Abstract
Heterotrophic plants provide evolutionarily independent, natural experiments in the genomic consequences of radically altered nutritional regimes. Here, we have sequenced and annotated the plastid genome of the endangered mycoheterotrophic orchid Hexalectris warnockii. This orchid bears a plastid genome that is ∼80% the total length of the leafy, photosynthetic Phalaenopsis, and contains just over half the number of putatively functional genes of the latter. The plastid genome of H. warnockii bears pseudogenes and has experienced losses of genes encoding proteins directly (e.g., psa/psb, rbcL) and indirectly involved in photosynthesis (atp genes), suggesting it has progressed beyond the initial stages of plastome degradation, based on previous models of plastid genome evolution. Several dispersed and tandem repeats were detected, that are potentially useful as conservation genetic markers. In addition, a 29-kb inversion and a significant contraction of the inverted repeat boundaries are observed in this plastome. The Hexalectris warnockii plastid genome adds to a growing body of data useful in refining evolutionary models in parasites, and provides a resource for conservation studies in these endangered orchids.
Keywords: orchid, heterotroph, plastome, pseudogene, evolution, chloroplast
Introduction
Plants that parasitize other plants or mycorrhizal fungi provide unique opportunities to study the genomic consequences of radically altered nutritional lifestyles and associated changes in selective regimes (Wolfe et al. 1992; Barrett et al. 2014; Wicke et al. 2016). In particular, plants that have become obligate parasites upon fungi for nutritional needs represent case studies of convergent evolution. Transitions to this lifestyle have occurred an estimated minimum of 30× in the orchid family alone, mostly due to their complete, parasitic dependence upon mycorrhizal fungi early in development, called “initial mycoheterotrophy” (Freudenstein and Barrett 2010; Merckx and Freudenstein 2010). Furthermore, many of these plants are rare or endangered (Freudenstein 1999; Merckx et al. 2013), and in many cases represent “ecological indicators” of undisturbed habitat, or may serve as “umbrella species” for conservation efforts (Taylor et al. 2013).
What happens to the genomes of organisms that have undergone such drastic changes in nutritional mode, from autotrophy to heterotrophy? Representative plastid genomes have been sequenced from plant lineages containing heterotrophs, allowing researchers to construct models of plastid genome degradation, including pseudogene formation (functional losses), physical gene losses, and increased substitution rates as a result of relaxed selective pressures on photosynthetic function (Wicke et al. 2011, 2016; Barrett and Davis 2012; Barrett et al. 2014; Graham et al. 2017). However, sampling gaps exist in these models, underscoring the need for more thorough representation of plant lineages containing nonphotosynthetic members, each representing an independent trajectory of plastome degradation.
One such lineage is the North American orchid genus Hexalectris Raf. Members of this genus are hypothesized to obtain most or all nutrients, including carbon, from their symbiotic mycorrhizal fungi (Taylor et al. 2003; Kennedy et al. 2011), a situation called mycoheterotrophy. Hexalectris contains ten currently recognized species, many of which are rare and restricted to highly specific habitats (Catling and Engel 1993; Catling 2004; Kennedy and Watson 2010). Hexalectris warnockii Ames and Correll, or the Texas purple-spike, is an endangered member of the genus restricted to Texas, Arizona, and Mexico, where it grows in shaded oak-juniper-pinyon canyons near seasonally dry creek beds, or on calcareous soils under juniper scrub (IUCN Red List: Endangered D; Goedeke et al. 2015). It is known from ∼24 sites in United States, including: Big Bend National Park, northeastern Texas (Dallas area), the Edwards Plateau, and Arizona; in Mexico it is found at a site in Coahuila and another at the southern tip of Baja California Sur (Catling 2004).
Here, we have sequenced, assembled, and annotated the plastid genome of Hexalectris warnockii. The goals of this study are: 1) to use genomic criteria—that is, extensive loss of photosynthesis-related genes—to determine if H. warnockii is nonphotosynthetic (fully mycoheterotrophic) or retains photosynthetic capability (partially mycoheterotrophic); 2) to compare the plastid genome of H. warnockii to those from members of other heterotrophic plant lineages; and 3) to provide a genomic resource for the development of plastid markers to facilitate studies of genetic diversity in populations of this endangered species.
Materials and Methods
Floral tissue of H. warnockii was collected from Brewster County, TX. A voucher specimen was deposited at The Miami University Willard Sherman Turrell Herbarium (Accession: Kennedy and Freeman #33). We extracted DNA using a CTAB protocol (Doyle and Doyle 1987), yielding 17.4 ng/μl based on a Qubit Fluorometer reading (ThermoFisher Scientific, Waltham, MA). Illumina libraries were prepared by shearing total genomic DNA to 350–400 bp fragments on a Covaris E220 ultrasonicator (Covaris, Woburn, MA), followed by the protocol of Glenn et al. (2016). Library concentrations and fragment sizes were calculated on an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA), pooled with 19 other libraries, and sequenced on two lanes of an Illumina Hiseq2000 for paired-end, 100-bp reads.
We carried out adapter removal and quality trimming with Trimmomatic v.0.36 (Bolger et al. 2014), using a 3-bp sliding window and a minimum PHRED score of 20 (1:100 error rate). The plastome was assembled from cleaned reads using NOVOPlasty v.2.6.3 (Dierckxsens et al. 2017), which uses a reference sequence as an initial seed (here, rbcL from the leafy, photosynthetic orchid Phalaenopsis equestris, GenBank# JF719062) and builds a circularized plastome. Reads were mapped with high stringency to the draft plastome produced by NOVOplasty in Geneious v.8.1 to check for assembly errors (http://www.geneious.com, last accessed May 1, 2018; Kearse et al. 2012; 98% similarity, allowing gaps up to 100 bp). The plastome was annotated initially in DOGMA (Wyman et al. 2004). Start/stop codons, exon/intron boundaries, inverted repeat (IR) boundaries, and putative loss-of-function pseudogenes were verified and adjusted by aligning the plastome to protein coding and RNA genes from P. equestris (GenBank accession JF719062), Phoenix dactylifera (Arecaceae, GU811709), and Heliconia collinsiana (Heliconiaceae, JX08866), as was done in Barrett et al. (2014).
The annotated H. warnockii plastome was aligned with that of P. equestris using the progressiveMAUVE (Darling et al. 2010) plugin for Geneious v. 8.1, which identifies syntenic regions between two or more genomes, thus allowing detection of genomic rearrangements. Putatively functional genes (with open reading frames or lacking drastic modifications in the case of RNA genes), pseudogenes (putative functional losses, i.e., those with interrupted reading frames or nontriplet insertions or deletions), and physical gene losses were recorded and compared with the plastome of the leafy, photosynthetic P. equestris. We also compared plastome size and functional gene content for a number of full mycoheterotrophs, partial mycoheterotrophs, holoparasites, hemiparasites, and other leafy, autotrophic species.
Genomic repeat type and abundance were calculated in REPuter (Kurtz et al. 2001), specifying a minimum length of 20 bp (for forward, reverse, palindromic, and reverse-complementary repeats), a Hamming distance of 3, and a maximum e-value of 1.0 × 10−3. Tandem repeats were identified using the Phobos plugin for Geneious (Mayer 2010), specifying 2–50 bp motif length, a minimum total length of 10 bp, and allowing only perfect repeats. All results were plotted in R (R Core Development Team 2013) or PAST v.3.8 (Hammer et al. 2001). A linearized plastome map was created in OGDraw (Lohse et al. 2013).
Results and Discussion
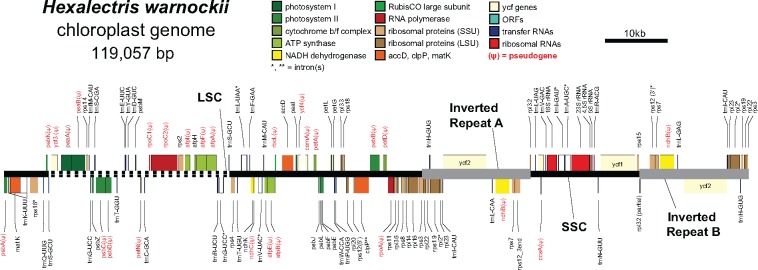
Illumina paired-end sequencing of H. warnockii yielded a total of 38,633,900 reads (after trimming), with an average insert size of 350 bp. Coverage depth of the finished plastome was 712.2×, representing 2.19% of the total read pool. The 119,057 bp plastome has a quadripartite structure as is typical for angiosperms (fig. 1), with a Large Single Copy region (LSC; 66,903 bp), Small Single Copy region (SSC; 17,490), and an Inverted Repeat (IR; 17,332) (table 1 and fig. 1). The H. warnockii plastome is thus 29,902 bp smaller than the leafy orchid P. equestris, or ∼79.9% the total size of the latter (148,959 bp, representing a typical orchid plastome size). The largest physical reduction in the H. warnockii plastome was in the LSC region, which was 27.6% smaller than that of P. equestris due to several large deletions. There is a contraction of the inverted repeat (IR) in H. warnockii, representing a 33% difference in total IR length relative to P. equestris. This contraction resulted in the following genes, typically found in the IR, becoming part of the SSC: 16S rRNA, trnIGAU, trnAUGC, 23S rRNA, 4.5S rRNA, 5S rRNA, trnRACG, trnNGUU, and the 5′ portion of ycf1. Total GC content is 36.9% after removing one copy of the IR, and similar to that of Phalaenopsis at 36.7%.
Fig. 1.
—Map of the plastid genome of Hexalectris warnockii. LSC, large single copy region; SSC, small single copy region; gray, inverted repeat; dashed line in LSC = a 29-kb inversion. Red text indicates the presence of a pseudogene (ψ). “*” and “**” denote genes with one or two introns, respectively.
Table 1.
Features of the Fully Mycoheterotrophic Hexalectris warnockii Plastid Genome Relative to That of the Leafy, Autotrophic Phalaenopsis equestris (GenBank accession JF719062)
| Hexalectris warnockii | Phalaenopsis equestris | % of Phalaenopsis | |
|---|---|---|---|
| Total length (bp) | 119,057 | 148,959 | 79.9 |
| Large single copy (LSC) | 66,903 | 85,967 | 77.8 |
| Inverted repeat (IR) | 17,332 | 25,846 | 67.1 |
| Small single copy (SSC) | 17,490 | 11,300 | 154.8 |
| protein coding genes (CDS) | 38 | 69 | 55.1 |
| Pseudogenes (ψ) | 25 | 3 | 833.3 |
| Transfer RNA genes (tRNA) | 30 | 30 | 93.3 |
| Ribosomal RNA genes (rRNA) | 4 | 4 | 100.0 |
| Putatively functional | 72 | 103 | 69.9 |
| Total genes and pseudogenes | 97 | 106 | 91.5 |
We identified 45 dispersed repeats across the genome passing our filters in REPuter: two were forward-compliment, 16 forward–forward, 22 palindromic, and five forward–reverse (table 2). We identified 419 tandem repeats with minimum motif lengths of 10 bp (table 2 and supplementary table S1, Supplementary Material online). The most abundant of these were hexanucleotide repeats (141) followed by pentanucleotide repeats (99). We identified three dinucleotide repeats, 17 trinucleotide repeats, and 50 tetranucleotide repeats. Thus, there are several options for the development of potentially variable satellite markers in H. warnocki, which will be useful in determining patterns of plastid genomic diversity across populations of this endangered orchid. Alignment with MAUVE detected a major genomic inversion of a ∼29-kb region of the LSC relative to P. equestris with breakpoints spanning trnSGCU and trnSGGA; the entire collinear block detected by MAUVE contains 29 genes (fig. 1).
Table 2.
Numbers of Dispersed and Tandem Repeats Detected with REPuter and Phobos, Respectively
| Dispersed Repeats | |
|---|---|
| Repeat Type | Number |
| Forward-compliment | 2 |
| Forward–forward | 16 |
| Palindromic | 22 |
| Forward–reverse | 5 |
|
Tandem repeats | |
| Motif Length (bp) | Number |
| 2 | 3 |
| 3 | 17 |
| 4 | 50 |
| 5 | 99 |
| 6 | 141 |
| 7 | 50 |
| 8 | 19 |
| 9 | 15 |
| >9 | 25 |
Note.—Tandem repeat sequences are listed in supplementary table S1, Supplementary Material online.
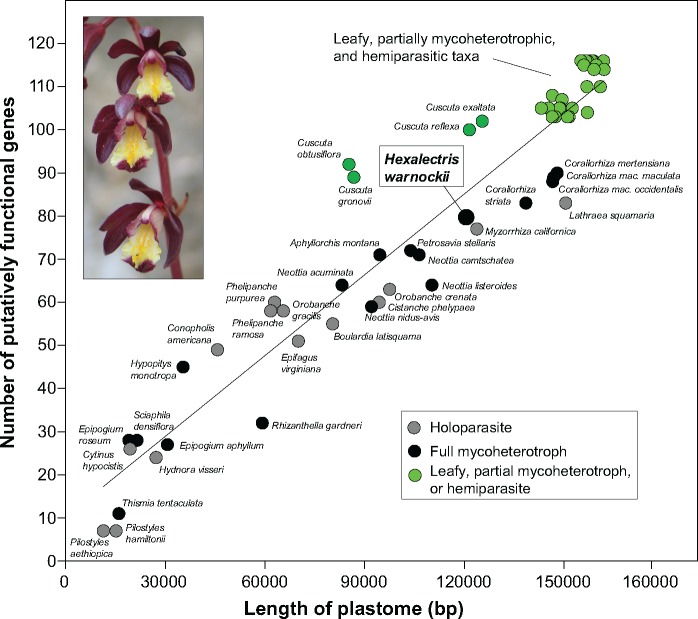
The plastome of H. warnockii encodes 72 putatively functional genes (protein-coding, tRNA, and rRNA), compared with 103 in P. equestris, a 31.1% difference in functional gene content, composed of pseudogenes (i.e., functional losses, 25 in H. warnockii relative to P. equestris s) and physical gene losses (11 in H. warnockii relative to P. equestris). The total plastome size reduction in H. warnockii is largely due to the deletion of regions containing photosynthesis-related genes (fig. 1), thus also reducing the gene count. Plastome size in H. warnockii is comparable to that of the mycoheterotrophic orchid Corallorhiza striata var. vreelandii at 137,505 bp (Barrett and Davis 2012), and to the holoparasite Myzorhiza californica at 120,840 bp (Wicke et al. 2013; see fig. 2). Overall there is a strong positive correlation between the number of putatively functional genes and plastome length among heterotrophic angiosperms (fig. 2; Pearson correlation r = 0.953, P < 0.0001); thus, physical gene loss is at least in part driving a reduction in plastome size.
Fig. 2.
—A comparison of plastid genome size and the number of putatively functional genes for sequenced holoparasites (gray), full mycoheterotrophs (black), and selected autotrophs (including partial mycoheterotrophs and hemiparasites; green), with trophic status based on whether or not photosynthetic genes are putatively functional in each. Inset: Hexalectris warnockii in flower (photo: A. Kennedy; Big Bend National Park, Brewster Co., TX).
Genes that are either functionally or physically lost conform to the models of Barrett and Davis (2012), Wicke et al. (2016) and Graham et al. (2017), and include: photosynthesis-related genes [Photosystem I and I subunits (psa, psb), Cytochrome subunits (pet), RuBisCO Large Subunit (rbcL), Photosystem Assembly Factors (ycf3, ycf4, also called paf1 and paf2, respectively; Wicke et al. 2011); subunits of the plastid-encoded RNA Polymerase (rpo); and subunits of the ATP synthase complex (atp)]. There are also substantial functional and physical losses among subunits of the NAD(P)H Dehydrogenase complex (ndh; all physically lost except ndhK, ψndhB, and ψndhC), but this is common in other orchids including Phalaenopsis, perhaps due to the tendency of orchids to occupy low-light environments (Lin et al. 2017).
Losses in subunits of these functional gene categories conform to “stage 4” of the model of plastome degradation by Barrett and Davis (2012), and are also in line with a recent mechanistic model of plastome evolution (Wicke et al. 2016). Functional loss of five out of six ATP Synthase subunit genes is significant, in that many parasitic lineages early in the process of plastome degradation tend to have preserved reading frames for atp genes despite having experienced major losses in photosynthesis-related and rpo genes (Barrett et al. 2014; Wicke et al. 2016; Braukmann et al. 2017; Graham et al. 2017). Thus, H. warnockii may have entered a new phase in plastome evolution following a period of evolutionary stasis, based on the “punctuated burst” model of plastome evolution put forth by Naumann et al. (2016).
The IR is hypothesized to function in plastid genome structural stability, but studies from highly rearranged genomes are equivocal (Palmer 1985; Lam et al. 2015; Lim et al. 2016). Here, a 29-kb LSC inversion is found in conjunction with a drastic reduction of the IR (fig. 1). Repeats have been shown in parasitic Orobanchaceae to be associated with plastome structural rearrangements and shifts in IR boundaries (Wicke et al. 2013); thus additional sampling of Hexalectris spp. and related genera will allow for explicit tests among repeat content, structural rearrangements, and substitution rates.
The ancestor of Hexalectris may have been evolving under relaxed selective pressure for up to 32 Myr, based on a stem-node age estimate of Hexalectris, which also includes members of the closely related genera Basiphyllaea and Bletia (Sosa et al. 2016). Hexalectris warnockii is consistently placed as sister to the remaining members of genus Hexalectris in previous studies (Kennedy and Watson 2010; Sosa et al. 2016); thus it is unknown whether this species has undergone an independent transition to full mycoheterotrophy, or if this condition is shared by all species in the genus. Regardless, plastome degradation has been occurring in H. warnockii for an estimated 24 Myr, when the first divergence occurred within Hexalectris (Sosa et al. 2016). Sequencing of additional members of Hexalectris, and the closely related members of tribe Bletiinae (Basiphyllaea, Bletia) will allow fine-scale reconstruction of plastid genome degradation, and testing of the hypothesis of a single origin of full mycoheterotrophy/loss of photosynthesis in Hexalectris. Furthermore, sampling of multiple individuals per species may uncover substantial variation in plastomes across the geographic range of each species, as has been recently demonstrated in the fully mycoheterotrophic orchid Corallorhiza striata (Barrett et al. 2018).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Big Bend National Park (US Department of Interior) for permission and assistance in collecting material; this research was supported by the West Virginia University Program to Stimulate Competitive Research Grant to C.F.B. We thank two anonymous reviewers for suggestions that improved the article.
Literature Cited
- Barrett CF, Davis JI.. 2012. The plastid genome of the mycoheterotrophic Corallorhiza striata (Orchidaceae) is in the relatively early stages of degradation. Am J Bot. 99(9):1513–1523. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Freudenstein JV, Li J, Mayfield-Jones DR, Perez L.. 2014. Investigating the path of plastid genome degradation in an early-transitional clade of heterotrophic orchids, and implications for heterotrophic angiosperms. Mol Biol Evol. 31(12):3095–3112. [DOI] [PubMed] [Google Scholar]
- Barrett CF, Wicke S, Sass C.. 2018. Dense infraspecific sampling reveals rapid and independent trajectories of plastome degradation in a heterotrophic orchid complex. New Phytol. 218:1192–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B.. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braukmann TWA, Broe MB, Stefanović S, Freudenstein JV.. 2017. On the brink: the highly reduced plastomes of nonphotosynthetic Ericaceae. New Phytol. 216(1):254–266. [DOI] [PubMed] [Google Scholar]
- Catling PM. 2004. A synopsis of the genus Hexalectris in the United States and a new variety of Hexalectris revoluta. Native Orchid Conf J. 1:5–25. [Google Scholar]
- Catling PM, Engel VS.. 1993. Systematics and distribution of Hexalectris spicata var. arizonica (Orchidaceae). Lindleyana 8:119–125. [Google Scholar]
- Darling AE, Mau B, Perna NT.. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5(6):e11147.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierckxsens N, Mardulyn P, Smits G.. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL.. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15. [Google Scholar]
- Freudenstein JV. 1999. A new species of Corallorhiza (Orchidaceae) from West Virginia, USA. Novon 9(4):511–513. [Google Scholar]
- Freudenstein JV, Barrett CF.. 2010. Mycoheterotrophy and diversity in Orchidaceae. In: Seberg O, Petersen G, Barfod A, Davis JI, editors. Diversity, phylogeny and evolution in the monocotyledons. The Proceedings of the Fourth International Conference on Monocot Systematics. Aarhus (Denmark): Aarhus University Press. p. 25–37.
- Glenn TC, et al. . 2016. Adapterama I: Universal stubs and primers for thousands of dual-indexed Illumina libraries (iTru & iNext). BioRxiv 049114. doi: https://doi.org/10.1101/049114. [DOI] [PMC free article] [PubMed]
- Goedeke T, Sharma J, Treher A, Frances A, Poff K.. 2015. Hexalectris warnockii. The IUCN Red List of Threatened Species.
- Graham SW, Lam VK, Merckx VS.. 2017. Plastomes on the edge: the evolutionary breakdown of mycoheterotroph plastid genomes. New Phytol. 214(1):48–55. [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DAT, Ryan PD.. 2001. PAST: paleontological Statistics Software Package for Education and Data Analysis. Palaeontol Electronica 4:1–9. [Google Scholar]
- Kearse M, et al. . 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AH, Taylor DL, Watson LE.. 2011. Mycorrhizal specificity in the fully mycoheterotrophic Hexalectris Raf. (Orchidaceae: epidendroideae). Mol Ecol. 20(6):1303–1316. [DOI] [PubMed] [Google Scholar]
- Kennedy AH, Watson LE.. 2010. Species delimitations and phylogenetic relationships within the fully myco-heterotrophic Hexalectris (Orchidaceae). Syst Bot. 35(1):64–76. [Google Scholar]
- Kurtz S, et al. . 2001. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 29(22):4633–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam VK, Soto Gomez M, Graham SW.. 2015. The highly reduced plastome of mycoheterotrophic Sciaphila (Triuridaceae) is colinear with its green relatives and is under strong purifying selection. Genome Biol Evol. 7(8):2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GS, Barrett CF, Pang CC, Davis JI.. 2016. Drastic reduction of plastome size in the mycoheterotrophic Thismia tentaculata relative to that of its autotrophic relative Tacca chantrieri. Am J Bot. 103(6):1129–1137. [DOI] [PubMed] [Google Scholar]
- Lin CS, et al. . 2017. Concomitant loss of NDH complex-related genes within chloroplast and nuclear genomes in some orchids. Plant J. 90(5):994–1006. [DOI] [PubMed] [Google Scholar]
- Lohse M, Drechsel O, Kahlau S, Bock R.. 2013. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C. 2010. Phobos 3.3.11. Available from: http://www.rub.de/ecoevo/cm/cm_phobos.htm, last accessed May 1, 2018.
- Merckx V, Freudenstein JV.. 2010. Evolution of mycoheterotrophy in plants: a phylogenetic perspective. New Phytol. 185(3):605–609. [DOI] [PubMed] [Google Scholar]
- Merckx VSFT, Smets EF, Specht CD.. 2013. Biogeography and conservation of mycoheterotrophic plants In: Merckx V, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 103–156. [Google Scholar]
- Naumann J, et al. . 2016. Detecting and characterizing the highly divergent plastid genome of the nonphotosynthetic parasitic plant Hydnora visseri (Hydnoraceae). Genome Biol Evol. 8(2):345–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. 1985. Comparative organization of chloroplast genomes. Annu Rev Genet. 19:325–354. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: A language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. Available from: http://www.R-project.org/, last accessed May 1, 2018.
- Sosa V, Cameron KM, Angulo DF, Hernandez-Hernandez T.. 2016. Life form evolution in epidendroid orchids: ecological consequences of the shift from epiphytism to terrestrial habit in Hexalectris. Taxon 65(2):235–248. [Google Scholar]
- Taylor DL, Bruns TD, Szaro TM, Hodges SA.. 2003. Divergence in mycorrhizal specialization within Hexalectris spicata (Orchidaceae), a nonphotosynthetic desert orchid. Am J Bot. 90(8):1168–1179. [DOI] [PubMed] [Google Scholar]
- Taylor DL, et al. . 2013. Progress and prospects for the ecological genetics of mycoheterotrophs. In: Merckx V, editor. Mycoheterotrophy: the biology of plants living on fungi. New York: Springer; p. 245–266. [Google Scholar]
- Wicke S, et al. . 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 25(10):3711–3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, et al. . 2016. Mechanistic model of evolutionary rate variation en route to a nonphotosynthetic lifestyle in plants. Proc Natl Acad Sci U S A. 113(32):9045–9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicke S, Schneeweiss GM, dePamphilis CW, Müller KF, Quandt D.. 2011. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 76(3–5):273–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Morden CW, Palmer JD.. 1992. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci U S A. 89(22):10648–10652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL.. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20(17):3252–3255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.