.1128/genomeA.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andrews S. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- 28.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 29.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crusoe MR, Alameldin HF, Awad S, Boucher E, Caldwell A, Cartwright R, Charbonneau A, Constantinides B, Edvenson G, Fay S, Fenton J, Fenzl T, Fish J, Garcia-Gutierrez L, Garland P, Gluck J, González I, Guermond S, Guo J, Gupta A, Herr JR, Howe A, Hyer A, Härpfer A, Irber L, Kidd R, Lin D, Lippi J, Mansour T, McA'Nulty P, McDonald E, Mizzi J, Murray KD, Nahum JR, Nanlohy K, Nederbragt AJ, Ortiz-Zuazaga H, Ory J, Pell J, Pepe-Ranney C, Russ ZN, Schwarz E, Scott C, Seaman J, Sievert S, Simpson J, Skennerton CT, Spencer J, Srinivasan R, Standage D, Stapleton JA, Steinman SR, Stein J, Taylor B, Trimble W, Wiencko HL, Wright M, Wyss B, Zhang Q, Zyme E, Brown CT. 2015. The khmer software package: enabling efficient nucleotide sequence analysis. F1000Res 4:900. doi: 10.12688/f1000research.6924.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. German Conf Bioinformatics 99:45–56. [Google Scholar]
- 32.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiyama M, Ito H, Hata Y, Ono E, Ito T. 2010. Complete nucleotide sequences of avian metapneumovirus subtype B genome. Virus Genes 41:389–395. doi: 10.1007/s11262-010-0518-z. [DOI] [PubMed] [Google Scholar]
- 34.Giraud P, Bennejean G, Guittet M, Toquin D. 1986. Turkey rhinotracheitis in France: preliminary investigations on a ciliostatic virus. Vet Rec 119:606–607. [PubMed] [Google Scholar]
- 35.Bäyon-Auboyer MH, Jestin V, Toquin D, Cherbonnel M, Eterradossi N. 1999. Comparison of F, G and N-based RT-PCR protocols with conventional virological procedures for the detection and typing of turkey rhinotracheitis virus. Arch Virol 144:1091–1109. doi: 10.1007/s007050050572. [DOI] [PubMed] [Google Scholar]
- 36.Yun B, Zhang Y, Liu Y, Guan X, Wang Y, Qi X, Cui H, Liu C, Zhang Y, Gao H, Gao L, Li K, Gao Y, Wang X. 2016. TMPRSS12 is an activating protease for subtype B avian metapneumovirus. J Virol 90:11231–11246. doi: 10.1128/JVI.01567-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The complete genome sequence of aMPV-B Hungary/657/4 has been deposited in GenBank under accession number MN729604. Raw data were deposited in the SRA under accession number SRR10518066, BioSample number SAMN13354755, and BioProject number PRJNA590745.