Abstract
Background
Staphylococcus aureus bloodstream infections (BSI) cause significant morbidity and mortality due to the frequent antibiotic resistance, toxin and adhesin production of the bacterium. These characteristics differ significantly in methicillin resistant (MRSA) and methicillin sensitive S. aureus (MSSA) and also among isolates of different MRSA clones, contributing to the outcome of S. aureus bacteraemia.
Methods
In this study, all MRSA BSI isolates from Semmelweis University, Budapest, Hungary, isolated between 2011–2016 and the same number of matched MSSA (overall 306 isolates) were characterised in terms of antibiotic susceptibility, virulence genes, clonality and their association with all-cause 30-day mortality. Effect of patient related variables, such as age, gender and comorbidities were also investigated.
Results
ST22-MRSA-IV and ST5-MRSA-II were the most prevalent clones in our study. SCCmec I isolates showed the highest resistance rates and SCCmec II carried most virulence genes. Infections caused by SCCmec IV isolates were associated with the highest mortality rate (42.2%), despite the similar comorbidity rates of the different patient groups. All-cause 30-day mortality was 39.9% in the MRSA and 30.7% in the MSSA group. Increased teicoplanin MIC was associated with high mortality rate. Resistance to ciprofloxacin, erythromycin and clindamycin was common in MRSA, whereas MSSA isolates were more sensitive to all antibiotics with the exception of doxycycline. All MRSA isolates were sensitive to glycopeptides and linezolid; resistance to rifampicin and sulfamethoxazole-trimethoprim was low. MRSA isolates carried more adhesion genes, superantigens were more frequent in MSSA. Panton-Valentine leukocidin was found in 2.3% of the isolates.
Conclusion
This study provides insight into the clonal composition and associated mortality of BSI S. aureus isolates in Hungary. The results suggest that the outcome of the infection is determined by the antibiotic resistance, genotype of the bacterium, and patient-related factors; rather than the virulence factors carried by the bacteria.
Keywords: Staphylococcus aureus, Bloodstream infection, MRSA, MSSA, Clonality
Background
Bloodstream infections (BSI) are the most severe form of Staphylococcus aureus infections [1]. Frequent antibiotic resistance, toxin and adhesin production of the bacterium result in significant morbidity and mortality.
Nevertheless, not all S. aureus isolates are the same. Antibiotic resistance and virulence of methicillin resistant (MRSA) and methicillin sensitive (MSSA) S. aureus differ significantly, contributing to the variable outcome of S. aureus BSI. Moreover, isolates of different MRSA clones also vary significantly in their antibiotic susceptibility, virulence and speed of replication, however, the impact of a specific clone on the clinical outcome of the infection is less studied.
Staphylococcus aureus express many cell-surface associated adhesins termed ‘microbial surface components recognizing adhesive matrix molecules’ (MSCRAMMs), allowing the bacterium to bind to extracellular matrix proteins (ECM) of the host, contributing to invasion and infection. Polysaccharide intercellular adhesin (PIA), also referred as poly N-acetylglucosamine (PNAG) mediates bacterial adhesion and is important part of staphylococcal biofilm. PIA is synthetized by N-acetylglucosamyl transferase, the product of icaA gene [2]. Collagen binding protein (CNA) has an important role in the pathogenesis of S. aureus, enhancing the adherence of the bacterium to connective tissue, and thus allow to cuase cause wound, skin and soft tissue infections [3]. Staphylococcal Protein A (SpA) is produced by the vast majority of clinical S. aureus strains and by binds to Fc and Fab domains of IgG antibodies, thus supresses immune response [4].
It is well-established that MRSA isolates are often multi-resistant towards antibiotics of different classes, while a significant proportion of MSSA strains are sensitive to non-β-lactam antibiotics. The virulence of the pathogen and the outcome of the infection are, however, difficult to compare between MRSA and MSSA isolates. Most studies report increased mortality rate in patients with MRSA infections [5]. Some other investigations debate this, suggesting that adjustment to confounding factors, such as comorbidities, age and severity of illness, and the delayed initiation of effective therapy may nullify the impact of resistance on outcome [6]. As toxin gene frequency may be as high in MSSA isolates as in MRSA, infections caused by MSSA should be taken seriously [7].
The genotype of the isolate may also have a role in the severity and outcome of the infection. In Hungary, during the 1990s the most prevalent S. aureus lineage was the ST239-MRSA-III (Hungarian/Brazilian) clone, replaced by the ST228-MRSA-I (South-German) and the ST5-MRSA-II (New York-Japan) clone from the beginning of the 2000s [8, 9]. An ESCMID survey on dominant clones of BSI S. aureus isolates in the European region in 2011 described ST22-EMRSA-15 being abundant in Hungary [10]. However, to our knowledge, this is the first comprehensive study describing antibiotic resistance, virulence factors and current clones of S. aureus BSI isolates from Hungary.
The objectives of this study were: [1] to compare antibiotic susceptibility, prevalence of virulence factors, genotype and mortality of patients with MRSA and MSSA strains from blood-stream infections over a 6-year period and [2] to gain insight into the S. aureus population currently causing bloodstream infection at our tertiary level health care clinic in Budapest, Hungary.
Methods
Strain collection
All non-duplicated blood stream infection (BSI) MRSA strains—isolated between January 2011 and December 2016, at the Institute of Laboratory Medicine, Semmelweis University, Budapest, Hungary—were included. Our laboratory serves a 2200-bed teaching hospital, with 135 000–140 000 inpatients admitted annually. Each year, the same number of MSSA BSI isolates, representing the same gender and age distribution of population and hospital wards were enrolled (from a much larger pool) to be compared to the MRSA strains. In total, 306 S. aureus BSI isolates (153 MRSA and 153 MSSA strains) were analysed. The isolates were non-epidemic strains. Patient data collected for each isolate included gender, age, comorbidities, current chemotherapy and steroid therapy, and all-cause 30-day mortality. Charlson comorbidity index was determined for each patient.
The statistical analysis of the results was carried out by Pearson’s Chi square test and Mann–Whitney U test using Microsoft Office Excel Analysis ToolPak. P-values less than 0.05 were considered statistically significant.
Identification and antibiotic susceptibility testing
The identification of S. aureus strains was carried out by standard and MALDI-TOF MS analysis (Bruker Corporation, USA). Genotypic identification was based on the detection of nucA, mecA and mecC genes by PCR [11, 12]. The ATCC 33591 and ATCC BAA-2312 strains were applied as positive controls for the mecA and mecC PCR, respectively.
Antibiotic susceptibility to oxacillin, erythromycin, clindamycin, gentamicin, tobramycin, amikacin, doxycyline, sulfamethoxazole-trimethoprim, rifampicin, linezolid and ciprofloxacin was tested by disc diffusion method, and MRSA isolates were additionally tested for vancomycin and teicoplanin susceptibility by broth microdilution according to the European Committee of Antibiotic Susceptibility Testing (EUCAST) guidelines [13].
Molecular analysis of virulence genes
Presence of the genes hla, hlb, hlg, hlg-v, spa, lukS-PV/lukF-PV, icaA, cna, sea, seb, sec, tst, eta, etb was detected by PCR [14–20]. For the detection of sea, eta and etb, we used newly designed primers (Table 1).
Table 1.
Primers used for detection of sea, eta and etb genes in S. aureus strains
| Gene | Primer sequence (5′-3′) | Annealing temperature (°C) | Amplicon size (bp) |
|---|---|---|---|
| sea for | TTATCAATGTGCGGGTGGTA | 54 | 265 |
| sea rev | CCTCTGAACCTTCCCATCAA | ||
| eta for | AAAAACCATGCAAAAGCAGAA | 54 | 372 |
| eta rev | ACCTGCACCAAATGGTTCTT | ||
| etb for | CAGCGCAGAAGAAATCAGAA | 54 | 609 |
| etb rev | CCGCCTTTACCACTGTGAAT |
Genotyping
Pulsed field gel electrophoresis (PFGE) was performed after SmaI digestion for 151 MRSA and 153 MSSA strains according to a previously published method [21]. SCCmec typing was performed for all MRSA isolates by PCR as described previously [22, 23]. Multi locus sequence typing (MLST) was carried out on a subset of representative isolates of the most prevalent PFGE pulsotypes and SCCmec types identified in our study and MLST sequence types were assigned through the MLST database [24].
Results
Patient characteristics
Most of our BSI samples in the study period originated from patients admitted to the internal medicine unit (42.2%), intensive care unit (18.0%), haematology (15.7%), with less isolates recovered from cardiology (10.3%), surgery ward (6.5%), transplant clinic (3.9%) and pulmonology ward (2.9%) of the University.
The baseline characteristics and Charlson comorbidity index (CCI) of the patients are shown in Table 2. Chronic liver disease and chemotherapy was more frequent in MSSA patients, whereas more of MRSA patients had surgery in the previous 30 days or endocarditis, however, Charlson comorbidity index did not differ significantly in the two groups (Table 2).
Table 2.
Characteristics of S. aureus BSI patients
| MSSA | MRSA | P | |||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Diabetes | 48 | 31.4 | 57 | 37.3 | 0.2785 |
| Chronic liver disease | 33 | 21.6 | 13 | 8.5 | 0.0014 |
| Chronic kidney disease | 26 | 17.0 | 21 | 13.7 | 0.4279 |
| Solid tumor | 39 | 25.5 | 25 | 16.3 | 0.0491 |
| Haematology malignancy | 20 | 13.1 | 15 | 9.8 | 0.3691 |
| Chemotherapy | 21 | 13.7 | 4 | 2.6 | 0.0006 |
| Steroid treatment | 14 | 9.2 | 9 | 5.9 | 0.2783 |
| Surgery in previous 30 days | 34 | 22.2 | 54 | 35.3 | 0.0115 |
| Endocarditis | 3 | 2.0 | 12 | 7.8 | 0.0172 |
| Charlson comorbidity index (mean) | 4.65 | 4.36 | 0.72634 | ||
The dominance of male gender among MRSA infections was statistically significant (61.4% males versus 38.6% females, p = 0.044). CCI was significantly higher in female patients (4.92 vs 4.24 in males, p = 0.0164).
Age range of the MRSA patients was 0–98 years (median = 68 years). MSSA patients were selected to match these figures as close as possible (0–94 years, median = 64 years).
Antibiotic resistance of MRSA and MSSA strains
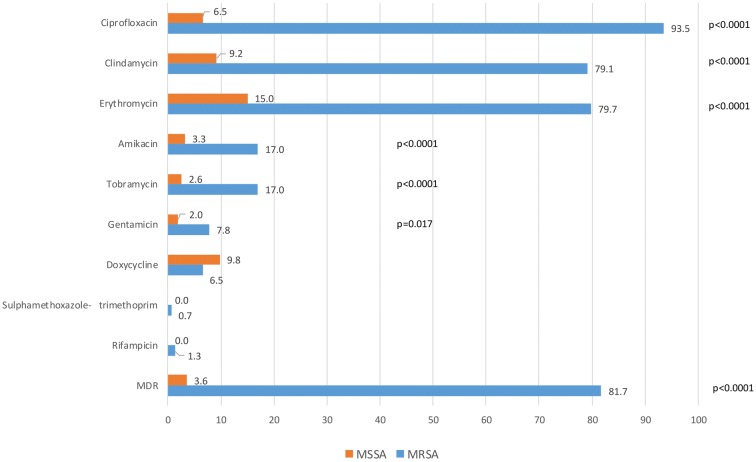
Resistance rates of the MRSA isolates were significantly higher towards ciprofloxacin, erythromycin, clindamycin, amikacin, tobramycin and gentamicin compared to MSSA isolates. The resistance rates of MSSA isolates were the highest to erythromycin and doxycycline (Fig. 1). Almost all isolates were sensitive to sulfamethoxazole-trimethoprim and rifampicin.
Fig. 1.
Antibiotic resistance of MRSA and MSSA isolates (Resistance rate (%) and multidrug resistance rate (MDR) (%))
The majority of MRSA isolates was multidrug-resistant (81.7%), i.e. resistant to at least three different antibiotic classes. Most prevalent resistance phenotype was resistance to β-lactams, erythromycin, clindamycin and ciprofloxacin. On the other hand, 75.8% of MSSA isolates were susceptible to all tested antibiotics and only 3.6% of them were multidrug-resistant (Fig. 1).
All MRSA and MSSA isolates were sensitive to vancomycin, teicoplanin and linezolid. Glycopeptide MICs ranged < 0.5–2 mg/L in each year. Vancomycin MIC was 2 mg/L in 7.8% of isolates. While in 2011–2012 less than 42% of the isolates had MIC ≥ 1 mg/L, in 2013–2015 more than 60% of the strains had MIC ≥ 1 mg/L. In 2016, their prevalence decreased to 51.7%. Vancomycin MIC50 values also increased from 0.5 mg/L in 2011–2012 to 1 mg/L in 2013–2016. Six point five percent of the isolates had teicoplanin MIC = 2 mg/L.
Virulence factors of MRSA and MSSA isolates
Among the examined virulence genes, toxic shock syndrome toxin and exfoliative toxin A and B encoding genes (tst, eta, etb) were detected in 1.3% of all strains. LukS-PV/lukF-PV gene was found in 2.3% of the isolates (Table 3). Out of the 14 studied virulence genes, cna, sea, ica and hlb were significantly more prevalent in MRSA, whereas tst, eta, sec and hlgv were significantly more frequent in MSSA. Superantigens were more frequent in MSSA isolates, while adhesins were more frequent in MRSA isolates (Tables 2 and 3). LukS-PV/lukF-PV positivity rate was 3.3% and 1.3% in MRSA vs MSSA, respectively. The prevalence of this gene changed significantly during the 6 years of the study: in was 13% in MRSA isolates in 2011, but never exceeded 4% in the later years.
Table 3.
Prevalence of virulence factors in MRSA and MSSA isolates
| Virulence genes | MRSA | MSSA | All | p values | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| Superantigens | |||||||
| tst | 0 | 0.0 | 4 | 2.6 | 4 | 1.3 | 0.044 |
| eta | 0 | 0.0 | 4 | 2.6 | 4 | 1.3 | 0.044 |
| etb | 1 | 0.7 | 3 | 2.0 | 4 | 1.3 | 0.3141 |
| sea | 30 | 19.6 | 17 | 11.1 | 47 | 15.4 | 0.0390 |
| seb | 58 | 37.9 | 49 | 32.0 | 107 | 35.0 | 0.2806 |
| sec | 12 | 7.8 | 25 | 16.3 | 37 | 12.1 | 0.0223 |
| Cytotoxins | |||||||
| lukS-PV/lukF-PV | 5 | 3.3 | 2 | 1.3 | 7 | 2.3 | 0.2513 |
| hla | 111 | 72.5 | 118 | 77.1 | 229 | 74.8 | 0.3564 |
| hlb | 106 | 69.3 | 75 | 49.0 | 181 | 59.2 | 0.0003 |
| hlg | 89 | 58.2 | 84 | 54.9 | 173 | 56.5 | 0.5642 |
| hlg-v | 31 | 20.3 | 93 | 60.8 | 124 | 40.5 | < 0.0001 |
| Adhesins | |||||||
| icaA | 122 | 79.7 | 85 | 55.6 | 207 | 67.6 | < 0.0001 |
| spa | 150 | 98.0 | 152 | 99.3 | 302 | 98.7 | 0.3141 |
| cna | 110 | 71.9 | 45 | 29.4 | 155 | 50.7 | < 0.0001 |
Italic values indicate statistically significant associations. (p < 0.05)
MRSA strains carried a median of six virulence genes. The most frequent virulence type in MRSA was positivity for hla, hlb, hlg, ica, spa, cna, and sea or seb (11.1% and 14.4% of the isolates, respectively). Isolates were highly diverse; we identified 57 different virulence gene combinations in MRSA isolates. MSSA strains carried less virulence factors (median of 5). Most frequent virulence type in MSSA was hla, hlb, hlg, hlgv, ica, spa positivity.
Clonality of the isolates
PFGE divided MRSA strains into 3 main pulsotypes, while MSSA strains proved to be much more diverse, no dominant clone could be identified (Fig. 2a, b).
Fig. 2.
PFGE patterns of the MRSA strains (a) and the MSSA strains (b)
The vast majority of the MRSA isolates in our study belonged to SCCmec type IV (66.7%). SCCmec II accounted for 23.5%, SCCmec I for 9.2% of the strains. One isolate belonged to SCCmec type V, whereas SCCmec types III and VI were not found. SCCmec type IV isolates were significantly more frequent in females (78.0% vs 59.6% in males, p = 0.0188).
MLST analysis was carried out on 12 representative MRSA isolates from the most frequent PFGE pulsotypes and SCCmec types, representing all 6 years of the study. All eight tested SCCmec IV, PFGE type A isolates belonged to the ST22 clone. Three SCCmec II, PFGE type B isolates were typed: two belonged to ST5 and one to ST225. Our representative SCCmec type I, pulsotype C isolate belonged to ST1.
Although SCCmec IV isolates dominated among the MRSA, these showed lower resistance to most antibiotics compared to SCCmec I and II isolates. Especially SCCmec I was associated with high resistance rates to aminoglycosides and doxycycline. Furthermore, the highest vancomycin MICs were observed also among these latter SCCmec types (Table 4).
Table 4.
Antibiotic resistance rates and high vancomycin MIC in different SCCmec types
| SCCmec I n = 14 |
SCCmec II n = 36 |
SCCmec IV n = 102 |
MRSA all n = 153 |
|||||
|---|---|---|---|---|---|---|---|---|
| R | % | R | % | R | % | R | % | |
| Erythromycin | 13 | 92.9 | 33 | 91.7 | 75 | 73.5 | 122 | 79.7 |
| Clindamycin | 13 | 92.9 | 32 | 88.9 | 75 | 73.5 | 121 | 79.1 |
| Gentamicin | 6 | 42.9 | 3 | 8.3 | 3 | 2.9 | 12 | 7.8 |
| Tobramycin | 7 | 50.0 | 12 | 33.3 | 7 | 6.9 | 26 | 17.0 |
| Amikacin | 7 | 50.0 | 12 | 33.3 | 7 | 6.9 | 26 | 17.0 |
| Ciprofloxacin | 11 | 78.6 | 35 | 97.2 | 97 | 95.1 | 143 | 93.5 |
| Sulfamethoxazole-trimethoprim | 0 | 0.0 | 0 | 0.0 | 1 | 1.0 | 1 | 0.7 |
| Doxycycline | 3 | 21.4 | 1 | 2.8 | 5 | 4.9 | 10 | 6.5 |
| Rifampicin | 1 | 7.1 | 0 | 0.0 | 1 | 1.0 | 2 | 1.3 |
| Vancomycin MIC = 2 mg/L | 3 | 21.4 | 4 | 11.1 | 4 | 3.9 | 10 | 6.5 |
SCCmec II isolates had the highest number of virulence genes. Panton-Valentine leukocidin was found only in SCCmec I and II isolates (Table 5). Interestingly, our single SCCmec V isolate did not carry any of the tested virulence factors.
Table 5.
Prevalence of virulence genes in different SCCmec types
| SCCmec I n = 14 |
SCCmec II n = 36 |
SCCmec IV n = 102 |
MRSA all n = 153 |
|||||
|---|---|---|---|---|---|---|---|---|
| R | % | R | % | R | % | R | % | |
| tst | 0 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0% |
| eta | 0 | 0.0 | 0 | 0 | 0 | 0 | 0 | 0% |
| etb | 0 | 0.0 | 1 | 2.8 | 0 | 0 | 1 | 0.7% |
| sea | 6 | 42.9 | 11 | 30.6 | 13 | 12.7 | 30 | 19.6% |
| seb | 5 | 35.7 | 12 | 33.3 | 41 | 40.2 | 58 | 37.9% |
| sec | 0 | 0.0 | 4 | 11.1 | 8 | 7.8 | 12 | 7.8% |
| pvl | 2 | 14.3 | 3 | 8.3 | 0 | 0.0 | 5 | 3.3% |
| hla | 10 | 71.4 | 29 | 80.6 | 72 | 70.6 | 111 | 72.5% |
| hlb | 8 | 57.1 | 23 | 63.9 | 75 | 73.5 | 106 | 69.3% |
| hlg | 5 | 35.7 | 22 | 61.1 | 62 | 60.8 | 89 | 58.2% |
| hlg-v | 10 | 71.4 | 19 | 52.8 | 2 | 2.0 | 31 | 20.3% |
| icaA | 11 | 78.6 | 28 | 77.8 | 83 | 81.4 | 122 | 79.7% |
| spa | 14 | 100.0 | 36 | 100.0 | 100 | 98.0 | 150 | 98.0% |
| cna | 5 | 35.7 | 12 | 33.3 | 93 | 91.2 | 110 | 71.9% |
| Median of n of virulence genes | 5.5 | 7 | 6 | 6 | ||||
Differences in mortality
Overall 30-day mortality was 35.3% in our BSI S. aureus cases, with higher rates in BSI cases caused by MRSA (39.9% vs 30.7% in MSSA, respectively, p < 0.0001), although CCI did not differ significantly in the 2 groups.
Females had significantly higher CCI than males, this can attribute to the higher mortality rate in this group (38.7% vs 33.2% of fatalities in females and males, respectively (p < 0.001)). Mortality increased with age: it was 20.0% in age group 0–49 years, 28.0% in 50–64 years, 40.2% in 65–79 years and 55.8% in patients older than 80 years.
Higher vancomycin MIC did not influence mortality risk. On the other hand, although we have found only 10 isolates with teicoplanin MIC of 2 mg/L, mortality was 70% in this group.
The number of carried virulence genes, the presence of specific virulence factors and antibiotic resistance to other drugs besides glycopeptides did not influence mortality.
Association of genotype, CCI and mortality are shown in Table 6. Interestingly, patients infected with SCCmec IV isolates had higher mortality, than patients infected with SCCmec I and II MRSA strains, however, these differences were not statistically significant. CCI of SCCmec I group and SCCmec II group do not differ significantly when compared to CCI of SCCmec IV group.
Table 6.
Association of S. aureus genotype, CCI and mortality rate
| MRSA-SCCmec I | MRSA-SCCmec II | MRSA-SCCmec IV | MSSA | |
|---|---|---|---|---|
| Charlson comorbidity index | 4.29 | 3.97 | 4.46 | 4.65 |
| Mortality rate (%) | 28.6 | 36.1 | 42.2 | 30.7 |
Discussion
Differences between MRSA and MSSA
Staphylococcus aureus is a significant and prevalent pathogen, however, the importance of methicillin resistance in the virulence of the bacterium and in the outcome of the infection is still not completely clear. According to a recent study from the USA, MRSA bacteraemia is associated with a higher risk of readmission for bacteraemia recurrence, increased mortality, and longer hospitalization [25]. Most studies support the concept that MRSA BSI is associated with poorer outcome [5], while some other researchers debate this, and report mortality comparable to that in MSSA BSI [6, 26]. Some studies even suggest that MSSA strains may cause more severe infections, which might be related to higher prevalence of virulence genes in MSSA or to the greater fitness cost associated with SCCmec cassettes in MRSA [27]. In our study, we observed higher mortality rates in patients with MRSA infections than those with MSSA. Female gender, older age, infection with SCCmec IV isolate and teicoplanin MIC = 2 mg/L were additional risk factors for mortality.
Higher antibiotic resistance of MRSA isolates may be an explanation for high mortality rates, as inappropriate empirical antibiotic therapy is described to be more frequent in patients with MRSA bacteraemia [28]. In our study, we found significantly higher resistance rates to several antibiotics and also more frequent multidrug resistance rate in MRSA than in MSSA as well.
In addition, we found different patterns of virulence genes in MRSA and MSSA isolates. Particularly adhesion factors (cna and ica) were significantly more prevalent in MRSA, meanwhile genes encoding for superantigens (especially sea and eta and tst) were more prevalent in MSSA isolates (Table 3). We found a low overall prevalence of pvl. On average, MRSA isolates carried more virulence genes than MSSA isolates. However, the number of the carried virulence genes or the presence of specific virulence genes did not influence 30-day all-cause mortality. Our findings suggest that the outcome of the infection is related to the antibiotic resistance and clonality of the bacterium and to patient-related factors, such as age and gender, rather than the virulence factors of the bacteria.
Antibiotic susceptibility
MRSA rates
In our laboratory, the MRSA rate among BSI S. aureus isolates varied between 27.5% and 40.7% during the investigated 6-years period. Similarly to a number of countries worldwide, MRSA prevalence decreased in Hungary in the recent years [29]. According to the surveillance data from the National Public Health Institute of Hungary, the proportion of MRSA strains among invasive S. aureus samples in Hungary increased from 20.2% to 30.1% between 2005 and 2010 (p < 0.001), then decreased to 24.2% by 2012 (p < 0.001). Since 2012, the prevalence of oxacillin resistance remained stable around 24% (data published in Hungarian) [30]. As our laboratory serves mostly university clinics, different patient population and increased disease severity may be responsible for the higher MRSA rates compared to the national average.
Glycopeptide susceptibility
The gradual increase of the number of MRSA isolates with high glycopeptide MIC values, referred as ‘vancomycin MIC creep’ in the literature is controversial; many studies report an increase of MICs, while others did not confirm these findings [31]. In our study, all MRSA isolates were sensitive to glycopeptides, however, vancomycin MIC seemed to creep higher from 2011 until 2015, with a slight decrease in 2016. The possibility of gradual increase in vancomycin MIC requires special attention, as it might lead to the development of resistant strains, and poorer clinical outcome was reported in patients infected with isolates exhibiting higher glycopeptide MIC values [31]. In our study, elevated vancomycin MIC was not associated with increased 30-day mortality, but patients with teicoplanin MIC = 2 mg/L had higher mortality than those with low teicoplanin MIC values.
Rifampicin resistance was very low in MRSA in our study, and none of the MSSA isolates were resistant to this drug, similar to data from other European countries [29]. Almost all of our isolates have retained susceptibility towards sulfamethoxazole-trimethoprim, in concordance with reports from Europe and other locations worldwide [32, 33].
Virulence
The prevalence of virulence genes in BSI S. aureus isolates varies highly according to geographical region and patient population.
Several studies have found low prevalence of PVL in S. aureus isolates from BSIs, and described it to be more closely associated with skin and soft tissue infections [27, 34]. However, for instance, pvl in BSI was more prevalent in Romania (22.4%) [35]. In our study, we found low pvl rate (2.3% for all samples). In a recent study on BSI MSSA, decreasing prevalence of pvl and other virulence genes during recent years was observed [27]. This is in concordance with our findings in MRSA isolates: in 2011, 13.0% of our MRSA isolates were positive for pvl, however, in the following years pvl frequency never exceeded 4% in our MRSA isolates. Tst was found exclusively in MSSA. Number of carried virulence genes and presence of specific virulence factors did not influence the outcome of the infection.
Clonality
It is well demonstrated that successful S. aureus clones are invading and replacing their competitors, changing the clonal map over time. In North and South America, and in Japan, USA 300 MRSA clone is becoming more prevalent, while in Europe and Asia ST239 Hungarian-Brazilian strains are being replaced by ST22-MRSA-IV (also known as EMRSA-15) [36]. According to a South-German study from 2016, this strain appeared in 2001 and became rapidly more common in their samples, accounting for nearly 80% of the MRSA strains in 2013 [35]. It was described as the most prevalent sequence type in NICU patient in Italy [37]. It has been causing nosocomial infections in the UK and in Ireland since the beginning of the 2000s [38] and was also described outside Europe, for example in Kuwait [39]. However, there are considerable differences in antibiotic susceptibility and virulence factors between variants of ST22-MRSA-IV clone. For example, UK-EMRSA-15/”Middle Eastern Variant” is generally susceptible to antibiotics and is characterized by the presence of tst1 gene, whereas our isolates completely lack this gene [37]. Another variant of ST22-MRSA-IV clone is positive for PVL [35], however, all of our SCCmec IV strains were PVL negative. SCCmec IV was previously considered as a usually community-acquired MRSA, however, it became widespread and successful in hospital settings, too [40]. In Hungary, ST22-MRSA-IV has been the most frequently described clone among BSI isolates since 2008, its prevalence was nearly 60% in 2013, however, its prevalence is decreasing in the recent years (personal communication from Ákos Tóth, National Public Health Center of Hungary). In our study, the majority (66.7%) of the BSI MRSA isolates belonged to the ST22-SCCmec IV type.
Our SCCmec II isolates belonged to clonal complex 5 (CC5). ST5-MRSA-II and its MLST single locus variant, ST225-MRSA-II were both found. ST5-MRSA-II (New York –Japan or Rhine-Hesse clone) has high worldwide prevalence [40]. It was the most prevalent MRSA type in the 2000s in Hungary, until its replacement by EMRSA-15 [9]. A representative of our PFGE type C isolates belonged to ST1-MRSA-I, a non-epidemic clone, which was found in a low number of patients, for example, in Croatia and in Italy [41, 42].
In accordance with other studies, we have found high genotypic diversity in MSSA strains, no predominant clone was identified among those isolates (Fig. 2b) [27, 43].
Role of SCCmec in the resistance, virulence and mortality of infected patients
In our study, SSCmec I and II isolates were associated with the highest rates of antibiotic resistance, while SCCmec IV was associated with low resistance, except for ciprofloxacin (Table 4). According to the literature, SCCmec types I-II-III are more likely to exhibit high glycopeptide MIC values and vancomycin hetero-resistance, than SCCmec IV isolates [44], our findings are in concordance with this (Table 4).
SCCmec II isolates carried the highest number of virulence genes. All our pvl positive MRSA isolates belonged to SCCmec type I or II (Table 5). Earlier studies had associated Panton-Valentine leukocidin positivity with CA-MRSA, but later on pvl genes emerged in various MRSA clones via PVL bacteriophages [45].
Although SCCmec type I isolates had higher resistance rates to antibiotics and SCCmec type II strains had the most virulence genes, interestingly, infections caused by SCCmec type IV isolates had the highest mortality, whereas their CCI value did not differ significantly (Table 6). As described by Recker et al., bacterial phenotype and genotype are highly predictive for adverse infection outcome, and have stronger impact on mortality than other factors, such as patient age, gender or comorbidities [46]. Our findings support that the genotype of the bacterium has a major influence on the outcome of the infection. This underlines the importance of having up-to-date knowledge on the clonal types circulating at a given location.
Our study has the following limitations: it focuses on strains originating from a single centre, only a subset of the isolates was analysed by MLST, severity of the illness at presentation and antibiotic treatment of the patients were not analysed.
Conclusions
In conclusion, antibiotic resistance and virulence of MRSA and MSSA isolates differ significantly. In our population, higher 30-day mortality was associated with BSI caused by MRSA and by strains with high teicoplanin MIC value. ST22-MRSA-IV was the dominant clone in our samples, and caused higher mortality than ST5-MRSA-II and ST1-MRSA-I strains. MSSA isolates had significantly lower antibiotic resistance rates and somewhat lower virulence gene prevalence than MRSA isolates. However, 30-day mortality of patients with MSSA BSI was still high, thus MSSA infections should be treated with appropriate care.
Staphylococcus aureus BSI continues to present a major clinical challenge. Further studies are required to gain a better understanding of differences in the antibiotic resistance, virulence and genotype of the bacterium and the impact of these factors on disease outcome.
Acknowledgements
The authors are thankful to Professor Barna Vásárhelyi (Head of the Laboratory Medicine Institute) for critically reviewing the manuscript. We are very grateful for Natasa Pesti for her help in the laboratory work. Parts of this study were presented at the 18th International Congress of the Hungarian Society for Microbiology, Budapest, July 2019.
Authors’ contributions
AH contributed to the acquisition, analysis and interpretation of data and was a major contributor in writing the manuscript. OD and KK designed the study, analysed data, and revised the manuscript. JST, EJ, JP, MI and EF worked on data acquisition and analysis. All authors read and approved the final manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
None.
Ethics approval and consent to participate
Ethical approval and informed consent were not required for this study as isolates were collected as part of routine patient care and no identifiable patient data was accessed for the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andrea Horváth, Email: horvath.andrea1@med.semmelweis-univ.hu.
Orsolya Dobay, Email: dobay.orsolya@med.semmelweis-univ.hu.
Judit Sahin-Tóth, Email: sahintothjudit@gmail.com.
Emese Juhász, Email: juhasz.emese@med.semmelweis-univ.hu.
Júlia Pongrácz, Email: pongracz.julia@med.semmelweis-univ.hu.
Miklós Iván, Email: ivan.miklos@med.semmelweis-univ.hu.
Enikő Fazakas, Email: fazakaseniko93@gmail.com.
Katalin Kristóf, Email: kristof.katalin@med.semmelweis-univ.hu.
References
- 1.Lakhundi S, Zhang K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev. 2018;31(4):e00020-18. doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin MH, Shu JC, Lin LP, Chong KY, Cheng YW, Du JF, et al. Elucidating the crucial role of poly N-acetylglucosamine from Staphylococcus aureus in cellular adhesion and pathogenesis. PLoS ONE. 2015;10(4):e0124216. doi: 10.1371/journal.pone.0124216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madani A, Garakani K, Mofrad MRK. Molecular mechanics of Staphylococcus aureus adhesin, CNA, and the inhibition of bacterial adhesion by stretching collagen. PLoS ONE. 2017;12(6):e0179601. doi: 10.1371/journal.pone.0179601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim HK, Falugi F, Thomer L, Missiakas DM, Schneewind O. Protein A suppresses immune responses during Staphylococcus aureus bloodstream infection in guinea pigs. MBio. 2015;6(1):e02369-14. doi: 10.1128/mBio.02369-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang JT, Hsu LY, Lauderdale TL, Fan WC, Wang FD. Comparison of outcomes among adult patients with nosocomial bacteremia caused by methicillin-susceptible and methicillin-resistant Staphylococcus aureus: a retrospective cohort study. PLoS ONE. 2015;10(12):e0144710. doi: 10.1371/journal.pone.0144710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Hal SJ, Jensen SO, Vaska VL, Espedido BA, Paterson DL, Gosbell IB. Predictors of mortality in Staphylococcus aureus Bacteremia. Clin Microbiol Rev. 2012;25(2):362–386. doi: 10.1128/CMR.05022-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imani Fooladi AA, Ashrafi E, Tazandareh SG, Koosha RZ, Rad HS, Amin M, et al. The distribution of pathogenic and toxigenic genes among MRSA and MSSA clinical isolates. Microb Pathog. 2015;81:60–66. doi: 10.1016/j.micpath.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 8.Conceicao T, Aires-de-Sousa M, Fuzi M, Toth A, Paszti J, Ungvari E, et al. Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin Microbiol Infect. 2007;13(10):971–979. doi: 10.1111/j.1469-0691.2007.01794.x. [DOI] [PubMed] [Google Scholar]
- 9.Horvath A, Dobay O, Kardos S, Ghidan A, Toth A, Paszti J, et al. Varying fitness cost associated with resistance to fluoroquinolones governs clonal dynamic of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 2012;31(8):2029–2036. doi: 10.1007/s10096-011-1536-z. [DOI] [PubMed] [Google Scholar]
- 10.Grundmann H, Schouls LM, Aanensen DM, Pluister GN, Tami A, Chlebowicz M, et al. The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill. 2014 doi: 10.2807/1560-7917.es2014.19.49.20987. [DOI] [PubMed] [Google Scholar]
- 11.Laub K, Tothpal A, Kardos S, Dobay O. Epidemiology and antibiotic sensitivity of Staphylococcus aureus nasal carriage in children in Hungary. Acta Microbiol Immunol Hung. 2017;64(1):51–62. doi: 10.1556/030.64.2017.001. [DOI] [PubMed] [Google Scholar]
- 12.Paterson GK, Larsen AR, Robb A, Edwards GE, Pennycott TW, Foster G, et al. The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J Antimicrob Chemother. 2012;67(12):2809–2813. doi: 10.1093/jac/dks329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.EUCAST European Comittee on Antimicrobial Susceptibility Testing—Clinical Breakpoints for bacteria.
- 14.Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, et al. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. 2002;70(2):631–641. doi: 10.1128/IAI.70.2.631-641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monday SR, Bohach GA. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J Clin Microbiol. 1999;37(10):3411–3414. doi: 10.1128/JCM.37.10.3411-3414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aarestrup FM, Larsen HD, Eriksen NH, Elsberg CS, Jensen NE. Frequency of alpha- and beta-haemolysin in Staphylococcus aureus of bovine and human origin. A comparison between pheno- and genotype and variation in phenotypic expression. APMIS. 1999;107(4):425–430. doi: 10.1111/j.1699-0463.1999.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 17.Lina G, Piemont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29(5):1128–1132. doi: 10.1086/313461. [DOI] [PubMed] [Google Scholar]
- 18.Campbell SJ, Deshmukh HS, Nelson CL, Bae IG, Stryjewski ME, Federspiel JJ, et al. Genotypic characteristics of Staphylococcus aureus isolates from a multinational trial of complicated skin and skin structure infections. J Clin Microbiol. 2008;46(2):678–684. doi: 10.1128/JCM.01822-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38(3):1032–1035. doi: 10.1128/JCM.38.3.1032-1035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker K, Friedrich AW, Lubritz G, Weilert M, Peters G, Von Eiff C. Prevalence of genes encoding pyrogenic toxin superantigens and exfoliative toxins among strains of Staphylococcus aureus isolated from blood and nasal specimens. J Clin Microbiol. 2003;41(4):1434–1439. doi: 10.1128/JCM.41.4.1434-1439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker J, Borrow R, Goering RV, Egerton S, Fox AJ, Oppenheim BA. Subtyping of methicillin-resistant Staphylococcus aureus isolates from the North-West of England: a comparison of standardised pulsed-field gel electrophoresis with bacteriophage typing including an inter-laboratory reproducibility study. J Med Microbiol. 1999;48(3):297–301. doi: 10.1099/00222615-48-3-297. [DOI] [PubMed] [Google Scholar]
- 22.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrob Agents Chemother. 2007;51(9):3374–3377. doi: 10.1128/AAC.00275-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Staphylococcus aureus MLST site https://pubmlst.org/saureus/.
- 25.Inagaki K, Lucar J, Blackshear C, Hobbs CV. Methicillin-Susceptible and Methicillin-Resistant Staphylococcus aureus bacteremia - Nationwide Estimates of 30-day Readmission, In-hospital Mortality, Length of Stay, and Cost in the US. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2019. [DOI] [PubMed]
- 26.Tom S, Galbraith JC, Valiquette L, Jacobsson G, Collignon P, Schonheyder HC, et al. Case fatality ratio and mortality rate trends of community-onset Staphylococcus aureus bacteraemia. Clin Microbiol Infect. 2014;20(10):O630–O632. doi: 10.1111/1469-0691.12564. [DOI] [PubMed] [Google Scholar]
- 27.Deasy EC, Brennan GI, Tecklenborg SC, Umeh C, Coleman DC, Shore AC. A molecular epidemiological investigation of methicillin-susceptible Staphylococcus aureus causing bloodstream infections in Ireland, 2006–2017. Clin Microbiol Infect. 2019;38(5):927–936. doi: 10.1007/s10096-019-03523-0. [DOI] [PubMed] [Google Scholar]
- 28.Wi YM, Rhee JY, Kang CI, Chung DR, Song JH, Peck KR. Clinical predictors of methicillin-resistance and their impact on mortality associated with Staphylococcus aureus bacteraemia. Epidemiol Infect. 2018;146(10):1326–1336. doi: 10.1017/S0950268818001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.European Centre for Disease Prevention and Control—Surveillance of antimicrobial resistance in Europe 2017 [https://ecdc.europa.eu/en/publications-data/surveillance-antimicrobial-resistance-europe-2017.
- 30.Hungarian National Antibiotic Surveillence Data [http://www.oek.hu/oek.web?to=2479&nid=505&pid=1&lang=hun.
- 31.Diaz R, Afreixo V, Ramalheira E, Rodrigues C, Gago B. Evaluation of vancomycin MIC creep in methicillin-resistant Staphylococcus aureus infections-a systematic review and meta-analysis. Clin Microbiol Infect. 2018;24(2):97–104. doi: 10.1016/j.cmi.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 32.Tissot-Dupont H, Gouriet F, Oliver L, Jamme M, Casalta JP, Jimeno MT, et al. High-dose trimethoprim-sulfamethoxazole and clindamycin for Staphylococcus aureus endocarditis. Int J Antimicrob Agents. 2019;54(2):143–148. doi: 10.1016/j.ijantimicag.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Paul M, Bishara J, Yahav D, Goldberg E, Neuberger A, Ghanem-Zoubi N, et al. Trimethoprim-sulfamethoxazole versus vancomycin for severe infections caused by meticillin resistant Staphylococcus aureus: randomised controlled trial. BMJ. 2015;350:h2219. doi: 10.1136/bmj.h2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saeed K, Gould I, Esposito S, Ahmad-Saeed N, Ahmed SS, Alp E, et al. Panton-Valentine leukocidin-positive Staphylococcus aureus: a position statement from the International Society of Chemotherapy. Int J Antimicrob Agents. 2018;51(1):16–25. doi: 10.1016/j.ijantimicag.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Monecke S, Jatzwauk L, Muller E, Nitschke H, Pfohl K, Slickers P, et al. Diversity of SCCmec elements in Staphylococcus aureus as observed in South-Eastern Germany. PLoS ONE. 2016;11(9):e0162654. doi: 10.1371/journal.pone.0162654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planet PJ, Narechania A, Chen L, Mathema B, Boundy S, Archer G, et al. Architecture of a Species: phylogenomics of Staphylococcus aureus. Trends Microbiol. 2017;25(2):153–166. doi: 10.1016/j.tim.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 37.Geraci DM, Giuffre M, Bonura C, Graziano G, Saporito L, Insinga V, et al. A Snapshot on MRSA Epidemiology in a Neonatal Intensive Care Unit Network, Palermo, Italy. Front Microbiol. 2016;7:815. doi: 10.3389/fmicb.2016.00815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson AP, Aucken HM, Cavendish S, Ganner M, Wale MC, Warner M, et al. Dominance of EMRSA-15 and -16 among MRSA causing nosocomial bacteraemia in the UK: analysis of isolates from the European Antimicrobial Resistance Surveillance System (EARSS) J Antimicrob Chemother. 2001;48(1):143–144. doi: 10.1093/jac/48.1.143. [DOI] [PubMed] [Google Scholar]
- 39.Udo EE, Boswihi SS, Al-Sweih N. High prevalence of toxic shock syndrome toxin-producing epidemic methicillin-resistant Staphylococcus aureus 15 (EMRSA-15) strains in Kuwait hospitals. New Microbes New Infect. 2016;12:24–30. doi: 10.1016/j.nmni.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner NA, Sharma-Kuinkel BK, Maskarinec SA, Eichenberger EM, Shah PP, Carugati M, et al. Methicillin-resistant Staphylococcus aureus: an overview of basic and clinical research. Nat Rev Microbiol. 2019;17(4):203–218. doi: 10.1038/s41579-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budimir A, Deurenberg RH, Bosnjak Z, Stobberingh EE, Cetkovic H, Kalenic S. A variant of the Southern German clone of methicillin-resistant Staphylococcus aureus is predominant in Croatia. Clin Microbiol Infect. 2010;16(8):1077–1083. doi: 10.1111/j.1469-0691.2009.03042.x. [DOI] [PubMed] [Google Scholar]
- 42.Campanile F, Bongiorno D, Borbone S, Stefani S. Hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) in Italy. Annals Clin Microbiol Antimicrob. 2009;8:22. doi: 10.1186/1476-0711-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perez-Montarelo D, Viedma E, Larrosa N, Gomez-Gonzalez C, de Ruiz-Gopegui E, Munoz-Gallego I, et al. Molecular epidemiology of Staphylococcus aureus bacteremia: association of molecular factors with the source of infection. Front Microbiol. 2018;9:2210. doi: 10.3389/fmicb.2018.02210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee HY, Chen CL, Liu SY, Yan YS, Chang CJ, Chiu CH. Impact of molecular epidemiology and reduced susceptibility to glycopeptides and daptomycin on outcomes of patients with methicillin-resistant Staphylococcus aureus bacteremia. PLoS ONE. 2015;10(8):e0136171. doi: 10.1371/journal.pone.0136171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu Q, Cheng H, Yuan W, Zeng F, Shang W, Tang D, et al. Panton-Valentine leukocidin (PVL)-positive health care-associated methicillin-resistant Staphylococcus aureus isolates are associated with skin and soft tissue infections and colonized mainly by infective PVL-encoding bacteriophages. J Clin Microbiol. 2015;53(1):67–72. doi: 10.1128/JCM.01722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Recker M, Laabei M, Toleman MS, Reuter S, Saunderson RB, Blane B, et al. Clonal differences in Staphylococcus aureus bacteraemia-associated mortality. Nat Microbiol. 2017;2(10):1381–1388. doi: 10.1038/s41564-017-0001-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
None.