To the Editor: Traditional approaches to respiratory virus surveillance may not identify novel pathogens in time to implement crucial public health interventions.1 The Seattle Flu Study is a multi-institutional, community-wide pandemic surveillance platform that was established in November 2018.2 Persons reporting symptoms of respiratory illness provided informed consent for testing to identify influenza and other respiratory pathogens (see the Supplementary Appendix, available with the full text of this letter at NEJM.org). In one study group, persons enrolled online and were sent kits, by rapid-delivery services, for home collection of a midnasal swab; samples were returned by mail. After identification of the first case of Covid-19 in Washington State, the samples that were collected were also tested for SARS-CoV-2. After March 4, 2020, a human subjects institutional review board determined that results could be reported to public health authorities and to participants, who were notified under a public health surveillance exemption.
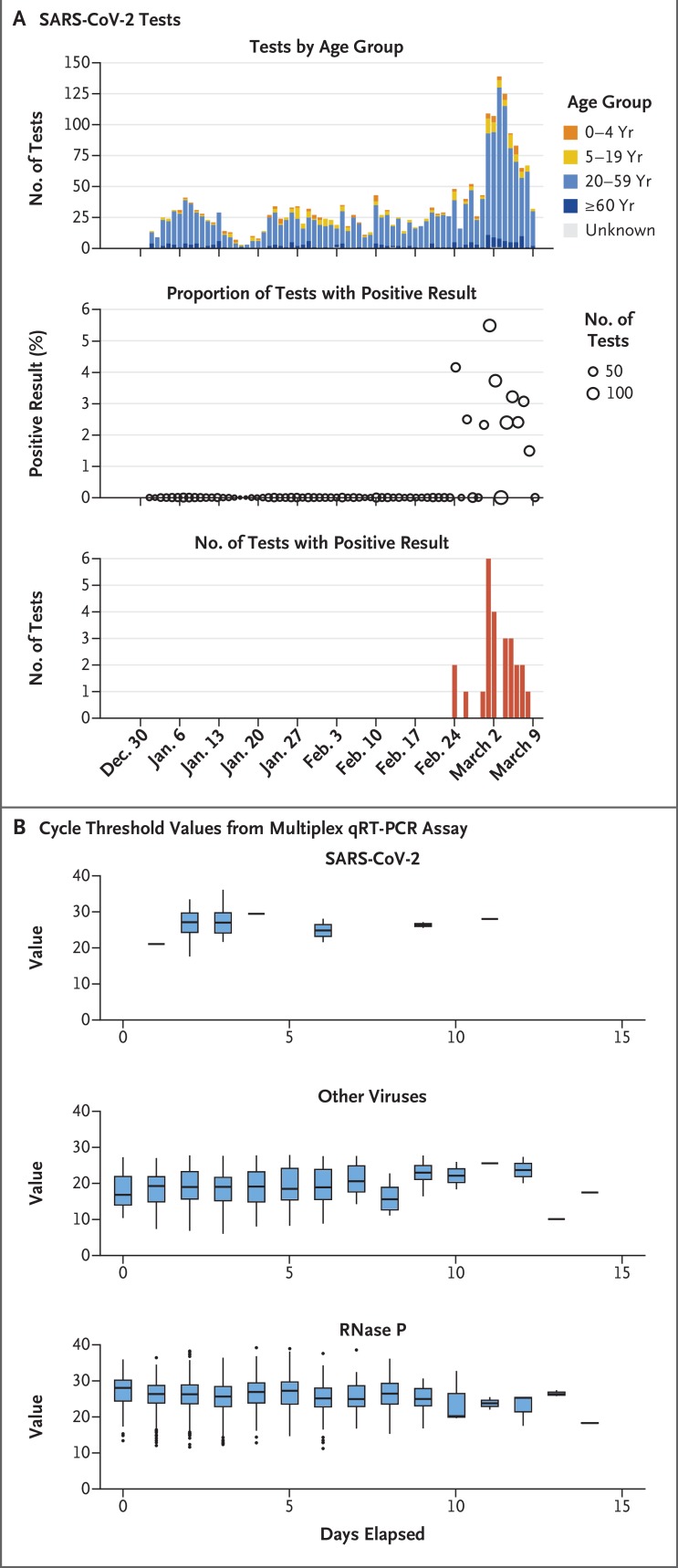
From January 1 through March 9, 2020, a total of 3524 participants provided specimens after online enrollment. Of these, 2353 (66.8%) completed all the study procedures, including testing; SARS-CoV-2 was detected in 25 (1.1%), of whom 2 were children (Figure 1A). Enrollment increased markedly in late February, most likely motivated by public concern about Covid-19 (Figure 1A). Some of the collection kits (1316 of 2353 [55.9%]) were sent to participants for same-day delivery, with a median delivery time of 2.3 hours (interquartile range, 1.7 to 3.1). The median number of days from swab collection to receipt of the specimen by the laboratory was 2 days (interquartile range, 2 to 4). Semiquantitative viral loads from home-collected midnasal swabs were stable over time for SARS-CoV-2, other respiratory viruses, and RNase P, a human cellular marker (Figure 1B).
Figure 1. Covid-19 Case Detection through the Seattle Flu Study.
Panel A shows SARS-CoV-2 tests over time, further stratified according to age group; detection of positive SARS-CoV-2 test results over time and as a percentage of the total number of tests run; and Covid-19–positive case counts over time. Panel B shows cycle threshold values from a laboratory-developed multiplex quantitative reverse-transcriptase polymerase-chain-reaction (qRT-PCR) assay as a function of days elapsed from swab collection to freezing or nucleic acid extraction for virus detection, shown separately for SARS-CoV-2, other respiratory viruses, and RNase P, a human cellular marker. The horizontal line in each box represents the median, the lower and upper boundaries of the boxes the interquartile range, and the ends of the vertical lines 1.5 times the interquartile range. Individual points represent outliers (>1.5 times the interquartile range).
In this early phase of the pandemic, only 7 of the 25 persons with Covid-19 (28%) reported seeking clinical care. The most common reported symptoms were fatigue, myalgia, fever, cough, and chills. The median duration of symptoms before swab collection was 4 days; 9 participants (36%) had had symptoms for less than 2 days at time of swab collection. Coinfection with another respiratory virus was present in 4 cases (16%).
The first Covid-19 case detected through the Seattle Flu Study, in a specimen collected on February 24, 2020, was the first documented U.S. case of community transmission at the time.3 These results initiated assessment of the spread of the virus in the Seattle region, which in turn accelerated public health efforts to mitigate the emerging pandemic.4 As the Covid-19 pandemic progresses, widespread implementation of simple methods that are scalable and require minimal interaction for collection of samples from persons who may not seek clinical care is critical for early detection of community cases. Looking beyond the current crisis, we envision ubiquitous, community-based sampling for respiratory illnesses as essential infrastructure for early detection and mitigation of future pandemics.
Supplementary Appendix
Disclosure Forms
This letter was published on May 1, 2020, at NEJM.org.
Footnotes
Supported by Gates Ventures.
Disclosure forms provided by the authors are available with the full text of this letter at NEJM.org.
References
- 1.Fink S, Baker M. “It’s just everywhere already”: how delays in testing set back the U.S. coronavirus response. New York Times. March 10, 2020. (https://www.nytimes.com/2020/03/10/us/coronavirus-testing-delays.html).
- 2.Chu HY, Boeckh M, Englund JA, et al. The Seattle Flu Study: a multi-arm community-based prospective study protocol for assessing influenza prevalence, transmission, and genomic epidemiology. March 6, 2020. (https://www.medrxiv.org/content/10.1101/2020.03.02.20029595v1) (preprint). [DOI] [PMC free article] [PubMed]
- 3.Bedford T, Greninger AL, Roychoudhury P, et al. Cryptic transmission of SARS-CoV-2 in Washington State. April 16, 2020. (https://www.medrxiv.org/content/10.1101/2020.04.02.20051417v1) (preprint). [DOI] [PMC free article] [PubMed]
- 4.Klein D, Hagedorn B, Kerr C, Hu H, Bedford T, Famulare M. Working paper — model-based estimates of COVID-19 burden in King and Snohomish counties through April 7, 2020. March 10, 2020. (https://institutefordiseasemodeling.github.io/COVID-public/reports/Working%20paper%20%E2%80%93%20model-based%20estimates%20of%20COVID-19%20burden%20in%20King%20and%20Snohomish%20counties%20through%20April%207.pdf).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.