Abstract
Bacteria can withstand killing by bactericidal antibiotics through phenotypic changes mediated by their pre-existing genetic repertoire. These changes can be exhibited transiently by a large fraction of the bacterial population, giving rise to tolerance, or displayed by a small subpopulation, giving rise to persistence. Apart from undermining the use of antibiotics, tolerant and persistent bacteria foster the emergence of antibiotic-resistant mutants. Persister formation has been attributed to alterations in the abundance of particular proteins, metabolites, and signaling molecules including toxin-antitoxin modules, adenosine triphosphate (ATP), and guanosine (penta) tetraphosphate, respectively. Here, we report that persistent bacteria form as a result of slow growth alone, despite opposite changes in the abundance of such proteins, metabolites, and signaling molecules. Our findings argue that transitory disturbances to core activities, which are often linked to cell growth, promote a persister state regardless of the underlying physiological process responsible for the change in growth.
Introduction
Persister bacteria are small subpopulations of a clonal population that survive exposure to high concentrations of an antibiotic (1, 2). Genetically identical to the original susceptible population, persistent bacteria prevent the eradication of bacterial infections and favor the emergence of antibiotic-resistant mutants (3–7). The occurrence of persisters also hinders pathogen elimination by the host immune system. For example, upon internalization by macrophages, the facultative intracellular pathogen Salmonella enterica serovar Typhimurium forms a slow-replicating subpopulation that is both resistant to killing by ampicillin and protected from clearance by macrophages (8). Because antibiotic persistence has been reported in phylogenetically diverse bacterial species (9, 10), the underlying basis for this process is likely to be conserved across bacteria.
In Escherichia coli, persistence was initially attributed to a signaling pathway that relied on the RelA and SpoT proteins (11, 12), the synthase and a combined synthase and hydrolase of the alarmone guanosine (penta) tetraphosphate [(p)ppGpp], respectively (13). Stochastically synthesized, (p)ppGpp indirectly activates the Lon protease, which degrades antitoxins from toxin-antitoxin (TA) modules. This allows the released toxins to inhibit core cellular processes, such as protein synthesis, resulting in tolerance to multiple antibiotics (11). Similarly, Salmonella has been reported to form persisters within 30 min of phagocytosis by murine macrophages through a pathway that depends on acidification of the Salmonella-containing vacuole, the synthesis of (p)ppGpp, and the resulting expression of genes encoding toxins from TA modules (14). In support of this model, mutants unable to synthesize (p)ppGpp or lacking the genes for particular TA modules are defective in persister formation, and this is also the case for wild-type Salmonella if phagosome acidification is inhibited (14). In addition, during growth in Luria-Bertani (LB) medium, serine hydroxamate [(Shx), a serine analog that induces (p)ppGpp production], or a mildly acidic pH, a condition that partially mimics the environment Salmonella encounters inside an acidic macrophage phagosome (15), promotes formation of persisters and expression of TA modules (14), an event often associated with the release of TA toxins (16).
However, TA modules were eventually shown to be dispensable for persister formation in both E. coli (17, 18) and Staphylococcus aureus (19). Moreover, the original report ascribing the formation of E. coli persisters to (p)ppGpp-mediated stochastic induction of TA modules was retracted (11, 20). Thus, an alternative model emerged suggesting a causal connection between a reduction in adenosine triphosphate (ATP) amounts and persistence. The latter model was supported by the low ATP amounts present in E. coli and S. aureus persisters (18, 19). That persister formation requires (p)ppGpp, TA modules and acidified LB broth in Salmonella (14) but not in the related Gram negative species E. coli, which shares this property with the distantly-related Gram positive S. aureus (17–19), raised the possibility of persister formation actually differing across bacterial species, and prompted us to reexamine persister formation in Salmonella.
Here, we report that growth rate is the ultimate indicator of persister formation in Salmonella, and most likely, in other bacterial species. That is, we establish that (p)ppGpp, TA modules and exposure to an acidified medium are all dispensable for persister formation in different media, demonstrating that Salmonella behaves like E. coli and S. aureus with regards to persister formation. We identify conditions in which antibiotic tolerance is achieved despite high ATP amounts, which rules out a low ATP concentration as being essential for this property. Our model accounts for the inextricable association between growth and the activity of core cellular processes, which are both the source of biosynthetic activity and the target of antibiotics. Moreover, it suggests potential avenues to eliminate persister organisms.
Results
Low cytoplasmic Mg2+ induces Salmonella tolerance to antibiotics independently of (p)ppGpp and TA modules
Three properties – slow growth, increased (p)ppGpp amounts, and low ATP abundance – have been implicated in persister formation and antibiotic tolerance (11, 18, 19, 21–23). Given that low cytoplasmic Mg2+ triggers slow growth, increased (p)ppGpp amounts, and low ATP abundance (24, 25), we wondered whether it also promotes persister formation or antibiotic tolerance. Thus, we examined bacterial survival to bactericidal antibiotics prior to and after a drop in cytoplasmic Mg2+ concentration that reduces growth, increases (p)ppGpp, and decreases ATP (i.e., 2 and 5 h in defined media containing 10 μM Mg2+, respectively) (25). Wild-type Salmonella grown in defined media with 10 μM Mg2+ for 5 h were ~10,000-fold less effectively killed by the cell wall inhibitor cefotaxime or the DNA gyrase inhibitor ciprofloxacin than were cells grown for 2 h in low Mg2+ (Fig. 1A and B). Because the media was at neutral pH, these results indicate that a mildly acidic pH is not required for tolerance to cefotaxime or ciprofloxacin in Salmonella.
Fig 1. Effects of low cytosolic Mg2+ on Salmonella antibiotic tolerance.
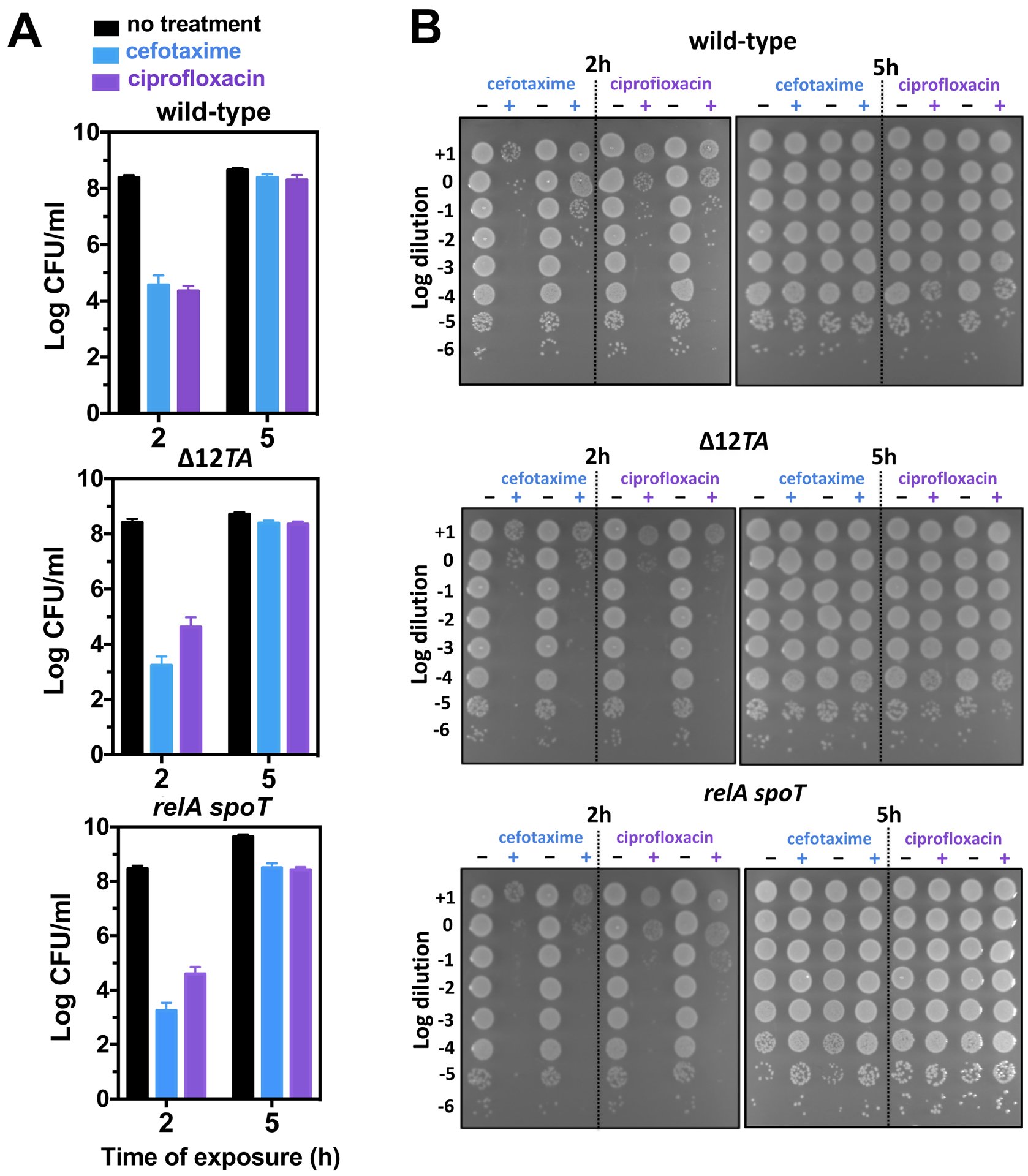
(A) Quantification of surviving wild-type (14028s), Δ12TA (MP1422), and relA::Tn10 spoT (MP342) Salmonella following 2.5 h exposure to cefotaxime (200 μg/mL) or ciprofloxacin (2 μg/mL) following 2 or 5 h of growth in low Mg2+ medium. Error bars represent standard deviations, derived from at least 8 biological samples and 3 independent experiments. Note log scale of y axis. (B) Representative dilution series of wild-type (14028s), Δ12TA (MP1422), and relA::Tn10 spoT (MP342) Salmonella following 2.5 h exposure to cefotaxime or ciprofloxacin following 2 or 5 h of growth in low Mg2+ medium. Each spot represents a 5 μL aliquot.
A Salmonella relA spoT double mutant, which is unable to produce (p)ppGpp (13), exhibited wild-type tolerance to cefotaxime and ciprofloxacin (Fig. 1A), and this was also the case for an engineered strain lacking all twelve bona fide TA modules (26) designated Δ12TA (Fig. 1B). Complete genome sequencing of the Δ12TA strain and its wild-type parental strain revealed the presence of a single genetic change in addition to the engineered mutations (see Materials and Methods). This mutation, consisting of a 125 bp deletion between the intergenic regions of the rrlB and rrfB genes, is not predicted to impact antibiotic tolerance. The wild-type parental strain and the Δ12TA mutant grow at similar rates (fig. S1A). These results indicate that (p)ppGpp and the 12 investigated TA modules were dispensable for antibiotic tolerance triggered by low cytoplasmic Mg2+ in Salmonella.
Acidic pH, (p)ppGpp, and TA modules are dispensable for persister formation in LB medium
The results presented in the previous section imply that either growth in defined media of low Mg2+ induces tolerance by a different mechanism from that taking place in acidified LB broth (14), or that tolerance promoted in LB broth does not actually require an acidic pH, (p)ppGpp and TA modules as reported (14). Therefore, we re-examined persister formation following growth in LB medium using the same experimental conditions and parental strain as those of the previous report (14).
The relA spoT double mutant and the Δ12TA mutant formed persisters in 5 to 8-fold lower amounts than wild-type Salmonella following a 24 h exposure to cefotaxime or ciprofloxacin (Fig. 2A). Moreover, a mildly acidic pH, which was previously reported to increase the levels of persisters by three orders of magnitude (14), did not affect the frequency of persisters (Fig. 2B). Furthermore, mutants lacking individual TA modules implicated in macrophage-induced persister formation (14) exhibited wild-type persistence when grown in LB broth (Fig. 2C). (Please note that the number of surviving bacteria reflects the number of persisters, as opposed to cells that acquired resistant mutations, because similar numbers of surviving cells were retrieved after a second antibiotic exposure (fig. S1B).) These results demonstrate that (p)ppGpp and TA modules are not essential for antibiotic persistence in Salmonella, and that an acidic pH does not increase the frequency of persisters formed in LB. Thus, they are in contrast to both the previous report on Salmonella (14) and a model in which E. coli persisters emerge through stochastic (p)ppGpp synthesis (11, 17, 19).
Fig 2. Effects toxin-antitoxin modules, (p)ppGpp biosynthetic and degradation genes, or acidic pH on Salmonella persistence.
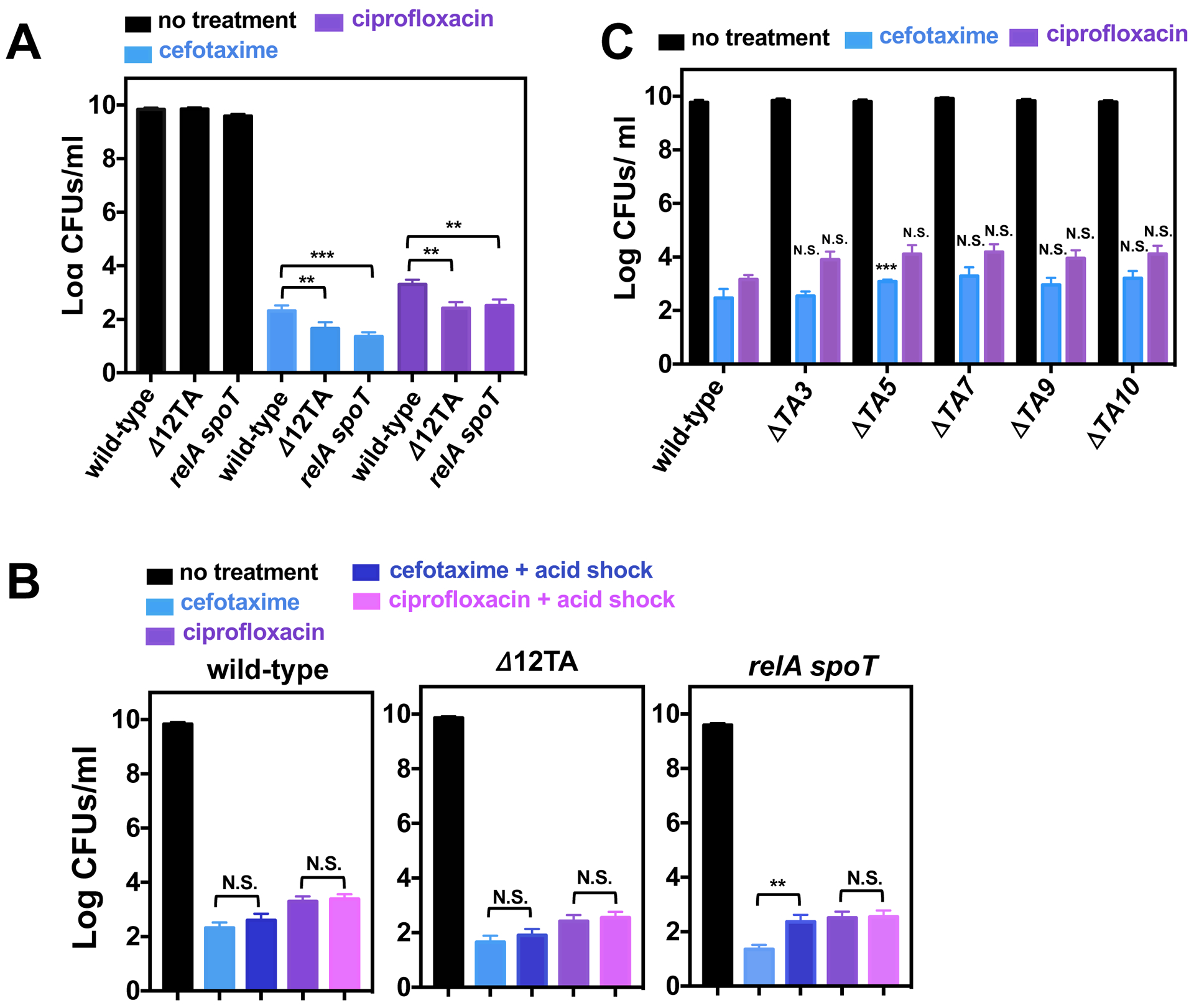
(A) Quantification of surviving wild-type (14028s), Δ12TA (MP1422), and relA::Tn10 spoT (MP342) Salmonella following 24 h exposure to cefotaxime or ciprofloxacin in LB medium. (B) Quantification of surviving wild-type (14028s), Δ12TA (MP1422), and relA::Tn10 spoT (MP342) Salmonella following acid shock (30 min in LB pH 4.5) prior to a 24 h exposure to cefotaxime or ciprofloxacin in LB medium. (C) Quantification of surviving wild-type (14028s), ΔTA3 (MP1454), ΔTA5 (MP1455), ΔTA7 (MP1456), ΔTA9 (MP1457), and ΔTA10 (MP1458) Salmonella following a 24 h exposure to cefotaxime or ciprofloxacin in LB medium. Error bars represent standard deviations (N = 8 biological replicates). Note log scale of y axis. For (A) and (B), two-tailed t-test between populations at the edge of brackets. For (C), two-tailed t-tests paired with wild-type populations: **p<0.01, ***p<0.001 and N.S. for no significance.
Triggering (p)ppGpp synthesis with serine hydroxamate (Shx) promotes transient antibiotic tolerance but does not affect persister frequency
The serine analog Shx was reported to promote persister formation in stationary phase cultures of Salmonella (14). This observation is puzzling for two reasons: first, Shx induces amino acid starvation by hindering charging of seryl-tRNA, which inhibits growth (27) and promotes (p)ppGpp synthesis through the RelA protein (28). However, we found that the (p)ppGpp-lacking relA spoT mutant forms persisters at a similar frequency as the wild-type strain (Fig. 2A). And second, logarithmically growing bacteria have active ribosomes that respond to Shx by synthesizing (p)ppGpp through the RelA protein (28). Therefore, Shx should promote tolerance in logarithmically growing bacteria, but not in stationary phase cultures because the latter, already nutrient starved, have high amounts of (p)ppGpp and reduced translation rates (27, 29–31).
As hypothesized, addition of Shx to bacteria in the stationary phase altered neither growth (Fig. 3A) nor tolerance (Fig. 3B and fig. S2). This is in contrast to Shx treatment of logarithmically growing Salmonella, which elicited a temporary growth arrest (Fig. 3A) during which bacteria were immune to killing by cefotaxime (Fig. 3B). We note that Shx did not increase persister frequency when added to either stationary or logarithmic growing bacteria (Fig. 3B and fig. S2).
Fig 3. Effect of Shx on bacterial growth and antibiotic tolerance.
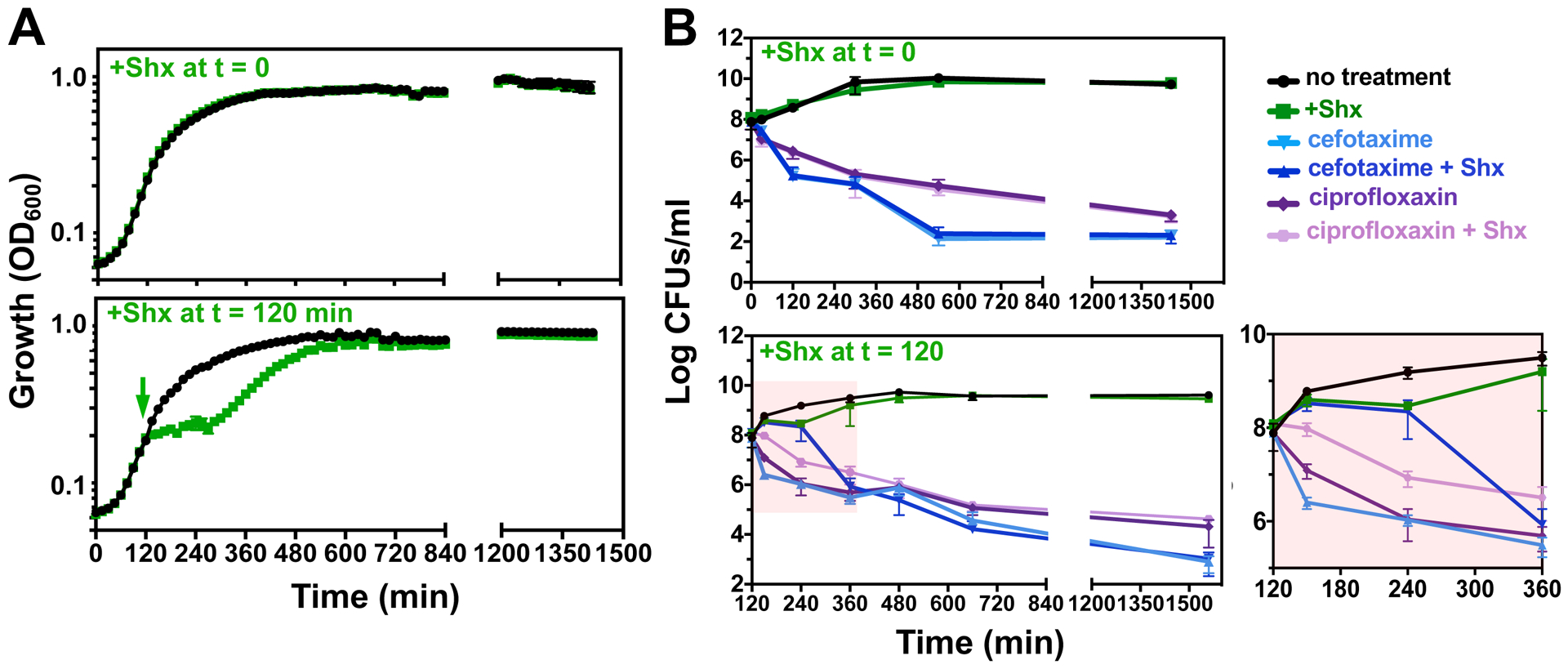
(A) Growth curves of wild-type Salmonella (14028s) in LB medium in the absence (black lines) or presence (green lines) of Shx (100 μg/mL). Shx was added for 30 min prior to outgrowth (top panel) or at 120 min of growth (bottom panel). (B) Concentration of viable bacteria (Log colony forming units (CFU)/mL) in cultures of wild-type Salmonella (14028s) gown as described in (A). Where indicated, bacteria were exposed to cefotaxime (200 μg/mL) or ciprofloxacin (2 μg/mL) for either 24 h (top panel) or 22 h (bottom panel). Error bars represent standard deviations (N = 6 biological replicates). Rightmost figure shows enlarged portion of left part of figure shaded in pink.
(In contrast to the complete inhibition of killing by cefotaxime (Fig. 3B), Shx only slowed down killing by ciprofloxacin when compared to the buffer control (Fig. 3B). This behavior likely reflects the inhibitory effects of (p)ppGpp on the initiation of chromosome replication (32) and pre-existing replication forks, which may be liable to the effects of the gyrase inhibitor ciprofloxacin.)
Altogether, the results presented above demonstrate that the transient growth inhibition caused by Shx in logarithmically-growing organisms increases antibiotic tolerance but does not impact persister frequency.
Growth inhibition promotes antibiotic tolerance despite increased ATP abundance
Wild-type Salmonella grown in 10 μM Mg2+ Mg2+ for 5 h is 10,000-fold more antibiotic tolerant (Fig. 1A and B) and harbors 15 times less ATP (25) than when grown in the same media for 2 h. These data support the inverse correlation between antibiotic tolerance and ATP concentration reported for E. coli (18, 22) and S. aureus (19). To independently test this correlation, we examined the behavior of wild-type Salmonella in which ATP abundance was decreased upon expression of the soluble subunit of the F1Fo ATPase using plasmid pATPase (33).
pATPase conferred high tolerance to both cefotaxime and ciprofloxacin (Fig. 4A and fig. S3A) whereas the vector control had no effect (Fig. 4A and fig. S2A). These results further support the inverse correlation between ATP abundance and survival to bactericidal antibiotics. However, they are also compatible with antibiotic tolerance resulting from a reduction in bacterial growth rate (34, 35), as opposed to ATP abundance per se. This is because bacteria are growing logarithmically at 2 h but linearly at 5 h (25), and also because a reduction in ATP abundance decreases growth rate (36) (fig. S1A).
Fig 4. Effects of ATP depletion and inhibition of protein synthesis on antibiotic tolerance.
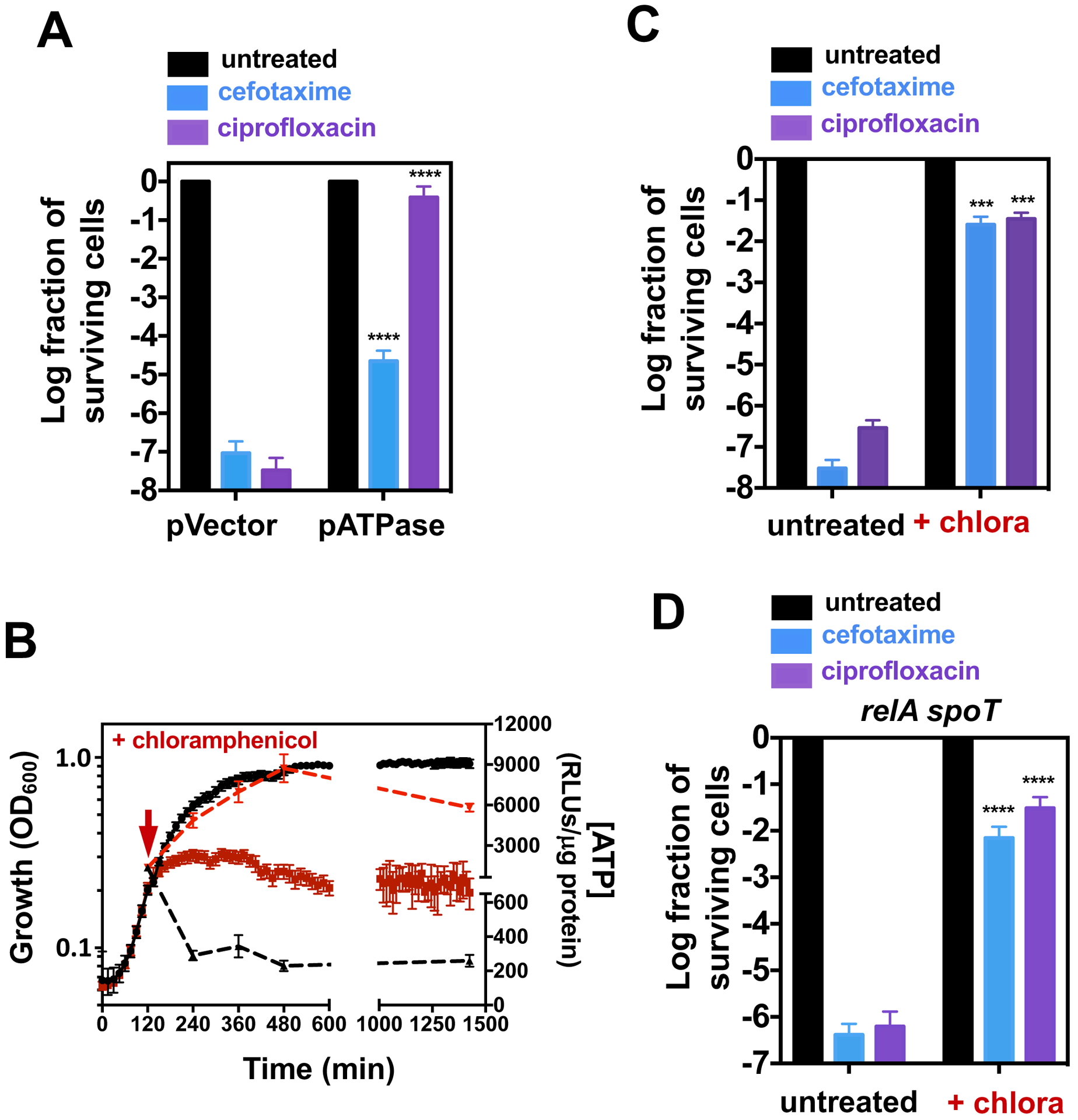
(A) Fraction of surviving wild-type Salmonella (14028s) carrying pATPase or the pVector control following 24 h exposure to cefotaxime (200 μg/mL) or ciprofloxacin (2 μg/mL). (B) Bacterial growth (left y-axis, solid lines) and ATP levels (right y-axis, dashed lines) of wild-type Salmonella (14028s) exposed to chloramphenicol at 120 min (red lines) or not exposed to chloramphenicol (black lines). Fraction of surviving (C) wild-type (14028s) or (D) relA::Tn10 spoT (MP342) Salmonella following cefotaxime or ciprofloxacin exposure in the presence or absence of chloramphenicol (50 μg/mL). Error bars represent standard deviations (N = 8 biological replicates). For (A), (C) and (D), note log scale of y axis. Wilcoxon rank-sum test between pVector and pATPase (A) or untreated and untreated populations (C) and (D): ***p<0.001, ****p<0.0001.
To determine whether the distinct antibiotic tolerance displayed by wild-type Salmonella harvested at 2 and 5 h in 10 μM Mg2+ reflected differences in growth rate versus ATP abundance, we investigated bacteria exposed to the bacteriostatic protein synthesis inhibitor chloramphenicol. Because protein synthesis is the activity that consumes most ATP in a cell (37), chloramphenicol treatment promoted a dramatic increase in ATP amounts (Fig. 4B) (38) that was accompanied by growth stasis (Fig. 4B) and a decrease in (p)ppGpp abundance (39).
Chloramphenicol rendered wild-type Salmonella highly tolerant to cefotaxime and ciprofloxacin (Fig. 4C and fig. S3B). This tolerance was still observed in the relA spoT mutant (Fig. 4D and fig. S3C), indicative that it is not mediated by (p)ppGpp. Thus, the pharmacological inhibition of protein synthesis leads to tolerance in Salmonella as observed in E. coli exposed to tetracycline (22), also a protein synthesis inhibitor. These data unequivocally demonstrate that bacteria exhibit high antibiotic tolerance despite having high amounts of ATP. We conclude that a reduction in bacterial growth rate promotes antibiotic tolerance.
To further explore the notion that conditions leading to growth slowdown or stoppage promote antibiotic tolerance, we examined the behavior of nutritional auxotrophs deprived of their required nutrients. First, we determined that a tryptophan auxotroph (trpEDCBA) is virtually immune to cefotaxime and ciprofloxacin in a medium lacking tryptophan (Fig. 5A). This mutant retained high cefotaxime tolerance in a relA spoT background (relA spoT trpA) (Fig. 5B and fig. S4B), indicating independence from (p)ppGpp. Second, the tolerance phenotype of the trpEDCBA mutant is due to growth stasis (as opposed to the tryptophan biosynthetic enzymes impacting tolerance by a mechanism unrelated to tryptophan synthesis), because the trpEDCBA mutant displayed antibiotic sensitivity when grown in the presence of tryptophan (Fig. 5A). And third, a mutant unable to catabolize the sugar mannose (i.e., manA) was rendered antibiotic tolerant when placed in media with mannose as the sole carbon source (Fig. 5A). This mutant lost its antibiotic tolerance when grown in glucose or in complex medium (Fig. 5A and fig. S4A). These results show that a variety of conditions promote antibiotic tolerance by slowing down bacterial growth.
Fig 5. Relationship between growth and antibiotic tolerance in nutritional auxotrophs and relA spoT strains.
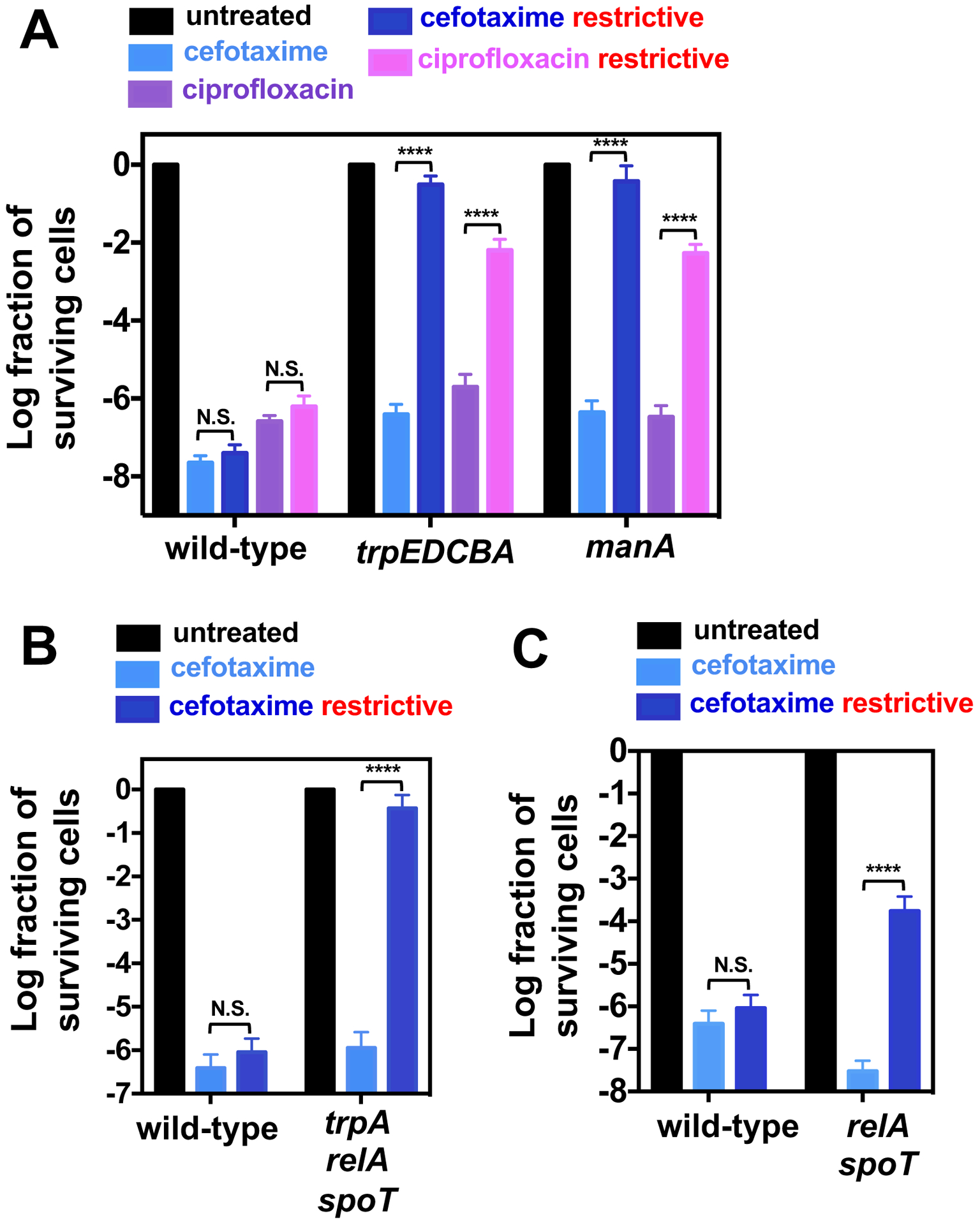
(A) Fraction of surviving wild-type (14028s), manA (MP50) and trpEDCBA::Tn10 (AS301) Salmonella following 24 h treatment with either cefotaxime (200 μg/mL) or ciprofloxacin (2 μg/mL) under growth-permissive and growth-restrictive conditions (see Materials and Methods). Fraction of surviving (B) wild-type (14028s) and relA::Tn10 spoT trpA::kan (MP1494), or (C) wild-type (14028s) and relA::Tn10 spoT (MP342) Salmonella following 24 h treatment with cefotaxime (200 μg/mL) under growth-permissive and growth-restrictive conditions (see Materials and Methods). Error bars represent standard deviations (N = 8 biological replicates). Note log scale of y axis. Wilcoxon rank-sum test between populations at the edge of brackets: ****p<0.0001 and N.S. for no significance.
(p)ppGpp inhibits antibiotic tolerance during restricted growth
(p)ppGpp plays a critical role during nutritional downshifts by promoting reallocation of cellular resources, which ensures balanced growth (27, 29, 30). Specifically, (p)ppGpp inhibits cellular processes favoring bacterial growth until the corresponding precursors become available. In downshift experiments, precursor availability is typically provided by the biosynthetic capability of the cell. Thus, although transiently inhibitory, (p)ppGpp does promote growth in the absence of certain nutrients, as evidenced by the fact that a relA spoT mutant displays multiple auxotrophies (13).
Given that growth inhibition promotes antibiotic tolerance in wild-type Salmonella, we postulated that a relA spoT mutant should display increased antibiotic tolerance under conditions in which growth is restricted. As hypothesized, the relA spoT mutant survived cefotaxime treatment better than wild-type Salmonella during growth in defined medium without amino acid supplementation (Fig. 5C and fig. S4C). These results identify a condition in which (p)ppGpp is actually detrimental to antibiotic tolerance.
Discussion
We herein report that a slowdown or stoppage in bacterial growth is a direct driver of antibiotic tolerance. Using an array of growth conditions and genetic contexts, we determined that antibiotic tolerance was inversely correlated with bacterial growth in Salmonella (Fig. 1, 3–5) (34, 35). In addition, we established that (p)ppGpp, TA modules, and acidic pH were all dispensable for persister formation during axenic growth in LB medium (Fig. 2 and Fig. 3). These results are in full agreement with those reported by others with E. coli (17, 18) and S. aureus (19).
The association between growth rate and antibiotic tolerance is supported by several independent findings. On the one hand, mutations in essential genes were historically isolated by enriching mutants that resisted killing by antibiotics because such mutants failed to replicate under particular conditions – i.e. in the absence of an essential nutrient, cellular precursor, or at non-permissive temperatures (40–43). On the other hand, multi-copy number plasmids expressing genes specifying proteins with unrelated biochemical functions render E. coli antibiotic tolerant by reducing bacterial growth (34, 35). These genes specify the chaperone DnaJ, the lipopolysaccharide-modifying PmrC, the transhydrogenase PntA, the sulfoquinovose isomerase YihS, the formate transport channel FocA, the transcriptional factor Zur, and YqjE, a putative protein of unknown function. Hence, a bacterium may become tolerant to antibiotics when the toxin of a TA module is active and reduces bacterial growth (23, 44, 45).
In the context of infection, naturally auxotrophic pathogens (46) may become tolerant when their required nutrients are scarce in host tissues. For instance, M. tuberculosis is unable to synthesize a number of nutrients, relying on dedicated transporters to obtain amino acids such as asparagine and aspartate from the host (47, 48). Because it occupies a variety of nutritional microenvironments during infection (49), M. tuberculosis replication may be hindered when experiencing limitation for asparagine, aspartate or another essential nutrient, rendering subpopulations of this pathogen antibiotic tolerant. Hence, although macrophage internalization leads to the formation of Salmonella persisters that are both antibiotic tolerant and resistant to killing by host immune cells (8, 14, 50, 51), we ascribe the antibiotic tolerance to the emergence of a Salmonella subpopulation exhibiting a much lower growth rate (8).
Because it slows down growth (36) (fig. S3A), a reduction in ATP amounts can lead to tolerance (18, 19, 22) (Fig. 4A and fig. S3A). However, a reduction in ATP amounts is not a causa sui. That is to say, ATP is critical in several cellular activities and alterations in ATP levels will inevitably result from changes in other physiological processes such as a metabolic switch, dissipation of membrane potential, etc (23, 52). Notably, the activity of ribosomal RNA (rRNA) P1 promoters – a phenotypic marker used to identify E. coli cells harboring low cytosolic ATP levels (18) – is also regulated by growth rate (31, 53, 54). Critically, the effects of ATP and growth rate on the rRNA P1 promoters can be uncoupled (55, 56). Therefore, low rRNA P1 transcription may reflect low ATP and/or slow growth.
It is presently unclear why others found a requirement for (p)ppGpp, TA modules and an acidic pH for persister formation in Salmonella (14) despite using the same experimental conditions and parental strain described in this paper. In fact, we established that (p)ppGpp actually reduces antibiotic tolerance under certain conditions (Fig. 5C). It is also puzzling that others reported a 100-fold increase in survival to antibiotics upon Shx treatment of bacteria in the stationary phase (14), when (p)ppGpp is abundant (27, 29–31). In light of the findings reported in this paper, it should be noted that (p)ppGpp does not impact persister formation in Mycobacterium smegmatis, a close relative of M. tuberculosis (57). Because of the disparate results obtained with Salmonella grown in laboratory media, and also because others found TA modules to be dispensable for persister formation in mice (51, 58), we chose not to re-examine the proposed requirement for (p)ppGpp, and TA modules in persister formation triggered when Salmonella is phagocytized by macrophages (14)
How, then, does a halt or decrease in bacterial growth rate promote the high degree of antibiotic tolerance reported in a broad range of bacterial species (9, 10)? Antibiotics target core cellular processes (2), and these processes are essential for growth. These cellular processes are normally coordinated through feedback mechanisms that ensure congruent macromolecular synthesis, balanced growth and survival (59–62). Therefore, a variety of conditions that inhibit bacterial growth may give rise to a persistence state by transiently disrupting housekeeping processes. Given the nature of various core processes, and the feedback mechanisms to which they are subjected during growth inhibition, it can be anticipated that variable frequencies of persisters may be recovered from exposure of bacteria to antibiotics that target distinct cellular processes (Fig. 1–5). For instance, although growth inhibition triggered by (p)ppGpp renders cells immediately immune to cell wall inhibitors, it will confer partial protection to DNA gyrase inhibitors, likely because active replication forks will not undergo inhibition (32) (Fig. 3B).
We propose that a condition will give rise to persistence if and only if it does not compromise a pre-existing feedback mechanism(s) and display bistability to allow the emergence of subpopulations (1, 2). That is, persistence normally arises from non-productive interactions among core cellular components, which are often enhanced when a bacterium experiences stress. In other words, there is no evidence of a dedicated “genetic program” driving formation of antibiotic persistent cells, which is in contrast to the role that specific genes play in genetic resistance to antibiotics (63).
The proposed model provides a parsimonious explanation for persistence, a phenomenon found not only among bacteria, but also in eukaryotes such as the phenotypically distinct cancer cells that are able to survive chemotherapy by entering into a quiescent state (64–70). In the context of bacterial persisters, our model accommodates the conclusions of a large number of studies, each ascribing a distinct underlying cause to persistence (18, 19, 21, 44, 45). Furthermore, it explains why certain antibiotic combinations increase bacterial survival (Fig 4B to 4D) (22, 71) whereas others (e.g., compounds that inhibit transcription and induce unspecific proteolysis) kill persister cells (72). That is, the latter drug combination likely disrupts a feedback control by degrading a combination of essential core proteins that cannot be resynthesized due to inhibition of transcription.
Materials and Methods
Microbial strains, plasmids and growth conditions
Microbial strains and plasmids used in this study are presented in Table S1. All S. enterica serovar Typhimurium strains are derived from strain 14028s (73) and were constructed by lambda-red-mediated recombination (74) followed by phage P22- mediated transduction as described (75). Bacterial strains used in transformation or recombination were grown in LB medium at 30°C or 37°C (74). For selection of recombinant mutants, LB medium was supplemented with ampicillin (100 μg/mL), chloramphenicol (20 μg/mL), tetracycline (20 μg/mL), kanamycin (50 μg/mL), apramycin (80 μg/mL), gentamycin (18 μg/mL), and/or spectinomycin (100 μg/mL). For antibiotic tolerance experiments, growth medium was supplemented with ampicillin (20 μg/mL), chloramphenicol (50 μg/mL), cefotaxime (200 μg/mL), ciprofloxacin (2 μg/mL), serine hydroxamate (Shx, 100 μg/mL). Antibiotic tolerance experiments were conducted in complex Luria Bertani (LB, Miller) liquid medium, or the following defined liquid medium formulations. For Mg2+ starvation experiments, cells were grown in MOPS minimal medium (76) lacking calcium and containing 10 mM and 2 mM K2HPO4 or 10 μM MgCl2 and 500 μM K2HPO4, 30 mM glucose, 0.1% casamino acids. For experiments involving tryptophan auxotrophs, cells were grown in MOPS minimal medium lacking calcium and containing 5 mM MgCl2, 2 mM K2HPO4, 30 mM glucose, amino acid mixture 1 (AA1MIX: 800 μM of arginine; 500 μM of methionine, histidine, valine, threonine and leucine; 250 μM of tyrosine and glycine), and the presence (permissive to growth) or absence (restrictive to growth) of 800 μM tryptophan. For experiments involving mannose catabolic mutant, cells were grown in MOPS minimal medium lacking calcium and containing 5 mM MgCl2, 2 mM K2HPO4, AA1MIX, 800 μM tryptophan, and 30 mM of either glucose (permissive to growth) or mannose (restrictive to growth). For experiments with strains containing relA spoT mutations cells were grown in LB medium (permissive to growth) or N-minimal medium (restrictive to growth) (24) with amino acid mixture 2 (AA2MIX: 100 μM methionine, histidine, valine, threonine, leucine, isoleucine) and 37 mM glycerol as the sole carbon source. In experiments involving strains harboring plasmid pATPase (33), cells were grown in MOPS minimal medium containing 5 mM MgCl2, 2 mM K2HPO4, 30 mM glucose, and 0.1% casamino acids. Expression of the ATPase was accomplished by addition of 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG). For all physiological experiments, bacterial were grown in 50 ml conical tubes containing 6 ml of culture medium. Cultures were incubated at 37°C in an orbital shaker (330 rpms).
Determination of colony forming units (CFU)
Following growth or exposure to cefotaxime or ciprofloxacin, cells were transferred to 5 ml microcentrifuge tubes and collected by centrifugation (13,500 × rpms for 20 min at 4°C). Cells were washed with 1.2 ml of ice-cold phosphate buffered saline, transferred to 1.5 ml microcentrifuge tubes and collected by centrifugation (14,000 × rpms for 20 min at 4°C). Finally, cells were resuspended in 500 μl LB medium, diluted and plated onto LB agar (1.5% w/v) plates.
Determination of minimal inhibitory concentration (MIC)
Two antibiotics with distinct modes of action were used in tolerance and persistence experiments: the inhibitor of cell wall biosynthesis cefotaxime, and the gyrase inhibitor ciprofloxacin. The MICs for ciprofloxacin and cefotaxime were determined in LB and MOPS minimal medium (containing 10 mM MgCl2, 2 mM K2HPO4) by serial dilution of antibiotic in 13 mm test tubes. Cultures were initiated in 1 ml of medium at a concentration of ≈ 1 × 107 CFU/ml and were allowed to grow at 37°C in an orbital shaker (330 rpms) for 24 h. The MIC was determined as the lowest concentration of antibiotic that prevented bacterial growth. In LB medium, the MIC for cefotaxime and ciprofloxacin were 25 μg/ml and 0.374 μg/ml, respectively. In MOPS medium MICs for cefotaxime and ciprofloxacin were 12.5 μg/ml and 0.187 μg/ml, respectively.
Mg2+ starvation and antibiotic exposure
Following overnight growth in MOPS medium containing 10 mM Mg2+ and 2 mM K2HPO4, cells were collected by centrifugation (14,000 × rpms for 1 min at room temperature), washed twice in MOPS medium containing no Mg2+ or phosphate and inoculated (1:100) in MOPS medium containing 10 μM MgCl2 and 500 μM K2HPO4. Cultures were grown for 2 or 5 h, aliquoted into separate tubes and grown for additional 2.5 h in the presence or absence of antibiotics.
Acid-shock treatment and antibiotic exposure
Stationary phase bacteria from overnight LB cultures were collected by centrifugation (14,000 × rpms for 1 min at room temperature), resuspended in fresh acidified LB (pH 4.5) or neutral (pH 7.2, control), and incubated for 30 min at 37°C. Cells were collected by centrifugation (14,000 × rpms for 1 min at room temperature), resuspended in fresh LB (pH 7.2) and diluted 1:200 into fresh LB (pH 7.2) containing antibiotics. Cultures were allowed to grow for 24 h prior to collection and estimation of CFU/ml.
Serine hydroxamate treatment and antibiotic exposure
Stationary phase bacteria from overnight cultures were collected by centrifugation (14,000 × rpms for 1 min at room temperature), resuspended in fresh LB containing 100 μg/ml Shx or LB alone (control) and incubated for 30 min at 37°C. (100 μg/ml of Shx is sufficient to induce a large increase in (p)ppGpp synthesis (25), inhibit growth (Fig. 4A) and elicit the global changes in transcription known as the stringent response (27)). Following incubation, cells were collected by centrifugation (14,000 × rpms for 1 min at room temperature), resuspended in fresh LB and diluted 1:200 in fresh LB containing antibiotics. For logarithmically growing cells, Shx was added to cultures following 120 min of growth. Following Shx addition, cultures were allowed to grow for an additional 15 min. Cultures were then subjected to a 24 h exposure to cefotaxime or ciprofloxacin prior to estimation of CFU/ml.
Growth restriction of tryptophan auxotroph and antibiotic exposure
Following overnight growth in minimal medium containing tryptophan, cells were diluted 1:200 into fresh medium and allowed to grow for 2 h. 3 ml of culture were collected on a 0.22 μM pore-size filter (Millipore), placed in a glass filter holder assembly with funnel, using a vacuum pump. Cells on the filter were vacuum-washed with 10 ml of dH2O and filters were placed on a 50 ml conical tube containing 6 ml of fresh medium containing either glucose or mannose as the sole carbon source. Cells were allowed to grow for an additional 35 min to a concentration of 2–20 × 107 CFU/ml prior to the addition of antibiotics. Cultures were subjected to a 24 h exposure to cefotaxime or ciprofloxacin prior to estimation of CFU/ml.
Growth restriction of mannose catabolic mutant and antibiotic exposure
Following overnight growth in minimal medium containing glucose as sole carbon source, cells were diluted 1:200 into fresh medium and allowed to grow for 2 h. 3 ml of culture were collected on a 0.22 μM pore-size filter (Millipore), placed in a glass filter holder assembly with funnel, using a vacuum pump. Cells on the filter were vacuum-washed with 10 ml of dH2O and filters were placed on a 50 ml conical tube containing 6 ml of fresh medium either containing or lacking tryptophan. Cells were allowed to grow for an additional 35 min to a concentration of 2–20 × 107 CFU/ml prior to the addition of antibiotics. Cultures were subjected to a 24 h exposure to cefotaxime or ciprofloxacin prior to estimation of CFU/ml.
Growth restriction of relA spoT strains and antibiotic exposure
Following overnight grown LB medium, cells were diluted 1:200 into fresh LB medium and allowed to grow for 2 h. 3 ml of culture were quickly collected on a 0.22 μM pore-size filter (Millipore), placed in a glass filter holder assembly with funnel, using a vacuum pump. Cells on the filter were vacuum-washed with 15 ml of dH2O and filters were placed on a 50 ml conical tube containing 6 ml of fresh LB medium or defined minimal medium. Cells were allowed to grow for an additional 35 min to a concentration of 2–20 × 107 CFU/ml prior to the addition of antibiotics. Cultures were subjected to a 24 h exposure to cefotaxime or ciprofloxacin prior to estimation of CFU/ml.
Expression of pATPase and antibiotic exposure
Following overnight grown in minimal medium, cells were diluted 1:200 into fresh medium and allowed to grow for 2 h. After the addition of IPTG (1 mM), cultures were allowed to grow for and additional 35 min prior to the addition of antibiotics. Cultures were subjected to a 24 h exposure to cefotaxime or ciprofloxacin prior to estimation of CFU/ml.
Secondary antibiotic exposure
Stationary phase bacteria from overnight LB cultures were collected by centrifugation (14,000 × rpms for 1 min at room temperature), resuspended in fresh LB and incubated at 37°C for 30 min. Cells were then diluted 1:200 into fresh LB in the presence of absence of antibiotics. Following 24 h of antibiotic exposure, cells were collected by centrifugation (14,000 × rpms for 20 min at 4°C). Cells were washed with 1.2 ml of ice-cold phosphate buffered saline, transferred to 1.5 ml microcentrifuge tubes and collected by centrifugation (14,000 × rpms for 20 min at 4°C). Finally, cells were resuspended in 500 μl LB medium, diluted and plated onto LB agar (1.5% w/v) plates. Cells were diluted 1:100 into fresh LB medium and allowed to grow overnight. Stationary phase bacteria from overnight cultures were subjected to a second round of antibiotic exposure as described above.
Construction of plasmid pKD4-TetR
Phusion® High-Fidelity DNA Polymerase (New England BioLabs) was used in reactions with primers W2990 and W2991 and genomic DNA derived from Salmonella strain MS7953 (phoP::Tn10) (73). Polymerase chain reaction (PCR) product was ligated into pKD4 (77) digested with BglII and AfeI using NEBuilder® HiFi DNA Assembly Cloning Kit (New England BioLabs). Plasmid was transformed into EC100D cells (Epicentre). The integrity of constructs was verified by DNA sequencing and the ability to render cells resistant to tetracycline.
Construction of plasmid pKD4-SpmR
Phusion® High-Fidelity DNA Polymerase (New England BioLabs) was used in reactions with primers W2992 and W3083 plasmid pSIM19 (72) as template. PCR product was ligated into pKD4 (77) digested with BglII and AfeI using NEBuilder® HiFi DNA Assembly Cloning Kit (New England BioLabs). Plasmid was transformed into EC100D cells (Epicentre). The integrity of constructs was verified by DNA sequencing and the ability to render cells resistant to spectinomycin.
Construction of plasmid pKD4-GenR
Phusion® High-Fidelity DNA Polymerase (New England BioLabs) was used in reactions with primers W2994 and W3084 and plasmid pSAM_Gent_Pbad (a gift from Dr. Barbara Kazmierczak) as template. PCR product was ligated into pKD4 (77) digested with BglII and AfeI using NEBuilder® HiFi DNA Assembly Cloning Kit (New England BioLabs). Plasmid was transformed into EC100D cells (Epicentre). The integrity of constructs was verified by DNA sequencing and the ability to render cells resistant to gentamycin.
Construction of plasmid pKD4-AprR
Phusion® High-Fidelity DNA Polymerase (New England BioLabs) was used in reactions with primers W2996 and W3082 and plasmid pMCS-7 (78) as template. PCR product was ligated into pKD4 (75) digested with BglII and AfeI using NEBuilder® HiFi DNA Assembly Cloning Kit (New England BioLabs). Plasmid was transformed into EC100D cells (Epicentre). The integrity of constructs was verified by DNA sequencing and the ability to render cells resistant to apramycin.
Recombineering protocol
Oligonucleotide sequences used in this study are presented in Table S2. S. enterica strain 14028s (71) harboring plasmid pSIM6 (74) was grown overnight in LB medium supplemented with 100 μg/ml of ampicillin at 30°C and 250 rpm. Cells were diluted (1:100) in 30 ml of the same medium and grown for approximately 2.5 h (OD600 ~0.35–0.4). The culture flask then grown in a water bath at 42°C and 250 rpm for 20 min (final OD600 ~0.6–0.75). Cells were immediately transferred to a 50 ml conical tube, collected by centrifugation (7,000 rpm for 2.5 min at 4°C) and resuspended in 40 ml of ice-cold dH2O. Cells were collected again by centrifugation and this washing procedure was repeated a second time. Finally, cells were resuspended in 150 μl of ice-cold dH2O. Homologous recombination was obtained by electroporating 70 μl of cell suspension with 10 μl of purified linear DNA fragments.
Construction of strain relA spoT trpA strain
The ΔtrpA::kan mutation from a Salmonella NR-42835 E11 strain from the BEi NR mutant library (79) was transferred to Salmonella strain MP342 (relA::Tn10 spoT) (25) using P22-mediated transduction (75).
Construction of manA mutant
A ΔmanA::cat mutation was generated by recombineering using purified linear DNA fragment generated from a PCR reaction using primers 12460 and 12461 plasmid pDK3 (77). The location of each insertion was verified by PCR using primers 12462 and 12190, which flank the recombination points, and by the ability to utilize mannose as the sole carbon source. ΔmanA::cat mutation was subsequently moved into S. enterica strain 14028s by P22-mediated transduction (75) to produce strain MP50.
Construction of strains containing deletions of single and multiple TA modules
Single ΔTA strains were generated by recombineering using purified linear DNA fragments generated from PCR reactions using primers listed in Table S2 and plasmids pDK3, pKD4 (77), pKD4-TetR, pKD4-SpmR, pKD4-GenR or pKD4-AprR as template. The location of each insertion was verified by polymerase chain reaction (PCR) using primers listed in Table S2. As the expression of recombineering functions can be mutagenic (80), these mutations were individually transferred into S. enterica strain 14028s by P22-mediated transduction (75) to produced single deletion strains. To generate s strains containing multiple deletions, six deletions, each containing a distinct antibiotic resistant marker, were sequentially moved into S. enterica strain 14028s by P22-mediated transduction. Antibiotic markers were removed using plasmid pCP20, which expresses the FLP recombinase (77), which acts on the FTR sites flanking the genes specifying the proteins conferring antibiotic resistance. This procedure was repeated a second time to delete the 12 TA modules.
Verification of strain lacking 12 TA modules
To verify the genetic integrity of strain MP1422, harboring deletions of multiple TA modules, genomic DNA was extracted using DNeasy kit (Qiagen). Purified DNA was submitted for library construction and high-throughput sequencing at the Yale Center for Genomic Analysis. Sequence fragments were assembled using Geneious Software (Biomatters) and S. enterica strain 14028s genome (NCBI ref. seq.: NC_016856.1) as reference. We identified a total of 10 polymorphisms relative to the 14028s strain. Sequencing of PCR products flanking the polymorphic regions from the S. enterica 14028s parental strain established that 9 of these nucleotide differences were actually errors generated in the original sequence of 14028s (79). Thus, strain sequence of MP1422 (NCBI SRA accession: PRJNA539960) contained a single mutation, consisting of a 125 bp deletion between the intergenic regions of rrlB and rrfB (i.e. a deletion between bp positions 4,369,814 and 4,369,939).
Estimation of intracellular ATP
Intracellular ATP was estimated as described (33). Briefly, 150 μl of cell cultures were aliquoted into PCR tubes, immediately heated at 80° C for 10 min, and subsequently placed on ice. At the same time, 1 ml of each of these cultures was transferred to a 1.5 ml tube and placed on ice. Using a multichannel pipette, 30–50 μl of heat-inactivated (80° C for 10 min) cells were transferred to 96-well black plates (Corning) containing 150 μl of BacTiter-Glo Microbial Cell Viability Assay Solution (Promega). Samples were mixed by pipetting, and their luminescence was measured in a Synergy H1 Reader (BioTek). Cells from the 1 ml of culture aliquots were collected by centrifugation (4° C at 14,000 rpm for 5 min), resuspended in 250 μl of 0.5% SDS, and lysed by boiling at 100° C for 10 min. The concentrations of proteins in the cell lysates were estimated using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Protein concentrations in these samples were used to calculate the protein amount present in the luminescence reactions. ATP levels were calculated by normalizing luminescence values of each sample by their protein content.
Supplementary Material
Fig. S1. Determination of of growth rates and of the emerge of antibiotic resistant mutants in bacterial cultures.
Fig. S2. Effect of Shx treatment on the frequency of persisters.
Fig. S3. Effects of ATP depletion and inhibition of protein synthesis on antibiotic tolerance.
Fig. S4. Effect of growth inhibition on antibiotic tolerance in Salmonella.
Table S1. Microbial strains and plasmids used in this study.
Table S2. Oligonucleotides sequences used in this study.
Acknowledgments:
The authors would like to thank Jennifer Aronson for useful discussions and comments, Dr. Barbara Kazmierczak (Yale University) for providing plasmid pSAM_Gent_Pbad, Dr. Christine Jacobs-Wagner (Yale University) for providing plasmid pMCS-7, Dr. Xiang Zhan (Penn State University) for discussions about statistical analyses, and Dr. Sophie Helaine (Imperial College London) for providing the sequence information of toxin-antitoxin modules described in her paper (14).
Funding: This research was supported by NIH grant AI49561 to EAG.
Footnotes
Competing Interests: The authors declare that they have no competing interests.
Data and Materials Availability: The sequence of strain MP1422 was deposited into NCBI SRA database (accession ID number PRJNA539960). All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Brauner A, Fridman O, Gefen O, Balaban NQ, Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Lewis K, Persister cells. Annu. Rev. Microbiol 64, 357–372 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Roth JR, Anderson DI, Amplification-mutagenesis—how growth under selection contributes to the origin of genetic diversity and explains the phenomenon of adaptive mutation. Res. Microbiol 155, 342–351 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Hersh MN, Ponder RG, Hastings PJ, Rosenberg SM, Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress. Res. Microbiol 155, 352–359 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Miller C, Thomsen LE, Gaggero C, Mosseri R, Ingmer H, Cohen SN, SOS response induction by beta-lactams and bacterial defense against antibiotic lethality. Science 305, 1629–1631 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Cirz RT, Chin JK, Andes DR, de Crécy-Lagard V, Craig WA, Romesberg FE, Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3, e176 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ, Antibiotic tolerance facilitates the evolution of resistance. Science 355, 826–830 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Abshire KZ, Neidhardt FC, Growth rate paradox of Salmonella typhimurium within host macrophages. J. Bacteriol 175, 3744–3748 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayrapetyan M, Williams T, Oliver JD, The relationship between the viable but nonculturable state and antibiotic persister cells. J. Bacteriol 200, e00249–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen NR, Lobritz MA, Collins JJ, Microbial persistence and the road to drug resistance. Cell Host Microbe. 13, 632–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maisonneuve E, Castro-Camargo M, Gerdes K, (p) ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 154, 1140–1150. (2013). [DOI] [PubMed] [Google Scholar]
- 12.Korch SB, Henderson TA, Hill TM, Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol 50, 1199–1213 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M, Residual guanosine 3’,5’-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem 266, 5980–5990 (1991). [PubMed] [Google Scholar]
- 14.Helaine S, Cheverton AM, Watson KG, Faure LM, Matthews SA, Holden DW, Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 343, 204–208 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranda CMA, Swanson JA, Loomis WP, Miller SI, Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. U. S. A 89, 10079–10083 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi Y, Inouye M, Regulation of growth and death in Escherichia coli by toxin-antitoxin systems. Nat. Rev. Microbiol 9, 779–790 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Goormaghtigh F, Fraikin N, Putrinš M, Hallaert T, Hauryliuk V, Garcia-Pino A, Sjödin A, Kasvandik S, Udekwu K, Tenson T, Kaldalu N, Van Melderen L, Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio 9, e00640–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan Y, Brown Gandt A, Rowe SE, Deisinger JP, Conlon BP, Lewis K, ATP-dependent persister formation in Escherichia coli. MBio. 8, e02267–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K, Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol 1, 16051 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Maisonneuve E, Castro-Camargo M, Gerdes K, (p) ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell 172, 1135 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Harms A, Fino C, Sørensen MA, Semsey S, Gerdes K, Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. MBio. 8, e01964–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwan BW, Valenta JA, Benedik MJ, Wood TK, Arrested protein synthesis increases persister-like cell formation. Antimicrob Agents Chemother. 57, 1468–1473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng HY, Soo VW, Islam S, McAnulty MJ, Benedik MJ, Wood TK, Toxin GhoT of the GhoT/GhoS toxin/antitoxin system damages the cell membrane to reduce adenosine triphosphate and to reduce growth under stress. Environ. Microbiol 16, 1741–1754 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Lee EJ, Groisman EA, Control of a Salmonella virulence locus by an ATP-sensing leader messenger RNA. Nature 486, 271–275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pontes MH, Yeom J, Groisman EA, Reducing ribosome biosynthesis promotes translation during low Mg2+ stress. Mol. Cell 64, 480–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobato-Márquez D, Moreno-Córdoba I, Figueroa V, Díaz-Orejas R, García-del Portillo F, Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Sci. Rep 5, 9374 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ, Transcription profiling of the stringent response in Escherichia coli. J. Bacteriol 190, 1084–1096 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH, Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Barker MM, Gaal T, Gourse RL, Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol 305, 689–702 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Murray HD, Schneider DA, Gourse RL, Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12, 125–134 (2003). [DOI] [PubMed] [Google Scholar]
- 31.Sarmientos P, Sylvester JE, Contente S, Cashel M, Differential stringent control of the tandem E. coli ribosomal RNA promoters from the rrnA operon expressed in vivo in multicopy plasmids. Cell 32,1337–1346 (1983). [DOI] [PubMed] [Google Scholar]
- 32.Levine A, Vannier F, Dehbi M, Henckes G, Séror SJ, The stringent response blocks DNA replication outside the ori region in Bacillus subtilis and at the origin in Escherichia coli. J. Mol. Biol 219, 605–613 (1991). [DOI] [PubMed] [Google Scholar]
- 33.Pontes MH, Groisman EA, Protein synthesis controls phosphate homeostasis. Genes Dev. 32, 79–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vázquez-Laslop N, Lee H, Neyfakh AA, Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol 188, 3494–3497 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhury N, Kwan BW, Wood TK, Persistence increases in the absence of the alarmone guanosine tetraphosphate by reducing cell growth. Sci. Rep 6, 20519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koebmann BJ, Westerhoff HV, Snoep JL, Nilsson D, Jensen PR, The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol 184, 3909–3916 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stouthamer AH, A theoretical study on the amount of ATP required for synthesis of microbial cell material. Antonie Van Leeuwenhoek. 39, 545–565 (1973). [DOI] [PubMed] [Google Scholar]
- 38.Bagnara AS, Finch LR, Relationships between intracellular contents of nucleotides and 5-phosphoribosyl 1-pyrophosphate in Escherichia coli. Eur. J. Biochem 36, 422–427 (1973). [DOI] [PubMed] [Google Scholar]
- 39.Lund E, Kjeldgaard KO, Metabolism of guanosine tetraphosphate in Escherichia coli. Eur. J. Biochem 28, 316–326 (1972). [DOI] [PubMed] [Google Scholar]
- 40.Davis BD, The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc. Natl. Acad. Sci. U. S. A 35, 1–10 (1949). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dewey DC, Work E, Diaminopimelic acid and lysine: diaminopimelic acid decarboxylase Nature 169, 533–534 (1952). [DOI] [PubMed] [Google Scholar]
- 42.Bukhari AI, Taylor AL, Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J. Bacteriol 105, 844–854 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eidlic L, Neidhardt FC, Protein and nucleic acid synthesis in two mutants of Escherichia coli with temperature-sensitive aminoacyl ribonucleic acid synthetases. J. Bacteriol 89, 706–711 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dörr T, Vulić M, Lewis K, Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 8, e1000317 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwan BW, Lord DM, Peti W, Page R, Benedik MJ, Wood T, The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ. Microbiol 17, 3168–3181 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Kwaik YA, Bunann D, Microbial quest for food in vivo: ‘nutritional virulence’ as an emerging paradigm. Cell. Microbiol 15, 882–890 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Gouzy A, Larrouy-Maumus G, Wu TD, Peixoto A, Levillain F, Lugo-Villarino G, Guerquin-Kern JL, de Carvalho LP, Poquet Y, Neyrolles O, Mycobacterium tuberculosis nitrogen assimilation and host colonization require aspartate. Nat. Chem. Biol 9, 674–676 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouzy A, Larrouy-Maumus G, Bottai D, Levillain F, Dumas A, Wallach JB, Caire-Brandli I, de Chastellier C, Wu TD, Poincloux R, Brosch R, Guerquin-Kern JL, Schnappinger D, Sório de Carvalho LP, Poquet Y, Neyrolles O, Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 10, e1003928 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sassetti CM, Rubin EJ, Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A 200, 12989–12994 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser P, Regoes RR, Dolowschiak T, Wotzka SY, Lengefeld J, Slack E, Grant AJ, Ackermann M, Hardt WD, Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol. 12, e1001793 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, Schmidt A, Bumann D, Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell. 158, 722–733 (2014). [DOI] [PubMed] [Google Scholar]
- 52.Verstraeten N, Knapen WJ, Kint CI, Liebens V, Van den Bergh B, Dewachter L, Michiels JE, Fu Q, David CC, Fierro AC, Marchal K, Beirlant J, Versées W, Hofkens J, Jansen M, Fauvart M, Michiels J, Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Schaechter M, Maaloe O, Kjeldgaard NO, Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J. Gen. Microbiol 19, 592–606 (1958). [DOI] [PubMed] [Google Scholar]
- 54.Gaal T, Gourse RL, Guanosine 3’-diphosphate 5’-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 87, 5533–5537 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petersen C, Møller LB, Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J. Biol. Chem 275, 3931–3935 (2000). [DOI] [PubMed] [Google Scholar]
- 56.Schneider DA, Gourse RL, Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J. Biol. Chem 279, 8262–8268 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Bhaskar A, De Piano C, Gelman E, McKinney JD, Dhar N, Elucidating the role of (p) ppGpp in mycobacterial persistence against antibiotics. IUBMB Life 70, 836–844 (2018). [DOI] [PubMed] [Google Scholar]
- 58.De la Cruz MA, Zhao W, Farenc C, Gimenez G, Raoult D, Cambillau C, Gorvel JP, Méresse S S1, A toxin-antitoxin module of Salmonella promotes virulence in mice. PLoS Pathog. 9, e1003827 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 59.Bremer H, Dennis PP “Modulation of chemical composition and other parameters of the cell by growth rate” in Escherichia coli and Salmonella: Cellular and molecular biology, Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, Eds. (ASM Press, 1996), vol. 2, pp. 1553–1569. [Google Scholar]
- 60.Helmstetter CE, “Timing of synthetic activities in the cell cycle” in Escherichia coli and Salmonella: Cellular and molecular biology, Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, Eds. (ASM Press, 1996), vol. 2, pp. 1627–1639. [Google Scholar]
- 61.Messer W, Weigel C “Initiation of chromosome replication” in Escherichia coli and Salmonella: Cellular and molecular biology, Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, Eds. (ASM Press, 1996), vol. 2, pp. 1579–1601. [Google Scholar]
- 62.Atkinson DE, Biological feedback control at the molecular level. Science 150, 851–857 (1965). [DOI] [PubMed] [Google Scholar]
- 63.Johnson PJ, Levin BR, Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genet. 9, e1003123 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA, Functional coupling of p38-induced up-regulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res. 66, 1702–1711 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ranganathan AC, Ojha S, Kourtidis A, Conklin DS, Aguirre-Ghiso JA, Dual function of pancreatic endoplasmic reticulum kinase in tumor cell growth arrest and survival. Cancer Res. 68, 3260–3268 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Terai H, Kitajima S, Potter DS, Matsui Y, Quiceno LG, Chen T, Kim TJ, Rusan M, Thai TC, Piccioni F, Donovan KA, Kwiatkowski N, Hinohara K, Wei G, Gray NS, Fischer ES, Wong KK, Shimamura T, Letai A, Hammerman PS, Barbie DA, ER stress signaling promotes the survival of cancer “persister cells” tolerant to EGFR tyrosine kinase inhibitors. Cancer Res. 78, 1044–1057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DY, Chen EI, Lyden D, Bissell MJ, The perivascular niche regulates breast tumour dormancy. Nat. Cell Biol 15, 807–817 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M, A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell 141, 583–594 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Drug M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, Evans L, Ji W, Hsu CH, Thurley K, Wei S, Zhou A, Koduru PR, Posner BA, Wu LF, Altschuler SJ, Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells Nat. Commun, 7, 10690 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Recasens A, Munoz L, Targeting cancer cell dormancy. Trends Pharmacol Sci. 40, 128–141 (2019). [DOI] [PubMed] [Google Scholar]
- 71.Ocampo PS, Lázár V, Papp B, Arnoldini M, Abel zur Wiesch P, Busa-Fekete R, Fekete G, Pál C, Ackermann M, Bonhoeffer S, Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob. Agents Chemother 58, 4573–4582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K, Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fields PI, Swanson RV, Haidaris CG, Heffron F, Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U. S. A 83, 5189–5193 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Datta S, Costantino N, Court DL, A set of recombineering plasmids for gram- negative bacteria. Gene 379, 109–115 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Davis RW, Botstein D, Roth JR, A Manual for Genetic Engineering Advanced Bacterial Genetics. (Cold Springer Harbor, New York, ed. 3, 1982). [Google Scholar]
- 76.Neidhardt FC, Bloch PL, Smith DF, Culture medium for enterobacteria. J. Bacteriol 119, 736–747 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Datsenko KA, Wanner BL, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A 83, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thanbichler M, Iniesta AA, Shapiro L, A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35, e137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, Canals R, Reynolds MM, Bogomolnaya L, Shields C, Cui P, Guo J, Zheng Y, Endicott-Yazdani T, Yang HJ, Maple A, Ragoza Y, Blondel CJ, Valenzuela C, Andrews-Polymenis H, McClelland M, Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One 9, e99820 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Murphy KC, Campellone KG, Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol 4, 11 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jarvik T, Smillie C, Groisman EA, Ochman H, Short-term signatures of evolutionary change in the Salmonella enterica serovar typhimurium 14028 genome. J. Bacteriol 192, 560–567 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hanahan D, “Techniques for transformation of E. coli” in DNA Cloning: A Practical Approach Glover DM, Ed. (IRL Press, 1985), pp. 109 −135. [Google Scholar]
- 83.Soncini FC, Véscovi EG, Groisman EA, Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol 177, 4364–4371 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Determination of of growth rates and of the emerge of antibiotic resistant mutants in bacterial cultures.
Fig. S2. Effect of Shx treatment on the frequency of persisters.
Fig. S3. Effects of ATP depletion and inhibition of protein synthesis on antibiotic tolerance.
Fig. S4. Effect of growth inhibition on antibiotic tolerance in Salmonella.
Table S1. Microbial strains and plasmids used in this study.
Table S2. Oligonucleotides sequences used in this study.


