Abstract
Microfluidic platforms can lead to miniaturisation, increased throughput and reduced reagent consumption, particularly when the processes are automated. Here, a programmable microcontroller is used for automation of a microfluidic platform configured to electrochemically determine the levels of 8 proteins simultaneously in complex liquid samples. The platform system is composed of a programmable Arduino microcontroller that controls inexpensive valve actuators, pump, magnetic stirrer and electronic display. The programmable microcontroller results in repeatable timing for each step in a complex assay protocol, such as sandwich immunoassays. Application of the platform is demonstrated using a multiplexed electrochemical immunoassay based on capture at the electrode surface of magnetic particles labelled with horseradish peroxidase and detection antibody. The multiplexed assay protocol is completed in less than 30 mins and results in detection of eight proteins associated with prostate cancer. The approach presented can be used to automate and simplify high-throughput screening campaigns, such as detection of multiple biomarkers in patient samples.
Keywords: microfluidic, automation, multiplex, electrochemical immunoassay, prostate cancer
Microfluidic platforms can provide increased throughput and reduced reagent consumption in analysis. However, most research reports of analysis based on microfluidic platforms utilise manual or semi-automated hardware. Introduction of programmed control of platform systems can improve the repeatability of timing in assay protocols. Programmed control over microfluidics can deliver highly integrated, clinical diagnostic devices for personalised treatment and healthcare use [1–5].
In this communication we report on control of microfluidic flow and direction using inexpensive mass-produced components: the Arduino Uno microcontroller, high torque servos, a commercial syringe pump and electronically commutated motors. Use of a microcontroller, instead of desktop or laptop, is a route towards miniaturisation, integration and automation of immunoassay protocols, identified as goals previously [6, 7]. The microcontroller can implement custom protocols and sequencing events in a rapid and simple manner using high-level programming language C/C + +.
Prostate-specific antigen (PSA)-based testing has significantly advanced prostate cancer (PCa) screening since its introduction [5]. However, PSA prognostic predictions can be challenged by PCa epidemiology alteration [8]. A multiplexed magnetic-bead based immunoassay for detection of proteins that may be useful in PCa diagnosis and staging is therefore used to demonstrate the applicability of the automated microfluidic platform. Multiplexing biomarkers increases predictive power compared to single-plexing in any cancer [8–12], but particularly relevant to PCa given the recognised inadequate PSA predictive values [13]. The panel of proteins selected for initial testing of the platform is PSA, VEGF, ERG, IGF-I, IGFBP-3, CD-14, PEDF and GOLM-1 (acronyms defined in Experimental).
Placing normal metabolism and ageing aside, PSA concentrations in serum are generally elevated in the blood of a patient with advanced stages of the disease [14].
VEGF is not specific to PCa, but is noted by links towards angiogenesis in various solid tumours, including PCa. Because PCa cannot continue to grow without an expansion of blood vessels, VEGF may help to stage the disease [15]. ERG is over-expressed in metastatic PCa samples, assumed to be from a recurrent gene fusion that is seen in approximately 50% of all PSA screening in the United States [16]. IGF-I and IGFBP-3 are reported as having an inverse expression phenomenon in PCa staging, with IGF-1 over-expressed and IGFPB-3 under-expressed in patient samples. It is proposed that IGFBP-3 might limit bio-availability of IGF-I and regulate apoptosis [17, 18]. CD-14 is an inflammatory protein present in critically ill patients. The main function of CD-14 is its presence in innate immune responses towards microbial infection. Hence, it has been associated with increased rates of infection, sepsis, or death, linking it to advanced stages of PCa [19, 20]. PEDF is either an angiogenesis inhibitor or inducer. In the case of PCa, it is an expected inhibitor of angiogenesis, showing decreased levels in serum while the body attempts to stop the spread of cancer [21, 22]. GOLM-1 is a protein originating from the Golgi apparatus. It is proposed to be responsible for molecular alterations associated with the Golgi apparatus that take place during prostate carcinogenesis [23].
Detection of these proteins is therefore selected to test platform automation, to confirm platform performance prior to application to patient sample testing. The automated microfluidic platform design, Figure 1, was selected to incorporate simple push button control to switch between programs, such as assay runs and system flushing protocols, allowing multiple types of automated immunoassay experiments to be activated once the program is uploaded to the platform. The platform also incorporates independently controlled, electrically actuated, servo-valves for fluid control. The assembled platform requires rudimentary instruction to use. In a clinical setting this versatility could allow for seamless integration into point-of-care detection and decisions in medicine.
Fig. 1.
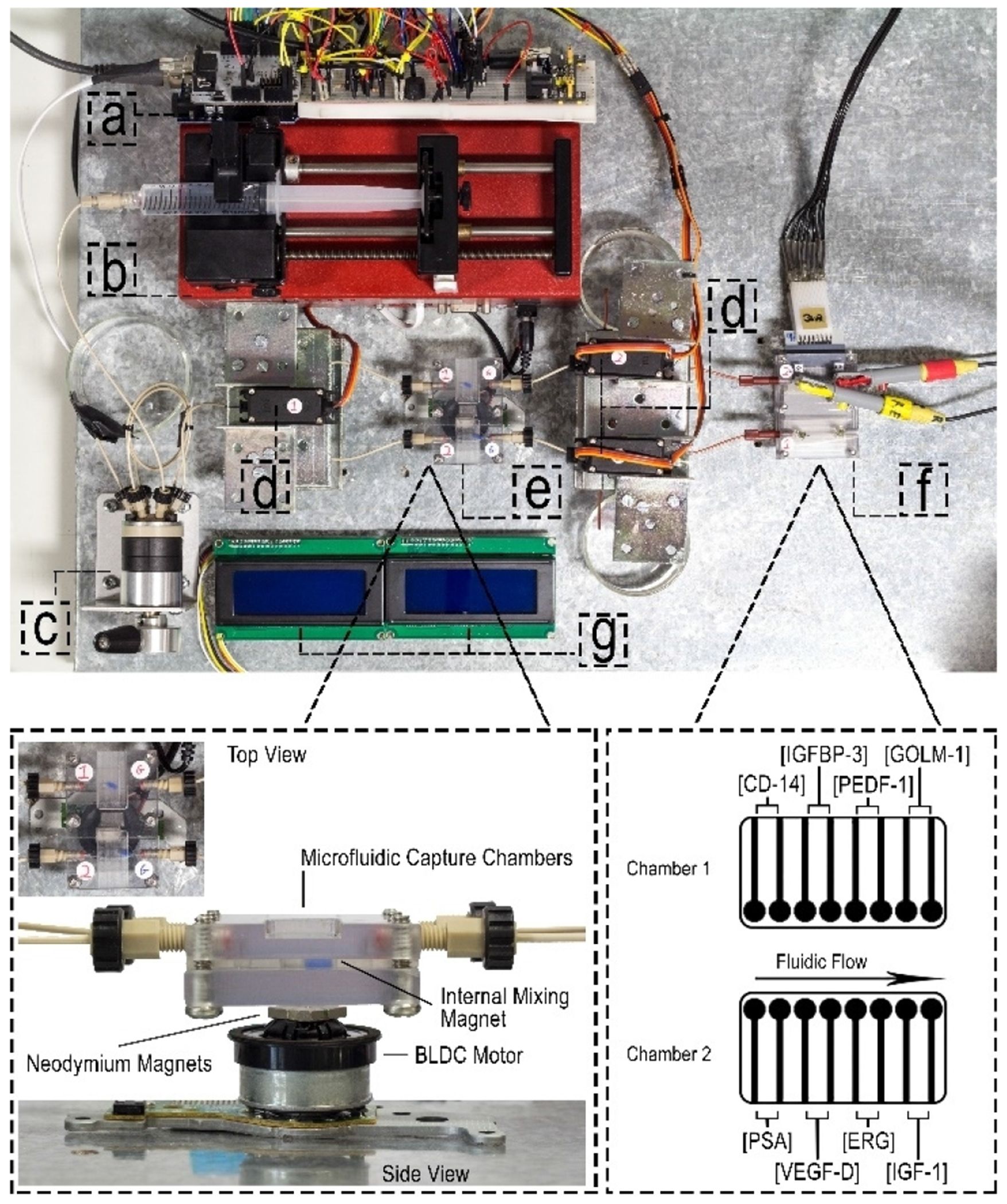
Photograph of automated microfluidic platform: (a) Arduino Uno microcontroller, (b) syringe pump, (c) sample injector, (d) servo-actuated valves, (e) capture chambers and magnetic stirrer, (f) detection chambers, (g) LCD displays.
The microfluidic platform consists of a microcontrolled syringe pump [24], three microcontrolled servo-valves [24], a microcontrolled fabricated magnetic stirrer [25], two capture chambers, two detection chambers, two liquid crystal displays (LCDs) and an Arduino Uno microcontroller (Figure 1). The LCDs visual display output is defined by 20 × 4 character displays and are programmed to update the system operator on tasks being, and to be, performed. The code used to control the system is encoded in C/C++ programming language and uploaded through the Arduino IDE software. The main function of the C/C++ program is to actuate the servos, operate magnetic stirring and initiate the syringe pump at specified flow rates, all at defined times.
The automated microfluidic platform is used to perform an electrochemical enzyme-amplified immunoassay for multiplexed detection of the proteins, with the protocol steps detailed in Table 1 of supporting information. This protocol is a modified version of that previously reported [26]. Briefly, capture chambers permit capture of biomarker proteins from serum samples (5X diluted calf serum, an excellent surrogate for human serum [10, 12]) by magnetic particles massively labelled with horseradish peroxidase (HRP) and secondary antibody (Ab2-HRP-MB) [27]. The capture chambers are first flooded with 0.05% PBS-T20 to minimise non-specific binding. The capture chambers are then filled with Ab2-HRP-MB followed by antigen standard solution containing protein mixture to be assayed. After a 10 min incubation period, assisted by magnetic stirrer [25], the contents of the capture chambers are directed, by valve and pump actuation, towards detection chambers and left to incubate for 10 min (Scheme 1). Screen printed carbon sensors modified with primary antibodies recognise and isolate proteins captured by the magnetic bead conjugate. Once these steps are complete, a CHI 1030 potentiostat connected to an 8-electrode array in the first detection chamber (see supporting information, Figure 1) is used to detect the amperometric signals achieved by introduction of 1 mM hydroquinone (HQ) in PBS and 0.1 mM H2O2. Catalytic reduction of peroxide is achieved by HRP with mediation of electron flow to the electrode by HQ. Amperometric signals are then developed, using the above approach, for the 8-electrode array in detection chamber 2.
Scheme 1.
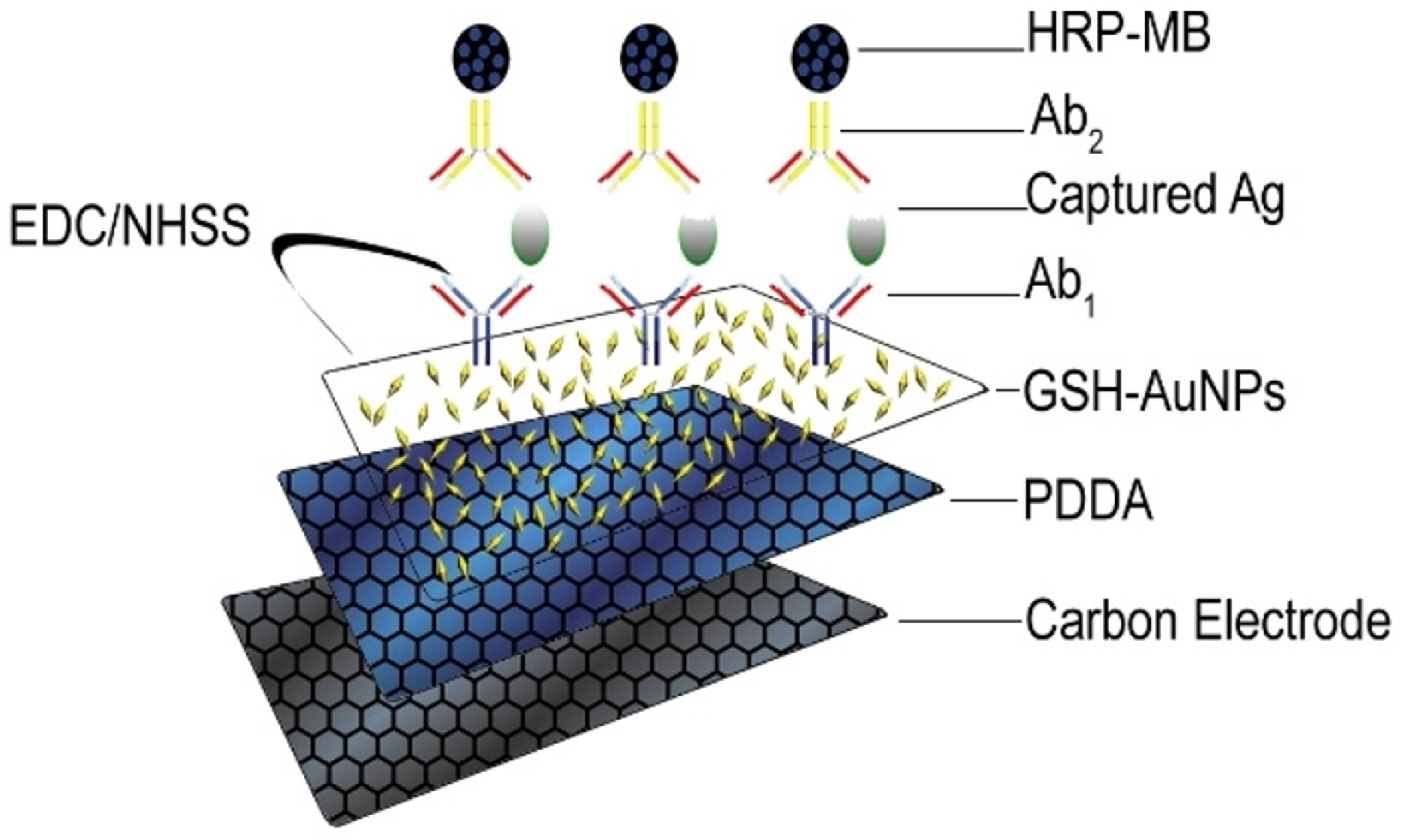
Schematic showing the sandwich assay process involved in the detection of a protein as antigen (Ag).
The proteins are detected, in duplicate, using two 8-electrode arrays, one in each detection chamber. Chamber 1 detects IGFBP-3, CD-14, PEDF-1, GOLM-1, while chamber 2 detects IGF-1, PSA, VEGF-D, ERG (see Figure 1, for detailed protocol see Table 1 of supporting information). Under these circumstances automation increases throughput and saves operator time and resources, permitting detection of 4 proteins in duplicate in each chamber. The use of separate chambers can be used to overcome potential cross-reactivity. For example, IGFBP-3 has the largest overall carrying capacity and highest affinity for IGF-I out of all known IGFBPs [17]. By utilising separate chambers, the potential of IGFBP-3 and IGF-I to cross react is eliminated. In the protocol, the immunoassay steps (Table 1 supporting information) are uploaded to the microcontroller, consequently only 4 injections for loading of reagents and samples are required by the operator (see steps 5–10).
The assay requires two 10 min incubation periods in each chamber in parallel, with the detection step taking less than 10 mins. This assay time totals far less than that for conventional ELISA (>5 hours) [28] and is similar to that reported using a manual on-line capture assay platform (~30 min) [27].
This approach provided simultaneous assays with detection limits (DLs, measured as three times the average standard deviation plus the zero protein control) of 140 pgml−1 for PSA, 90 pgml−1 for VEGF-D, 15 pgml−1 for ERG, 13 pgml−1 for IGF-1, 130 pgml−1 for CD-14, 150 pgml−1 for IGFBP-3, 90 pgml−1 for PEDF-1 and 15 pgml−1 for GOLM-1 in serum. These preliminary results show that amperometric peak currents increase in a semi-logarithmic trend over the detected ranges of 0.14 to 34.2 ng ml−1 for PSA, 0.09 to 23.8 ngml−1 for VEGF-D, 0.015 to 3.9 ngml−1 for ERG, 0.013 to 3.4 ngml−1 for IGF-1, 0.13 to 32.5 ngml−1 for CD-14, 0.15 to 38.7 ngml−1 for IGFBP-3, 0.09 to 11.2 ngml−1 for PEDF-1 and 0.015 to 1.95 ngml−1 for GOLM-1 (Figure 2). These DLs compare well to similar research involving a 2 protein multiplex using a manual on-line capture assay platform [26].
Fig. 2.
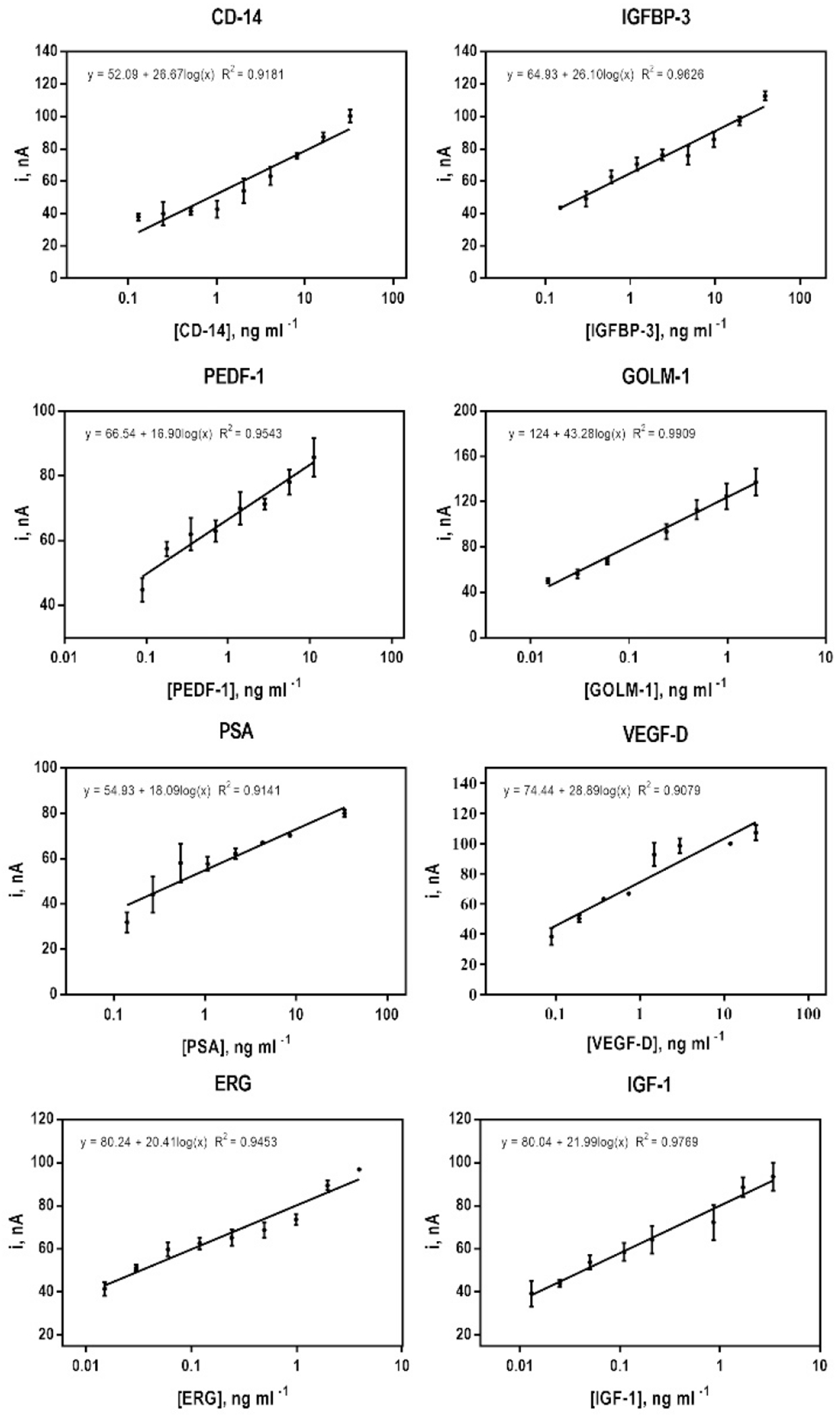
Immunoarray calibration plots using amperometric responses for PSA, VEGF, ERG, IGF-I, IGFBP-3, CD-14, PEDF and GOLM-1 at −0.2 V vs Ag/AgCl developed by injecting 1 mM HQ and 0.1 mM H2O2 after capturing analyte Ab2-HRP-MB-Ag bioconjugates on modified carbon electrode surfaces contained within the microfluidic device detection chambers. Protein standards were diluted 5X in calf serum, to mimic a complex sample matrix. Error bars represent the range (n=2) for the two electrodes in each immunoarray designed to detect the protein.
The high sensitivity of the assay can allow for high dilution of the sample to help with very distinct clinical thresholds of protein biomarkers, but can also be traded-off for shorter assay times [5, 29].
The automated assay protocol provides a repeatable, multiplexed, rapid electrochemical assay platform that can be implemented for high-throughput detection of multiple proteins in patient samples that may be useful in PCa diagnosis and staging. The platform brings immunoarray diagnostics closer to point-of-care technology.
Gold(III) chloride trihydrate, HEPES, sodium borohydride, reduced glutathione (GSH), Amicon Ultra centrifugal filter units (MWCO 50 kDa), hydrogen peroxide solution (H2O2 30 (w/w) in H2O), ammonium metavanadate, phosphate buffer solution with 0.05% Tween-20 (PBS-T20), sodium phosphate dibasic, sodium phosphate monobasic monohydrate, sodium chloride, potassium chloride, hydroquinone (HQ), poly(diallyldimethylammonium chloride) (PDDA), 1-(3-(dimethylamino)propyl)-3-ethylcarbodiimide hydrochloride (EDC), N-hydroxysulfo-succinimide (NHSS), bovine serum albumin (BSA), and bovine calf serum was purchased from Sigma and used as received. Biotinylated peroxidase, streptavidin dynabeads T1 and dyna mag 2 was purchased from Fisher and used as received. PDMS, Sylgard 184 was purchased from Dow Corning and used with a ratio of 1:10 PDMS per master mold. Erythroblast transformation specific related gene (ERG) and golgi membrane protein 1 (GOLM-1) were from OriGene Technologies. Prostate-specific antigen (PSA), insulin-like growth factor I (IGF-I), IGF-binding protein-3 (IGFBP-3), cluster of differentiation 14 (CD-14), vascular endothelial growth factor (VEGF), and pigment epithelium-derived factor (PEDF) proteins were from R&D Systems. Rheodyne 7725i injector was purchased from Sigma and used as received. Screen-printed carbon arrays consisting of eight 700 micrometer diameter sensors were from Kanichi Research (UK). MG996R servos were from TowerPro and the RS232 Shield V2 were purchased from LinkSprite.
Biotinylated Ab2 and biotinylated HRP labels were attached onto the 1 mm diameter streptavidin-coated super- paramagnetic beads (MBs) as previously described [30].
Monolayer films of 5 nm glutathione-decorated gold nanoparticles (GSH–AuNPs) [31] were deposited on the electrode array on an underlayer of adsorbed PDDA as previously reported [30]. Ab1 were attached onto GSH–AuNPs on the electrode array via EDC–NHSS coupling overnight. The electrode array was washed and incubated with 2% BSA in PBS for 1 h to block non-specific binding. A fresh AuNP-antibody array is inserted into the detection module for each assay.
Supplementary Material
Acknowledgements
This work was supported financially by US-Ireland Program (Grant Number EB014586) administered by the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and funded by NIH and Science Foundation Ireland under the US/Ireland programme (Grant Number 13/US/B2546). The authors are grateful to Dr. Colleen Krause and Dr. Brunah Otieno for sharing details on the original non-automated microfluidic immunoarray.
Footnotes
Supporting information for this article is available on the WWW under https://doi.org/10.1002/elan.201800632
References
- [1].Whitesides GM, Nature 2006,442, 368–373. [DOI] [PubMed] [Google Scholar]
- [2].Volpatti LR, Yetisen AK, Trends Biotechnol. 2014, 32, 347–350. [DOI] [PubMed] [Google Scholar]
- [3].Au AK, Bhattacharjee N, Horowitz LF, Chang TC, Folch A, Lab Chip 2015,15,1934–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mark D, Haeberle S, Roth G, von Stetten F, Zengerle R, Chem. Soc. Rev 2010,39,1153. [DOI] [PubMed] [Google Scholar]
- [5].Rusling JF, Anal. Chem 2013, 85, 5304–5310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Patabadige DEW, Jia S, Sibbitts J, Sadeghi J, Sellens K, Culbertson CT, Anal. Chem 2016, 88, 320–338. [DOI] [PubMed] [Google Scholar]
- [7].Mercer C, Leech D, J. Chem. Educ 2018, 95, 1221–1225. [Google Scholar]
- [8].Bensalah K, Lotan Y, Karam JA, Shariat SF, Prostate Cancer Prostatic Dis. 2008,11, 112–120. [DOI] [PubMed] [Google Scholar]
- [9].Xiao Z, Prieto D, Conrads TP, Veenstra TD, Issaq HJ, Mol. Cell. Endocrinol 2005, 230, 95–106. [DOI] [PubMed] [Google Scholar]
- [10].Rusling JF, V Kumar C, Gutkind JS, Patel V, Analyst 2010,135, 2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chikkaveeraiah BV, Bhirde AA, Morgan NY, Eden HS, Chen X, ACS Nano 2012, 6, 6546–6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hanash SM, Pitteri SJ, Faca VM, Nature 2008, 452, 571–579. [DOI] [PubMed] [Google Scholar]
- [13].Lilja H, Ulmert D, Vickers AJ, Nat. Rev. Cancer 2008, 8, 268–278. [DOI] [PubMed] [Google Scholar]
- [14].Hassan MI, Kumar V, Singh TP, Yadav S, Chem. Biol. Drug Des 2007, 70, 261–267. [DOI] [PubMed] [Google Scholar]
- [15].Yu L, Deng L, Li J, Zhang Y, Hu L, Gynecol. Oncol 2013,128, 391–396. [DOI] [PubMed] [Google Scholar]
- [16].Park K, Tomlins SA, Mudaliar KM, Chiu Y-L, Esgueva R, Mehra R, Suleman K, Varambally S, Brenner JC, MacDonald T, Neoplasia 2010,12, 590–IN21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Meinbach DS, Lokeshwar BL, Urol. Oncol. Semin. Orig. Investig 2006,24, 294–306. [DOI] [PubMed] [Google Scholar]
- [18].Chan JM, Stampfer MJ, Giovannucci E, Ma J, Pollak M, Growth Horm. IGF Res 2000, 10, S32–S33. [DOI] [PubMed] [Google Scholar]
- [19].Butkus de Aguiar B, Girardi I, D’Avila Paskulin D, de Franca E, Dornelles C, Suparregui Dias F, Bonorino C, Sampaio Alho C, Immunol. Invest 2008, 37, 752–769. [DOI] [PubMed] [Google Scholar]
- [20].Zhang A-Q, Yue C-L, Gu W, Du J, Wang H-Y, Jiang J, PLoS One 2013, 8, e71237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dawson DW, Science, 1999, 285, 245–248. [DOI] [PubMed] [Google Scholar]
- [22].Ji D, Li M, Zhan T, Yao Y, Shen J, Tian H, Zhang Z, Gu J, Carcinogenesis 2013, 34, 1265–1272. [DOI] [PubMed] [Google Scholar]
- [23].Wei S, A Dunn T, Isaacs WB, De Marzo AM, Luo J, Prostate 2008, 68, 1387–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mercer C, Bennett R, Conghaile PÓ, Rusling JF, Leech D, Sensors Actuators B Chem. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Mercer C, Leech D, J. Chem. Educ 2017, 94, 816–818. [Google Scholar]
- [26].Otieno BA, Krause CE, Latus A, Chikkaveeraiah BV, Faria RC, Rusling JF, Biosens. Bioelectron 2014, 53, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Otieno BA, Krause CE, Latus A, Chikkaveeraiah BV, Faria RC, Rusling JF, Biosens. Bioelectron 2014, 53, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Findlay JWA, Smith WC, Lee JW, Nordblom GD, Das I, DeSilva BS, Khan MN, Bowsher RR, J. Pharm. Biomed. Anal 2000, 21, 1249–1273. [DOI] [PubMed] [Google Scholar]
- [29].Krause CE, Otieno BA, Latus A, Faria RC, Patel V, Gutkind JS, Rusling JF, ChemistryOpen 2013, 2, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, Abraham MT, Dey DK, Gutkind JS, Rusling JF, Anal. Chem 2012, 84, 6249–6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mani V, Chikkaveeraiah BV, Patel V, Gutkind JS, Rusling JF, ACS Nano 2009, 3, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


