Abstract
The emergence and rapid global spread of SARS-CoV-2 mark the third such identification of a novel coronavirus capable of causing severe, potentially fatal disease in humans in the 21st century. As noted by Andersen et al. (Nature Medicine), the sequencing of proximal zoonotic ancestors to SARS-CoV-2 has aided in the identification of alleles that may contribute to the virus’ virulence in humans.
The emergence and rapid global spread of SARS-CoV-2 mark the third such identification of a novel coronavirus capable of causing severe, potentially fatal disease in humans in the 21st century. As noted by Andersen et al. (Nature Medicine), the sequencing of proximal zoonotic ancestors to SARS-CoV-2 has aided in the identification of alleles that may contribute to the virus’ virulence in humans.
Main Text
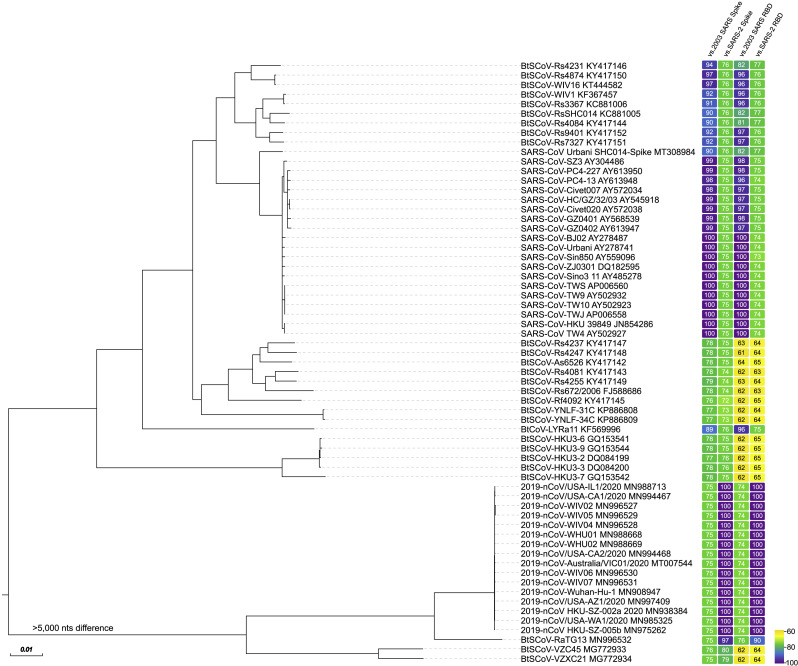
Three novel coronaviruses that are capable of causing severe disease have emerged in human populations in the 21st century. The 2003 severe acute respiratory coronavirus (SARS-CoV) and the 2012 Middle East respiratory coronavirus (MERS-CoV) foreshadowed the emergence potential of zoonotic coronaviruses. In December 2019, a strain of coronavirus that was 22% different from the 2003 SARS-CoV, later named severe acute respiratory syndrome strain 2 (SARS-CoV-2), emerged in Wuhan, China, and the resulting pandemic has caused over 3.2 million confirmed infections and over 225,000 deaths in nearly 5 months (as of April 29, 2020). The advent of high-throughput sequencing technologies has simplified the tracking of viral sequence diversity and evolution in both human and animal populations. Metagenomic surveillance of bat populations in areas near population centers in China has led to the identification of numerous civet and bat coronavirus strains closely related to the 2003 SARS-CoV. Moreover, strains like BatCoV-RaTG13 and pangolin GD/P2S1 share 96.2% and <90%, respectively, genome identity with SARS-CoV-2 (Lam et al., 2020, Zhang et al., 2020, Zhou et al., 2020) (Figure 1 ). Recently, Andersen et al. (2020) outlined the two most notable genetic features of SARS-CoV-2 that likely contribute to its virulence in humans: (1) a receptor-binding domain (RBD) that is optimized for binding to the human angiotensin-converting enzyme 2 (hACE2) molecule as the viral receptor and (2) the presence of a polybasic (furin) cleavage site at the S1-S2 boundary in the spike protein. The authors describe how these features contribute to virulence in other betacoronaviruses and then sketch two possibilities by which they may, through natural selection processes, have arisen in the coronavirus currently infecting humans worldwide.
Figure 1.
Genome Phylogeny and Spike and Receptor-Binding Domain Identity of Representative Group 2b Betacoronaviruses
The genome, spike, and RBD sequences of selected group 2b betacoronaviruses were aligned and phylogenetically compared. Sequences were aligned using free end gaps with the Blosum62 cost matrix in Geneious Prime. The phylogenetic tree was constructed from the multiple genome sequence alignment using the neighbor-joining method based on 100 replicates, also in Geneious Prime. The GenBank accession number follows each sequence name. Spike and RBD amino acid sequence identities from their respective alignments are represented by color-coded boxes to the right of each tree position, with colors ranging from yellow (~60% similarity) to purple (~100% similarity), shown in the scale in the lower right. Identities are represented as versus 2003 SARS-CoV spike (listed in figure as “2003 SARS Spike”), versus SARS-CoV-2 spike (“SARS-2 Spike”), versus 2003 SARS-CoV RBD (“2003 SARS RBD”), and versus SARS-CoV-2 RBD (“SARS-2 RBD”). The phylograms and alignments were exported from Geneious and then rendered for publication using EvolView (www.evolgenius.info) and Adobe Illustrator CC 2020.
Of the 14 residues of the RBD of 2003 SARS-CoV known to interact with hACE2 (Li et al., 2005), six residues are more critical for RBD-hACE2 binding and are host range determinants for SARS-CoV-like viruses (Wan et al., 2020). Interestingly, SARS-CoV and SARS-CoV-2 differ at 8/14 of these residues, including 5/6 critical interacting residues, and in vitro and structural studies indicate that SARS-CoV-2 has affinity for ACE2 molecules with high homology to hACE2 (Wan et al., 2020). However, while computational analyses indicate that this interaction has high affinity, the RBD sequence is clearly different from those shown to be optimal for hACE2 binding, suggesting that this binding interface is a product of a natural selection process on hACE2 or a human-like animal ACE2.
The other distinctive genetic feature SARS-CoV-2 possesses that potentially mediates virulence in humans is a polybasic (i.e., furin) cleavage site at the S1-S2 junction in the spike amino acid sequence. This site allows cleavage by proteases such as furin and is another factor that can determine viral infectivity and host range (Nao et al., 2017). While such cleavage sites have not been detected in other lineage B betacoronaviruses, they have been identified in betacoronaviruses in lineages A and C (in HCoV-HKU1 and MERS-like CoVs, respectively). Moreover, the O-linked glycans likely associated with the polybasic site may alter immunogenicity in response to herd immunity within natural animal hosts, which is likely not necessary in naive human populations (Bagdonaite and Wandall, 2018). Thus, the functional significance of the polybasic cleavage site awaits characterization.
In light of social media speculation about possible laboratory manipulation and deliberate and/or accidental release of SARS-CoV-2, Andersen et al. theorize about the virus’ probable origins, emphasizing that the available data argue overwhelmingly against any scientific misconduct or negligence (Andersen et al., 2020). As has been previously described, the SARS-CoV-genome contains over 1,200 nucleotide changes as compared with RaTG13, its closest relative. Moreover, the RaTG13 S glycoprotein is 97% identical at the amino acid level to the SARS-CoV-2 S glycoprotein (Figure 1), and it encodes an RBD that is not optimized for hACE2 interaction (Wan et al., 2020). Anderson cites these genetic and biological data as strong evidence against deliberate generation, and the arguments are compelling. It is noteworthy that many early COVID-19 cases had not visited the Huanan wet market, suggesting that either the index cases occurred earlier and were not identified or that these sites were not major sites of epidemic expansion. How, then, did the virus emerge? Anderson et al. cite multiple lines of strong evidence that argue, instead, in favor of various mechanisms of natural selection, either in an animal host before the virus was transmitted to humans or in humans after the zoonotic transmission event(s). These possibilities will be reviewed below. Nevertheless, speculation about accidental laboratory escape will likely persist, given the large collections of bat virome samples stored in labs in the Wuhan Institute of Virology, the facility’s proximity to the early outbreak, and the operating procedures at the facility (Zeng et al., 2016). Transparency and open scientific investigation will be essential to resolve this issue, noting that forensic evidence of natural escape is currently lacking, and other explanations remain reasonable.
Given the high correlation of many, but not all, of the early cases of COVID-19 disease in Wuhan with the Huanan wet market, it is possible that an animal reservoir of the virus was present at that location, and genome evolution analyses have suggested an earlier time of origin (Zhang et al., 2020). This scenario would have allowed for the establishment of earlier human-to-human transmission networks independent of the open market. The BtCoV-RaTG13 virus is the closest currently characterized relative to SARS-CoV-2, and it encodes 7/14 changes in the S glycoprotein RBD. More distantly related coronavirus genome sequences have also been identified in illegally imported Malayan pangolins (Lam et al., 2020), and while these strains encode 8/14 changes in the RBD interface residues, they do retain 6/6 of the most critical ACE2-interacting RBD residues with SARS-CoV-2 (Lam et al., 2020, Zhang et al., 2020). The presence of highly related viral sequences in diverse species argues strongly for natural selection being the major driving force for the optimization of the SARS-CoV-2 spike RBD among these related viruses. While a more homologous zoonotic relative has yet to be identified that shares the polybasic site with SARS-CoV-2, the sheer diversity of coronavirus sequences that have been identified in bat populations in China and worldwide indicates that zoonotic reservoirs are drastically under-sampled and under-characterized. Clearly, additional studies into the diversity of zoonotic coronavirus strains are essential for global public health preparedness, for the development of countermeasures, and to clarify the origins of SARS-CoV-2.
Anderson et al. also argue that it is possible that a progenitor coronavirus jumped to humans prior to acquiring its polybasic site and key hACE2 interaction residues, acquiring these features through undetected human-to-human transmission events prior to the first documented cases of COVID-19 disease that triggered human surveillance systems (Wu et al., 2020, Zhou et al., 2020). In support, antibodies targeting the group 2b SARS-like coronaviruses can be detected in people living and working near or in bat hibernacula in China, suggesting frequent exposures in a rural setting. As SARS-CoV-2 infections are frequently asymptomatic or mild, initial exposures would easily have allowed for extended silent transmission events in rural settings prior to the emergence of a strain that could support sustained human-to-human transmission, especially when brought into an urban setting.
As emphasized by the authors, retroactive mapping of the paths of emergence of human pathogens is critical, especially in light of the global emergency fomented by the current pandemic. The presence of abundant sources of coronaviruses in zoonotic populations and the continuing and advancing encroachment of humans into animal habitats argue that emergence events will only become more common in future years. Indeed, prior to 2003, only two human coronaviruses were known: HCoV-OC43 and HCoV-229E, which cause mild, cold-like disease. After 2003, heightened surveillance retroactively identified two additional human coronaviruses, HCoV-NL63 and HCoV-HKU1. Nearly 10 years separated the documented emergence of SARS-CoV in 2003 and MERS-CoV in 2012, and just under 8 years have now separated the emergences of MERS-CoV and SARS-CoV-2. These patterns suggest that the global ecology has shifted and now favors the continued emergence of zoonotic coronaviruses, resulting in micro-outbreaks, continued low-level epidemics, or global pandemics.
In summary, Andersen et al. have outlined many of the key elements of the SARS-CoV-2 spike protein that could be mediating its extraordinary global expansion and summarize how the virus may have emerged from zoonotic populations (Andersen et al., 2020). The authors do not discuss the potential role for other less defined virulence determinants in the spike protein that alter host signaling networks and cytokine levels that may be associated with disease or transmission frequency. The virus, which is similar to yet distinct from the two previous zoonotic coronaviruses from the 21st century, SARS-CoV and MERS-CoV, marks the third emergence of a coronavirus that is capable of causing severe disease within the last 20 years. Novel bat coronaviruses have also emerged in swine populations in the past few years. As the pace of coronavirus emergence appears to be accelerating, these data not only underscore a common event in nature but also emphasize the urgency to develop vaccines and therapeutics with broad efficacy. Thus, studies characterizing the SARS-CoV-2 neutralizing epitopes and identifying broadly cross-neutralizing epitopes are clear priorities for immunotherapeutic and vaccine countermeasure design. This work should be performed with due caution, ensuring that putative enhancing epitopes are likewise identified and avoided in the course of vaccine design to minimize the risk of potentiating disease. Additionally, T cell epitopes should be identified across outbred populations to determine the key correlates of protective immunity. A key priority in combating the current pandemic and constructing readiness programs for future emergence events is the development of broadly effective medical countermeasures and therapeutics that can be stockpiled as insurance against future viral emergence events to prevent the human loss and economic and social catastrophe of global pandemics.
References
- Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26:450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28:443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T.-Y., Shum M.H.-H., Zhu H.-C., Tong Y.-G., Ni X.-B., Liao Y.-S., Wei W., Cheung W.Y.-M., Li W.-J., Li L.-F. Identifying SARS-CoV-2 Related Coronaviruses in Malayan Pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. Published online March 26, 2020. [DOI] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Nao N., Yamagishi J., Miyamoto H., Igarashi M., Manzoor R., Ohnuma A., Tsuda Y., Furuyama W., Shigeno A., Kajihara M. Genetic Predisposition To Acquire a Polybasic Cleavage Site for Highly Pathogenic Avian Influenza Virus Hemagglutinin. MBio. 2017;8 doi: 10.1128/mBio.02298-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020;94:94. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L.P., Gao Y.T., Ge X.Y., Zhang Q., Peng C., Yang X.L., Tan B., Chen J., Chmura A.A., Daszak P., Shi Z.L. Bat Severe Acute Respiratory Syndrome-Like Coronavirus WIV1 Encodes an Extra Accessory Protein, ORFX, Involved in Modulation of the Host Immune Response. J. Virol. 2016;90:6573–6582. doi: 10.1128/JVI.03079-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Probable Pangolin Origin of SARS-CoV-2 Associated With the COVID-19 Outbreak. Curr. Biol. 2020;30:1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]