Abstract
Human lysine demethylase KDM5A is a chromatin-modifying enzyme associated with transcriptional regulation, because of its ability to catalyze removal of methyl groups from methylated lysine 4 of histone H3 (H3K4me3). Amplification of KDM5A is observed in many cancers, including breast cancer, prostate cancer, hepatocellular carcinoma, lung cancer, and gastric cancer. In this study, we employed alanine scanning mutagenesis to investigate substrate recognition of KDM5A and identify the H3 tail residues necessary for KDM5A-catalyzed demethylation. Our data show that the H3Q5 residue is critical for substrate recognition by KDM5A. Our data also reveal that the protein–protein interactions between KDM5A and the histone H3 tail extend beyond the amino acids proximal to the substrate mark. Specifically, demethylation activity assays show that deletion or mutation of residues at positions 14–18 on the H3 tail results in an 8-fold increase in the , compared to wild-type 18mer peptide, suggesting that this distal epitope is important in histone engagement. Finally, we demonstrate that post-translational modifications on this distal epitope can modulate KDM5A-dependent demethylation. Our findings provide insights into H3K4-specific recognition by KDM5A, as well as how chromatin context can regulate KDM5A activity and H3K4 methylation status.
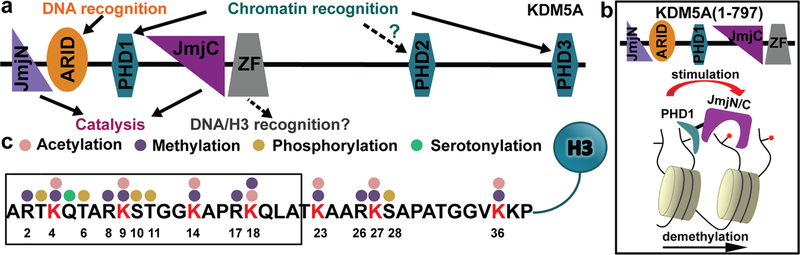
By regulating chromatin structure and accessibility, post-translational modifications (PTMs) on histone proteins control several cellular processes, such as transcription, cell differentiation, and DNA damage repair.1,2 These modifications are dynamic, because they are installed, read, and removed by specialized proteins. Histone lysine demethylases (KDMs) are enzymes that demethylate lysine residues in histone proteins. The KDM5 subfamily of the histone demethylases specifically remove methylation from lysine 4 in histone H3 (H3K4me1/2/3), which is a mark that is associated with active transcription. KDM5 family members share a conserved multidomain architecture composed of the jumonji N and C domains (JmjN and JmjC), which together comprise the catalytic domain, a DNA binding ARID domain, a zinc-finger domain (ZF), and two-to-three plant homeo- domain (PHD) reader domains (Figure 1a).
Figure 1.
Domain architecture in KDM5A and H3 substrate sequence. (a) KDM5A domain architecture, (b) proposed model for allosteric stimulation of demethylation by its PHD1 domain (the product of KDM5A-catalyzed demethylation binds to the PHD1 and stimulates catalytic activity, allowing feed-forward regulation), and (c) sequence and select post-translational modifications of the H3 tail.
There are four paralogs of KDM5 enzymes in humans: KDM5A—KDM5D. KDM5A has three PHD reader domains (Figure 1a). The PHD1 domain binds preferentially to the product of KDM5A demethylation (H3K4me0) and allosterically enhances demethylase activity (Figure 1b).3,4 Function of the PHD2 is yet to be determined, while the PHD3 domain recognizes H3K4me3 marks and is postulated to recruit KDM5A to its substrate.5 While PHD1 is required for efficient demethylation in cells, the deletion of the PHD2 and PHD3 domains does not abrogate demethylase activity, and a KDM5A construct that lacks these domains (KDM5A1—797) has been utilized for the in vitro characterization of this protein (Figure 1b).3,4,6,7
The presence of multiple methylated lysine residues in the H3 tail (Figure 1c) raises questions about the molecular basis for H3K4-specific demethylation by KDM5A.8–11 Currently, there is limited information regarding the molecular details of KDM5A substrate recognition. While KDM5 enzymes have been previously crystallized,12–18 none of these structures contain histone substrate. The co-crystal structure of a KDM5 plant homologue with a 10mer H3 peptide provided insights into substrate engagement by the plant enzyme,19 which substantially differs from human KDM5 enzymes, because it lacks ARID and PHD1 domains.
In this study, we identified the H3 residues necessary for substrate recognition, as well as interactions between KDM5A and its peptide substrate at sites distant from the methylated H3K4 residue. To our knowledge, this is the first time that interactions between KDM5A and distal residues of H3 peptide are identified as significant for peptide binding and modulation of KDM5A activity.
RESULTS AND DISCUSSION
Contributions of the Lysine Proximal Residues to K4- Specific Demethylation by KDM5A
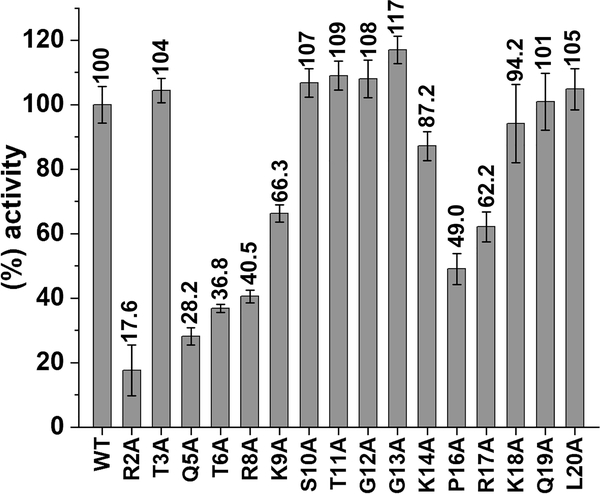
To probe KDM5A-H3 substrate interactions, we investigated how alanine mutations in a H3 N-terminal 21mer (aa A1-A21) peptide substrate affect demethylase activity (Figure 2). The activity of KDM5A1–797 was most significantly impaired toward peptides harboring mutations in the first 9 residues, with the exception of the T3A mutant peptide (R2A, Q5A, T6A, R8A, and K9A peptides) (Figure 2). Single alanine substitution of the C-terminal half of the peptide (aa 10–20) had no significant effect on KDM5A1–797 activity, apart from P16A and R17A mutants, which retained only half of the activity of their wild-type (WT) counterpart.
Figure 2.
Alanine scanning mutagenesis of histone H3 tail in KDM5A-catalyzed demethylation. KDM5A1–797 activity for 21mer H3K4me3 WT and mutant peptides. KDM5A1–797 activity was normalized to the 21mer H3K4me3 WT peptide reaction (considered as 100%). Results are means ± standard error of the mean (SEM) of two independent experiments.
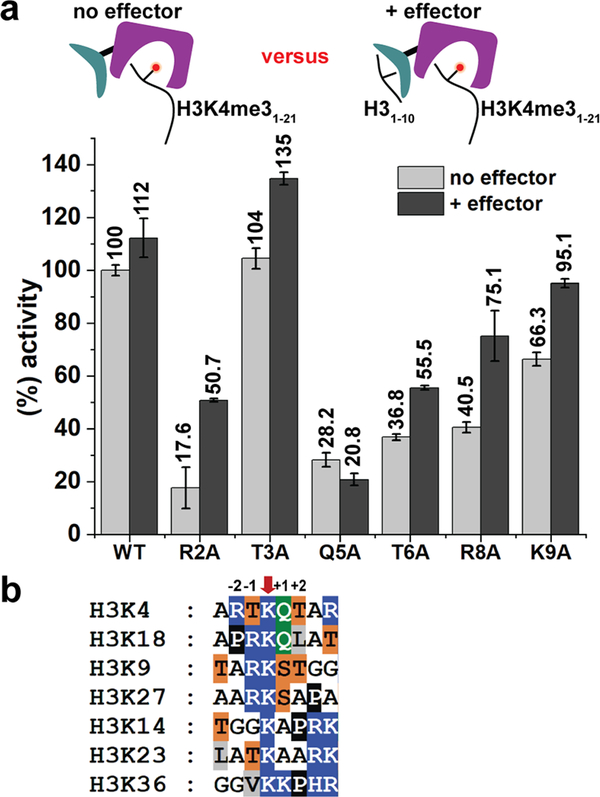
As the first 10 residues of the H3 tail are also recognized by the PHD1 domain, and peptide binding to PHD1 stimulates demethylation activity, it is possible that the observed reduction in demethylation is due to impaired engagement of the mutant peptide by the PHD1 domain, diminishing allosteric stimulation. To test this, we preincubated KDM5A1–797 with unmodified H3K4 peptide (effector peptide) at a concentration where the PHD1 is saturated with effector peptide (20 times the previously determined KD value), allowing for maximal allosteric stimulation,3,4 and compared the activity for wild-type (WT) and mutant H3K4me3 peptide substrates (see Figure 3a). In the case of R2A, T6A, and R8A mutant substrates, the activity was only partially rescued in the presence of the saturating PHD1 effector peptide (Figure 3a). These findings suggest that the observed reduced demethylation of these mutant substrates is a consequence of both their decreased engagement by the catalytic domain and reduced allosteric stimulation (Figure 3a). The most prominent (~3-fold) rescue in demethylation in the presence of the effector peptide was observed with R2A mutant peptide. Indeed, our previous studies point to decreased engagement of R2A peptide by the PHD1 domain.4 In contrast, almost complete rescue of K9A demethylation suggests the minimal role of this residue in substrate recognition. Notably, very low KDM5A1–797 activity toward the Q5A substrate peptide was observed (~20%). This reduction in activity could not be rescued by the presence of the effector peptide, suggesting that the Q5 position is critical for substrate recognition and engagement by the catalytic domain. Previous experiments with a predicted Q5 interacting mutant in the paralog demethylase KDM5B support the importance of Q5-enzyme interaction for demethylation.19
Figure 3.
Effect of N-terminal H3 tail mutations on KDM5A activity. (a) Impact of the PHD1 effector ligand on demethylase activity. KDM5A1–797 activity for 21mer H3K4me3 WT and mutant peptides in the absence and presence of 20 μM effector (unmodified 10mer peptide). Error bars represent the standard error of the mean of two independent experiments. (b) Sequence alignment of H3 tail methylated lysine residues. Amino acids are color coded as follows: blue, R/K/H; orange, T/S; green, Q; white, A/G; black, P; and gray, L/V/I.
In order to determine how these substrate mutants affect catalysis, we obtained apparent Michaelis-Menten (M-M) kinetic constants under conditions where the PHD1 domain was saturated with effector peptide. While the measured kcat was essentially the same for WT and mutant peptide substrates, the apparent KM values were significantly different (see Table 1, as well as Table S2 and Figure S1 in the Supporting Information). Specifically, the value for Q5A (415.2 ± 71.4 μM) was >30-fold higher than that for the WT peptide (12.5 ± 0.9 μM), while the values for R2A, T6A, and R8A were, respectively, ~12-fold, ~8-fold, and ~4-fold higher. We further probed the T6 interaction by mutating this residue to a serine and a valine. We found that the T6S peptide behaved similar to the WT substrate while substitution of T6 by valine resulted in a 3-fold increase in the KM value. These results highlight the importance of the hydroxyl group of T6 in the KDM5A-substrate interaction. Taken together, our findings suggest that R2, Q5, T6, and R8 all contribute to substrate recognition by KDM5A, with Q5 being the most important residue for substrate engagement. Interestingly, a comparison of our findings to those observed for the plant homologue JMJ14 indicates species-specific substrate recognition in H3K4 demethylases. In the JMJ14 study, mutation of the residues that interact with H3R2 abrogated JMJ14 activity while H3T6 had no direct interaction with JMJ14.19 Such differences between KDM5s and the plant homologue are not surprising, given the differences in domain architecture, as well as 50%—60% similarity of their catalytic domains.
Table 1.
Apparent Michaelis—Menten (M-M) Kinetic Parameters for 21mer H3K4me3 Peptides in the Presence of Saturating (20 μM) Effector Peptide (Unmodified H3 10mer, aa 1–10)a
| 21mer H3K4me3 | (× 10−6 M) | kcat (s−1) | kcat/ (M−1 s−1) |
|---|---|---|---|
| WT | 12.5 ± 0.9 | 0.052 ± 0.001 | 4126.4 ± 300.6 |
| R2A | 153.6 ± 11.6 | 0.069 ± 0.002 | 451.8 ± 35.7 |
| T3A | 9.0 ± 0.6 | 0.056 ± 0.001 | 6232.4 ± 413.9 |
| Q5A | 415.2 ± 71.4 | 0.056 ± 0.003 | 134.4 ± 24.4 |
| T6A | 96.5 ± 15.7 | 0.075 ± 0.004 | 772.7 ± 131.6 |
| R8A | 55.1 ± 7.6 | 0.054 ± 0.002 | 980.7 ± 142.5 |
| K9A | 21.9 ± 2.8 | 0.061 ± 0.002 | 2791.3 ± 373.5 |
| T6S | 14.7 ± 1.5 | 0.070 ± 0.002 | 4756.4 ± 507.0 |
| T6V | 39.4 ± 6.8 | 0.050 ± 0.003 | 1258.5 ± 228.5 |
Results are means ± SEM of two independent experiments.
A sequence alignment of the H3 tail lysine residues that undergo methylation (K9, K14, etc.) (Figure 3b), together with our data, provides further insights into KDM5A specificity for H3K4. For the various methylated lysines, Arg in the −2 (H3R2) position is replaced by a small aliphatic residue (alanine, glycine, or proline), while Thr in the +2 (H3T6) position is replaced by an aliphatic residue in all instances except K9. However, K9 differs from K4 at the —2 (H3R2) and +1 (H3Q5) positions; these substitutions are sufficient to render H3K9me3 an unfavorable substrate for KDM5A.7 Finally, in addition to K4, the only H3 lysine that has a glutamine at position +1 (H3Q5 position) is K18. Since methylation of K18 is a newly detected mark with unknown function,8,10 we tested whether KDM5A1—797 can demethylate K18. When H3K18me3 (aa 12—32) peptide was incubated with KDM5A1—797, no demethylation was detected (see Figure S3 in the Supporting Information). The observation that the adjacent glutamine is not sufficient for demethylation further supports recognition of an extended H3 region by KDM5A.
KDM5A Recognizes an H3 Epitope Distal to K4
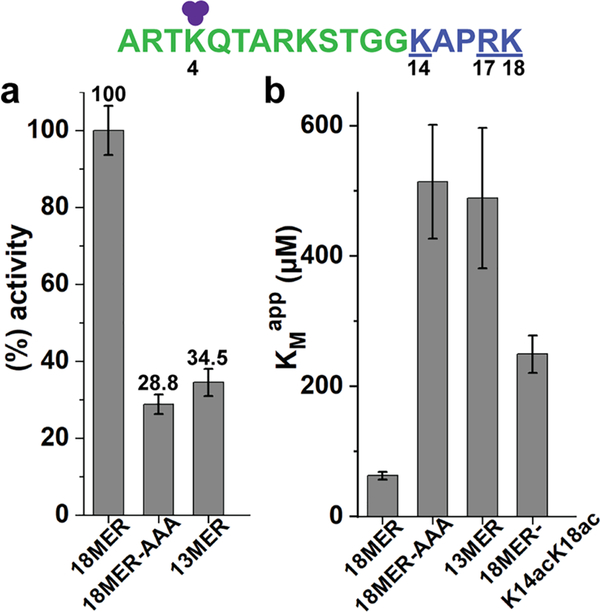
In addition to the N-terminal half of the 21mer H3K4me3 peptide substrate (aa A1-K9), we observed a significant reduction of KDM5A activity when P16 was substituted by alanine (~50%, Figure 2). Since the PHD1 domain engages only the first 10 residues of H3,3,4 and mutations in downstream positions are not expected to affect peptide engagement by PHD1, the decreased activity for P16A suggests that structural rigidity at position 16 may be important for substrate engagement. Furthermore, alanine substitution at the next position (R17A) decreased KDM5A activity to 62% (Figure 2). These findings led us to hypothesize that (i) the basic residues surrounding P16 (K14, R17, and K18) may engage in interactions with KDM5A, and (ii) their effect on substrate binding may be cumulative.
To test this hypothesis, we utilized peptide substrates that either lacked the K14-K18 basic patch (13mer peptide) or the three basic residues were mutated to alanines (K14A/R17A/ K18A triple mutant, 18mer-AAA), and compared them to 18mer WT peptide substrate (Figure 4a). Both the deletion and alanine substitution of this basic patch substantially impaired demethylation of these substrates (~28%−35%, compared to the 18mer WT peptide). Further kinetic characterization showed that, when this basic patch is removed, either by deletion or mutagenesis, the KMapp value increases 8-fold, compared to the 18mer WT peptide (see Table 2, as well as Table S3 in the Supporting Information, and Figure 4b, as well as Figure S2 in the Supporting Information). These results support our hypothesis that the K14-K18 basic patch is engaged by KDM5A and that P16 provides required structural rigidity for the basic patch-KDM5A interaction. The observed difference in KMapp also explains the preference of KDM5 enzymes toward longer peptide substrates.3,4
Figure 4.
Recognition of H3 K14-K18 basic patch by KDM5A. (a) KDM5A1–797 activity for H3K4me3 WT and basic patch mutant/ deletion peptides normalized to the 18mer WT peptide reaction (considered as 100%). (b) Apparent KM values for the various basic patch peptides. Error bars represent the standard error of the mean of two independent experiments. (purple circles represent methyl groups, green characters denote 13mer, and blue characters (underlined) denote basic patch residues).
Table 2.
Apparent Michaelis—Menten (M-M) Kinetic Parameters for H3K4me3 WT and Mutant Peptides of Various Lengthsa
| H3K4me3 | (× 10−6 M) | kcat (s−1) | kcat/ (M−1 s−1) |
|---|---|---|---|
| 21mer | 24.9 ± 3.3 | 0.057 ± 0.002 | 2289.3 ± 312.3 |
| 18mer | 62.3 ± 5.6 | 0.078 ± 0.002 | 1248.8 ± 117.2 |
| 13mer | 488.7 ± 107.5 | 0.111 ± 0.009 | 227.6 ± 53.7 |
| 18mer-K14A/R17A/K18A | 514.1 ± 87.6 | 0.108 ± 0.007 | 210.9 ± 38.6 |
| 18mer-K14acK18ac | 249.1 ± 28.5 | 0.101 ± 0.004 | 404.6 ± 48.7 |
Results are means ± SEM of two independent experiments.
Acetylation of K14 and K18 is associated with active transcription, and a previous study identified a positive crosstalk between these modifications and K4me3.8 Hence, we proceeded to test how acetylation of K14 (K14ac) and K18 (K18ac) affects KDM5A substrate engagement and activity. Utilizing an 18mer peptide with acetylated lysines 14 and 18 (18mer-K14acK18ac), we found that acetylation of the basic patch increased the value of 4-fold, relative to WT (see Table 2, as well as Table S3, and Figure 4b, as well as Figure S2). This finding reveals that post-translational modifications installed on the K14-K18 basic patch can modulate KDM5A activity. Interestingly, K14ac was shown to inhibit K4me2 demethylation by histone demethylase KDM1A (LSD1).20
CONCLUSION
Our investigations have defined substrate requirements for demethylation by human histone demethylase KDM5A. Specifically, we showed that engagement of H3Q5 is crucial for substrate binding, while H3R2, H3T6, and H3R8 further contribute to H3K4me3-specific recognition by KDM5A. These findings provide a rationale for selectivity of KDM5A toward the H3K4me3 mark over other methylated lysine residues in H3. Interestingly, we found that a basic patch on the histone tail downstream of H3K4 (aa K14-K18) is also involved in interactions with the enzyme. We demonstrated that post-translational modifications of this distal epitope can alter demethylase activity. Our study uncovered extended interactions between the catalytic domain of KDM5A and the H3 tail and revealed a distal basic epitope on H3 that modulates KDM5A demethylation. Combined with the allosteric regulation achieved through the PHD1 domaineffector peptide interaction, these findings provide important insights into how chromatin context can regulate catalytic activity of KDM5A and, consequently, H3K4 methylation status.
Future studies with modified nucleosomes, rather than peptide substrates, will further contribute to understanding the role of H3 residues in H3K4me3 demethylation by KDM5A. While this study focuses on H3 tail recognition by the catalytic domain of KDM5A, the roles of auxiliary domains and interacting partners of the demethylase on regulation of the catalytic activity warrant further investigation.
Supplementary Material
Acknowledgments
Funding
This research was supported by the National Institutes of Health (Grant No. GM114044) and the Cancer League and UCSF Helen Diller Family Comprehensive Cancer Center Award (to D.G.F.).
Footnotes
The authors declare no competing financial interest.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.biochem.9b01036.
Experimental procedures, sequences of H3 peptides, MM curves, KM comparison graph and expanded M-M tables (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.biochem.9b01036
Accession Codes
UniProtKB ID for human KDM5A: P29375
Contributor Information
Nektaria Petronikolou, Department of Cellular and Molecular Pharmacology, University of California, San Francisco, California 94158, United States.
James E. Longbotham, Department of Cellular and Molecular Pharmacology, University of California, San Francisco, California 94158, United States
Danica Galonić Fujimori, Department of Cellular and Molecular Pharmacology and Department of Pharmaceutical Chemistry, University of California, San Francisco, California 94158, United States.
REFERENCES
- (1).Kouzarides T (2007) Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- (2).Allis CD, and Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat. Rev. Genet 17, 487–500. [DOI] [PubMed] [Google Scholar]
- (3).Longbotham JE, Chio CM, Dharmarajan V, Trnka MJ, Torres IO, Goswami D, Ruiz K, Burlingame AL, Griffin PR, and Fujimori DG (2019) Histone H3 binding to the PHD1 domain of histone demethylase KDM5A enables active site remodeling. Nat. Commun 10, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Torres IO, Kuchenbecker KM, Nnadi CI, Fletterick RJ, Kelly MJ, and Fujimori DG (2015) Histone demethylase KDM5A is regulated by its reader domain through a positive-feedback mechanism. Nat. Commun 6, 6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, and Allis CD (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, and Helin K (2007) RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128, 1063–1076. [DOI] [PubMed] [Google Scholar]
- (7).Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument- Bromage H, Tempst P, Gilliland DG, Zhang Y, and Kaelin WG Jr. (2007) The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128, 889–900. [DOI] [PubMed] [Google Scholar]
- (8).Schwammle V, Sidoli S, Ruminowicz C, Wu X, Lee CF, Helin K, and Jensen ON (2016) Systems Level Analysis of Histone H3 Post-translational Modifications (PTMs) Reveals Features of PTM Crosstalk in Chromatin Regulation. Mol. Cell. Proteomics 15, 2715–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, and Hunt DF (2006) Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J. Biol. Chem 281, 559–568. [DOI] [PubMed] [Google Scholar]
- (10).Luense LJ, Wang X, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, and Berger SL (2016) Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenet. Chromatin 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhao Y, and Garcia BA (2015) Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harbor Perspect. Biol 7, a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Vinogradova M, Gehling VS, Gustafson A, Arora S, Tindell CA, Wilson C, Williamson KE, Guler GD, Gangurde P, Manieri W, Busby J, Flynn EM, Lan F, Kim HJ, Odate S, Cochran AG, Liu Y, Wongchenko M, Yang Y, Cheung TK, Maile TM, Lau T, Costa M, Hegde GV, Jackson E, Pitti R, Arnott D, Bailey C, Bellon S, Cummings RT, Albrecht BK, Harmange JC, Kiefer JR, Trojer P, and Classon M (2016) An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells. Nat. Chem. Biol 12, 531–538. [DOI] [PubMed] [Google Scholar]
- (13).Liang J, Labadie S, Zhang B, Ortwine DF, Patel S, Vinogradova M, Kiefer JR, Mauer T, Gehling VS, Harmange JC, Cummings R, Lai T, Liao J, Zheng X, Liu Y, Gustafson A, Van der Porten E, Mao W, Liederer BM, Deshmukh G, An L, Ran Y, Classon M, Trojer P, Dragovich PS, and Murray L (2017) From a novel HTS hit to potent, selective, and orally bioavailable KDM5 inhibitors. Bioorg. Med. Chem. Lett 27, 2974–2981. [DOI] [PubMed] [Google Scholar]
- (14).Horton JR, Liu X, Gale M, Wu L, Shanks JR, Zhang X, Webber PJ, Bell JSK, Kales SC, Mott BT, Rai G, Jansen DJ, Henderson MJ, Urban DJ, Hall MD, Simeonov A, Maloney DJ, Johns MA, Fu H, Jadhav A, Vertino PM, Yan Q, and Cheng X (2016) Structural Basis for KDM5A Histone Lysine Demethylase Inhibition by Diverse Compounds. Cell Chem. Biol 23, 769–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Horton JR, Engstrom A, Zoeller EL, Liu X, Shanks JR, Zhang X, Johns MA, Vertino PM, Fu H, and Cheng X (2016) Characterization of a Linked Jumonji Domain of the KDM5/JARID1 Family of Histone H3 Lysine 4 Demethylases. J. Biol. Chem 291, 2631–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Horton JR, Woodcock CB, Chen Q, Liu X, Zhang X, Shanks J, Rai G, Mott BT, Jansen DJ, Kales SC, Henderson MJ, Cyr M, Pohida K, Hu X, Shah P, Xu X, Jadhav A, Maloney DJ, Hall MD, Simeonov A, Fu H, Vertino PM, and Cheng X (2018) Structure-Based Engineering of Irreversible Inhibitors against Histone Lysine Demethylase KDM5A. J. Med. Chem 61, 10588–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Johansson C, Velupillai S, Tumber A, Szykowska A, Hookway ES, Nowak RP, Strain-Damerell C, Gileadi C, Philpott M, Burgess-Brown N, Wu N, Kopec J, Nuzzi A, Steuber H, Egner U, Badock V, Munro S, LaThangue NB, Westaway S, Brown J, Athanasou N, Prinjha R, Brennan PE, and Oppermann U (2016) Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat. Chem. Biol 12, 539–545. [DOI] [PubMed] [Google Scholar]
- (18).Bavetsias V, Lanigan RM, Ruda GF, Atrash B, McLaughlin MG, Tumber A, Mok NY, Le Bihan YV, Dempster S, Boxall KJ, Jeganathan F, Hatch SB, Savitsky P, Velupillai S, Krojer T, England KS, Sejberg J, Thai C, Donovan A, Pal A, Scozzafava G, Bennett JM, Kawamura A, Johansson C, Szykowska A, Gileadi C, Burgess-Brown NA, von Delft F, Oppermann U, Walters Z, Shipley J, Raynaud FI, Westaway SM, Prinjha RK, Fedorov O, Burke R, Schofield CJ, Westwood IM, Bountra C, Muller S, van Montfort RL, Brennan PE, and Blagg J (2016) 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors. J. Med. Chem 59, 1388–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Yang Z, Qiu Q, Chen W, Jia B, Chen X, Hu H, He K, Deng X, Li S, Tao WA, Cao X, and Du J (2018) Structure of the Arabidopsis JMJ14-H3K4me3 Complex Provides Insight into the Substrate Specificity of KDM5 Subfamily Histone Demethylases. Plant Cell 30, 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Wu M, Hayward D, Kalin JH, Song Y, Schwabe JW, and Cole PA (2018) Lysine-14 acetylation of histone H3 in chromatin confers resistance to the deacetylase and demethylase activities of an epigenetic silencing complex. eLife 7, No. e37231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.