Abstract
Myocardial infarction (MI) is a leading cause of morbidity and mortality around the world. A major goal of regenerative medicine is to replenish the dead myocardium after MI. Although several strategies have been used to regenerate myocardium, stem cell therapy remains a major approach to replenish the dead myocardium of an MI heart. Accumulating evidence suggests the presence of resident cardiac stem cells (CSCs) in the adult heart and their endocrine and/or paracrine effects on cardiac regeneration. However, CSC isolation and their characterization and differentiation toward myocardial cells, especially cardiomyocytes, remains a technical challenge. In the present study, we provided a simple method for the isolation, characterization, and differentiation of CSCs from the adult mouse heart. Here, we describe a density gradient method for the isolation of CSCs, where the heart is digested by a 0.2% collagenase II solution. To characterize the isolated CSCs, we evaluated the expression of CSCs/cardiac markers Sca-1, NKX2–5, and GATA4, and pluripotency/stemness markers OCT4, SOX2, and Nanog. We also determined the proliferation potential of isolated CSCs by culturing them in a Petri dish and assessing the expression of the proliferation marker Ki-67. For evaluating the differentiation potential of CSCs, we selected seven- to ten-days cultured CSCs. We transferred them to a new plate with a cardiomyocyte differentiation medium. They are incubated in a cell culture incubator for 12 days, while the differentiation medium is changed every three days. The differentiated CSCs express cardiomyocyte-specific markers: actinin and troponin I. Thus, CSCs isolated with this protocol have stemness and cardiac markers, and they have a potential for proliferation and differentiation toward cardiomyocyte lineage.
Keywords: Sca-1, OCT4, Nanog, Ki-67, NKX2–5, GATA4 actinin, troponin I
SUMMARY:
The overall goal of this article is to standardize the protocol for the isolation, characterization, and differentiation of cardiac stem cells (CSCs) from the adult mouse heart. Here, we describe a density gradient centrifugation method to isolate murine CSCs and elaborated methods for CSC culture, proliferation, and differentiation into cardiomyocytes.
INTRODUCTION
Ischemic heart disease, including myocardial infarction (MI), is a major cause of death around the world1. Stem cell therapy for regenerating dead myocardium remains a major approach to improve the cardiac function of an MI heart2–5. Different types of stem cells have been used to replenish dead myocardium and to improve the cardiac function of an MI heart. They can be broadly categorized into embryonic stem cells6 and adult stem cells. In adult stem cells, various types of stem cells have been used, such as bone marrow-derived mononuclear cells7,8, mesenchymal stem cells derived from bone marrow9,10, adipose tissue11,12, and umbilical cord13, and CSCs14,15. Stem cells can promote cardiac regeneration through endocrine and/or paracrine actions16–20. However, a major limitation of stem cell therapy is obtaining an adequate number of stem cells that can proliferate and/or differentiate toward a specific cardiac lineage21,22. Autologous and allogenic transplantation of stem cells is an important challenge in stem cell therapy9. CSCs could be a better approach for cardiac regeneration because they are derived from the heart and they can be more easily differentiated into cardiac lineages than non-cardiac stem cells. Thus, it reduces the risk of teratoma. In addition, the endocrine and paracrine effects of CSCs, such as exosomes and miRNAs derived from the CSCs, could be more effective than other types of stem cells. Thus, CSCs remains a better option for cardiac regeneration 23,24.
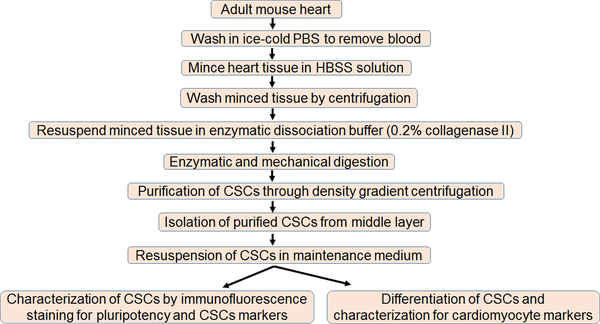
Although CSCs are a better candidate for cardiac regeneration in an MI heart due to their cardiac origin, a major limitation with CSCs is less yield due to the lack of an efficient isolation method. Another limitation could be the impaired differentiation of CSCs toward cardiomyocytes lineage2,25–27. To circumvent these limitations, it is important to develop an efficient protocol for CSC isolation, characterization, and differentiation towards cardiac lineage. There is no single acceptable marker for CSCs and a specific cell-surface marker-based isolation of CSCs yields less CSCs. Here, we standardize a simple gradient centrifugation approach to isolate CSCs from the mouse heart that is cost-effective and results in an increased yield of CSCs. These isolated CSCs can be selected for specific cell-surface markers by fluorescence-activated cell sorting. In addition to CSC isolation, we provided a protocol for CSC culture, characterization, and differentiation towards cardiomyocyte lineage. Thus, we present an elegant method to isolate, characterize, culture, and differentiate CSCs from adult mouse hearts (Figure 5).
Figure 5: Schematic representation of the different steps of CSC isolation, culture, and differentiation.
We used surgically removed hearts from four to five adult mice for the isolation of CSCs. The stepwise process of CSC isolation, culture, and differentiation toward cardiomyocytes lineages are presented.
PROTOCOL:
The housing, anesthesia, and sacrifice of mice were performed following the approved IACUC protocol of the University of Nebraska Medical Center.
-
Materials
-
1.1
Use 10- to 12-week-old C57BL/6J black male mice, kept in-house at the institutional animal facility, for the isolation of CSCs. CSCs can also be isolated from non-pregnant female mice.
-
1.2
Sterilize all the necessary surgical instruments, including surgical scissors, fine surgical scissors, curved shank forceps, and a surgical blade, by autoclaving them before the euthanasia of the mouse.
-
1.3
Prepare CSC isolation buffers in a sterilized condition and store them at 4 °C on ice. These buffers include polysucrose and a sodium diatrizoate solution adjusted to a density of 1.077 g/mL, incomplete Dulbecco’s modified Eagle’s Medium, 1x Ham’s balanced salt solution (HBSS), and cell culture-grade 1x phosphate-buffered saline (PBS). Prepare a 1% collagenase II stock solution (filtered and sterilized) and a 0.2% working solution in HBSS.
-
1.4
Keep the tissue culture materials in the sterilized condition in the biosafety cabinet, including a 10-mm Petri dish, a 6-well plate, a 24-well plate, T-25 and T-75 culture flasks, 15-mL and 50-mL conical tubes, and disposable serological pipettes with 40-μm and 100-μm cell strainers with 0.22-μm filters.
-
1.5
Sterilize the 10-μL, 200-μL, and 1,000-μL pipette tips by autoclave.
-
1.6
Use powder-free nitrile gloves and 70% ethanol to maintain a sterile condition throughout the experiment.
-
1.7
Use 10% bleach as a disinfectant.
-
1.8
Use commercially available CSC maintenance medium and cardiomyocytes differentiation medium for the culture and differentiation of CSCs, respectively.
-
1.1
-
Isolation Method of Cardiac Stem Cells
-
2.1
Euthanize four to five adult male (or non-pregnant female) mice using a CO2 chamber. Fix each mouse’s arms on a separate dissection cardboard with pins or sticky tapes.
-
2.2
Spray 70% ethanol on the mouse to get rid of outside germs, and maintain the sterilized condition during the harvesting of the heart. Make an incision with scissors in the middle of the abdomen and cut the skin from the abdomen up to the thorax in a straight line. Remove the skin from both sides of the middle cut to clear the abdominal area of skin and hairs. Using scissors and tweezers cut the diaphragm below the ribcage.
-
2.3
Open the thoracic cavity of the mouse by cutting the ribcage inside the hood. Expose the heart and remove the blood near the heart. Wash the surrounding area of the heart with ice-cold PBS to get rid of any blood in the vicinity of the heart.
-
2.4
Dissect the heart using surgical scissors and tweezers. Place the heart in a 100-mm Petri dish containing 10 mL of ice-cold PBS.
-
2.5
Palpitate the heart using curved shank forceps and remove the residual blood inside the heart.
-
2.6
Use the whole heart for the isolation of CSCs.
-
2.7
Transfer all four or five whole hearts to a 50-mL conical tube containing 10 mL of ice-cold HBSS. Keep the tube on ice until further processing.
-
2.8
Wipe inside and outside of a biosafety cabinet with 70% ethanol to create a sterilized environment. Sterilize the heart-containing tube and other required materials with 70% ethanol. Place the heart-containing tube into the biosafety cabinet for further processing.
-
2.9
Transfer the hearts to a 100-mm Petri dish containing 10 mL of ice-cold HBSS. Wash the hearts again by palpating to remove any residual blood inside the heart. Change the HBSS solution after each wash to ensure the complete removal of the blood.
-
2.10
Cut the hearts into small pieces using fine surgical scissors or a surgical blade in a 100-mm Petri dish containing 5 – 7 mL of HBSS solution. Hold each heart with a pair of forceps and use a pair of surgical fine scissors or a surgical blade to cut the heart into 2- to 4-mm pieces. Change the HBSS solution frequently to ensure blood-free HBSS. Mince the hearts in the HBSS solution.
-
2.11
Centrifuge the minced tissue at 500 x g at 4 °C for 5 min. Discard the supernatant and keep the pellet.
-
2.12
Resuspend the pellet in 5 – 6 mL of 0.2% collagenase II solution in a 50-mL conical tube. The volume of collagenase solution should be almost 1.5- to 2-fold to the volume of the pellet.
Note: For the digestion of tissue, 0.2% collagenase I and 2.5 U/mL dispase solution can replace 0.2% collagenase II. Use an almost equal volume of dispase solution to the collagenase I solution.
-
2.13
Thoroughly mix the pellet with 0.2% collagenase II solution by agitating the tube or by rocking the tube on a shaker at 75 x g for 45–60 min at 37 °C. Shake the tube vigorously by hand after every 20 – 30 min to enhance the lysis.
-
2.14
Triturate the lysed tissue mix using a 5mL serological pipette and 1-mL pipette tip to dissociate the tissue lump into the single-cell suspension.
Note: To get the maximum recovery of the CSCs, triturate the lysis mix for at least 5 min. If the tissue lumps are relatively big, cut the tip of a 1-mL pipette tip with scissors or a surgical blade and use it for trituration.
-
2.15
Stop the enzymatic lysis of the tissue by adding commercially available CSC maintenance medium. Add almost 2 – 3x as much maintenance medium as the volume of the lysed tissue in step 2.14.
-
2.16
Pass the digested cell suspension through a 100-μm cell strainer to remove any undigested tissue pieces.
-
2.17
Repeat step 2.16 using a 40-μm cell strainer.
-
2.18
Separate the cells through density gradient centrifugation. For the separation of cells, gently overlay an equal volume of the filtrate over polysucrose and sodium diatrizoate solution. For example, first, add 20 mL of polysucrose and sodium diatrizoate solution in a 50-mL conical tube and then, add 20 mL of filtrate from step 2.17 over the polysucrose and sodium diatrizoate solution.
-
2.18.1
Note Prewarm the polysucrose and sodium diatrizoate solution at 37 °C before use with the cells. Add the filtrate from step 2.17 over the polysucrose and sodium diatrizoate solution very carefully and slowly to avoid any mixing of the two solutions. This is considered to be a crucial step.
-
2.18.1
-
2.19
Centrifuge the tube containing both solutions at 500 x g for 20 min at room temperature using a swing bucket centrifuge. Set the centrifuge at a lower acceleration and deceleration speed to avoid mixing the two solutions and for proper separation of the CSCs. This is an important step in CSC isolation. Put the tube containing the gradient mixture very slowly without disturbance inside the centrifuge and set it for centrifugation.
-
2.20
Take out the buffy coat along with extra solution from the upper layer using either a 1-mL pipette or a plastic Pasteur pipette in a 15-mL sterilized tube. This buffy layer will contain the CSCs.
Note: Take extra precaution during the pipetting of the buffy coat not to take out the polysucrose and sodium diatrizoate solution layer because it is toxic to CSCs.
-
2.21
Add an equal volume of CSC maintenance medium to the cell suspension from step 2.20 and mix it properly to neutralize the residual polysucrose and sodium diatrizoate solution, if present.
-
2.22
Centrifuge the above cell suspension at 500 x g for 5 min at 4 °C. Discard the supernatant.
-
2.23
Resuspend the pellet in 7 – 10 mL of incomplete DMEM medium and centrifuge it at 500 x g for 5 min at 4 °C to get rid of any residual polysucrose and sodium diatrizoate solution.
-
2.24
Collect the pellet, which contains the purified CSCs.
-
2.25
Resuspend the pellet in 1 mL of CSC maintenance medium, count the CSC number, and use the pellet for culture. To count the CSC number, use Trypan blue staining and a hemocytometer. With four male mice hearts, the yield of CSCs is ~1.8 million.
-
2.26
Seed the CSCs in a 6-well culture plate coated with a 0.02% gelatin solution containing 0.5% fibronectin. Use 2 mL of complete maintenance medium for seeding. Incubate the culture plate in an incubator at 37 °C with 5% CO2.
-
2.27
Replace the medium of culture CSCs with complete CSC maintenance medium every day until the third day. After that, change the medium on alternate days.
-
2.28
Determine the growth of the CSCs by observing them under a microscope.
-
2.1
-
Culture Maintenance and Passage of Cardiac Stem Cells
-
3.1
Culture the isolated CSCs in a commercially available maintenance medium. Replace the culture medium with fresh maintenance medium on alternate days. When the CSC culture has reached confluency, transfer the CSCs to a new plate coated with gelatin and fibronectin, as mentioned in step 2.26. Detach the cultured CSCs by enzymatic cell dissociation buffer. Follow the standard protocol of cell dissociation from the culture plate. At this stage, CSCs are considered at passage 0 (P0) stage.
-
3.2
Culture P0 CSCs in a new gelatin and fibronectin-coated plate in complete CSC maintenance medium at 37 °C in a 5% CO2 incubator, allow them to be confluent, and then follow step 3.1 to get passage 1 (P1) CSCs. Store P1 CSCs in liquid nitrogen or use them for experiments.
-
3.3
Seed 0.5 million cells in a 6-well plate or 1 million cells in T-25 to maintain the CSC culture at its initial stage.
-
3.4
Passage the CSCs when they become 85% – 90% confluent. CSCs can be cultured in the maintenance medium up to four to six weeks.
Note: Keep the initial seeding density of the CSCs relatively high (40% – 50%) because, at a lower seeding density, the CSCs may change their morphology to flat and elongated.
-
3.1
-
Characterization of Cardiac Stem Cells
-
4.1
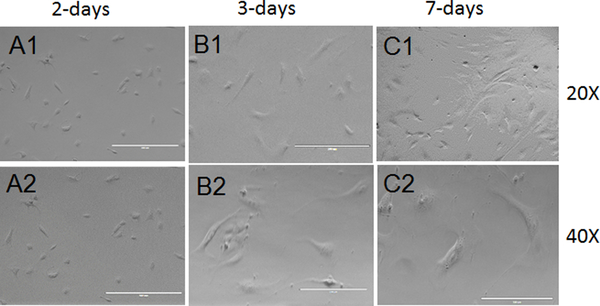
To characterize the cultured CSCs, observe them under a phage-contrast or a bright-field microscope. Place the CSC-containing plate on the objective of a fluorescent microscope. Set the magnification fairly low (10X) to focus the cells. Use phase-contrast aperture to observe the cells. Increase the magnification to 20X by changing the objective lens and image the CSCs. Increase the objective to 40X and image the CSCs. In two days, the CSCs appear almost round in shape, but the size of the cell increases with time, and in seven days, the CSCs appear almost cylindrical in shape (Figures 1A - 1C).
-
4.2
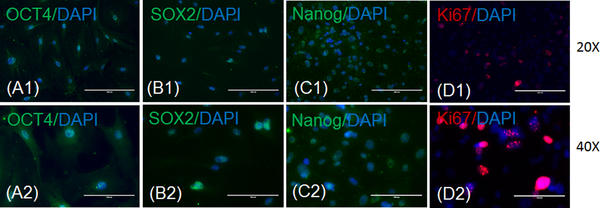
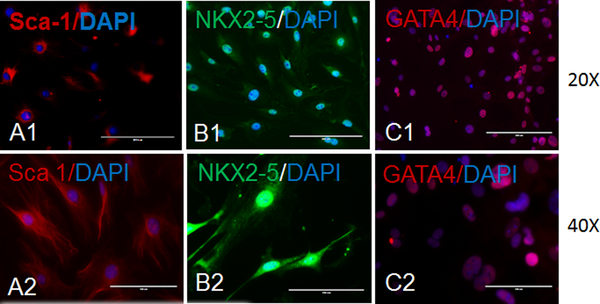
Perform immunostaining of the CSCs with markers of pluripotency/stemness (OCT4, SOX2, or Nanog), proliferation (Ki-67) (Figures 2A - 2D), and cardiac origin/progenitor cells (Sca-1, NKX2–5, or GATA4) (Figures 3A - 3C).
-
4.2.1
For the immunostaining of the CSCs, seed 10,000 CSCs in a 24-well culture plate.
-
4.2.2
The next day (after 18 – 24 h), fix the CSCs in 300 μL of 4% paraformaldehyde for 10 min.
-
4.2.3
Wash the CSCs with 500 μL of ice-cold PBS, 3x with 5-min intervals.
-
4.2.4
Permeabilize the CSCs with 300 μL of 0.2% Triton X-100 diluted in PBS for 10 min.
-
4.2.5
Wash the CSCs with 500 μL of ice-cold PBS, 3x with 5-min intervals.
-
4.2.6
Perform the blocking of the CSCs with 300 μL of 1% BSA diluted in PBST (PBS + 0.1% Tween 20) or 300 μL of 10% normal chicken serum diluted in PBST for 1 h.
-
4.2.7
Incubate the CSCs in 200 μL of primary antibody diluted in blocking solution overnight at 4 °C. Dilute the primary antibody as recommended by the datasheet of antibodies or as per standardization.
-
4.2.8
Wash the CSCs with 500 μL of ice-cold PBS, 3x with 5-min intervals.
-
4.2.9
Incubate the CSCs in 200 μL of secondary antibody diluted in blocking solution (see step 4.2.6) for 1 h at room temperature. Dilute the secondary antibody as recommended by the datasheet of antibodies or as per standardization.
-
4.2.10
Wash the CSCs with 500 μL of ice-cold PBS, 3x with 5-min intervals.
-
4.2.11
Counterstain the CSCs with 200 μL of DAPI (1 μg/mL) diluted in PBS for 5 min.
-
4.2.12
Wash the CSCs 1x with 500 μL of ice-cold PBS. Then, add 300 μL of ice-cold PBS in each well.
-
4.2.13
Perform imaging using a fluorescence microscope. Follow the instructions of fluorescent imaging, such as to avoid direct light on the plate. Keep the culture plate under the microscope. Focus the cells at different magnifications. Observe the cells under phase-contrast and/or different excitation filters and save the images.
-
4.2.14
Store the plate in the dark at 4 °C.
-
4.2.1
-
4.1
-
Differentiation of Cardiac Stem Cells into Cardiomyocyte
-
5.1
For the differentiation of CSCs, seed 500,000 CSCs in a 6-well plate with 200 μL of prewarm (37 °C) commercially available CSC maintenance medium.
-
5.2
Incubate the plate containing the CSCs in a CO2 incubator at 37 °C. Allow the CSCs to attach and proliferate until sub-confluent growth. If the medium turns yellow, replace the culture medium with fresh maintenance medium.
-
5.3
At 85% – 90% confluency, replace the maintenance medium with 2 mL of prewarm (37 °C) cardiomyocytes differentiation medium. Incubate the plate in a CO2 incubator at 37 °C for up to 2 – 3 weeks.
-
5.4
Change the differentiation medium every 2 – 3 d. Observe the CSCs’ morphology under a microscope every time during the medium change to ensure the good culture conditions.
-
5.5
After 12 d of CSC culture in differentiation medium, image the CSCs for differentiation markers following a standard immunostaining protocol (see steps 4.2.1 – 4.2.14). Save the fluorescent images for the expression of cardiomyocytes markers (Figures 4A - 4B).
-
5.1
Figure 1: Morphology of cultured CSCs.
These panels show phase-contrast imaging of cultured CSCs, namely representative CSCs after 2 days of culturing in maintenance medium at (A1) 20X and (A2) 40X objectives, representative CSCs after 3 days of culturing in maintenance medium at (B1) 20X and (B2) 40X objectives, and representative CSCs after 7 days of culturing in maintenance medium at (C1) 20X and (C2) 40X objectives. The scale bars are 100 μm for the 40X magnifications and 200 μm for the 20X magnifications.
Figure 2: Characterization of CSCs for pluripotency markers.
These panels show representative fluorescence imaging of cultured CSCs for markers of pluripotency (OCT4, SOX2, and Nanog) and proliferation (Ki-67). Panels A show expressions of OCT4 (green) and DAPI (blue) in CSCs at (A1) 20X and (A2) 40X magnifications. Panels B show expressions of SOX2 (green) and DAPI (blue) in CSCs at (B1) 20X and (B2) 40X magnifications. Panels C show expressions of Nanog (green) and DAPI (blue) in CSCs at (C1) 20X and (C2) 40X magnifications. Panels D show expressions of Ki-67 (red) and DAPI (blue) in CSCs at (D1) 20X and (D2) 40X magnifications. The scale bars are 100 μm for the 40X magnifications and 200 μm for the 20X magnifications.
Figure 3: Characterization of CSCs for cardiac markers.
These panels show representative fluorescence imaging of cultured CSCs for cardiac markers. Panels A show expressions of Sca-1 (red) and DAPI (blue) in CSCs at (A1) 20X and (A2) 40X magnifications. Panels B show expressions of NKX2–5 (green) and DAPI (blue) in CSCs at (B1) 20X and (B2) 40X magnifications. Panels C show expressions of GATA4 (red) and DAPI (blue) in CSCs at (C1) 20X and (C2) 40X magnifications. The scale bars are 100 μm for the 40X magnifications and 200 μm for the 20X magnifications.
Figure 4: Characterization of CSC differentiation toward cardiomyocyte lineage.
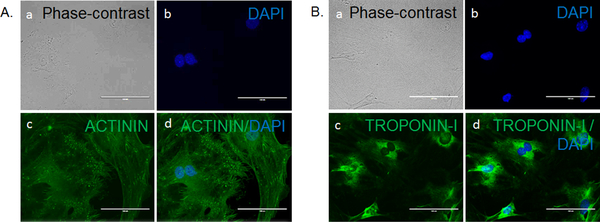
These panels show representative phase-contrast and fluorescence imaging of 12-days differentiated CSCs. (A) This panel shows the expression of cardiomyocyte marker actinin in differentiated CSCs, with (a) a phase-contract image, (b) a fluorescence image of DAPI (staining nucleus), (c) a fluorescence image of actinin (green), and (d) a merged image of panels a - c. The magnifications are 40X. (B) This panel shows the expression of the cardiomyocyte marker troponin I in differentiated CSCs, with (a) a phase-contract image, (b) a fluorescence image of DAPI (staining nucleus), (c) a fluorescence image of troponin I (green), and (d) a merged image of panels a - c. The magnifications are 40X. The scale bars are 100 μm for the 40X magnifications and 200 μm for the 20X magnifications.
REPRESENTATIVE RESULTS:
In the present study, we isolated CSCs from 10- to 12-week-old C57BL/6J male mice hearts. Here, we have presented a simple method for CSC isolation and characterization using markers of pluripotency. We also presented an elegant method for CSC differentiation and the characterization of CSCs that differentiated toward cardiomyocytes lineage. We observed a spindle shape morphology of 2- to 3-days-cultured CSCs under a phase-contrast microscope (Figures 1A and 1B). We found a change in the morphology of CSCs at 7 days of culture in maintenance medium, during which time the stem cells become elongated (Figure 1C). We characterized CSCs for markers of pluripotency and found that they express OCT4, SOX2, and Nanog (Figures 2A - 2C). We also found that CSCs proliferate in culture medium and express the proliferation marker Ki-67 (Figure 2D). To characterize the cardiac origin of CSCs, we determined the expression of cardiac markers Sca-1, NKX2–5, and GATA4 in CSCs. We found an expression of cardiac markers in the cultured CSCs (Figures 3A - 3C). To determine the differentiation of CSCs toward cardiomyocyte lineage, we cultured CSCs in cardiomyocyte differentiation medium for 12 days. After 12 days, we imaged the differentiated CSCs for the cardiomyocyte markers actinin and troponin I. We observed that cardiomyocyte markers were expressed in differentiated CSCs (Figures 4A - 4B). Overall, these results demonstrate that we successfully isolated CSCs from mouse heart and that these CSCs can proliferate and differentiate toward cardiomyocytes lineage.
DISCUSSION:
The critical steps of this CSC isolation protocol are as follows. 1) A sterilized condition must be maintained for extraction of the hearts from the mice. Any contamination during the heart extraction may compromise the quality of the CSCs. 2) The blood must be completely removed before mincing the heart, which is done by several washes of the whole heart and the heart pieces with HBSS solution. 3) The heart pieces must be completely lysed into a single-cell suspension with collagenase solution. 4) The polysucrose and sodium diatrizoate gradient solution for the separation of the cells must be prewarmed. 5) The medium for the CSC culture must be prewarmed. 6) The confluence of the CSCs during the seeding in a culture dish should be high because, in low confluency, CSCs may change their morphology. 7) The CSC number should be scored after the isolation from the heart and before the plating into a culture dish. The number of CSCs indicates the efficiency of the isolation from the heart. The counting of the CSCs is important for the seeding of the CSCs in the culture plate.
We used adult mice to isolate CSCs, which has been used by other investigators in rodent models23,28. CSCs can be isolated from neonatal rodent hearts29,30 and even from the biopsy of a human heart31. The previously reported CSC isolation protocols have some limitations, such as less yield, a complex isolation procedure, a long duration for differentiation, and less cardiomyocyte differentiation potential32–34. The CSC isolation protocol presented here is simple: it takes less time and is cost-effective and more efficient in CSC yield. Moreover, the isolated CSCs are Sca-1+ve (a well-accepted marker for mouse CSCs) (Figures 1A1 – 3C2). For the differentiation of CSCs, other protocols need three to four weeks35,36. However, this protocol requires 12 days (Figures 4A – 4B). The gradient centrifugation-based CSC isolation presented here yields ~1.8 million CSCs from four male mice hearts.
Although the yield of CSCs is high, a limitation of this protocol is the uniformity and purity of the isolated CSCs. Because there is no single acceptable marker for CSCs, it is difficult to use a single-cell-surface marker to select CSCs. However, CSCs isolated by this protocol can be used to obtain a uniform population of CSCs based on a specific marker or several markers of CSCs, using fluorescence-activated cell sorting. Thus, this method of CSC isolation is cost-effective with a high yield and has the option to get a uniform CSC population based on specific cell-surface marker(s).
Supplementary Material
ACKNOWLEDGMENTS:
This work is supported, in parts, by the National Institutes of Health grants HL-113281 and HL116205 to Paras Kumar Mishra.
Footnotes
DISCLOSURES:
The authors have nothing to disclose.
REFERENCES:
- 1.Benjamin EJ et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 135, e146–e603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen PK, Rhee JW, Wu JC Adult Stem Cell Therapy and Heart Failure, 2000 to 2016: A Systematic Review. The Journal of the American Medical Association Cardiology. 1, 831–841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emmert MY et al. Safety and efficacy of cardiopoietic stem cells in the treatment of post-infarction left-ventricular dysfunction - From cardioprotection to functional repair in a translational pig infarction model. Biomaterials. 122, 48–62 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Silvestre JS, Menasche P. The Evolution of the Stem Cell Theory for Heart Failure. EBioMedicine. 2, 1871–1879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terzic A, Behfar A. Stem cell therapy for heart failure: Ensuring regenerative proficiency. Trends in Cardiovascular Medicine. 26, 395–404 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yamada S. et al. Embryonic stem cell therapy of heart failure in genetic cardiomyopathy. Stem Cells. 26, 2644–2653 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadek HA, Martin CM, Latif SS, Garry MG, Garry DJ Bone-marrow-derived side population cells for myocardial regeneration. Journal of Cardiovascular Translational Research. 2, 173–181 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vrtovec B. et al. Effects of intracoronary CD34+ stem cell transplantation in nonischemic dilated cardiomyopathy patients: 5-year follow-up. Circulation Research. 112, 165–173 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Hare JM et al. Comparison of allogeneic vs autologous bone marrow-derived mesenchymal stem cells delivered by transendocardial injection in patients with ischemic cardiomyopathy: the POSEIDON randomized trial. The Journal of American Medical Association. 308, 2369–2379 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guijarro D. et al. Intramyocardial transplantation of mesenchymal stromal cells for chonic myocardial ischemia and impaired left ventricular function: Results of the MESAMI 1 pilot trial. International Journal of Cardiology. 209, 258–265 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Bobi J. et al. Intracoronary Administration of Allogeneic Adipose Tissue-Derived Mesenchymal Stem Cells Improves Myocardial Perfusion But Not Left Ventricle Function, in a Translational Model of Acute Myocardial Infarction. Journal of the American Heart Association. 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki E, Fujita D, Takahashi M, Oba S. & Nishimatsu H. Adipose tissue-derived stem cells as a therapeutic tool for cardiovascular disease. World Journal of Cardiology. 7, 454–465 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao LR et al. Intracoronary infusion of Wharton’s jelly-derived mesenchymal stem cells in acute myocardial infarction: double-blind, randomized controlled trial. BMC Medicine. 13, 162 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson DL et al. A strong regenerative ability of cardiac stem cells derived from neonatal hearts. Circulation. 126, S46–53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazakov A. et al. C-kit(+) resident cardiac stem cells improve left ventricular fibrosis in pressure overload. Stem Cell Research. 15, 700–711 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Ong SG et al. Cross talk of combined gene and cell therapy in ischemic heart disease: role of exosomal microRNA transfer. Circulation. 130, S60–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sahoo S, Losordo DW Exosomes and cardiac repair after myocardial infarction. Circulation Research. 114, 333–344 (2014). [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z. et al. Pretreatment of Cardiac Stem Cells With Exosomes Derived From Mesenchymal Stem Cells Enhances Myocardial Repair. Journal of the American Heart Association. 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ibrahim AG, Cheng K, Marban E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports. 2, 606–619 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emanueli C, Shearn AI, Angelini GD, Sahoo S. Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vascular Pharmacology. 71, 24–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menasche P. Cardiac cell therapy: lessons from clinical trials. Journal of Molecular and Cellular Cardiology. 50, 258–265 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Trounson A, McDonald C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell. 17, 11–22 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Takamiya M, Haider KH, Ashaf M. Identification and characterization of a novel multipotent sub-population of Sca-1(+) cardiac progenitor cells for myocardial regeneration. PLoS One. 6, e25265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cambria E. et al. Translational cardiac stem cell therapy: advancing from first-generation to next-generation cell types. NPJ Regenerative Medicine. 2, 17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruyneel AA, Sehgal A, Malandraki-Miller S, Carr C. Stem Cell Therapy for the Heart: Blind Alley or Magic Bullet? Journal of Cardiovascular Translational Research. 9, 405–418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garbern JC, Lee RT Cardiac stem cell therapy and the promise of heart regeneration. Cell Stem Cell. 12, 689–698 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh H, Ito H, Sano S. Challenges to success in heart failure: Cardiac cell therapies in patients with heart diseases. Journal of Cardiology. 68, 361–367 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Smith AJ et al. Isolation and characterization of resident endogenous c-Kit+ cardiac stem cells from the adult mouse and rat heart. Nature Protocols. 9, 1662–1681 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Rutering J. et al. Improved Method for Isolation of Neonatal Rat Cardiomyocytes with Increased Yield of C-Kit+ Cardiac Progenitor Cells. Journal of Stem Cell Research and Therapy. 5, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saravanakumar M, Devaraj H. Distribution and homing pattern of c-kit+ Sca-1+ CXCR4+ resident cardiac stem cells in neonatal, postnatal, and adult mouse heart. Cardiovascular Pathology. 22, 257–263 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Monsanto MM et al. Concurrent Isolation of 3 Distinct Cardiac Stem Cell Populations From a Single Human Heart Biopsy. Circulation Research. 121, 113–124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidyasekar P, Shyamsunder P, Santhakumar R, Arun R, Verma RS A simplified protocol for the isolation and culture of cardiomyocytes and progenitor cells from neonatal mouse ventricles. European Journal of Cell Biology. 94, 444–452 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Dergilev KV et al. Comparison of cardiac stem cell sheets detached by Versene solution and from thermoresponsive dishes reveals similar properties of constructs. Tissue Cell. 49, 64–71 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Zaruba MM, Soonpaa M, Reuter S, Field LJ Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 121, 1992–2000 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H. et al. Isolation and characterization of a Sca-1+/CD31- progenitor cell lineage derived from mouse heart tissue. BMC Biotechnology. 14, 75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smits AM et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nature Protocols. 4, 232–243 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.