Abstract
Background: The Glycemia Reduction Approaches in Diabetes: A Comparative Effectiveness (GRADE) study has enrolled a racially and ethnically diverse population with type 2 diabetes, performed extensive phenotyping, and randomly assigned the participants to one of four second-line diabetes medications. The continuous glucose monitoring (CGM) substudy has been added to determine whether there are racial/ethnic differences in the relationship between average glucose (AG) and hemoglobin A1c (HbA1c). CGM will also be used to compare time in target range, glucose variability, and the frequency and duration of hypoglycemia across study groups.
Methods: The observational CGM substudy will enroll up to 1800 of the 5047 GRADE study participants from the four treatment groups, including as many as 450 participants from each of 4 racial/ethnic minority groups to be compared: Hispanic White, non-Hispanic White, non-Hispanic African American, and non-Hispanic Other. CGM will be performed for 2 weeks in proximity to a GRADE annual visit, during which an oral glucose tolerance test will be performed and HbA1c and glycated albumin measured. Indicators of interindividual variation in red blood cell turnover, based on specialized erythrocyte measurements, will also be measured to explore the potential causes of interindividual HbA1c variations.
Conclusions: The GRADE CGM substudy will provide new insights into whether differences exist in the relationship between HbA1c and AG among different racial/ethnic groups and whether glycemic profiles differ among frequently used diabetes medications and their potential clinical implications. Understanding such differences is important for clinical care and adjustment of diabetes medications in patients of different races or ethnicities.
Keywords: Type 2 diabetes, Glycated hemoglobin, Interracial differences, Continuous glucose monitoring, Average glucose
Background
Interindividual differences in the relationship among average glucose (AG) levels, glycated hemoglobin, and other measures of glycemic control, including interracial and interethnic differences, have been suggested1–6 but remain controversial.7–10 Most studies to date, with rare exceptions,11,12 have not collected reliable measures of average glycemia or explored plausible mechanisms for the putative differences. Interindividual variability in red blood cell (RBC) turnover and genetic variation in hemoglobin glycation13–17 have been proposed as explanations for any interindividual differences in the relationship between AG and glycated hemoglobin levels. Understanding whether such putative differences exist among racial and ethnic groups is important because of the potential to overtreat or undertreat subgroups of patients based on the incorrect translation of hemoglobin A1c (HbA1c) into AG levels, which could in turn result in excess hypoglycemia or increased risk for long-term micro- and macrovascular complications, respectively.
The Glycemia Reduction Approaches in Diabetes: A Comparative Effectiveness (GRADE) Study has recruited participants with type 2 diabetes (T2D) with a large representation of African Americans and Hispanic Americans as well as non-Hispanic Whites (NHWs).18 The study is primarily directed at comparing glycemic effects, based on HbA1c, of four of the most commonly used diabetes medicines when added to metformin. We describe herein a continuous glucose monitoring (CGM) Substudy added to GRADE which, with its diverse population, will definitively address whether the relationship between AG and HbA1c differs among racial and ethnic populations. In addition, the random assignment to different diabetes therapies in GRADE provides the opportunity to examine potential differential effects of diabetes medications on glucose profiles, including their relative risk for hypoglycemia and postprandial hyperglycemia. Putative differences in glucose profiles between diabetes medications beyond mean glycemia achieved have also been suggested as a risk factor for long-term complications,19 but comparative effectiveness studies with random treatment assignment and reliable measurements of glycemic profiles have been extremely limited.
Methods
Population and setting
The design of the GRADE study has been described in detail.18 In brief, the GRADE study, a pragmatic, parallel design clinical trial, recruited participants ≥30 years of age with diabetes duration <10 years. At the time of randomization eligible participants were treated with at least 1000 mg of metformin per day for the preceding 8 weeks, but with no other diabetes medications, and had HbA1c levels between 6.8% and 8.5%. Eligible participants were randomly assigned to the sulfonylurea glimepiride, the DPP-4 inhibitor sitagliptin, the GLP-1 receptor agonist liraglutide, or the long-acting insulin analog glargine. Recruitment ended in August 2017 with a cohort of 5047. Participant race is self-reported according to seven categories and ethnicity as Hispanic or not. The CGM study will be an observational substudy within the GRADE clinical trial.
CGM substudy cohort
Approximately 1800 participants of the GRADE parent study will be recruited to enroll in the CGM substudy, including up to 450 members of each of the four racial and ethnic minorities of interest that will provide adequate power to allow comparisons. We expect that this selection will also provide similar distributions across the treatment groups. The four racial/ethnic groups considered are Non-Hispanic African American (NHAA), Hispanic White, NHW and Non-Hispanic Other (NHO), where the NHO group includes American Indians, Asian Americans, and Pacific Islanders, and participants who report race/ethnicity as “other.”
Measurements
Continuous glucose monitoring
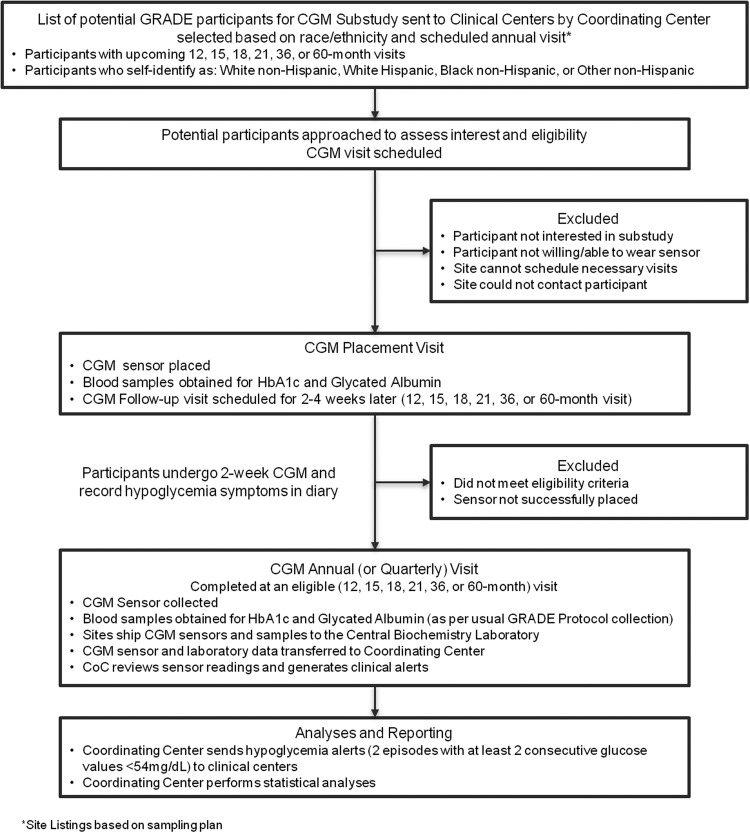
CGM will be performed for 2 weeks on the selected subcohort in close proximity (about 2–4 weeks prior) to the time of a scheduled annual year 1, 3, or 5 visit (Fig. 1: Study Overview) to have information on metabolic progression over time. Owing to practical scheduling challenges, a small number of “year one” participants will be studied after the first year annual visit at months 15, 18, or 21.
FIG. 1.
Continuous glucose monitoring substudy overview.
The Abbott Freestyle Libre™ Pro CGM Professional model (Abbott Diabetes Care, Alameda, CA), was selected for its ease of use (small and lack of need for calibration) and the masked nature of the glucose data collected (results not visible to the participants to avoid influencing diabetes management). Staff training will be provided centrally at CGM substudy start-up for all sites with a didactic and hands-on demonstration and practice. The selection of staff to insert the sensor will be based on local guidelines and scope of practice and all staff engaged in the CGM substudy will be required to review the manual of procedures and complete a certification quiz.
The CGM sensor will be inserted on the back of either upper arm, activated with the CGM reader and worn for 10 to 14 days. At the time of sensor placement, study staff will review with participants an instruction sheet that covers care of the CGM sensor, sensor removal, diary completion, and how to return the sensor to the GRADE clinical site. The sensor is removed either at the time of the follow-up visit if it occurs on day 14 of CGM use, or by the participant at home depending on the timing of the completion of monitoring (Fig. 1). If the sensor is dislodged before the 14-day study period is completed and the participant is willing, a second sensor will be placed within the study period. The data from the two sensors will be combined. Ten or more days of CGM readings will be considered a complete study for inclusion in the main analyses. If participants have experienced an acute illness likely to affect glycemia or have been treated with systemic glucocorticoids during the CGM collection, the GRADE Coordinating Center is alerted and those results will be censored. All sensors are read by the GRADE Central Biochemical Laboratory (CBL) at the Advanced Research and Diagnostic Laboratory, Department of Laboratory Medicine and Pathology, University of Minnesota in Minneapolis, Minnesota.
During the 14-day monitoring period, each participant will keep a diary of any symptomatic episodes consistent with hypoglycemia, including the date, time, and treatment of the episode and the date of sensor removal/dislodgement. CGM results will be blinded to both the participant and the clinical site staff during the monitoring period; however, after the CGM results have been analyzed, alert levels (defined as two episodes, each with two or more consecutive CGM values ≤54 mg/dL) (with readings recorded every 15 min so an episode is at least 30 min in duration) will be reported to the centers so that appropriate adjustments in lifestyle or medication may be considered.
Besides AG, other features of interest will be the time spent in the designated hyperglycemic ranges of >180 and >250 mg/dL, time spent in the target range of 70–180 mg/dL, and the time spent in the designated hypoglycemic ranges of <70 and <54 mg/dL. In addition, we will measure the number of episodes of hypoglycemia (<70 and <54 mg/dL), where an episode is defined as two events, each of which has two or more consecutive glucose levels <54 mg/dL. Glycemic variability will be captured with derived measures such as the mean amplitude of glycemic excursion and the continuous overall net glycemic action, the mean of daily differences, coefficients of variation, and standard deviations.20
Laboratory measurements
Blood samples for HbA1c and glycated albumin21 will be obtained at the time of the CGM insertion and approximately 2 weeks later when the CGM is completed. These blood samples will also be used to assess the mean RBC ages and turnover among participants. Blood samples obtained subsequently may be analyzed similarly. All measurements are performed in the GRADE CBL. Glycated hemoglobin is measured in EDTA whole blood using the Tosoh HPLC Glycohemoglobin Analyzer, which is an automated high-performance liquid chromatography method (Tosoh Medics, Inc., San Francisco, CA). Calibration of this method is evaluated utilizing standard values derived by the National Glycohemoglobin Standardization Program (NGSP). The interassay coefficients of variation in the GRADE Central Laboratory are 1.16% at a HbA1c value of 5.34% and 0.55% at a HbA1c value of 10.11%. Glycated albumin will be measured using a multienzyme, stepwise assay (Asahi Kasei, Inc.,Tokyo, Japan) in serum and expressed as the percentage of albumin that is glycated. The interassay coefficient of variation (CV) of glycated albumin in the GRADE Central Laboratory is 4.4% at a mean concentration of 0.45 g/dL and 2.8% at a mean concentration of 1.64 g/dL; the interassay CV of albumin is 2.4% at a mean concentration of 4.45 g/dL and 1.9% at a mean concentration of 3.86 g/dL.
Estimation of red blood cell age and turnover
Two approaches will be used to estimate each participant's RBC age and turnover and their potential extraglycemic influence on HbA1c. First, CGM and HbA1c will be combined with a mechanistic model of hemoglobin glycation to provide an indirect estimate of the subject's mean RBC age (eMRBC).22 Second, single-RBC volume and hemoglobin concentration will be measured (ADVIA 2120i, Siemens AG or CELL-DYN Sapphire; Abbott Diagnostics, Abbott Park, IL). These measurements provide quantitative estimates of RBC age and turnover.23,24 Each measurement of RBC turnover will be used to identify participants whose RBC turnover is relatively faster or slower than the norm and in whom the AG/HbA1c relationship may be altered accordingly.
Statistical analyses
Initial analyses will compare the characteristics of the racial/ethnic groups, including the distribution of HbA1c and the distribution of CGM results (including AG) with an adjustment for six pairwise comparisons. A linear model will regress the HbA1c on AG, calculated from CGM, with a class effect for racial/ethnic group and AG by group interaction. A 2-degrees of freedom joint test of the equality of intercepts and slopes will investigate differences among groups in the HbA1c to AG regression lines with an adjustment for six pairwise comparisons. The principal comparison of NHW versus NHAA will also be conducted secondarily for males and females with a test of homogeneity. Similar analyses will be performed using glycemic measures derived from the oral glucose tolerance test, and with glycated albumin instead of HbA1c, and employing measures of HbA1c and AG corrected for variation attributed to estimated mean RBC age.
Additional analyses will compare these relationships among the four treatment groups under the intention-to-treat principle, using all available data, and additional terms for racial group and interactions of AG with treatment group.
Cox proportional hazards models stratified by the year of the CGM assessment (1, 3, or 5), with and without adjustment for AG, HbA1c, and other risk factors (such as sex and age) will assess the association between the level of glucose variability and selected incident GRADE outcomes observed after the CGM evaluation. Outcomes of interest will include the time-to-occurrence of the GRADE primary, secondary, and tertiary metabolic outcomes18 and time to an episode of severe hypoglycemia (defined as requiring assistance), as well as time to microvascular complications (such as micro- and macroalbuminuria defined as an albumin excretion: urine creatinine ratio >30 and 300 mg/g, respectively) and the risk of cardiovascular disease (CVD).
Multiple comparisons conducted across subgroups will be adjusted for multiplicity using the Holm procedure.25
Power calculations and sample size
The sample size for the study was based on an assessment of the power to detect a difference in intercepts of 4 mg/dL and slopes of 1 mg/dL/% between racial/ethnic groups (NHW vs. NHAA) when regressing HbA1c on AG, the primary aim. The calculations employed estimates from the A1c-Derived Average Glucose (ADAG) study,11 allowing for nonhomogeneous (homoscedastic) random errors over the range of HbA1c. These quantities were then employed in the expression for the noncentrality parameter for the 2 df chi-square test of the difference in slopes and intercepts to calculate sample size and power.
A sample size of 86 per racial/ethnic group within each treatment group would provide ∼90% power to detect the difference described above in intercepts and slopes for the comparison of 2 racial/ethnic groups adjusting for 6 pairwise comparisons. Allowing for possible model misspecification through a 1.2 variance inflation factor, and 10% missing data yields a sample size of 112 participants in each of the 4 racial/ethnic groups within each of the four treatment groups, for a total sample size of 1800 participants, 450 within each racial group evenly divided among the four treatment groups (112 each) and 450 within each treatment group evenly divided among the four racial groups (112 each).
This sample size will provide excellent power (>90%) to detect a meaningful difference between two racial groups within one of the four treatment groups. The power to determine whether glycemic variability contributes to or affects the outcomes in GRADE, including microvascular and CVD events, is a function of the effect sizes (i.e., hazard ratios) and the number of participants who have the event. The latter will be limited.
Conclusions
Many epidemiologic studies over the past 20 years have demonstrated higher HbA1c levels in African Americans than Whites.26 Until the mid-2000s, the observed differences in HbA1c by race were universally attributed to health disparities, that is, differences in health care in different population groups. Subsequent studies that compared HbA1c by race and statistically adjusted for sociodemographic and clinical factors, and controlled for access to care and quality of care, were unable to explain the observed differences in HbA1c by race.27,28 More recently, differences in HbA1c have been demonstrated in African American and White adults who were selected to have the same fasting and postchallenge glucose levels.4,29–31 Still, unmeasured differences in diet and physical activity between African Americans and Whites might have resulted in chronic differences in glucose levels and thus in HbA1c. To address the limited measurements of actual mean glycemia in prior studies, a recent study of African American and White patients with type 1 diabetes (T1D) used CGM to provide a complete assessment of AG levels.12 It demonstrated that HbA1c was 0.8% higher in African Americans than Whites. Approximately one-half of the difference was explained by differences in glycemia, but the remaining ∼0.4% difference was not explained by average glycemia. In contrast, no significant racial differences were found in the relationship between mean glucose and glycated albumin or fructosamine.
Racial differences in single nucleotide polymorphisms that are associated with HbA1c, but operate through nonglycemic mechanisms, have been proposed as an explanation for the differences in HbA1c between African Americans and Whites.32 To date, however, no polymorphisms have been identified to explain more than a small proportion of the observed difference in HbA1c by race. Racial differences in red cell turnover have also been invoked to explain the racial differences in HbA1c,33 although there is no direct evidence of racial differences in red cell turnover to date, owing in part to the difficulty in its measurement.16
CGM provides frequent real-time reliable measurements of glucose levels that allows calculation of AG. Other key features added by CGM glucose analysis beyond AG levels include the ability to quantitate hypoglycemia and glucose variability. While the GRADE CGM substudy is measuring a single 2-week period of glucose control, this period of CGM has been shown to be a good reflection of 30–90 days of CGM in most individuals.34
Most trials to date studying the added value of using CGM have focused on individuals with T1D who are using pump therapy or multiple daily insulin injections.12,35 Only a few studies have used CGM to characterize glucose metrics in T2D patients or examined the value of CGM in T2D management.36 No studies to date have characterized or compared CGM metrics or glucose profiles in a large number of T2D patients randomized to commonly used T2D medications, as will be done in the GRADE CGM substudy. A key component of diabetes management today is focused on minimizing hypoglycemia in both T1D and T2D, since hypoglycemia has been clearly demonstrated to be potentially dangerous, as well as disruptive to the quality of life and costly to the health care system.37,38 While some T2D medications carry a low risk of hypoglycemia, sulfonylureas and insulin have been associated with an increased risk of hypoglycemia. CGM is the only practical way to compare rates of hypoglycemia 24 h/day, and capture all the episodes of hypoglycemia whether they are symptomatic or asymptomatic.
Glucose variability, the other key glucose profile metric not captured by an HbA1c, has been associated with an increased risk for hypoglycemia, diabetes complications, and reduced quality of life.19,39 Few studies have been performed in T2DM, and the GRADE CGM substudy will greatly expand the characterization of glucose variability across four major diabetes treatment regimens. Similarly, the relationship between microvascular and macrovascular complications and glucose variability is based on a few observational studies.40,41 GRADE is measuring renal microvascular disease and cardiovascular risk factors, allowing us to determine if these outcome measures are related to glucose variability.
In summary, the GRADE CGM substudy will provide a large and diverse population of patients with T2D to determine whether, and the extent to which, the relationship between AG and HbA1c differs among racial and ethnic groups. The additional measurement of glycated albumin and RBC profiling should help to distinguish the contribution of abnormalities in RBC turnover from differences in glycation. This substudy will also provide new insights into whether glycemic profiles and measures of time in target range, hypoglycemia, and glucose variability differ among frequently used diabetes medications.
Acknowledgments
GRADE Research Group (May 30, 2019):
Designations: Principal Investigator (PI); Co-Principal Investigator (Co-PI); Co-Investigator (Co-I); Study Coordinator (SC); Recruitment/Retention Coordinator (RC); Research Staff (RS).
Current Clinical Centers:
Albert Einstein College of Medicine: Jill P. Crandall (PI); Melissa Diane McKee (PI, past); Janet Brown-Friday (SC); Entila Xhori (RC, past); Keisha Ballentine-Cargill (RS); Sally Duran (RS); Jennifer Lukin (RS); Stephanie Beringher (RS, past); Susana Gonzalez de la torre (RS, past).
Atlanta VA Medical Center: Lawrence Phillips (PI); Elizabeth Burgess (Co-I); Darin Olson (Co-I); Mary Rhee (Co-I); Peter Wilson (Co-I); Tasha Stephanie Raines (SC); Julie Costello (SC); Chona Gullett (SC); Maxine Maher-Albertelli (SC); Folayan Morehead (SC); Radhika Mungara (SC); Saranjit Person (SC); Louise Savoye (SC); Mabil Sibymon (SC); Sridhar Tanukonda (SC); Carol Ann White (SC); Leah Holloway (SC, past); Cynthia Adams (RC, past); April Ross (RC, past).
Baylor College of Medicine: Ashok Balasubramanyam (PI); Erica Gonzalez (SC); Charlyne Wright (RS, past).
Baylor Research Institute: Priscilla Hollander (PI); Erin Roe (Co-I); Analyn Uy (SC); Polly Burt (SC, past); Lorie Estrada (RS); Kris Chionh (RS, past).
Case Western Reserve University/Cleveland VA/MetroHealth Medical Center: Faramarz Ismail-Beigi (PI); Corinna Falck-Ytter (Co-PI); Laure Sayyed Kassem (Co-PI); Ajay Sood (Co-PI, past); Margaret Tiktin (Co-I, SC); Bethany Cramer (SC); Jacalyn Iacoboni (SC); Maria V. Kononets (SC); Tanya Kulow (SC); Cynthia Newman (SC); Katherine A. Stancil (SC); Cristina Sanders (SC, past); Lisa Tucker (SC, past); Amanda Werner (SC, past); Adrienne Krol (RS); Gloria McPhee (RS); Christine Patel (RS); Linda Colosimo (RS, past).
Columbia University Medical Center: Robin Goland (PI); James Pring (SC); Patricia Kringas (SC, past); Jessica Tejada (RC); Camille Hausheer (RC, past); Harvey Schneier (RS); Kelly Gumpel (RS, past); Amanda Kirpitch (RS, past).
Duke University Medical Center: Jennifer B. Green (PI); Hiba AbouAssi (Co-I); Ranee Chatterjee (Co-I); Mark N. Feinglos (Co-I); Jennifer English Jones (SC, RC); Shubi A. Khan (SC, RC); Jeanne B. Kimpel (SC); Ronna P. Zimmer (SC, past); Mary Furst (RC, past); Barbara M. Satterwhite (RS); Connie R. Thacker (RS); Kathryn Evans Kreider (RS, past).
Indiana University: Kieren J. Mather (PI); Amale Lteif (Co-I, past); Tonya Hamilton (SC); Nick Patel (SC, past); Gabriela Riera (RC); Marcia Jackson (RC, past); Vivian Pirics (RC, past); Devin Howard (RS); Danielle Aguillar (RS, past); Sloan Hurt (RS, past).
International Diabetes Center: Richard Bergenstal (PI); Anders Carlson (Co-I); Thomas Martens (Co-I); Mary Johnson (SC); Renae Hill (SC); Jamie Hyatt (SC); Connie Jensen (SC); Marcia Madden (SC); Dianna Martin (SC); Holly Willis (SC); Wanda Konerza (RS); Rebecca Passi (RS); Kathleen Kleeberger (RS, past).
Kaiser Permanente Northwest: Stephen Fortmann (PI); Michael Herson (Co-I); Karen Mularski (Co-I); Harry Glauber (Co-I, past); James Prihoda (Co-I, past); Britt Ash (SC); Christina Carlson (SC); Phyllis Anne Ramey (SC); Emily Schield (SC); Britta Torgrimson-Ojerio (SC); Kathy Arnold (SC, past); Bryan Kauffman (SC, past); Elease Panos (SC, past); Samantha Sahnow (RC); Kristi Bays (RS); Jennifer Cook (RS); Jennifer Gluth (RS); Debra Sasaki (RS); Katrina Schell (RS); Jennifer Criscola (RS, past); Camille Friason (RS, past); Suzi Jones (RS, past); Sergey Nazarov (RS, past).
Kaiser Permanente of Georgia: Joshua Barzilay (PI); Negah Rassouli (Co-PI); Rachel Puttnam (Co-I); Michelle Curtis (SC); Kia Stokes (SC); Bonita Hollis (SC, past); Cynthia Sanders-Jones (SC, past); Roslin Nelson (RC); Zakiah El-Haqq (RS, past); Abby Kolli (RS, past); Tu Tran (RS, past).
Massachusetts General Hospital: Deborah Wexler (PI); Mary Larkin (Co-I); James Meigs (Co-I); Amy Dushkin (SC); Gianna Rocchio (SC); Brittany Chambers (SC, past); Mike Yepes (SC, past); Barbara Steiner (RC); Hilary Dulin (RC, past); Melody Cayford (RS); Andrea DeManbey (RS); Lindsey Gurry (RS); Mallory Hillard (RS); Kimberly Martin (RS); Christine Stevens (RS); Nopporn Thangthaeng (RS); Raquel Kochis (RS, past); Elyse Raymond (RS, past); Valerie Ripley (RS, past).
MedStar Health Research Institute/MedStar Baltimore: Jean Park (PI); Vanita Aroda (PI, past); Adline Ghazi (Co-PI); Amy Loveland (SC); Maria Hurtado (SC); Alexander Kuhn (SC); Florence Mofor (SC).
Miami VA Healthcare System/University of Miami: Hermes J. Florez (PI); Willy Marcos Valencia (PI); Jennifer Marks (Co-PI, past); Lisset Oropesa-Gonzalez (SC); Ana K. Riccio Veliz (SC); Ramfis Nieto-Martinez (SC); Miriam Gutt (RC).
Oregon Health & Science University: Andrew Ahmann (PI); Diana Aby-Daniel (Co-I); Farahnaz Joarder (Co-I); Victoria Morimoto (Co-I); Carol Sprague (Co-I); Daisuke Yamashita (Co-I); Nancy Cady (SC); Patricia Kirchhoff (SC); Nadia Rivera-Eschright (SC); Joseph Adducci (RC); Brianna Morales Gomez (RC); Alina Goncharova (RC, past).
Pacific Health Research and Education Institute/VA Pacific Islands: Sophia H. Hox (PI); Helen Petrovitch (Co-PI); Michael Matwichyna (SC); Victoria Jenkins (SC, past); Nina O. Bermudez (RS); Renée R. Ishii (RS).
Pennington Biomedical Research Center: Daniel S. Hsia (PI); William T. Cefalu (PI, past); Frank L. Greenway (Co-I); Celeste Waguespack (Co-I); Erin King (SC); Natalie Haynes (SC, past); Amy Thomassie (SC, past); Brandi Bourgeois (RC, past); Claire Hazlett (RS).
San Diego VA Medical Center: Robert Henry (PI); Sunder Mudaliar (Co-I); Schafer Boeder (Co-I, past); Jeremy Pettus (Co-I, past); Elsa Diaz (SC); Catherine DeLue (SC, past); Erick Castro (RC, past); Sylvia Hernandez (RC, past).
Southwestern American Indian Center: Jonathan Krakoff (PI); Jeffrey M. Curtis (Co-I); Tina Killean (SC); Erica Joshevama (RC); Enrique Diaz (RS); Denelle Martin (RS); Tracey Karshner (RS, past).
St. Luke's-Roosevelt Hospital: Jeanine Albu (Co-PI); F. Xavier Pi-Sunyer (Co-PI); Sylvaine Frances (SC); Carol Maggio (SC, past); Emily Ellis (RC); Joseph Bastawrose (RC, past); Xiuqun Gong (RS).
SUNY Downstate Medical Center/New York Hospital-Queens: Mary Ann Banerji (PI); Phyllis August (Co-I); Daniel Lorber (Co-I); Necole M. Brown (SC, RC); Debra H. Josephson (SC); Lorraine L. Thomas (SC, RC); Mari Tsovian (SC, RC); Ajini Cherian (SC, RC, past); Marlo H. Jacobson (RS); Motria M. Mishko (RS).
The University of North Carolina Diabetes Care Center: M. Sue Kirkman (PI); Katherine Bergamo (Co-I); John B. Buse (Co-I); Jean Dostou (Co-I); Laura Young (Co-I); April Goley (Co-I, past); Jeffrey Kerr (Co-I, past); Joseph F. Largay (Co-I, past); Sonia Guarda (SC); Juanita Cuffee (SC, past); Dawn Culmer (SC, past); Rachael Fraser (RC); Hope Almeida (RC, past); Samantha Coffer (RC, past); Elizabeth Debnam (RC, past); Lauren Kiker (RC, past); Sarah Morton (RC, past); Kim Josey (RS); Gail Fuller (RS, past).
University of Alabama Birmingham: W. Timothy Garvey (PI); Andrea Cherrington (Co-PI); Dana Golson (SC); Olivia Griffith (SC); Mary Catherine Robertson (SC); April Agne (RC); Steve McCullars (RC).
University of Cincinnati/Cincinnati VA Medical Center: Robert M. Cohen (PI); Jacqueline Craig (SC); Kimberly Kersey (SC, RC); M. Colleen Rogge (SC); Carla Wilson (SC); Kathryn Burton (SC, past); Sonia Lipp (RC, past); Mary Beth Vonder Meulen (RC, past).
University of Colorado-Denver/VA: Neda Rasouli (PI); Emily Schroeder (Co-I); Stephanie Steiner (SC); Chelsea Baker (SC, RS); Chantal Underkofler (SC); Sara Douglass (SC, past).
University of Iowa: William Sivitz (PI); Erin Cline (SC); Laura Knosp (SC); Jennifer McConnell (SC, past); Tamara Lowe (RC).
University of Michigan: William H. Herman (PI); Rodica Pop-Busui (Co-PI); Meng H. Tan (Co-I); Catherine Martin (SC); Andrea Waltje (SC, RC); Lynn Goodhall (SC, past); Rebecca Eggleston (RC, past); Shihchen Kuo (RS); Stephanie Bule (RS, past); Nancy Kessler (RS, past); Elizabeth LaSalle (RS, past).
University of Minnesota: Elizabeth R. Seaquist (PI); Anne Bantle (Co-I); Anjali Kumar (Co-I); Bruce Redmon (Co-I); John Bantle (Co-I, past); Tasma Harindhanavudhi (Co-I, past); Mary Coe (SC); Michael Mech (SC); Abdisa Taddese (RC); Lesia Lesne (RS); Shannon Smith (RS).
University of Nebraska Medical Center/Omaha VA: Cyrus Desouza (PI); Lisa Kuechenmeister (Co-I); Vijay Shivaswamy (Co-I); Ana Laura Morales (SC); Maria Grace Rodriguez (SC); Kris Seipel (SC); Alissa Alfred (SC, past); Jenna Eggert (RS); Grace Lord (RS); William Taylor (RS); Renee Tillson (RS).
University of New Mexico: David S. Schade (PI); Allen Adolphe (Co-PI); Mark Burge (Co-PI); Elizabeth Duran-Valdez (SC); Janae Martinez (RC); Doris Hernandez McGinnis (RS); Benjamin Pucchetti (RS, past); Elizabeth Scripsick (RS, past).
University of Texas Health Science Center: Ralph A. DeFronzo (PI); Eugenio Cersosimo (Co-PI); Muhammad Abdul-Ghani (Co-I); Curtis Triplitt (Co-I); Hector Verastiqui (SC); Rosa Irene Garza (SC); Kathryn Wright (RC, past); Curtiss Puckett (RS).
University of Texas-Southwestern Medical Center: Philip Raskin (PI); Chanhaeng Rhee (Co-I, past); Soma Abraham (SC); Lin Fan Jordan (SC); Serey Sao (SC); Luisa Morton (SC, past); Oralenda Smith (SC, past); Laura Osornio Walker (RC, past); Laura Schnurr-Breen (RC, past); Rosa Ayala (RS); Robert Brian Kraymer (RS); Daytheon Sturgess (RS, past).
VA Puget Sound Healthcare System/University of Washington: Kristina M. Utzschneider (PI); Steven E. Kahn (Co-PI); Lorena Alarcon-Casas Wright (Co-I); Edward J. Boyko (Co-I); Elaine C. Tsai (Co-I); Dace L. Trence (Co-I, past); Basma N. Fattaleh (SC); Brenda K. Montgomery (SC, past); Karen M. Atkinson (RS); Tessa Concepcion (RS, past); Alexandra Kozedub (RS); Cameron Moak (RS, past); Samantha Rhothisen (RS, past).
Vanderbilt University: Tom A. Elasy (PI); Stephanie Martin (SC); Laura Shackelford (RC, RS); Rita Goidel (RS); Nina Hinkle (RS); Janie Lipps Hogan (RS); Cynthia Lovell (RS); Janet Myers (RS).
Washington University: Janet B. McGill (PI); Maamoun Salam (Co-I); Sarah Kissel (SC, RC); Toni Schweiger (SC, RC); Carol Recklein (SC, past).
Yale University/Fair Haven Community Health Center: William Tamborlane (PI); Patricia Gatcomb (SC); Anne Camp (Co-I); Barbara Gulanski (Co-I); Silvio Inzucchi (Co-I); Kim Pham (Co-I); Michele Alguard (SC, RC); Katarzyna Lessard (SC); Magalys Perez (SC); Elizabeth Magenheimer (RC); Abmaridel Montoza (RC).
Study Units:
NIH/NIDDK (Sponsor): Henry B. Burch (Project Scientist); Andrew Bremer (Program Scientist, past); Barbara Linder (Program Official); Judith Fradkin (Director, past).
Chairman's Office, Massachusetts General Hospital, Harvard Medical School: David M. Nathan (Study Chair, Study Co-PI).
Executive Committee: David M. Nathan (Study Chair, Study Co-PI); John Lachin (U01 Contact PI, Study Co-PI); John B. Buse (Co-I); Steven E. Kahn (Co-I); Heidi Krause-Steinrauf (Co-I, Project Director); Mary Larkin (Co-I); Margaret Tiktin (Co-I, SC); Deborah Wexler (PI); Henry B. Burch (Program Scientist); Barbara Linder (Program Official); Andrew Bremer (Program Scientist, past).
Coordinating Center, The George Washington University Biostatistics Center: John Lachin (U01 Contact PI, Study Co-PI); Heidi Krause-Steinrauf (Co-I); Naji Younes (Co-I); Michael Backman (RS); Ionut Bebu (RS); CJ Buys (RS); Anna Fagan Murphy (RS); Yuping Gao (RS); Michaela Gramzinski (RS); Stephanie Hall (RS); Elizabeth Legowski (RS); Alyssa Arey (RS, past) Joel Bethepu (RS, past); Claire Lund (RS, past); Pam Mangat Dhaliwal (RS, past); Paula McGee (RS, past); Emily Mesimer (RS, past); Lisa Ngo (RS, past).
Central Biochemical Laboratory, University of Minnesota Advanced Research and Diagnostic Laboratory: Michael Steffes (PI); Jesse Seegmiller (Co-I); Amy Saenger (Co-I, past); Valerie Arends (SC); Deanna Gabrielson (SC, past).
Drug Distribution Center, VA Cooperative Studies Program Clinical Research Pharmacy Coordinating Center: Todd Conner (PI); Stuart Warren (PI, past); Jolene Day (RS); Alexandra Scrymgeour (RS).
ECG Reading Center, EPICARE, Wake Forest University: Elsayed Z. Soliman (PI); Zhu-Ming Zhang (Co-I, past); Charles Campbell (SC); Julie Hu (SC); Lisa Keasler (SC); Susan Hensley (SC, past); Yabing Li (RS).
Economic Evaluation and Assessment Center:
University of Michigan: William Herman (PI); Catherine Martin (SC); Andrea Waltje (SC, RC); Shihchen Kuo (RS); Rada Mihalcea (RS); Veronica Perez-Rosas (RS); Lisa Prosser (RS); Kenneth Resnicow (RS); Wen Ye (RS).
Centers for Disease Control and Prevention: Hui Shao (RS); Ping Zhang (RS).
Neurocognitive Coordinating Center, Columbia University Medical Center: Jose Luchsinger (PI); Danurys Sanchez (SC).
QWB Reading Center, University of California San Diego Health Services Research Center: Erik Groessl (PI); Helen Chong (SC); Naomi Hillery (RS).
Collaborators:
Collaborating Investigators (Recruitment Site): Ivan Abdouch (University of Nebraska Medical Center/Omaha VA); Paula Brantley (Pennington Biomedical Research Center (LSU)); Frances E. Broyles (SIBCR—VA Puget Sound); Gay Canaris (University of Nebraska Medical Center/Omaha VA); Paul Copeland (Massachusetts General Hospital); Jeri J. Craine (SIBCR—VA Puget Sound); Warren L. Fein (SIBCR—VA Puget Sound); Melissa S. Lee (SUNY Downstate Medical Center); Rebecca Meiners (Pennington Biomedical Research Center—LSU); Vaughn Meiners (Pennington Biomedical Research Center—LSU); Hollis O'Neal (Pennington Biomedical Research Center—LSU); James E. Park (SIBCR—VA Puget Sound); Edward Sledge Jr. (Pennington Biomedical Research Center—LSU); Jeanne Steppel-Resnick (Massachusetts General Hospital); Alexander Turchin (Massachusetts General Hospital).
Continuous Glucose Monitoring Substudy: John Higgins (Massachusetts General Hospital).
Contributor Information
Collaborators: the GRADE Research Group, Jill P. Crandall, Melissa Diane McKee, Janet Brown-Friday, Entila Xhori, Keisha Ballentine-Cargill, Sally Duran, Jennifer Lukin, Stephanie Beringher, Susana Gonzalez de la torre, Lawrence Phillips, Elizabeth Burgess, Darin Olson, Mary Rhee, Peter Wilson, Tasha Stephanie Raines, Julie Costello, Chona Gullett, Maxine Maher-Albertelli, Folayan Morehead, Radhika Mungara, Saranjit Person, Louise Savoye, Mabil Sibymon, Sridhar Tanukonda, Carol Ann White, Leah Holloway, Cynthia Adams, April Ross, Ashok Balasubramanyam, Erica Gonzalez, Charlyne Wright, Priscilla Hollander, Erin Roe, Analyn Uy, Polly Burt, Lorie Estrada, Kris Chionh, Faramarz Ismail-Beigi, Corinna Falck-Ytter, Laure Sayyed Kassem, Ajay Sood, Bethany Cramer, Jacalyn Iacoboni, Maria V. Kononets, Tanya Kulow, Cynthia Newman, Katherine A. Stancil, Cristina Sanders, Lisa Tucker, Amanda Werner, Adrienne Krol, Gloria McPhee, Christine Patel, Linda Colosimo, Robin Goland, James Pring, Patricia Kringas, Jessica Tejada, Camille Hausheer, Harvey Schneier, Kelly Gumpel, Amanda Kirpitch, Jennifer B. Green, Hiba AbouAssi, Ranee Chatterjee, Mark N. Feinglos, Jennifer English Jones, Shubi A. Khan, Jeanne B. Kimpel, Ronna P. Zimmer, Mary Furst, Barbara M. Satterwhite, Connie R. Thacker, Kathryn Evans Kreider, Kieren J. Mather, Amale Lteif, Tonya Hamilton, Nick Patel, Gabriela Riera, Marcia Jackson, Vivian Pirics, Devin Howard, Danielle Aguillar, Sloan Hurt, Richard Bergenstal, Anders Carlson, Thomas Martens, Mary Johnson, Renae Hill, Jamie Hyatt, Connie Jensen, Marcia Madden, Dianna Martin, Holly Willis, Wanda Konerza, Rebecca Passi, Kathleen Kleeberger, Stephen Fortmann, Michael Herson, Karen Mularski, Harry Glauber, James Prihoda, Britt Ash, Christina Carlson, Phyllis Anne Ramey, Emily Schield, Britta Torgrimson-Ojerio, Kathy Arnold, Bryan Kauffman, Elease Panos, Samantha Sahnow, Kristi Bays, Jennifer Cook, Jennifer Gluth, Debra Sasaki, Katrina Schell, Jennifer Criscola, Camille Friason, Suzi Jones, Sergey Nazarov, Joshua Barzilay, Negah Rassouli, Rachel Puttnam, Michelle Curtis, Kia Stokes, Bonita Hollis, Cynthia Sanders-Jones, Roslin Nelson, Zakiah El-Haqq, Abby Kolli, Tu Tran, Deborah Wexler, James Meigs, Amy Dushkin, Gianna Rocchio, Brittany Chambers, Mike Yepes, Barbara Steiner, Hilary Dulin, Melody Cayford, Andrea DeManbey, Lindsey Gurry, Mallory Hillard, Kimberly Martin, Christine Stevens, Nopporn Thangthaeng, Raquel Kochis, Elyse Raymond, Valerie Ripley, Jean Park, Vanita Aroda, Adline Ghazi, Amy Loveland, Maria Hurtado, Alexander Kuhn, Florence Mofor, Hermes J. Florez, Willy Marcos Valencia, Jennifer Marks, Lisset Oropesa-Gonzalez, Ana K. Riccio Veliz, Ramfis Nieto-Martinez, Miriam Gutt, Andrew Ahmann, Diana Aby-Daniel, Farahnaz Joarder, Victoria Morimoto, Carol Sprague, Daisuke Yamashita, Nancy Cady, Patricia Kirchhoff, Nadia Rivera-Eschright, Joseph Adducci, Brianna Morales Gomez, Alina Goncharova, Sophia H. Hox, Helen Petrovitch, Michael Matwichyna, Victoria Jenkins, Nina O. Bermudez, Renée R. Ishii, Daniel S. Hsia, William T. Cefalu, Frank L. Greenway, Celeste Waguespack, Erin King, Natalie Haynes, Amy Thomassie, Brandi Bourgeois, Claire Hazlett, Robert Henry, Sunder Mudaliar, Schafer Boeder, Jeremy Pettus, Elsa Diaz, Catherine DeLue, Erick Castro, Sylvia Hernandez, Jonathan Krakoff, Jeffrey M. Curtis, Tina Killean, Erica Joshevama, Enrique Diaz, Denelle Martin, Tracey Karshner, Jeanine Albu, F. Xavier Pi-Sunyer, Sylvaine Frances, Carol Maggio, Emily Ellis, Joseph Bastawrose, Xiuqun Gong, Mary Ann Banerji, Phyllis August, Daniel Lorber, Necole M. Brown, Debra H. Josephson, Lorraine L. Thomas, Mari Tsovian, Ajini Cherian, Marlo H. Jacobson, Motria M. Mishko, M. Sue Kirkman, Katherine Bergamo, John B. Buse, Jean Dostou, Laura Young, April Goley, Jeffrey Kerr, Joseph F. Largay, Sonia Guarda, Juanita Cuffee, Dawn Culmer, Rachael Fraser, Hope Almeida, Samantha Coffer, Elizabeth Debnam, Lauren Kiker, Sarah Morton, Kim Josey, Gail Fuller, W. Timothy Garvey, Andrea Cherrington, Dana Golson, Olivia Griffith, Mary Catherine Robertson, April Agne, Steve McCullars, Jacqueline Craig, Kimberly Kersey, M. Colleen Rogge, Carla Wilson, Kathryn Burton, Sonia Lipp, Mary Beth Vonder Meulen, Neda Rasouli, Emily Schroeder, Stephanie Steiner, Chelsea Baker, Chantal Underkofler, Sara Douglass, William Sivitz, Erin Cline, Laura Knosp, Jennifer McConnell, Tamara Lowe, Rodica Pop-Busui, Meng H. Tan, Catherine Martin, Andrea Waltje, Lynn Goodhall, Rebecca Eggleston, Shihchen Kuo, Stephanie Bule, Nancy Kessler, Elizabeth LaSalle, Elizabeth R. Seaquist, Anne Bantle, Anjali Kumar, Bruce Redmon, John Bantle, Tasma Harindhanavudhi, Mary Coe, Michael Mech, Abdisa Taddese, Lesia Lesne, Shannon Smith, Cyrus Desouza, Lisa Kuechenmeister, Vijay Shivaswamy, Ana Laura Morales, Maria Grace Rodriguez, Kris Seipel, Alissa Alfred, Jenna Eggert, Grace Lord, William Taylor, Renee Tillson, David S. Schade, Allen Adolphe, Mark Burge, Elizabeth Duran-Valdez, Janae Martinez, Doris Hernandez McGinnis, Benjamin Pucchetti, Elizabeth Scripsick, Ralph A. DeFronzo, Eugenio Cersosimo, Muhammad Abdul-Ghani, Curtis Triplitt, Hector Verastiqui, Rosa Irene Garza, Kathryn Wright, Curtiss Puckett, Philip Raskin, Chanhaeng Rhee, Soma Abraham, Lin Fan Jordan, Serey Sao, Luisa Morton, Oralenda Smith, Laura Osornio Walker, Laura Schnurr-Breen, Rosa Ayala, Robert Brian Kraymer, Daytheon Sturgess, Kristina M. Utzschneider, Steven E. Kahn, Lorena Alarcon-Casas Wright, Edward J. Boyko, Elaine C. Tsai, Dace L. Trence, Basma N. Fattaleh, Brenda K. Montgomery, Karen M. Atkinson, Tessa Concepcion, Alexandra Kozedub, Cameron Moak, Samantha Rhothisen, Tom A. Elasy, Stephanie Martin, Laura Shackelford, Rita Goidel, Nina Hinkle, Janie Lipps Hogan, Cynthia Lovell, Janet Myers, Janet B. McGill, Maamoun Salam, Sarah Kissel, Toni Schweiger, Carol Recklein, William Tamborlane, Patricia Gatcomb, Anne Camp, Barbara Gulanski, Silvio Inzucchi, Kim Pham, Michele Alguard, Katarzyna Lessard, Magalys Perez, Elizabeth Magenheimer, Abmaridel Montoza, Henry B. Burch, Andrew Bremer, Barbara Linder, Judith Fradkin, John Lachin, Steven E. Kahn, Henry B. Burch, Naji Younes, Michael Backman, CJ Buys, Anna Fagan Murphy, Yuping Gao, Michaela Gramzinski, Stephanie Hall, Elizabeth Legowski, Alyssa Arey, Joel Bethepu, Pam Mangat Dhaliwal, Paula McGee, Emily Mesimer, Lisa Ngo, Michael Steffes, Jesse Seegmiller, Amy Saenger, Deanna Gabrielson, Todd Conner, Stuart Warren, Jolene Day, Alexandra Scrymgeour, Elsayed Z. Soliman, Zhu-Ming Zhang, Charles Campbell, Julie Hu, Lisa Keasler, Susan Hensley, Yabing Li, William Herman, Rada Mihalcea, Veronica Perez-Rosas, Lisa Prosser, Kenneth Resnicow, Wen Ye, Hui Shao, Ping Zhang, Jose Luchsinger, Danurys Sanchez, Erik Groessl, Helen Chong, Naomi Hillery, Ivan Abdouch, Paula Brantley, Frances E. Broyles, Gay Canaris, Paul Copeland, Jeri J. Craine, Warren L. Fein, Melissa S. Lee, Rebecca Meiners, Vaughn Meiners, Hollis O'Neal, James E. Park, Edward Sledge, Jr., Jeanne Steppel-Resnick, Alexander Turchin, and John Higgins
Author Disclosure Statement
All authors affirm that authorship is merited based on the ICMJE authorship criteria. Mary E. Larkin contributed to the design, writing, and critical review of this article. David M. Nathan contributed to the design, interpretation of data and results, obtainment of funds, supervision and management of research, writing, and critical review of this article. Ionut Bebu contributed to the design, analysis, writing, and critical review of this article. Heidi Krause-Steinrauf contributed to the design, writing, and critical review of this article. William H. Herman contributed to the design, supervision, and management of research, and critical review of this article. John M. Higgins contributed to the design, interpretation of data and results, writing, and critical review of this article.
Margaret Tiktin contributed to the acquisition of data, and critical review of this article. Robert M. Cohen contributed to the design, interpretation of data and results, supervision and management of research, and critical review of this article. Claire Lund contributed to the design, supervision and management of research, writing, and critical review of this article. Richard M. Bergenstal contributed to the design, acquisition of data, interpretation of data, and critical review of this article. Mary L. Johnson contributed to the acquisition of data, supervision and management of research, writing, and critical review of this article. Valerie Arends contributed to the acquisition of data, supervision and management of the research, and critical review of this article.
Mary E. Larkin is the guarantor of this work, and as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. David M. Nathan performed an investigator-initiated study of a point-of-care A1c device funded by the sponsor (Alere, Inc.), now Abbott. Robert M. Cohen owns stock in Bristol Myers Squibb. Richard M. Bergestal has received research support, consulted, or has been on the scientific advisory board for Abbott Diabetes Care, DexCom, Medtronic, Onduo, Roche, Senseonics, and United Healthcare. His diabetes technology research is partly funded by NIDDK UC4DK108611. Richard M. Bergenstal's employer, nonprofit HealthPartners Institute, contracts for his services and no personal income goes to Richard M. Bergenstal. Mary L. Johnson has received research support and/or consulted for Abbott, Dexcom, Johnson & Johnson, Medtronic, NIDDK, Novo Nordisk, Roche, and Sanofi. Her employer, nonprofit International Diabetes Center/HealthPartners Institute/Park Nicollet, contracts for her services and she receives no personal income from these activities. Mary E. Larkin, Heidi Krause-Steinrauf, William H. Herman, John M. Higgins, Margaret Tiktin, Claire Lund, and Valerie Arends: No competing financial interests exist.
Funding Information
The CGM Substudy is supported by grant support from the National Institute of Diabetes and Digestive and Kidney Diseases, NIH and with material support from Asahi Kasei Pharma Corporation, Siemens Healthcare Diagnostics, Inc., and Abbott, Inc. This research was supported by a grant from Abbott Laboratories. The planning of GRADE was supported by a U34 planning grant from the NIDDK, NIH (U34-DK-088043). The American Diabetes Association supported the initial planning meeting for the U34 proposal. GRADE is supported by a U01 grant (U01-DK-098246). Educational materials have been provided by the National Diabetes Education Program. Material support in the form of donated medications and supplies has been provided by Becton Dickinson, Bristol-Myers Squibb, Merck, Novo-Nordisk, Roche Diagnostics, and Sanofi.
References
- 1. Yudkin JS, Forrest RD, Jackson CA, et al. : Unexplained variability of glycated haemoglobin in non-diabetic subjects not related to glycaemia. Diabetologia 1990;33:208–215 [DOI] [PubMed] [Google Scholar]
- 2. Gould BJ, Davie SJ, Yudkin JS: Investigation of the mechanism underlying the variability of glycated haemoglobin in non-diabetic subjects not related to glycaemia. Clin Chim Acta 1997;260:49–64 [DOI] [PubMed] [Google Scholar]
- 3. Cohen RM, Holmes YR, Chenier TC, et al. : Discordance between hemoglobin A1c and fructosamine: evidence for a glycosylation gap and its relation to nephropathy in longstanding type 1 diabetes. Diabetes Care 2003;26:163–167 [DOI] [PubMed] [Google Scholar]
- 4. Herman WH, Ma Y, Uwaifo G, et al. ; Diabetes Prevention Program Research Group: Differences in A1C by race and ethnicity among patients with impaired glucose tolerance in the Diabetes Prevention Program. Diabetes Care 2007;30:2453–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chalew SA, McCarter RJ, Thomas J, et al. : A comparison of the glycosylation gap and hemoglobin glycation index in patients with diabetes. J Diabetes Complications 2005;19:218–222 [DOI] [PubMed] [Google Scholar]
- 6. Kilpatrick ES, Rigby AS, Atkin SL: Variability in the relationship between mean plasma glucose and HbA1c: implications for the assessment of glycemic control. Clin Chem 2007;53:897–901 [DOI] [PubMed] [Google Scholar]
- 7. Lachin JM, Genuth S, Nathan DM, et al. : The hemoglobin glycation index is not an independent predictor of the risk of microvascular complications in the Diabetes Control and Complications Trial. Diabetes 2007;56:1913–1921 [DOI] [PubMed] [Google Scholar]
- 8. Sacks DB, Nathan DM, Lachin JM: Gaps in the glycation gap hypothesis. Clin Chem 2011;57:150–152 [DOI] [PubMed] [Google Scholar]
- 9. Parirnello CM, Sharrett AR, Maruther NM, et al. : Racial differences in and prognostic value of biomarkers of hyperglycemia. Diabetes Care 2016;39:589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Selvin E: Are there clinical implications of racial differences in HbA1c? A difference, to be a difference, must make a difference. Diabetes Care 2016;39:1462–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nathan DM, Kuenen J, Borg R, et al. : A1C-derived average glucose study group translating the A1C assay into estimated average glucose values. Diabetes Care 2008;31:1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bergenstal RM, Gal RL, Connor CG, et al. ; T1D Exchange Racial Differences Study Group. Racial differences in the relationship of glucose concentrations and hemoglobin A1c levels. Ann Intern Med 2017;167:95–102 [DOI] [PubMed] [Google Scholar]
- 13. Lindsell CJ, Franco RS, Smith EP, et al. : A method for the continuous calculation of the age of labeled red blood cells. Am J Hematol 2008;83:454–457 [DOI] [PubMed] [Google Scholar]
- 14. Cohen RM, Ciraolo P, Palascak MB, et al. Red cell lifespan heterogeneity in hematologically normal people sufficient to alter the apparent HbA1c. Blood 2008;112:4284–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khera PK, Smith EP, Lindsell CJ, et al. Use of an oral stable isotope label to confirm variation in red blood cell mean age that influences HbA1c interpretation. Am J Hematol 2015;90:50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Franco RS: The measurement and importance of red cell survival. Am J Hematol 2008;84:109–114 [DOI] [PubMed] [Google Scholar]
- 17. Soranzo N, Sanna S, Wheeler E, et al. : Common variants at 10 genomic loci influence hemoglobin A1(C) levels via glycemic and nonglycemic pathways. Diabetes 2010;59:3229–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. GRADE Study Research Group. Rationale and design of the glycemia reduction approaches in diabetes: a comparative effectiveness study. Diabetes Care 2013;36:2254–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hirsch IB: Glycemic variability and diabetes complications: does it matter? Of course it does! Diabetes Care 2015;38:1610–1614 [DOI] [PubMed] [Google Scholar]
- 20. Danne T, Nimri R, Battelino T, et al. : International consensus on use of continuous glucose monitoring. Diabetes Care 2017;40:1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kouzuma T, Uemastu Y, Usami T, Imamura S: Study of glycated amino acid elimination reaction for an improved enzymatic glycated albumin measurement method. Clin Chim Acta 2004;346:135–143 [DOI] [PubMed] [Google Scholar]
- 22. Malka R, Nathan DM, Higgins JM: Mechanistic modeling of hemoglobin glycation and red blood cell kinetics enables personalized diabetes monitoring. Sci Transl Med 2016;8:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgins JM, Mahadevan L: Physiological and pathological population dynamics of circulating human red blood cells. Proc Nat Acad Sci USA 2010;107:20587–20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patel HH, Patel HR, Higgins JM: Modulation of red blood cell population dynamics is a fundamental homeostatic response to disease. Am J Hematol 2015;90:422–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Holm S: A simple sequentially rejective multiple test procedure. Scand J Stat 1979;75, 383–386 [Google Scholar]
- 26. Kirk JK, D'Agostino RB, Jr, Bell RA, et al. : Disparities in HbA1c levels between African-American and non-Hispanic white adults with diabetes: a meta-analysis. Diabetes Care 2006;29:2130–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Heisler M, Faul JD, Hayward RA, et al. : Mechanisms for racial and ethnic disparities in glycemic control in middle-aged and older Americans in the health and retirement study. Arch Intern Med 2007. 24;167:1853–1860 [DOI] [PubMed] [Google Scholar]
- 28. Adams AS, Trinacty CM, Zhang F, et al. : Medication adherence and racial differences in A1C control. Diabetes Care 2008;31:916–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Viberti G, Lachin J, Holman R, et al. : ADOPT Study Group. A Diabetes Outcome Progression Trial (ADOPT): baseline characteristics of Type 2 diabetic patients in North America and Europe. Diabet Med 2006;23:1289–1294 [DOI] [PubMed] [Google Scholar]
- 30. Wolffenbuttel BH, Herman WH, Gross JL, et al. : Ethnic differences in glycemic markers in patients with type 2 diabetes. Diabetes Care 2013;36:2931–2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ziemer DC, Kolm P, Weintraub WS, et al. : Glucose-independent, black-white differences in hemoglobin A1c levels: a cross-sectional analysis of 2 studies. Ann Int Med 2010;152:770–777 [DOI] [PubMed] [Google Scholar]
- 32. Grimsby JL, Porneala BC, Vassy JL, et al. ; MAGIC Investigators. Race-ethnic differences in the association of genetic loci with HbA1c levels and mortality in U.S. adults: the third National Health and Nutrition Examination Survey (NHANES III). BMC Med Genet 2012;13:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mortensen HB, Christophersen C: Glucosylation of human haemoglobin a in red blood cells studied in vitro. Kinetics of the formation and dissociation of haemoglobin A1c. Clin Chim Acta 1983;134:317–326 [DOI] [PubMed] [Google Scholar]
- 34. Riddlesworth TD, Beck RW, Gal RL, et al. : Optimal sampling duration for continuous glucose monitoring to determine long-term glycemic control. Diabetes Technol Ther 2018;20:314–316 [DOI] [PubMed] [Google Scholar]
- 35. Juvenile Diabetes Research Foundation Continuous Glucose Monitoring Study Group: Continuous glucose monitoring and intensive treatment of type 1 diabetes. N Engl J Med 2008;359:1464–1476 [DOI] [PubMed] [Google Scholar]
- 36. Carlson AL, Mullen DM, Bergenstal RM: Clinical use of continuous glucose monitoring in adults with type 2 diabetes. Diabetes Technol Ther 2017;19:S4–S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Frier BM: Hypoglycaemia in diabetes mellitus: epidemiology and clinical implications. Nat Rev Endocrinol 2014:10:711–722 [DOI] [PubMed] [Google Scholar]
- 38. Lipska KJ, Ross JS, Wang Y, et al. : National trends in US hospital admissions for hyperglycemia and hypoglycemia among Medicare beneficiaries, 1999 to 2011. JAMA Intern Med 2014;174:1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qu Y, Jacober SJ, Zhang Q, et al. : Rate of hypoglycemia in insulin-treated patients with type 2 diabetes can be predicted from glycemic variability data. Diabetes Technol Ther 2012;14:1008–1012 [DOI] [PubMed] [Google Scholar]
- 40. Siegelaar SE, Kerr L, Jacober SJ, et al. : A decrease in glucose variability does not reduce cardiovascular event rates in type 2 diabetic patients after acute myocardial infarction: a reanalysis of the HEART2D study. Diabetes Care 2011;34:855–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Monnier L, Colette C: Glycemic variability: can we bridge the divide between controversies? Diabetes Care 2011;34:1058–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]