Abstract
Purple Broccoli (Brassica oleracea L. var italica) attracts growing attention as a functional food. Its purple coloration is due to high anthocyanin amounts. Light represents a critical parameter affecting anthocyanins biosynthesis. In this study, ‘Purple Broccoli’, a light-responding pigmentation cultivar, was assessed for exploring the mechanism underlying light-induced anthocyanin biosynthesis by RNA-Seq. Cyanidin, delphinidin and malvidin derivatives were detected in broccoli head samples. Shading assays and RNA-seq analysis identified the flower head as more critical organ compared with leaves. Anthocyanin levels were assessed at 0, 7 and 11 days, respectively, with further valuation by RNA-seq under head-shading and light conditions. RNA sequences were de novo assembled into 50,329 unigenes, of which 38,701 were annotated against four public protein databases. Cluster analysis demonstrated that anthocyanin/phenylpropanoid biosynthesis, photosynthesis, and flavonoid biosynthesis in cluster 8 were the main metabolic pathways regulated by light and had showed associations with flower head growth. A total of 2,400 unigenes showed differential expression between the light and head-shading groups in cluster 8, including 650 co-expressed, 373 specifically expressed under shading conditions and 1,377 specifically expressed under normal light. Digital gene expression (DGE) analysis demonstrated that light perception and the signal transducers CRY3 and HY5 may control anthocyanin accumulation. Following shading, 15 structural genes involved in anthocyanin biosynthesis were downregulated, including PAL, C4H, 4CL, CHS, CHI, F3H and DFR. Moreover, six BoMYB genes (BoMYB6-1, BoMYB6-2, BoMYB6-3, BoMYB6-4, BoMYBL2-1 and BoMYBL2-2) and three BobHLH genes (BoTT8_5-1, BoTT8_5-2 and BoEGL5-3) were critical transcription factors controlling anthocyanin accumulation under light conditions. Based on these data, a light-associated anthocyanin biosynthesis pathway in Broccoli was proposed. This information could help improve broccoli properties, providing novel insights into the molecular mechanisms underpinning light-associated anthocyanin production in purple vegetables.
Keywords: Broccoli, Transcriptome, Anthocyanins, Light induction
Introduction
Broccoli (Brassica oleracea L. var. italica) represents a nutritious plant whose flower heads are rich in vitamins A, B2 and C, minerals, and antioxidant phytochemicals such as glucoraphanin (Moreno et al., 2010). Purple broccoli serves as a functional food with significant anthocyanin production in the head, leaves and seeds. In Brassica species, purple/red color conferred by anthocyanin accumulation is tightly associated with the induction of structural genes and transcription factors (Song et al., 2018). A previous study showed that insertion of Harbinger transposon of BoMYB2 results in its upregulation, which then together with BobHLHs upregulates the downstream genes of anthocyanin biosynthesis in purple cauliflower (Chiu et al., 2010). In red cabbage, TT8 and MYB2 upregulate structural genes in the anthocyanin pathway (Yuan, Chiu & Li, 2009) as well as the structural genes F3’H, DFR and ANS in kale (Zhang et al., 2012; Neugart, Krumbein & Zrenner, 2016). In addition, deletion or substitution in the coding sequences of BoMYBL2-1 leads to purple color accumulation of cabbage (Song et al., 2018). In Kohlrabi, BoPAP1 promotes purple coloration in a light dependent manner as well as BoCHS while BoTT8 and BoPAP2 are light-independent (Zhang et al., 2015). Some candidate genes controlling color formation have also been mapped in Brassica crops. In broccoli, a major locus (qPH.C01-2) and two minor loci (qPH.C01-4 and qPH.C01-5) for purple sepal trait of the flower head were mapped onto chromosome C01 (Yu et al., 2019). In ornamental kale, single genes for pink or purple leaf traits were mapped onto chromosome C03 ( Zhu et al., 2016) and C09 (Liu et al., 2017), respectively. The change in color of inner and outer flower head (from green to purple) in broccoli is induced by light. However, the mechanism underlying purple color formation remains largely undefined in broccoli.
Anthocyanins comprise the largest subclass of hydrosoluble pigments conferring red, orange, purple, and blue colorations to flowers, fruits, seeds, and vegetables in plants (Cominelli et al., 2008; Espley et al., 2007; Li et al., 2017). Anthocyanins play important roles in furnishing flowers and fruits for attracting pollinations and dispersers, protecting plants from various biotic and abiotic stressors (Harborne & Williams, 2000; Ahmed et al., 2014). In addition, the health benefits of anthocyanins attract growing attention due to their antioxidant activities (Pojer et al., 2013). Genetic parameters, developmental stages and environmental conditions control anthocyanin accumulation, including high-light, UV-light, cold temperature, nutrient availability and infection (Dixon & Paiva, 1995; Chalker-Scott, 1999). Based on the above, a comprehensive understanding of regulation of anthocyanin biosynthesis should be investigated to obtain anthocyanin-rich foods via breeding and/or environmental control.
Anthocyanin biosynthesis has been assessed in various species such as Arabidopsis, petunia, tobacco, and fruit plants (Liu, Osbourn & Ma, 2015). The genes of the anthocyanin biosynthetic pathway are well-conserved across species (Holton & Cornish, 1995; Zhang, Butelli & Martin, 2014; Guo et al., 2014). Anthocyanin synthesis starts with phenylalanine, and is catalyzed stepwise by phenylalanine ammonia lyase (PAL), 4-coumarate-CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxyl enzyme (F3H), flavonoid 3′-hydroxylase (F3’H), dihydroflavonol reductase (DFR) and anthocyanin synthase (ANS) or leucoanthocyanidin dioxygenase (LDOX).
These structural genes are transcriptionally controlled by the MBW complex composed of R2R3-MYB, basic Helix-Loop-Helix (bHLH) and WD repeat protein (WDR). In Eudicots, R2R3-MYB transcription factors (TFs) are primarily implicated as positive regulators to initiate and activate the MBW complex in leaves under stress and young flowers/fruits. It was reported for the first time in maize that R2R3-MYB TFs C1 regulates anthocyanin synthesis (Paz-Ares et al., 1986), and co-action of bHLH proteins and Leaf color (Lc) was found recently (Ludwig et al., 1989). AtMYB75/PAP1, AtMYB90/PAP2, AtMYB112, AtMYB113 and AtMYB114 contribute to anthocyanin production in Arabidopsis (Gonzalez et al., 2008; Lotkowska et al., 2015). In petunia, AN1 and AN2 contribute to anthocyanin synthesis and vacuolar acidification (Koes, Verweij & Quattrocchio, 2005); meanwhile, PyMYB10 and PyMYB10.1 bind to bHLH for enhancing anthocyanin production in pears (Feng et al., 2015). However, some MYB TFs, such as AtMYB3/4/6/60 (Jin et al., 2000) and AtMYBL2 (Matsui, Umemura & Ohme-Takagi, 2008), FaMYB1 and FcMYB1 (Aharoni et al., 2001; Zhang et al., 2018), VvMYB4/C2 (Matus, Aquea & Arce-Johnson, 2008), and MdMYB16/17/111 (Lin-Wang et al., 2011), exert inhibitory effects. These negative regulators interact with the bHLH protein, thereby competing with R2R3-MYB activators. The bHLH proteins controlling anthocyanin production have been described in multiple species, including Arabidopsis (GL3, EGL3 and TT8) (Payne, Zhang & Lloyd, 2000; Zhang et al., 2003; Nesi et al., 2000), petunia (PhAN1, PhJAF13) (Spelt et al., 2000; Quattrocchio et al., 1998), apple (MdbHLH3/MdbHLH33) (Espley et al., 2007), grape (VvMYC1) (Hichri et al., 2010) and peach (PpbHLH3/PpbHLH33) (Ravaglia et al., 2013). The various transcriptional modulators control anthocyanin production as well as their modifications and translocation into vacuoles via glutathione S-transferases (GSTs), the ATP-binding cassette (ABC) and multidrug and toxic compound extrusion (MATE) proteins ( Wang et al., 2017).
Multiple parameters including genetic, developmental and environmental factors control anthocyanin biosynthesis. Light indexes, including intensity and quality, represent critical factors affecting anthocyanin accumulation (Albert et al., 2009). In lettuce and turnip, UV-A and UV-B increase anthocyanin contents via upregulation of DFR and CHS in the anthocyanin biosynthetic pathway (Zhou et al., 2007; Park et al., 2007). In the presence of light, photoreceptors are activated, including PHYs (PHYA to E) that absorb red/far-red light; CRYs (CRY1 to 3) and PHOTs (PHOT1 and 2) that sense blue/UV-A light, and UVR8 that absorbs UV-B (Zoratti et al., 2014), interacting with the ubiquitin E3 ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC1) that controls the degradation of target transcription factors, including ELONGATED HYPOCOTYL5 (HY5). HY5 is associated with induced CHS, CHI and flavonoid production under light and UV-B radiation conditions in Arabidopsis (Shin, Park & Choi, 2007). It also binds the MYB75/12/111 promoter to increase their expression and modulate anthocyanin biosynthesis (Stracke et al., 2010; Shin et al., 2013; Nguyen et al., 2015).
The present work aimed to assess the global transcription of regulatory, structural and hormone signal transduction genes which might positively or negatively regulate broccoli’s anthocyanin biosynthetic pathway. The light sensitivity of pigment biosynthesis makes the broccoli ‘Long Jing’ an optimal plant for evaluating anthocyanin synthesis and explore the underpinning mechanisms under light conditions. Indeed, anthocyanin accumulation accompanied with broccoli head growth and development under natural light conditions. However, broccoli heads show reduced coloration under shading conditions. Here, the ‘Long Jing’ cultivar was employed for analyzing light-associated anthocyanin biosynthesis and the underlying mechanisms by RNA-seq. Based on the association of shading time with anthocyanin amounts, anthocyanin accumulation was assessed at 0 d, 7 d (highest production rate) and 11 d (peak amounts). The present findings provide insights into the molecular mechanisms underpinning light-associated anthocyanin production in broccoli, facilitating genetic engineering for increasing anthocyanin amounts in vegetables.
Materials & Methods
Plant materials and RNA preparation
The purple broccoli cultivar ‘Long Jing’ was assessed as the experimental material. After flower head formation, a total of seven stages were defined as 0, 3, 5, 7, 9, 11, and 14 days. Flower heads were collected for phenotype observation and anthocyanin level measurement at each developmental stage of the head under light and dark (shading using a sunshade net over the whole heads or leaves) conditions. In addition, light and darkness treated heads at 0, 7 and 11 days, and darkness treated leaves at 7 days were collected for RNA-Seq from the flower heads of the purple cultivar, respectively. Three biological replicate specimens were obtained. All specimens underwent snap freezing in liquid nitrogen and storage at −80 °C. To assess cells displaying purple coloration, head flowers underwent transverse sectioning by hand and analysis under a Zeiss Axioscope photo microscope.
Quantification of total anthocyanin amounts
Total anthocyanin amounts were determined by the pH differential method (Lee, Durst & Wrolstad, 2005) with some modifications. Briefly, 100 mg of fresh flowers were soaked in 500 µl acidified methanol (1% v/v formic acid), sonicated (15 min × 3 times), and centrifuged for 5 min at 4,000 rpm. The precipitate was extracted twice. All supernatants were collected and stored at −20 °C. Then, 200 µl samples were mixed with 800 µl KCl (pH 1) and 800 µl NaAc (pH 4.5), respectively. After incubation at room temperature for 20 min in the dark, optical density was read at 510 and 700 nm. The following equation was employed to derive anthocyanin amounts: total anthocyanin amounts (mg C3G/g FW) = A × MW × DF × V /(ε × 1 × m), where A = (A510 nm–A700 nm)pH 1.0–(A510 nm–A700 nm)pH 4.5; A510 and A700 are absorbance values at 510 and 700 nm, respectively; MW = 449.2; DF = 5; V is the total volume of supernatants; ε = 26900; m is fresh sample weight. Five replicates were assessed per biological specimen.
Qualitative analysis of anthocyanin compounds by high- performance liquid chromatography (HPLC)
For HPLC, specimens underwent freeze-drying and pulverization in liquid nitrogen. Then, 100 mg of each specimen was thoroughly mixed for 5 min in 2 ml of 5% formic acid (v/v) in ultrapure water, and submitted to sonication (20 min). Upon centrifugation (12,000 rpm, 10 min), the supernatants underwent filtration with 0.45 µm PTFE filters (Toyo Roshi Kaisha, Japan). Anthocyanin amounts were assessed on an Agilent 1200 series HPLC (Agilent Technologies, USA), on a reverse phase Kromasil C18 column (5 µm; C18 80A, 250 × 4.60 mm; Kromasil) at 40 °C. Samples were injected at 10 µl, and a flow rate of 1 ml min−1 was adopted. Mobile phases A (1.6% formic acid) and B (methanol containing 0.01% formic acid) were employed as eluents: 0–5 min, 85% A+15% B; 5–10 min, 80% A+20% B; 10–20 min, 72% A+28% B; 20–35 min, 40% A+60% B; 35–35.1 min, 100% B; 35.1–50 min, 100% B; 50–50.1 min, 85% A+15% B; 50.1–60 min, 85% A+15% B. Detection was performed at 530 nm. Individual anthocyanins were quantitated (mg g-1 dry weight) via comparisons of areas under their HPLC peaks with those of known standards (delphinidin-3-O-galactoside, delphinidin-3-O-glucoside, cyanidin-3-O-galactoside, cyanidin-3-O-glucoside, malvidin-3-O-galactoside and malvidin-3-O-glucoside).
RNA purification and library generation for transcriptomics
Total RNA was extracted from head flowers in purple broccoli at different times under both light conditions with mirVana miRNA Isolation Kit (Ambion) as directed by the manufacturer. RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies). Specimens with RNA Integrity Number (RIN) ≥7 were further evaluated. The libraries were generated with TruSeq Stranded mRNA LT Sample Prep Kit (Illumina, USA) as instructed by the manufacturer.
Transcriptome sequencing, de novo assembly and functional annotation
The obtained libraries underwent sequencing by the PE strategy on a HiSeqTM 2500 or Illumina HiSeq X Ten; cDNA fragments approximated 125 or 150 bp. The raw reads obtained underwent pre-processing with Trimmomatic; those with ploy-N or showing low quality were excluded, leaving clean reads. These clean reads underwent assembly into contigs and de novo assembly into transcripts using Trinity (version: 2.4) (Grabherr et al., 2011) by the paired end method. The longest transcripts were selected as unigenes for further assessment. Original sequencing data were deposited in SRA (Short Read Archive; accession number PRJNA560282).
Unigene quantification, assessment of differentially expressed genes (DEGs) and gene annotation
Fragments per kilobase of transcript per Million (FPKM) and read counts for each unigene were assessed with Bowtie 2 and eXpress. DEG identification was carried out with the DESeq functions estimate Size Factors and negative binomial Test. P < 0.05 and fold Change >2 or <0.5 were thresholds for determining significant differential expression. Hierarchical clustering of DEGs was carried out to assess the transcripts’ expression patterns. The assembled unigenes were assessed with R according to hypergeometric distribution.
GO and KEGG pathway analyses of DEGs
The assembled unigenes underwent annotation by alignment in protein databases, including the NCBI non redundant (NR), SwissProt (http://www.expasy.ch/sprot), and Clusters of orthologous groups for eukaryotic complete genomes (COG) databases (https://www.ncbi.nlm.nih.gov/COG/) with Blastx (E<10−5). Proteins best matching the unigenes were employed for assigning functions. Based on the SwissProt annotation, GO classification was carried out using the Blast2GO software (Du et al., 2010; Conesa et al., 2005). Unigenes were mapped to the KEGG database (http://www.genome.jp/kegg) for determining the associated metabolic pathways (Kanehisa et al., 2008).
Real-time quantitative reverse transcription-PCR
To confirm the results of Illumina analysis, qRT-PCR was performed for multiple genes. Total RNA isolation from specimens collected in various developmental stages under light or dark conditions was carried out. Reverse-transcription used PrimeScript RT Master Mix Perfect Real Time Kit (Takara) as directed by the manufacturer. Finally, qRT-PCR was carried out on a QuantStudio 5 Real-Time PCR System (Fisher Scientific, USA) with SYBR Premix Ex Taq (TaKaRa, Japan) in triplicate. Data were normalized to Actin 2 amounts, and the comparative CT method was employed for analysis.
Results
Phenotypic characterization of purple broccoli responding to light during anthocyanin production
The color of purple broccoli cultivar ‘Long Jing’ head is affected by pigment types and contents in flower buds as well as light during the developmental stages. Firstly, pigment types in broccoli heads were evaluated. Anthocyanin amounts were assessed by HPLC multistage tandem mass spectrometry (Fig. 1A). Peaks 1, 2, 3, 4, 5 and 6 were identified as delphinidin-3-O-galactoside, delphinidin-3-O-glucoside, cyanidin-3-O-galactoside, cyanidin-3-O-glucoside, malvidin-3-O-galactoside and malvidin-3-O-glucoside, respectively.
Figure 1. Accumulation of anthocyanins under head-shading, leaves-shading and normal light (CK) treatment in Broccoli.
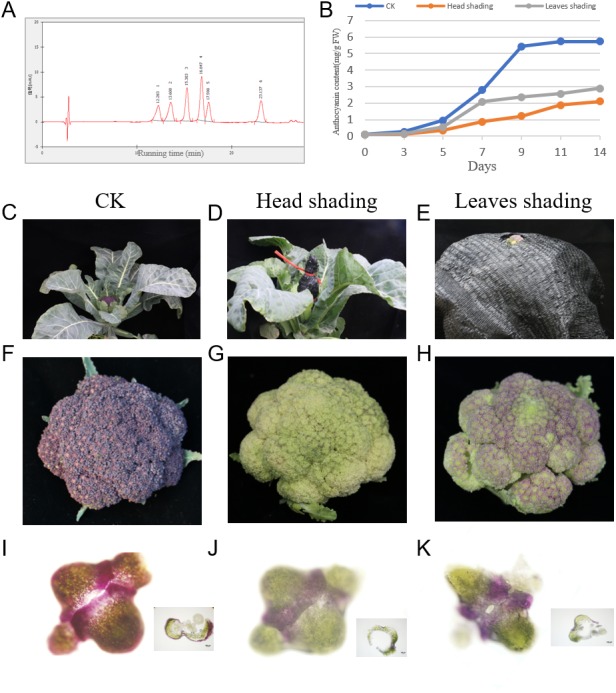
(A) Mass spectrometry assessment of anthocyanin levels by HPLC; (B) Relative anthocyanin amounts during head flower development; (C–E) Various shading treatments; (F–H) Respective head phenotypes under various treatments; (I–K) Anthocyanin levels in various parts of purple head flowers revealed by hand section.
To study light-response factors in anthocyanin production in the broccoli cultivar ‘Long Jing’, we shaded the whole head and leaves using a sunshade net during the developmental stages of the head from 0 d to 14 d, with light conditions employed as a control treatment. In comparison with controls, flower buds grown under head- and leaves-shading all faded, with head shading exerting more pronounced effects (Fig. 1C). The relative amounts of total anthocyanin were measured, and the head-shading treatment group showed lower levels compared with the leaf-shading treatment group, and both of these groups had lower values than the control group (Fig. 1B; Table S1). In addition, decrease in relative anthocyanin levels was more pronounced under head-shading (2.04 mg g−1 FW) compared with leaf-shading (1.47 mg g−1FW), indicating that shading during the head development in broccoli significantly affected the relative contents of total anthocyanin.
Quantitative analysis was further conducted to identify the key developmental stage under light during anthocyanin production. Spectra were obtained at 200–600 nm, and chromatograms at 520 nm (Fig. 1B). In the control group, relative anthocyanin amounts rose during head development (0 d to 11 d), showed a peak growth rate at 7 d, and then remained steady after 11 d, while in shaded plants they increased slowly during head’s developmental stages from 0 d to 14 d. Therefore, 0 d, 7 d and 11 d were considered critical times for response to light during anthocyanin production.
Microscopic examination of sections of flower buds from head shading, leaf shading and control plants revealed that the prominent purple color extended to more bud tissue cells, with anthocyanins accumulating closer to the outer layer of buds (Fig. 1D), in accordance with previously published data on purple Graffiti cauliflower (Chiu & Li, 2012). Compared with the control treatment, head and leaves shading conditions resulted in lighter purple pigments in upper epidermal layers. The above results suggested light had a pivotal function in anthocyanin production, and shading treatment significantly repressed anthocyanin accumulation during head development in broccoli.
Organ responses to light during anthocyanin biosynthesis
In order to assess differences in organ response to light during anthocyanin production in broccoli, RNA-Seq was performed under head-shading, leaves-shading and normal night conditions at the fastest point (7 d), respectively. The flowers were collected to construct nine libraries for transcriptomics in three biological replicates.
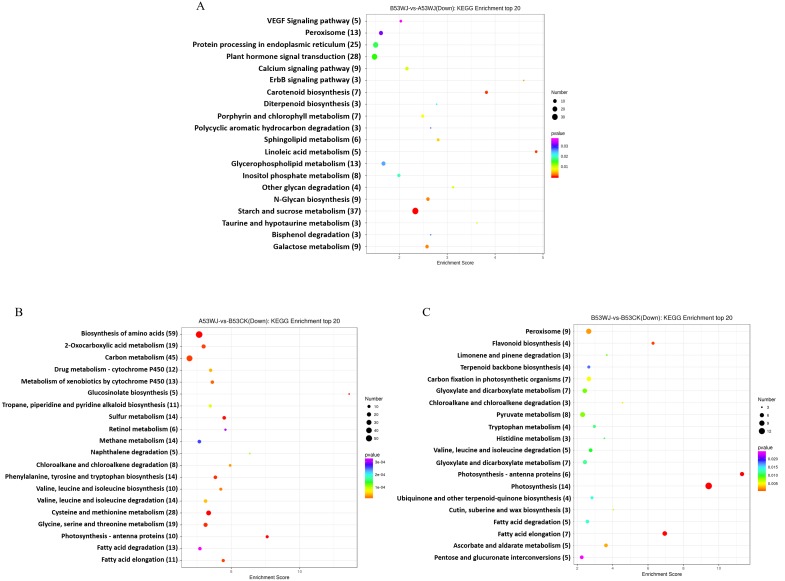
Comparing the expression amounts of differently expressed genes (DEGs) under head-shading and leaves-shading treatment, a total of 2,223 and 2,558 DEGs were up- and down-regulated, respectively. To identify the functions of these down-regulated DEGs, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were carried out (Du et al., 2010; Conesa et al., 2005; Kanehisa et al., 2008). Thirty GO terms were found as enriched biological processes based on the DEGs (Table S2; Fig. S1): “nucleus”, “response to abscisic acid”, “DNA-binding transcription factor activity”, “response to water deprivation” and “sequence-specific DNA binding”, which were the major GO terms. The significantly enriched pathways were identified using KEGG analysis (Fig. 2A). Among the significantly enriched pathways, “Starch and sucrose metabolism”, “Plant hormone signal transduction” and “Protein processing in endoplasmic reticulum” were the major public pathway-related database.
Figure 2. KEGG function classification of DEGs in B53WJ-vs-A53WJ (A), A53WJ-vs-B53CK (B) and B53WJ-vs-B53CK (C).
B53WJ-vs-A53WJ refers to the number of DEGs at 7 d between head-shading and leaf-shading treatments; A53WJ -VS-B53CK refers to the number of DEGs at 7 d between leaf-shading and light treatments; B53WJ -VS-B53CK refers to the number of DEGs at 7 d between head-shading and light treatments.
Comparing the expression amounts of differently expressed genes (DEGs) under shading and normal light treatment, a total of 378 and 660 DEGs were up- and down-regulated under head-shading treatment, respectively. For DEGs under leaf-shading treatment, we found that a total of 1532 and 1628 DEGs were up- and down- regulated, respectively (Tables S3 and S4; Fig. S2 and S3). In GO analysis, DEGs encoding proteins related to response to “light stimulus”, “chloroplast and photosystem I” were down-regulated under both head- and leaf-shading treatments. In KEGG analysis, DEGs were grouped in 20 functional classes. Under leaf-shading treatment, DEGs were significantly enriched in “amino acid biosynthesis”, “carbon metabolism”, “sulfur metabolism”, “cysteine and methionine metabolism”, and “photosynthesis-antenna proteins” (Fig. 2B), while downregulated DEGs with previously described functions were associated with “photosynthesis”, “peroxisome” and “flavonoid biosynthesis”, indicating such pathways/processes might be affected by head shading (Fig. 2C). The KEGG analysis showed the critical pathways in response to light. The above results indicated that photosynthesis and anthocyanin biosynthetic process were markedly inhibited by head shading while photosynthesis and carbon-nitrogen-sulfur metabolism were significantly repressed by leaf shading by transcriptional regulation in response to light.
According to flower head phenotypes, pigment synthesis and associated DEGs, the head might constitute the main organ showing a response to light during anthocyanin biosynthesis, in accordance with previous data on chrysanthemum in which the capitulum was the key organ responding more to light compared with the leaf during anthocyanin production (Hong et al., 2015).
Overall transcriptomic analysis under head shading and control treatments
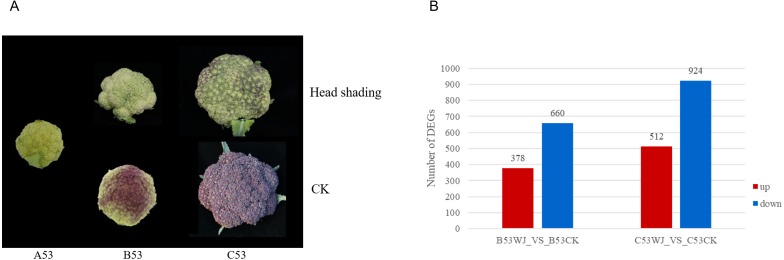
To assess how light induces anthocyanin production in broccoli, RNA-Seq was performed for three triplicate groups at 0 d, 7 d and 11 d under head-shading and normal light treatments, respectively (Fig. 3A). Averagely 20 million clean reads were produced per sample, including 81∼85% which were mapped to the Brassica oleracea genome (Table S5). Cumulatively, 90231 transcripts were detected across all six treatments with an FPKM ≥1. The full annotation and the expression levels of all genes (FPKM values) are found in Tables S6 and S7.
Figure 3. Differentially expressed genes (DEGs) in the broccoli head treated with head-shading and normal light conditions.
(A) Head-shading treatment and corresponding head flower phenotypes during head flower development at 0 d (A53), 7 d (B53) and 11 d (C53); (B) Changes in gene expression at 7 d and 11 d. B53WJ -VS-B53CK refers to the number of DEGs at 7 d between the head-shading and light treatments. C53WJ-VS-C53CK refers to the amounts of DEGs at 11 d between the head-shading and light treatments.
Thousands of genes are activated in broccoli in response to light
To identify genes responding to light, differential gene expression analyses were performed at 0 d, 7 d and 11 d under both head-shading and control treatments. Transcriptomics revealed 1461 (7 versus 0 day) and 5132 (11 versus 0 day) DEGs under control treatment, and 1752 (7 versus 0 day) and 1859 (11 vs. 0 day) DEGs under head-shading treatment. The number of shared DEGs increased over time, probably due to the total number of DEGs increasing between treatments. Relative to the control treatment, 378 upregulated and 660 downregulated DEGs (P < 0.05) were detected at 7 d, while 512 upregulated and 924 downregulated DEGs were found at 11 days, respectively (Fig. 3B).
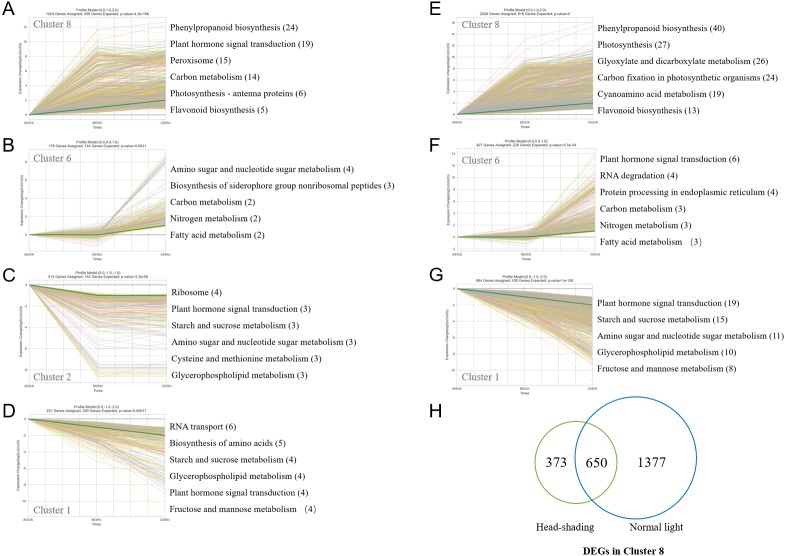
To gain a deeper understanding regarding the associated biological processes, transcripts were assigned to nine clusters per treatment group (Figs. S4 and S5). Then, KEGG analysis was performed for identifying biological pathways enriched in clusters of comparably regulated genes (Figs. 4A and 4B). Cluster 1, 6 and 8 were under both light and head-shading treatments. Cluster 1 encompassed genes downregulated throughout the study, including those controlling Plant hormone signal transduction, Starch and sucrose metabolism, Glycero-phospholipid metabolism and Fructose and mannose metabolism. Cluster 6 comprised genes positively regulated throughout the entire study, including those associated with Carbon metabolism, Nitrogen metabolism and Fatty acid metabolism. Cluster 8 contained genes significantly upregulated from 0 d to 7 d, many of which were involved in Phenylpropanoid biosynthesis, Photosynthesis, and Flavonoid biosynthesis. These findings indicated that cluster 8 DEGs were expressed in early phases of light induction and played roles in regulating anthocyanin biosynthesis. As many as 2400 DEGs showed differential expression between the light and shading libraries in cluster 8, in which 373 and 1377 individual DEGs showed specific expression under head-shading and normal light conditions, respectively, and 650 genes were co-expressed under both conditions (Fig. 4C; Table S8, S9 and S10). Thus, we focused on the 650 co-expressed and 373 specifically expressed under head-shading, which most likely represented light-induced genes.
Figure 4. Clustering of differentially expressed genes with marked expression level differences and KEGG pathway analysis after head-shading treatment (A–D) and light treatment (E–G); (H) Overlap of DEGs in cluster 8 in response to light.
Different expression patterns of anthocyanin biosynthesis structural genes
The expression profiling of structural genes was performed to assess their roles in anthocyanin biosynthetic pathway after shading. In this study, fifteen such genes, e.g., PAL, C4H, 4CL, CHS, CHI, F3H and DFR were regulated by light (Fig. 5). Most of the genes had comparable expression profiles; their expression amounts were low in the A53CK sample at 0 d, and gradually increased in the B53CK sample exposed to normal light for 7 d, peaking in the C53CK sample treated by normal light for 11 d. The expression levels of all transcripts were suppressed in the head-shading treatment group, corroborating the reduced anthocyanin amounts in plants under head-shading. Most genes showed higher expression levels in the C53WJ group treated by head-shading for 11 d compared with the B53WJ group treated by head-shading for 7 d, except assembly18039 (PAL), assembly 39669 (C4H) and assembly 52832 (CHS), which showed higher expression levels in the B53WJ group. Thus, these genes were considered critical structural genes associated with the effects of light on anthocyanin production.
Figure 5. The flavonoid biosynthetic pathway associated with anthocyanin biosynthesis.
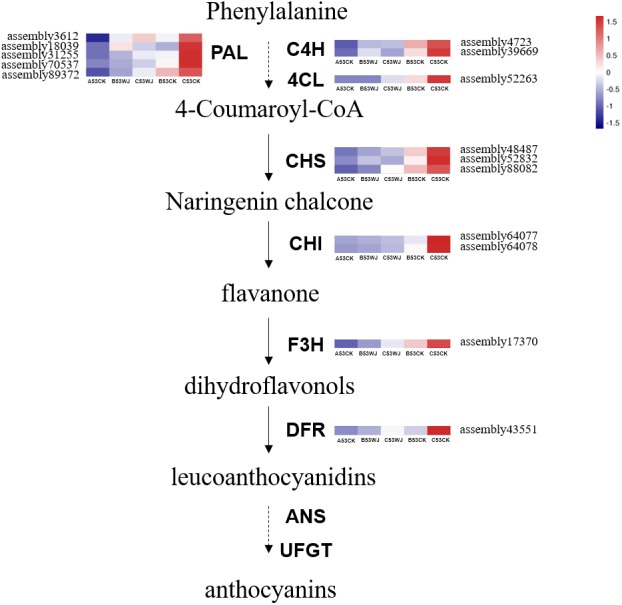
The expression patterns of various structural genes contributing to anthocyanin production in A53CK, B53WJ, C53WJ, B53CK and C53CK are arranged from left to right. A53CK, B53WJ, C53WJ, B53CK and C53CK indicate samples treated by head-shading for 0 d, 7 d and 11 d, and normal light for 7 d and 11 d, respectively. The color scale reflected the log-transformed FPKM values.
Expression of genes associated with light signal perception and transduction
Photoreceptors (PHYA, PHYB, PHYC, PHYD, PHYE), cryptochromes (CRY1, CRY2, CRY3), phototropins (PHOT1, PHOT2) and UV RESISTENCE LOCUS8 (UVR8) are four classes of photoreceptors contributing to light response (Chaves et al., 2011; Christie, 2007; Jenkins, 2014). Compared with head shading samples, flower head response to light was mediated by three CRY3 photoreceptors, including assembly 37255, assembly 86925 and assembly 18166 (Table 1), which were all downregulated under head-shading treatment. Under light conditions, the expression levels of assembly 37255 and assembly 18166 gradually increased and showed highest values in C53CK under light treatment, while assembly 86925 showed the highest value in B53CK under light treatment.
Table 1. Genes expression related to photoreceptors and light signal transduction.
| Accession | Gene description (blast NR) | FPKM value | ||||
|---|---|---|---|---|---|---|
| A53CK | B53WJ | C53WJ | B53CK | C53CK | ||
| photoreceptors | ||||||
| assembly37255 | CRY3 | 0.16 | 0.41 | 0.21 | 3.05 | 5.01 |
| assembly86925 | CRY3 | 0.00 | 0.87 | 0.00 | 3.32 | 2.12 |
| assembly18166 | CRY3 | 0.00 | 0.57 | 0.17 | 2.18 | 3.48 |
| genes related to light signal transduction | ||||||
| assembly42644 | HY5, TED 5 | 5.57 | 8.08 | 22.31 | 17.71 | 58.64 |
Notes.
- CRY
- Cryptochrome
- HY5
- HYPOCOTYL 5
HY5 induces photomorphogenesis under all light conditions and exerts direct regulatory effects on light-responsive genes (Chattopadhyay et al., 1998). Here, we found BoHY5 (assembly 42644) was downregulated under head-shading treatment in comparison with light conditions. Assembly 42644 showed rapid upregulation in the B53CK group, peaking in C53CK, with reduced amounts under head-shading treatment.
DEGs Encoding Transcription Factors and their interaction with Hormone-Related Genes
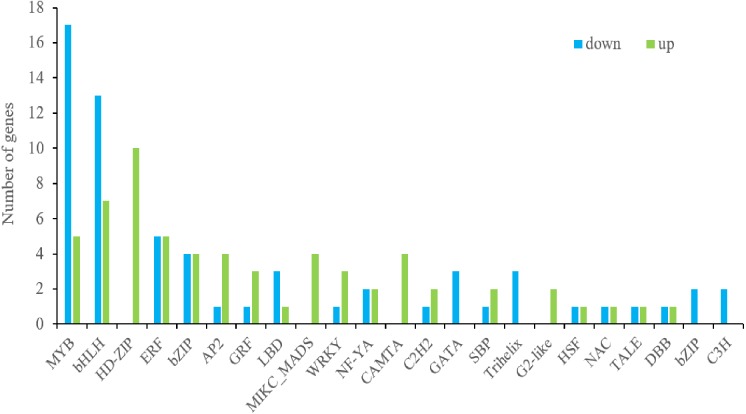
To assess the complex network of signaling pathways in light-induced anthocyanin biosynthesis, we further compared the expression profiles of transcription factors. A total of 133 genes were assigned to the MapMan “transcription factors” bin and more than half were down-regulated (Fig. 6). Members of the GATA, Trihelix, bZIP and C3H families were downregulated and HD-ZIP, MIKC_MADS, CAMTA, G2-like families were upregulated, while other TF families were regulated in both directions. Of these TFs, MYB and bHLH constituted the largest family with 22 and 20 members, followed by the HD-ZIP, ERF, bZIP, AP2, GRF, LBD, MIKC_MADS, WRKY, NF-YA and CAMTA families, with 10, 10, 8, 5, 4, 4, 4, 4, 4, 4 members, respectively. Most of the MYB and bHLH TFs were downregulated, including MYB90, MYB114, EGL3 and TT8, which are key regulators of anthocyanin biosynthesis.
Figure 6. Statistical column diagram showing transcription factors between up- and down-regulated differentially expressed genes (DEGs).
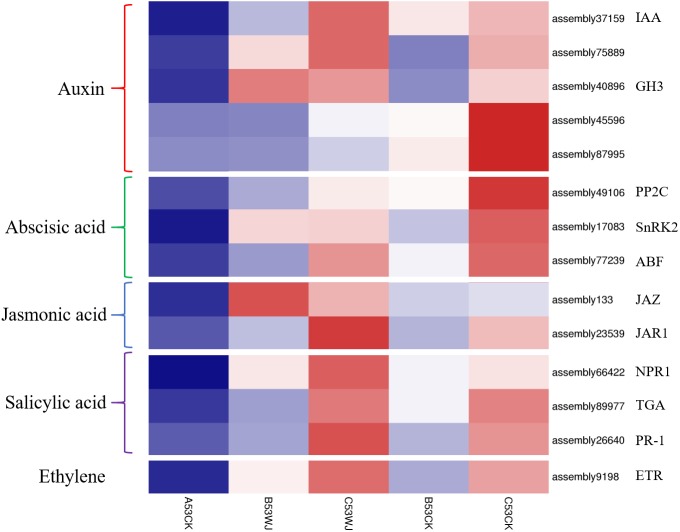
The interaction partners of these TFs activated as molecular responses are key components of signal transduction pathways that take place during anthocyanin biosynthesis. To investigate the functions of plant hormones in light-induced anthocyanin biosynthesis, the expression patterns of genes involved in plant hormone response as receptors and response factors were assessed by heat map analysis (Fig. 7). The results showed that multiple genes involved in abscisic acid, auxin, salicylic acid, ethylene and jasmonic acid signaling pathway were mostly upregulated in samples treated for 11 d in comparison with those treated for 0 d and 7 d.
Figure 7. Heat map representation of the expression patterns of DEGs related to the phytohormone biosynthesis and signaling pathways during light-responsive reactions.
A53CK, B53WJ, C53WJ, B53CK and C53CK indicate samples treated by head-shading for 0 d, 7 d and 11 d, and normal light for 7 d and 11 d, respectively. The color scale reflected the log-transformed FPKM values.
In the auxin transduction pathway, the expressions levels of genes encoding auxin influx carrier/auxin-responsive protein IAA (AUX/IAA) (assembly 37159 and assembly 75889) and auxin responsive GH3 gene family (GH3) (assembly 40896) peaked in the C53WJ sample treated by head-shading for 11 d. Meanwhile, the other two GH3 genes (assembly45596 and assembly 87995) were significantly up-regulated in the C53CK sample treated by normal light for 11 d. In the abscisic acid transduction pathway, genes encoding protein phosphatase 2C (PP2C) (assembly 49106), serine/threonine protein kinase SRK2n (SnRK2) genes (assembly17083) and ABRE binding factors (ABF) (assembly 77239) were downregulated under head-shading conditions in comparison with normal light conditions. In the jasmonic acid transduction pathway, the transcriptional level of jasmonate ZIM (JAZ) domain-containing gene (assembly133) was highest in the B53WJ sample treated by headed-shading for 7 d, while gene encoding jasmonic acid resistant 1 (JAR1) (assembly23539) was highly expressed in the C53WJ sample treated by head-shading for 11 d. In the salicylic acid (SA) signaling pathway, SA receptors Non-Expresser of Pathogenesis Related Gene 1 (NPR1) genes (assembly 66422), TGA factor (assembly 89977) and pathogenesis-related 1 (PR-1; assembly 26640) genes were upregulated in the C53WJ sample treated by head-shading for 11 d. These results indicated that gene expression patterns in plant hormone signal transduction pathways showed alterations in broccoli during the shading treatment.
Different expression patterns of anthocyanin biosynthetic regulatory genes
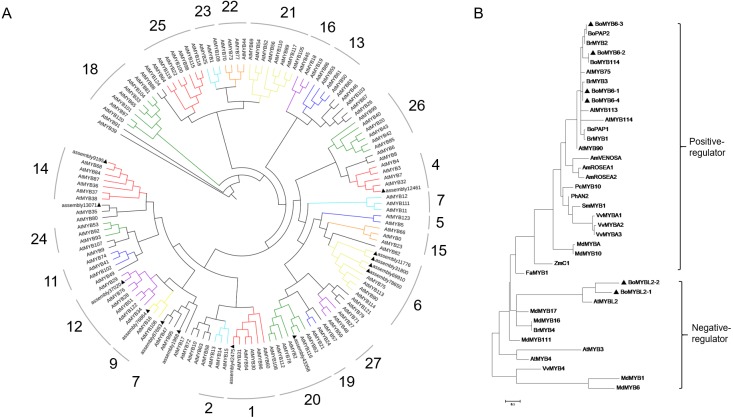
To classify and assess the potential functions and evolutionary characteristics of BoMYB proteins in broccoli, a phylogenetic tree was generated for the 125 AtMYBs of Arabidopsis. Thirteen broccoli R2R3-MYBs were clustered together with AtMYBs and grouped into 8 MYB sub-categories (Fig. 8). In the present study, there were one BoMYB in subfamily 1 (S1; assembly 42475), one in S4 (assembly 12461), four in S6 (assembly 69910, assembly 31800, assembly 11776 and assembly 78650), two in S7 (assembly 87693 and assembly 1968), one in S9 (assembly 76864), one in S12 (assembly 37020), one in S14 (assembly 9195), one in S20 (assembly 43358), and one in unknown class (assembly 43358). Then, phylogenetic analysis was carried out to identify probable candidate MYBs involved in anthocyanin regulation in purple broccoli. Notably, S6 molecules were found to modulate flavonoid and/or anthocyanin metabolic pathways. The first S6 group genes, i.e., assembly 69910, assembly 31800, assembly 11776 and assembly 78650, were renamed BoMYB6-1, BoMYB6-2, BoMYB6-3 and BoMYB6-4, respectively, while assembly 75153 and assembly 75155 were renamed BoMYBL2-1 and BoMYB2-2, respectively. Phylogenetic analysis indicated that BoMYB6-1, BoMYB6-2, BoMYB6-3 and BoMYB6-4 were assigned to the positive-regulator group, while BoMYBL2-1 and BoMYB2-2 belonged to the negative-regulator category.
Figure 8. Phylogenetic relationship of BoMYBs with other R2R3-MYBs.
The UPGMA method was employed for tree generation using 1,000 bootstrap values with Mega 6.0. Putative modulatory functions of respective R2R3-MYB proteins are shown. (A) Evolutionary relationships of 125 AtMYBs and BoMYBs; (B) Phylogenetic relationships of BoMYBs controlling anthocyanin production and those of other species. Triangles indicate the putatively encoded BoMYB TFs.
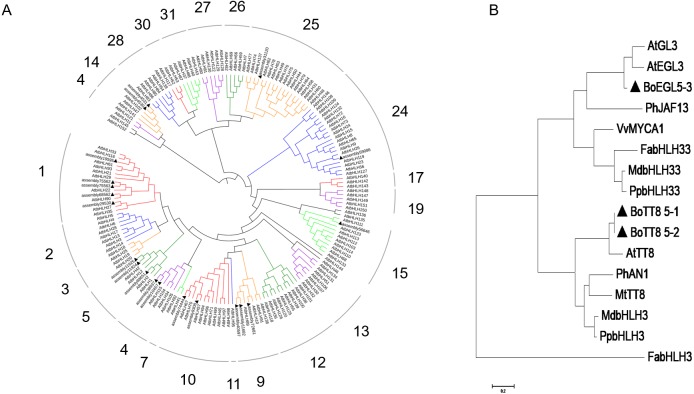
Using the same methods, a total of twenty broccoli BobHLHs and 152 Arabidopsis bHLHs were employed to generate a phylogenetic tree, and divided into eight subfamilies, including 1, 4, 5, 10, 14, 15, 24 and 25. As shown in Fig. 9, subfamily 5 was associated with flavonoid and/or anthocyanin biosynthesis, including three BobHLHs (assembly 11924, assembly 11925 and assembly 88651, renamed BoTT8_5-1, BoTT8_5-2 and BoEGL5-3, respectively). Phylogenetic analysis showed BobHLH clustering with Arabidopsis anthocyanin bHLH regulator, BoTT8_5-1 and BoTT8_5-2 showed a more distant association with Arabidopsis AtTT8; BoEGL5-3 had more distant associations with Arabidopsis AtGL3 and AtEGL3. These results suggested that BoTT8_5-1, BoTT8_5-2 and BoEGL5-3 played roles in regulating anthocyanin synthesis.
Figure 9. Phylogenetic associations of BobHLH with other bHLHs.
The UPGMA method was employed for tree generation using 1,000 bootstrap values with Mega 6.0. Putative modulatory functions of respective BobHLH proteins are shown. (A) Evolutionary relationships of 152 AtbHLHs and BoHLHs; (B) Phylogenetic relationships of BoHLHs controlling anthocyanin production in subfamily 5 and those of other species. Triangles indicate the putatively encoded bHLH TF.
Validation of the expression of genes of the Anthocyanin Biosynthetic Pathway
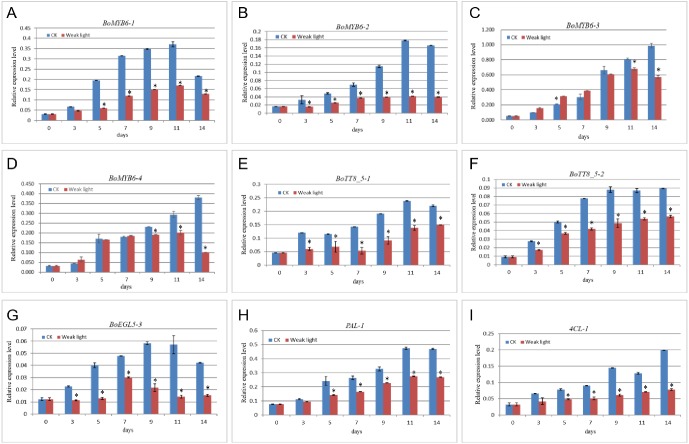
To confirm gene expression data revealed by transcriptomics, seven regulatory genes (BoMYB6-1, BoMYB6-2, BoMYB6-3, BoMYB6-4, BoTT8_5-1, BoTT8_5-2 and BoEGL5-3) and two structural genes (PAL1, 4CL-1) contributing to anthocyanin production in broccoli underwent amplification from specimens obtained in head’s developmental stages under shading and light conditions, respectively, by RT-qPCR (Fig. 10), with Actin2 employed as a reference gene. All 9 genes displayed identical expression trends obtained by RNA-seq. Most of the transcription factors assessed also showed identical expression trends as determined by transcriptomics. Moreover, the light and shading groups were significantly different (P < 0.05).
Figure 10. Validation of the expression of genes associated with anthocyanin biosynthesis in broccoli.
∗ represents significance at the 0.05 level.
Discussion
Broccoli head is the key light-response receptor in anthocyanin production
The purple broccoli has abundant flavonoids and other bioactive molecules in addition to glucosinolate-derived isothiocyanates, vitamins and minerals, indicating a great nutritional value for this plant (Moreno et al., 2010). In the present study, delphinidin-3-O-galactoside, delphinidin-3-O-glucoside, cyanidin-3-O-galactoside, cyanidin-3-O-glucoside, malvidin-3-O-galactoside and malvidin-3-O-glucoside were identified in broccoli head, in accordance with previously published data on broccoli sprouts, purple Graffiti cauliflower and red cabbage (Moreno et al., 2010; Chiu et al., 2010; Yuan, Chiu & Li, 2009). Light represents a predominant environmental stimulus controlling plant anthocyanin production (Liu et al., 2018). As shown above, shading reduced anthocyanin production in broccoli. Under light conditions, anthocyanins were produced in a rapid manner (Fig. 1). In this study, the relative contents of total anthocyanins under head-shading were more reduced than those obtained under leaves shading and normal light conditions. This was similar to anthocyanin accumulation in chrysanthemum under head-shading and leaf shading conditions as reported by Hong et al. (2015).
Usually, more focus is placed on fresh broccoli heads which provide economic benefits directly; however, purple broccoli varieties with purple or green leaves are variable in leaf pigmentation. In order to illustrate this mechanism, RNA-seq was performed under head-shading and leaves-shading treatment. Downregulated DEGs, GO and KEGG analyses were carried out. GO analyses showed that, “nucleus”, “response to abscisic acid”, “DNA-binding transcription factor activity”, “response to water deprivation” and “sequence-specific DNA binding” were the major GO terms. Moreover, KEGG analyses showed that, “Starch and sucrose metabolism”, “Plant hormone signal transduction” and “Protein processing in endoplasmic reticulum” were the major public pathway-related database. ABA played positive role in modulating anthocyanin biosynthesis (Carvalho, Carvalho & Duque, 2010). Starch degradation and Sucrose-specific contribute to anthocyanin biosynthesis, while MdSnRK1.1 interacts with MdJAZ18 to induce sucrose-induced anthocyanin and proanthocyanidin biosynthesis in apple (Liu et al., 2017), however IAA might play an crucial role in anthocyanin accumulation regardless of sugar and starch in ornamental kale (Ren et al., 2019). Endoplasmic reticulum likely function in the biosynthesis and transport of anthocyanin pigments (Wagner, 1987). Therefore, the downregulated genes in hormone signaling, starch and sucrose metabolism, endoplasmic reticulum and transcription factors might affect the anthocyanin accumulation under head-shading treatment in broccoli.
Although anthocyanin biosynthesis is well characterized, the associated light-response receptors in broccoli are less clear. Photosynthesis was significantly repressed under both head and leaves-shading conditions, suggesting that shading affects plants during photosynthesis. The DEGs in “photosynthesis”, “peroxisome”, “flavonoid biosynthesis” were significantly repressed by head shading while “amino acid biosynthesis”, “carbon metabolism”, “sulfur metabolism”, “cysteine and methionine metabolism”, and “photosynthesis-antenna proteins” were significantly suppressed by leaves shading via transcriptional regulation. We can infer that under leaves-shading treatment, carbon fixation and carbohydrate production were affected by less light indirectly leading to less anthocyanin contents (Shao et al., 2014). Under head-shading conditions, some photoreceptors and anthocyanin biosynthesis-associated genes downregulated directly leading to decreased anthocyanin contents in response to light. Therefore, the head might be the more critical role as light-response receptor in anthocyanin production.
Light-induced anthocyanin biosynthesis is mediated by signal transduction pathways in ‘Long Jing’
Plants use many photoreceptors for coordinating responses to environmental light (Ma et al., 2019). When broccoli flowers under head-shading conditions were compared with those under normal light conditions, we found three CRY3 genes were downregulated, in agreement with the tendency of anthocyanin production in broccoli head flower (Fig. 1B). These finding indicate that CRY3 may be an important photoreceptor in broccoli head flower, with critical functions in regulating anthocyanin production (Opseth et al., 2015). Li et al. (2017) and Li et al. (2018) characterized the eggplant photomorphogenic factors CRY3 was upregulated by light. In the current study, HY5 was identified among DEGs during head flower development and its transcription levels quickly rose under light conditions but were reduced under head-shading treatment. In Turnip (Brassica rapa), the upregulation of BrHY5 further induced BrPAP1 expression to produce more anthocyanins under sunlight (Yang et al., 2017). Shin et al. (2013) has reported that HY5 induced anthocyanin accumulation by directly binding the promoter of MYB75/PAP1 transcription factor in Arabidopsis. In apple, MdHY5 also bound on the 5′ upstream region of MdMYBA in a yeast system (Peng et al., 2013). This suggests HY5 might represent an inducer of broccoli head flower coloration by binding the promoter of MYB TFs under light conditions (Stracke et al., 2010; Shin et al., 2013; Nguyen et al., 2015).
Transcription factors are involved in light-induced broccoli coloration
A set of TFs are considered to regulate anthocyanin biosynthesis at the transcriptional level, including R2R3-MYB, bHLH and WD40, as well as members of several other TF families. In this study, 133 transcription factors were differently expressed in response to light which were classified into 23 families (Fig. 6). Among them, HY5 (Shin et al., 2013), MYB90 (Bac-Molenaar et al., 2015), MYB114 (Gonzalez et al., 2008), MYBL2 (Dubos et al., 2008), EGL3 (Nemie-Feyissa et al., 2015), TT8 (Li et al., 2017) and WRKY26 (Amato et al., 2017), WRKY70 (Li et al., 2006) were recorded in the study, which were known for their response to anthocyanin biosynthesis. As is well known, HY5 and MYB could triggered expression of light-inducible genes CHS after light exposure (Chattopadhyay et al., 1998). In our dataset, the differently expressed R2R3MYB TFs BoMYB6-1, BoMYB6-2, BoMYB6-3, BoMYB6-4 as well as bHLH TFs BoTT8_5-1, BoTT8_5-2, BoEGL5-3, were categorised into subfamily 6 and 5 respectively, which were found to modulate flavonoid and/or anthocyanin metabolic pathways (Figs. 8 and 9). In ornamental cabbage, R2R3MYB TFs BoPAP2, bHLH TFs BoTT8, BoEGL3.1 and BoMYC1.2 and WDR genes BoTTG1 were identified as candidates involved in the anthocyanin biosynthesis pathway (Jin et al., 2018). Ectopic expression of the R2R3 MYB TFs Pr-D allele in cauliflower induced tissue-specific anthocyanin accumulation (Chiu et al., 2010). In the turnip seedlings, PAP1 transcript levels simultaneously depend on light spectra and hypocotyl localization, suggesting that MYBs have critical functions in light spectrum- and location-dependent transduction of light signals (Wang et al., 2012). In turnip, BrTT8, interacting with BrPAP1 and BrTTG1, plays a positive role in anthocyanin biosynthesis via regulating expression of LBGs (BrDFR, BrANS1, BrANS2, and BrUFGT) in response to light (Yang et al., 2017). In addition, many other differently expressed TFs response to light were detected in our datasets, especially some MYB genes, MADS-box and NAC, suggesting their potential roles in regulation of anthocyanin accumulation (Jaakola, Poole & Jones, 2010; Wang et al., 2018). As a negative regulator, loss of BoMYBL2-1 expression led to the establishment of purple color in cabbages (Song et al., 2018). Moreover, MYBL2 expression was inhibited by high light-induced stress, which triggered strong accumulation of anthocyanins in Arabidopsis (Dubos et al., 2008). At the present study, we screened numerous differently expressed TFs, including 10 HD-ZIPs, 10 ERFs, 8 bZIPs, 5 AP2, 4 WRKYs, 4 GRFs, 4 LBDs, 4 MIKC_MADS, 4 NF-YAs and 4 CAMTAs families. Also, further studies are needed to ascertain their roles in anthocyanin biosynthesis in response to light in broccoli.
Plant hormones are involved in light-induced broccoli coloration
Previous studies have shown that phytohormones controlling anthocyanin accumulation can be affected by light (Loreti et al., 2008; Zhang et al., 2016a; Zhang et al., 2016b). Endogenous application of auxins can inhibit the expression of anthocyanin-related genes (Deikman & Hammer, 1995). In this study, the expressions levels of IAAs and one GH3 genes were higher in the C53WJ sample treated by head-shading for 11 d. However, two GH3s were upregulated in the C53CK sample treated by normal light for 11 d. These distinct expression patterns indicate that auxin signaling has different functions in the regulation of anthocyanin biosynthesis in broccoli through various transduction pathways. In purple ornamental cabbage, Jin et al. (2019) suggested that ABA might increase the intensity of purple pigmentation of the inner leaves. Carvalho, Carvalho & Duque (2010) provided evidence that ABA plays a positive role in modulating anthocyanin biosynthesis in hormone mutants after exogenous application (Carvalho, Carvalho & Duque, 2010). We observed significant upregulation of genes encoding ABA-responsive elements, such as PP2C, SnRK2 and ABF in the normal light treatment (Fig. 7). Thus, we speculate that these ABA signaling factors might promote the expression of anthocyanin-related genes. Wang et al. (2019) indicated that ethylene acts as a negative regulator in red light-regulated anthocyanin biosynthesis in cabbage. Ethylene suppresses the anthocyanin biosynthesis via binding to ETRs in Arabidopsis (Jeong et al., 2010) and peel (Ma et al., 2019). Similarly, we observed that the expression levels of ETR were higher under the head-shading treatments. Previous studies have shown ethylene treatment significantly lowers anthocyanin accumulation, while SA alleviates these effects in canola plants (Brassica napus L.) (Tirani, Nasibi & Kalantari, 2013). ABA, JA and SA pre-treatments could increase anthocyanin accumulation in turnip (Brassica rapa ssp. rapa) and Brassica juncea L. (Thiruvengadam et al., 2016; Sharma et al., 2018). Horváth et al. (2007) reported exogenously applied SA increases the accumulation of anthocyanin in UV-B exposed T. aestivum. Endogenous application of jasmonate can also increase the production of anthocyanin (Memelink). However, we found that all genes encoding JAZ, JAR1 and NPR1 in jasmonic acid signalling pathway as well as TGA and PR-1 in the salicylic acid signaling pathway were upregulated under head-shading treatment, which implies that SA and JA play negative roles in light-induced anthocyanin accumulation in broccoli. In general, some form of hormonal cross-talk may be present in pigment accumulation of broccoli flower head.
Conclusions
Overall, comprehensive gene expression analysis was performed for exploring the mechanisms underlying light-associated anthocyanin accumulation in broccoli, and the following regulatory sequence was proposed. Under light conditions, the expression levels of CRY3 and HY5 contributing to light signal perception and transduction are closely associated with anthocyanin production in broccoli head flower. HY5 activates the downstream MYBs related to anthocyanin biosynthesis. Strikingly, the above results revealed that BoMYB6-1, BoMYB6-2, BoMYB6-3, BoMYB6-4, BoMYBL2-1, BoMYBL2-2, BoTT8_5-1, BoTT8_5-2 and BoEGL5-3 affect anthocyanin production and regulate structural genes such as PAL, C4H, 4CL, CHS, CHI, F3H and DFR. In addition, HD-ZIP, ERF, bZIP, AP2, GRF, LBD, MIKC_MADS, WRKY, NF-YA, CAMTA transcription factors and abscisic acid, auxin, salicylic acid, ethylene, jasmonic acid signaling pathway involved in the anthocyanin biosynthesis in response to light. This study provides novel insights into the functions of these genes in regulating light-associated anthocyanin production.
Supplemental Information
Funding Statement
This work was supported by the National Key R&D Program of China (2017YFD0101805), the Shanghai Agriculture Applied Technology Development Program, China (Grant No. G2016060105), and the Youth Talent Development Plan of Shanghai Municipal Agricultural System, China (Grant No. 20180116). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Chunqing Liu performed the experiments, analyzed the data, prepared figures and/or tables, authored or reviewed drafts of the paper, and approved the final draft.
Xueqin Yao and Zhujie Xie conceived and designed the experiments, authored or reviewed drafts of the paper, and approved the final draft.
Guangqing Li conceived and designed the experiments, prepared figures and/or tables, and approved the final draft.
Lei Huang analyzed the data, prepared figures and/or tables, and approved the final draft.
Data Availability
The following information was supplied regarding data availability:
Data is available in Table S1 and at NCBI SRA: PRJNA560282.
References
- Aharoni et al. (2001).Aharoni A, De Vos CHR, Wein M, Sun Z, Greco R, Kroon A, Mol JN, O’Connell AP. The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant Journal. 2001;28:319–332. doi: 10.1046/j.1365-313X.2001.01154.x. [DOI] [PubMed] [Google Scholar]
- Ahmed et al. (2014).Ahmed NU, Park JI, Jung HJ, Yang TJ, Hur Y, Nou IS. Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene. 2014;550(1):46–55. doi: 10.1016/j.gene.2014.08.013. [DOI] [PubMed] [Google Scholar]
- Albert et al. (2009).Albert NW, Lewis DH, Zhang H, Irving LJ, Jameson PE, Davies K. Light induced vegetative anthocyanin pigmentation in Petunia. Journal of Experimental Botany. 2009;60(7):2191–2202. doi: 10.1093/jxb/erp097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato et al. (2017).Amato A, Cavallini E, Zenoni S, Finezzo L, Begheldo M, Ruperti B, Tornielli GB. A grapevine TTG2-Like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front Plant Science. 2017;7:1979. doi: 10.3389/fpls.2016.01979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bac-Molenaar et al. (2015).Bac-Molenaar JA, Fradin EF, Rienstra JA, Vreugdenhil D, Keurentjes JJB. GWA mapping of anthocyanin accumulation reveals balancing selection of MYB90 in Arabidopsis thaliana. PLOS ONE. 2015;10(11):e0143212. doi: 10.1371/journal.pone.0143212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho, Carvalho & Duque (2010).Carvalho RF, Carvalho SD, Duque P. The plant-specific SR45 protein negatively regulates glucose and ABA signaling during early seedling development in Arabidopsis. Plant Physiology. 2010;154(2):772–783. doi: 10.1104/pp.110.155523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker-Scott (1999).Chalker-Scott L. Environmental significance of anthocyanins in plant stress responses. Photochemistry and Photobiology. 1999;70(1):1–9. doi: 10.1111/j.1751-1097.1999.tb01944.x. [DOI] [Google Scholar]
- Chattopadhyay et al. (1998).Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. The Plant Cell. 1998;10(5):673–683. doi: 10.1105/tpc.10.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves et al. (2011).Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, Brettel K, Essen LO, van der Horst GT, Batschauer A, Ahmad M. The cryptochromes: blue light photoreceptors in plants and animals. Annual Review of Plant Biology. 2011;62:335–364. doi: 10.1146/annurev-arplant-042110-103759. [DOI] [PubMed] [Google Scholar]
- Chiu & Li (2012).Chiu LW, Li L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta. 2012;236(4):1153–1164. doi: 10.1007/s00425-012-1665-3. [DOI] [PubMed] [Google Scholar]
- Chiu et al. (2010).Chiu LW, Zhou X, Burke S, Wu X, Prior RL, Li L. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiology. 2010;154(3):1470–1480. doi: 10.1104/pp.110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie (2007).Christie JM. Phototropin blue-light receptors. Annual Review of Plant Biology. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- Cominelli et al. (2008).Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C. Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. Plant Physiology. 2008;165:886–894. doi: 10.1016/j.jplph.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Conesa et al. (2005).Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Deikman & Hammer (1995).Deikman J, Hammer PE. Induction of anthocyanin accumulation by cytokinins in Arabidopsis thaliana. Plant Physiology. 1995;108:47–57. doi: 10.1104/pp.108.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon & Paiva (1995).Dixon RA, Paiva NL. Stress-induced phenylpropanoid metabolism. The Plant Cell. 1995;7(7):1085–1097. doi: 10.1105/tpc.7.7.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du et al. (2010).Du Z, Zhou X, Ling Y, Zhang ZH, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research. 2010;38:W64–W70. doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos et al. (2008).Dubos C, Le Gourrierec J, Baudry A, Huep G, Lanet E, Debeaujon I, Routaboul JM, Alboresi A, Weisshaar B, Lepiniec L. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant Journal. 2008;55(6):940–953. doi: 10.1111/j.1365-313X.2008.03564.x. [DOI] [PubMed] [Google Scholar]
- Espley et al. (2007).Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant Journal. 2007;49:414–427. doi: 10.1111/j.1365-313x.2006.02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng et al. (2015).Feng S, Sun S, Chen X, Wu S, Wang D, Chen X. PyMYB10 and PyMYB10.1 interact with bHLH to enhance anthocyanin accumulation in pears. PLOS ONE. 2015;10(11):e0142112. doi: 10.1371/journal.pone.0142112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez et al. (2008).Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant Journal. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Grabherr et al. (2011).Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, Di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature Biotechnology. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo et al. (2014).Guo N, Cheng F, Wu J, Liu B, Zheng S, Liang J, Wang X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genomics. 2014;15:426. doi: 10.1186/1471-2164-15-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborne & Williams (2000).Harborne JB, Williams CA. Advances in flavonoid research since 1992. Phytochemistry. 2000;55(6):481–504. doi: 10.1016/S0031-9422(00)00235-1. [DOI] [PubMed] [Google Scholar]
- Hichri et al. (2010).Hichri I, Heppel SC, Pillet J, Leon C, Czemmel S, Delrot S, Lauvergeat V, Bogs J. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Molecular Plant. 2010;3(3):509–523. doi: 10.1093/mp/ssp118. [DOI] [PubMed] [Google Scholar]
- Holton & Cornish (1995).Holton T, Cornish E. Genetics and biochemistry of anthocyanin biosynthesis. The Plant Cell. 1995;7:1071–1083. doi: 10.2307/3870058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong et al. (2015).Hong Y, Tang X, Huang H, Zhang Y, Dai S. Transcriptomic analyses reveal species-specific light-induced anthocyanin biosynthesis in chrysanthemum. BMC Genomics. 2015;16:202. doi: 10.1186/s12864-015-1428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth et al. (2007).Horváth E, Pál M, Szalai G, Páldi E, Janda T. Exogenous 4-hydroxybenzoic acid and salicylic acid modulate the effect of short-term drought and freezing stress on wheat plants. Biologia Plantarum. 2007;51:480–487. doi: 10.1007/s10535-007-0101-1. [DOI] [Google Scholar]
- Jaakola, Poole & Jones (2010).Jaakola L, Poole M, Jones MO. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiology. 2010;153:1619–1629. doi: 10.1104/pp.110.158279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins (2014).Jenkins GI. The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell. 2014;26(1):21–37. doi: 10.1105/tpc.113.119446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong et al. (2010).Jeong SW, Das PK, Jeoung SC, Song JY, Lee HK, Kim YK, Kim WJ, Park YIl, Yoo SD, Choi SB, Park YL. Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiology. 2010;154:1514–1531. doi: 10.1104/pp.110.161869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin et al. (2000).Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO Journal. 2000;19(22):6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin et al. (2019).Jin SW, Rahim MA, Jung HJ, Afrin KS, Kim HT, Park JI, Kang JG, Nou IS. Abscisic acid and ethylene biosynthesis-related genes are associated with anthocyanin accumulation in purple ornamental cabbage (Brassica oleracea var. acephala) Genome. 2019;62(8):513–526. doi: 10.1139/gen-2019-0038. [DOI] [PubMed] [Google Scholar]
- Jin et al. (2018).Jin SW, Rahim MA, Kim HT, Park JI, Kang JG, Nou IS. Molecular analysis of anthocyanin-related genes in ornamental cabbage. Genome. 2018;61(2):111–120. doi: 10.1139/gen-2017-0098. [DOI] [PubMed] [Google Scholar]
- Kanehisa et al. (2008).Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36:480–484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes, Verweij & Quattrocchio (2005).Koes R, Verweij W, Quattrocchio F. Flavonoids: a colorful model for the regulation and evolution of biochemical pathways. Trends Plant Science. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lee, Durst & Wrolstad (2005).Lee J, Durst RW, Wrolstad RE. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. Journal of AOAC International. 2005;88:1269–1278. doi: 10.3200/JACH.54.2.108-115. [DOI] [PubMed] [Google Scholar]
- Li et al. (2006).Li J, Brader G, Kariola T, Palva ET. WRKY70 modulates the selection of signaling pathways in plant defense. Plant Journal. 2006;46(3):477–491. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- Li et al. (2018).Li J, He YJ, Zhou L, Liu Y, Jiang M, Ren L, Chen H. Transcriptome profiling of genes related to light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) before purple color becomes evident. BMC Genomics. 2018;19(1):201. doi: 10.1186/s12864-018-4587-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li et al. (2017).Li J, Ren L, Gao Z, Jiang M, Liu Y, Zhou L, He Y, Chen H. Combined transcriptomic and proteomic analysis constructs a new model for light-induced anthocyanin biosynthesis in eggplant (Solanum melongena L.) Plant Cell Environment. 2017;40(12):3069–3087. doi: 10.1111/pce.13074. [DOI] [PubMed] [Google Scholar]
- Lin-Wang et al. (2011).Lin-Wang K, Micheletti D, Palmer J, Volz R, Lozano L, Espley R, Hellens RP, Chagnè D, Rowan DD, Troggio M, Iglesias I, Allan AC. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant, Cell & Environment. 2011;34:1176–1190. doi: 10.1111/j.1365-3040.2011.02316.x. [DOI] [PubMed] [Google Scholar]
- Liu, Osbourn & Ma (2015).Liu J, Osbourn A, Ma P. MYB transcription factors as regulators of phenylpropanoid metabolism in plant. Molecular Biology. 2015;8(5):689–708. doi: 10.1016/j.molp.2015.03.012. [DOI] [PubMed] [Google Scholar]
- Liu et al. (2017).Liu XJ, An XH, Liu X, Hu DG, Wang XF, You CX, Hao YJ. MdSnRK1.1 interacts with MdJAZ18 to regulate sucrose-induced anthocyanin and proanthocyanidin accumulation in apple. Journal of Experimental Botany. 2017;68(11):2977–2990. doi: 10.1093/jxb/erx150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu et al. (2018).Liu Y, Tikunov Y, Schouten RE, Marcelis LFM, Visser RGF, Bovy A. Anthocyanin biosynthesis and degradation mechanisms in Solanaceous vegetables: a review. Front Chemistry. 2018;6:52. doi: 10.3389/fchem.2018.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti et al. (2008).Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytologist. 2008;179(4):1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- Lotkowska et al. (2015).Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B. The arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiology. 2015;169(3):1862–1880. doi: 10.1104/pp.15.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig et al. (1989).Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proceedings of the National Academy of Sciences of the United States of America. 1989;86(18):7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma et al. (2019).Ma CQ, Liang B, Chang B, Yan J, Liu L, Wang Y, Yang Y, Zhao Z. Transcriptome profiling of anthocyanin biosynthesis in the peel of ’Granny Smith’ apples (Malus domestica) after bag removal. BMC Genomics. 2019;20(1):353. doi: 10.1186/s12864-019-5730-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui, Umemura & Ohme-Takagi (2008).Matsui K, Umemura Y, Ohme-Takagi M. AtMYBL2, a protein with a single MYB domain, acts as a negative regulator of anthocyanin biosynthesis in Arabidopsis. Plant Journal. 2008;55(6):954–967. doi: 10.1111/j.1365-313x.2008.03565.x. [DOI] [PubMed] [Google Scholar]
- Matus, Aquea & Arce-Johnson (2008).Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology. 2008;8:1–15. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno et al. (2010).Moreno DA, Perez-Balibrea S, Ferreres F, Gil-Izquierdo A, Garcia-Viguera C. Acylated anthocyanins in broccoli sprouts. Food Chemistry. 2010;123:358–363. doi: 10.1016/j.foodchem.2010.04.044. [DOI] [Google Scholar]
- Nemie-Feyissa et al. (2015).Nemie-Feyissa D, Heidari B, Blaise M, Lillo C. Analysis of interactions between heterologously produced bHLH and MYB proteins that regulate anthocyanin biosynthesis: quantitative interaction kinetics by Microscale Thermophoresis. Phytochemistry. 2015;111:21–26. doi: 10.1016/j.phytochem. [DOI] [PubMed] [Google Scholar]
- Nesi et al. (2000).Nesi N, Debeaujon I, Jond C, Pelletier G, Caboche M, Lepiniec L. The TT8 gene encodes abasic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. The Plant Cell. 2000;12(10):1863–1878. doi: 10.2307/3871198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugart, Krumbein & Zrenner (2016).Neugart S, Krumbein A, Zrenner R. Influence of light and temperature on gene expression leading to accumulation of specific flavonol glycosides and hydroxycinnamic acid derivatives in Kale (Brassica oleracea var. sabellica) Frontiers in plant science. 2016;7:326. doi: 10.3389/fpls.2016.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen et al. (2015).Nguyen NH, Jeong CY, Kang GH, Yoo SD, Hong SW, Lee H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant Journal. 2015;84:1192–1205. doi: 10.1111/tpj.13077. [DOI] [PubMed] [Google Scholar]
- Opseth et al. (2015).Opseth L, Holefors A, Rosnes AKR, Lee Y, Olsen JE. FTL2 expression preceding bud set corresponds with timing of bud set in Norway spruce under different light quality treatments. Environmental and Experimental Botany. 2015;121:121–131. doi: 10.1016/j.envexpbot.2015.05.016. [DOI] [Google Scholar]
- Park et al. (2007).Park JS, Choung MG, Kim JB, Hahn BS, Kim JB, Bae SC, Roh KH, Kim YH, Cheon CI, Sung MK, Cho KJ. Genes up-regulated during red coloration in UV-B irradiated lettuce leaves. Plant Cell Reports. 2007;26:507–516. doi: 10.1007/s00299-006-0255-x. [DOI] [PubMed] [Google Scholar]
- Payne, Zhang & Lloyd (2000).Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics. 2000;156(3):1349–1362. doi: 10.0000/PMID11063707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares et al. (1986).Paz-Ares J, Wienand U, Peterson PA, Saedler H. Molecular cloning of the c locus of Zea mays: a locus regulating the anthocyanin pathway. EMBO Journal. 1986;5(5):829–833. doi: 10.1002/j.1460-2075.1986.tb04291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng et al. (2013).Peng T, Saito T, Honda C, Ban Y, Kondo S, Liu JH, Hatsuyama Y, Moriguchi T. Screening of UV-B-induced genes from apple peels by SSH: possible involvement of MdCOP1-mediated signaling cascade genes in anthocyanin accumulation. Physiologia Plantarum. 2013;148(3):432–444. doi: 10.1111/ppl.12002. [DOI] [PubMed] [Google Scholar]
- Pojer et al. (2013).Pojer E, Mattivi F, Johnson D, Stockley CS. The case for anthocyanin consumption to promote human health: a review. Comprehensive Reviews in Food Science and Food Safety. 2013;12:483–508. doi: 10.1111/1541-4337.12024. [DOI] [PubMed] [Google Scholar]
- Quattrocchio et al. (1998).Quattrocchio F, Wing JF, Woude KVD, Mol JNM, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant Journal. 1998;13(4):475–488. doi: 10.1046/j.1365-313X.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Ravaglia et al. (2013).Ravaglia D, Espley R, Henry-Kirk R, Andreotti C, Ziosi V, Hellens RP. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biology. 2013;13:68. doi: 10.1186/1471-2229-13-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren et al. (2019).Ren J, Liu Z, Chen W, Xu H, Feng H. Anthocyanin Degrading and Chlorophyll Accumulation Lead to the Formation of Bicolor Leaf in Ornamental Kale. International Journal of Molecular Science. 2019;20(3):603. doi: 10.3390/ijms20030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao et al. (2014).Shao Q, Wang H, Guo H, Zhou A, Huang Y, Sun Y, Li M. Effects of shade treatments on photosynthetic characteristics; chloroplast ultrastructure; and physiology of Anoectochilus roxburghii. PLOS ONE. 2014;9(2):e85996. doi: 10.1371/journal.pone.0085996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma et al. (2018).Sharma A, Kumar V, Yuan H, Kanwar MK, Bhardwaj R, Thukral AK, Zheng B. Jasmonic acid seed treatment stimulates insecticide detoxification in brassica juncea L. Frontiers in Plant Science. 2018;9:1609. doi: 10.3389/fpls.2018.01609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin et al. (2013).Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, Choi G, Park YI. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Letters. 2013;587:1543–1547. doi: 10.1016/j.febslet.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Shin, Park & Choi (2007).Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant Journal. 2007;49:981–994. doi: 10.1111/j.1365-313x.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- Song et al. (2018).Song H, Yi H, Lee M, Han CT, Lee J, Kim H, Park JI, Nou IS, Kim SJ, Hur Y. Purple Brassica oleracea var. capitata F. rubra is due to the loss of BoMYBL2-1 expression. BMC Plant Biology. 2018;18:82. doi: 10.1186/s12870-018-1290-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt et al. (2000).Spelt C, Quattrocchio F, Mol JN, Koes R. Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. The Plant Cell. 2000;12(9):1619–1632. doi: 10.1105/tpc.12.9.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke et al. (2010).Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. The Arabidopsis bzip transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-b radiation. Plant, Cell & Environment. 2010;33:88–103. doi: 10.1111/j.1365-3040.2009.02061.x. [DOI] [PubMed] [Google Scholar]
- Thiruvengadam et al. (2016).Thiruvengadam M, Baskar V, Kim S, Chung I. Effects of abscisic acid, jasmonic acid and salicylic acid on the content of phytochemicals and their gene expression profiles and biological activity in turnip (Brassica rapa ssp. rapa) Plant Growth Regulation. 2016;80:377–390. doi: 10.1007/s10725-016-0178-7. [DOI] [Google Scholar]
- Tirani, Nasibi & Kalantari (2013).Tirani MM, Nasibi F, Kalantari KM. Interaction of salicylic acid and ethylene and their effects on some physiological and biochemical parameters in canola plants (Brassica napus L.) Photosynthetica. 2013;51:411–418. doi: 10.1007/s11099-013-0041-2. [DOI] [Google Scholar]
- Wagner (1987).Wagner GJ. The possible role of endoplasmic reticulum in the biosynthesis and transport of anthocyanin pigments. Plant Vacuoles. 1987;134:393–400. doi: 10.1007/978-1-4684-5341-6_51. [DOI] [Google Scholar]
- Wang et al. (2019).Wang FH, Ahammed GJ, Li GY, Bai PT, Jiang Y, Wang SX, Chen SC. Ethylene is involved in red light-induced anthocyanin biosynthesis in cabbage (Brassica oleracea) International Journal of Agriculture & Biology. 2019;21:955–963. doi: 10.17957/IJAB/15.0980. [DOI] [Google Scholar]
- Wang et al. (2017).Wang ZG, Du H, Zhai R, Song LY, Ma FW, Xu LF. Transcriptome analysis reveals candidate genes related to color fading of ‘Red Bartlett’ (Pyrus communis L.) Frontiers in Plant Science. 2017;8:455. doi: 10.3389/fpls.2017.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2018).Wang YC, Wang N, Xu HF, Jiang SH, Fang HC, Su MY, Zhang ZY, Zhang TL, Chen XS. Auxin regulates anthocyanin biosynthesis through the Aux/IAA-ARF signaling pathway in apple. Horticulture Research. 2018;5:59. doi: 10.1038/s41438-018-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang et al. (2012).Wang Y, Zhou B, Sun M, Li Y, Kawabata S. UV-A light induces anthocyanin biosynthesis in a manner distinct from synergistic blue + UV-B light and UV-A/blue light responses in different parts of the hypocotyls in turnip seedlings. Plant Cell Physiology. 2012;53(8):1470–1480. doi: 10.1093/pcp/pcs088. [DOI] [PubMed] [Google Scholar]
- Yang et al. (2017).Yang JF, Chen YZ, Kawabata S, Li YH, Wang Y. Identification of light-independent anthocyanin biosynthesis mutants induced by ethyl methane sulfonate in turnip tsuda (brassica rapa) International Journal of Molecular Sciences. 2017;18(7):1288. doi: 10.3390/ijms18071288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu et al. (2019).Yu HF, Wang JS, Sheng XG, Zhao ZQ, Shen YS, Branca F, Gu HH. Construction of a high-density genetic map and identification of loci controlling purple sepal trait of flower head in Brassica oleracea L. italica. BMC Plant Biology. 2019;19(1):228. doi: 10.1186/s12870-019-1831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Chiu & Li (2009).Yuan Y, Chiu L, Li L. Transcriptional regulation of anthocyanin biosynthesis in red cabbage. Planta. 2009;230(6):1141–1153. doi: 10.1007/s00425-009-1013-4. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2012).Zhang B, Hu Z, Zhang Y, Li Y, Zhou S, Chen G. A putative functional MYB transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (Brassica Oleracea var. acephala f. tricolor) Plant Cell Report. 2012;31(2):281–289. doi: 10.1007/s00299-011-1162-3. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2003).Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130(20):4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2016a).Zhang HN, Li WC, Wang HC, Shi SY, Shu B, Liu LQ, Wei YZ, Xie JH. Transcriptome profiling of light-regulated anthocyanin biosynthesis in the pericarp of litchi. Frontiers in Plant Science. 2016a;7:963. doi: 10.3389/fpls.2016.00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang et al. (2016b).Zhang N, Qi Y, Zhang HJ, Wang X, Li H, Shi Y, Guo YD. Genistein: a novel anthocyanin synthesis promoter that directly regulates biosynthetic genes in red cabbage in a light-dependent way. Front Plant Science. 2016b;7:1804. doi: 10.3389/fpls.2016.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Butelli & Martin (2014).Zhang Y, Butelli E, Martin C. Engineering anthocyanin biosynthesis in plants. Current Opinion in Plant Biology. 2014;19:81–90. doi: 10.1016/j.pbi.2014.05.011. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2015).Zhang YJ, Hu ZL, Zhu MK, Zhu ZG, Wang ZJ, Tian SB, Chen GP. Anthocyanin accumulation and molecular analysis of correlated genes in purple kohlrabi (brassica oleracea var. gongylodes L.) Journal of Agricultural and Food Chemistry. 2015;63(16):4160–4169. doi: 10.1021/acs.jafc.5b00473. [DOI] [PubMed] [Google Scholar]
- Zhang et al. (2018).Zhang YT, Jiang L, Li YL, Chen Q, Ye Y, Zhang Y, Luo Y, Sun B, Wang XR, Tang HR. Effect of red and blue light on anthocyanin accumulation and differential gene expression in strawberry (Fragaria × ananassa) Molecules. 2018;23(4):820. doi: 10.3390/molecules23040820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou et al. (2007).Zhou B, Li Y, Xu Z, Yan H, Homma S, Kawabata S. Ultraviolet a-specific induction of anthocyanin biosynthesis in the swollen hypocotyls of turnip (brassica rapa) Journal of Experimental Botany. 2007;58(7):1771–1781. doi: 10.1093/jxb/erm036. [DOI] [PubMed] [Google Scholar]
- Zhu et al. (2016).Zhu PF, Cheng MM, Feng X, Xiong Y, Liu C, Kang YH. Mapping of Pi, a gene conferring pink leaf in ornamental kale (Brassica oleracea L. var. acephala DC) Euphytica. 2016;207:377–385. doi: 10.1007/s10681-015-1555-4. [DOI] [Google Scholar]
- Zoratti et al. (2014).Zoratti L, Karppinen K, Luengo Escobar A, Häggman H, Jaakola L. Light-controlled flavonoid biosynthesis in fruits. Front Plant Science. 2014;5:534. doi: 10.3389/fpls.2014.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following information was supplied regarding data availability:
Data is available in Table S1 and at NCBI SRA: PRJNA560282.