Summary
The phenotypic and functional dichotomy between IRF8+ type 1 and IRF4+ type 2 conventional dendritic cells (cDC1s and cDC2s, respectively) is well accepted; it is unknown how robust this dichotomy is under inflammatory conditions, when additionally monocyte-derived cells (MCs) become competent antigen-presenting cells (APCs). Using single-cell technologies in models of respiratory viral infection, we found that lung cDC2s acquired expression of the Fc receptor CD64 shared with MCs and of IRF8 shared with cDC1s. These inflammatory cDC2s (inf-cDC2s) were superior in inducing CD4+ T helper (Th) cell polarization while simultaneously presenting antigen to CD8+ T cells. When carefully separated from inf-cDC2s, MCs lacked APC function. Inf-cDC2s matured in response to cell-intrinsic Toll-like receptor and type 1 interferon receptor signaling, upregulated an IRF8-dependent maturation module, and acquired antigens via convalescent serum and Fc receptors. Because hybrid inf-cDC2s are easily confused with monocyte-derived cells, their existence could explain why APC functions have been attributed to MCs.
Keywords: dendritic cell, monocyte, transcription factor, IRF8, virus, CD64, inf-cDC2, inflammation, type 1 interferon, Fc receptor, convalescent serum
Graphical Abstract
Highlights
-
•
Type I interferon drives differentiation of inf-cDC2s that closely resemble MCs
-
•
Inf-cDC2s prime CD4+ and CD8+ T cells, whereas MCs lack APC function
-
•
Inf-cDC2s internalize antibody-complexed antigen via Fc receptors
-
•
IRF8 controls maturation gene module in inf-cDC2s
The dichotomy between type 1 and 2 conventional DCs under steady-state conditions is well defined. Bosteels et al. demonstrate that, upon inflammation, cDC2s acquire a hybrid inf-cDC2 phenotype, sharing phenotype, gene expression, and function with cDC1s and monocyte-derived cells, to optimally boost CD4 and CD8 immunity via Fc receptors.
Introduction
Type 1 and type 2 conventional dendritic cells (cDC1s and cDC2s, respectively) and plasmacytoid DCs (pDCs) are defined by expression of cell surface markers and develop from well-known common DC and pre-cDC progenitors through the action of lineage-defining transcription factors (TFs) (Bosteels and Scott, 2020, Grajales-Reyes et al., 2015, Guilliams et al., 2014a, Guilliams et al., 2016, Murphy et al., 2016, Schlitzer et al., 2015). The TFs interferon regulatory factor 8 (IRF-8) and Batf3 drive the development of chemokine receptor XCR1-expressing cDC1s, which have the capacity to present and cross-present antigens to CD8+ T cells and are a copious source of interleukin-12 (IL-12) (Bagadia et al., 2019, Bajaña et al., 2016, Durai et al., 2019, Edelson et al., 2010, Everts et al., 2016, Grajales-Reyes et al., 2015, Sichien et al., 2016). On the other hand, IRF4 drives the development and terminal differentiation of the CD11b+CD172a+-expressing cDC2 lineage, which is more specialized in polarizing CD4+ T helper (Th) cell responses and providing help to B cells (Bajaña et al., 2012, Gao et al., 2013, Krishnaswamy et al., 2017, Kumamoto et al., 2013, Vander Lugt et al., 2014, Naessens et al., 2020, Satpathy et al., 2013, Schlitzer et al., 2013, Tussiwand et al., 2015, Williams et al., 2013).
Many studies have dissected the antigen-presenting functions of cDC1s and cDC2s by sorting by unique surface marker expression (Ballesteros-Tato et al., 2010, GeurtsvanKessel et al., 2008, Helft et al., 2012, Lehmann et al., 2017) or conditional inactivation of Irf8 or Irf4 in Itgax-expressing cells to target cDC1s or cDC2s, respectively (Baptista et al., 2019, Van der Borght et al., 2018, Deckers et al., 2017, Ivanov et al., 2016, Luda et al., 2016, Mayer et al., 2017, Persson et al., 2013). However, the selectivity and validity of these approaches mainly derives from studies that have measured cell surface markers and IRF expression in the steady state without properly questioning how expression is altered by inflammation. Moreover, as soon as tissue inflammation is present, Ly6ChiCD11b+CD172a+ monocytes enter antigen-exposed barrier sites and lymph nodes (LNs). Monocytes can then rapidly upregulate the expression of Major Histocompatibility Complex class II (MHCII) and CD11c while downregulating expression of Ly6C. These cells are known by various names, like monocyte-derived cells (MCs), monocyte-derived DCs (moDCs), or inflammatory DCs, and can be easily confused with CD11b+CD172a+ cDC2s, particularly because they also express CD11b, CD172a, and intermediate amounts of IRF4 (León et al., 2007, Menezes et al., 2016, Naik et al., 2006, Plantinga et al., 2013). Although MafB lineage tracing studies suggest that MCs are much more related to macrophages than DCs, they perform well in ex vivo antigen presentation assays, leading to their classification as professional antigen-presenting cells (APCs) and their designation as moDCs (Cheong et al., 2010, Kool et al., 2008a, Kool et al., 2008b, León et al., 2007, Sallusto and Lanzavecchia, 1994, Wu et al., 2016). Although cDCs readily migrate to draining nodes, MCs are usually less migratory.
It is now assumed that MCs and macrophages can be readily discriminated from cDCs based on their surface expression of the high-affinity Fc gamma receptor CD64, by staining with the MAR-1 clone of the anti-FcεRI antibody, or by expression of Tyrosine-protein kinase Mer (MerTK) and CD88 (Gautier et al., 2012, Hammad et al., 2010, Nakano et al., 2015, Plantinga et al., 2013, Tamoutounour et al., 2012, Tamoutounour et al., 2013, Tang et al., 2019). However, CD64 has been reported to also identify a subset of kidney cDCs in the steady state (Schraml et al., 2013). Understanding which APCs express Fc receptors is important because uptake of antigen via convalescent serum or immune complexes is an effective way of targeting antigen to APCs during an ongoing primary or recall immune response (Guilliams et al., 2014b, Lehmann et al., 2017).
We found significant overlap in marker and TF expression in cDCs and MCs. Bona fide inflammatory cDC2s (inf-cDC2s) acquired characteristics traditionally defining cDC1 and macrophages in a type I interferon (IFN)-dependent manner. By also acquiring shared functions such as IL-12 production and Fc receptor-mediated antigen uptake, inf-cDC2s optimally primed CD4+ and CD8+ T cell-mediated immunity to respiratory virus infection.
Results
CD26+CD64+ MAR-1+ DCs Accumulate in Tissues and LNs of Virus-Infected Mice
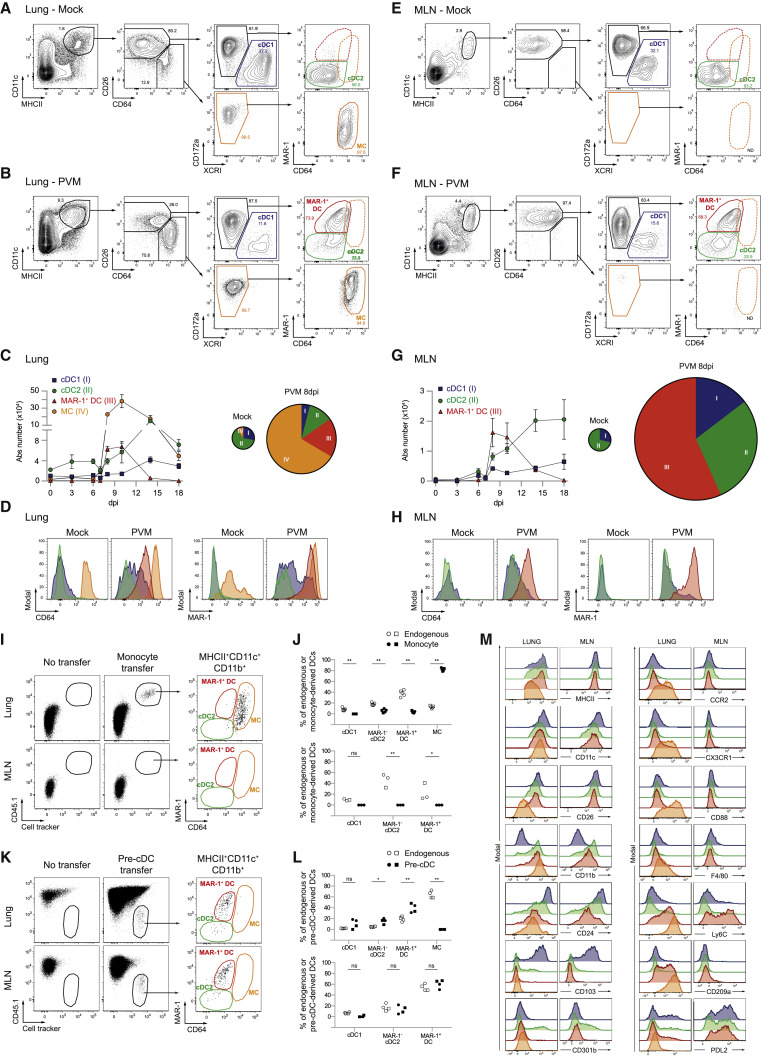
DC subsets and CD11c+MHCII+ MCs were studied in naive (mock-infected) lungs and lungs of mice infected with the single-stranded RNA virus pneumonia virus of mice (PVM), a virus closely related to human respiratory syncytial virus (RSV), which causes a severe acute respiratory distress syndrome (ARDS)-like disease (Vandersarren et al., 2017). Bona fide cDCs were separated from MCs by surface staining for CD26 and CD64, respectively, whereas XCR1 and CD172a (Sirpα) were used to separate cDC1s from cDC2s, respectively (Guilliams et al., 2016). We additionally stained cells with the antibody MAR-1 raised against FcεRI, also known to bind CD64 and FcγRIV on DCs and shown previously to mark inflammatory DCs (Grayson et al., 2007, Hammad et al., 2010, Tang et al., 2019).
In mock-infected mice, CD26+ XCR1+ cDC1s and CD172a+ cDC2s made up around one- and two-thirds of the lung cDC population, respectively, whereas CD26loCD172a+CD64hi MCs were barely recovered in the MHCII+CD11c+ cell population (Figure 1 A; summarized in Figure 1C). At 8 days post infection (dpi) with PVM (Figure 1B), when the viral load is highest in this model, total lung MHCII+CD11c+ cells had expanded greatly. The proportion of cDC1s and cDC2s in this cell fraction had decreased, whereas the proportion of MCs was significantly increased (Figure 1C), and expression of MAR-1 was upregulated (Figure 1D). Another DC population appeared, expressing CD26 and CD172a like cDC2s but was marked by expression of CD64 and MAR-1 (Figures 1B and 1D), which we termed CD26+CD64+MAR-1+ DCs. The intensity of CD64 staining on CD26+CD64+ DCs was between that of cDCs and that of MCs (Figure 1D), but without use of CD26, it would be very difficult to separate these cells from MCs. Following viral clearance, CD26+CD64+MAR-1+ DCs were no longer identifiable in the lung, but cDC and MC numbers remained elevated at 18 dpi (Figure 1C).
Figure 1.
CD26+MAR-1+CD64+ DCs Are Induced after Pneumovirus Infection
(A and B) Gating strategy of lung DC subsets pre-gated on live CD3−CD19− non-autofluorescent cells in mock-infected controls (A) or 8 dpi with PVM (B). cDC1s are shown in blue, cDC2s in green, MAR-1+ DCs in red, and MCs in orange gates. These colored gates are maintained throughout the manuscript.
(C) Kinetics of different lung DC subset numbers upon PVM infection (left panel) and pie charts depicting relative distribution of DC subsets in the lung upon mock or 8 dpi PVM infection (right panel) (see also Figure S1A). Error bars indicate ± SEM.
(D) Surface expression of CD64 and MAR-1 on different DC subsets in the lung 8 dpi with PVM.
(E and F) Gating strategy of migratory MHCIIhi DC subsets in MLNs pre-gated on live CD3−CD19− cells in mock-infected controls (E) or 8 dpi with PVM (F).
(G) Kinetics of different migratory DC subset numbers in MLNs upon PVM infection (left panel) and pie charts depicting relative distribution of DC subsets in MLNs upon mock or 8 dpi PVM infection (right panel) (see also Figure S1B).
(H) Surface expression of CD64 and MAR-1 on different DC subsets in MLN 8 dpi with PVM.
(I) CD45.1.2 monocytes were sorted from eFl450+ cell tracker-labeled BM and transferred intravenously (i.v.) into CD45.2 Ccr2−/− recipient mice at 1 dpi with IAV. Four days later, donor cells were identified in the lungs and MLNs (see also Figures S1C–S1H).
(J) Distribution of different DC subsets in the endogenous population (white) and adoptively transferred monocyte-derived population (black) in the lungs (top panel) and MLNs (bottom panel).
(K) Pre-cDCs were sorted from eFl450+ CellTrace™ Violet (CTV)-labeled BM from FLT3L-treated CD45.2 WT mice and transferred i.v. into CD45.1.2 WT mice at 1 dpi with IAV. Four days later, donor cells were identified in the lungs and MLNs (see also Figures S1C–S1F).
(L) Distribution of different DC subsets in the endogenous population (white) and adoptively transferred pre-cDC-derived population (black) in the lungs (top panel) and MLNs (bottom panel).
(M) Histograms showing surface expression of commonly used DC and macrophage markers on different DC subsets in the lungs and MLNs 8 dpi with PVM.
(A–H) Data are representative of at least 2 or 3 independent experiments with 4–6 mice per group. The size of the pie chart is proportional to the absolute number of DCs in the lungs (C) or MHCIIhi DCs in MLNs (G).
(I–L) Data pooled from 2 independent experiments (circles and squares, n = 5 in total), analyzed with a two-way ANOVA with Sidak correction for multiple comparisons. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant.
The same gating strategy was applied to the migratory MHCIIhi DCs in lung draining mediastinal LNs (MLNs) (Figures 1E and 1F). In mock-infected mice, cDC2s made up the highest fraction of MLN migratory DCs (Figures 1E and 1G). At 8 dpi, there was a robust increase in cDC1s and cDC2s and a substantial population of CD26+CD64+ DCs, representing more than half of the total DC population in the MLNs. These CD26+MHCII+CD11c+ cells expressed CD64 and MAR-1 at an intensity comparable with lung equivalents (Figures 1F, 1D, and 1H) and were no longer detectable in MLNs following recovery from viral infection (Figure 1G). Despite their accumulation in the lungs, CD26loCD64hi MCs did not accumulate in MLNs (Figures 1E and 1F). Similar changes occurred in the lungs and LNs of mice infected with mild influenza A virus (IAV; strain X31) but with more rapid kinetics (Figures S1A and S1B). Thus, respiratory viral infection induces a population of CD26+ DCs that express CD64 and MAR-1, usually found on MCs and macrophages.
CD26+CD64+MAR-1+ DCs Are Bona Fide cDC2s
We next questioned whether the CD26+CD64+MAR-1+ DC population was of monocyte or pre-cDC origin. Monocytes were sorted from the bone marrow (BM) of CD45.1 wild-type mice and transferred into virus-infected CD45.2 Ccr2 −/− mice. Four days after monocyte transfer, when viral titers are maximal in the IAV model (GeurtsvanKessel et al., 2008), the transferred cells were present exclusively in the lung MC gate (Figure 1I). MCs could not be recovered in MLNs, showing lack of migratory capacity (Figures 1I and 1J). Conversely, sorted CD45.2 BM pre-cDCs gave rise to cDC1s and cDC2s 4 days after transfer, but not to the CD64hiMAR-1+ MC population (Figures 1K and 1L). Pre-cDCs also gave rise to the MAR-1+CD64+ DC population in the lung and MLNs, demonstrating these cells to be genuine conventional DCs with a characteristic capacity to migrate to LNs (Figures 1K and 1L).
Conventional DCs typically depend on Flt3 ligand, and virus-infected Flt3l −/− mice had reduced numbers of all cDC subsets, including MAR-1+ DCs (Figures S1C–S1F). Granulocyte-macrophage colony-stimulating factor (GM-CSF) acting on Csf-2r has been shown to play a cell-intrinsic pro-survival role in resident cDC homeostasis in vivo (Greter et al., 2012). In wild-type (WT):Csf2rb −/− chimeras, Csf2rb signaling was, however, not intrinsically required for generation of MAR-1+ DCs and MCs (Figure S1I). Dependency on the chemokine receptor CCR2 is widely used to define monocyte descent (Serbina and Pamer, 2006), but it has been shown that subtypes of tissue CD11b+cDC2s also depend on CCR2 (Nakano et al., 2017, Scott et al., 2015). In WT:Ccr2 −/− chimeras infected with the virus, cDC1s derived equally well from both donors, whereas MCs were generated preferentially from the WT donor component. MAR-1+CD11b+ cDCs had an intermediate dependence on CCR2 (Figures S1G and S1H).
Compared with MCs, pre-cDC-derived MAR-1+ DCs expressed higher amounts of MHCII and CD26 protein (Figure 1M). In addition, CX3CR1 and CD88 were exclusively expressed by MCs (Nakano et al., 2015). Ly6C has often been used to distinguish cDC2s from MCs (GeurtsvanKessel et al., 2008, Grayson et al., 2007, Hammad et al., 2009). However, both inf-cDC2s and MCs expressed this marker (Figure 1M). In the lungs, CD209a Dendritic cell-specific ICAM-3-grabbing non-integrin (DC-SIGN) was expressed by inf-cDC2s and MCs despite earlier reports of MC exclusivity (Cheong et al., 2010). CD301b expression was only found on a fraction of cDC2s (Gao et al., 2013, Kumamoto et al., 2013).
Thus, in virally infected tissues, CD26+CD64+MAR-1+ DCs are pre-cDC-derived cells that express CD172a and CD11b but not XCR1, CD103, or CD88, identifying them as a subset of cDC2s with great potential to migrate to draining MLNs under conditions of inflammation. We therefore propose to call these cells inf-cDC2s.
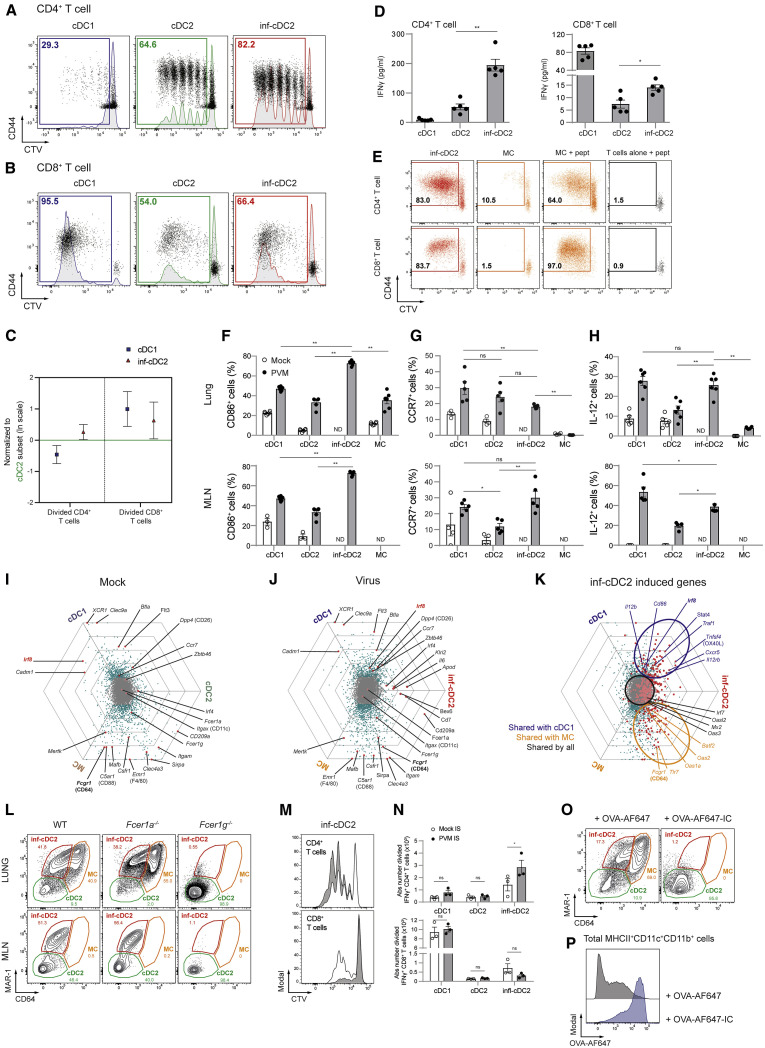
Inf-cDC2s Present Antigens to CD4+ and CD8+ T Cells
We created mice transgenic for a T cell receptor (TCR) recognizing a Major Histocompatibility Complex class I (MHCI)-restricted (N339–347) or MHCII-restricted (M37-47), PVM-derived T cell epitope by CD8+ and CD4+ T cells, respectively (Figures S1K–S1P; Vandersarren et al., 2017, Walsh et al., 2013). After 4 days of co-culture, cDC2s retrieved from MLNs at 8 dpi were more efficient in inducing virus-specific CD4+ T cell proliferation compared with cDC1s (Figure 2 A). Inf-cDC2s induced the highest degree of CD4+ T cell proliferation (Figures 2A–2C), whereas cDC1s were superior in inducing proliferation of CD8+ T cells. Inf-cDC2s performed better than their classical cDC2 counterparts in stimulating CD8+ T cell immunity (Figures 2B and 2C). IFN-γ was produced in much larger quantities when CD4+ T cells were co-cultured with inf-cDC2s compared with cDC2s or cDC1s (Figure 2D). In contrast, IFN-γ production by CD8+ T cells was higher upon stimulation with cDC1s compared with cDC2s, but again, inf-cDC2s were more potent in inducing IFN-γ production from dividing CD8+ T cells (Figure 2D). MCs that were sorted from the lungs and carefully separated from inf-cDC2s could not induce proliferation of naive CD4+ or CD8+ T cells whereas lung inf-cDC2s readily did (Figure 2E). However, when exogenous preprocessed peptide was added to MCs, they readily induced proliferation (Figure 2E). Thus, inf-cDC2s, which represent the dominant DC population in lung-draining LNs at the peak of PVM infection, can optimally prime CD4+ and CD8+ T cell responses to virus.
Figure 2.
Activated inf-cDC2s Share Characteristics with cDC1 and MC Subsets
(A and B) Proliferation profile of CTV-labeled CD4+ (A) and CD8+ (B) PVM-specific TCR transgenic T cells cocultured for 4 days with different migratory cDC subsets sorted from 4–6 pooled MLNs 8 dpi with PVM (see also Figures S1K–S1P).
(C) Number of divided CD4+ and CD8+ PVM TCR transgenic T cells cocultured for 4 days with different migratory cDC subsets sorted from MLNs 8 dpi with PVM relative to the cDC2 subset, which is set to 0. Error bars represent 95% confidence intervals. Data were pooled from 3 independent experiments (n = 5–7).
(D) IFN-γ measured in supernatants of cocultured CD4+ and CD8+ PVM TCR transgenic T cells for 4 days with different MLN-derived cDC subsets sorted 8 dpi with PVM. Data are representative of 3 independent experiments. Error bars indicate ± SEM. Mann-Whitney U test; ∗p < 0.05, ∗∗p < 0.01.
(E) Proliferation profile of CTV-labeled CD4+ and CD8+ PVM TCR transgenic T cells cocultured for 4 days with T cells alone, inf-cDC2s, or MCs sorted from 3 pooled lungs 8 dpi with PVM. For peptide controls, 1 μg/mL of CD4+ (M37–M47) or CD8+ (N339–N347) immunodominant epitope was added ex vivo.
(F–H) Percentage of CD86-expressing (F) and CCR7-expressing (G) and IL-12-producing (H) lung or migratory DC subsets in MLNs upon mock infection (white circles) or 10 dpi with PVM (black dots). For IL-12 staining, samples were restimulated ex vivo for 6 h. Data are representative of 2 independent experiments with 3–6 mice per group, analyzed with a two-way ANOVA with Sidak correction for multiple comparisons. Error bars indicate ± SEM. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant. (see also Figure S1J).
(I–K) To visualize DEG between DC subsets sorted from the lungs 4 dpi after mock (I) and IAV infection (J) and those induced in inf-cDC2s (K), each gene was plotted in a hexagonal triwise diagram in which the direction of a point represents the relative higher expression in one or two populations, whereas the distance from the origin represents the magnitude of expression. Genes that are 32-fold or more differentially expressed are plotted on the outer grid line. Genes represented by gray dots in the center of the triwise plot are not differentially expressed (see also Figures S2A–S2D).
(L) MAR-1 and CD64 staining of live CD3−CD19−MHCII+CD11c+CD172a+ cells 8 dpi with PVM in the lungs and MLNs of WT, Fcer1a−/−, and Fcer1g−/− mice.
(M) Proliferation profile of CTV-labeled CD4+ and CD8+ PVM TCR transgenic T cells cocultured for 4 days with different migratory cDC subsets sorted from 4 pooled MLNs 8 dpi with PVM from mice that received mock (white) or PVM (gray) IS i.t. 6 dpi.
(N) Absolute number IFNγ+ CD4+ and CD8+ PVM TCR transgenic T cells cocultured for 4 days with different migratory cDC subsets sorted from 4 pooled MLNs 8 dpi with PVM from mice that received mock (white) or PVM (gray) IS i.t. 6 dpi. Two-way ANOVA with Sidak correction for multiple comparisons. Error bars indicate ± SEM. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant. (see also Figures S3A and S3B).
(O and P) MAR-1 and CD64 staining of live CD3−CD19−MHCII+CD11c+CD11b+ cells (O) 8 dpi with PVM in lungs of mice treated i.t. at 6 dpi with 50 μg Alexa Fluor 647 (AF647)-labeled OVA administered alone (left panel) or as OVA-AF647-IgG2c-IC (right panel) and the respective uptake of OVA-AF647 by these cells (P) (see also Figures S3C and S3D).
Inf-cDC2s Share Characteristics with cDC1s and MCs
In PVM-infected mice, inf-cDC2s expressed significantly higher amounts of the DC maturation marker CD86 in the lungs and MLNs compared with cDC1s, cDC2s, and MCs (Figure 2F). Upon activation, antigen-bearing cDCs migrate to MLNs in a CCR7-dependent manner (Hintzen et al., 2006, Vermaelen et al., 2001). Surface CCR7 expression was induced by infection on cDC1s, cDC2s and inf-cDC2s but not MCs (Figure 2G), and CCR7 was required for migration of inf-cDC2s to MLNs (data not shown). In steady-state mice, IL-12 is typically produced by the cDC1 subset (Everts et al., 2016). Upon virus infection, however, lung and MLN inf-cDC2s gained enhanced capacity to produce IL-12 compared with cDC2s after 6 h of ex vivo restimulation, to a degree comparable with cDC1s (Figure 2H), explaining why inf-cDC2s were good inducers of IFN-γ production in CD4+ and CD8+ T cells (Figure 2D; Figure S1J).
We next performed a RNA microarray analysis on sorted lung APC subsets to study which other features might be shared between inf-cDC2s, cDC1s, and MCs (Figure S2A). In mock-infected mice, Xcr1, Clec9a, Cadm1, and Irf8 were highly specific for cDC1s and Mertk, Emr1 (F4/80), and Fcgr1 (CD64) for MCs (Figure 2I). No cDC2-specific genes could be defined because most genes were shared with cDC1s (Dpp4 [CD26] and Zbtb46) or MCs (Sirpa, Itgam, Csfr1, and Clec4a3). Upon infection, a small fraction of the genes encoding for (surface) markers remained cDC1 specific (e.g., Xcr1, Cadm1, Clec9a, Gcsam, Zdhhc2, Tlr3, and Itgae) or MC specific (e.g., Mertk, Mafb, and Emr1) (Figure 2J; Figures S2B–S2D). As for cDC2s in the steady state, there were no genes specific to inf-cDC2s. All genes that were upregulated in inf-cDC2s compared with steady-state cDC2s were shared with cDC1s (e.g., Irf8, Il12b, Il15, Cd80, Cd86, and Tnfrsf4), MCs (e.g., Fcgr1 and Tlr7), or both subsets (Figure 2K; Figures S2B–S2D). Thus, inf-cDC2s induced upon virus infection were activated cDC2s that acquired a hybrid phenotypic, functional, and transcriptomic identity with characteristics of cDC1s and MCs (Figures 2A–2K; Figures S2A–S2D).
Inf-cDC2s Internalize Antigen via Activating Fc Receptors
Although Fcgr1 and Fcer1g were clearly shared between inf-cDC2s and MCs (Figures 2I and 2J), we did not detect Fcer1a, coding for the high-affinity IgE receptor FcεRIα recognized by the MAR-1 antibody on mast cells and basophils. In addition, MAR-1 staining was intact in inf-cDCs of Fcer1a −/− virus-infected mice (Figure 2L) and abolished in Fcer1g −/− mice (Figure 2L). Thus, as reported previously (Tang et al., 2019), MAR-1 recognizes an unknown epitope of an activating Fc receptor on inf-cDC2s and MCs. Because activating Fc receptors like CD64 and CD16b (FcγRIV) can target antigens to macrophages and DCs via binding surface immunoglobulin G (IgG) or uptake of immune complexes (Guilliams et al., 2014b, Lehmann et al., 2017), we next tested whether this would be a specialized function of inf-cDC2s. Heat-inactivated convalescent immune serum (IS) from PVM-infected mice (PVM-IS, obtained at 15 dpi) or control serum from mock-infected mice was transferred to another cohort of infected mice at 6 dpi, and DC subsets were sorted from MLNs 36 h later. Compared with mock serum, PVM-IS transfer led to enhanced capacity of inf-cDC2s to present antigen to CD4+ T cells, which led to higher T cell division (Figure 2M) and increased numbers of IFN-γ producing T cells (Figure 2N). This effect was not seen when cDC2s were sorted from Fcer1g−/− mice (Figures S3A and S3B). Transfer of convalescent PVM-IS did not enhance the capacity of cDC1s or MAR-1- cDC2s to present viral antigen.
Immune complexes (ICs) were made from fluorescently labeled ovalbumin (OVA)-Alexa Fluor 647 mixed with two mouse OVA-specific IgG2c monoclonal antibodies (mAbs) and injected intratracheally at 6 dpi. Strikingly, 48 h later, all CD64 and MAR-1 staining had disappeared from the total pool of lung cDC2s as well as MCs when ICs were injected (Figure 2O). This was due to Fc receptor-mediated internalization by inf-cDC2s because the amount of internalized OVA-Alexa Fluor 647 in MHCII+CD11c+CD11b+ cells comprising cDC2s and MCs was much higher following injection of OVA-Alexa Fluor 647-IC compared with OVA-Alexa Fluor 647 alone (Figure 2P). Intracellular FcgR staining allowed us to discriminate the different DC subsets, and acquisition of CD64 and MAR-1 by inf-cDC2s licensed these cells to more efficiently internalize OVA-Alexa Fluor 647 (AF647)-ICs compared with OVA-AF647 alone (Figures S3C and S3D). Thus, inf-cDC2s induced by respiratory virus infection acquire antigens via convalescent serum components and Fc receptors to primarily boost CD4+ Th cell immunity.
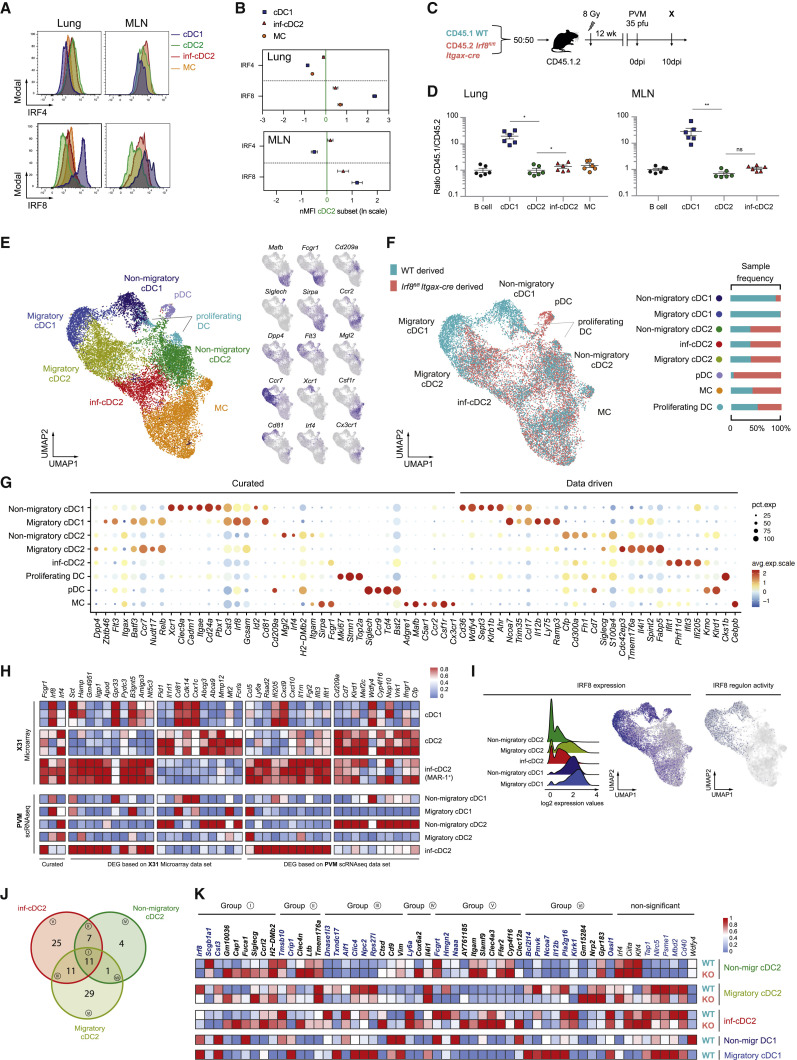
Inf-cDC2s Develop Normally in the Absence of IRF4 and IRF8
IRF8 drives cDC1 development, whereas IRF4 drives cDC2s. In line with the cDC2 origin of inf-DCs, IRF4 was expressed at baseline and minimally altered by infection (Figures 3A and 3B; Figure S2F). Lung inf-cDC2s upregulated IRF8 relative to their MAR-1− cDC2 counterparts, and this was even more marked after migration of inf-cDC2s to the MLN, where the intensity of IRF8 staining almost reached that of cDC1s (Figure 3A and 3B; Figure S2G). We next questioned whether IRF8 was needed to drive inf-cDC2s development. Irf8 fl/fl Itgax-cre mice lack cDC1, potentially affecting viral clearance and confounding comparisons with WT mice because of immunopathology. We therefore generated CD45.1 WT:CD45.2 Irf8 fl/fl Itgax-cre mixed BM chimeric mice (Figure 3C) which were infected with PVM 12 weeks later. At 10 dpi, cDC1s in the lungs and MLNs were almost exclusively derived from the WT CD45.1 BM compartment (Figure 3D). In contrast, cDC2s and inf-cDC2s were derived equally well from the WT and Irf8−/− BM compartments, although inf-cDC2s had a slightly higher WT:Irf8 fl/fl Itgax-cre ratio compared with cDC2s (Figure 3D). Lack of IRF4 in CD11c+ cells of Irf4 fl/fl Itgax-cre mice resulted in a reduction in cDC2s in the lungs and a severe reduction in migratory cDC2s in MLNs in the steady state (Bajaña et al., 2016). Although lung inf-cDC2s were not affected by loss of IRF4 in infected mice, there was less migration of CCR7+ cDC2s and inf-cDC2s in Irf4 fl/fl Itgax-cre mice to MLNs (Figures S4A–S4E). A subset of CD301b(MGL2)+CD24+cDC2s known to induce Th2 cell responses (Gao et al., 2013, Kumamoto et al., 2013) was lower in the lungs and MLNs of Irf4 fl/fl Itgax-cre mice; however, this was a minor population of all cDC2s (Figure S4F).
Figure 3.
IRF8 Controls the Gene Network in inf-cDC2s
(A) Expression of IRF4 (top panel) and IRF8 (bottom panel) by different DC subsets in the lungs and MLNs 10 dpi with PVM.
(B) Normalized Median Fluoresent Intensity (MFI) for IRF4 and IRF8 relative to the cDC2 subset, which is set to 0 for DC subsets in the lungs (top panel) or the different migratory cDC subsets in MLNs (bottom panel) 10 dpi with PVM. Error bars represent 95% confidence intervals. Data are representative of 3 independent experiments with 4–6 mice per group.
(C) Schematic representation of CD45.1 WT:CD45.2 Irf8fl/flItgax-cre BM chimeras.
(D) Normalized CD45.1/CD45.2 ratio relative to B cells of DC subsets in the lungs (left panel) and migratory cDCs in MLNs (right) 10 dpi with PVM. Data are representative of 2 independent experiments with 4–6 mice per group, analyzed with a one-way ANOVA with Sidak correction for multiple comparisons. Error bars indicate ± SEM. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant.
(E) UMAP plot of scRNA-seq data of pooled, sorted, live CD3-CD19-SiglecF-CD11c+MHCII+ cells from lungs of CD45.1 WT:CD45.2 Irf8fl/flItgax-cre chimeric mice 10 dpi with PVM, showing assigned clusters, and UMAPs showing expression of key annotation markers by color (gray, low expression; blue, high expression) (see also Figures S5A–S5C).
(F) UMAP plot overlay (left panel) similar to (E) but with the colors representing origin of the WT (teal) or Irf8fl/flItgax-cre (red) compartment and sample frequency per cluster (right panel).
(G) Dot plot heatmap showing expression of selected (curated) and top DEG (data driven) per cluster. The dot size represents the percentage of cells expressing the gene, and the color represents the average expression of that gene within a cluster.
(H) Heatmap showing relative expression of the top DEG retrieved from a microarray dataset of cDC2s and inf-cDC2s sorted from IAV-infected lungs 4 dpi, plotted on lung inf-cDC2s, migratory and non-migratory cDC1 and cDC2 clusters of an scRNA-seq dataset 10 dpi with PVM, and vice versa. Relative expression was calculated by transforming the normalized expression values to a 0-1 scale for each gene separately.
(I) Histogram showing Irf8 RNA expression profiles of different lung cDC clusters derived from the WT compartment of WT:Irf8fl/flItgax-cre chimeras 10 dpi with PVM (left panel), UMAP showing Irf8 expression (center panel), and a UMAP plot showing Irf8 regulon activity as predicted by SCENIC (right panel) on lung cells derived from the WT and Irf8−/− compartments. Cells in which the Irf8 regulon is active (i.e., regulon activity exceeds a regulon-specific area under the curve [AUC] threshold) are shown in blue.
(J) Venn diagram detailing overlap between different cDC2 subsets of Irf8-dependent DEG reaching a log-fold change of ± 0.25.
(K) Heatmap of normalized expression of Irf8-dependent genes in cDC2 subsets derived from lungs of CD45.1 WT:CD45.2 Irf8fl/flItgax-cre chimeric mice 10 dpi with PVM. Genes with a log fold-change lower than −0.25 (i.e., intrinsically induced by IRF8) are shown in bold blue, and genes with a log fold-change higher than 0.25 (i.e., intrinsically suppressed by IRF8) are shown in bold black. Group names are based on the cDC2 subsets in which the log fold threshold was reached (J).
Inf-cDC2s Express Genes Shared with All DC Subsets and MCs
Single-cell RNA sequencing (scRNA-seq) analysis was performed on sorted lineage−CD11c+MHCII+ cells obtained from the lungs of WT:Irf8 fl/fl Itgax-cre chimeric mice 10 dpi with PVM, according to expression of congenic markers. An unsupervised Uniform Manifold Approximation and Projection (UMAP) was performed on 9,446 WT CD45.1 and 9,142 Irf8 fl/fl Itgax-cre CD45.2 cells derived from chimeric mice (Figure 3E). To account for non-IRF8-related differences because of the congenic background, the same analysis was performed on 10,362 WT CD45.1 and 10,142 non-transgenic CD45.2 cells sorted from the lungs of infected WT:Irf8 fl/fl Itgax-cre − chimeric mice (Figures S5B and S5C). Cells were pooled into a single dataset, and their origin was visualized in a dual color-coded UMAP (Figure 3F; Figure S5C).
Annotation of cell clusters was based on expression of curated and data-driven genes (Figures 3G and 3H). pDCs were defined as Siglech + Tcf4 + Ccr9 + Bst2 + and cDCs as Flt3 + Dpp4 + Zbtb46 +. The cDCs were further subdivided into “non-migratory” (Ccr7 lo Nudt17 lo) cDC1s (Cd81 + Gcsam + Xcr1 + Irf8 +) and cDC2s (Sirpa + S100a4 + CD209a +) and “migratory” (Ccr7 hi Nudt17 hi profile) cDC1s (Cd81 + Gcsam + Laptmb4 + Ncoa7 +; the latter two genes were chosen because of downregulation of Xcr1 mRNA) and cDC2s (S100a4 + Anxa3 + Cdc42ep3 +). A small group of cDCs highly expressed genes associated with cell proliferation (Mki67 + Stmn1 +), confirming reports that cDCs can proliferate in the lungs (Cabeza-Cabrerizo et al., 2019). Approximately one-third of lung MHCII+CD11c+ cells at 10 dpi with PVM in chimeric mice were MCs (expression of Mafb, Mertk, and Fcgr1 and lack of expression of Flt3, Zbtb46, and Dpp4; Figures 3E and 3F). In the cDC2 compartment, inf-cDC2s were identified based on expression of Fcgr1 and other genes (e.g., Irf8 and Irf4). Although, in flow cytometry analysis, up to 60% of all cDCs were inf-cDC2s (Figures 1C and 1G), scRNA-seq revealed only a representation of approximately 20% (Figure 3E). This most likely stems from differences in the kinetics of mRNA and protein expression of marker genes, and inf-cDC2s might be masked by downregulation or delayed expression of Fcgr1 mRNA. The most likely cluster where inf-cDC2s are to be expected is the migratory cDC2 cluster because this cluster contains the highest number of Irf8 mRNA+ cells.
To compare the differentially expressed genes (DEG) between cDC2s and inf-cDC2s defined by viral infection, we mapped the top DEG between sorted inf-cDC2s and cDC2s of lungs of IAV-infected mice onto cDC2 clusters of the scRNA-seq data of PVM-infected mice and vice versa (Figure 3H). The top DEG of the inf-cDC2s subset clearly overlapped between both models of viral respiratory infection (Figure 3H), illustrating a common signature of inf-cDC2s. Compared with non-migratory cDC2s and migratory cDC2s, the most discriminative genes in inf-cDC2s were related to a type I IFN signature (e.g., Stat1, Iigp1, Rsad2, ifi205, Isg20, Ifit1, and Ifit3), genes encoding for pro-inflammatory cytokines (e.g., Ccl5, Cxcl9, and Cxcl10), and genes promoting Th1 cell differentiation (e.g., Il12b), whereas genes involved in Th2 cell development appeared to be downregulated (e.g., Irf4, Klf4, and Mgl2) (Figures 3G, 3H, and 3K). In line with the flow cytometry data, cDC1s were exclusively derived from the WT compartment (Figure 3F). MCs and the entire cDC2 lineage, including inf-cDC2s, developed equally well from both compartments. Sorted pDCs were almost completely derived from the IRF8-deficient compartment. We reported earlier that IRF8-deficient pDCs have higher expression of CD11c, IRF4, and MHCII, which explains why IRF8-deficient pDCs are present in the pool of CD11c+MHCII+ cells of these WT:Irf8 fl/fl Itgax-cre chimeras (Sichien et al., 2016). In WT:WT control mixed BM chimeras, all cell populations were equally derived from both compartments (Figure S5).
IRF8 Drives Maturation in inf-cDC2s
The precise gene expression module depending on IRF8 in cDCs has been difficult to elucidate because cDC1s do not develop in the absence of IRF8 (Murphy et al., 2016, Sichien et al., 2016). However, the finding that IRF8 was turned on in inf-cDC2 without affecting development gave us the unique opportunity to study the IRF8-dependent genes in cDC2s. We quantified IRF8 regulon activity in cells of chimeric mice by applying the single-cell regulatory network inference and clustering (SCENIC) workflow (Aibar et al., 2017), which predicted a high-confidence regulon governed by IRF8 in the cDC1 lineage. This IRF8 regulon activity was also observed during cDC2 maturation and migration upon PVM infection, supported by Irf8 mRNA expression in cells with IRF8 regulon activity (Figure 3I). We then compared DEG between Irf8 −/− and WT cells in each cluster of cDC2s (i.e., non-migratory cDC2s, migratory cDC2s, and inf-cDC2s). After removal of 9 genes that were differentially expressed because of the congenic background in chimeric mice, a small set of 88 genes was obtained (Figures 3J and 3K). Most of the IRF8-dependent genes were involved in co-stimulation, T cell differentiation (e.g., promotes Il12b and suppresses Irf4, Klf4, and Ccrl2), and antigen processing and presentation (e.g., Siglecg, H2-DMb2, Cst3, Fcgrt, and Nlrc5). In addition to Irf8, other transcription factors associated with cDC1 development and function were adopted by migratory cDC2s, like Id2, Batf3, IL12b, Tap1, Tap2, Tapbp, Tapbpl, B2m, Psme genes, Swap70, Nfil, Nlrc5, and Rab43 (Jaiswal et al., 2013, Kretzer et al., 2016; Figures 3G and 3K). Genes typically associated with cDC2 function (Irf4, Ciita, H2-DMb2, and Siglecg) were upregulated in Irf8−/− cells (Figure 3K).
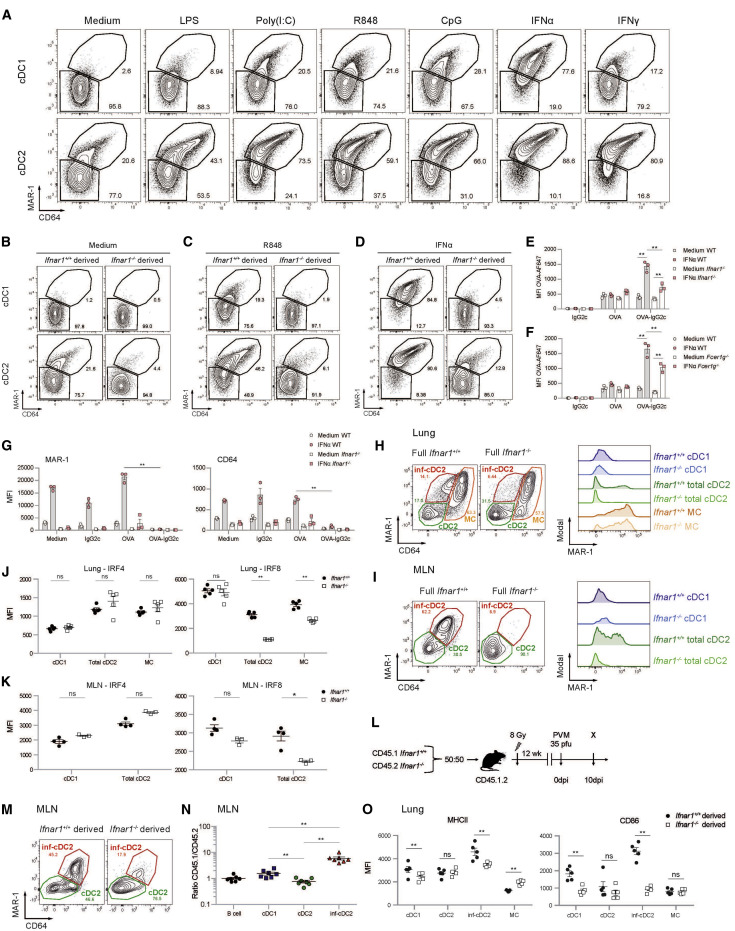
Inf-cDC2s Are Induced in a Cell-Intrinsic, Type I IFN-Dependent Manner
PVM and IAV are RNA viruses known to contain TLR3 and TLR7 agonists, whereas the fusion protein of PVM is a known TLR4 agonist. To study the signals leading to inf-cDC2 development, we set up Flt3L BM cultures (Kirkling et al., 2018), which were exposed to agonists of TLR3 (poly(I:C)), TLR4 Lipopolysaccharide (LPS), TLR7 (R848), and TLR9 (CpG motifs). Because Ingenuity Pathway Analysis (IPA) of the transcriptional profile of inf-cDC2s suggested that Interferon alpha/beta receptor 1 (IFNAR1), STAT1, IRF7, and IRF8 were potential upstream regulators of the distinct gene expression profile (Figure S2E), cells were also exposed to type 1 and type 2 IFN. After 9 days of culture, XCR1+CD172a− cDC1s and XCR1−CD172a+ cDC2s were clearly distinguishable. Without addition of TLR ligands, some cDC2s already expressed MAR-1 and low amounts of CD64. Addition of LPS, poly(I:C), R848, and CpG motifs induced much higher MAR-1 and CD64 on cDC2s, and these cells now resembled inf-cDC2s. Upregulation of MAR-1 on cDC1s was also seen after exposure to poly(I:C) and imiquimod, although the shift was bigger in cDC2s. Type 1 IFN induced CD64 and MAR-1 on cDC2s and cDC1s, whereas IFN-γ induced CD64 and MAR-1 exclusively on cDC2s (Figure 4 A).
Figure 4.
MAR-1, CD64, and IRF8 Are Induced in a Type I IFN-Dependent Manner in cDC2s
(A) On day 8, Flt3L bone marrow dendritic cells (BMDCs) were stimulated with LPS (200 ng/mL), poly(I:C) (1,000 ng/mL), R848 (10 ng/mL), CpG (100 ng/mL), IFNα (200 ng/mL), or IFNγ (200 ng/mL). cDCs were harvested 20 h later and analyzed for expression of MAR-1 and CD64.
(B–D) Flt3L culture of a 50:50 mix of CD45.1 WT (Ifnar1+/+) and CD45.2 Ifnar1−/− DCs that was unstimulated (B) or stimulated with R848 (C) or IFNα (D) and harvested 20 h later (see also Figures S3E and S3F).
(E–G) Uptake of OVA-AF647 (10 μg/mL) added alone or as OVA-AF647-IgG2c-IC to Flt3L culture of a 50:50 mix of CD45.1 WT and CD45.2 Ifnar1−/− (E) or Fcer1g−/− (F) type 2 cDCs and surface expression (MFI) of MAR-1 and CD64 (G) on Flt3L (un)stimulated WT and Ifnar1−/− BM-derived type 2 cDCs 20 h after addition of OVA-AF647 (10 μg/mL) alone or as OVA-AF647-IgG2c-IC. Data are representative of 3 independent experiments with 3 mice per group, analyzed with a two-way ANOVA with Sidak correction for multiple comparisons. Error bars indicate ± SEM. ∗∗p < 0.01 (see also Figures S3G and S3H).
(H and I) Flow cytometry plots pre-gated on live CD3−CD19−CD172a+ DCs in the lungs (H) and migratory DCs in MLNs (I) of WT (Ifnar1+/+) and Ifnar1−/− mice 10 dpi with PVM. Histograms show expression of MAR-1 on the different DC subsets (see also Figures S2H–S2J).
(J and K) Expression of IRF4 and IRF8 shown as MFI by DC subsets in the lungs (J) and MLNs (K) of WT (Ifnar1+/+) and Ifnar1−/− mice 4 dpi with IAV. Data are representative of 2 independent experiments with 4–5 mice per group and were analyzed with a two-way ANOVA with Sidak correction for multiple comparisons. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant.
(L) Schematic representation of CD45.1 WT:CD45.2 Ifnar1−/− BM chimeras.
(M) Flowcytometry plots pre-gated on WT (Ifnar1+/+) or Ifnar1−/−−derived live CD3−CD19− migratory CD172a+ DCs in MLNs of CD45.1 WT:CD45.2 Ifnar1−/− chimeric mice 10 dpi with PVM.
(N) Normalized CD45.1/CD45.2 ratio relative to B cells of migratory cDCs in the MLN 10 dpi with PVM. Data are representative of 2 independent experiments with 5–7 mice per group, analyzed with a one-way ANOVA with Sidak correction for multiple comparisons. Error bars indicate ± SEM. ∗∗p < 0.01.
(O) Expression of MHCII and CD86 shown as MFI by DC subsets in the lungs of CD45.1 WT:CD45.2 Ifnar1−/− chimeric mice 10 dpi with PVM. Data are representative of 2 independent experiment with 5–7 mice per group and were analyzed with a two-way ANOVA with Sidak correction for multiple comparisons. ∗∗p < 0.01; ns, not statistically significant.
Mixed cultures set up from 50% CD45.1 WT and 50% CD45.2 Ifnar1 −/− BM cells stimulated by medium or TLR7 ligation revealed that spontaneous upregulation of MAR-1 on cDC2s was only seen in the WT compartment but not in the Ifnar1 −/− compartment of these mixed cultures, suggesting autocrine production of type 1 IFN and cell-intrinsic IFNAR1 signaling (Figure 4B). Likewise, TLR7-induced upregulation of MAR-1 and CD64 on the WT fraction of cells was almost completely absent in the Ifnar1 −/− fraction, suggesting a role of autocrine type 1 IFN in upregulation of activating Fc receptors on cDCs driven by TLR ligation (Figure 4C). Similar results were found for the other TLR ligands (data not shown). Type 1 IFN induced upregulation of CD64 and MAR-1 in the WT fraction but not in the Ifnar1 −/− fraction.
Upregulation of activating Fc receptors in vitro increased the capacity of cDC1s and cDC2s to take up OVA-AF647-ICs compared with OVA-AF647 alone (Figures 4E and 4F; Figures S3G and S3H). This increased uptake was intrinsically dependent on type I IFN signaling and expression of the Fc common gamma chain (Figures 4E and 4F). Moreover, the uptake of OVA-AF647-ICs was accompanied by loss of surface expression of MAR-1 and CD64 because of internalization (Figure 4G).
To validate these findings in vivo, Ifnar1 −/− and WT mice were infected with PVM and analyzed for DC subsets at 10 dpi. In the absence of type I IFN signaling, inf-cDC2s were hardly detectable in the lungs, whereas MAR-1+ MCs were intact (Figure 4H). This was mostly seen in MLNs, in which migratory inf-cDC2s were completely lacking (Figure 4I). Similar findings were seen in IAV-infected mice at 4 dpi (Figures S2H and S2I). Moreover, we found that IRF8 expression was reduced in lung and MLN cDC2s and MCs but not in cDC1s of virus-infected Ifnar1 −/− mice compared with WT controls (Figures 4J and 4K), indicating that type 1 IFN signaling acts as an upstream inducer of IRF8 selectively in cDC2s and MCs. No changes were observed in IRF4 expression in virus-infected WT and Ifnar1 −/− mice (Figures 4J and 4K).
In mixed WT:Ifnar1 −/− BM chimeras (Figure 4L) infected with PVM, induction of inf-cDC2s also intrinsically relied on IFNAR1 signaling in vivo. In MLNs at 10 dpi, inf-cDC2s were almost exclusively derived from the WT component of chimeric mice (Figures 4M and 4N), whereas cDC1s were equally derived from WT and Ifnar1 −/− cells. The increased maturation state of lung inf-cDC2s, reflected by enhanced expression of co-stimulatory molecules compared with cDC2s, was also intrinsically dependent on IFNAR1 signaling (Figure 4O).
Taken together, these data show that TLR-driven activation of type I IFN production causes cell-intrinsic IFNAR1 signaling, inducing activation of migratory cDC2s that acquire the phenotype of inf-cDC2s in virally infected mice.
Inf-cDC2s Are Found Abundantly in Other Models of Inflammation and Infection
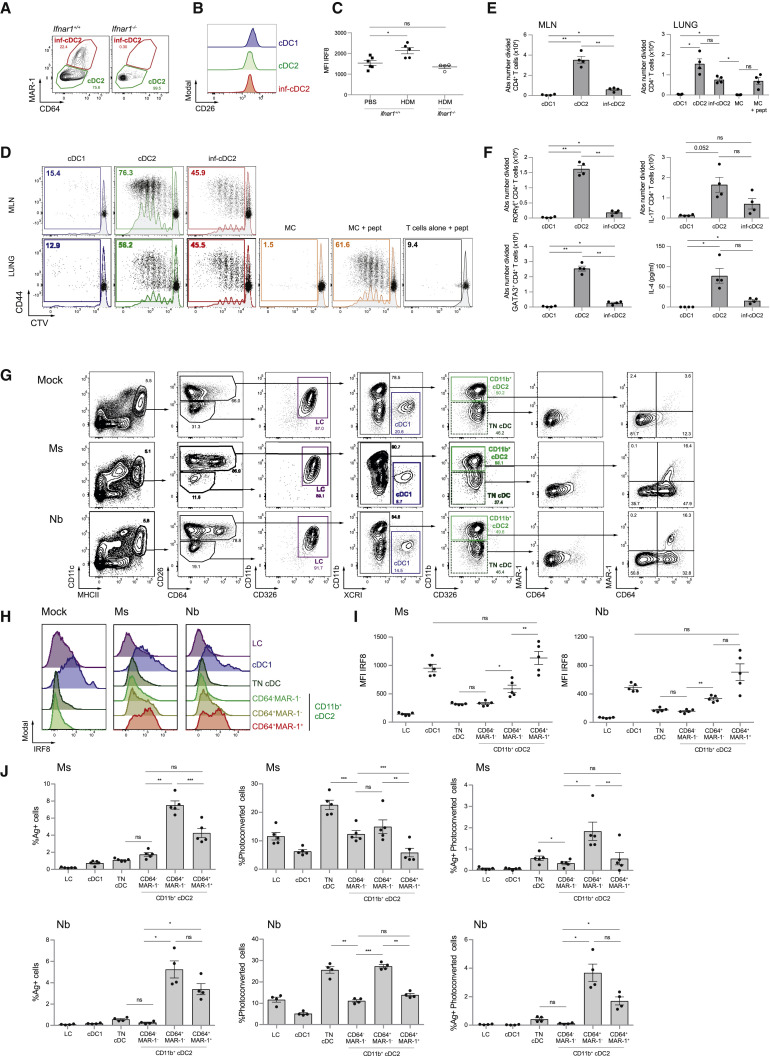
We reported previously, using a Th2 cell-driven house dust mite (HDM) mouse model of asthma, that MAR-1+ CD64+ MHCII+ CD11c+ cells present Der p 1 antigens to ex vivo allergen-specific T cells and prime for asthma upon adoptive transfer to naive mice. However, because MAR-1 and CD64 were considered markers of monocyte-derived macrophages at that time, and based on the dependency on CCR2, we called these cells inflammatory monocyte-derived DCs (Hammad et al., 2010, Plantinga et al., 2013). Given the phenotype of inf-cDC2s in viral lung infections, we repeated this experiment with intratracheal (i.t.) administration of 100 μg of HDM. As reported, MAR-1+ CD64int CD172a+ DCs were found to accumulate in MLNs 3 days after HDM administration in WT mice but not Ifnar1 −/− mice (Figure 5 A). These MAR-1+ DCs expressed the same amount of the common cDC marker CD26 as cDC1s and MAR-1− cDC2s, arguing for their pre-cDC origin (Figure 5B). Like in viral infection, the total cDC2 population in MLNs acquired intermediate IRF8 expression (Figure 5C) upon HDM administration, but this was not seen in Ifnar1 −/− mice. Selected pro-Th2 cell cytokines known to affect DCs, like Thymic stromal lymphopoietin (TSLP) and IL-33, were unable to induce inf-cDC2s in vitro (Figure S3F). HDM sensitization in combination with LPS and beta-glucans promotes neutrophilic inflammation (Hadebe et al., 2015), and mixed Th2-Th17 cell pulmonary immunity is also found in severe asthmatics. Inf-cDC2s were capable of priming naive Der p 1-specific (1-Der) T cells and induce RORγt and IL-17A expression, but less efficiently than CD64− cDC2s (Figures 5D–5F). Sorted lung MCs were again unable to present endogenously acquired antigen to 1-Der T cells ex vivo, but when preprocessed exogenous Der p 1 peptide was added, they readily induced proliferation.
Figure 5.
Inf-cDC2s Are Found in Other Models of Inflammation and Infection
(A) Flowcytometry plots pre-gated on live CD3−CD19− migratory CD172a+ DCs in MLNs of WT (Ifnar1+/+) and Ifna1r−/− mice 3 days after 100 μg HDM administration i.t.
(B) CD26 expression by the different DC subsets in MLNs of WT mice 3 days after 100 μg HDM administration i.t.
(C) Expression of IRF8, shown as MFI, by the total cDC2 subset in MLNs of WT (Ifnar1+/+) and Ifnar1−/− 3 days after 100 μg HDM administration i.t. The experiment had 4–6 mice per group and was analyzed with a one-way ANOVA with Sidak correction for multiple comparisons. ∗p < 0.05; ns, not statistically significant.
(D) Proliferation profile of CTV-labeled Der p 1-specific CD4+ TCR transgenic T cells cocultured for 4 days with the different DC subsets sorted from MLNs (top row) or lungs (bottom row) 72 h after i.t. challenge with HDMs (75 μg) in combination with LPS (300 ng) and beta-glucans (30 μg). For the peptide control, 1 μg/mL of Der p 1 peptide was added ex vivo.
(E and F) Absolute number total divided (E) and RORγt+, IL-17+, and GATA3+ (F) Der p 1-specific TCR transgenic CD4+ T cells and IL-4 measured in supernatants of coculture as in (D). One-way ANOVA with Sidak correction for multiple comparisons. Error bars indicate ± SEM. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant.
(G) Gating strategy of DC subsets in ELNs 48 h after i.d. injection of the ear pinna with mock, nonviable Ms or Nb L3 larvae; pre-gated on live CD3−B220−Ly6G−Ly6C− cells.
(H and I) Expression of IRF8 by different DC subsets in ELNs 48 h after i.d. immunization of the ear pinna with Ms or Nb or mock immunization, shown in representative histograms (H) and as MFI (I). Data are representative of 2 independent experiment with 5 mice per group and were analyzed with a one-way ANOVA with Sidak correction for multiple comparisons. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant.
(J) Proportion of CTFR-Ms+/Nb+ and/or photoconverted DC subsets in ELNs 48 h after i.d. immunization and violet laser exposure of the ear pinna. Data are representative of 2 independent experiments with 4 Kaede mice per group and were analyzed with a one-way ANOVA with Sidak correction for multiple comparisons. ∗p < 0.05, ∗∗p < 0.01; ns, not statistically significant.
To study whether inf-cDC2s were also found in other tissues, we examined the distinct DC populations isolated from mouse ear draining LNs (ELNs) 48 h after intradermal (i.d.) injection in the ear pinna of nonviable freeze-thawed Nippostrongylus brasiliensis L3 larvae (Nb) or heat-killed Mycobacterium smegmatis (Ms) as models for skin type 2 parasitic and type 1 mycobacterial infection, respectively (Connor et al., 2017). An increase in MHCIIhi DCs in ELNs was seen 48 h after Ms and Nb infection, mainly caused by influx of skin CD11b+ and triple-negative (CD11b−CD103−CD326−) CD26+ XCR1− cDC2s (Figure 5G and data not shown). Again, no CD26loCD64hi MCs could be identified in the actively migrating MHCIIhi DC population in draining LNs (Figure 5G). Unique to the skin, a CD26loMHCIIhi population in ELNs was identified as Langerhans cells (LCs) expressing CD11b and CD326 but not CD64. Conventional CD26+ DCs were further subdivided in XCR1+ cDC1s, CD11b+ cDC2s, and TN cDCs. CD11b+ cDC2s acquired expression of CD64 and MAR-1 (Figure 5H) upon immunization, the degree of which correlated with intracellular IRF8 expression (Figure 5I). In CD64+MAR-1+ skin inf-cDC2s in the Ms and Nb model, the intensity of IRF8 was at least as high as in cDC1s, potentially because a subset of CD326+ skin cDC1s express lower IRF8 compared with other tissue cDC1s (Figure 5I; Tussiwand et al., 2012).
To evaluate active migration to ELNs, we immunized Kaede mice, which ubiquitously express a violet light-sensitive photoconvertible fluorescence protein, with CellTrace™ Far Red (CTFR)-labeled Nb and Ms and studied migration at 48 h (Tomura et al., 2008; Figure 5J). In keeping with a previous report (Connor et al., 2014), CD11b+ cDC2s accounted for the majority of Ms and Nb antigen transported to the ELN. Photoconverted skin DCs were found exclusively in the MHCIIhi fraction of ELNs. MAR-1+ and/or CD64+ cDC2s of the skin were a bona fide migratory DC population.
Thus, inf-cDC2s that express IRF8 and develop in an IFNAR1-dependent manner can be found in multiple models of infection and inflammation in and outside of the lung.
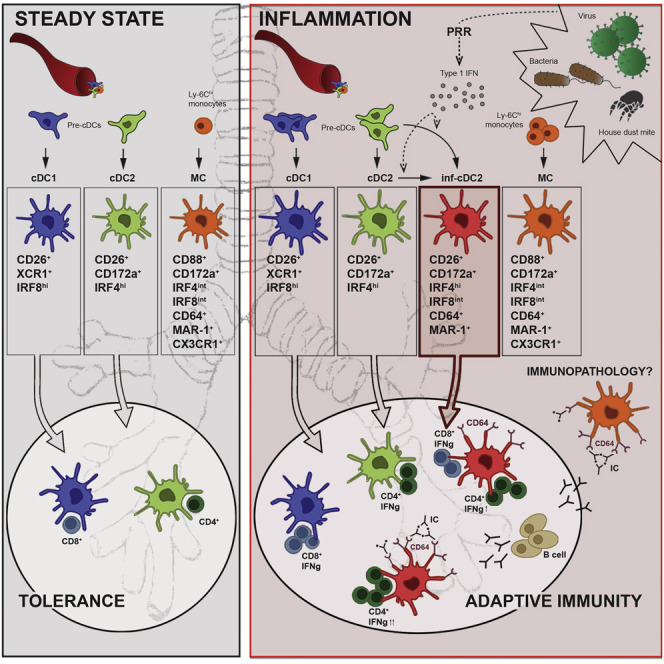
Discussion
Here we have shown that what are known as “monocyte-derived inflammatory DCs” are a mixture of monocyte-derived MCs that have little migratory and APC potential and bona fide pre-cDC-derived, CD26-expressing inf-cDC2s that depend on Flt3L but not on GM-CSF. Inf-cDC2s had upregulated expression of cytokines, chemokines, costimulatory molecules, and Fc receptors in an IFNAR1-dependent manner. Given these hybrid features of inf-cDC2s, sharing expression of activating Fc receptors with MCs and macrophages and the potential to upregulate IRF8 like cDC1s, we suspect that they have often contaminated other populations of APCs. Because most investigators have not used CD26 to precisely define cDCs in the CD11b+CD11c+MHCII+ pool of cells, the mistaken identity of contaminating inf-cDC2 cells might be the main reason why in vivo monocyte-derived DCs have been shown to be excellent APCs with migratory capacity in models of allergy, vaccination, infection, and cancer (Cheong et al., 2010, Kool et al., 2012, León et al., 2007, Min et al., 2018, Nakano et al., 2009, Plantinga et al., 2013, Tanaka et al., 2007). When we carefully excluded inf-cDC2s from monocyte-derived “DCs,” the latter were no longer able to process and present native antigens to naive CD4+ and CD8+ T cells.
The argument that moDCs are important for immunopathology and Th1 cell-mediated immunity has also heavily relied on use of Ccr2 −/− mice, which have defects in monocyte egress from the BM, moDC and MC recruitment, and Th1 cell induction (Iijima et al., 2011, De Koker et al., 2017, Menezes et al., 2016, Nakano et al., 2009, Sheng et al., 2017, Tamoutounour et al., 2012). Our data now show that inf-cDC2s are IL-12-producing mature APCs that resemble moDCs and also depend on CCR2 for migrating to the lungs, at least in the setting of respiratory viral infection. These data are in line with reports describing CCR2 expression on circulating Ly6ChiSiglecH+ pre-cDC2s and dependence of CD11b+ cDC2s on CCR2 in mucosal tissues such as the gut and lungs (Nakano et al., 2017, Schlitzer et al., 2015, Scott et al., 2015). Use of the CCR2 model and the presence of CD64+MAR-1+-expressing DCs in MLNs have also misled us to suggest that moDCs were drivers of Th2 cell immunity in the HDM model of allergy (Plantinga et al., 2013), an interpretation we would now like to revise. By using CD26 as a defining marker, it is very clear that the moDCs we proposed to migrate to LNs to induce Th2 cell priming after high-dose HDM exposure were, in fact, inf-cDC2s (Plantinga et al., 2013), and we additionally demonstrate how these emerge in a type 1 IFN-dependent manner. These findings also realign our work with that of others proposing that lung inflammatory Ly6C+ moDCs are more related to macrophages and have a poor capacity to migrate to lung LNs or prime T cell responses (Mesnil et al., 2012, Nakano et al., 2013, Wu et al., 2016).
It has been proposed that CD209 (DC-SIGN) is a unique marker for moDCs that identifies DCs with the highest degree of maturity and potential to cross-present antigens (Cheong et al., 2010, Menezes et al., 2016). When carefully separating MCs from inf-cDC2s, in our hands, CD209 mRNA was only found on cDC2s. Cell surface CD88 and CX3CR1 are potentially much better and stable positive discriminators of MCs, particularly in combination with CD26 to identify cDCs (Nakano et al., 2015). We no longer recommend exclusive use of CD64 or MAR-1 to separate cDCs from MCs (Cabeza-Cabrerizo et al., 2019, Guilliams et al., 2014b, Guilliams et al., 2016, Plantinga et al., 2013, Tamoutounour et al., 2012).
Mutant Irf8 (BHX2), Irf8 fl/fl Itgax-cre, and Irf8 fl/fl Zbtb46-cre mice have been used or proposed as models to interrogate the in vivo functions of cDC1s, whereas Irf4 fl/fl Itgax-cre mice have been used to probe cDC2 function (Ainsua-Enrich et al., 2019, Bajaña et al., 2012, Bajaña et al., 2016, Baptista et al., 2019, Krishnaswamy et al., 2017, Persson et al., 2013, Schlitzer et al., 2013). Because IRF8 is induced in inf-cDC2s and regulates a functional module, including antigen-presentation capacity, chemokine production, and cytokine production, we caution to draw the conclusion that a particular phenotype of Irf8 mutant mice would be exclusively due to defects of cDC1s. It will be very important to better understand how IRF8 expression is regulated in cDC2s and whether this is different compared with cDC1s.
Our work has implications for understanding (cross)-presentation and uptake of opsonized antigens through use of convalescent serum. In the steady state, cDC1s have a higher capacity to cross-present cell-bound and particulate antigens to CD8+ T cells, and cDC2s are better at inducing CD4+ T cell responses (Burgdorf et al., 2007, Dudziak et al., 2007, Hildner et al., 2008). However, many studies have shown that CD11b+ DCs and moDCs (MCs) can also stimulate CD8 immunity, particularly in vitro and after cytokine activation in vivo (Briseño et al., 2016, Cheong et al., 2010, Desch et al., 2014, Larson et al., 2016). As an example, we and others have shown that cDC1s prime CD8+ T cell immunity to influenza virus infection (GeurtsvanKessel et al., 2008, Helft et al., 2012), whereas others have claimed that CD11b+ cDC2s are more proficient (Ainsua-Enrich et al., 2019, Ballesteros-Tato et al., 2010). Here, using PVM-specific TCR transgenic mice, we show that cDC1s and bona fide CD11b+ inf-cDC2s from virus-infected mice are almost equally potent in stimulating CD8+ T cells ex vivo and that inf-cDC2s simultaneously stimulate CD4+ T cell responses, inducing polarization of IFN-γ-producing cells. Contrary to dogma, MCs were unable to present viral antigens to naive CD8+ or CD4+ T cells. Therefore, the role of tissue-residing MCs might rather be found in orchestration of the peripheral effector response or inflammation in the presence of antibody-complexed antigen. It will be important to examine whether convalescent serum can activate MCs to produce pro-inflammatory cytokines and chemokines. This is a pertinent question for patients with severe viral infections causing acute lung injury and ARDS (such as COVID-19 and influenza). It has been proposed that convalescent serum be given during the acute lung injury phase, but if this leads to activation of Fc receptor-bearing MCs, it could be detrimental (Casadevall and Pirofski, 2020).
Triggering of activating Fc receptors by opsonized antigens or ICs can lead to enhanced APC function of moDCs (Regnault et al., 1999), whereas inhibitory Fc receptors (FcRIIb) suppress APC function (Kalergis and Ravetch, 2002). However, most studies that interrogated expression of Fc receptors on steady-state cDCs in vivo only demonstrated inhibitory Fc receptors on cDCs and pDCs and extremely low expression of activating FcγRI (CD64), FcγRIII, or FcγRIV receptors (summarized in Guilliams et al., 2014b, Lehmann et al., 2017, Tamoutounour et al., 2012). Here we show that bona fide inf-cDC2s upregulate activating Fc receptors in a TLR- and type I IFN-dependent manner and proficiently use these activating receptors for enhanced presentation of opsonized viral antigens to CD4+ T cells upon transfer of convalescent serum, whereas CD8+ T cell responses are not altered. Fc receptor-mediated uptake can divert antigens to a more acidic endosomal compartment conducive to MHCII antigen processing. Mouse IgG2c-antigen IC injection in vivo led to rapid Fc receptor-mediated internalization of antigen in inf-cDC2s. More mechanistic studies are needed, but this ligand-induced internalization could be one of the main reasons why these cells have been ignored as major players in the uptake of ICs in vivo, a function that has traditionally been attributed to cDC1 in the steady state but could be an induced function on inf-cDC2s (den Haan and Bevan, 2002).
In conclusion, in respiratory viral infection and allergy, inf-cDC2s are generated in a type I IFN-dependent manner to optimally prime CD4+ and CD8+ T cell immunity. It will be essential to revisit the role of APCs in various inflammation models because the function of inf-cDC2s may have been wrongly attributed to cDC1s or MCs.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse FceR1 alpha Monoclonal antibody (Armenian hamster, clone MAR-1), Biotin conjugated | Thermo Fisher Scientific | Cat#13-5898-85; RRID:AB_466784 |
| Anti-mouse CD3e Monoclonal antibody (Armenian hamster, clone 145-2C11), PE-Cy5 conjugated | Thermo Fisher Scientific | Cat#15-0031-82; RRID:AB_468690 |
| Anti-mouse CD3e Monoclonal antibody (Armenian hamster, clone 145-2C11), BUV737 conjugated | BD Biosciences | Cat#564618; RRID:AB_2738868 |
| Anti-mouse CD3e Monoclonal antibody (Armenian hamster, clone 145-2C11), Biotin conjugated | Thermo Fisher Scientific | Cat#13-0031-85; RRID:AB_466320 |
| Anti-mouse CD3 Monoclonal antibody (Rat, clone 17A2), AF700 conjugated | Thermo Fisher Scientific | Cat#56-0032-82; RRID:AB_529507 |
| Anti-mouse CD19 Monoclonal antibody (Rat, clone eBio1D3(1D3)), PE-Cy5 conjugated | Thermo Fisher Scientific | Cat#15-0193-83; RRID:AB_657673 |
| Anti-mouse CD19 Monoclonal antibody (Rat, clone eBio1D3(1D3)), AF700 conjugated | Thermo Fisher Scientific | Cat#56-0193-82; RRID:AB_837083 |
| Anti-mouse CD19 Monoclonal antibody (Rat, clone eBio1D3(1D3)), Biotin conjugated | Thermo Fisher Scientific | Cat#13-0193-85; RRID:AB_657658 |
| Anti-mouse Ly-6G Monoclonal antibody (Rat, clone 1A8), BUV395 conjugated | BD Biosciences | Cat#563978; RRID:AB_2716852 |
| Anti-mouse Ly-6G Monoclonal antibody (Rat, clone 1A8), Biotin conjugated | BioLegend | Cat#127604; RRID:AB_1186108 |
| Anti-mouse Ly-6C Monoclonal antibody (Rat, clone AL-21), AF700 conjugated | BD Biosciences | Cat#561237; RRID:AB_10612017 |
| Anti-mouse Ly-6C Monoclonal antibody (Rat, clone HK1.4), AF647 conjugated | BioLegend | Cat#128010; RRID:AB_1236550 |
| Anti-mouse CD326 Monoclonal antibody (Rat, clone G8.8), BV711 conjugated | BioLegend | Cat#118233; RRID:AB_2632775 |
| Anti-mouse CD11c Monoclonal antibody (Armenian hamster, clone N418), PE-Cy7 conjugated | Thermo Fisher Scientific | Cat#25-0114-82; RRID:AB_469590 |
| Anti-mouse CD11c Monoclonal antibody (Armenian hamster, clone N418), eFluor 450 conjugated | Thermo Fisher Scientific | Cat#48-0114-82; RRID:AB_1548654 |
| Anti-mouse CD11c Monoclonal antibody (Armenian hamster, clone HL3), BV786 conjugated | BD Biosciences | Cat#563735; RRID:AB_2738394 |
| Anti-mouse I-A/I-E (MHCII) Monoclonal antibody (Rat, clone M5/114.15.2), APC-eFluor 780 conjugated | Thermo Fisher Scientific | Cat#47-5321-82; RRID:AB_1548783 |
| Anti-mouse I-A/I-E (MHCII) Monoclonal antibody (Rat, clone M5/114.15.2), FITC conjugated | Thermo Fisher Scientific | Cat#11-5321-85; RRID:AB_465233 |
| Anti-mouse I-A/I-E (MHCII) Monoclonal antibody (Rat, clone M5/114.15.2), FITC conjugated | Thermo Fisher Scientific | Cat#56-5321-82; RRID:AB_494009 |
| Anti-mouse CD103 Monoclonal antibody (Armenian hamster, clone 2E7), PE conjugated | Thermo Fisher Scientific | Cat#12-1031-83; RRID:AB_465800 |
| Anti-mouse CD103 Monoclonal antibody (Armenian hamster, clone 2E7), Pacific Blue conjugated | BioLegend | Cat#121418; RRID:AB_2128619 |
| Anti-mouse CD11b Monoclonal antibody (Rat, clone M1/70), BV605 conjugated | BD Biosciences | Cat#563015; RRID:AB_2737951 |
| Anti-mouse CD11b Monoclonal antibody (Rat, clone M1/70), V450 conjugated | BD Biosciences | Cat#560455; RRID:AB_1645266 |
| Anti-mouse CD11b Monoclonal antibody (Rat, clone M1/70), BUV737 conjugated | BD Biosciences | Cat#564443; RRID:AB_2738811 |
| Anti-mouse CD64 Monoclonal antibody (Mouse, clone X54-5/7.1), AF647 conjugated | BD Biosciences | Cat#558539; RRID:AB_647120 |
| Anti-mouse CD64 Monoclonal antibody (Mouse, clone X54-5/7.1), PE conjugated | BD Biosciences | Cat#558455; RRID:AB_647241 |
| Anti-mouse CD64 Monoclonal antibody (Mouse, clone X54-5/7.1), BV711 conjugated | BioLegend | Cat#139311; RRID:AB_2563846 |
| Anti-mouse CD64 Monoclonal antibody (Mouse, clone X54-5/7.1), PE-Cy7 conjugated | BioLegend | Cat#139313; RRID:AB_2563903 |
| Anti-mouse CD24 Monoclonal antibody (Rat, clone M1/69), PE conjugated | Thermo Fisher Scientific | Cat#48-0242-82; RRID:AB_1311169 |
| Anti-mouse XCR1 Monoclonal antibody (Mouse, clone ZET), BV660 conjugated | BioLegend | Cat#148220; RRID:AB_2566410 |
| Anti-mouse XCR1 Monoclonal antibody (Mouse, clone ZET), PE conjugated | BioLegend | Cat#148204; RRID:AB_2563843 |
| Anti-mouse CD172a Monoclonal antibody (Rat, clone P84), PerCP-eFluor 710 conjugated | Thermo Fisher Scientific | Cat#46-1721-82; RRID:AB_10804639 |
| Anti-mouse CD172a Monoclonal antibody (Rat, clone P84), PE-Cy7 conjugated | BioLegend | Cat#144008; RRID:AB_2563546 |
| Anti-mouse CD172a Monoclonal antibody (Rat, clone P84), APC conjugated | BD Biosciences | Cat#560106; RRID:AB_1645218 |
| Anti-mouse CD26 Monoclonal antibody (Rat, clone H194-112), FITC conjugated | BD Biosciences | Cat#559652; RRID:AB_397295 |
| Anti-mouse CD26 Monoclonal antibody (Rat, clone H194-112), APC conjugated | BioLegend | Cat#137807; RRID:AB_10663403 |
| Anti-mouse MerTK Monoclonal antibody (Rat, clone 2B10C42), APC conjugated | BioLegend | Cat#151508; RRID:AB_2650739 |
| Anti-mouse Siglec-F Monoclonal antibody (Rat, clone E50-2440), PE conjugated | BD Biosciences | Cat#552126; RRID:AB_394341 |
| Anti-mouse Siglec-F Monoclonal antibody (Rat, clone E50-2440), BV786 conjugated | BD Biosciences | Cat#740956; RRID:AB_2740581 |
| Anti-mouse CCR2 Monoclonal antibody (Rat, clone SA203G11), BV421 conjugated | BioLegend | Cat#150605; RRID:AB_2571913 |
| Anti-mouse CCR2 Monoclonal antibody (Rat, clone 475301), PE conjugated | R&D Systems | Cat#FAB5538P; RRID:AB_10718414 |
| Anti-mouse CD45.1 Monoclonal antibody (Mouse, clone A20), BV605 conjugated | BioLegend | Cat#110738; RRID:AB_2562565 |
| Anti-mouse CD45.2 Monoclonal antibody (Mouse, clone 104), AF700 conjugated | Thermo Fisher Scientific | Cat#56-0454-82; RRID:AB_657752 |
| Anti-mouse CD45.2 Monoclonal antibody (Mouse, clone 104), PerCP-Cy5.5 conjugated | BD Biosciences | Cat#552950; RRID:AB_394528 |
| Anti-mouse CD80 Monoclonal antibody (Rat, clone 16-10A1), PerCP-Cy5.5 conjugated | BD Biosciences | Cat# 560526, RRID:AB_1727514 |
| Anti-mouse CD86 Monoclonal antibody (Rat, clone PO3), PE-Cy7 conjugated | BioLegend | Cat#105116; RRID:AB_493600 |
| Anti-mouse CD88 Monoclonal antibody (Rat, clone 20/70), PE conjugated | BioLegend | Cat#135806; RRID:AB_2243735 |
| Anti-mouse CD88 Monoclonal antibody (Rat, clone 20/70), PerCP-Cy5.5 conjugated | BioLegend | Cat#135813; RRID:AB_2750209 |
| Anti-mouse CD197 (CCR7) Monoclonal antibody (Rat, clone 4B12), PE-Cy7 conjugated | Thermo Fisher Scientific | Cat#25-1971-82; RRID:AB_469652 |
| Anti-mouse B220 Monoclonal antibody (Rat, clone RA3-6B2), BUV395 conjugated | BD Biosciences | Cat#563793; RRID:AB_2738427 |
| Anti-mouse IRF4 Polyclonal antibody (Goat, clone M-17), unconjugated | Santa Cruz Biotechnology | Cat#sc-6059; RRID:AB_2127145 |
| Anti-mouse IRF8 Monoclonal antibody (Rat, clone V3GYWCH), PerCP-eFluor 710 conjugated | Thermo Fisher Scientific | Cat#46-9852-82; RRID:AB_2573904 |
| Anti-mouse IRF8 Monoclonal antibody (Rat, clone V3GYWCH), APC conjugated | Thermo Fisher Scientific | Cat#17-9852-82; RRID:AB_2573318 |
| Anti-goat Polyclonal antibody (Donkey), AF647 conjugated | Thermo Fisher Scientific | Cat#A-21447; RRID:AB_2535864 |
| Anti-mouse IL-12 Monoclonal antibody (Rat, clone C15.6), APC conjugated | BD Biosciences | Cat#554479; RRID:AB_395420 |
| Anti-mouse CD3e Monoclonal antibody (Armenian hamster, clone 145-2C11), APC-Cy7 conjugated | BD Biosciences | Cat#557596; RRID:AB_396759 |
| Anti-mouse CD4 Monoclonal antibody (Rat, clone RM4-5), BV605 conjugated | BD Biosciences | Cat#563151; RRID:AB_2687549 |
| Anti-mouse CD8 Monoclonal antibody (Rat, clone 53-6.7), PerCP-Cy5.5 conjugated | Thermo Fisher Scientific | Cat#45-0081-82; RRID:AB_1107004 |
| Anti-mouse CD44 Monoclonal antibody (Rat, clone IM7), redFluor 710 conjugated | Tonbo Biosciences | Cat#80-0441; RRID:AB_2621985 |
| Anti-mouse IFN-g Monoclonal antibody (Rat, clone XMG1.2), PE-eFluor 610 conjugated | Thermo Fisher Scientific | Cat#61-7311-82; RRID:AB_2574662 |
| Anti-mouse CD209a Monoclonal antibody (Mouse, clone MMD3), eFluor 660 conjugated | Thermo Fisher Scientific | Cat#50-2094-82; RRID:AB_11219065 |
| Anti-mouse CD301b (Mgl2) Monoclonal antibody (Rat, clone URA-1), PerCP-Cy5.5 conjugated | BioLegend | Cat#146809; RRID:AB_2563391 |
| Anti-mouse CD273 (PD-L2) Monoclonal antibody (Rat, clone TY25), BUV395 conjugated | BD Biosciences | Cat#565102; RRID:AB_2739068 |
| Anti-mouse F4/80 Monoclonal antibody (Rat, clone BM8), BV785 conjugated | BioLegend | Cat#123141; RRID:AB_2563667 |
| Anti-mouse CX3CR1 Monoclonal antibody (Mouse, clone SA011F11), BV785 conjugated | BioLegend | Cat#149029; RRID:AB_2565938 |
| Anti-mouse TCR V beta 6 Monoclonal antibody (Rat, clone RR4-7), FITC conjugated | BD Biosciences | Cat#557004; RRID:AB_647180 |
| Anti-mouse TCR V beta 12 Monoclonal antibody (Mouse, clone MR11-1) FITC conjugated | BD Biosciences | Cat#557004; RRID:AB_647180 |
| Anti-mouse OVA IgG2c (Mouse, hybridoma clone 23-9 and 27-6) | Provided by Dr. Karasuyama and Argenx | This paper |
| Bacterial and Virus Strains | ||
| PVM strain J3666 | Gift from Andrew Easton (Cook et al., 1998, Vandersarren et al., 2017) | N/A |
| H3N2 Influenza strain X31 | Medical Research Council (GeurtsvanKessel et al., 2008, Neyt et al., 2016) | N/A |
| Mycobacterium smegmatis (MC2155) | Malaghan Institute of Medical Research (Connor et al., 2014) | N/A |
| Nippostrongylus brasiliensis L3 larvae | Malaghan Institute of Medical Research (Camberis et al., 2013) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Fc block (clone 2.4G2) | Bioceros | N/A |
| Streptavidin, PE-CF594 conjugated | BD Biosciences | Cat#562284 |
| Cell proliferation dye eFl450 | Thermo Fisher Scientific | Cat#65-0842-90 |
| β-mercaptoethanol | Sigma-Aldrich | Cat#3148 |
| DNase I | Roche | Cat#04 536 282 001 |
| Liberase TL | Roche | Cat#05 401 020 001 |
| PBS | GIBCO | Cat#10010-015 |
| EDTA | Westburg | Cat#51234 |
| Fetal Bovine Serum | Bodinco | Cat#S181G |
| BSA | Sigma-Aldrich | Cat#A8806 |
| Gentamicin | Thermo Fisher Scientific | Cat#15710-049 |
| GlutaMAX | Thermo Fisher Scientific | Cat#35050-038 |
| MEM alpha | Thermo Fisher Scientific | Cat#22571-020 |
| RPMI | Thermo Fisher Scientific | Cat#21875-059 |
| FoxP3 Transcription factor staining buffer kit | Thermo Fisher Scientific | Cat#00-5523-00 |
| Optiprep | Axis Shield | Cat#1114542 |
| PVM M37-47 (PMFQTSLPKNS) | Pepscan | N/A |
| PVM N339-347 (GAPRNRELF) | Pepscan | N/A |
| Der p 1 peptide (GCGSCWAFSGVAATESA YLAYR) |
Pepscan | N/A |
| Recombinant Human/Mouse Flt3L | VIB Protein Service Facility | N/A |
| Recombinant Mouse GM-CSF | VIB Protein Service Facility | N/A |
| Recombinant Mouse IFNα | Provided by Dr. Roosmarijn E. Vandenbroucke, VIB-Ghent University, Belgium | Meister et al., 1986 |
| Recombinant Mouse IFNγ | R&D systems | Cat#485-MI-100 |
| Recombinant Mouse TSLP | Thermo Fisher Scientific | Cat#14-8498-80 |
| Recombinant Mouse IL-33 | R&D systems | Cat#3626-ML |
| Recombinant Mouse IL-25 | R&D systems | Cat#1399-IL |
| Ultrapure E. coli LPS | Invivogen | Cat#tlrl-3pelps |
| R848 (resiquimod) | Invivogen | Cat#tlrl-r848-5 |
| Poly(I:C) LMW | Invivogen | Cat#tlrl-picw |
| CpG class B ODN1826 | Invivogen | Cat#tlrl-1826-5 |
| AF647 Succinimidyl Ester | Thermo Fisher Scientific | Cat#A-20006 |
| CellTracker Deep Red | Thermo Fisher Scientific | Cat#C34565 |
| EndoFit Ovalbumine | Invivogen | Cat#vac-pova |
| House dust mite extract | Greer Laboratories | Cat#XPB82D3A2.5 |
| Beta-glucan (Candida albicans SC5314) | Provided by Dr. David L. Williams, East Tennessee State University, USA | Hadebe et al., 2015 |
| Critical Commercial Assays | ||
| Live/Dead eFluor 506 | Thermo Fisher Scientific | Cat#65-0866-18 |
| Live/Dead eFluor 780 | Thermo Fisher Scientific | Cat#65-0865-14 |
| Bio-Plex Pro Mouse Cytokine 23-plex Assay | Bio-Rad Laboratories | Cat#M60009RDPD |
| MagniSort Streptavidin negative selection beads | Thermo Fisher Scientific | Cat#MSNB-6002-74 |
| RNeasy Micro Plus kit | QIAGEN | Cat#74034 |
| Deposited Data | ||
| Murine RNA-sequencing data | This paper | GSE149619 |
| Murine Micro-array data | This paper | GSE149619 |
| Experimental Models: Cell Lines | ||
| OP9-GFP | Gift from Tom Taghon | Van de Walle et al., 2011 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6j SPF | Janvier Labs or Jackson Laboratory | N/A |
| Mouse: PVM M37-47 CD4 TCR Tg | This paper | N/A |
| Mouse: PVM N339–347 CD8 TCR Tg | This paper | N/A |
| Mouse: 1-Der: Der p 1-specific CD4 TCR Tg | In house generated | Plantinga et al., 2013 |
| Mouse: Irf8loxP/loxP | In house generated | Sichien et al., 2016 |
| Mouse: Irf4loxP/loxP | Provided by Dr. W. Agace, University of Lund, Sweden | Persson et al., 2013 |
| Mouse: Fcer1g−/− :B6.129P2-Fcer1gtm1RavN12 | Taconic | Model 583 |
| Mouse: Kaede mice: B6.Cg-Tg(CAG-tdKaede)15Utr | Provided by Dr. Tatyana Chtanova, Garvan Institute of Medical Research, Australia | Tomura et al., 2008 |
| Mouse: Ifnar1−/− | Provided by Dr. Roosmarijn E. Vandenbroucke, VIB-Ghent University, Belgium | Grine et al., 2015 |
| Mouse: Ccr2−/−:B6(C)-Ccr2tm1.1Cln/J | Jackson Laboratories | Stock No: 027619 |
| Mouse: Flt3L−/−:C57BL/6-Flt3ltm1Imx/TacMmjax | Jackson Laboratories | Stock No: 37395 |
| Mouse: Itgax-cre:B6.Cg-Tg(Itgax-cre)1-1Reiz/J | Jackson Laboratories | Stock No: 008068 |
| Mouse: CD45.1:B6.SJL-PtprcaPepcb/BoyJ | Jackson Laboratories | Stock No: 002014 |
| Mouse: Csf2rb−/−: B6.129S1-Csf2rbtm1Cgb/J | Jackson Laboratories | Stock No: 005940 |
| Mouse: Fcer1a−/−:B6.C.129S2-Fcer1atm1Knt/J | Jackson Laboratories | Stock No: 005629 |
| Software and Algorithms | ||
| FlowJo v11 | FlowJo | https://www.flowjo.com |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com; RRID: SCR_002798 |
| Ingenuity Pathway Analysis | QIAGEN Bioinformatics | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ |
| Adobe Illustrator | Adobe | https://www.adobe.com/ |
Resource Availability
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Bart N. Lambrecht (bart.lambrecht@ugent.be).
Data and Code Availability
Cell type-specific expression patterns of individual genes together with the gene-barcode matrices and annotation matrices can be accessed via an online browser tool (http://bioit2.irc.ugent.be/cdc2/). The accession number for the raw RNA-sequencing data reported in this paper is GSE149619.
Experimental Model and Subject Details
In vivo Animal Studies
The following mice were used in this study; female C57BL/6 mice (aged 6-10 weeks) were purchased from Janvier (France). Fcer1g −/− mice were purchased from Taconic (USA). PVM M37-47 CD4+ and N339–347 specific CD8+ TCR transgenic mice were generated in house (Vandersarren et al., 2017, Vanheerswynghels et al., 2018, Walsh et al., 2013) (Figure S2). CD45.1, CD45.1.2, Der p 1-specific (1-Der) CD4+ TCR Tg, PVM CD4+ TCR Tg Rag2 −/−, PVM CD8+ PVM TCR Tg Rag2 −/−, FcεRIa −/−, Ccr2 −/− , Flt3l −/− , Csf2rb -/ , - Irf4 fl/fl and Irf8 fl/fl Itgax-cre mice (Caton et al., 2007, Persson et al., 2013, Plantinga et al., 2013, Sichien et al., 2016) were bred in house in specific pathogen-free conditions at the animal facility of Ghent University. Itgax-cre mice with germline deletion were detected by PCR and excluded from further analysis. Ifnar1 −/− mice were kindly donated by Prof. Dr. Roos Vandenbroucke (Grine et al., 2015). Mice for the Ms and Nb skin infection models were bred and housed at the Malaghan Institute of Medical Research animal facility under specific pathogen-free conditions and age- and sex-matched mice were used within experiments. C57BL/6J were originally obtained from the Jackson Laboratory (Bar Harbor, ME). B6.Cg-Tg(CAG-tdKaede)15Utr mice (Ando et al., 2002, Tomura et al., 2008) were bred from pairs obtained from Dr. Tatyana Chtanova (Garvan Institute of Medical Research, Australia). All experiments were approved by the independent animal ethical committee “Ethische Commissie Dierproeven – faculteit Geneeskunde en Gezondheids-wetenschappen Universiteit Gent.” Experimental protocols for the Ms and Nb skin infection models were approved by the Victoria University of Wellington Animal Ethics Committee and performed in accordance with institutional guidelines.
Generation of MHC class I-restricted, N339-347 and MHC class II-restricted, M37-47 PVM-specific TCR transgenic mice
The generation of the PVM-TCR transgenic mice was done as described recently (Vanheerswynghels et al., 2018). Briefly, splenocytes retrieved from C57BL/6 mice 16 days after sublethal i.t. infection (35 pfu) with PVM (J3666 strain), were depleted for either CD8+ or CD4+ cells through negative selection by MACS, and then restimulated with either the CD4+ or CD8+ immunodominant PVM epitope corresponding to amino acids 37-47 in the matrix protein (M37-47) (Vandersarren et al., 2017) or 339-347 in the nucleoprotein (N339-347) (Walsh et al., 2013) respectively. Next, CD4+ or CD8+ cells were positively selected by MACS from these enriched cell pools and immortalized by fusion with BW5147 cells. From specificity-selected monoclonal T cell clones (IL2+CD69+ in DC:T cell co-culture), the Vα and Vβ of the CD4+ TCR were subcloned in the p783 (containing a transcriptional CD8+ silencer element) and p428 expression vector respectively. For the CD8+ TCR both the Vα and Vβ were subcloned in the p428 expression vector. The TCR constructs were micro-injected in fertilized Rag2 −/− oocytes. Validation of the TCR transgenic mice was done by genotyping using primers from the TCR PCR panel corresponding to the construct (TCR Vα and Vβ), analysis of T cells in peripheral blood (Figures S2B and S2E) and in vitro evaluation of TCR specificity in a DC-T cell co-culture (Figures S2A and S2D). Finally, adoptive transfer of TCR transgenic T cells into PVM infected wild-type acceptor mice provided in vivo validation (Figures S2C and S2F).
Method Details
BM Chimeric Mice
BM was prepared by crushing femurs and tibias in phosphate buffer saline (PBS) followed by filtering through a sterile 70 μm cell strainer. BM cells were suspended in RPMI-1640 medium (Sigma-Aldrich) supplemented with 50% FCS and 10% DMSO (Sigma-Aldrich) and stored at −150°C. BM chimeras were generated by lethally irradiating (8 Gy dose) CD45.2+ or CD45.1.2+ C57BL/6 female recipient mice aged 6-8 weeks. Five hours later, mice received 2 x 106 mixed BM cells of gender- and age-matched CD45.1+ WT and CD45.2+ transgenic donor mice (either Csf2rb −/−, Ccr2 −/−, Irf8 fl/fl Itgax-cre Tg, Irf8 fl/fl Itgax-cre Wt or Ifnar1 −/−), at a 50-50 ratio. Chimeric mice were subjected to infection 12 weeks upon reconstitution.
Infection, immunizations and in vivo treatments
Mice were anesthetized with isoflurane (2 l/min, 2%–3%) and infected intranasally with 105 TCID50 H3N2 X31 influenza A virus or mock virus (allantoic fluid of uninfected eggs); all diluted in 50 μl PBS (GeurtsvanKessel et al., 2008, Neyt et al., 2016). Mouse-passaged stocks of PVM strain J3666 were grown as described (Cook et al., 1998). Anesthetized mice were infected intratracheally with a previously in vivo titrated sub-lethal dose of 35 pfu PVM virus (J3666) diluted in 80 μl PBS (Vandersarren et al., 2017). Weight loss was monitored daily. For HDM (Greer Laboratories) experiments, 100 μg HDM diluted in 80 μl PBS was administered i.t. as described previously (Plantinga et al., 2013) or 75 μg HDM (Greer Laboratories, USA) in combination with 300 ng LPS (Invivogen) and beta-glucans (Provided by Dr. David L. Williams (Hadebe et al., 2015)).
For skin immunizations, mice were anesthetized and injected intradermally (i.d.) with 4x106 CFU heat-killed Ms (Connor et al., 2014) or 300 non-viable Nb into the ear pinna (Camberis et al., 2013). For photoconversion experiments, mice were anesthetized and each side of the ear was exposed to a 130mV 405nm violet laser for 10 s, while all other parts of the head were protected by a face shield. Immediately after photoconversion, fluorescently labeled antigens were injected intradermally as mentioned before.
Preparation of immunogens
M. smegmatis (Ms, mc2155) was grown in Luria-Bertani broth overnight at 37°C. Bacteria were washed three times in PBS containing 0.05% Tween 80 and heat-killed at 75°C for 1 hour. N. brasiliensis (Nb) infective L3 larvae were prepared, washed in sterile PBS and killed by three freeze-thaw cycles as described (Camberis et al., 2013). For fluorescent labeling of pathogens, non-viable Ms or Nb were labeled with CellTrace™ Far Red (ThermoFisher Scientific) according to the manufacturer’s instructions.
Immune serum transfer
Whole blood from 8-10 weeks old PVM convalescent female C57BL/6 mice was obtained at 15dpi with 35 pfu or from mock infected controls by terminal bleeding. Whole blood was allowed to clot at room temperature (RT) for 30 minutes, then centrifuged for 10 minutes at 1000 g at RT. Next, collected serum was heat-inactivated at 56°C for 30 minutes and stored at −20°C. Serum was diluted 1:2 in PBS (ThermoFisher Scientific) and administered i.t. in a volume of 80 μl at 6dpi with PVM.
OVA and OVA immune complexes
Ovalbumine (Invivogen) was fluorescently labeled with AF647 Succinimidyl Ester (ThermoFisher Scientific) according to the manufacturer’s instructions. Immune complexes were pre-formed at RT for 30 minutes by using 2 different monoclonal anti-OVA IgG2c antibodies in a 1:1 molar ratio with OVA-AF647. For the anti-OVA IgG2c antibodies, mRNA from hybridomas (clone 23-9 and 27-6; mIgG1) gifted by Dr. H. Karasuyama, was reverse transcribed and variable domains were amplified, sequenced and recloned in mouse IgG2c constant domains. HEK293-E produced antibodies were purified using MabSelectSure LX affinity and Superdex 200 gelfiltration (GE Life Sciences). At 6dpi with PVM, 50μg OVA-AF647 alone or as an immune complex was administered i.t. in a volume of 80 μl PBS.
Flt3L Bone marrow cultures
Single BM cell suspensions were suspended in tissue culture medium (TCM; RPMI-1640 medium (Thermo Fisher Scientific) containing 10% fetal calf serum (FCS, Bodinco), 1.1 mg/ml β-Mercaptoethanol (Sigma-Aldrich), 2 mM L-alanyl-L-glutamine dipeptide (Thermo Fisher Scientific) and 56 μg/ml Gentamicin (Thermo Fisher) Scientific) supplemented with 250 ng/ml Flt3L (PSF, VIB Protein Service Facility). Cells were plated at 2x106 cells per well in 2 ml of TCM in 24-well plates and cultured at 37°C in a humidified atmosphere at 5% CO2 and kept for 3 days. OP9 cells were cultured in OP9 culture medium (MEMα medium (Thermo Fisher) containing 20% FCS (Bodinco), 2 mM L-alanyl-L-glutamine dipeptide (Thermo Fisher Scientific) and 56 μg/ml Gentamicin (Thermo Fisher Scientific)) (Van de Walle et al., 2011). On day 3 of differentiation, half of the volume (i.e., 1 ml) of BM cells in TCM was transferred to a single well containing a monolayer of OP9 cells in 1ml fresh OP9 medium supplemented with 250 ng/ml Flt3L in 24-well plates (Kirkling et al., 2018). On day 8 of differentiation, half of the medium was refreshed (i.e., 1ml 50:50 OP9:TCM medium with 250 ng/ml Flt3L) and supplemented with LPS (200 ng/ml; Invivogen), Poly (I:C) (1000 ng/ml; Invivogen), R848 (1000 mg/ml; Invivogen,), CpG (100 ng/ml; Invivogen), IFNα (200 ng/ml; Donated by Prof. Dr. Vandenbroucke (Meister et al., 1986)), IFNγ (200 ng/ml; R&D systems), GM-CSF (300 ng/ml; PSF, VIB Protein Service Facility), IL-33 (200 ng/ml; R&D systems), TSLP (200 ng/ml; Thermo Fisher Scientific) and IL-25 (200 ng/ml; R&D systems). For OVA-uptake assay 10 μg/ml OVA-AF647 either alone or as IC was added immediately after adding the stimuli. The cultures were harvested 20h later.
Tissue sampling and processing
Mice were sacrificed at time points indicated by intraperitoneal (i.p.) injection of sodium pentobarbital. The lungs were then mechanically disrupted by GentleMACS dissociation (Miltenyi Biotec) (Lung program 01_01) in 3 ml RPMI 1640 (GIBCO) containing 20 μg/ml Liberase and 10 U/ml Dnase (Roche), followed by digestion for 30 min at 37°C and final GentleMacs homogenization (Lung program 02_01). Single cell suspensions were prepared by digestion in collagenase/DNase (Roche) solution; lungs were digested for 30 minutes at 37°C, MLNs were digested 15 minutes at 37°C. Next, the cell suspension was passed through a 100 μm filter and red blood cells were lysed using ammonium chloride lysis buffer (10 mM KHCO3, 155 mM NH4Cl, 0,1 mM EDTA in MilliQ water). Auricular LNs were digested with 100μg/ml Liberase TL and 100μg/ml DNase I (Roche) for 30 minutes at 37°C before passing through a 70 μm cell strainer.
Flow cytometry and cell sorting
Single cell suspensions were incubated with a mix of fluorescently labeled monoclonal antibodies (Ab) for 30 minutes at 4°C. To reduce non-specific binding, 2.4G2 Fc receptor Ab was added. Dead cells were removed from analysis, using fixable viability dye eFluor506 (eBioscience). For intracellular staining, cells were fixed using a fixation/permeabilization kit (Thermo Fisher Scientific) according to the manufacturer’s protocol. For cell restimulation, 5 x 106 cells were incubated with 1 ml 1x Cell Stimulation Cocktail (eBioscience) for 6h at 37°C and fixed using a fixation/permeabilization kit (Thermo Fisher Scientific). In order to monitor individual cell divisions of the T cells, cells were stained with Cell proliferation dye eFl450 (CTV, Thermo Fisher Scientific) according to the manufacturer’s protocol.
Before acquisition, photomultiplier tube voltages were adjusted to minimize fluorescence spillover. Single-stain controls were prepared with UltraComp eBeads (Thermo Fisher Scientific) following the manufacturer’s instructions, and were used to calculate a compensation matrix. Sample acquisition was performed on a LSR Fortessa or FACSymphony cytometer equipped with FACSDiva software (BD biosciences). Final analysis and graphical output were performed using FlowJo software (Tree Star, Inc.) and GraphPad Prism 6 (GraphPad Software, Inc.). For sorting of DC subsets and T cells, cells were stained as described and cell sorting was performed on a FACSAria II and III (BD biosciences).
Pre-cDC transfer
Pre-cDC transfer was done as previously described (Scott et al., 2015). Briefly, to expand pre-cDCs, CD45.2 wild-type mice were injected every other day with 10 μg Flt3L (PSF, VIB). Mice were killed 8 days after the first treatment and bone marrow was isolated. Red blood cells were lysed by ammonium chloride osmotic lysis buffer. Cells were labeled with eFl450 cell proliferation dye (Thermo Fisher Scientific) and pre-cDCs were sorted as CD45+, Lin- (CD3, CD19, MHC II, CD49b, CD11b, B220), CD11cint, Sirpαint. 8 × 105 cells were injected i.v. into CD45.1/2 wild-type mice 1dpi. Lung and MLN cells were analyzed 4 days later (5dpi).
Monocyte transfer
Monocyte transfer was done as previously described (Bain et al., 2013). Bone marrow cells were isolated from CD45.1 wild-type mice. Red blood cells were lysed by using osmotic lysis buffer. Cells were labeled with eFl450 cell proliferation dye (CTV, Thermo Fisher Scientific) and monocytes were sorted as Lin- (CD3, CD19, MHCII, SiglecF, Ly6G), CD11c-, c-kit-, CD11bhi and Ly6Chi. 8 × 105 cells were injected i.v. into CD45.2 Ccr2 −/− mice 1dpi. Lung and MLN cells were analyzed 4 days later (5dpi).
DC–CD4+ and CD8+ T cell co-culture
Migratory DCs harvested from the MLN and lung 8 dpi with PVM or 72h after challenge with HDM, LPS and beta-glucans, were enriched by gradient selection (Bosteels et al., 2018) and sorted by distinct subset. Naive CD44-CD62L+Lin-(CD11c, MHCII, CD19) CD4+ and CD8+ T cells specific for the H2-IAb-restricted M37-47 (Vandersarren et al., 2017) and the H-2b-restricted N339–347 (Walsh et al., 2013) PVM-epitope respectively or H2-IAb-restricted Der p 1 peptide (Plantinga et al., 2013), were isolated and sorted from spleens and LN of Rag2 −/− TCR Tg mice and labeled with Cell proliferation dye eFluor450 (CTV, Thermo Fisher Scientific) according to the manufacturer’s instructions. 5000 sorted DCs were co-cultured with sorted T cells in a 1:10 DC:T cell ratio in sterile tissue culture medium (TCM; RPMI (GIBCO) containing 10% fetal calf serum (Bodinco), 1.1 mg/ml β-Mercaptoethanol (Sigma-Aldrich), 2 mM L-alanyl-L-glutamine dipeptide (Thermo Fisher Scientific) and 56 μg/ml Gentamicin (Thermo Fisher Scientific)) on ice. CTV dilution and T cell activation were evaluated by flow cytometry after 4 days of incubation at 37°C, 5% CO2 and 20% O2. For peptide controls, 5 μg/ml PVM M37-47, 1 ug/ml PVM N339-347 or (Pepscan) or 5 ug/ml Der p 1 peptide (Pepscan) were added to the cultures. Cytokine production by the PVM-specific or 1-Der T cells was measured in a Bio-Plex system (Biorad) on the co-culture supernatants.
Micro-array
10000 cells of each of the DC subsets were isolated from the lung and MLN 4dpi as described above. RNA was obtained with an RNEasy Plus Micro Kit according to the manufacturer’s instructions (QIAGEN) and RNA integrity was assessed with a Bioanalyser 2100 (Agilent). With a WT Expression Kit (Ambion) 50 ng of total RNA per sample ‘spiked’ with bacterial poly(A) RNA positive control (Affymetrix) was converted to double-stranded cDNA in a reverse-transcription reaction. Samples were fragmented and labeled with biotin in a terminal labeling reaction according to the Affymetrix WT Terminal Labeling Kit. A mixture of fragmented biotinylated cDNA and hybridization controls (Affymetrix) was hybridized on a GeneChip Mouse Gene 1.0 ST Array (Affymetrix), followed by staining and washing in a GeneChip fluidics station 450 according to the manufacturer’s procedures (Affymetrix). For analysis of raw probe signal intensities, chips were scanned with a GeneChip scanner 3000 (Affymetrix). Samples were subsequently analyzed with software of the R project for statistical computing (Bioconductor). All samples passed quality control, and the robust multiarray average procedure was used for normalization of data within arrays (probe-set summarization, background correction and log2 transformation) and between arrays (quantile normalization). In a subsequent step, probe sets that either mapped to multiple genes or had low variance were filtered out. The final analysis of the obtained data was performed by using Ingenuity Pathway Analysis (IPA, QIAGEN).
Single Cell RNA Sequencing
Sorting and RNA Isolation
Single cell suspensions were obtained as described earlier from the pooled lungs of 8 WT:Irf8 fl/fl Itgax-CreTg or 6 WT: Irf8 fl/fl Itgax-CreWt. chimeras at 10dpi with PVM. Prior to FACS purification, cells were stained with biotinylated CD3eCD19and Ly6G, followed by MagniSort Streptavidin negative selection beads. Enrichment by negative selection prior to sorting was done according to the manufacturer’s instructions (Thermo Fisher Scientific). 72000 SiglecF-CD3-CD19-CD11c+MHCII+ CD45.1 and CD45.2 live cells from lungs 10 dpi with PVM of WT(CD45.1):Irf8fl/fl Itgax-Cre Tg(CD45.2) and control WT(CD45.1):Irf8fl/fl Itgax-Cre Wt(CD45.2) chimeric mice were FACS-purified. Cells were sorted into PBS 0.04% BSA, spun down and resuspended in PBS with 0.04% BSA at an estimated final concentration of 1000 cells/μl. Cellular suspensions (target recovery of 10000 cells) were loaded on a GemCode Single-Cell Instrument (10x Genomics, Pleasanton) to generate single-cell Gel Bead-in-EMulsion (GEMs). Single-cell RNA-Seq libraries were prepared using GemCode Single-Cell 3ʹGel Bead and Library Kit (10x Genomics) according to the manufacturer’s instructions and as previously described (Scott et al., 2018).
Single Cell RNA Sequencing Analysis
Sequencing libraries were loaded on an HiSeq4000 (Illumina, San Diego, Ca) with custom sequencing settings (28/8/0/98 - 2.1pM loading concentration). Sequencing was performed at the VIB Nucleomics Core (VIB, Leuven). Demultiplexing of the raw data and mapping to the mouse genome mm10 was done by the 10X CellRanger software (version 2.0.0 and version 2.0.2; cellranger). The aggregation of the WT (CD45.1) and transgenic (CD45.2 either Irf8fl/fl Itgax-CreTg or Itgax-CreWt) samples was done using ‘cellranger aggr’ (10x’s CellRanger software).
Preprocessing Data
Preprocessing of the data was done by the scran and scater R package according to workflow proposed by the Marioni lab (Lun et al., 2016). Outlier cells were identified based on 3 metrics (library size, number of expressed genes and mitochondrial proportion) and cells were tagged as outliers when they were 3 median absolute deviation (MADs) away from the median value of each metric across all cells. Detecting highly variable genes, finding clusters and creating tSNE plots was done using the Seurat pipeline. Marker genes per identified subpopulation were found using the findMarker function of the Seurat pipeline. Additional low quality (low UMI counts, high % of mitochondrial genes) and contaminating cells (potential doublets, lymphocytes, non-immune cells) were also removed from the analysis (Figure S4A).
Single-cell regulatory network inference and clustering using SCENIC
We performed SCENIC (Aibar et al., 2017) by starting from the raw counts and following the proposed workflow using the default parameters. The co-expression network was generated using GENIE3 rather than GRNBoost2. AUC, which identifies and scores gene regulatory networks or regulons in single cells, was calculated using AUCell as previously described (Aibar et al., 2017). The better the targets of a regulon match the highly expressed genes of a certain cell, the higher the AUC value of that regulon in that particular cell.
Quantification and Statistical Analysis
In all experiments, data are presented as mean ± SEM. Statistical tests were selected based on appropriate assumptions with respect to data distribution and variance characteristics. One-way ANOVA with Sidak correction for multiple testing was used for the statistical analysis of differences between more than two groups. Two-way ANOVA with Sidak post-test was used for the statistical analysis of differences between more than two groups and with 2 different independent variables. Statistical significance was defined as p < 0.05. Sample sizes were chosen according to standard guidelines. Number of animals is indicated as ‘‘n.’’ Of note, sizes of the tested animal groups were also dictated by availability of the transgenic strains and litter sizes, allowing littermate controls. Statistical details of experiments can be found in the figure legends. The investigator was not blinded to the mouse group allocation.
Acknowledgments
We thank the technicians of the Hammad, Lambrecht, and Guilliams lab and the VIB Nucleomics and Flow cores. B.N.L. is supported by a European Research Council (ERC) advanced grant, a concerted research initiative grant (GOA) from Ghent University, and an Excellence of Science (EOS) research grant. M.G. is supported by Fonds Wetenschappelijk Onderzoek Vlaanderen (FWO), a GOA from Ghent University, and an ERC consolidator grant. Y.S. is supported by the FWO and is a Marylou Ingram Scholar. C.L.S. is supported by the FWO and an ERC starting grant. C.B., K.N., N.D., and V.B. are supported by grants from FWO. D.S., M.L., and E.L. are supported by Flanders Innovation & Entrepreneurship (VLAIO). This research was further funded by a Health Research Council of New Zealand Independent Research Organisation grant (to the Malaghan Institute) and a Health Research Council of NZ project grant (to F.R.).
Author contributions
Conceptualization, B.N.L., C.B., K.N., and M.G.; Methodology, C.B., K.N., D.S., and C.L.S.; Software, N.V., L.M., and Y.S.; Validation, S.-C.T., J.U.M., and F.R.; Formal Analysis, C.B., N.V., L.M., and Y.S.; Investigation, C.B., K.N., and J.U.M.; Resources, M.L. and E.L.; Data Curation, C.B., K.N., and N.V.; Writing – Original Draft, C.B., K.N., C.L.S., M.G., and B.N.L.; Writing – Review & Editing, C.B., J.U.M., C.L.S., M.G., and B.N.L.; Visualization, C.B., K.N., and N.V.; Supervision, M.J.v.H., Y.S., F.R., H.H., M.G., and B.N.L.; Project Administration, C.B., K.N., M.V., N.V., N.D., V.B., and J.U.M.; Funding Acquisition, C.B. K.N., F.R., H.H., M.G., and B.N.L.
Declaration of Interests
The authors declare no competing interests.
Published: May 8, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.04.005.
Contributor Information
Martin Guilliams, Email: martin.guilliams@ugent.vib.be.
Bart N. Lambrecht, Email: bart.lambrecht@ugent.be.
Supplemental Information
References
- Aibar S., González-Blas C.B., Moerman T., Huynh-Thu V.A., Imrichova H., Hulselmans G., Rambow F., Marine J.C., Geurts P., Aerts J. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods. 2017;14:1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsua-Enrich E., Hatipoglu I., Kadel S., Turner S., Paul J., Singh S., Bagavant H., Kovats S. IRF4-dependent dendritic cells regulate CD8+ T-cell differentiation and memory responses in influenza infection. Mucosal Immunol. 2019;12:1025–1037. doi: 10.1038/s41385-019-0173-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando R., Hama H., Yamamoto-Hino M., Mizuno H., Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagadia P., Huang X., Liu T.-T., Durai V., Grajales-Reyes G.E., Nitschké M., Modrusan Z., Granja J.M., Satpathy A.T., Briseño C.G. An Nfil3-Zeb2-Id2 pathway imposes Irf8 enhancer switching during cDC1 development. Nat. Immunol. 2019;20:1174–1185. doi: 10.1038/s41590-019-0449-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain C.C., Scott C.L., Uronen-Hansson H., Gudjonsson S., Jansson O., Grip O., Guilliams M., Malissen B., Agace W.W., Mowat A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013;6:498–510. doi: 10.1038/mi.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S., Roach K., Turner S., Paul J., Kovats S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J. Immunol. 2012;189:3368–3377. doi: 10.4049/jimmunol.1102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaña S., Turner S., Paul J., Ainsua-Enrich E., Kovats S. IRF4 and IRF8 Act in CD11c+ Cells To Regulate Terminal Differentiation of Lung Tissue Dendritic Cells. J. Immunol. 2016;196:1666–1677. doi: 10.4049/jimmunol.1501870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A., León B., Lund F.E., Randall T.D. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat. Immunol. 2010;11:216–224. doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista A.P., Gola A., Huang Y., Milanez-Almeida P., Torabi-Parizi P., Urban J.F., Jr., Shapiro V.S., Gerner M.Y., Germain R.N. The Chemoattractant Receptor Ebi2 Drives Intranodal Naive CD4+ T Cell Peripheralization to Promote Effective Adaptive Immunity. Immunity. 2019;50:1188–1201.e6. doi: 10.1016/j.immuni.2019.04.001. [DOI] [PubMed] [Google Scholar]
- Bosteels C., Scott C.L. Transcriptional regulation of DC fate specification. Mol. Immunol. 2020;121:38–46. doi: 10.1016/j.molimm.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosteels C., Lambrecht B.N., Hammad H. Isolation of conventional murine lung dendritic cell subsets. Curr. Protoc. Immunol. 2018;120:3.7B.1–3.7B.16. doi: 10.1002/cpim.39. [DOI] [PubMed] [Google Scholar]
- Briseño C.G., Haldar M., Kretzer N.M., Wu X., Theisen D.J., Kc W., Durai V., Grajales-Reyes G.E., Iwata A., Bagadia P. Distinct Transcriptional Programs Control Cross-Priming in Classical and Monocyte-Derived Dendritic Cells. Cell Rep. 2016;15:2462–2474. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorf S., Kautz A., Böhnert V., Knolle P.A., Kurts C. Distinct pathways of antigen uptake and intracellular routing in CD4 and CD8 T cell activation. Science. 2007;316:612–616. doi: 10.1126/science.1137971. [DOI] [PubMed] [Google Scholar]
- Cabeza-Cabrerizo M., van Blijswijk J., Wienert S., Heim D., Jenkins R.P., Chakravarty P., Rogers N., Frederico B., Acton S., Beerling E. Tissue clonality of dendritic cell subsets and emergency DC poiesis revealed by multicolor fate mapping of DC progenitors. Sci. Immunol. 2019;4:eaaw1941. doi: 10.1126/sciimmunol.aaw1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camberis M., Prout M., Tang S.C., Forbes-Blom E., Robinson M., Kyle R., Belkaid Y., Paul W., Le Gros G. Evaluating the in vivo Th2 priming potential among common allergens. J. Immunol. Methods. 2013;394:62–72. doi: 10.1016/j.jim.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L.A. The convalescent sera option for containing COVID-19. J. Clin. Invest. 2020;130:1545–1548. doi: 10.1172/JCI138003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8- dendritic cells in the spleen. J. Exp. Med. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong C., Matos I., Choi J.H., Dandamudi D.B., Shrestha E., Longhi M.P., Jeffrey K.L., Anthony R.M., Kluger C., Nchinda G. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor L.M., Tang S.-C., Camberis M., Le Gros G., Ronchese F. Helminth-conditioned dendritic cells prime CD4+ T cells to IL-4 production in vivo. J. Immunol. 2014;193:2709–2717. doi: 10.4049/jimmunol.1400374. [DOI] [PubMed] [Google Scholar]
- Connor L.M., Tang S.-C., Cognard E., Ochiai S., Hilligan K.L., Old S.I., Pellefigues C., White R.F., Patel D., Smith A.A.T. Th2 responses are primed by skin dendritic cells with distinct transcriptional profiles. J. Exp. Med. 2017;214:125–142. doi: 10.1084/jem.20160470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook P.M., Eglin R.P., Easton A.J. Pathogenesis of pneumovirus infections in mice: detection of pneumonia virus of mice and human respiratory syncytial virus mRNA in lungs of infected mice by in situ hybridization. J. Gen. Virol. 1998;79:2411–2417. doi: 10.1099/0022-1317-79-10-2411. [DOI] [PubMed] [Google Scholar]
- Deckers J., Sichien D., Plantinga M., Van Moorleghem J., Vanheerswynghels M., Hoste E., Malissen B., Dombrowicz D., Guilliams M., De Bosscher K. Epicutaneous sensitization to house dust mite allergen requires interferon regulatory factor 4-dependent dermal dendritic cells. J. Allergy Clin. Immunol. 2017;140:1364–1377.e2. doi: 10.1016/j.jaci.2016.12.970. [DOI] [PubMed] [Google Scholar]
- Desch A.N., Gibbings S.L., Clambey E.T., Janssen W.J., Slansky J.E., Kedl R.M., Henson P.M., Jakubzick C. Dendritic cell subsets require cis-activation for cytotoxic CD8 T-cell induction. Nat. Commun. 2014;5:4674. doi: 10.1038/ncomms5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Chae G.P. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- Durai V., Bagadia P., Granja J.M., Satpathy A.T., Kulkarni D.H., Davidson J.T., 4th, Wu R., Patel S.J., Iwata A., Liu T.-T. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat. Immunol. 2019;20:1161–1173. doi: 10.1038/s41590-019-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 2010;207:823–836. doi: 10.1084/jem.20091627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B., Tussiwand R., Dreesen L., Fairfax K.C., Huang S.C.C., Smith A.M., O’Neill C.M., Lam W.Y., Edelson B.T., Urban J.F., Jr. Migratory CD103+ dendritic cells suppress helminth-driven type 2 immunity through constitutive expression of IL-12. J. Exp. Med. 2016;213:35–51. doi: 10.1084/jem.20150235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Nish S.A., Jiang R., Hou L., Licona-Limón P., Weinstein J.S., Zhao H., Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., Immunological Genome Consortium Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Willart M.A., van Rijt L.S., Muskens F., Kool M., Baas C., Thielemans K., Bennett C., Clausen B.E., Hoogsteden H.C. Clearance of influenza virus from the lung depends on migratory langerin+CD11b- but not plasmacytoid dendritic cells. J. Exp. Med. 2008;205:1621–1634. doi: 10.1084/jem.20071365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α(+) conventional DC clonogenic progenitor. Nat. Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson M.H., Cheung D., Rohlfing M.M., Kitchens R., Spiegel D.E., Tucker J., Battaile J.T., Alevy Y., Yan L., Agapov E. Induction of high-affinity IgE receptor on lung dendritic cells during viral infection leads to mucous cell metaplasia. J. Exp. Med. 2007;204:2759–2769. doi: 10.1084/jem.20070360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M., Helft J., Chow A., Hashimoto D., Mortha A., Agudo-Cantero J., Bogunovic M., Gautier E.L., Miller J., Leboeuf M. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36:1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grine L., Dejager L., Libert C., Vandenbroucke R.E. Dual Inhibition of TNFR1 and IFNAR1 in Imiquimod-Induced Psoriasiform Skin Inflammation in Mice. J. Immunol. 2015;194:5094–5102. doi: 10.4049/jimmunol.1403015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Ginhoux F., Jakubzick C., Naik S.H., Onai N., Schraml B.U., Segura E., Tussiwand R., Yona S. Dendritic cells, monocytes and macrophages: a unified nomenclature based on ontogeny. Nat. Rev. Immunol. 2014;14:571–578. doi: 10.1038/nri3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Bruhns P., Saeys Y., Hammad H., Lambrecht B.N. The function of Fcγ receptors in dendritic cells and macrophages. Nat. Rev. Immunol. 2014;14:94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- Guilliams M., Dutertre C.A., Scott C.L., McGovern N., Sichien D., Chakarov S., Van Gassen S., Chen J., Poidinger M., De Prijck S. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity. 2016;45:669–684. doi: 10.1016/j.immuni.2016.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan J.M.M., Bevan M.J. Constitutive versus activation-dependent cross-presentation of immune complexes by CD8(+) and CD8(-) dendritic cells in vivo. J. Exp. Med. 2002;196:817–827. doi: 10.1084/jem.20020295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadebe S., Kirstein F., Fierens K., Chen K., Drummond R.A., Vautier S., Sajaniemi S., Murray G., Williams D.L., Redelinghuys P. Microbial Ligand Costimulation Drives Neutrophilic Steroid-Refractory Asthma. PLoS ONE. 2015;10:e0134219. doi: 10.1371/journal.pone.0134219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., Chieppa M., Perros F., Willart M.A., Germain R.N., Lambrecht B.N. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 2009;15:410–416. doi: 10.1038/nm.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., Plantinga M., Deswarte K., Pouliot P., Willart M.A.M., Kool M., Muskens F., Lambrecht B.N. Inflammatory dendritic cells--not basophils--are necessary and sufficient for induction of Th2 immunity to inhaled house dust mite allergen. J. Exp. Med. 2010;207:2097–2111. doi: 10.1084/jem.20101563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J., Manicassamy B., Guermonprez P., Hashimoto D., Silvin A., Agudo J., Brown B.D., Schmolke M., Miller J.C., Leboeuf M. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Invest. 2012;122:4037–4047. doi: 10.1172/JCI60659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S. Batf3 Deficiency Reveals a Critical Role for CD8 + Dendritic Cells in Cytotoxic T Cell Immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hintzen G., Ohl L., del Rio M.-L., Rodriguez-Barbosa J.-I., Pabst O., Kocks J.R., Krege J., Hardtke S., Förster R. Induction of tolerance to innocuous inhaled antigen relies on a CCR7-dependent dendritic cell-mediated antigen transport to the bronchial lymph node. J. Immunol. 2006;177:7346–7354. doi: 10.4049/jimmunol.177.10.7346. [DOI] [PubMed] [Google Scholar]
- Iijima N., Mattei L.M., Iwasaki A. Recruited inflammatory monocytes stimulate antiviral Th1 immunity in infected tissue. Proc. Natl. Acad. Sci. USA. 2011;108:284–289. doi: 10.1073/pnas.1005201108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov S., Scallan J.P., Kim K.W., Werth K., Johnson M.W., Saunders B.T., Wang P.L., Kuan E.L., Straub A.C., Ouhachi M. CCR7 and IRF4-dependent dendritic cells regulate lymphatic collecting vessel permeability. J. Clin. Invest. 2016;126:1581–1591. doi: 10.1172/JCI84518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal H., Kaushik M., Sougrat R., Gupta M., Dey A., Verma R., Ozato K., Tailor P. Batf3 and Id2 have a synergistic effect on Irf8-directed classical CD8α+ dendritic cell development. J. Immunol. 2013;191:5993–6001. doi: 10.4049/jimmunol.1203541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalergis A.M., Ravetch J.V. Inducing tumor immunity through the selective engagement of activating Fcgamma receptors on dendritic cells. J. Exp. Med. 2002;195:1653–1659. doi: 10.1084/jem.20020338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkling M.E., Cytlak U., Lau C.M., Lewis K.L., Resteu A., Khodadadi-Jamayran A., Siebel C.W., Salmon H., Merad M., Tsirigos A. Notch Signaling Facilitates In Vitro Generation of Cross-Presenting Classical Dendritic Cells. Cell Rep. 2018;23:3658–3672.e6. doi: 10.1016/j.celrep.2018.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Koker S., Van Hoecke L., De Beuckelaer A., Roose K., Deswarte K., Willart M.A., Bogaert P., Naessens T., De Geest B.G., Saelens X. Inflammatory monocytes regulate Th1 oriented immunity to CpG adjuvanted protein vaccines through production of IL-12. Sci. Rep. 2017;7:5986. doi: 10.1038/s41598-017-06236-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M., Soullié T., van Nimwegen M., Willart M.A.M., Muskens F., Jung S., Hoogsteden H.C., Hammad H., Lambrecht B.N. Alum adjuvant boosts adaptive immunity by inducing uric acid and activating inflammatory dendritic cells. J. Exp. Med. 2008;205:869–882. doi: 10.1084/jem.20071087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M., Pétrilli V., De Smedt T., Rolaz A., Hammad H., van Nimwegen M., Bergen I.M., Castillo R., Lambrecht B.N., Tschopp J. Cutting edge: alum adjuvant stimulates inflammatory dendritic cells through activation of the NALP3 inflammasome. J. Immunol. 2008;181:3755–3759. doi: 10.4049/jimmunol.181.6.3755. [DOI] [PubMed] [Google Scholar]
- Kool M., Fierens K., Lambrecht B.N. Alum adjuvant: some of the tricks of the oldest adjuvant. J. Med. Microbiol. 2012;61:927–934. doi: 10.1099/jmm.0.038943-0. [DOI] [PubMed] [Google Scholar]
- Kretzer N.M., Theisen D.J., Tussiwand R., Briseño C.G., Grajales-Reyes G.E., Wu X., Durai V., Albring J., Bagadia P., Murphy T.L., Murphy K.M. RAB43 facilitates cross-presentation of cell-associated antigens by CD8α+ dendritic cells. J. Exp. Med. 2016;213:2871–2883. doi: 10.1084/jem.20160597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy J.K., Gowthaman U., Zhang B., Mattsson J., Szeponik L., Liu D., Wu R., White T., Calabro S., Xu L. Migratory CD11b + conventional dendritic cells induce T follicular helper cell–dependent antibody responses. Sci. Immunol. 2017;2:eaam9169. doi: 10.1126/sciimmunol.aam9169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumamoto Y., Linehan M., Weinstein J.S., Laidlaw B.J., Craft J.E., Iwasaki A. CD301b+ dermal dendritic cells drive T helper 2 cell-mediated immunity. Immunity. 2013;39:733–743. doi: 10.1016/j.immuni.2013.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson S.R., Atif S.M., Gibbings S.L., Thomas S.M., Prabagar M.G., Danhorn T., Leach S.M., Henson P.M., Jakubzick C.V. Ly6C(+) monocyte efferocytosis and cross-presentation of cell-associated antigens. Cell Death Differ. 2016;23:997–1003. doi: 10.1038/cdd.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann C.H.K., Baranska A., Heidkamp G.F., Heger L., Neubert K., Lühr J.J., Hoffmann A., Reimer K.C., Brückner C., Beck S. DC subset-specific induction of T cell responses upon antigen uptake via Fcγ receptors in vivo. J. Exp. Med. 2017;214:1509–1528. doi: 10.1084/jem.20160951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- León B., López-Bravo M., Ardavín C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity. 2007;26:519–531. doi: 10.1016/j.immuni.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Luda K.M., Joeris T., Persson E.K., Rivollier A., Demiri M., Sitnik K.M., Pool L., Holm J.B., Melo-Gonzalez F., Richter L. IRF8 Transcription-Factor-Dependent Classical Dendritic Cells Are Essential for Intestinal T Cell Homeostasis. Immunity. 2016;44:860–874. doi: 10.1016/j.immuni.2016.02.008. [DOI] [PubMed] [Google Scholar]
- Vander Lugt B., Khan A.A., Hackney J.A., Agrawal S., Lesch J., Zhou M., Lee W.P., Park S., Xu M., DeVoss J. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat. Immunol. 2014;15:161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- Lun A.T.L., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J.U., Demiri M., Agace W.W., MacDonald A.S., Svensson-Frej M., Milling S.W. Different populations of CD11b+ dendritic cells drive Th2 responses in the small intestine and colon. Nat. Commun. 2017;8:15820. doi: 10.1038/ncomms15820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister A., Uzé G., Mogensen K.E., Gresser I., Tovey M.G., Grütter M., Meyer F. Biological activities and receptor binding of two human recombinant interferons and their hybrids. J. Gen. Virol. 1986;67:1633–1643. doi: 10.1099/0022-1317-67-8-1633. [DOI] [PubMed] [Google Scholar]
- Menezes S., Melandri D., Anselmi G., Perchet T., Loschko J., Dubrot J., Patel R., Gautier E.L., Hugues S., Longhi M.P. The Heterogeneity of Ly6Chi Monocytes Controls Their Differentiation into iNOS+ Macrophages or Monocyte-Derived Dendritic Cells. Immunity. 2016;45:1205–1218. doi: 10.1016/j.immuni.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesnil C., Sabatel C.M., Marichal T., Toussaint M., Cataldo D., Drion P.V., Lekeux P., Bureau F., Desmet C.J. Resident CD11b(+)Ly6C(-) lung dendritic cells are responsible for allergic airway sensitization to house dust mite in mice. PLoS ONE. 2012;7:e53242. doi: 10.1371/journal.pone.0053242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J., Yang D., Kim M., Haam K., Yoo A., Choi J.H., Schraml B.U., Kim Y.S., Kim D., Kang S.J. Inflammation induces two types of inflammatory dendritic cells in inflamed lymph nodes. Exp. Mol. Med. 2018;50:e458. doi: 10.1038/emm.2017.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.L., Grajales-Reyes G.E., Wu X., Tussiwand R., Briseño C.G., Iwata A., Kretzer N.M., Durai V., Murphy K.M. Transcriptional Control of Dendritic Cell Development. Annu. Rev. Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naessens T., Morias Y., Hamrud E., Gehrmann U., Budida R., Mattsson J., Baker T., Skogberg G., Israelsson E., Thörn K. Human Lung Conventional Dendritic Cells Orchestrate Lymphoid Neogenesis During COPD. Am. J. Respir. Crit. Care Med. 2020 doi: 10.1164/rccm.201906-1123OC. Published online April 7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Metcalf D., van Nieuwenhuijze A., Wicks I., Wu L., O’Keeffe M., Shortman K. Intrasplenic steady-state dendritic cell precursors that are distinct from monocytes. Nat. Immunol. 2006;7:663–671. doi: 10.1038/ni1340. [DOI] [PubMed] [Google Scholar]
- Nakano H., Lin K.L., Yanagita M., Charbonneau C., Cook D.N., Kakiuchi T., Gunn M.D. Blood-derived inflammatory dendritic cells in lymph nodes stimulate acute T helper type 1 immune responses. Nat. Immunol. 2009;10:394–402. doi: 10.1038/ni.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Burgents J.E., Nakano K., Whitehead G.S., Cheong C., Bortner C.D., Cook D.N. Migratory properties of pulmonary dendritic cells are determined by their developmental lineage. Mucosal Immunol. 2013;6:678–691. doi: 10.1038/mi.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Moran T.P., Nakano K., Gerrish K.E., Bortner C.D., Cook D.N. Complement receptor C5aR1/CD88 and dipeptidyl peptidase-4/CD26 define distinct hematopoietic lineages of dendritic cells. J. Immunol. 2015;194:3808–3819. doi: 10.4049/jimmunol.1402195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H., Lyons-Cohen M.R., Whitehead G.S., Nakano K., Cook D.N. Distinct functions of CXCR4, CCR2, and CX3CR1 direct dendritic cell precursors from the bone marrow to the lung. J. Leukoc. Biol. 2017;101:1143–1153. doi: 10.1189/jlb.1A0616-285R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyt K., GeurtsvanKessel C.H., Lambrecht B.N. Double-negative T resident memory cells of the lung react to influenza virus infection via CD11c(hi) dendritic cells. Mucosal Immunol. 2016;9:999–1014. doi: 10.1038/mi.2015.91. [DOI] [PubMed] [Google Scholar]
- Persson E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., Agace W.W. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Plantinga M., Guilliams M., Vanheerswynghels M., Deswarte K., Branco-Madeira F., Toussaint W., Vanhoutte L., Neyt K., Killeen N., Malissen B. Conventional and monocyte-derived CD11b(+) dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 2013;38:322–335. doi: 10.1016/j.immuni.2012.10.016. [DOI] [PubMed] [Google Scholar]
- Regnault A., Lankar D., Lacabanne V., Rodriguez A., Théry C., Rescigno M., Saito T., Verbeek S., Bonnerot C., Ricciardi-Castagnoli P., Amigorena S. Fcgamma receptor-mediated induction of dendritic cell maturation and major histocompatibility complex class I-restricted antigen presentation after immune complex internalization. J. Exp. Med. 1999;189:371–380. doi: 10.1084/jem.189.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., Kc W., Wu X., Thomas S.R., Lee W.L., Turkoz M. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 2013;14:937–948. doi: 10.1038/ni.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W.S., See P., Shin A., Wasan P.S. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R., Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 2015;16:718–728. doi: 10.1038/ni.3200. [DOI] [PubMed] [Google Scholar]
- Schraml B.U., van Blijswijk J., Zelenay S., Whitney P.G., Filby A., Acton S.E., Rogers N.C., Moncaut N., Carvajal J.J., Reis e Sousa C. Genetic tracing via DNGR-1 expression history defines dendritic cells as a hematopoietic lineage. Cell. 2013;154:843–858. doi: 10.1016/j.cell.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Scott C.L., Bain C.C., Wright P.B., Sichien D., Kotarsky K., Persson E.K., Luda K., Guilliams M., Lambrecht B.N., Agace W.W. CCR2(+)CD103(-) intestinal dendritic cells develop from DC-committed precursors and induce interleukin-17 production by T cells. Mucosal Immunol. 2015;8:327–339. doi: 10.1038/mi.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C.L., T’Jonck W., Martens L., Todorov H., Sichien D., Soen B., Bonnardel J., De Prijck S., Vandamme N., Cannoodt R. The Transcription Factor ZEB2 Is Required to Maintain the Tissue-Specific Identities of Macrophages. Immunity. 2018;49:312–325.e5. doi: 10.1016/j.immuni.2018.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina N.V., Pamer E.G. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat. Immunol. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- Sheng J., Chen Q., Soncin I., Ng S.L., Karjalainen K., Ruedl C. A Discrete Subset of Monocyte-Derived Cells among Typical Conventional Type 2 Dendritic Cells Can Efficiently Cross-Present. Cell Rep. 2017;21:1203–1214. doi: 10.1016/j.celrep.2017.10.024. [DOI] [PubMed] [Google Scholar]
- Sichien D., Scott C.L., Martens L., Vanderkerken M., Van Gassen S., Plantinga M., Joeris T., De Prijck S., Vanhoutte L., Vanheerswynghels M. IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity. 2016;45:626–640. doi: 10.1016/j.immuni.2016.08.013. [DOI] [PubMed] [Google Scholar]
- Tamoutounour S., Henri S., Lelouard H., de Bovis B., de Haar C., van der Woude C.J., Woltman A.M., Reyal Y., Bonnet D., Sichien D. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 2012;42:3150–3166. doi: 10.1002/eji.201242847. [DOI] [PubMed] [Google Scholar]
- Tamoutounour S., Guilliams M., Montanana Sanchis F., Liu H., Terhorst D., Malosse C., Pollet E., Ardouin L., Luche H., Sanchez C. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Tanaka A., Minoguchi K., Samson K.T.R., Oda N., Yokoe T., Tazaki T., Yamamoto Y., Yamamoto M., Ohta S., Adachi M. Inhibitory effects of suplatast tosilate on the differentiation and function of monocyte-derived dendritic cells from patients with asthma. Clin. Exp. Allergy. 2007;37:1083–1089. doi: 10.1111/j.1365-2222.2006.02616.x. [DOI] [PubMed] [Google Scholar]
- Tang X.-Z., Jung J.B., Allen C.D.C. A case of mistaken identity: The MAR-1 antibody to mouse FcεRIα cross-reacts with FcγRI and FcγRIV. J. Allergy Clin. Immunol. 2019;143:1643–1646.e6. doi: 10.1016/j.jaci.2018.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomura M., Yoshida N., Tanaka J., Karasawa S., Miwa Y., Miyawaki A., Kanagawa O. Monitoring cellular movement in vivo with photoconvertible fluorescence protein “Kaede” transgenic mice. Proc. Natl. Acad. Sci. USA. 2008;105:10871–10876. doi: 10.1073/pnas.0802278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Lee W.L., Murphy T.L., Mashayekhi M., Kc W., Albring J.C., Satpathy A.T., Rotondo J.A., Edelson B.T., Kretzer N.M. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tussiwand R., Everts B., Grajales-Reyes G.E., Kretzer N.M., Iwata A., Bagaitkar J., Wu X., Wong R., Anderson D.A., Murphy T.L. Klf4 expression in conventional dendritic cells is required for T helper 2 cell responses. Immunity. 2015;42:916–928. doi: 10.1016/j.immuni.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Borght K., Scott C.L., Martens L., Sichien D., Van Isterdael G., Nindl V., Saeys Y., Boon L., Ludewig B., Gillebert T.C., Lambrecht B.N. Myocarditis Elicits Dendritic Cell and Monocyte Infiltration in the Heart and Self-Antigen Presentation by Conventional Type 2 Dendritic Cells. Front. Immunol. 2018;9:2714. doi: 10.3389/fimmu.2018.02714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandersarren L., Bosteels C., Vanheerswynghels M., Moon J.J., Easton A.J., Van Isterdael G., Janssens S., Lambrecht B.N., van Helden M.J. Epitope mapping and kinetics of CD4 T cell immunity to pneumonia virus of mice in the C57BL/6 strain. Sci. Rep. 2017;7:3472. doi: 10.1038/s41598-017-03042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanheerswynghels M., Toussaint W., Schuijs M., Vanhoutte L., Killeen N., Hammad H., Lambrecht B.N. The generation and use of allergen-specific TCR transgenic animals. Methods Mol. Biol. 2018;1799:183–210. doi: 10.1007/978-1-4939-7896-0_15. [DOI] [PubMed] [Google Scholar]
- Vermaelen K.Y., Carro-Muino I., Lambrecht B.N., Pauwels R.A. Specific migratory dendritic cells rapidly transport antigen from the airways to the thoracic lymph nodes. J. Exp. Med. 2001;193:51–60. doi: 10.1084/jem.193.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Walle I., De Smet G., Gärtner M., De Smedt M., Waegemans E., Vandekerckhove B., Leclercq G., Plum J., Aster J.C., Bernstein I.D. Jagged2 acts as a Delta-like Notch ligand during early hematopoietic cell fate decisions. Blood. 2011;117:4449–4459. doi: 10.1182/blood-2010-06-290049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh K.B., Sidney J., Welch M., Fremgen D.M., Sette A., Oldstone M.B.A. CD8+ T-cell epitope mapping for pneumonia virus of mice in H-2b mice. J. Virol. 2013;87:9949–9952. doi: 10.1128/JVI.00339-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.W., Tjota M.Y., Clay B.S., Vander Lugt B., Bandukwala H.S., Hrusch C.L., Decker D.C., Blaine K.M., Fixsen B.R., Singh H. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat. Commun. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Briseño C.G., Durai V., Albring J.C., Haldar M., Bagadia P., Kim K.-W., Randolph G.J., Murphy T.L., Murphy K.M. Mafb lineage tracing to distinguish macrophages from other immune lineages reveals dual identity of Langerhans cells. J. Exp. Med. 2016;213:2553–2565. doi: 10.1084/jem.20160600. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cell type-specific expression patterns of individual genes together with the gene-barcode matrices and annotation matrices can be accessed via an online browser tool (http://bioit2.irc.ugent.be/cdc2/). The accession number for the raw RNA-sequencing data reported in this paper is GSE149619.