Abstract
Alcoholism remains a prevalent health concern throughout the world. Previous studies have identified transcriptomic patterns in the brain associated with alcohol dependence in both humans and animal models. But none of these studies have systematically investigated expression within the unique cell types present in the brain. We utilized single nucleus RNA sequencing (snRNA-seq) to examine the transcriptomes of over 16 000 nuclei isolated from the prefrontal cortex of alcoholic and control individuals. Each nucleus was assigned to one of seven major cell types by unsupervised clustering. Cell type enrichment patterns varied greatly among neuroinflammatory-related genes, which are known to play roles in alcohol dependence and neurodegeneration. Differential expression analysis identified cell type-specific genes with altered expression in alcoholics. The largest number of differentially expressed genes (DEGs), including both protein-coding and non-coding, were detected in astrocytes, oligodendrocytes and microglia. To our knowledge, this is the first single cell transcriptome analysis of alcohol-associated gene expression in any species and the first such analysis in humans for any addictive substance. These findings greatly advance the understanding of transcriptomic changes in the brain of alcohol-dependent individuals.
Introduction
Alcohol abuse is involved in over 200 pathologies and health conditions (e.g. alcohol dependence, liver cirrhosis, cancers and injuries) and creates substantial social and economic burdens (1,2). To develop more effective therapeutic strategies, we must first understand how alcohol affects the body at the cellular and molecular level. Previous studies of transcriptomic responses in human alcoholics (3–6) relied on RNA extracted from brain regions using tissue homogenates comprised of multiple cell types. This approach likely masks differences in gene expression patterns among specific cells, as well as heterogeneity from cell-to-cell variation within a given cell type.
Single cell RNA sequencing (scRNA-seq) has recently gained attention in cell and molecular biology research for its ability to profile novel cell types and measure cell-to-cell variation in gene expression. To our knowledge, transcriptomic responses to chronic alcohol exposure or any other abused drug have not been studied at the cellular level in the human brain. We hypothesized that cell type-specific gene expression patterns associated with alcoholism will identify novel alcohol targets that were previously missed by bulk analysis of tissue homogenates, as has been shown for other neuropathologies (7,8). Using single nucleus RNA-seq (snRNA-seq), a popular scRNA-seq alternative for analyzing frozen brain tissue (9–18), we profiled the transcriptomes of 16 305 nuclei extracted from frozen prefrontal cortex (PFC) samples of four control and three alcohol-dependent individuals. The PFC is involved in executive function and is an important substrate in the reward circuitry associated with development of alcohol dependence (19). The PFC has also been the focus of many transcriptomic studies (3,4,20,21) and was a logical choice for our initial work. Using the approach presented here, we discovered novel cell type-specific transcriptome changes associated with alcohol abuse in the human PFC.
Results and Discussion
Like most organs in the body, the brain consists of a diverse array of cell types. In order to systematically assess the roles of different cells in alcohol dependence, we examined gene expression in the PFC of alcohol-dependent versus control donors at the single cell level (Supplementary Material, Table S1). For this specific approach, we utilized droplet-based snRNA-seq technology to prepare libraries from postmortem tissue samples. The dataset was comprised of transcriptomes of 16 305 nuclei from seven donors (four control donors and three donors with alcohol dependence) with approximately 30 000–35 000 total genes detected in each sample (Supplementary Material, Table S2). We visualized the data using uniform manifold approximation and projection (UMAP) (22). Despite variation in the number of transcript counts per nucleus among some of the samples from the different donors (Supplementary Material, Table S2), the nuclei did not separate by the donor or batch from which they were isolated (Fig. 1A and Supplementary Material, Fig. S1), suggesting that samples with fewer nuclei and transcripts detected had experienced unbiased undersampling, which is accounted for in the normalization steps (Methods). Therefore, we included all samples for further analysis in this study [see Supplementary Material, Fig. S3 for the impact of removing the sample with the lowest UMI/nucleus, 777C, on the differential expression (DE) analysis].
Figure 3.
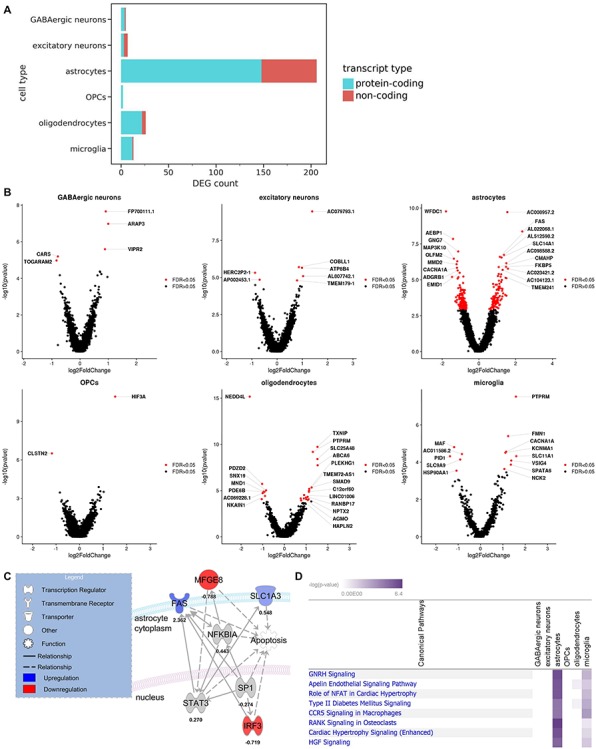
DEGs associated with alcoholism were detected in every neural cell type. (A) Breakdown of DEGs by cell type and transcript type (FDR < 0.05). (B) Volcano plots of top DEGs in each cell type (FDR < 0.05). (C) An IPA molecular network including four neuroinflammation-associated DEGs in astrocytes (FDR < 0.25). Solid lines indicate direct relationships, while dashed lines indicate indirect relationships. (D) Log2 fold changes and adjusted P-values of genes were processed by IPA with FDR < 0.25 as the cutoff for indicating significant DEGs. The top canonical pathways are shown. Negative log (P-values) are derived from Fisher’s exact test.
Figure 1.
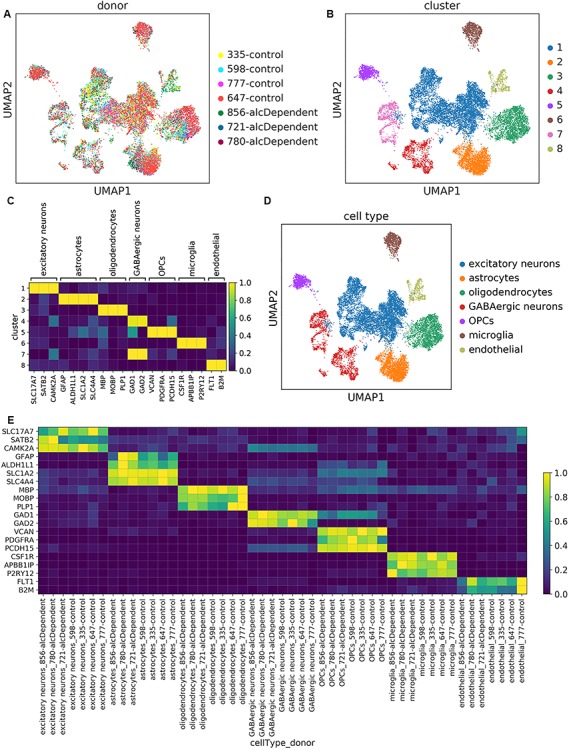
Unsupervised clustering captured expected cell types in every sample. (A) UMAP plots of the 16 305 nuclei in our dataset, colored by donor or (B) transcriptomically distinct clusters determined from unsupervised clustering. (C) Scaled mean expression of known cell type markers in the different clusters. For each gene, expression was scaled from 0.0 to 1.0 to maintain a balanced colormap. (D) UMAP plot of nuclei colored by cell type assignment. (E) Scaled mean expression of marker genes for each cell type in each donor.
Unsupervised clustering of nuclei
In order to identify transcriptomically distinct groups of nuclei, we performed unsupervised, graph-based clustering, yielding eight clusters (Fig. 1B). We then annotated the clusters using the following markers of major brain cell types that are consistently detected in single cell/nucleus transcriptomics studies (7,8,20,23,24): excitatory neurons, astrocytes, oligodendrocytes, inhibitory/GABAergic neurons, oligodendrocyte progenitor cells (OPCs), microglia, and endothelial cells (Fig. 1C and D). Two clusters corresponded with GABAergic neurons, while all other clusters corresponded to a single cell type. We confirmed that the cell types and signatures were conserved among samples from all of the donors by examining marker expression in each donor cell-type combination (Fig. 1E). Proportions of the different cell types were comparable to other snRNA-seq studies (8,23) with neurons (particularly excitatory neurons) being over-represented relative to non-neuronal cell types (Supplementary Material, Fig. S2). Furthermore, we did not find differences in cell type proportions between alcoholics and controls (Supplementary Material, Fig. S2). This is consistent with previous findings (3).
While some other single cell transcriptomics studies have attempted to define novel cell states and subtypes, our goal was to evaluate gene expression differences between alcoholics and controls in known, established cell types. Therefore, we performed the clustering using a low-resolution parameter (Materials and Methods) rather than using subclustering.
Neuroinflammatory signaling
The neuroimmune system is critical for the pathogenesis of neurological diseases such as Alzheimer’s disease and multiple sclerosis (25) and is also involved in regulating alcohol abuse and dependence (26). To date, however, single cell/nucleus RNA-seq studies have not specifically evaluated neuroimmune gene expression among cell types. Of the 85 human genes directly listed in the Neuroinflammation Signaling Pathway in the Ingenuity Pathway Analysis (IPA) database, 79 were detected in our snRNA-seq dataset, and 68 were detected in more than 20 nuclei. The expression of these genes among the different cell types is displayed in Fig. 2A. Expression of some genes was fairly pervasive across cell types (e.g. HMGB1) while other genes exhibited relative enrichment in one or two specific cell types (e.g. SNCA in excitatory neurons, BCL2 in astrocytes, P2RX7 in oligodendrocytes, GRIA1 in GABAergic neurons, S100B in OPCs, TLR2 in microglia (Fig. 2A–C) and CTNNB1 in endothelial cells). Nearly half of the genes whose expression was highest in astrocytes are involved with interferon signaling: BCL2 (27), IRF3, HMGB1, TICAM1 and TRAF3 (28).
Figure 2.
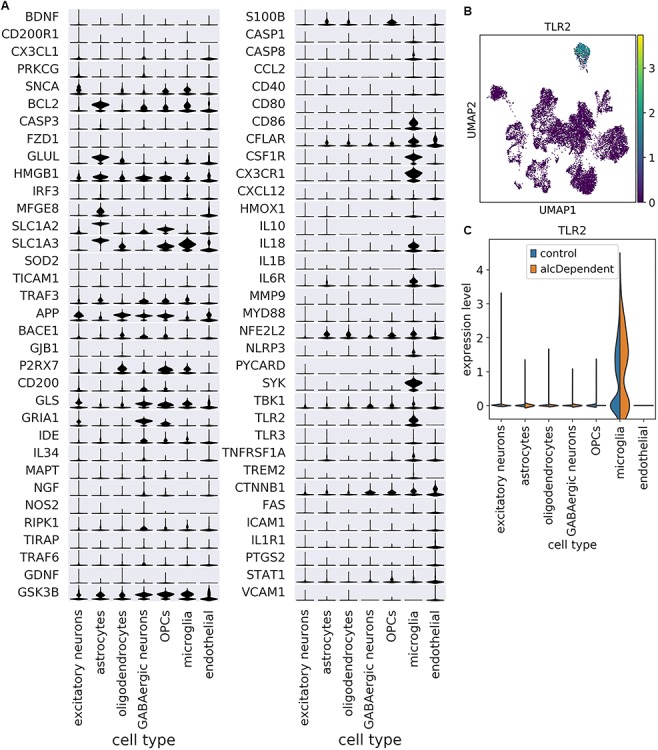
Cell type enrichment patterns vary among neuroimmune genes. (A) Scaled expression of neuroimmune genes among cell types. Genes are ordered by which cell type had the highest mean expression. Scaling was done for each gene across all cells. (B) UMAP plot depicting expression of TLR2 as an example of a gene that is enriched in a specific cell type (microglia). (C) TLR2 expression is enriched in microglia among nuclei from both controls and alcoholics.
While mean expression of a given gene may be higher in a specific cell type, that does not imply that expression in other cell types is statistically negligible or biologically unimportant. For example, expression of FAS is lower in astrocytes relative to endothelial cells; however, FAS is known to be active in both astrocytes (29) and endothelial cells (30).
DE and pathway analysis
Prior to DE analysis, we pooled transcript counts within each cell type and each donor, creating ‘pseudo-bulk’ transcriptomes. This approach has been employed by several single cell transcriptomics studies (31–34) since it provides robustness to varying numbers and library sizes of cells/nuclei among replicates (donors acting as replicates in our case), provides strong type I error control and allows the use of bulk RNA-seq DE tools that have been optimized and validated over the course of many years. After pseudo-bulking, DESeq2 was used to test for differentially expressed genes (DEGs), with batch as a covariate. We identified a total of 916 DEGs at FDR < 0.25 and 253 at FDR < 0.05. There were large differences in DEG counts among the different cell types, with endothelial cells having 0 DEGs and astrocytes having the most DEGs (Fig. 3A and Supplementary Material, Fig. S3). Recent mouse RNA-seq studies on isolated glial cell types also detected more alcohol-related DEGs in astrocytes than in total brain homogenate (20) and microglia (21). These findings support key roles for astrocytes in response to chronic alcohol that were not detected in other studies lacking cell type resolution.
DEGs corresponding to both protein-coding and non-coding transcripts were present in every cell type that had more than two total DEGs (Fig. 3A and B). Bulk and scRNA-seq studies often exclude non-coding RNAs from their analyses (3,8,35). However, the importance of ncRNAs is becoming increasingly apparent, not only in alcohol dependence (36) but in brain physiology in general (37), and these RNAs were therefore included in our data. Interestingly, the top DEG in astrocytes was AC008957.2, a lncRNA that is antisense to the gene encoding SLC1A3, a glutamate uptake transporter that plays important roles in the neurocircuitry of addiction (38–40). SLC1A3 was also upregulated in astrocytes, albeit at a lower significance level (FDR = 0.07).
In addition to non-coding genes, protein-coding genes of interest were also identified. For example, four DEGs involved in neuroinflammation (SLC1A3, FAS, MFGE8, IRF3) were found in astrocytes (Fig. 3C) (FDR < 0.25). IPA revealed that all four are associated with apoptosis, a component of the neuroimmune response. SLC1A3 (also called ‘GLAST’) was downregulated with alcohol consumption in mouse astrocytes (20,21) but upregulated in the human alcoholic brain (39). FAS and MFGE8, however, showed similar regulation between mouse and human astrocytes (21).
We also used IPA to evaluate DEGs (FDR < 0.25) in all cell types. Only astrocytes, microglia and oligodendrocytes showed significant pathways (P < 0.05, Fisher’s exact test) containing multiple DEGs. The top overall pathway was GNRH signaling (Fig. 3D). One of the top DEGs in this pathway was CACNA1A, which was downregulated in astrocytes and upregulated in microglia.
A notable limitation of our DE analysis was the sample size of our dataset (seven donors). A small sample size in a single cell genomics study does affect statistical power, but does not preclude the possibility of drawing meaningful inferences of cell type gene expression in the human brain (9,41). That said, our goal is to increase the sample size for future single cell studies to better identify important expression patterns, particularly for genes that have smaller effect sizes. Additionally, this study and future single cell studies do not aim to supplant previous bulk RNA-seq studies, some of which have large sample sizes and high statistical power, but instead to complement them by adding cell type specificity.
Comparison with previous bulk RNA-seq datasets
We next determined which cell types were most similar or dissimilar to bulk data in terms of alcohol-associated expression changes. We compared our DE results to that from a published bulk RNA-seq analysis of the PFC from 65 alcoholics and 73 control donors (3). Hierarchical clustering revealed that all non-microglia cell types were more similar to bulk than they were to microglia in terms of DE patterns between alcoholics and controls (Fig. 4A). Other studies have also found that microglial expression, in general, is particularly underrepresented in bulk transcriptomes, relative to that of other brain cell types (8,42). Among the 33 DEGs in microglia, only the gene with the highest log fold change, SLC11A1, had been detected as a significant hit in the bulk data (Fig. 4B). This suggests that microglia-specific DEGs must have a particularly large effect size in order to be detected in bulk DE analysis.
Figure 4.
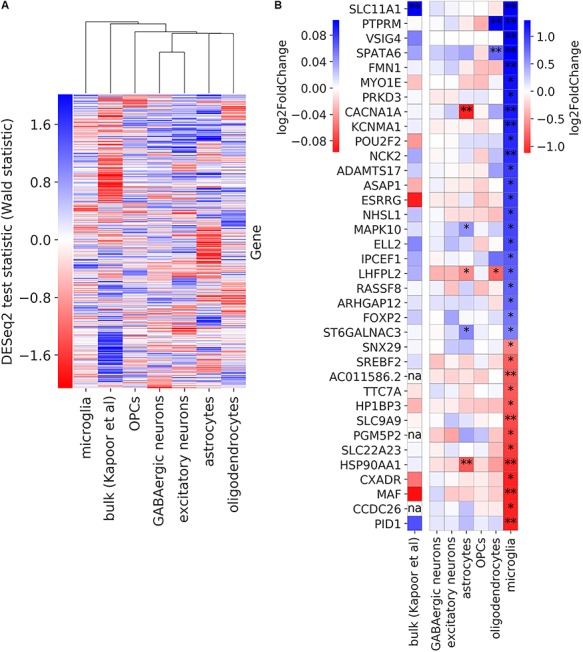
Comparison of alcohol dependence-associated DE from human bulk RNA-seq and snRNA-seq data. (A) Gene expression changes in alcoholics for each cell type as well as bulk data from a previous study (3). Groups were hierarchically clustered using average as the linkage method. (B) Log fold change of expression in alcohol-dependent donors compared with controls for genes that are differentially expressed (FDR < 0.25) in microglia. A single asterisk indicates DE at FDR < 0.25, while a double asterisk indicates FDR < 0.05. The three genes with ‘na’ are non-protein-coding genes, which had not been included in the bulk study.
Aside from utilizing different technologies (snRNA-seq versus bulk RNA-seq), another major difference between the present study and the previous bulk study is the inclusion of donors of both sexes in the latter. We focused on a single sex in the present study since introducing an additional biological variable would not be appropriate for a study of this scale. Future, large-scale single cell genomics studies could be designed to assess similarities and differences between the sexes in terms of cell type-resolved expression patterns of alcohol dependence.
GWAS enrichment meta-analysis
Genome-wide association studies (GWAS) have identified genetic variants associated with numerous diseases to prioritize candidate genes for further study, thus generating hypotheses about mechanisms of pathogenesis. In addition, GWAS have confirmed the polygenic nature of complex diseases, particularly for psychiatric disorders including alcohol dependence (3). When comparing data from the Psychiatric Genomics Consortium (PGC) alcohol dependence GWAS (43) with the DE results for each cell type in this study, we found significant enrichment in astrocytes (P = 0.031) (Supplementary Material, Table S3). GWAS enrichment analysis can be influenced by gene size and density at the loci. The larger locus will have more SNPs and can influence the association testing. This problem was circumvented by including the gene size and gene density as a covariate in the model, and the association remained significant after correcting for technical covariates. These observations are consistent with the DE analysis in a bulk tissue analysis where the PGC GWAS genes were significantly enriched in the network and module related to immunity (3). These observations establish a link between genetic risk factors and astrocytic transcriptional changes in alcohol dependence, adding functional significance to risk variants identified by genetic studies.
Conclusions
This study presents the first single cell transcriptomic dataset of the human alcoholic brain. Among 16 305 nuclear transcriptomes, we detected major neural cell types from seven donors: three alcoholics and four controls. Every cell type displayed relative enrichment of different genes linked to neuroinflammation, a process associated with excessive alcohol use. We detected DEGs between alcoholics and controls in every neural cell type. Many more DEGs were found in astrocytes, oligodendrocytes and microglia relative to neurons, indicating that glial cells should be given particular attention in future studies of alcohol-associated gene expression.
Materials and Methods
Case selection and postmortem tissue collection
Diagnosis of alcohol dependence was based on DSM-IV criteria. All donors were required to meet the following criteria to be considered in this study: no head injury at time of death, lack of developmental disorder, no recent cerebral stroke, no history of other psychiatric or neurological disorders and no history of intravenous or polydrug abuse. Alcoholic and control donors were selected to be as close as possible in terms of postmortem interval (PMI), pH of tissue and cause of death (Supplementary Material, Table S1). Fresh frozen tissue samples were collected as previously described (3).
Isolation of nuclei from frozen postmortem brain tissue
Nuclei were isolated from tissue based on a modified version of Luciano Martelotto’s Customer-Developed Protocol provided on the 10X Genomics website (44). All procedures were carried out on ice, and all centrifugation steps were at 500 g for 5 min at 4°C. PFC tissue (~50 mg) was homogenized in 5 ml of lysis buffer [Nuclei EZ Lysis Buffer (Sigma, NUC101), 0.2 U/μl RNase inhibitor (NEB, M0314), 1× protease inhibitor (Sigma, 4 693 132 001)] in 7 ml dounce ~ 30 times until no visible tissue pieces remained. An aliquot of homogenate (500 μl) was snap-frozen on dry ice and stored at −80°C for later RNA integrity assessment. The remaining homogenate was incubated on ice for 5–10 min and then centrifuged. The supernatant was removed, and nuclei were resuspended in 2 ml of lysis buffer without protease inhibitor. The suspension was incubated for another 5 min on ice and centrifuged. Supernatant was removed, and nuclei were resuspended in 2 ml of Wash & Resuspension buffer (PBS with 1% BSA and 0.2 U/μl of RNase inhibitor). Nuclei were centrifuged and the supernatant was removed. Nuclei were then resuspended in Wash & Resuspension buffer containing 10 μg/ml of DAPI and filtered into a FACS tube with a 35 μm strainer cap. Nuclei were then sorted into a tube containing 10X Genomics (v3.0) reverse transcription (RT) master mix (20 μl of RT reagent, 3.1 μl of template switch oligo, 2 μl of reducing agent B and 10 μl of water). Nuclei-RT mix was topped off to 71.7 μl using water, and then 8.3 μl of RT enzyme C was added. It should be noted that the sample 647 was prepared in another batch using a slightly different protocol, the main difference being that nuclei were sorted into an empty tube, concentrated by centrifuging and removing all but 100 μl of supernatant and then resuspended before being combined with the RT master mix.
RNA integrity assessment
The Qiagen RNeasy Lipid Tissue Mini Kit (Qiagen, 74 804) was used to extract RNA from tissue homogenates using the manufacturer’s instructions. The RNA was quantified using a NanoDrop 1000 (Thermo Fisher) and assayed for quality using an Agilent 2100 TapeStation (Agilent Technologies). Homogenate for sample 647 had not been saved.
Droplet-based snRNA-seq
Nuclei-RT mix (75 μl) was transferred to a Chromium B Chip, and libraries were prepared using the 10X Genomics 3′ Single Cell Gene Expression protocol. Paired-end sequencing of the libraries was conducted using a NovaSeq 6000 with an S2 chip (100 cycles).
Alignment to reference genome
A GENCODE v30 GTF file was modified to create a ‘pre-mRNA’ GTF file so that pre-mRNAs would be included as counts in the subsequent analysis. Cellranger’s (v3.0.2) mkref command was then used to create a pre-mRNA reference from the GTF file and a FASTA file of the GRCh38.p12 genome. FASTQ files of the snRNA-seq libraries were then aligned to the pre-mRNA reference using the cellranger count command, producing gene expression matrices. The matrices for the different samples were concatenated into a single matrix using the cellranger aggr command with normalization turned off, so that the raw counts would remain unchanged at this point.
Filtering and normalization
The filtered matrix produced by cellranger was loaded into scanpy (v1.4.3) (45). Nuclei with more than 20% of UMI (transcript) counts attributed to mitochondrial genes were removed, and then all mitochondrial genes were removed from the dataset. Normalization was conducted based on the recommendations from multiple studies that compared several normalization techniques (35,46,47). In brief, three steps were performed: (1) preliminary clustering of cells by constructing a nearest network graph and using scanpy’s implementation of Louvain community detection, (2) calculating size factors using the R package scran (v1.10.2) (48) and (3) dividing counts by the respective size factor assigned to each cell. Normalized counts were then transformed by adding a pseudocount of 1 and taking the natural log.
Assigning nuclei to transcriptomically distinct clusters
Genes that are highly variable within every sample were identified using scanpy’s highly_variable_genes function with sample identity used for the batch_key argument. This type of approach helps select genes that distinguish cell types from each other (9). Normalized expression of every gene was then centered to a mean of zero. Principal component (PC) analysis was performed on the highly variable genes, and the top 50 PCs were used to construct a nearest neighbor graph. Nuclei were then assigned to clusters using the Louvain algorithm with the resolution set to 0.1.
Differential gene expression analysis
DE testing was performed separately on each cell type. We adapted a transcript count summation strategy (31) (also called ‘pseudo-bulking’). First, nuclei corresponding to the given cell type were selected from the full dataset. Second, raw counts were summed in order to produce a ‘pseudo-bulk’ transcriptome for each donor. Third, DEGs between the alcohol-dependent and control donors were detected using DESeq2 (v1.24.0) (49) with batch as a covariate. Additional covariates were not included given the sample size and the fact that including donor covariates inflates the DEG count (e.g. adding age, PMI and RIN covariates, the number of total DEGs at FDR < 0.05 was 1492), likely increasing type I error.
Pathway analyses of DE
Qiagen’s IPA software was used to identify canonical pathways associated with DEGs in each cell type. A cutoff of FDR < 0.25 was used to separate DEGs from non-DEGs.
GWAS enrichment meta-analysis
Summary statistics for meta-analysis of alcohol dependence GWAS was obtained from Psychiatric Genetics Consortium’s web resources (50). MAGMA (v1.24.0) (51) was used to perform gene-based association analysis for the GWAS summary statistics. The output of gene-based analysis was used to perform the cell-type enrichment analysis for the GWAS genes. The enrichment scores were calculated on the ranked absolute values of log2 fold change for DEGs in alcoholics for each cell type. Gene size (per cluster), gene density (per cluster) and sample size were included as covariates for the enrichment analysis.
Data Availability
Data is available on the Gene Expression Omnibus (GEO) with accession ID GSE141552.
Supplementary Material
Acknowledgements
The authors thank The University of Texas at Austin’s Genomic Sequencing and Analysis Facility (GSAF) and its members for preparing the cDNA libraries and the Center for Medical Genomics at Indiana University School of Medicine for sequencing the libraries. We are also grateful to the New South Wales Tissue Resource Center at the University of Sydney for providing human brain samples; the Center is supported by the National Health and Medical Research Council of Australia, Schizophrenia Research Institute and National Institute on Alcohol Abuse and Alcoholism (NIH/NIAAA R24AA012725).
Conflicts of Interest. We have no competing interests to declare for this study.
Funding
National Institutes of Health (U01-AA020926 to R.D.M., R01-AA012404 to R.D.M.); E.B. receives support from two internal fellowships at The University of Texas at Austin: the Provost’s Graduate Excellence Fellowship and the F.M. Jones and H.L. Bruce Endowed Graduate Fellowship in Addiction Science.
References
- 1. Alcohol Facts and Statistics (2011) Alcohol Facts and Statistics https://www.niaaa.nih.gov/alcohol-health/overview-alcohol-consumption/alcohol-facts-and-statistics(accessed July 16, 2019).
- 2. Sacks J.J., Gonzales K.R., Bouchery E.E., Tomedi L.E. and Brewer R.D. (2015) 2010 national and state costs of excessive alcohol consumption. Am. J. Prev. Med., 49, e73–e79. [DOI] [PubMed] [Google Scholar]
- 3. Kapoor M., Wang J.-C., Farris S.P., Liu Y., McClintick J., Gupta I., Meyers J.L., Bertelsen S., Chao M., Nurnberger J. et al. (2019) Analysis of whole genome-transcriptomic organization in brain to identify genes associated with alcoholism. Transl. Psychiatry, 9, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Farris S.P., Arasappan D., Hunicke-Smith S., Harris R.A. and Mayfield R.D. (2015) Transcriptome Organization for chronic alcohol abuse in human brain. Mol. Psychiatry, 20, 1438–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Augier E., Barbier E., Dulman R.S., Licheri V., Augier G., Domi E., Barchiesi R., Farris S., Nätt D., Mayfield R.D. et al. (2018) A molecular mechanism for choosing alcohol over an alternative reward. Science, 360, 1321–1326. [DOI] [PubMed] [Google Scholar]
- 6. Rao X., Thapa K.S., Chen A.B., Lin H., Gao H., Reiter J.L., Hargreaves K.A., Ipe J., Lai D., Xuei X. et al. (2019) Allele-specific expression and high-throughput reporter assay reveal functional variants in human brains with alcohol use disorders. bioRxiv, 514992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nagy C., Maitra M., Tanti A., Suderman M., Théroux J.-F., Mechawar N., Ragoussis J. and Turecki G. (2019) Single-nucleus RNA sequencing shows convergent evidence from different cell types for altered synaptic plasticity in major depressive disorder. bioRxiv, 384479. [Google Scholar]
- 8. Mathys H., Davila-Velderrain J., Peng Z., Gao F., Mohammadi S., Young J.Z., Menon M., He L., Abdurrob F., Jiang X. et al. (2019) Single-cell transcriptomic analysis of Alzheimer’s disease. Nature, 570, 332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Del-Aguila J.L., Li Z., Dube U., Mihindukulasuriya K.A., Budde J.P., Fernandez M.V., Ibanez L., Bradley J., Wang F., Bergmann K. et al. (2019) A single-nuclei RNA sequencing study of Mendelian and sporadic AD in the human brain. Alzheimers Res. Ther., 11, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abdelmoez M.N., Iida K., Oguchi Y., Nishikii H., Yokokawa R., Kotera H., Uemura S., Santiago J.G. and Shintaku H. (2018) SINC-seq: correlation of transient gene expressions between nucleus and cytoplasm reflects single-cell physiology. Genome Biol., 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bakken T.E., Hodge R.D., Miller J.M., Yao Z., Nguyen T.N., Aevermann B., Barkan E., Bertagnolli D., Casper T., Dee N. et al. (2018) Equivalent high-resolution identification of neuronal cell types with single-nucleus and single-cell RNA-sequencing. bioRxiv, 239749. [Google Scholar]
- 12. Sathyamurthy A., Johnson K.R., Matson K.J.E., Dobrott C.I., Li L., Ryba A.R., Bergman T.B., Kelly M.C., Kelley M.W. and Levine A.J. (2018) Massively parallel single nucleus transcriptional profiling defines spinal cord neurons and their activity during behavior. Cell Rep., 22, 2216–2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lake B.B., Ai R., Kaeser G.E., Salathia N.S., Yung Y.C., Liu R., Wildberg A., Gao D., Fung H.-L., Chen S. et al. (2016) Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science, 352, 1586–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lacar B., Linker S.B., Jaeger B.N., Krishnaswami S., Barron J., Kelder M., Parylak S., Paquola A., Venepally P., Novotny M. et al. (2016) Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat. Commun., 7, 11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krishnaswami S.R., Grindberg R.V., Novotny M., Venepally P., Lacar B., Bhutani K., Linker S.B., Pham S., Erwin J.A., Miller J.A. et al. (2016) Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat. Protoc., 11, 499–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grindberg R.V., Yee-Greenbaum J.L., McConnell M.J., Novotny M., O’Shaughnessy A.L., Lambert G.M., Araúzo-Bravo M.J., Lee J., Fishman M., Robbins G.E. et al. (2013) RNA-sequencing from single nuclei. Proc. Natl. Acad. Sci. U. S. A., 110, 19802–19807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Habib N., Li Y., Heidenreich M., Swiech L., Avraham-Davidi I., Trombetta J.J., Hession C., Zhang F. and Regev A. (2016) Div-Seq: single-nucleus RNA-Seq reveals dynamics of rare adult newborn neurons. Science, 353, 925–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S.R., Aguet F., Gelfand E., Ardlie K. et al. (2017) Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods, 14, 955–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ball G., Stokes P.R., Rhodes R.A., Bose S.K., Rezek I., Wink A.-M., Lord L.-D., Mehta M.A., Grasby P.M. and Turkheimer F.E. (2011) Executive functions and prefrontal cortex: a matter of persistence? Front. Syst. Neurosci., 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Erickson E.K., Farris S.P., Blednov Y.A., Mayfield R.D. and Harris R.A. (2018) Astrocyte-specific transcriptome responses to chronic ethanol consumption. Pharmacogenomics J., 18, 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Erickson E.K., Blednov Y.A., Harris R.A. and Mayfield R.D. (2019) Glial gene networks associated with alcohol dependence. Sci. Rep., 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F. and Newell E.W. (2018) Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol., 37, 38–44. [DOI] [PubMed] [Google Scholar]
- 23. Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C., Duong T.E., Gao D., Chun J., Kharchenko P.V. et al. (2018) Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol., 36, 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McKenzie A.T., Wang M., Hauberg M.E., Fullard J.F., Kozlenkov A., Keenan A., Hurd Y.L., Dracheva S., Casaccia P., Roussos P. et al. (2018) Brain cell type specific gene expression and co-expression network architectures. Sci. Rep., 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian L., Ma L., Kaarela T. and Li Z. (2012) Neuroimmune crosstalk in the central nervous system and its significance for neurological diseases. J. Neuroinflammation, 9, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Warden A., Erickson E., Robinson G., Harris R.A. and Mayfield R.D. (2016) The neuroimmune transcriptome and alcohol dependence: potential for targeted therapies. Pharmacogenomics, 17, 2081–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rongvaux A., Jackson R., Harman C.C.D., Li T., West A.P., Zoete M.R., Wu Y., Yordy B., Lakhani S.A., Kuan C.-Y. et al. (2014) Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell, 159, 1563–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. GO_TYPE_I_INTERFERON_PRODUCTION http://software.broadinstitute.org/gsea/msigdb/cards/GO_TYPE_I_INTERFERON_PRODUCTION.html(accessed September 3, 2019).
- 29. Choi C., Park J.Y., Lee J., Lim J.H., Shin E.C., Ahn Y.S., Kim C.H., Kim S.J., Kim J.D., Choi I.S. et al. (1999) Fas ligand and Fas are expressed constitutively in human astrocytes and the expression increases with IL-1, IL-6, TNF-alpha, or IFN-gamma. J. Immunol. Baltim. Md 1950, 162, 1889–1895. [PubMed] [Google Scholar]
- 30. Sata M., Suhara T. and Walsh K. (2000) Vascular endothelial cells and smooth muscle cells differ in expression of Fas and Fas ligand and in sensitivity to Fas ligand-induced cell death: implications for vascular disease and therapy. Arterioscler. Thromb. Vasc. Biol., 20, 309–316. [DOI] [PubMed] [Google Scholar]
- 31. Lun A.T.L. and Marioni J.C. (2017) Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostat. Oxf. Engl., 18, 451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hay S.B., Ferchen K., Chetal K., Grimes H.L. and Salomonis N. (2018) The human cell atlas bone marrow single-cell interactive web portal. Exp. Hematol., 68, 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Messmer T., Meyenn F., Savino A., Santos F., Mohammed H., Lun A.T.L., Marioni J.C. and Reik W. (2019) Transcriptional heterogeneity in naive and primed human pluripotent stem cells at single-cell resolution. Cell Rep., 26, 815–824.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ernst C., Eling N., Martinez-Jimenez C.P., Marioni J.C. and Odom D.T. (2019) Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat. Commun., 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Luecken M.D. and Theis F.J. (2019) Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol., 15, e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayfield R.D. (2017) Emerging roles for ncRNAs in alcohol use disorders. Alcohol Fayettev. N, 60, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zampa F., Hartzell A.L., Zolboot N. and Lippi G. (2019) Non-coding RNAs: the gatekeepers of neural network activity. Curr. Opin. Neurobiol., 57, 54–61. [DOI] [PubMed] [Google Scholar]
- 38. Bell R.L., Hauser S.R., McClintick J., Rahman S., Edenberg H.J., Szumlinski K.K. and McBride W.J. (2016) Ethanol-associated changes in glutamate reward neurocircuitry: a minireview of clinical and preclinical genetic findings. Prog. Mol. Biol. Transl. Sci., 137, 41–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kashem M.A., Sultana N., Pow D.V. and Balcar V.J. (2019) GLAST (GLutamate and ASpartate transporter) in human prefrontal cortex; interactome in healthy brains and the expression of GLAST in brains of chronic alcoholics. Neurochem. Int., 125, 111–116. [DOI] [PubMed] [Google Scholar]
- 40. Spencer S. and Kalivas P.W. (2017) Glutamate transport: a new bench to bedside mechanism for treating drug abuse. Int. J. Neuropsychopharmacol., 20, 797–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Boldog E., Bakken T.E., Hodge R.D., Novotny M., Aevermann B.D., Baka J., Bordé S., Close J.L., Diez-Fuertes F., Ding S.-L. et al. (2018) Transcriptomic and morphophysiological evidence for a specialized human cortical GABAergic cell type. Nat. Neurosci., 21, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kelley K.W., Nakao-Inoue H., Molofsky A.V. and Oldham M.C. (2018) Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci., 21, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Walters R.K., Polimanti R., Johnson E.C., McClintick J.N., Adams M.J., Adkins A.E., Aliev F., Bacanu S.-A., Batzler A., Bertelsen S. et al. (2018) Trans-ancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat. Neurosci., 21, 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Customer Developed Protocols Customer Developed Protocols https://community.10xgenomics.com/t5/Customer-Developed-Protocols/ct-p/customer-protocols(accessed August 20, 2019).
- 45. Wolf F.A., Angerer P. and Theis F.J. (2018) SCANPY: large-scale single-cell gene expression data analysis. Genome Biol., 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Büttner M., Miao Z., Wolf F.A., Teichmann S.A. and Theis F.J. (2019) A test metric for assessing single-cell RNA-seq batch correction. Nat. Methods, 16, 43–49. [DOI] [PubMed] [Google Scholar]
- 47. Vieth B., Parekh S., Ziegenhain C., Enard W. and Hellmann I. (2019) A systematic evaluation of single cell RNA-seq analysis pipelines: library preparation and normalisation methods have the biggest impact on the performance of scRNA-seq studies. bioRxiv, 583013. [Google Scholar]
- 48. Lun A.T.L., Bach K. and Marioni J.C. (2016) Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol., 17, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Love M.I., Huber W. and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Psychiatric Genomics Consortium Downloads Psychiatric Genomics Consortium Downloads https://www.med.unc.edu/pgc/results-and-downloads/(accessed October 8, 2019).
- 51. Leeuw C.A., Mooij J.M., Heskes T. and Posthuma D. (2015) MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput. Biol., 11, e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is available on the Gene Expression Omnibus (GEO) with accession ID GSE141552.


