Abstract
Pancreatic cancer (PC) is one of the most malignant tumors. Despite considerable progress in the treatment of PC, the prognosis of patients with PC is poor. The aim of this study was to identify potential biomarkers for the diagnosis and prognosis of PC. First, the original data of three independent mRNA expression datasets were downloaded from the Gene Expression Omnibus and The Cancer Genome Atlas databases and screened for differentially expressed genes (DEGs) using the R software. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses of the DEGs were performed, and a protein-protein interaction (PPI) network was constructed to screen for hub genes. The hub genes were analyzed for genetic variations, as well as for survival, prognostic, and diagnostic value, using the cBioPortal and Gene Expression Profiling Interactive Analysis (GEPIA) databases and the pROC package. After screening for potential biomarkers, the mRNA and protein levels of the biomarkers were verified at the tissue and cellular levels using the Cancer Cell Line Encyclopedia, GEPIA, and the Human Protein Atlas. As a result, a total of 248 DEGs were identified. The GO terms enriched in DEGs were related to the separation of mitotic sister chromatids and the binding of the spindle to the extracellular matrix. The enriched pathways were associated with focal adhesion, ECM-receptor interaction, and phosphatidylinositol 3-kinase (PI3K)/AKT signaling. The top 20 genes were selected from the PPI network as hub genes, and based on the analysis of multiple databases, MCM2 and NUSAP1 were identified as potential biomarkers for the diagnosis and prognosis of PC. In conclusion, our results show that MCM2 and NUSAP1 can be used as potential biomarkers for the diagnosis and prognosis of PC. The study also provides new insights into the underlying molecular mechanisms of PC.
1. Introduction
Pancreatic cancer (PC) is one of the most common malignant tumors, with a 5-year survival rate of only 9% [1]. Currently, surgery is the most effective way to improve the survival rate of patients with PC. However, the prognosis of patients with PC is still very poor because the onset of PC is cryptic, symptoms are atypical, lymph node metastasis occurs early, the degree of malignancy is high, and the progress is rapid [2]. Therefore, early diagnosis and intervention are essential for reducing mortality and improving the clinical prognosis of patients with PC.
The main potential biomarkers of PC identified in the past two decades are CA19-9, DUPAN-2, CAM17.1, TPS, SPan-1, TAT1, POA, YKL-40, TUM2-PK, and matrix metalloproteinases [3]. Although CA19-9, which is considered a better biomarker for the diagnosis and prognosis of PC [4], is highly sensitive, its application in early diagnostic screening for PC is limited owing to a low specificity [5, 6]. Therefore, research to find effective biomarkers for PC continues.
Increasing evidence indicates that abnormal expression of and mutations in certain genes are closely related to the occurrence and development of PC. Abnormal expression of OPN and CISD2 has been shown to play a key role in the progression of PC [7, 8], while PAM4, S100A6, and SPARC have been identified as biomarkers of PC [9–11]. Therefore, further elucidation of the pathogenesis of PC at the genetic level may help identify new diagnostic and prognostic indicators. With the rapid development of sequencing technology, microarray analyses, based on high-throughput platforms, have been widely used in biomedical and clinical research for screening genetic variants [12, 13]. At present, there are many PC-related expression profile datasets of varying quality. However, most of the PC-related bioinformatics studies based on The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) did not perform quality control of the original data, nor did they verify their findings in other databases. Consequently, potential diagnostic and prognostic biomarkers showing compatibility across different transcriptomic platforms and patient cohorts have not been systematically investigated. In this study, we aimed to identify potential diagnostic and prognostic biomarkers for PC using the data available in the GEO and TCGA databases and validate the expression of these biomarkers using the Cancer Cell Line Encyclopedia (CCLE), International Cancer Genome Consortium (ICGC), cBioPortal, Gene Expression Profiling Interactive Analysis (GEPIA), and Human Protein Atlas databases. Our results will help develop novel therapeutic strategies to improve clinical outcomes and provide new insights into the pathogenesis of PC.
2. Materials and Methods
2.1. Gene Expression Profile Datasets
GEO (http://www.ncbi.nlm.nih.gov/geo) is a public repository of various high-throughput experimental data [14]. In this study, two mRNA expression profile datasets for PC (GSE15471 [15] and GSE16515 [16]), based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array), were downloaded from the GEO database. The GSE15471 dataset consisted of 39 PC tissues and 21 normal pancreatic tissues, and the GSE16515 dataset included 36 PC tissues and 16 normal pancreatic tissues. TCGA (http://www.cancergenome.nih.gov) is a large-scale cancer genetic information database [17] that provides information regarding the key genomic changes and clinical data for 33 cancers. An mRNA expression profile that contained data for 179 PC tissues and four normal pancreatic tissues was obtained from TCGA.
2.2. Data Preprocessing and Identification of Differentially Expressed Genes (DEGs)
The affy package [18] in the R software (version 3.6.1, http://r-project.org/) was used to read the raw data (CEL file) of the three datasets and then convert the original data format, fill the missing values, and apply background correction. The samples were then subjected to differential expression analysis, and the DEGs were exported using the limma package [19]. DEGs that satisfied the adj. P value < 0.05 and ∣log2FC∣ > 1 criteria were considered. Finally, the intersection of the DEGs from the three datasets was obtained using the FunRich software [20] (version 3.1.3, http://funrich.org/index.html).
2.3. Construction of the Protein-Protein Interaction (PPI) Network and Analysis of Important Modules
The Search Tool for the Retrieval of Interacting Genes database (version 11.0, http://string-db.org) [21] was used to construct a PPI network for DEGs, and interactions with a combined score of greater than 0.9 were considered statistically significant. The PPI network was then visualized using the Cytoscape software (version 3.7.1) [22]. To better extract valuable clues from important modules, the cytoHubba plugin [23] was used to select and sort 20 genes using the Maximum Correlation Criteria (MCC) algorithm. The NetworkAnalyst database (https://www.networkanalyst.ca/faces/home.xhtml) was used to display the coexpressed gene network of DEGs; cBioPortal (https://www.cbioportal.org) [24] was used to analyze the hub genes and their coexpressed genes in PC; the Cancer RNA-Seq Nexus (CRN) database (http://syslab4.nchu.edu.tw) [25] was used to further validate the expression of the hub genes.
2.4. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment Analyses of DEGs
GO covers three aspects of biology: cellular component (CC), molecular function (MF), and biological process (BP) [26]. KEGG is a database that analyzes high-level functions of biological systems at the molecular level [27]. To further analyze the functions of DEGs, GO and KEGG pathway enrichment analyses of DEGs were performed using the clusterProfiler package [28]. FDR < 0.05 was considered statistically significant.
2.5. GO and KEGG Pathway Enrichment Analyses of the Hub Genes
The Database for Annotation, Visualization, and Integrated Discovery (DAVID; https://david.ncifcrf.gov; version 6.8) provides a comprehensive set of gene and protein functional annotation information [29]. GO and KEGG pathway enrichment analyses of the hub genes were performed using the DAVID database, and FDR < 0.05 was considered statistically significant.
2.6. Screening for Biomarkers
To further screen for potential biomarkers for the diagnosis and prognosis of PC, a comprehensive analysis of the 20 hub genes was performed. We used cBioPortal to analyze genetic variations in the hub genes. The GEPIA database (http://gepia.cancer-pku.cn) [30] was used to analyze associations of hub gene expression with overall survival (OS) and disease-free survival (DFS) of patients with PC. To further investigate the diagnostic value of the hub genes for PC, receptor operating characteristic (ROC) curves were plotted using the pROC package [31]. Potential biomarkers for the diagnosis and prognosis of PC were investigated by analyzing genetic changes, verifying differential expression, and evaluating the survival, prognostic, and diagnostic value of the hub genes.
2.7. Multidimensional Verification of Biomarkers
To minimize the bias and improve the accuracy of the results of analysis, multiple online databases, including CCLE (https://portals.broadinstitute.org/ccle) [32], the Human Protein Atlas (https://www.Proteinatlas.org/) [33], and GEPIA, were used to determine the mRNA and protein expression levels of potential biomarkers at the tissue and cell levels. The therapeutic potential of the biomarkers was investigated by analyzing genetic correlations between the screened biomarkers and EGFR, ERBB2, and KRAS, which are important therapeutic targets in PC [34–36], using the GEPIA database and circlize package in R [37].
3. Results
3.1. DEGs Identified in the Three Datasets
The results of normalization of the sample data from the GSE15471 and GSE16515 datasets are presented in box plots (Fig. S1a and S1b). Samples in both datasets were at the same level, indicating high consistency. The results of the sample cluster analysis of the two datasets are shown in Fig. S1c and S1d, indicating that the sample quality was reliable. After data preprocessing, we extracted 2,759 and 1,629 DEGs from the GSE15471 and GSE16515 mRNA expression profiles, respectively, using the R software. The volcano plots of the upregulated and downregulated DEGs are shown in Figures 1(a) and 1(b). Figures 1(c) and 1(d) show the hierarchical clustering heatmaps of DEGs from the two mRNA expression profiles. A total of 5,134 DEGs were obtained from the mRNA expression profiles derived from TCGA database. The DEGs from the three datasets were then intersected, and 248 DEGs were found to overlap among the three datasets (Figure 2(a)).
Figure 1.
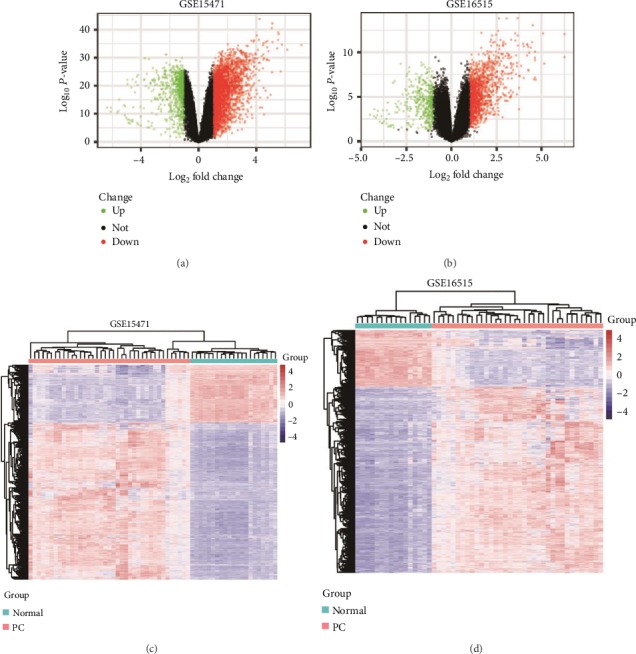
Differential expression analysis. (a) Volcano plot of DEGs in the GSE15471 dataset. (b) Volcano plot of DEGs in the GSE16515 dataset. Red dots indicate upregulated genes; green dots indicate downregulated genes; and black dots indicate unaltered genes. (c) Hierarchical clustering heatmap of DEGs in the GSE15471 dataset. (d) Hierarchical clustering heatmap of DEGs in the GSE16515 dataset. Blue represents downregulated genes, and red represents upregulated genes.
Figure 2.
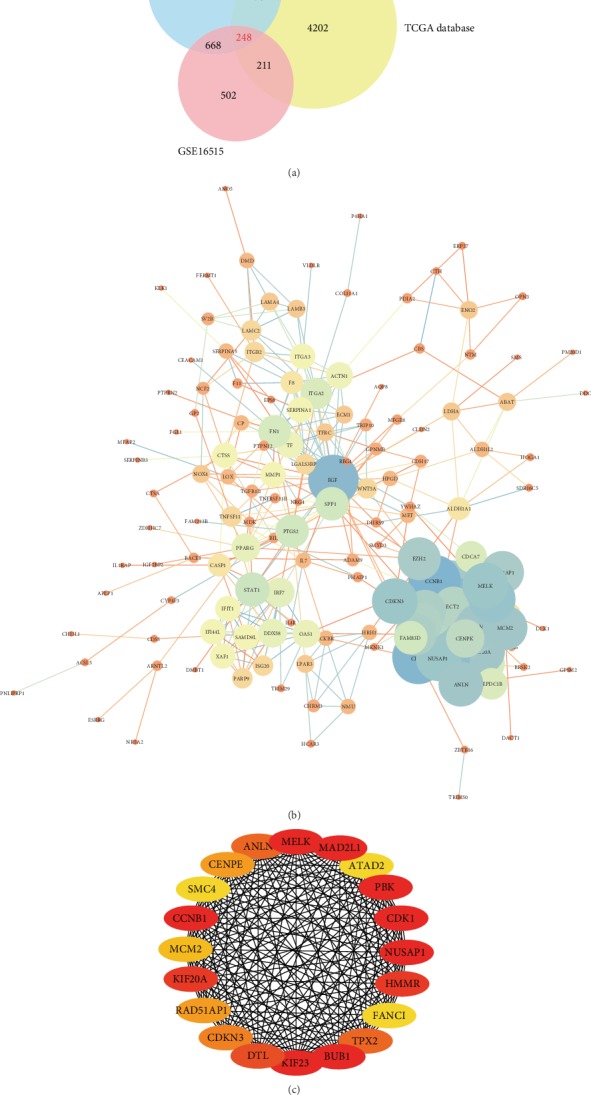
Venn diagram and the PPI network. (a) Venn diagram of the overlapping DEGs in the three datasets. (b) PPI network of DEGs. The size of the circle indicates the weight of the gene action; the line indicates the interaction between the genes, and the thickness of the line indicates the strength of the interaction. (c) Twenty hub genes. A darker color represents a higher score, and a lighter color represents a lower score.
3.2. Analysis of Important Modules in the PPI Network
The PPI network for the DEGs is presented in Figure 2(b). The top 20 genes, which were selected as the hub genes using the most relevant standard (MCC), included MELK, MAD2L1, ATAD2, PBK, CDK1, NUSAP1, HMMR, FANCI, TPX2, BUB1, KIF23, DTL, CDKN3, RAD51AP1, KIF20A, MCM2, CCNB1, SMC4, CENPE, and ANLN (Figure 2(c) and Table 1). The coexpression gene network of the DEGs is presented in Figure 3(a). The DEGs had a large weight ratio in the coexpression network, with a large number of related genes, indicating that the DEGs play a comprehensive and complex role in the pathogenesis of PC. Simultaneously, the hub genes and their coexpression gene network in PC (Figure 3(b)) were analyzed using cBioPortal. The results showed that the 20 hub genes were closely related to and interacted with their coexpressed genes. The heatmap of the 20 hub genes, constructed using the CRN database, is shown in Figure 3(c). These 20 hub genes were highly expressed in PC tissues but not in normal tissues.
Table 1.
Top 20 hub genes ranked using the Maximum Correlation Criteria algorithm.
| Rank | Gene symbol | Full name | Entrez ID | Score |
|---|---|---|---|---|
| 1 | BUB1 | BUB1 mitotic checkpoint serine/threonine kinase | 699 | 2.71E + 22 |
| 1 | CCNB1 | Cyclin B1 | 891 | 2.71E + 22 |
| 1 | CDK1 | Cyclin-dependent kinase 1 | 983 | 2.71E + 22 |
| 1 | KIF23 | Kinesin family member 23 | 9493 | 2.71E + 22 |
| 1 | MAD2L1 | Mitotic arrest deficient 2-like 1 | 4085 | 2.71E + 22 |
| 1 | MELK | Maternal embryonic leucine zipper kinase | 9833 | 2.71E + 22 |
| 1 | NUSAP1 | Nucleolar and spindle-associated protein 1 | 51203 | 2.71E + 22 |
| 1 | PBK | PDZ-binding kinase | 55872 | 2.71E + 22 |
| 9 | HMMR | Hyaluronan-mediated motility receptor | 3161 | 2.71E + 22 |
| 9 | KIF20A | Kinesin family member 20A | 10112 | 2.71E + 22 |
| 11 | DTL | Denticleless E3 ubiquitin protein ligase homolog | 51514 | 2.71E + 22 |
| 12 | ANLN | Anillin actin binding protein | 54443 | 2.71E + 22 |
| 12 | TPX2 | TPX2 microtubule nucleation factor | 22974 | 2.71E + 22 |
| 14 | CDKN3 | Cyclin-dependent kinase inhibitor 3 | 1033 | 2.71E + 22 |
| 15 | CENPE | Centromere protein E | 1062 | 2.71E + 22 |
| 15 | RAD51AP1 | RAD51-associated protein 1 | 10635 | 2.71E + 22 |
| 17 | MCM2 | Minichromosome maintenance complex component 2 | 4171 | 2.71E + 22 |
| 18 | ATAD2 | ATPase family AAA domain-containing 2 | 29028 | 2.71E + 22 |
| 18 | FANCI | FA complementation group I | 55215 | 2.71E + 22 |
| 18 | SMC4 | Structural maintenance of chromosomes 4 | 10051 | 2.71E + 22 |
Figure 3.
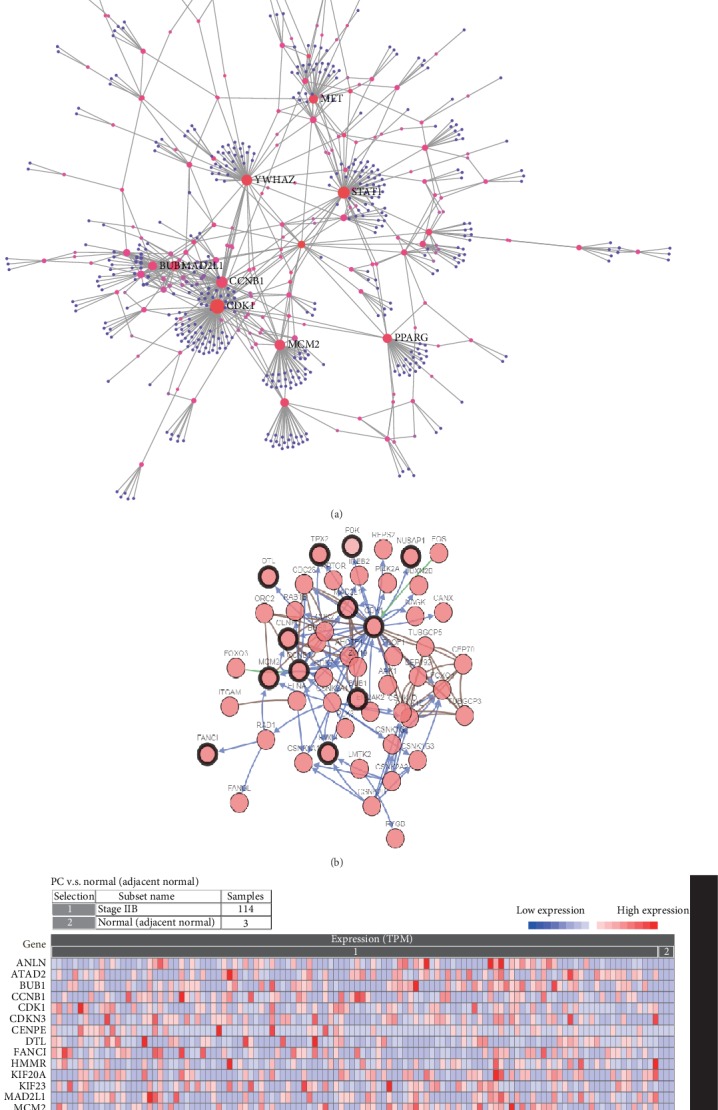
DEGs and hub gene coexpression networks. (a) Coexpression gene networks of DEGs. (b) Network of interactions between the hub genes and their coexpressed genes in PC. (c) Heatmap verifying the expression of the hub genes in the CRN database. Red indicates high expression of the gene, and blue indicates low expression of the gene.
3.3. GO and KEGG Pathways Enriched in DEGs
GO analysis showed that BP-related changes in DEGs were significantly enriched in mitotic sister chromatid separation, regulation of cell cycle phase transition, and positive regulation response to cytokine stimulation (Figure 4(a)). The changes in CC were mainly in the centromere region, spindle, and endoplasmic reticulum cavity (Figure 4(b)). The changes in MF were mainly enriched in the extracellular matrix binding and integrin binding (Figure 4(c)). The KEGG pathway enrichment analysis showed that the enriched pathways were associated with focal adhesion, ECM-receptor interaction, and the phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway (Figure 4(d)).
Figure 4.
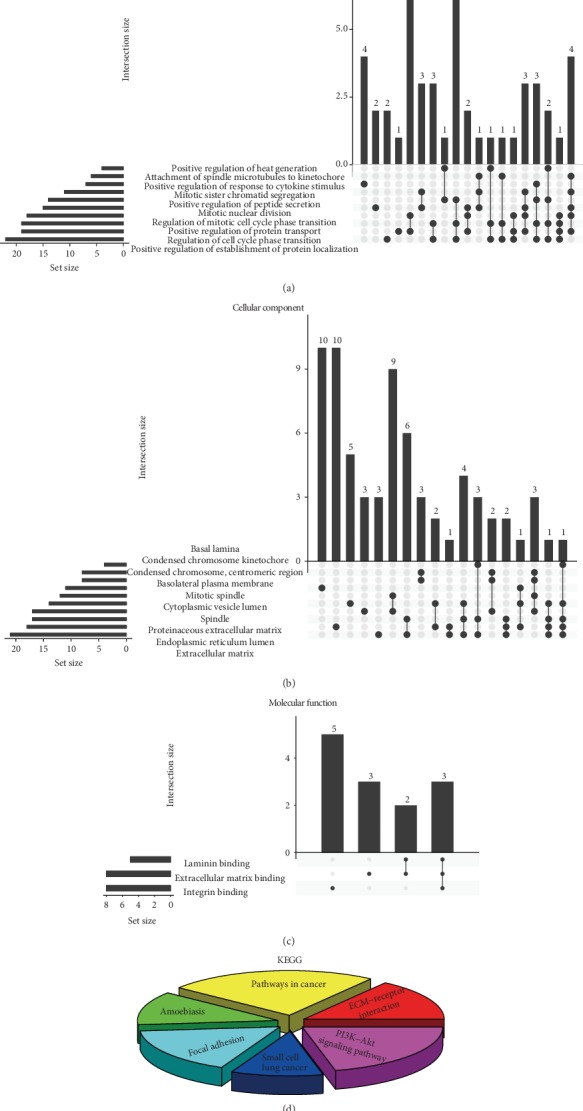
GO and KEGG pathway enrichment analyses of DEGs. (a) BP, (b) CC, (c) MF, and (d) KEGG analysis results.
3.4. GO and KEGG Pathways Enriched in the Hub Genes
The GO terms enriched in the hub genes were mainly related to the apoptosis process, cell cycle, chromosome, and histone kinase activity, while the KEGG pathways enriched in the hub genes were related to the cell cycle and p53 signaling pathway (Fig. S2).
3.5. Identification of MCM2 and NUSAP1 as Potential Biomarkers
To screen for potential biomarkers for the diagnosis and prognosis of PC, we first used cBioPortal to analyze whether genetic variations in the hub genes are involved in PC progression. As shown in Fig. S3, the 20 hub genes showed genetic variations in the PC samples. Among the genes, the highest genetic variation rate was found in ATAD2 (5%), including missense, start lost, initiator codon, frameshift, stop lost, and stop gained mutations, which suggested that these mutations might be involved in the occurrence and progression of PC. To investigate the prognostic value of the hub genes in PC, we performed a survival analysis using the GEPIA database. As shown in Figure 5, high mRNA levels of BUB1, CDK1, FANCI, KIF20A, HMMR, KIF23, MCM2, NUSAP1, and TPX2 were associated with a poor OS and were also closely related to a poor DFS of patients (Fig. S4). Thus, the results of the survival analysis suggested that these nine hub genes might be new indicators for predicting the prognosis of PC. The results obtained using ROC curve analysis showed that MCM2 (area under the curve (AUC) = 0.954) and NUSAP1 (AUC = 0.93) had the highest diagnostic value for PC (Figure 6). In summary, we identified MCM2 and NUSAP1 as potential biomarkers for the diagnosis and prognosis of PC using analysis of genetic variation, verification of differential expression, and evaluation of the survival, prognostic, and diagnostic value of the 20 hub genes.
Figure 5.
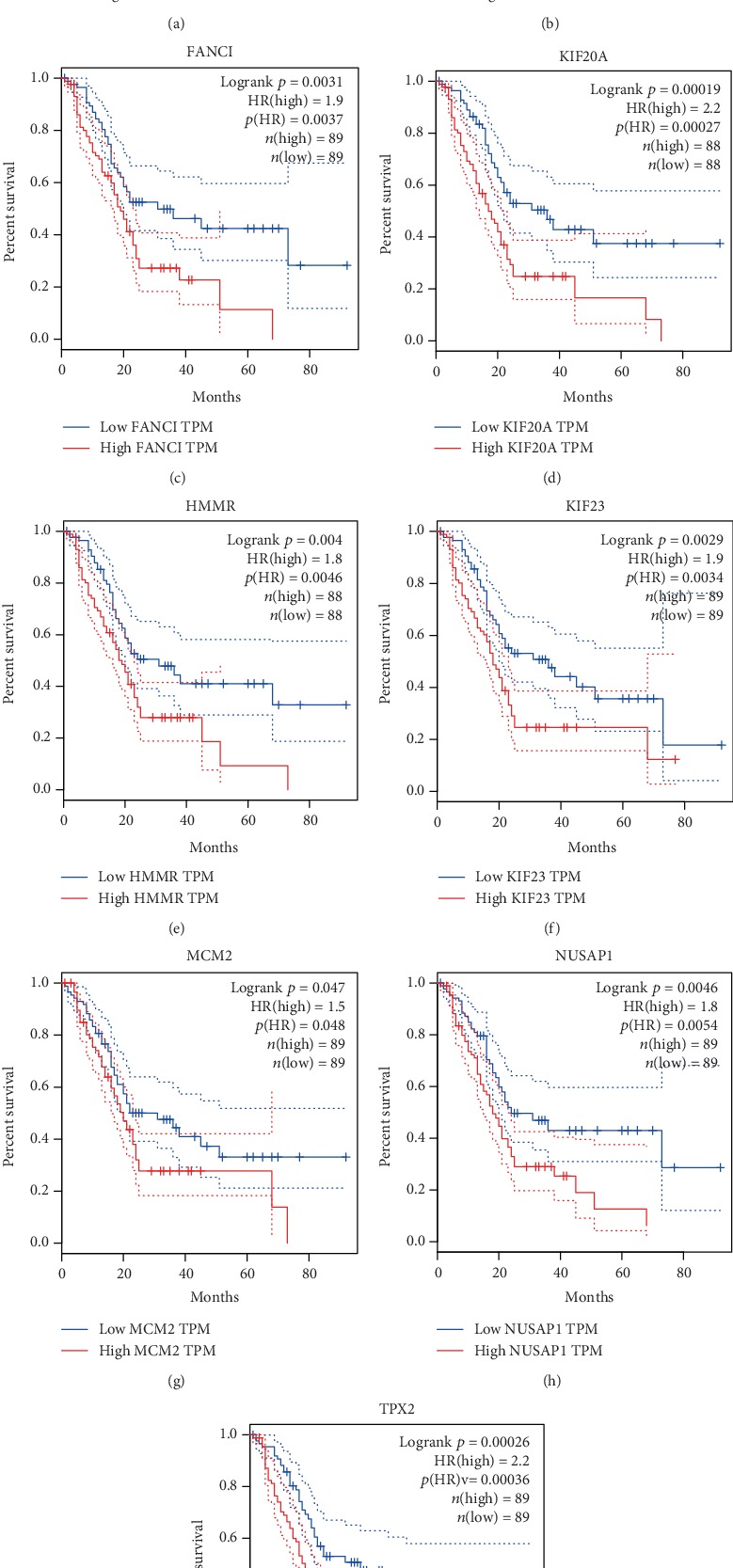
Association of hub gene expression with overall survival of patients. (a) BUB1, (b) CDK1, (c) FANCI, (d) KIF20A, (e) HMMR, (f) KIF23, (g) MCM2, (h) NUSAP1, and (i) TPX2. The solid line represents the survival curve, and the dashed line represents the 95% confidence interval. Patients with higher than the median value are indicated by the red line, and those with lower than the median value are indicated by the blue line. Log-rank P < 0.05 was considered statistically significant.
Figure 6.
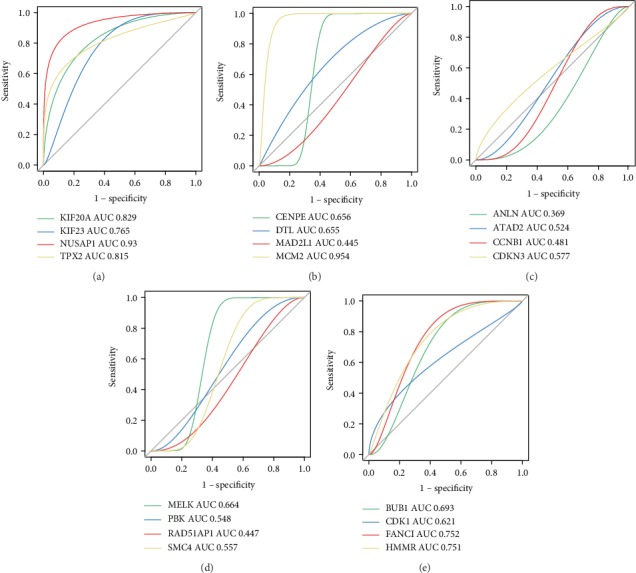
Analysis of the diagnostic value of the hub genes using ROC curve analysis. (a) KIF20A, KIF23, NUSAP1, and TPX2. (b) CENPE, DTL, MAD2L1, and MCM2. (c) ANLN, ATAD2, CCNB1, and CDKN3. (d) MELK, PBK, RAD51AP1, and SMC4. (e) BUB1, CDK1, FANCI, and HMMR. AUC represents the area under the ROC curve.
3.6. Multidimensional Verification of MCM2 and NUSAP1 as Potential Biomarkers
To ensure the accuracy of the analytical results, we used multiple databases to validate the mRNA and protein expression of MCM2 and NUSAP1 in PC at the tissue and cell levels. We used the GEPIA (Figure 7(a)), ICGC (Figure 7(b)), and TCGA (Figure 7(c)) databases to analyze the MCM2, NUSAP1, EGFR, ERBB2, and KRAS expression. The correlation coefficient (R) values are shown in Table 2. At the cell level, CCLE was used to analyze the expression of MCM2 and NUSAP1 in cell lines derived from different tissues. The results showed that MCM2 and NUSAP1 were expressed in various cell lines, but their mRNA levels were low in pancreatic cell lines (Figures 8(a) and 8(b)). We also analyzed the mRNA expression of MCM2 and NUSAP1 in 21 experimental PC cell lines and found that both were upregulated in most of the PC cell lines (Figures 8(c) and 8(d)). At the tissue level, the GEPIA-based analysis indicated that MCM2 and NUSAP1 were upregulated in PC tissues (Fig. S5). Further analysis using the Human Protein Atlas showed that MCM2 and NUSAP1 were poorly expressed at the mRNA and protein levels in normal pancreatic tissues compared with their expression in other tissues (Fig. S6). The MCM2 and NUSAP1 protein levels were higher in PC tissues than in normal pancreatic tissues (Figure 9).
Figure 7.
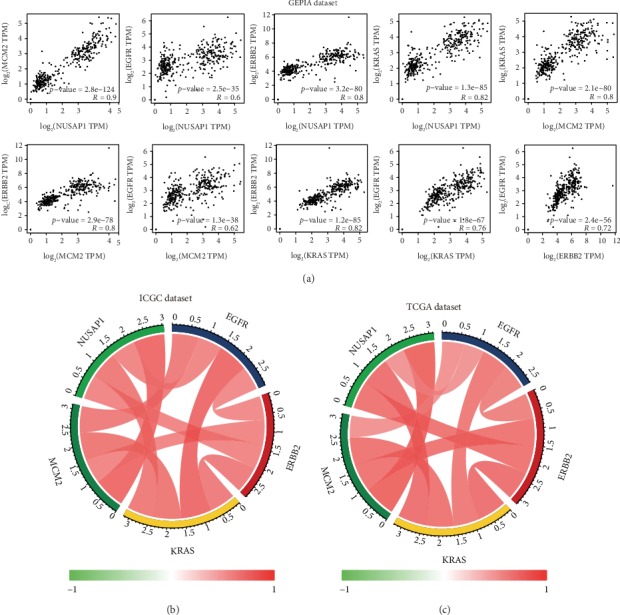
Intergene correlation analysis of expression levels based on (a) the GEPIA database, (b) the ICGC database, and (c) TCGA database.
Table 2.
Intergene correlation analysis of expression levels of MCM2, NUSAP1, EGFR, ERBB2, and KRAS.
| GEPIA dataset | ICGC dataset | TCGA dataset | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene 1 | Gene 2 | Correlation coefficient | Gene 1 | Gene 2 | Correlation coefficient | Gene 1 | Gene 2 | Correlation coefficient |
| MCM2 | NUSAP1 | 0.9 | MCM2 | NUSAP1 | 0.879 | MCM2 | NUSAP1 | 0.9 |
| EGFR | NUSAP1 | 0.6 | MCM2 | ERBB2 | 0.704 | MCM2 | ERBB2 | 0.8 |
| ERBB2 | NUSAP1 | 0.8 | MCM2 | KRAS | 0.78 | MCM2 | KRAS | 0.8 |
| KRAS | NUSAP1 | 0.82 | MCM2 | EGFR | 0.75 | MCM2 | EGFR | 0.62 |
| KRAS | MCM2 | 0.8 | NUSAP1 | ERBB2 | 0.705 | NUSAP1 | ERBB2 | 0.8 |
| ERBB2 | MCM2 | 0.8 | NUSAP1 | KRAS | 0.76 | NUSAP1 | KRAS | 0.82 |
| EGFR | MCM2 | 0.62 | NUSAP1 | EGFR | 0.692 | NUSAP1 | EGFR | 0.6 |
| KRAS | KRAS | 0.82 | KRAS | ERBB2 | 0.748 | KRAS | ERBB2 | 0.82 |
| EGFR | KRAS | 0.76 | KRAS | EGFR | 0.86 | KRAS | EGFR | 0.76 |
| EGFR | KRAS | 0.72 | ERBB2 | EGFR | 0.673 | ERBB2 | EGFR | 0.72 |
Figure 8.
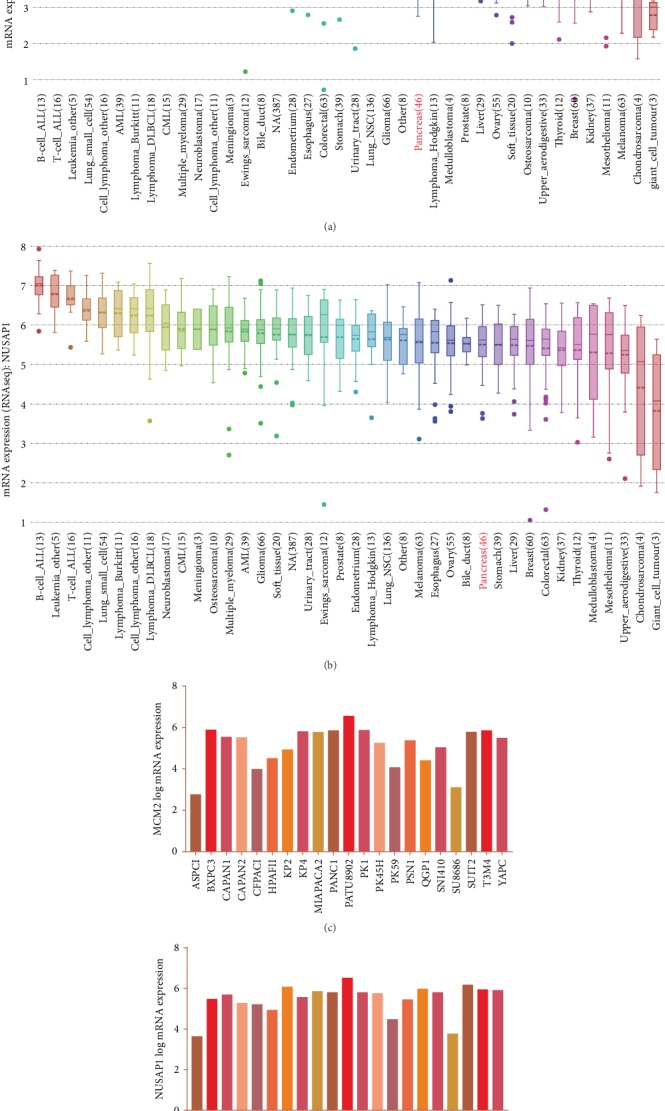
Expression of MCM2 and NUSAP1 at the cell level. (a) Expression of MCM2 in cell lines derived from different tissues. (b) Expression of NUSAP1 in cell lines derived from different tissues. (c) Expression of MCM2 in 21 commonly used PC cell lines. (d) Expression of NUSAP1 in 21 commonly used PC cell lines.
Figure 9.
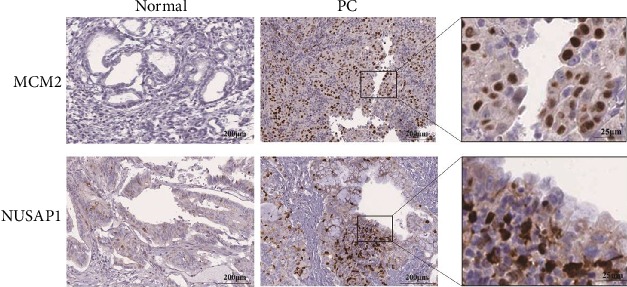
Representative immunohistochemical images of MCM2 and NUSAP1 expression in normal pancreatic and PC tissues.
4. Discussion
The lack of methods for early screening for and detection of PC leads to late detection of the disease and a high mortality rate in patients [38]. Hence, early diagnosis and treatment are pivotal for improving the clinical outcome of patients with PC. A growing body of evidence suggests that some of dysregulated genes in PC may be potential biomarkers for the diagnosis and prognosis of the disease [39, 40]. Therefore, we used bioinformatics tools to analyze PC-associated mRNA expression profiles and to identify potential biomarkers for diagnosis and prognosis of PC.
In this study, we downloaded three PC-related mRNA expression datasets from the GEO and TCGA databases and screened the data for DEGs. In total, 248 DEGs were identified, and it was found that the GO terms enriched in DEGs were mainly related to “mitotic mitosis”, “centromere region”, “mitotic spindle”, and “extracellular matrix binding”; the enriched pathways were mainly associated with “focus adhesion”, “ECM-receptor interactions”, and “PI3K/Akt signaling pathway”. ECM and focal adhesion have been shown to be important components of tumorigenesis and cancer progression [41–43]. Dysregulation of the cell cycle is the key factor in the malignant biological behaviors associated with the proliferation, invasion, and metastasis of PC cells [44, 45]. Zhang et al. [46] showed that LAMB3 affected the proliferation, invasion, and metastasis of PC by regulating the PI3K/Akt signaling pathway. Our data are consistent with the above findings and provide new insights into molecular mechanisms of pathogenesis of PC.
In addition, we constructed a PPI network for DEGs and selected the top 20 genes as the hub genes. The hub genes were analyzed using GO and KEGG pathway enrichment and were found to be mainly enriched in the GO terms “apoptotic process” and “cell cycle” and in the KEGG pathways related to the “cell cycle” and “p53 signaling pathway”. Previous studies have reported that the apoptotic process and cell cycle are closely related to the development and progression of PC [47, 48]. As one of the important signaling pathways in the body, the p53 signaling pathway has been shown to be involved in the development, invasion, and metastasis of various tumors [49, 50]. Studies have reported that targeting of DTL can induce cell cycle arrest and senescence and can therefore be used to treat liver cancer [51]. Downregulation of MELK [52], MAD2L1 [53], and CCNB1 [54] can also inhibit cell cycle progression of liver cancer. ANLN [55], BUB1 [56], CDK1 [57], SMC4 [58], CENPE [59], ATAD2 [60], TPX2 [61], KIF23 [62], CDKN3 [63], and KIF20A [64] are closely related to the development of various tumors. However, reports regarding the role of FANCI and RAD51AP1 in tumors are limited, and hence, these genes deserve further investigation.
To further screen for potential biomarkers for the diagnosis and prognosis of PC, we analyzed genetic variations, verified the differential expression, and evaluated the survival, prognostic, and diagnostic value of the hub genes. Finally, we identified MCM2 and NUSAP1 as potential biomarkers for the diagnosis and prognosis of PC. To minimize the bias, we used multiple databases to verify the mRNA and protein levels of MCM2 and NUSAP1 at the tissue and cell levels. The results showed that the mRNA and protein levels of MCM2 and NUSAP1 were significantly upregulated in PC cells and tissues. In addition, gene correlation analysis showed that the expression levels of MCM2, NUSAP1, EGFR, ERBB2, and KRAS significantly correlated, and thus, MCM2 and NUSAP1 are likely to become potential therapeutic targets for PC.
MCM2 is an important DNA replication initiation factor in humans. It is present in the nucleus and is highly expressed in proliferating cells. MCM2 is not expressed or is poorly expressed in quiescent or well-differentiated cells, suggesting that it can be used as a specific marker for proliferating cells. Studies have reported that MCM2 is a key molecule involved in the pathogenesis of non-small-cell lung cancer and may be a novel therapeutic target for lovastatin in the treatment of this cancer [65]. Proteomic analysis of clinical specimens of high-grade small cell lung cancer showed that MCM2 was associated with a poor prognosis in patients [66]. Increasing evidence has confirmed that MCM2 can be used as a biomarker for the diagnosis and prognosis of various cancers. Yang et al. [67] showed that MCM2 expression in gastric cancer tissues was significantly higher than that in the normal gastric mucosa, and the levels of expression positively correlated with the prognosis, suggesting that MCM2 can be used as a novel prognostic biomarker for gastric cancer. Torres-Rendon et al. [68] analyzed the MCM2 expression in clinical samples of oral squamous cell carcinoma using immunohistochemistry and demonstrated that MCM2 might be a useful prognostic marker for the disease. In another study, staining for MCM2 was directly performed on 183 liquid-based cytological samples. Studies have shown that the positive rate of MCM2 expression increased with the severity of cervical lesions and was related to the type of human papilloma virus, which indicates a potential application value of MCM2 in the diagnosis of cervical lesions [69]. All these results show that MCM2 is involved in the occurrence and progression of various tumors and can be a potential biomarker for their diagnosis and prognosis, which is consistent with the results of our analysis. However, there are no studies regarding the role of MCM2 in PC, and thus, MCM2 needs to be confirmed as a potential marker for the diagnosis and prognosis of PC.
NUSAP1 is a tubulin that is involved in the assembly of the spindle to ensure a normal cell cycle, thus playing an important role in mitosis [70]. An abnormal cell cycle is an important feature of tumor formation, and currently, the role of NUSAP1 in cancer is being actively investigated. Some studies have shown overexpression of NUSAP1 in renal cell carcinoma [71], colon cancer [72], glioma [73], and other malignant tumors and its significant association with tumor invasion and metastasis, as well as with a poor prognosis in patients. Thus, the results of this study are consistent with those regarding the role of NUSAP1 in other tumors. NUSAP1 expression in acute myeloid leukemia can block the cell cycle [74], whereas reduction in the NUSAP1 expression increased the killing effect of paclitaxel on oral epithelial squamous cell carcinoma cells [75]. In patients with liver cancer, the level of NUSAP1 expression is closely associated with the severity of the prognosis, whereas interference with NUSAP1 expression inhibits the growth of liver cancer cells [76]. Gulzar et al. [77] found that high levels of NUSAP1 expression were related to the growth characteristics of tumor cells in PC, thus making NUSAP1 a novel biomarker for PC recurrence after surgery. Studies have also confirmed that NUSAP1 has a high prognostic value for breast cancer [78]. Thus, the high prognostic value of NUSAP1 for various tumors is consistent with the results of this study. Although relevant research on the role of NUSAP1 in PC is lacking, we speculated that NUSAP1 may be a potential biomarker for PC.
Most bioinformatics studies have reported analysis of a single mRNA expression dataset. In this study, we selected three mRNA expression datasets from two databases, thereby increasing the sample size and confidence level. We used different bioinformatics methods to mine the data deeper and used multiple databases to perform multidimensional verification of MCM2 and NUSAP1 expression in PC, providing diverse perspectives. However, this study has certain limitations. First, a certain degree of heterogeneity was present in the datasets selected for this study, and only four normal samples were included in TCGA. Although we removed the batch data and performed quality control and standardization of the raw data, a larger sample size and a higher-quality dataset are still required to verify the reliability of the results. Second, our study involved a second round of mining and analysis of previously published datasets. Although some previous data were consistent with those of our analysis, further molecular biology experiments are required to verify the accuracy of our results. As a future research direction, validation of MCM2 and NUSAP1 as diagnostic and prognostic markers of PC is needed in a large number of clinical PC samples and PC cell lines.
5. Conclusions
In conclusion, using bioinformatics analysis of three mRNA expression datasets, a total of 20 hub genes were identified, which may play key roles in the occurrence and progression of PC. Analysis of the clinical significance of the 20 hub genes in PC revealed two abnormally regulated genes, MCM2 and NUSAP1, which were confirmed, using multidimensional verification, as potential biomarkers for the diagnosis and prognosis of PC. However, the results need to be validated in a larger number of clinical samples using additional experimental methods before using MCM2 and NUSAP1 as effective diagnostic and prognostic markers for PC.
Acknowledgments
This work has benefited from GEO and TCGA. We thank the GEO and TCGA network for their generous sharing of large amounts of data. We would like to thank FigureYa (Xiao Ya Hua Tu) for the figure technology support.
Contributor Information
Jinyong Hao, Email: haojinyong517@sina.com.
Qiqi Xie, Email: jieqq16@lzu.edu.cn.
Ruochen Zhao, Email: zhaorch16@lzu.edu.cn.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
Yajun Deng and Hanyun Ma contributed equally to this work.
Supplementary Materials
Fig. S1: data preprocessing. (a) Box plot of standardized expression data for the GSE15471 dataset. (b) Box plot of the standardized expression data for the GSE16515 dataset. The red symbol indicates a normal tissue sample, and the blue symbol indicates a sample from a patient with PC. The red diamond in each box represents the average level of gene expression in each sample. (c) Sample clustering map of the GSE15471 dataset. (d) Sample clustering map of the GSE16515 dataset. Blue represents downregulation of genes, and red represents upregulation of genes.
Fig. S2: GO and KEGG pathway enrichment analyses of the hub genes. (a) BP, (b) CC, (c) MF, and (d) KEGG analysis results.
Fig. S3: genetic variation analysis of the hub genes. (a) Changes in the hub gene copy number and a mutation panorama. (b) Mutation details of the hub genes.
Fig. S4: association of hub gene expression with disease-free survival (DFS) of patients with PC. (a) BUB1, (b) CDK1, (c) FANCI, (d) KIF20A, (e) HMMR, (f) KIF23, (g) MCM2, (h) NUSAP1, and (i) TPX2. The solid line represents the survival curve, and the dashed line represents the 95% confidence interval. Patients with higher than the median value are indicated by the red line, and those with lower than the median value are indicated by the blue line. Log-rank P < 0.05 was considered statistically significant.
Fig. S5: dot plots of MCM2 and NUSAP1 expression in different tumor and normal specimens. Each point represents a sample, with red representing tumor samples and green representing normal samples. (a) MCM2 expression. (b) NUSAP1 expression.
Fig. S6: mRNA and protein expression of MCM2 and NUSAP1 in normal human tissues, based on the Human Protein Atlas. (a) MCM2 mRNA. (b) NUSAP1 mRNA. (c) MCM2 protein. (d) NUSAP1 protein.
References
- 1.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2017. CA: a Cancer Journal for Clinicians. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Garrido-Laguna I., Hidalgo M. Pancreatic cancer: from state-of-the-art treatments to promising novel therapies. Nature Reviews Clinical Oncology. 2015;12(6):319–334. doi: 10.1038/nrclinonc.2015.53. [DOI] [PubMed] [Google Scholar]
- 3.Ray K. Pancreatic cancer: biomarkers for the early detection of PDAC. Nature Reviews Gastroenterology & Hepatology. 2017;14(9):504–505. doi: 10.1038/nrgastro.2017.111. [DOI] [PubMed] [Google Scholar]
- 4.Ballehaninna U. K., Chamberlain R. S. The clinical utility of serum CA 19-9 in the diagnosis, prognosis and management of pancreatic adenocarcinoma: an evidence based appraisal. Journal of gastrointestinal oncology. 2012;3(2):105–119. doi: 10.3978/j.issn.2078-6891.2011.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Useros J., Garcia-Foncillas J. Can molecular biomarkers change the paradigm of pancreatic cancer prognosis? BioMed Research International. 2016;2016:13. doi: 10.1155/2016/4873089.4873089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crawley A. S., O'Kennedy R. J. The need for effective pancreatic cancer detection and management: a biomarker-based strategy. Expert Review of Molecular Diagnostics. 2015;15(10):1339–1353. doi: 10.1586/14737159.2015.1083862. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y., Bai Y. S., Wang Q. CDGSH iron sulfur domain 2 activates proliferation and EMT of pancreatic cancer cells via Wnt/β-Catenin pathway and has prognostic value in human pancreatic cancer. Oncology Research. 2017;25(4):605–615. doi: 10.3727/096504016X14767450526417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koopmann J., Fedarko N. S., Jain A., et al. Evaluation of osteopontin as biomarker for pancreatic adenocarcinoma. Cancer Epidemiology, Biomarkers & Prevention. 2004;13(3):487–491. [PubMed] [Google Scholar]
- 9.Zihao G., Jie Z., Yan L., et al. Analyzing S100A6 expression in endoscopic ultrasonography-guided fine-needle aspiration specimens: a promising diagnostic method of pancreatic cancer. Journal of Clinical Gastroenterology. 2013;47(1):69–75. doi: 10.1097/MCG.0b013e3182601752. [DOI] [PubMed] [Google Scholar]
- 10.Gold D. V., Gaedcke J., Ghadimi B. M., et al. PAM4 enzyme immunoassay alone and in combination with CA 19-9 for the detection of pancreatic adenocarcinoma. Cancer. 2013;119(3):522–528. doi: 10.1002/cncr.27762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han W., Cao F., Chen M. B., et al. Prognostic value of SPARC in patients with pancreatic cancer: a systematic review and meta-analysis. PLoS One. 2016;11(1, article e0145803) doi: 10.1371/journal.pone.0145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Z. K., Zhang R. Y., Yong Y. L., et al. Identification of crucial genes based on expression profiles of hepatocellular carcinomas by bioinformatics analysis. PeerJ. 2019;7, article e7436 doi: 10.7717/peerj.7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang H., Zhang M., Xie Q., Yu J., Qi Y., Yue Q. Identification of diagnostic markers for major depressive disorder by cross-validation of data from whole blood samples. PeerJ. 2019;7, article e7171 doi: 10.7717/peerj.7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrett T., Wilhite S. E., Ledoux P., et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Research. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badea L., Herlea V., Dima S. O., Dumitrascu T., Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepato-Gastroenterology. 2008;55(88):2016–2027. [PubMed] [Google Scholar]
- 16.Pei H., Li L., Fridley B. L., et al. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell. 2009;16(3):259–266. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomczak K., Czerwinska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Współczesna Onkologia . 2015;19(1a):A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier L., Cope L., Bolstad B. M., Irizarry R. A. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 19.Ritchie M. E., Phipson B., Wu D., et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):p. e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pathan M., Keerthikumar S., Ang C. S., et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- 21.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chin C. H., Chen S. H., Wu H. H., Ho C. W., Ko M. T., Lin C. Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. Systems Biology. 2014;8(Supplentary 4):p. S11. doi: 10.1186/1752-0509-8-s4-s11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao J., Aksoy B. A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6(269):p. pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J. R., Sun C. H., Li W., et al. Cancer RNA-Seq Nexus: a database of phenotype-specific transcriptome profiling in cancer cells. Nucleic Acids Research. 2016;44(D1):D944–D951. doi: 10.1093/nar/gkv1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris M. A., Clark J., Ireland A., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Research. 2004;32(Database issue):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M. The KEGG database. ‘In Silico’ Simulation of Biological Processes: Novartis Foundation Symposium. 2002;247:91–101. [PubMed] [Google Scholar]
- 28.Yu G., Wang L. G., Han Y., He Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis G., Jr., Sherman B. T., Hosack D. A., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biology. 2003;4(5) doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 30.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Research. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):p. 77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barretina J., Caponigro G., Stransky N., et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su A. I., Wiltshire T., Batalov S., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(16):6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., Wang M., Yao X., et al. Development of a novel EGFR-targeting antibody-drug conjugate for pancreatic cancer therapy. Targeted Oncology. 2019;14(1):93–105. doi: 10.1007/s11523-018-0616-8. [DOI] [PubMed] [Google Scholar]
- 35.Skrypek N., Vasseur R., Vincent A., Duchene B., Van Seuningen I., Jonckheere N. The oncogenic receptor ErbB2 modulates gemcitabine and irinotecan/SN-38 chemoresistance of human pancreatic cancer cells via hCNT1 transporter and multidrug-resistance associated protein MRP-2. Oncotarget. 2015;6(13):10853–10867. doi: 10.18632/oncotarget.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang M. T., Fer N., Galeas J., et al. Blockade of leukemia inhibitory factor as a therapeutic approach to KRAS driven pancreatic cancer. Nature Communications. 2019;10(1):p. 3055. doi: 10.1038/s41467-019-11044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu Z., Gu L., Eils R., Schlesner M., Brors B. circlize implements and enhances circular visualization in R. Bioinformatics. 2014;30(19):2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 38.Malvezzi M., Carioli G., Bertuccio P., et al. European cancer mortality predictions for the year 2017, with focus on lung cancer. Annals of Oncology. 2017;28(5):1117–1123. doi: 10.1093/annonc/mdx033. [DOI] [PubMed] [Google Scholar]
- 39.Joergensen M. T., Brunner N., De Muckadell O. B. Comparison of circulating MMP-9, TIMP-1 and CA19-9 in the detection of pancreatic cancer. Anticancer Research. 2010;30(2):587–592. [PubMed] [Google Scholar]
- 40.Suh H., Pillai K., Morris D. L. Mucins in pancreatic cancer: biological role, implications in carcinogenesis and applications in diagnosis and therapy. American Journal of Cancer Research. 2017;7(6):1372–1383. [PMC free article] [PubMed] [Google Scholar]
- 41.Malik R., Lelkes P. I., Cukierman E. Biomechanical and biochemical remodeling of stromal extracellular matrix in cancer. Trends in Biotechnology. 2015;33(4):230–236. doi: 10.1016/j.tibtech.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eke I., Cordes N. Focal adhesion signaling and therapy resistance in cancer. Seminars in Cancer Biology. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 43.Okegawa T., Pong R. C., Li Y., Hsieh J. T. The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochimica Polonica. 2004;51(2):445–457. doi: 10.18388/abp.2004_3583. [DOI] [PubMed] [Google Scholar]
- 44.Usugi E., Ishii K., Hirokawa Y., et al. Antifibrotic agent pirfenidone suppresses proliferation of human pancreatic cancer cells by inducing G0/G1 cell cycle arrest. Pharmacology. 2019;103(5-6):250–256. doi: 10.1159/000496831. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y., Kim K. H., Lee I. S., et al. Apoptosis and G2/M cell cycle arrest induced by a timosaponin A3 from Anemarrhena asphodeloides Bunge on AsPC-1 pancreatic cancer cells. Phytomedicine. 2019;56:48–56. doi: 10.1016/j.phymed.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H., Pan Y. Z., Cheung M., et al. LAMB3 mediates apoptotic, proliferative, invasive, and metastatic behaviors in pancreatic cancer by regulating the PI3K/Akt signaling pathway. Cell Death & Disease. 2019;10(3):p. 230. doi: 10.1038/s41419-019-1320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jing W., Song N., Liu Y. P., et al. DNMT3a promotes proliferation by activating the STAT3 signaling pathway and depressing apoptosis in pancreatic cancer. Cancer Management and Research. 2019;11:6379–6396. doi: 10.2147/CMAR.S201610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Guo J., Fan H. MiR-155 regulates the proliferation and apoptosis of pancreatic cancer cells through targeting SOCS3. European Review for Medical and Pharmacological Sciences. 2019;23(12):5168–5175. doi: 10.26355/eurrev_201906_18181. [DOI] [PubMed] [Google Scholar]
- 49.Zhang H., Zhang X., Li X., et al. Effect of CCNB1 silencing on cell cycle, senescence, and apoptosis through the p53 signaling pathway in pancreatic cancer. Journal of Cellular Physiology. 2018;234(1):619–631. doi: 10.1002/jcp.26816. [DOI] [PubMed] [Google Scholar]
- 50.Dong Y. X., Pang Z. G., Zhang J. C., Hu J. Q., Wang L. Y. Long non-coding RNA GClnc1 promotes progression of colorectal cancer by inhibiting p53 signaling pathway. European Review for Medical and Pharmacological Sciences. 2019;23(13):5705–5713. doi: 10.26355/eurrev_201907_18308. [DOI] [PubMed] [Google Scholar]
- 51.Chen Y. C., Chen I. S., Huang G. J., et al. Targeting DTL induces cell cycle arrest and senescence and suppresses cell growth and colony formation through TPX2 inhibition in human hepatocellular carcinoma cells. OncoTargets and Therapy. 2018;11:1601–1616. doi: 10.2147/OTT.S147453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y., Li Y., Chen Y., et al. MicroRNA-214-3p inhibits proliferation and cell cycle progression by targeting MELK in hepatocellular carcinoma and correlates cancer prognosis. Cancer Cell International. 2017;17(1):p. 102. doi: 10.1186/s12935-017-0471-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Y., Bai W., Zhang J. MiR-200c-5p suppresses proliferation and metastasis of human hepatocellular carcinoma (HCC) via suppressing MAD2L1. Biomedicine & Pharmacotherapy. 2017;92:1038–1044. doi: 10.1016/j.biopha.2017.05.092. [DOI] [PubMed] [Google Scholar]
- 54.Chai N., Xie H. H., Yin J. P., et al. FOXM1 promotes proliferation in human hepatocellular carcinoma cells by transcriptional activation of CCNB1. Biochemical and Biophysical Research Communications. 2018;500(4):924–929. doi: 10.1016/j.bbrc.2018.04.201. [DOI] [PubMed] [Google Scholar]
- 55.Magnusson K., Gremel G., Rydén L., et al. ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer. 2016;16(1):p. 904. doi: 10.1186/s12885-016-2923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Z., Katsaros D., Shen Y., et al. Biological and clinical significance of MAD2L1 and BUB1, genes frequently appearing in expression signatures for breast cancer prognosis. PLoS One. 2015;10(8, article e0136246) doi: 10.1371/journal.pone.0136246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y. X., Zhu T., Zou T., et al. Prognostic and predictive values of CDK1 and MAD2L1 in lung adenocarcinoma. Oncotarget. 2016;7(51):85235–85243. doi: 10.18632/oncotarget.13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang C., Kuang M., Li M., Feng L., Zhang K., Cheng S. SMC4, which is essentially involved in lung development, is associated with lung adenocarcinoma progression. Scientific Reports. 2016;6(1):p. 34508. doi: 10.1038/srep34508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shan L., Zhao M., Lu Y., et al. CENPE promotes lung adenocarcinoma proliferation and is directly regulated by FOXM1. International Journal of Oncology. 2019;55(1):257–266. doi: 10.3892/ijo.2019.4805. [DOI] [PubMed] [Google Scholar]
- 60.Liu N., Funasaka K., Obayashi T., et al. ATAD2 is associated with malignant characteristics of pancreatic cancer cells. Oncology Letters. 2019;17(3):3489–3494. doi: 10.3892/ol.2019.9960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang M. Y., Liu X. X., Li H., Li R., Liu X., Qu Y. Q. Elevated mRNA levels of AURKA, CDC20 and TPX2 are associated with poor prognosis of smoking related lung adenocarcinoma using bioinformatics analysis. International Journal of Medical Sciences. 2018;15(14):1676–1685. doi: 10.7150/ijms.28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X. L., Ji Y. M., Song R., Li X. N., Guo L. S. KIF23 promotes gastric cancer by stimulating cell proliferation. Disease Markers. 2019;2019:9. doi: 10.1155/2019/9751923.9751923 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Chang S. L., Chen T. J., Lee Y. E., Lee S. W., Lin L. C., He H. L. CDKN3 expression is an independent prognostic factor and associated with advanced tumor stage in nasopharyngeal carcinoma. International Journal of Medical Sciences. 2018;15(10):992–998. doi: 10.7150/ijms.25065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shen T., Yang L., Zhang Z., et al. KIF20A affects the prognosis of bladder cancer by promoting the proliferation and metastasis of bladder cancer cells. Disease Markers. 2019;2019:9. doi: 10.1155/2019/4863182.4863182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X., Teng Y., Yang F., et al. MCM2 is a therapeutic target of lovastatin in human non-small cell lung carcinomas. Oncology Reports. 2015;33(5):2599–2605. doi: 10.3892/or.2015.3822. [DOI] [PubMed] [Google Scholar]
- 66.Fujii K., Miyata Y., Takahashi I., et al. Differential proteomic analysis between small cell lung carcinoma (SCLC) and pulmonary carcinoid tumors reveals molecular signatures for malignancy in lung cancer. Proteomics Clinical Applications. 2018;12(6, article e1800015) doi: 10.1002/prca.201800015. [DOI] [PubMed] [Google Scholar]
- 67.Yang C., Wen Y., Li H., et al. Overexpression of minichromosome maintenance 2 predicts poor prognosis in patients with gastric cancer. Oncology Reports. 2012;27(1):135–142. doi: 10.3892/or.2011.1473. [DOI] [PubMed] [Google Scholar]
- 68.Torres-Rendon A., Roy S., Craig G. T., Speight P. M. Expression of Mcm2, geminin and Ki67 in normal oral mucosa, oral epithelial dysplasias and their corresponding squamous-cell carcinomas. British Journal of Cancer. 2009;100(7):1128–1134. doi: 10.1038/sj.bjc.6604967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng J. Diagnostic value of MCM2 immunocytochemical staining in cervical lesions and its relationship with HPV infection. International Journal of Clinical and Experimental Pathology. 2015;8(1):875–880. [PMC free article] [PubMed] [Google Scholar]
- 70.Mills C. A., Suzuki A., Arceci A., et al. Nucleolar and spindle-associated protein 1 (NUSAP1) interacts with a SUMO E3 ligase complex during chromosome segregation. The Journal of Biological Chemistry. 2017;292(42):17178–17189. doi: 10.1074/jbc.M117.796045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fang L., Zhang M., Chen L., et al. Downregulation of nucleolar and spindle-associated protein 1 expression suppresses cell migration, proliferation and invasion in renal cell carcinoma. Oncology Reports. 2016;36(3):1506–1516. doi: 10.3892/or.2016.4955. [DOI] [PubMed] [Google Scholar]
- 72.Liu Z., Guan C., Lu C., et al. High NUSAP1 expression predicts poor prognosis in colon cancer. Pathology, Research and Practice. 2018;214(7):968–973. doi: 10.1016/j.prp.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 73.Zhu T., Xie P., Gao Y. F., et al. Nucleolar and spindle-associated protein 1 is a tumor grade correlated prognosis marker for glioma patients. CNS Neuroscience & Therapeutics. 2018;24(3):178–186. doi: 10.1111/cns.12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hatfield K. J., Reikvam H., Bruserud O. Identification of a subset of patients with acute myeloid leukemia characterized by long-term in vitro proliferation and altered cell cycle regulation of the leukemic cells. Expert Opinion on Therapeutic Targets. 2014;18(11):1237–1251. doi: 10.1517/14728222.2014.957671. [DOI] [PubMed] [Google Scholar]
- 75.Okamoto A., Higo M., Shiiba M., et al. Down-regulation of nucleolar and spindle-associated protein 1 (NUSAP1) expression suppresses tumor and cell proliferation and enhances anti-tumor effect of paclitaxel in oral squamous cell carcinoma. PLoS One. 2015;10(11, article e0142252) doi: 10.1371/journal.pone.0142252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Roy S., Hooiveld G. J., Seehawer M., et al. microRNA 193a-5p regulates levels of nucleolar- and spindle-associated protein 1 to suppress hepatocarcinogenesis. Gastroenterology. 2018;155(6):1951–1966.e26. doi: 10.1053/j.gastro.2018.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gulzar Z. G., McKenney J. K., Brooks J. D. Increased expression of NuSAP in recurrent prostate cancer is mediated by E2F1. Oncogene. 2013;32(1):70–77. doi: 10.1038/onc.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L., Yang L., Qiao F., et al. High levels of nucleolar spindle-associated protein and reduced levels of BRCA1 expression predict poor prognosis in triple-negative breast cancer. PLoS One. 2015;10(10, article e0140572) doi: 10.1371/journal.pone.0140572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: data preprocessing. (a) Box plot of standardized expression data for the GSE15471 dataset. (b) Box plot of the standardized expression data for the GSE16515 dataset. The red symbol indicates a normal tissue sample, and the blue symbol indicates a sample from a patient with PC. The red diamond in each box represents the average level of gene expression in each sample. (c) Sample clustering map of the GSE15471 dataset. (d) Sample clustering map of the GSE16515 dataset. Blue represents downregulation of genes, and red represents upregulation of genes.
Fig. S2: GO and KEGG pathway enrichment analyses of the hub genes. (a) BP, (b) CC, (c) MF, and (d) KEGG analysis results.
Fig. S3: genetic variation analysis of the hub genes. (a) Changes in the hub gene copy number and a mutation panorama. (b) Mutation details of the hub genes.
Fig. S4: association of hub gene expression with disease-free survival (DFS) of patients with PC. (a) BUB1, (b) CDK1, (c) FANCI, (d) KIF20A, (e) HMMR, (f) KIF23, (g) MCM2, (h) NUSAP1, and (i) TPX2. The solid line represents the survival curve, and the dashed line represents the 95% confidence interval. Patients with higher than the median value are indicated by the red line, and those with lower than the median value are indicated by the blue line. Log-rank P < 0.05 was considered statistically significant.
Fig. S5: dot plots of MCM2 and NUSAP1 expression in different tumor and normal specimens. Each point represents a sample, with red representing tumor samples and green representing normal samples. (a) MCM2 expression. (b) NUSAP1 expression.
Fig. S6: mRNA and protein expression of MCM2 and NUSAP1 in normal human tissues, based on the Human Protein Atlas. (a) MCM2 mRNA. (b) NUSAP1 mRNA. (c) MCM2 protein. (d) NUSAP1 protein.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


