Abstract
Inflammatory bowel disease (IBD) is a chronic and debilitating disorder that has few treatment options due to a lack of comprehensive understanding of its molecular pathogenesis. We used multiplexed mass spectrometry to collect high-content information on protein phosphorylation in two different mouse models of IBD. Because the biological function of the vast majority of phosphorylation sites remains unknown, we developed Substrate-based Kinase Activity Inference (SKAI), a methodology to infer kinase activity from phosphoproteomic data. This approach draws upon prior knowledge of kinase-substrate interactions to construct custom lists of kinases and their respective substrate sites, termed kinase-substrate sets that employ prior knowledge across organisms. This expansion as much as triples the amount of prior knowledge available. We then used these sets within the Gene Set Enrichment Analysis framework to infer kinase activity based on increased or decreased phosphorylation of its substrates in a dataset. When applied to the phosphoproteomic datasets from the two mouse models, SKAI predicted largely non-overlapping kinase activation profiles. These results suggest that chronic inflammation may arise through activation of largely divergent signaling networks. However, the one kinase inferred to be activated in both mouse models was mitogen-activated protein kinase-activated protein kinase 2 (MAPKAPK2 or MK2), a serine/threonine kinase that functions downstream of p38 stress-activated mitogen-activated protein kinase. Treatment of mice with active colitis with ATI450, an orally bioavailable small molecule inhibitor of the MK2 pathway, reduced inflammatory signaling in the colon and alleviated the clinical and histological features of inflammation. These studies establish MK2 as a therapeutic target in IBD and identify ATI450 as a potential therapy for the disease.
Keywords: mouse model, proteomics, kinases, gene set enrichment analysis, inflammation
INTRODUCTION
Inflammatory bowel disease (IBD) is characterized by perpetual and incurable inflammation of the gastrointestinal tract. IBDs, including Crohn’s disease (CD) and ulcerative colitis (UC), affect more than 5 million people worldwide, with a dramatic increase in the last 50 years [1]. IBD patients suffer from debilitating symptoms including abdominal pain, fever, rectal bleeding, severe diarrhea, and weight loss. Medical therapy with targeted inhibitors is effective for some patients, but most patients are not responsive and/or become resistant, making the search for new inhibitors of the utmost importance. Most available targeted therapies for IBD hit immunomodulatory molecules, like TNF-α, α4β7 integrin, and the cytokines IL-12 and IL-23 [2]. While less common in IBD, the use of small molecule kinase inhibitors is more established in oncology, and considerable effort has been devoted to developing drugs targeting these enzymes. Whether these drugs will be useful for IBD is unclear, given that little is known about the role that kinase signaling plays in the disease. Tofacitinib, which targets the JAK1/3 tyrosine kinases, was the first small molecule kinase inhibitor approved for treatment of IBD [3]. The inhibition of p38 mitogen-activated protein kinase (MAPK) seemed a promising treatment due to its increased activity in patients with IBD, however inhibitor studies were not effective [4].
Global proteomics and phosphoproteomics by mass spectrometry (MS) provide high-content, quantitative data to interrogate changes in protein expression and phosphorylation [5, 6]. Nevertheless, there is some uncertainty as to how to draw insights from large scale datasets where the biological functions and regulatory mechanisms of many individual analytes (i.e. phosphorylated proteins) are still unknown [7]. While phosphorylation site annotation with biological function is limited, prior knowledge of kinases and their respective substrate sites provides the opportunity to infer information regarding kinase activity through knowledge of substrate phosphorylation state [8, 9]. Indeed, multiple prior studies have used MS-based phosphoproteomics as a proxy for direct measurements of kinase activity. These techniques vary with respect to sources of prior knowledge of kinase-substrate interactions, data input, and the metrics used to infer activity [10, 11]. For example, Kinase Enrichment Analysis (KEA) integrates kinase-substrate interactions from a variety of online resources (from both computationally predicted interactions and the literature) and calculates the significance associated with each kinase based on the presence (but not quantity) of its substrates in the input list relative to the background kinase-substrate dataset [12]. Similarly, Kinase-Substrate Enrichment Analysis (KSEA) employs a variety of input data sources, but, in contrast to KEA, utilizes quantitative input data and defines enrichment through three different output metrics [13–15]. Drake et al. also synthesized kinase-substrate lists from a variety of sources,however chose an enrichment score metric analogous to Gene Set Enrichment Analysis (GSEA) [16]. Phosphosite-set Enrichment Analysis (PSEA), while enriching sets more general than individual kinases (e.g. pathways), similarly employed the GSEA algorithm with sets comprised of phosphorylation site specific prior knowledge [17]. PTM signature enrichment analysis (PTM-SEA) infers activity of phosphorylation signatures (which include kinases as well as pathways and perturbations, PTMsigDB) also using the GSEA algorithm [18]. Inference of kinase activities from phosphoproteomics (IKAP) employs a machine learning algorithm to calculate kinase activities, drawing on PhosphoSitePlus for prior knowledge [19]. Integrative Inferred Kinase Activity (INKA) analysis integrates both kinaseand substrate-centric information [20]. Alternatively, while not explicitly focused on inferring kinase activity, the motif-x algorithm operates on input data of phosphopeptide sequences (among others) and extracts statistically significant motifs, such as those belonging to a particular kinase [21, 22]. An extension of this motif-search approach, combined with prior knowledge of motifs associated with kinases, was used to study signaling downstream of the EGFRvIII mutation in glioblastoma [23].
While these techniques enable identification of kinase activities from phosphoproteomic data, many lack consideration of site-specific kinase-substrate interactions from studies across organisms. Moreover, and perhaps to a larger detriment, most approaches consider primarily human substrate sites and are not applicable to multiple organisms.
Targeted and global proteomic and phosphoproteomic data from a mouse model of colitis indicate that p21-activated kinase (PAK) and mechanistic target of rapamycin (mTOR) promote gastrointestinal inflammation [24, 25]. Here, we have further developed a user-friendly GSEA-based approach to infer changes in kinase activity from global phosphoproteomic data. We first drew upon and expanded kinase-substrate interactions available from PhosphoSitePlus by integrating information across organisms [8]. We then uniquely matched organism- and isoform-specificity of MS data output creating custom substrate sets for a large panel of kinases. Lastly, we outlined the use of GSEA as an algorithm to calculate enrichment. We applied this approach—called Substrate-based Kinase Activity Inference (SKAI)—to phosphoproteomic data from two mechanistically distinct mouse models of IBD, providing new hypotheses for kinases that may contribute to chronic inflammation. Using SKAI predictions as a foundation, we then validated the activation of MAPKAPK2 (MK2), a serine/threonine kinase in the p38 MAPK signaling pathways, during inflammation [26]. In patients with IBD, p38 activity is increased; this has been found to have a variety of effects including a recruitment and activation of immune cells such as lymphocytes and neutrophils [4]. However, inhibition of p38 MAPK has not been successful clinically. This highlights MK2 as particularly interesting in the context of not only our results, but also in the established biological understanding of IBD. Lastly, we performed a preclinical trial in mice to demonstrate that MK2 activation plays a central role in IBD and serves as a relevant target for future preclinical and clinical studies.
METHODS
Animal models
T cell transfer (TCT) was performed according to established methods [27]. Splenocytes were extracted from wildtype (WT) C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) and subsequently processed to isolate naïve T cells and regulatory T cells (Treg). These were injected into Rag1 null animals at 400 000 and 200 000 cells/animal, respectively. Animals were then monitored for colitis with direct screening by rigid colonoscopy. Optical measurements were performed using the Image 1 camera system (Karl-Storz, Tuttlingen, Germany) and UR-4MD HD video recorder (TEAC, Montebello, CA). Animals injected with naïve T cells become inflamed; Treg injected animals remain uninflamed.
TNFΔARE animals were maintained by backcrossing to C57BL/6 animals from the Jackson Laboratory. TNFΔARE mice used were as described in Kontoyiannis et al. [28].
All animal work was approved by the Institutional Care and Use Committee of Beth Israel Deaconess Medical Center under protocol number 078–2014, and 080–2017. Approved protocols conformed to the USDA Animal Welfare Act, PHS Policy on Humane Care and Use of Laboratory Animals, and the “Institute for Laboratory Animal Research (ILAR) Guide for the Care and Use of Laboratory Animals.”
Collection of MS datasets
Proteomic and phosphoproteomic data from a TCT mouse model were described and generated previously (MassIVE, accession no. MSV000081198) [25].
TNFΔARE tissue samples were collected and processed as in Lyons et al. [25], however from the ileum rather than the colon. Sample processing and MS measurements were performed as described in previous work [25]. These datasets were collected for this manuscript and have been made available in the Mass Spectrometry Interactive Virtual Environment (MassIVE) database, accession no. MSV000082217.
Phosphoproteomic scaled to total proteomic data (phosphor-/total) were calculated as a composite ratio of a phosphopeptide scaled by its corresponding (based on UniProt ID) total protein abundance where possible.
To ensure consistent nomenclature with the sets, we created an identifier for each phosphorylated peptide that concatenates its respective UniProt ID and phosphorylated site (UniProtID_SiteNumber). We utilize UniProt IDs as they provide a unique name for each analyte. Gene or protein names can be used as well, however this may require careful consideration if more than one analyte has the same gene name and site (depending on the post processing of MS results this may occur). Additionally, we note that for SKAI analyses, we excluded multiply phosphorylated peptides due to lack of clarity in interpretation.
Unsupervised clustering
Unsupervised hierarchical clustering of total proteomic, phosphoproteomic, and phosphoproteomic scaled to total proteomic data was conducted on data transformed as the log2 fold change (FC) relative to the median control value. Clustering was performed using MATLAB (MathWorks, Natick, MA, r2015b) with the Euclidean distance metric, average linkage method, and optimal leaf ordering.
Differential expression analysis
Differential expression of MS data between inflamed and control samples was conducted for total proteomic, phosphoproteomic, and phosphoproteomic scaled to total proteomic data using a Wilcoxon Rank Sum test. Within each dataset, multiple hypothesis correction was employed using the Benjamini Hochberg procedure. Analyses were conducted in MATLAB.
Principal component analysis
Principal component analyses (PCAs) of total proteomic, phosphoproteomic, and phosphoproteomic scaled to total proteomic data were carried out on data transformed as the log2 FC relative to the median control value. Analyses were conducted in MATLAB. Given use of the log2 FC transform, data were not centered in order to retain the control as the point of reference for the data.
Substrate-based kinase activity inference (SKAI)
SKAI employs both the kinase substrate and phosphorylation site datasets available from PhosphoSitePlus. Updated monthly, this study used the 2017–10-02 version. Each entry in the kinase substrate dataset provides an organism-specific phosphorylation site paired with a kinase capable of phosphorylating it and associated meta information. Identifiers from three naming systems are provided for both kinase and substrate: gene name, protein name, and UniProt ID. Additional metadata includes site-specific information such as the amino acid sequence surrounding the phosphorylation site and its corresponding site group ID. The latter allows for expansion of the kinase substrate dataset beyond the specific organisms and isoforms presented, detailed in this manuscript, as the site group ID uniquely identifies each site with homology across protein isoforms and organisms.
The phosphorylation site dataset is a collection of known sites of phosphorylation. Entries are organism specific. Similar to the kinase substrate dataset, metadata includes identifiers from three naming systems (protein, gene, and UniProt ID) as well as the site group ID. This dataset includes phosphorylation sites for which a kinase has not yet been identified, or perhaps not yet identified in a particular organism, and as a result contains a superset of the sites described in the aforementioned kinase substrate dataset.
We expanded the kinase-substrate pairings in the kinase substrate dataset to encompass all entries in the phosphorylation site dataset that have the same site group IDs. While the kinase and site group ID associations remain the same, this broadens the organisms and isoforms considered. For example, the literature the kinase-substrate set is derived from may have focused on, for example, a specific organism (e.g. human) or isoform. We assume that the kinase-substrate interaction is applicable to all substrate isoforms and organisms with a given site group ID (the validity of this assumption is supported by the conservation of relationships between organisms) [29]. Note, this process will likely result in the creation of duplicate pairings (i.e. pairings of the same site group ID, isoform, and organism); these are consolidated into one entry after the expansion.
The organism-specific kinase-substrate sets are assembled to satisfy the GSEA gene set format [30, 31]. They consist of a first row of kinases. We used protein name identifiers as they are more general than the UniProt IDs (e.g. they do not depend on an entry being reviewed), however other identifiers are available. A second row contains an optional description; we left this blank. Subsequent rows in each column contain the substrate sites corresponding to the cognate kinase. Kinase-substrate sets are organism-specific, such that the site numbering corresponds to the organism-specific substrate site. This is accomplished by selecting the subset of the expanded pairings that pertain to the organism of interest. We chose to use the organism-specific UniProt ID as these identifiers uniquely identified analytes in our phosphoproteomic datasets (UniProtID_SiteNumber). The last step is to remove any duplicate entries within a column that may have arisen as a result of the expansion or assembly of kinase-substrate sets. Sets for mouse, human, and rat are available formatted for immediate use with GSEA (data file S1).
We performed enrichment analysis via the GSEA algorithm and employed the GSEA software available at http://www.broad.mit.edu/gsea/ [30, 31]. To infer kinase activity, we used phosphoproteomic data or phosphoproteomic scaled to total proteomic data and replaced ‘gene sets’ with our expanded, organism-specific kinase-substrate sets.
As outlined in Subramanian et al., there exist a variety of parameter values within the GSEA algorithm [30]. We gave careful consideration to the ranking metric used. Given the small sample sizes often available from phosphoproteomic studies (e.g. n = 3), we used a nonparametric ranking metric. Specifically, we ran the GSEA algorithm using a pre-ranked list of the phosphorylated peptides in the dataset of interest (multiple polyphosphorylated peptides were excluded from analysis due to lack of clarity in interpretation). Specifically, we chose a nonparametric signal-to-noise value,
where m is the median and MAD is the median absolute deviation when comparing groups A versus B (e.g. inflamed to control). MAD has a minimum value of 0.2∗|m|(as recommended in the GSEA documentation). Additional parameters used in the analysis can be found in data file S9. Significantly enriched sets were defined as those having a false discovery rate (FDR) Q-value <0.25 and P-value <0.1.
ATI450 dosing studies
Dosing studies were carried out to ensure animals were dosed sufficiently with ATI450 (Aclaris Therapeutics, Wayne, PA) to induce MK2 pathway inhibition. Standard rodent chow was formulated with ATI450 at 172, 516, and 1458 ppm. This was fed to adult female BALB/c mice for 3 days ad libitum. Mice were subsequently challenged with a 50 μg intraperitoneal injection of Salmonella typhosa-derived LPS for 1 hour. Serum was then collected and TNFα concentration, a metric of MK2 pathway activity, quantified by a single-plex assay (Meso Scale Diagnostics, Rockville, MD). We chose an ATI450 concentration of 1000 ppm for our preclinical mouse studies. This is lower than the maximum dose employed while still within the range of maximum MK2 pathway inhibition; furthermore, this concentration has been effective in previous publications [32, 33].
Tissue processing, histology, and immunohistochemistry
To further interrogate the TCT model, colons were dissected and flushed with phosphate-buffered saline (PBS) and measured lengthwise. They were then opened longitudinally and one lengthwise section was divided into proximal and distal colon and flash frozen in liquid nitrogen. A matched lengthwise section was taken and placed in PBS for flow cytometry. The remaining piece was rolled and fixed overnight in 10% formalin for histology. Fixed tissue was processed and embedded into paraffin blocks and then cut into 5 μm sections using a Leica RM2235 microtome (Leica, Wetzlar, Germany). Hematoxylin and eosin (H&E) staining was preformed according to standard protocols. H&E stained sections were imaged using a BX61VS slide scanner (Olympus, Waltham, MA). To quantify crypt height, a line, approximated visually by histology, was drawn from top of the muscle layer to the lumen every five crypts across the length of the colon. For scatter plots, all measurements were evenly spaced across the distal and proximal regions separately using polyline measurements of the lengths of those sections. Averages were taken of all measurements in the distal and proximal regions separately.
For immunohistochemistry using anti-Ki-67 (D2H10) from cell signaling (Danvers, MA), tissue sections were deparaffinized and rehydrated and subjected to antigen retrieval by pressure cooker in Target Retrieval Solution, pH 6 (DAKO, Santa Clara, CA). Tissue sections were blocked with DAKO peroxidase block for 15 minutes at room temperature and DAKO serum-free protein block for 30 minutes at room temperature and then exposed to primary antibodies overnight at 4°C. Tissue sections were incubated with DAKO HRP rabbit secondary for 30 minutes at room temperature, detected with 3,3′-diaminobenzidine (DAB), and then counterstained with hematoxylin. To determine the number of positive Ki-67 cells, the number of positive stain cells was quantified per half crypt along the length of the whole distal colon.
Flow cytometry
Once all colons were removed and placed in PBS, tissues were homogenized in Dulbecco’s Modified Eagle Medium (DMEM) with 2 mg/ml collagenase type I and incubated for 1 hour at 37°C on a shaker. After incubation, samples were pushed through a 45 μm filter and centrifuged for 5 minutes at 500 g. Cells were stained with BioLegend (San Diego, CA) antibodies for 10 minutes: APC/Cy7 anti-mouse CD45 and BV-605 anti-mouse CD4. Cells were analysed on a 5-laser BD Aria sorter.
Signaling analysis
Frozen colonic tissue samples were thawed and lysed in 250 μl Bio-Plex lysis buffer (Bio-Rad, Hercules, CA) supplemented with Factor I, Factor II, 2 mM PMSF, and cOmplete protease inhibitor cocktail (Roche Diagnostics Deutschland GmbH, Mannheim, Germany). Protein lysates were quantified using a bicinchoninic acid assay (Thermo, Waltham, MA) and samples were equally loaded and run on 10% SDS-PAGE gels. Following transfer onto polyvinylidene fluoride membranes, membranes were blocked with Odyssey Blocking Buffer for 1 hour at room temperature and probed with primary antibodies from Cell Signaling Technology: MK2 (#3042), p-MK2 (Thr334, #3041), HSP27 (#95357), p-HSP27 (Ser82, #9709), and GAPDH (#5174). β-Tubulin antibody was from Sigma-Aldrich (St. Louis, MO). Following incubation, membranes were washed with 1X Tris-buffered saline and Tween-20 (TBST) and then incubated in secondary antibody. Membranes were washed in 1X TBST and analysed on an Odyssey CLx Infrared Imaging System (LI-COR, Lincoln, NE).
Luminex analysis was also performed on colonic tissue lysates. Measurements were carried on a Millipore MAGPIX® according to the manufacturer’s instructions, as described [24], using the following kits: MILLIPLEX MAP Mouse Cytokine/Chemokine Magnetic Bead Panel—Immunology Multiplex Assay (MCYTOMAG-70 K); Bio-Plex Pro Cell Signaling Akt panel, 8-plex (lq00006jk0k0rr); and Bio-Plex Pro Cell Signaling MAPK Panel, 9-plex (lq00000s6kl81s). Analytes whose signal was at or below background were excluded from analysis.
Statistical analysis of MK2 pathway inhibition and signaling studies
Statistical analysis of dosing studies, quantitative western blots, histology and immunohistochemistry quantification, flow cytometry, and Luminex analysis data was conducted in Prism 7 (GraphPad, San Diego, CA). Comparisons are made using a Wilcoxon Rank Sum test.
RESULTS
Expanded kinase-substrate sets as much as triple prior knowledge
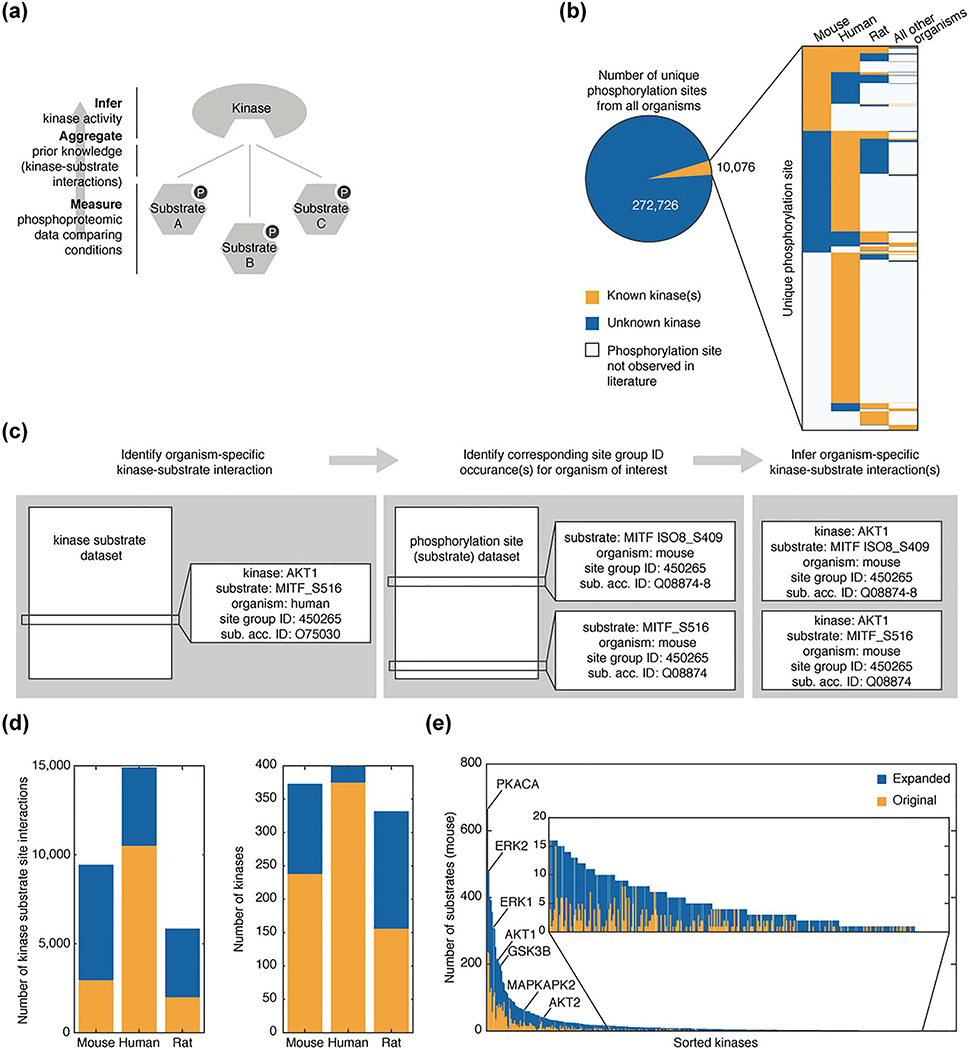
To utilize kinase-substrate relationships as a bottom-up approach to kinase activity inference from phosphoproteomic data (Fig. 1a), we first compiled a list of known kinase-substrate associations. There exist multiple sources of prior knowledge—determined both experimentally and in silico—of kinase-substrate relationships [10]. We chose PhosphoSitePlus because it is based on experimental data, is updated monthly, and identifies each phosphorylation site with a ‘site group ID’—a classifier that identifies homologous phosphorylation sites across isoforms and organisms [8].
Figure 1.
Expansion of organism-specific kinase substrate datasets. (a) Workflow diagram illustrating bottom-up approach to infer kinase activity from prior knowledge and phosphoproteomic data. (b) Analysis of the number of unique phosphorylation sites with (orange) and without (blue) at least one known associated kinase from the kinase substrate dataset and phosphorylation site dataset available from PhosphoSitePlus as of 2017–10-02. Inset presents the organism breakdown of substrate sites within the set of kinase associated sites. Substrate sites for which organism-specific phosphorylation is not observed in the literature are shown in white. (c) Example illustrating process to expand kinase substrate dataset for mice. (d) Comparison of organism-specific kinase-substrate sets for mouse, rat, and human within the original dataset (orange) and after the expansion (blue). (e) The number of mouse substrates associated with each kinase in the original dataset (orange) and after expansion (blue) are ordered based on substrate set size after expansion.
PhosphoSitePlus provides two sets of phosphorylation data: the full set of observed phosphorylation sites (phosphorylation site dataset) and the subset of phosphorylation sites with an identified kinase (kinase substrate dataset). Both these datasets are organism- and isoform-specific based on the literature result(s) from which they were identified. When considering the total number of unique phosphorylation sites in PhosphoSitePlus—identified using the site group ID—<4% were found to have at least one known kinase associated with them and many of the kinase-substrate relationships were known only for a single organism (Fig. 1b). Overcoming this organism-specific, prior knowledge is key to conducting robust kinase activity inference analysis as both identifiers used in global phosphoproteomics (UniProt IDs) and phosphorylation site positions are organismand isoform-specific. Nevertheless, many of the phosphorylation sites for a specific organism that were not yet associated with a kinase were associated with a kinase in another organism, allowing us to infer a kinase-substrate relationship for that site in the original organism. This was true for 17% of human sites and 59% of mouse sites (Fig. 1b).
To create the most inclusive prior knowledge kinase-substrate set, we expanded the original kinase substrate dataset utilizing organism-specific information available in the phosphorylation site dataset. Based on the conservation of kinase-substrate relationships between organisms [29], we assumed that a kinase-substrate relationship is generalizable to all organisms and substrate protein isoforms in which the substrate site is seen to have been phosphorylated in prior studies. One example of this expansion is the interaction of the RAC-alpha serine/threonine-protein kinase (AKT1) with its substrate microphthalmia-associated transcription factor, MITF. AKT1 was experimentally shown to phosphorylate MITF on residue Ser516 in humans [34]. Using the site group ID for this site (450265), we identified corresponding sites in mice within the phosphorylation site dataset, MITF isoform 8 (ISO8) site Ser409 and MITF site Ser516. The resultant mouse-specific expanded kinase-substrate set contains two additional interactions that are inferred by homology from this original human-specific entry within the kinase substrate dataset (Fig. 1c). We carried out this expansion for mouse, human, and rat interactions (Fig. 1d).
Following this expansion, kinase-substrate sets were formed for each kinase by collecting the substrate sites associated with the kinase (data file S1). Substrates were identified using UniProt IDs and phosphorylation site to ensure consistent nomenclature with output from global MS studies. For mouse kinase-substrate relationships, this expansion, on average, approximately tripled the number of substrates for each kinase, and did so fairly uniformly. It added new substrate annotations for kinases already highly annotated such as cAMP-dependent protein kinase catalytic subunit alpha (PKACA) and extracellular signal-regulated kinase 2 (ERK2) and for kinases with fewer known substrates (Fig. 1e). We also calculated the percentage overlap between kinase-substrate sets and found this to be minimal (Fig. S1 and Table S1.
There exist a breadth of computational approaches and metrics used to infer kinase activity from phosphoproteomic data [10]. We chose to use the GSEA algorithm as it is frequently used by both bench and computational scientists. We term our approach, implementing the expanded kinase-substrate sets within the GSEA algorithm to infer kinase activity from phosphoproteomic data, Substrate-based Kinase Activity Inference, SKAI. For a full description of kinase-substrate set generation and use of GSEA, see Methods.
Univariate and multivariate analysis of global data provides limited insights
Our overarching goal is to identify signal transduction pathways that are activating during chronic gastrointestinal inflammation. To this end, we analysed global total proteomic and phosphoproteomic data from the TCT model of colitis [27] and from the TNFΔARE model of ileitis (Crohn’s-like IBD) [28] (Fig. 2a). To account for changes in phosphorylation that result from changes in total protein abundance, we calculated a dataset of phosphoproteomic scaled to total proteomic data, which represents phosphoproteomic measurements that are normalized to their respective total proteomic measurements (Tables S2–S7). Importantly, while the TCT and TNFΔARE models both develop chronic inflammation of the intestinal tract, they develop inflammation in different regions (colon and ileum, respectively), which raises the interesting question of whether the pathways that are dysregulated are similar or different.
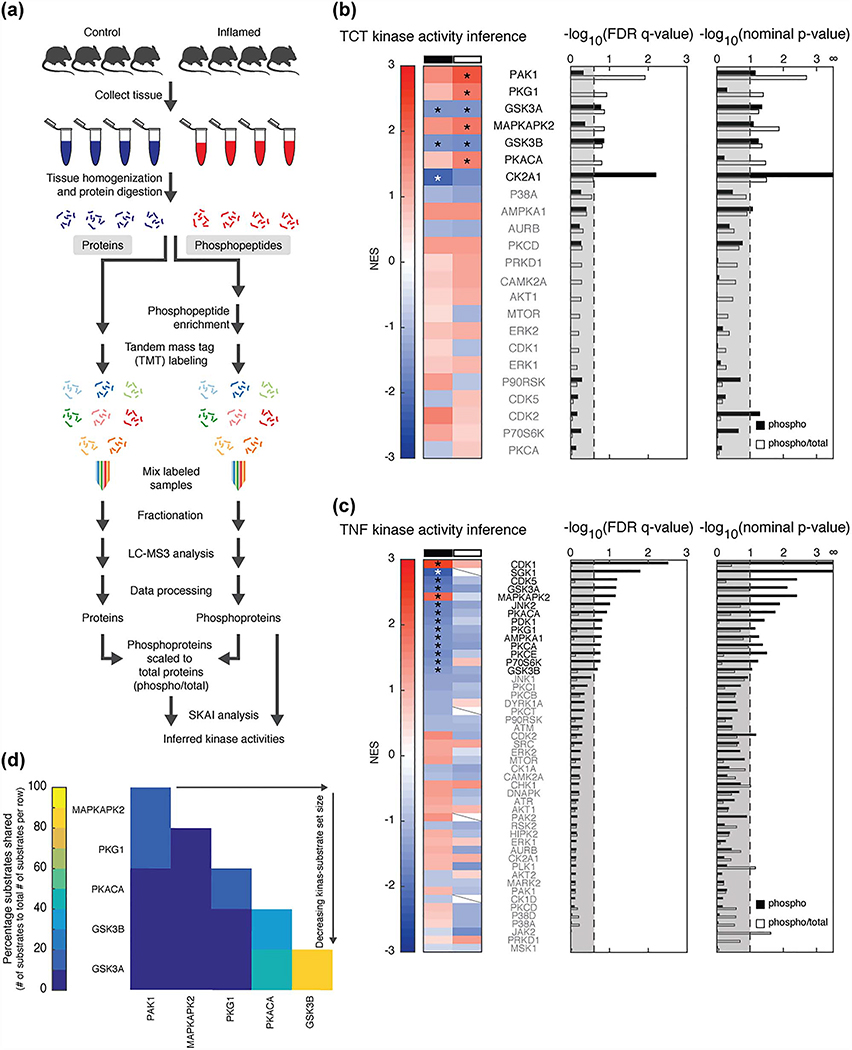
Figure 2.
SKAI reveals activated kinases. (a) MS workflow. (b and c) SKAI conducted on phosphoproteomic data (phospho, black) and phosphoproteomic scaled to total proteomic data (phospho/total, white) from (b) TCT and (c) TNFΔARE models. Enrichment compares the inflamed to control samples. Each line in the heat map represents the NES associated with a kinase (set size ≥5). Slashes indicate the kinase’s respective substrate set was not detected in the analysis due to insufficient (<5) substrates in the respective dataset. Bar graphs present FDR q-values and nominal p-values for each kinase; thresholds are at an FDR q-value <0.25 and nominal p-value <0.1. Kinases shown in bold and marked with an asterisk have results that meet these thresholds. (d) Substrate overlap in TCT SKAI results from analysis of phosphoproteomic scaled to total proteomic data.
We visualized each dataset (via unsupervised hierarchical clustering and PCA) to interrogate measurement repeatability and to investigate changes in global phosphopeptide signalling associated with the inflamed state. Biological replicates clustered together, while inflamed and control samples were well separated in principal component space (Figs.S2 and S3). We also carried out univariate differential expression analyses, however, due to limited annotation of phosphorylation signals, they did not lend themselves to actionable hypotheses regarding their effect (Tables S2–S7).
SKAI identifies kinases dysregulated during inflammation
To move beyond univariate analysis, we applied SKAI to the TCT and TNFΔARE phosphoproteomic datasets to demonstrate the technique’s ability to generate hypotheses in two established models of IBD. For each dataset, we calculated the kinase-substrate set enrichment score (normalized enrichment score, NES) for the relative quantification of phosphosites in the inflamed versus control tissues, using both phosphoproteomic and phosphoproteomic scaled to total proteomic data. We inferred differential activation of kinases in the two models based on the NES. In the TCT model, comparison of phosphopeptides in inflamed versus control data identified seven unique kinases with significant substrate set enrichment or de-enrichment (Fig. 2b, Table S8). We noted that PAK1 had a significant positive substrate set enrichment, consistent with our earlier work [25]. SKAI analysis of the TNFΔARE model resulted in more kinases identified overall than the TCT model, likely due to the larger size of the TNFΔARE dataset (Fig. 2c, Table S8). However, in this model only the phosphoproteomic data identified significantly enriched and de-enriched kinase-substrate sets; none were significant in the phosphoproteomic scaled to total proteomic data analysis. For comparison, analyses employing the unexpanded organism-specific kinase-substrate lists were also conducted, which resulted in fewer detected and significantly enriched kinases (Fig. S4).
In comparing the TCT and TNFΔARE analyses, we found minimal overlap. The activities of two kinases (GSK3A and GSK3B) were predicted to be decreased in both models, although these kinases share many substrates and therefore cannot be easily distinguished by SKAI (Fig. 2d). The only kinase predicted to have increased activity in both models was MAPKAPK2 (MK2), a serine/threonine kinase in the p38 stress-induced MAPK pathway (Table S9) [26]. In contrast to what we found with GSK3A and GSK3B, it is unlikely that the MK2 inference is due to the activity of another kinase. In phosphoproteomic scaled to total proteomic data from the TCT model, the MK2 substrate list had the most overlap with that of PAK1, but only 17% of MK2 substrates were shared by PAK1 (Fig. 2d).
MK2 pathway inhibition resolves inflammation
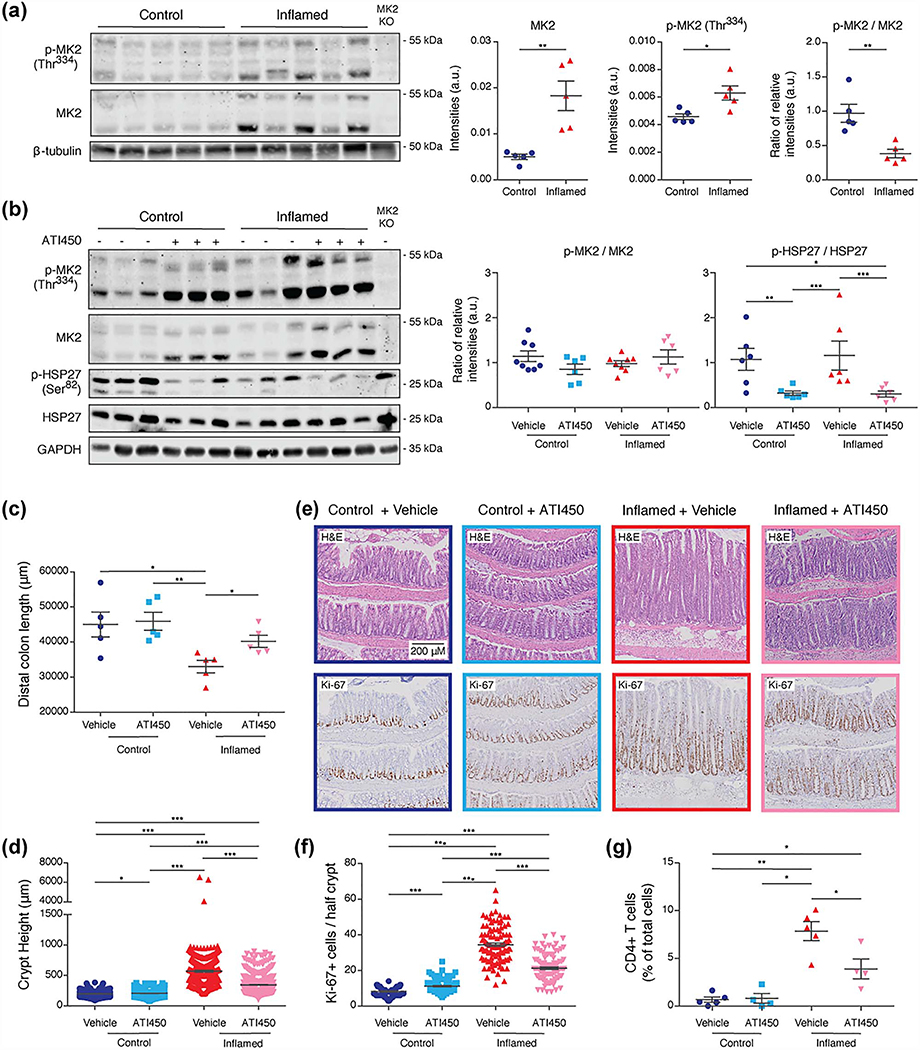
Due to the biological significance of this pathway in IBD [4] and the potential of MK2 as a therapeutic target, we sought to further investigate its role in colitis. We utilized the TCT model of colitis [27]. After adoptive transfer, we used colonoscopy to monitor animals for clinical signs of colitis, such as thickening of the mucosa and bleeding ulcers, which typically arose in 8–16 weeks (Fig. S5a). We found an increase in MK2 expression and phosphorylation on Thr334 (a sign of activation) relative to animals without inflammation (Fig. 3a). We also found increased MK2 phosphorylation in TNFARE animals with inflammation (Fig. S5b). These observations validate the prediction of MK2 activation made by SKAI.
Figure 3.
ATI450 treatment reduces inflammatory burden in animals with colitis. (a) Immunoblot analysis of MK2 and p-MK2 (Thr334) in lysates from colon tissue of animals with and without colitis. Graphs represent quantification of western blots, 5 mice/group. (b) Immunoblot analysis of MK2, p-MK2 (Thr334), HSP27, and p-HSP27 (Ser82) in lysates from colon tissue of ATI450-treated animals. Graphs represent quantification of western blots from 6 to 8 mice/group (representative blot shown). (c and d) Quantification of distal colon length (c) and crypt height (d) in animals treated with ATI450, 7 mice/group. (e and f) H&E staining and immunohistochemistry for Ki-67 in colon tissue from mice treated with ATI450. Images (e) are representative of 7 mice/group, each analysed separately. The number of Ki-67+ cells (f) from analysis of 100 crypts/mouse and 7 mice/group. (g) Flow cytometry analysis of CD4+ T cells in colon tissue from mice treated with ATI450, 4–5 mice/group. Error bars shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005 by Wilcoxon Rank Sum test.
To determine whether inhibition of MK2 could alleviate colitis in the TCT model, we treated inflamed (those with established colitis) and control animals with ATI450, an orally available small molecule inhibitor, and monitored the progression of inflammation via colonoscopy. Dosing was determined based on prior studies as well as a dosing study (Fig. S5c) [32, 33]. Following acute ATI450 treatment, we did not observe a change in the ratio of phosphorylated MK2 to total MK2, but we did observe a significant decrease in the phospho/total ratio of HSP27 (Ser82 in human and Ser86 in mouse), an MK2 substrate (Figs. 3b and S5d). These observations indicate that MK2 is activated during colitis and that treatment with ATI450 effectively blocks MK2 pathway activity. This is consistent with SKAI results; HSP27 (Ser86) was one of the substrates that contributed to the positive enrichment of MK2 (Table S9). Inflammation became more severe in animals fed standard mouse chow, while colitis—as assessed by colonoscopy—was reduced over the 2-week treatment in animals that received ATI450 (Fig. S5a). Following the endoscopy scoring system develop by Kodani et al. [35], all control animals presented with an endoscopy score of 0. Inflamed animals scored between 8 and 10, except when measured post-ATI450 treatment where they were found to have a score between 0 and 4 (Fig. S5e). Moreover, upon sacrifice at the end of the treatment period, we noted that ATI450 treatment reversed the colonic shortening phenotype that is common in mouse models of colitis (Fig. 3c).
One of the primary phenotypes associated with TCT-induced colitis is hyperplasia of the colonic crypts [36]. Treatment with ATI450 significantly reduced crypt height in animals with inflammation, while not affecting animals without inflammation (Fig. 3d and e). Consistent with the reduction in crypt height, we noted a significant reduction in the number of Ki-67-positive cells when animals with colitis were treated with ATI450 (Fig. 3f). In mice without colitis, ATI450 had a small, but significant, effect on the number of Ki-67-positive cells, albeit in the opposite direction (Fig. 3f). Consistent with a resolution of inflammation, we found that ATI450 treatment led to a significant decrease in the number of colonic T cells in animals that had inflammation (Fig. 3g). Altogether, these results demonstrate that inhibition of the MK2 pathway with ATI450 suppresses active colitis in the TCT model.
Pro-inflammatory signaling is reduced by ATI450
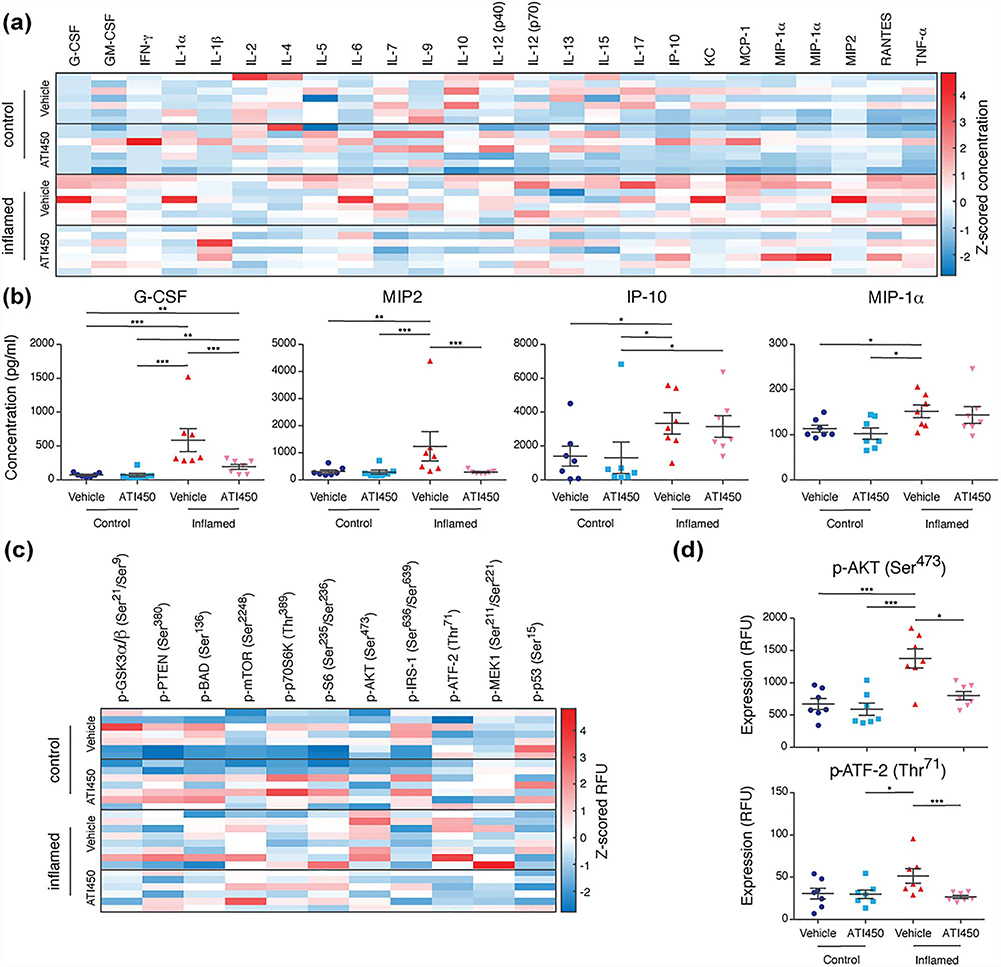
MK2 signals through downstream proteins regulate the expression of pro-inflammatory cytokines and chemokines [37]. To determine the effect of ATI450 on the expression of inflammatory signals, we used Luminex’s xMAP technology to measure the expression of 25 cytokines and chemokines in the colons of animals from our preclinical trial, many of which were significantly up-regulated during colitis (Figs. 4a and S6, Table S10). Consistent with prior studies linking MK2 to cytokine production, some of these, for example, G-CSF and MIP2, were significantly reduced after ATI450 treatment (Fig. 4b). There were some cytokines, such as IP-10 and MIP-1α, that were significantly up-regulated during inflammation, but were not affected by MK2 pathway inhibition, even though the colitis phenotype was obviously reduced (Fig. 4b).
Figure 4.
ATI450 treatment inhibits pro-inflammatory signaling.(a and b) Luminex analysis of cytokine abundance in distal colon tissue from control and inflamed mice treated with or without ATI450 for 2 weeks. Data are concentration Z-scores of all analytes (a) or concentration of G-CSF, MIP2, IP-10, or MIP-1α (b) with 7 mice/group (three technical replicates per mouse, averaged). (c and d) Luminex analysis of phosphoproteins in lysates of distal colon tissue from control and inflamed mice treated with or without ATI450 for 2 weeks.Data are raw fluorescence (RFU) Z-scores of all analytes (c) or relative amount of AKT (Ser473) and ATF-2 (Thr71) (d) with 7 mice/group (three technical replicates per mouse, averaged). Error bars shown as mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.005 by Wilcoxon Rank Sum test.
Although there are many known MK2 substrates, there is little known about the specific MK2 signaling cascade in colitis. To explore MK2 signaling during colitis, we again used Luminex’s xMAP technology, this time to measure phosphorylation of a panel of 11 cell signaling molecules (Figs. 4c and S7, Table S10). Consistent with our prior work on the TCT model, we found that mTOR phosphorylation on Ser2248 was up-regulated during colitis [24], however this was not significantly affected by MK2 pathway inhibition. Previously published work has shown that MK2 phosphorylates AKT on Ser473 [38]. Our analysis revealed a significant decrease in AKT (Ser473) phosphorylation after ATI450 treatment in animals with colitis, supporting the notion that AKT is a substrate for MK2 in this system (Fig. 4d). We also found that ATI450 treatment led to a decrease in phosphorylation of ATF-2 (Thr71), a transcription factor that is known to be phosphorylated directly by members of the stress-induced MAPK pathway (Fig. 4d) [39].
DISCUSSION
MS-based phosphoproteomics analysis provides a wealth of high-content information relating to protein regulation. Nevertheless, it can be difficult to determine the activation states of pathways and individual proteins given the large number of unannotated phosphorylation sites. Moreover, the activation state of specific kinases is difficult to quantify if they (and their regulatory phosphorylation sites) are not present in a given dataset. As a result, the collection of global phosphorylation data can fall short of its promise to identify dysregulated signaling in pathogenic states. Here, we present an approach for kinase activity inference, SKAI, that provides a refined methodology for the generation of experimentally testable hypotheses based on global phosphoproteomics. Specifically, we integrate information across organisms and provide expanded organism-specific kinase-substrate sets. By applying SKAI to global phosphoproteomic and phosphoproteomic scaled to total proteomic data from the TCT and TNFΔARE mouse models, we were able to acquire general insights into the computational approach as well as specific insights into the mouse models and new therapeutic opportunities for IBD.
SKAI employs the GSEA algorithm to infer kinase activity by using gene sets composed of bona fide phosphorylation substrates for protein kinases. We maximized these substrate lists by combining known kinase-substrate relationships across organisms (Fig. 1). McDonald et al. demonstrated that more that 80% of human kinase-substrate associations are conserved in mouse, where the remaining do not occur due to lack of conservation of the substrate [29]. Our expansion excludes organism-specific substrates not observed in studies of each respective organism. We therefore expect even high than 80% conservation of kinase-substrate associations.To provide further evidence for the validity of the cross-organism expansion, we turn to the kinase-substrate dataset from PhosphoSitePlus. Specifically, we considered the ground truth mouse kinase-substrate relationships identified in PhosphoSitePlus and those of other organisms. Of the mouse substrates paired with a kinase, we then looked to those which also have a kinase association with another organism. The percent conserved is calculated as the number of relationships that are the same between mouse and another organism relative to the number of substrates that have a kinase (the same or not) identified in mouse and another organism. This is determined to be 69%. We note that of the ground truth mouse kinase-substrate interactions, 56% do not have a corresponding relationship in another organism. This is anticipated to be due to experimental bias (e.g. wealth of human cell line data overwhelming that of mouse, which kinases were studied in prior work) rather than a biological implication. We hypothesize this is a likely cause for a lower percentage than the percent conserved reported in McDonald et al. While earlier approaches have noted the importance of having organism-specific kinase-substrate sets [18], our organism expansion that integrates information across studies is the first of its kind.
Analysis of phosphoproteomic data from animal studies receive the most significant benefit from this expansion—as much as three times the available prior knowledge—as they gain from knowledge of kinase interactions observed in a wealth of studies performed in human cell lines in vitro (Fig. 1). We further observed this benefit through comparison of kinase activity inference carried out using the original mouse kinase-substrate set of interactions and the results using the expanded set (Figs. 2 and S4). Not only did the expanded set facilitate detection of additional kinases, it also detected MK2, which we have successfully validated as a potential therapeutic target. Without the use of the expanded sets, MK2 would not have been detected or significant in both models (Figs. 2 and S4).
A key observation from our study is the potential for insights provided by SKAI analysis on both phosphoproteomic data and phosphoproteomic scaled to total proteomic data (Fig. 2). Each data type likely has its own merit and interrogates a different question based on the respective data assumptions. We hypothesize that results derived from phosphor-proteomic data are more tightly linked to kinase activity when the kinase is limiting, and that phosphoproteomic scaled to total proteomic data is more relevant in the case of a limiting substrate. This is just a model, however. While relevant biochemically, its accuracy within the context of tissue is unknown. Our data derive from MS analysis of bulk tissue, so it is plausible that when considering phosphoproteomic scaled to total proteomic data, the phosphorylated peptides originate from a tissue region that is distinct from the majority of the total protein. With this uncertainty, our approach has been to use both data types for generating hypotheses and then to follow-up experimentally (as presented here in the case of MK2). Overall, it is evident that both quantities can be biologically relevant—neither one is exclusively influential; this is further highlighted by the variety of use in the literature [15, 40].
One goal in creating SKAI was to provide a user-friendly approach for the broader community of signaling biology researchers studying a variety of model systems. Organism-specific, expanded kinase-substrate sets can be downloaded (data file S1) and directly implemented with existing GSEA software and minimal processing of MS data output is necessary to format substrate site identifiers. Investigators with no prior experience with programming have immediate access to this approach and can implement it in their existing workflow, which may already include pathway gene set enrichment through GSEA. Across biological systems, SKAI can provide an opportunity to delve into the wealth of information available via phosphoproteomic data, teasing out new mechanisms and consequences of phosphorylation state regulation.
One consideration when interpreting the results of SKAI is the potential overlap between kinase-substrate sets. We observed minimal overlap when considering the fraction of substrates shared between sets overall, but there was considerable overlap between some sets (Fig. 2d and Table S1). This overlap influenced the results of our analysis, as we identified negative enrichment of substrates for GSK3A and GSK3B, which share >80% of their substrates (Fig. 2). As such, we do not know whether either or both kinases contribute to the increased substrate phosphorylation observed in our datasets. However, MK2 was observed to only share a small fraction of substrates with other kinases, providing confidence in this result.
We chose to apply SKAI to datasets from two different mouse models of IBD in order to determine how different the global signaling network is in the two models and identify potential therapeutically tractable kinases. Initially, we found that the enrichment results for the two models were largely distinct (Fig. 2). Since our ultimate goal was to identify therapeutic targets, we were most interested in the kinases that were predicted by SKAI to be activated during colitis. In the TCT model, four kinases (PAK1, PKG1, MK2, and PKACA) were predicted to be activated in the phosphoproteomic scaled to total proteomic dataset (Fig. 2b). The identification of PAK1 is consistent with our prior work showing that this kinase plays an important role in maintaining colitis in the TCT model [25]. Collectively, PAK1, MK2, and PKACA are members of the MAPK pathway. While the individual kinases have not directly been associated with IBD outside of work within our lab [25], the relevance of the MAPK to IBD has been identified in prior work [41]. In the TNFΔARE model, only two kinases (CDK1 and MK2) were predicted to be activated, all in the phosphoproteomic dataset (Fig. 2c). Common between these two pathways is the cellular senescence pathway, indicating that perhaps there is a feedback mechanism at work to compensate for the inflammatory response seen in IBD. The majority of kinase activities in the TNFΔARE model were predicted to have a decrease in activity. While therefore of less therapeutic interest, we note that biologically, they are found across a range of pathways, including PI3K-AKT, FoxO, and cGMP-PKG. These at least offer additional starting points for future work that broadens further explorations of IBD. Whether the differences in activated kinases observed between the TNFΔARE and TCT models reflect the distinct location of the inflammation (small intestine versus colon) or the distinct mechanism of initiation (TNF-α over-expression versus T cell activation) is unclear. Moreover, of the five different kinases identified by SKAI to be significantly activated or de-activated in both mouse models (GSK3A, GSK3B, MK2, PKACA, and PKG1), none has a clear pre-established role in human IBD. It is important to note that SKAI is designed to be a hypothesis-generating analysis; the statistical thresholds are set to allow for false positives, but to minimize false negatives. The key is that SKAI identifies pathways to test for functional significance through additional experimentation.
We chose to focus our experimental validation studies on MK2, which was the only kinase predicted to be up-regulated in both models. MK2 is a serine/threonine kinase that is activated by p38 stress-induced MAPK via phosphorylation on multiple residues [42]. The p38 pathway plays a central role in inflammation by regulating the expression of pro-inflammatory cytokines [43]. In patients with IBD, there is an increase in p38 expression in both immune and non-immune cells, leading to the idea that p38 inhibitors could show efficacy in this disease [44]. Moreover, inhibition of p38 was found to suppress colitis in both dextran sodium sulfate (DSS)- and trinitrobenzenesulfonic acid (TNBS)-induced models [45, 46]. In our study, we were able to validate the activation of MK2 predicted by SKAI through quantitative western blotting for phosphorylation of Ser334, a direct p38α substrate (Figs. 3 and S5b). The finding that MK2 is activated in the TCT model is particularly interesting given that we previously showed that p38α activity is reduced in this model [25]. These observations suggest a disconnect between p38α and MK2 in this system. MK2 could be activated by another isoform of p38 (β or γ ) and it can also be phosphorylated by ERK, although SKAI did not predict activation of ERK1 or ERK2 in either of our datasets.
Because the p38 pathway plays an important role in inflammation, there was hope that inhibitors would show efficacy in the clinic. Nevertheless, systemic p38 inhibition in patients is associated with unacceptable side effects and tachyphylaxis [4, 47], possibly due to feedback activation of NF-κB, JNK, and MEK signaling [48]. This limitation had led to the search for inhibitors that target other part of the p38 signaling pathway. A selective, orally available small molecule inhibitor of the MK2 pathway, ATI450 (formerly CDD-450), inhibits inflammatory responses in animals and is well tolerated at doses up to 30× higher than the predicted Cmax exposure [33]. We therefore evaluated the efficacy of ATI450 in a preclinical model of IBD. We demonstrated its activity at multiple levels. This confirms its capacity to impact downstream signaling pathways (Fig. 4c), to down-regulate pro-inflammatory molecules (Fig. 4a) and to promote mucosal healing (Fig. 3c–g). Importantly, no significant side effects were observed in mice treated with ATI450 and MK2 KO mice are viable and fertile [33, 37], suggesting that MK2 pathway inhibition represents a viable therapeutic modality for IBD patients.
One of the interesting insights revealed by treatment of TCT animals with ATI450 concerns the regulation of ATF-2 and AKT. ATF-2 is a transcription factor regulated by cellular stress and our data suggest that it is downstream of MK2 (Fig. 4d). AKT activity is regulated by phosphorylation on Thr308 and Ser473, which are predominantly regulated by Pyruvate dehydrogenase kinase 1 (PDK1) and mTOR [49]. We had noted in our prior work that AKT Ser473 phosphorylation is up-regulated during colitis in the TCT model, but that inhibition of mTOR did not decrease this phosphorylation [24]. Here, we also found that AKT Ser473 phosphorylation is up-regulated during colitis, but that this phosphorylation is inhibited by ATI450 (Fig. 4d). This observation indicates that in the context of colitis, AKT is regulated primarily by MK2, rather than by mTOR.
CONCLUSION
We have developed an organism-specific GSEA-based approach (SKAI) to predict kinase activation state from global phosphoproteomic data. Unlike previous methods, we integrate information across organisms; this allows us to compile expanded organism-specific kinase-substrate sets. When applied to mouse models of IBD, SKAI revealed activation of MK2, which we validated as a therapeutic target through a preclinical study with a clinically relevant inhibitor. Our study demonstrates that therapeutically relevant information is present in global phosphoproteomic data beyond individual phosphorylation sites and that the activation of therapeutically relevant kinases can be gleaned from phosphorylation data even when the kinase itself is not measured.
Supplementary Material
INSIGHT, INNOVATION, INTEGRATION.
Analyzing phosphoproteomic changes during pathogenesis can reveal mechanisms of disease and therapeutic targets, but matching kinase inhibitors with the appropriate disease state requires a detailed understanding of the key functional changes within the phosphoproteome. The most effective way to measure global phosphorylation changes is through mass spectrometry. Translating mass spectrometry data to a functional global signaling state is difficult, however, in light of the lack of widespread functional annotation throughout the phosphoproteome and poor coverage of the relatively small number of annotated sites in any given mass spectrometry experiment. Computational approaches that overcome these limitations can help to identify therapeutic targets from global phosphoproteomic data
ACKNOWLEDMENTS
We are grateful for the provision of mice from Ken Lau at Vanderbilt University.
FUNDING
This work was supported by grants from the National Institutes of Health (U01-CA199252 and R01-CA195744 to K.M.H, U01CA215798 to D.A.L, K.M.H. and W.H., R35-ES028374 to M.B.Y.) and the Department of Defense (W81XWH-16-1-0042 to K.M.H.). S.D.S. was supported by a National Science Foundation Graduate Research Fellowship (Grant No. 1122374), the Barbara J. Weedon Fellowship, and the Alan L. McWhorter Fellowship. J.L. and L.S-L. were supported by postdoctoral fellowships from the Crohn’s and Colitis Foundation of America.
Footnotes
SUPPLEMENTARY DATA
Supplementary data is available at Integrative Biology Journal online.
REFERENCES
- 1.Molodecky NA, Soon IS, Rabi DM et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012;142:46–54 e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- 2.Neurath MF. Current and emerging therapeutic targets for IBD. Nat Rev Gastroenterol Hepatol 2017;14:269–78. [DOI] [PubMed] [Google Scholar]
- 3.Panes J, Sandborn WJ, Schreiber S et al. Tofacitinib for induction and maintenance therapy of Crohn’s disease: results of two phase IIb randomised placebo-controlled trials. Gut 2017;66:1049–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng YJ, Li YY. The role of p38 mitogen-activated protein kinase in the pathogenesis of inflammatory bowel disease. J Dig Dis 2011;12:327–32. [DOI] [PubMed] [Google Scholar]
- 5.Macek B, Mann M, Olsen JV. Global and site-specific quantitative phosphoproteomics: principles and applications. Annu Rev Pharmacol Toxicol 2009;49:199–221. [DOI] [PubMed] [Google Scholar]
- 6.Riley NM, Coon JJ. Phosphoproteomics in the age of rapid and deep proteome profiling. Anal Chem 2016;88:74–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Needham EJ, Parker BL, Burykin T et al. Illuminating the dark phosphoproteome. Sci Signal 2019;12:eaau8645. [DOI] [PubMed] [Google Scholar]
- 8.Hornbeck PV, Zhang B, Murray B et al. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res 2015;43:D512–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinkel H, Chica C, Via A et al. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res 2011;39:D261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirbel J, Cutillas P, Saez-Rodriguez J. Phosphoproteomicsbased profiling of kinase activities in cancer cells. Methods Mol Biol 2018;1711:103–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez-Armenta C, Ochoa D, Goncalves E et al. Benchmarking substrate-based kinase activity inference using phosphoproteomic data. Bioinformatics 2017;33: 1845–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lachmann A, Ma’ayan A. KEA: Kinase enrichment analysis. Bioinformatics 2009;25:684–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casado P, Rodriguez-Prados JC, Cosulich SC et al. Kinase-Substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci Signal 2013;6:rs6. [DOI] [PubMed] [Google Scholar]
- 14.Wiredja DD, Koyuturk M, Chance MR. The KSEA App: a webbased tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics 2017;33:3489–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ochoa D, Jonikas M, Lawrence RT et al. An atlas of human kinase regulation. Mol Systems Biol 2016;12:888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drake JM, Graham NA, Stoyanova T et al. Oncogene-specific activation of tyrosine kinase networks during prostate cancer progression. Proc Natl Acad Sci USA 2012;109:1643–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miraldi ER, Sharfi H, Friedline RH et al. Molecular network analysis of phosphotyrosine and lipid metabolism in hepatic PTP1b deletion mice. Integr Biol 2013;5:940–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krug K, Mertins P, Zhang B et al. A curated resource for Phosphosite-specific signature analysis. Mol Cell Proteomics 2019;18:576–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mischnik M, Sacco F, Cox J et al. IKAP: A heuristic framework for inference of kinase activities from Phosphoproteomics data. Bioinformatics 2016;32:424–31. [DOI] [PubMed] [Google Scholar]
- 20.Beekhof R, van Alphen C, Henneman AA et al. INKA, an integrative data analysis pipeline for phosphoproteomic inference of active kinases. Mol Systems Biol 2019;15:e8981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from largescale data sets. Nature Biotech 2005;23:1391–8. [DOI] [PubMed] [Google Scholar]
- 22.Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics 2011;Chapter 13:Unit 13.15–24. [DOI] [PubMed] [Google Scholar]
- 23.Joughin BA, Naegle KM, Huang PH et al. An integrated comparative phosphoproteomic and bioinformatic approach reveals a novel class of MPM-2 motifs upregulated in EGFRvIII-expressing glioblastoma cells. Mol Biosyst 2009;5:59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lyons J, Ghazi PC, Starchenko A et al. The colonic epithelium plays an active role in promoting colitis by shaping the tissue cytokine profile. PLoS Biol 2018;16:e2002417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyons J, Brubaker DK, Ghazi PC et al. Integrated in vivo multiomics analysis identifies p21-activated kinase signaling as a driver of colitis. Sci Signal 2018;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res 2005;15:11–8. [DOI] [PubMed] [Google Scholar]
- 27.Ostanin DV, Bao J, Koboziev I et al. T cell transfer model of chronic colitis: Concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol 2009;296: G135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kontoyiannis D, Pasparakis M, Pizarro TT et al. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: Implications for joint and gut-associated immunopathologies. Immunity 1999;10:387–98. [DOI] [PubMed] [Google Scholar]
- 29.McDonald M, Trost B, Napper S. Conservation of kinase-phosphorylation site pairings: Evidence for an evolutionarily dynamic phosphoproteome. PLoS ONE 2018;13: e0202036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mootha VK, Lindgren CM, Eriksson KF et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature Genet 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 32.Murali B, Ren Q, Luo X et al. Inhibition of the stromal p38MAPK/MK2 pathway limits breast cancer metastases and chemotherapy-induced bone loss. Cancer Res 2018;78: 5618–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C, Hockerman S, Jacobsen EJ et al. Selective inhibition of the p38alpha MAPK-MK2 axis inhibits inflammatory cues including inflammasome priming signals. J Exp Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang C, Zhao L, Su Q et al. Phosphorylation of MITF by AKT affects its downstream targets and causes TP53dependent cell senescence. Int J Biochem Cell Biol 2016;80: 132–42. [DOI] [PubMed] [Google Scholar]
- 35.Kodani T, Rodriguez-Palacios A, Corridoni D et al. Flexible colonoscopy in mice to evaluate the severity of colitis and colorectal tumors using a validated endoscopic scoring system. J Vis Exp 2013;80:e50843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McGuire VA, Gray A, Monk CE et al. Cross talk between the Akt and p38alpha pathways in macrophages downstream of Toll-like receptor signaling. Mol Cell Biol 2013;33: 4152–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh RK, Najmi AK, Dastidar SG. Biological functions and role of mitogen-activated protein kinase activated protein kinase 2 (MK2) in inflammatory diseases. Pharmacol Rep 2017;69:746–56. [DOI] [PubMed] [Google Scholar]
- 38.Gupta S, Campbell D, Derijard B et al. Transcription factor ATF2 regulation by the JNK signal transduction pathway. Science 1995;267:389–93. [DOI] [PubMed] [Google Scholar]
- 39.Suarez-Lopez L, Sriram G, Kong YW et al. MK2 contributes to tumor progression by promoting M2 macrophage polarization and tumor angiogenesis. Proc Natl Acad Sci USA 2018;115:E4236–E44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaudet S, Janes KA, Albeck JG et al. A compendium of signals and responses triggered by prodeath and prosurvival cytokines. Mol Cell Proteomics 2005;4:1569–90. [DOI] [PubMed] [Google Scholar]
- 41.Coskun M, Olsen J, Seidelin JB et al. MAP kinases in inflammatory bowel disease. Clin Chim Acta 2011;412:513–20. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Levy R, Leighton IA, Doza YN et al. Identification of novel phosphorylation sites required for activation of MAPKAP kinase-2. EMBO J 1995;14:5920–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cuenda A,Rousseau S.p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta 2007;1773:1358–75. [DOI] [PubMed] [Google Scholar]
- 44.Li YY, Yuece B, Cao HM et al. Inhibition of p38/Mk2 signaling pathway improves the anti-inflammatory effect of WIN55 on mouse experimental colitis. Lab Invest 2013;93: 322–33. [DOI] [PubMed] [Google Scholar]
- 45.Hollenbach E, Neumann M, Vieth M et al. Inhibition of p38 MAP kinase- and RICK/NF-kappaB-signaling suppresses inflammatory bowel disease. FASEB J 2004;18:1550–2. [DOI] [PubMed] [Google Scholar]
- 46.Hollenbach E, Vieth M, Roessner A et al. Inhibition of RICK/nuclear factor-kappaB and p38 signaling attenuates the inflammatory response in a murine model of Crohn disease. J Biol Chem 2005;280:14981–8. [DOI] [PubMed] [Google Scholar]
- 47.Dambach DM. Potential adverse effects associated with inhibition of p38alpha/beta MAP kinases. Curr Top Med Chem 2005;5:929–39. [DOI] [PubMed] [Google Scholar]
- 48.Jones DS, Jenney AP, Joughin BA et al. Inflammatory but not mitogenic contexts prime synovial fibroblasts for compensatory signaling responses to p38 inhibition. Sci Signal 2018;11:eaal1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manning BD, Toker A. AKT/PKB Signaling: Navigating the network. Cell 2017;169:381–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.