Abstract
With the growing threat of antimicrobial resistance worldwide, uncovering the molecular epidemiology is critical for understanding what is driving this crisis. We aimed to evaluate the prevalence of plasmid-mediated-quinolone-resistance (PMQR) and extended-spectrum beta-lactamase- (ESBL) producing gram-negative organisms among primigravid women with bacteriuria. We collected urine specimens from primigravid women attending their first antenatal visit at Gandhi Hospital during October 1, 2015 to September 30, 2016. We determined antimicrobial susceptibility and ESBL and quinolone resistance using VITEK-2. We performed polymerase chain reaction amplification on resistant isolates for detection of ESBL-encoding genes (TEM, SHV, CTX-M) and PMQR genes (qnrA, qnrB, qnrD, qnrS, aac (6’)-Ib-cr). Of 1,841 urine samples, 133 demonstrated significant bacterial growth with gram-negative bacilli accounting for 85% of isolates, including Escherichia coli (n = 79), Klebsiella pneumoniae (n = 29), Sphingomonas (n = 3), Enterobacter (n = 1), and Citrobacter (n = 1). We found 65% of E. coli isolates and 41% of K. pneumoniae isolates were ESBL positive. Of ESBL-positive isolates, the most common genes conferring resistance were TEM-1 (66.7%) followed by CTX-M-15 (33.3%). Fifty-seven percent of ESBL-positive E. coli also demonstrated resistance to quinolones with the most common PMQR genes being qnr-S (62.5%) and aac (6')-Ib-cr (37.5%). We did not find any resistance to quinolones among ESBL-positive K. pneumoniae isolates. Across different classes of antibiotics we found a strong clustering of multi-drug resistance in E. coli with over 45% of ESBL-positive isolates demonstrating resistance to at least three classes of antibiotics. This study emphasizes the high prevalence of plasmid-mediated ESBL and quinolone resistance in community-acquired urinary tract infections of primigravid women. The overall abundance of multi-drug-resistant isolates in this population is alarming and may present therapeutic challenges.
Introduction
Emergence of community-acquired multi-drug-resistant bacterial infections poses a grave public health threat. Urinary tract infections (UTIs) are a major proportion of community-acquired infections that have demonstrated increasing patterns of antimicrobial resistance. UTIs occur in 2–10% of pregnant women, which may be symptomatic or asymptomatic [1]. Regardless of symptoms, untreated or undertreated bacteriuria in pregnancy increases risk for adverse outcomes including preterm birth, low birth weight, and pyelonephritis, which can lead to excess maternal and neonatal morbidity and mortality [2–4]. Thus, screening and treating pregnant women for bacteriuria has become a routine part of prenatal care [5, 6]. Evaluating the bacteriological profiles of bacteriuria in pregnant women attending antenatal clinics provides an opportunity to study the prevalence of antimicrobial resistance in community-acquired uropathogens and determine appropriateness of empiric treatment options.
Cephalosporins and combination beta-lactam/beta-lactamase inhibitors are considered first-line therapy in the treatment of bacteriuria in pregnancy. Similarly, cephalosporins and fluoroquinolones are frequently used for treating community-acquired UTIs in non-pregnant adults due to their potency, broad spectrum of activity, oral bioavailability, and safety profile [7]. However, with increasing antibiotic resistance worldwide, the efficacy of these antibiotic treatment options may be threatened.
Extended-spectrum beta-lactamases (ESBLs) are a group of genetic mutations that confer resistance by hydrolysing penicillins, first-, second-, and third-generation cephalosporins, and aztreonam. They can be inhibited by beta-lactamase inhibitors. ESBLs are encoded by three major groups of genes: TEM, SHV, and CTX-M [8], and these enzymes are often found in Escherichia coli and Klebsiella pneumoniae [9]. Several different species of bacteria are capable of producing ESBLs, which were initially associated with healthcare-associated infections (HCAIs), but are increasingly being associated with community-acquired infections.
Fluoroquinolones are used to treat UTIs caused by both gram-positive and gram-negative bacteria. Wide usage of these antibiotics has led to resistance, especially among Enterobacteriaceae [10]. Fluoroquinolone resistance varies from 2.2% to 69% among community-acquired UTIs [11]. The emergence of plasmid-mediated quinolone resistance (PMQR) was first found in a strain of K. pneumonia in the USA in 1998 and shown to be due to a member of the pentapeptide repeat family of proteins qnr [12]. Qnr interacts with DNA gyrase and topoisomerase IV to prevent quinolone inhibition. In subsequent years, several distantly-related plasmid-mediated qnr determinants have been described in Enterobacteriaceae (qnrB, qnrC, qnrD, qnrS). The qnr genes are usually integrated into plasmids, which make them particularly susceptible to cross-species transmission [13]. In addition, these plasmids often harbor other antibiotic resistance genes such as ESBLs and favor the selection and dissemination of fluoroquinolone-resistant strains by chemically unrelated drug classes and vice versa [14].
There is limited information regarding the frequency of ESBL and fluoroquinolone-resistance genes in community-acquired infections in India. Therefore, this study aimed to uncover the epidemiology of PMQR and ESBL genes in gram-negative bacilli among primigravid women with bacteriuria attending the antenatal clinic for the first time.
Materials and methods
Study design and sampling
A cross-sectional observational study was conducted at Gandhi Medical College and Hospital, Hyderabad from October 1, 2015 to September 30, 2016. We collected urine specimens from primigravid women attending their first antenatal clinic visit at the outpatient department of Gandhi Hospital. We excluded multigravid and primigravid women who previously attended antenatal clinics to rule out the possibility of any HCAIs or colonization resulting from healthcare exposure. In addition, we administered surveys regarding community exposures at the time of enrollment that indicated recent antibiotic use was rare among study participants [15]. The study was approved by IRB committees from Gandhi Medical College and Hospital, Stanford University, and the Indian Council of Medical Research. We also obtained written informed consent from all study participants before specimen collection and interviews.
A structured case proforma was filled by a trained investigator during the antenatal visit. The questionnaire was based on the Demographic and Health Surveys tool AMR Module for Population-Based Surveys [16]. The questionnaire included their usual and current residence, occupation, husband’s occupation, household income, religion, caste, education level, dietary and hygiene practices, and recent non vitamin tablet consumption. As most of the women were unaware of the antimicrobial drug use, ingestion of any other tablet other than vitamin was the question asked for antimicrobial drug use.
Antimicrobial susceptibility testing
We performed phenotypic antimicrobial susceptibility testing and ESBL and quinolone resistance screening using a VITEK-2 system in accordance with CLSI guidelines [17].
Genotypic characterization of ESBL and PMQR genes
We targeted ESBL-producing and fluoroquinolone-resistant isolates for detection of ESBL-encoding genes (TEM, SHV, CTX-M) and PMQR-encoding genes (qnrA, qnrB, qnrD, qnrS, aac (6’)-Ib-cr), respectively. We isolated DNA from bacterial cells by using a boiling method. For polymerase chain reaction (PCR) amplification, we prepared master mix using 10 μl dNTPs, 2μl of each primer, 1 μl of Taq polymerase in 6.5 μl PCR buffer, and 22 μl of RNase-free water. We added DNA to the above mixture to a final volume of 50 μl [18], utilizing published primers (Tables 1 and 2) [19, 20]. We performed amplification in a thermocycler according to the following cycling parameters: initial denaturation step at 95°C for 5minutes, 35 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, extension at 72°C for 2 minutes, final extension step at 72°C for 10 minutes, and a hold at 4°C. We subjected amplified PCR products to agarose gel electrophoresis with 1.8% agarose gel using suitable molecular weight markers. We visualized the gel on a UV platform in a gel documentation system (Figs 1 and 2).
Table 1. Primers for polymerase chain reaction of ESBL genes.
| Primer | Orientation | Oligonucleotide sequence (5΄-3΄) | Size (bp) |
|---|---|---|---|
| CTX-M | Forward | 5'- GAAGGTCATCAAGAAGGTGCG -3' | 560bp |
| Reverse | 5'- GCATTGCCACGCTTTTCATAG- 3' | ||
| TEM | Forward | 5'—GAGACAATAACCCTGGTAAAT- 3' | 459 bp |
| Reverse | 5'- AGAAGTAAGTTGGCAGCAGTG- 3' | ||
| SHV | Forward | 5'-GTCAGCGAAAAACACCTTGCC- 3' | 383bp |
| Reverse | 5'-GTCTTATCGGCGATAAACCAG- 3' |
Table 2. Primers for polymerase chain reaction of PMQR genes.
| Primer | Orientation | Oligonucleotide sequence (5΄-3΄) | Size (bp) | Annealing Temperature |
|---|---|---|---|---|
| qnrA | Forward | CAGCAAGAGGATTTCTCACG | 630 bp | 58°C |
| Reverse | AATCCGGCAGCACTATTACTC | |||
| qnrB | Forward | GGCTGTCAGTTCTATGATCG | 488 bp | 59.1°C |
| Reverse | SAKCAACGATGCCTGGTAG | |||
| qnrD | Forward | CGAGATCAATTTACGGGGAATA | 581 bp | 57°C |
| Reverse | AACAAGCTGAAGCGCCTG | |||
| qnrS | Forward | GCAAGTTCATTGAACAGGGT | 428 bp | 55.6°C |
| Reverse | TCTAAACCGTCGAGTTCGGCG | |||
| aac (6’)-Ib-cr | Forward | TTGGAAGCGGGGACGGAM | 260 bp | 58°C |
| Reverse | ACACGGCTGGACCATA |
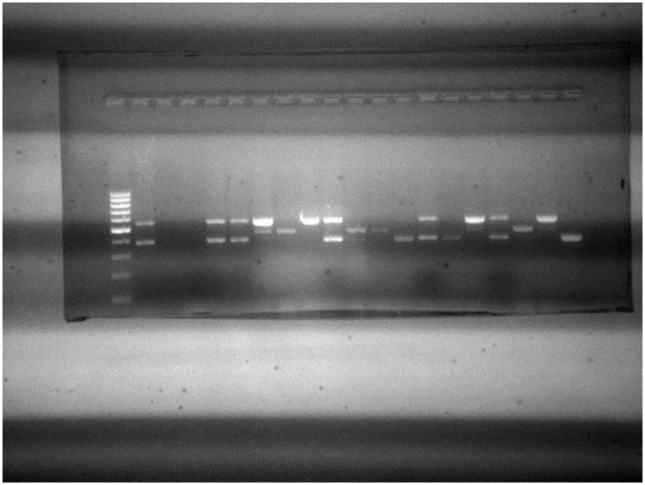
Fig 1. Gel electrophoresis detection of ESBL-producing genes among E. coli and K. pneumoniae isolates.
Results by lane: 1- ladder (100bp); 2- K. pneumoniae (ATCC 700603) CTX-M-15+SHV-38 genes; 3-E. coli (ATCC 25922); 4-Undetected; 5,6,10,14,17- CTX-M-15+SHV-38 genes; 13,15,20- SHV-38 gene; 7,16,19- CTX-M-15 +TEM-1 genes; 8,11,12,18- TEM-1 gene.
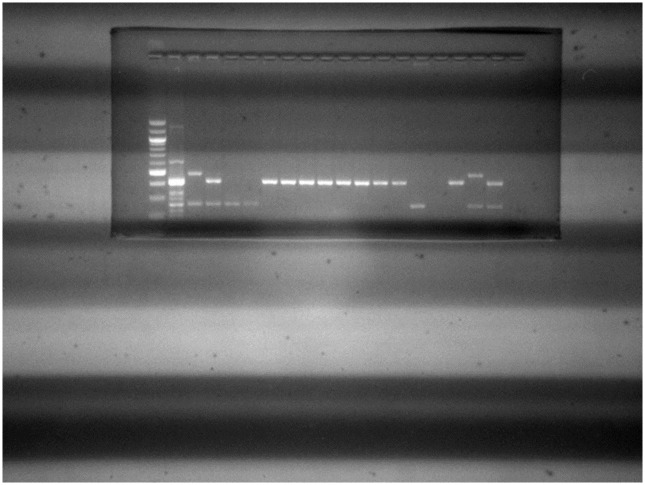
Fig 2. Gel electrophoresis detection of PMQR genes among E. coli and K. pneumoniae isolates.
Results by lane: 1- ladder (100bp); 2- ladder (50bp); 3- K. pneumonia qnrB+aac (6’)-lb-cr genes; 4- qnrS+aac (6’)-lb-cr genes; 5,6,15- aac (6’)-lb-cr genes; 7–14- qnrS gene; 16-Undetected.
DNA sequencing
We sequenced the purified PCR products with an ABI 3730XL sequencer (Applied Biosystems, USA). We analysed the nucleic acid sequences using the Basic Local Alignment Search Tool available at the National Centre for Biotechnology database. We submitted the nucleic acid sequences to Genbank (accession numbers for ESBLs: MH745708, MH745709, MH745710, MH745711, MH745712, MH745713, MH745714, MH745715, MH745716 and PMQRs: MK761221, MK761222, MK761223, MK761224, MK761225, and MK761226).
Results
Distribution of ESBL and quinolone resistance among isolates
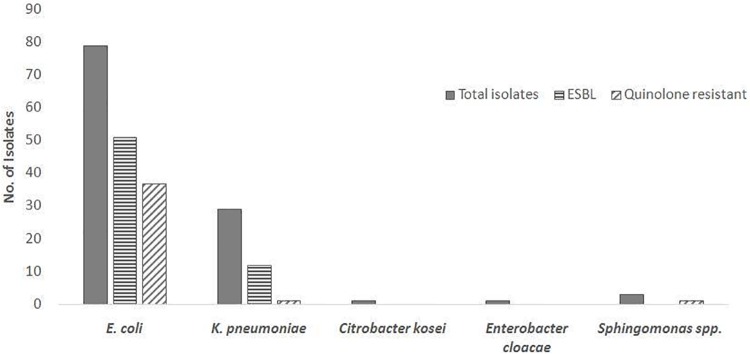
Of 1,841 urine samples, 133 had significant bacterial growth, defined as ≥105 CFU/mL. Gram-negative bacilli accounted for 85% (113) of the isolates, including E. coli (n = 79), K. pneumoniae (n = 29), Sphingomonas (n = 3), Enterobacter (n = 1), and Citrobacter (n = 1) (Fig 3).
Fig 3. Phenotypic distribution of ESBL and quinolone resistance among isolates.
High levels of ESBL and quinolone resistance were observed among E. coli isolates. K. pneumoniae isolates demonstrated less but still substantial resistance.
Based on VITEK-2 determinations, we detected ESBL positivity in 65% (51) of E. coli isolates and 41% (12) of K. pneumonia isolates. Quinolone resistance was observed in 47% (37) of E. coli isolates, whereas only one isolate of K. pneumoniae demonstrated resistance to quinolones.
Resistance patterns to other antibiotics
We evaluated for resistance against individual antimicrobial agents separated by ESBL determination. Among ESBL-positive E.coli isolates, we observed the highest resistance against nalidixic acid (86%), which can signify reduced susceptibility to fluoroquinolones. High levels of resistance were also noted for ciprofloxacin (57%), trimethoprim/sulfamethoxazole (55%), and gentamicin (33%). Multi-drug resistance (resistance to at least 3 classes of antibiotics) was noted in 45% of ESBL-positive E. coli isolates (Table 3).
Table 3. Antimicrobial resistance patterns of E. coli isolates by ESBL positivity.
| Antibiotic | ESBL E. coli (n = 51) | Non-ESBL E. coli (n = 28) | ||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Amoxicillin/Clavulanic acid | 28 (56%) | 18 (36%) | 4 (8%) | 26 (92.9%) | 2 (7.1%) | - |
| Cefuroxime | 10 (20%) | 1 (2%) | 39 (78%) | 25 (89.3%) | 1 (3.6%) | 2 (7.1%) |
| Ceftriaxone | 13 (26%) | 2 (4%) | 35 (70%) | 24 (88.9%) | 1 (3.7%) | 2 (7.4%) |
| Piperacillin/Tazobactam | 43 (86%) | 3 (6%) | 4 (8%) | 25 (96.2%) | - | 1 (3.8%) |
| Gentamicin | 34 (66.7%) | - | 17 (33.3%) | 28 (100%) | - | - |
| Nalidixic acid | 7 (14%) | - | 43 (86%) | 11 (39.3%) | - | 17 (60.7%) |
| Ciprofloxacin | 21 (41.2%) | 1 (2.0%) | 29 (56.9%) | 20 (71.4%) | 1 (3.6%) | 7 (25%) |
| Meropenem | 51 (100%) | - | - | 28 (100%) | - | - |
| Nitrofurantoin | 48 (96%) | 1 (2.0%) | 1 (2.0%) | 25 (89.3%) | 2 (7.1%) | 1 (3.6%) |
| Trimethoprim/sulfamethoxazole | 23 (45.1%) | - | 28 (54.9%) | 20 (76.9%) | - | 6 (23.1%) |
| Multi-drug resistant (≥3 classes) | 23 (45.1%) | 0 | ||||
Among ESBL-positive K. pneumoniae isolates, rates of resistance to other antibiotics was lower, though a substantial number of isolates demonstrated only intermediate susceptibility to nitrofurantoin (50%), a common treatment option for community-acquired UTIs. Resistance to other antimicrobial classes are shown in Table 4. All the ESBL-positive K. pneumoniae isolates were sensitive to nalidixic acid and ciprofloxacin, and only one isolate demonstrated multi-drug resistance (8%).
Table 4. Antimicrobial resistance patterns of K. pneumoniae isolates by ESBL positivity.
| Antibiotic | ESBL K. pneumoniae (n = 12) | Non-ESBL K. pneumoniae (n = 17) | ||||
|---|---|---|---|---|---|---|
| S | I | R | S | I | R | |
| Amoxicillin/Clavulanic acid | 11 (91.7%) | - | 1 (8.3%) | 17 (100%) | - | - |
| Cefuroxime | 3 (25%) | - | 9 (75%) | 17 (100%) | - | - |
| Ceftriaxone | 2 (16.7%) | - | 10 (83.3%) | 17 (100%) | - | - |
| Piperacillin/Tazobactam | 12 (100%) | - | - | 17 (100%) | - | - |
| Gentamicin | 11 (91.7%) | - | 1 (8.3%) | 17 (100%) | - | - |
| Nalidixic acid | 12 (100%) | - | - | 15 (88.2%) | - | 2 (11.8%) |
| Ciprofloxacin | 12 (100%) | - | - | 16 (94.1%) | - | 1 (5.9%) |
| Meropenem | 12 (100%) | - | - | 17 (100%) | - | - |
| Nitrofurantoin | 5 (41.7%) | 6 (50.0%) | 1 (8.3%) | 12 (70.6%) | 5 (29.4%) | - |
| Trimethoprim/sulfamethoxazole | 12 (100%) | - | - | 17 (100%) | - | - |
| Multi-drug resistant (≥3 classes) | 1 (8.3%) | 0 | ||||
Distribution of ESBL genes
We identified 63 phenotypically-confirmed ESBLs (51 E. coli and 12 K. pneumoniae), which were genotypically characterized for ESBL genes (CTX-M, SHV, and TEM). Among these isolates, the most common ESBL gene was TEM-1 in E. coli (62.7%) and K. pneumoniae (83.3%), followed by CTX-M-15 as the second most prevalent gene at 35.2% and 25%, respectively (Table 5). We found a co-occurrence of CTX-M-15 and TEM-1 genes in 23.8% of isolates and TEM-1 and SHV-38 genes in 4.8%. Overall 28.6% of isolates carried two resistance genes.
Table 5. Frequency of ESBL genes among ESBL-producing E. coli and K. pneumoniae.
| ESBL Gene | E. coli (n = 51) | K. pneumoniae (n = 12) |
|---|---|---|
| CTX-M-15 | 18 (35.2%) | 3 (25.0%) |
| TEM-1 | 32 (62.7%) | 10 (83.3%) |
| SHV-38 | 10 (19.6%) | 2 (16.6%) |
| TEM-1+SHV-38 | 3 (5.8%) | 0 |
| CTX-M-15+TEM-1 | 12 (23.5%) | 3 (25.0%) |
Distribution of PMQR genes
We identified 43 phenotypically-confirmed quinolone-resistant ESBL E. coli strains, and we genotypically characterized 32 for PMQR genes. The most common gene in the present study was qnrS at 62.5% followed by aac (6')-Ib-cr at 37.5% (Table 6). QnrA, qnrB, and qnrD were not detected. Since all of the ESBL-positive K. pneumoniae were sensitive to quinolones they were not tested for PMQR genes.
Table 6. Frequency of PMQR genes among quinolone-resistant, ESBL-positive E. coli.
| PMQR Gene | E. coli (n = 32) | Nalidixic acid resistance | Ciprofloxacin Resistance |
|---|---|---|---|
| qnrA | 0 (0%) | 0 | 0 |
| qnrB | 0 (0%) | 0 | 0 |
| qnrD | 0 (0%) | 0 | 0 |
| qnrS | 20 (62.5%) | 20 | 15 |
| aac (6')-Ib-cr | 12 (37.5%) | 9 | 9 |
| qnrS +aac (6')-Ib-cr | 2 (6.25%) | 9 | 4 |
Discussion
The aim of the study was to evaluate the prevalence of PMQR and ESBL-producing gram-negative organisms among primigravid women with bacteriuria. Pregnant women are an understudied population with regards to antimicrobial resistance, and very few studies have previously examined the prevalence of PMQR and ESBL genes among enterobacterial isolates from pregnant women [21]. However, the implications of the findings likely extend to uropathogen resistance profiles in the community given the absence of prior healthcare exposure in this population. The high overall rates of plasmid-mediated resistance in this study are cause for concern.
Since the early 2000s a change in epidemiology of ESBL-producing Enterobacteriaceae was observed with increasing reports of their occurrence in community-acquired infections [22]. We detected an overall prevalence of ESBL-producing E. coli isolates of 65% in this study with 48% phenotypic resistance to third-generation cephalosporins. This contrasts with studies published from 2011–2014 evaluating bacteriuria in pregnant women in India that found rates of E. coli resistance to cephalosporins as low as 0–14% [23, 24]. Similarly, a previous study of rectal E. coli isolates from pregnant women in India demonstrated 17–19% resistance to third-generation cephalosporins, with 86% of those isolates producing ESBLs, highlighting either the rapid spread of antibiotic resistance or substantial variations in local epidemiology [21].
The emergence of ESBL-producing Enterobacteriaceae was attributed to the spread of the CTX-M gene [22]. However, the most common ESBL gene in the present study was TEM-1, similar to findings from other studies in India [25–27]. Additionally, in our study a third of isolates carried more than one type of beta-lactamase gene, similar to prior reports that have demonstrated a frequent co-occurrence of ESBL genes [22, 27]. Overall, 84% percent of ceftriaxone-resistant isolates carried at least one ESBL gene. Several studies in India have shown Klebsiella spp. as the major ESBL producer [25, 26], though our study revealed greater ESBL positivity among E. coli, which has also been reflected in other studies [28].
We demonstrated a high level of quinolone resistance among ESBL-positive E. coli, supporting the co-transmission of resistance genes on plasmids. This has been seen in other studies in India where 61% of ciprofloxacin-resistant E. coli isolates demonstrated ESBL production [29]. Accordingly, multi-drug resistance was common in E. coli, though lower resistance was seen in K. pneumoniae isolates. As E. coli is one of the most common causes of bacteriuria in pregnant women as well as community-acquired UTIs, the clustering of resistance genes among E. coli isolates presents concerning therapeutic challenges.
Fluoroquinolone therapy is generally avoided in pregnant women because of safety concerns in developing foetuses, though it remains an important treatment for UTIs and other types of infections in non-pregnant individuals. Overall resistance to fluoroquinolones in this study both in ESBL and non-ESBL varied substantially between E. coli (45.1%) and K. pneumonia (3.6%). Qnr genes were detected in 86% of quinolone-resistant isolates. This is higher than another study from India that reported a prevalence of 50% among ciprofloxacin-resistant uropathogenic E. coli [29]. The total percentage of E. coli isolates harbouring qnr genes in this study, as shown in Table 3, is also significantly higher compared to Brazil, Europe, the USA, and elsewhere in Asia [19, 30–35].
In this study, we only detected qnrS and aac (6’)-Ib-cr genes in E. coli, with almost a quarter of isolates containing both genes. QnrA, qnrB, and qnrC were not found. Few studies in India reported PMQR genes among clinical isolates of Enterobacteriaceae, but the most prevalent gene reported has been aac (6’)-lb-cr [21, 29, 36]. However, regionally qnrS has been found to be a major antimicrobial resistant gene in environmental samples in India [36]. Studies on E. coli strains showing higher prevalence of qnrA and qnrB have mostly been among clinical isolates from HCAIs [31].
The increased frequency of ESBL and PMQR genes detected in this study carries significant implications for the management of community-acquired infections and further spread of these plasmid-mediated resistance mechanisms through horizontal gene transfer to other bacterial classes. Additionally, it raises the question about what is driving this trend. Overuse and misuse of antibiotics has changed the landscape of resistance, but this study population has not been directly influenced by the typical risk factors such as recent antibiotic use or healthcare contact/hospitalization [21, 37, 38]. After eliminating the effects of healthcare, presence of resistant bacteria has been found to be most closely linked to low socioeconomic status [15]. In fact, a review of community-acquired UTIs found that exposure to food, animal, and environmental sources may predict UTIs caused by ESBL-producing E. coli [39]. This finding suggests that there are environmental selective pressures that are distinct from the healthcare setting, and contamination of the environment by biocides and antibiotic residues are increasing resistance in community flora [40–42].
Indeed, in a series of studies conducted near the present research site, high levels of fluoroquinolones have been detected in water and environmental sources as a result of contamination by pharmaceutical manufacturing effluent [43, 44]. Environmental contamination in particular selects for antibiotic resistance genes on mobile genetic elements because it promotes increased uptake of foreign DNA, which contributes to bacterial resilience under selective pressures [45, 46]. The findings of this study provide concerning evidence that these environmental processes may be spreading into human pathogens.
In conclusion, our study emphasizes the high prevalence of plasmid-mediated ESBL and quinolone resistance in community-acquired UTIs. While lower resistance was found in K. pneumoniae isolates compared with E. coli, overall abundance of drug resistance among uropathogens in this population is alarming. The findings of this study limit empiric treatment options for community-acquired UTI, including among pregnant women. Early detection of multi-drug-resistant isolates in routine microbiology laboratories is critical to avoid treatment failure and the complications thereof. Future surveillance and characterization of plasmids carrying multi-drug-resistance determinants could improve understanding of the origin and evolution of gram-negative bacterial resistance and inform infection control efforts.
Supporting information
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Freeman Spogli Institute for International Studies, Stanford University http://dx.doi.org/10.13039/100006100 Dr.Manisha Rani one of the co-author was recruited as Research Assistant in this project and received salary. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Tomezsko J.E. and Sand P.K., Pregnancy and intercurrent diseases of the urogenital tract. Clin Perinatol, 1997. 24(2): p. 343–68. [PubMed] [Google Scholar]
- 2.Smaill F.M. and Vazquez J.C., Antibiotics for asymptomatic bacteriuria in pregnancy. Cochrane Database Syst Rev, 2015(8): p. CD000490 10.1002/14651858.CD000490.pub3 [DOI] [PubMed] [Google Scholar]
- 3.Delzell J.E. and Lefevre M.L., Urinary tract infections during pregnancy. Am Fam Physician, 2000. 61(3): p. 713–21. [PubMed] [Google Scholar]
- 4.Millar L.K. and Cox S.M., Urinary tract infections complicating pregnancy. Infect Dis Clin North Am, 1997. 11(1): p. 13–26. 10.1016/s0891-5520(05)70339-1 [DOI] [PubMed] [Google Scholar]
- 5.Force, U.S.P.S.T., Screening for asymptomatic bacteriuria in adults: U.S. Preventive Services Task Force reaffirmation recommendation statement. Ann Intern Med, 2008. 149(1): p. 43–7. 10.7326/0003-4819-149-1-200807010-00009 [DOI] [PubMed] [Google Scholar]
- 6.Nicolle L.E., et al. , Clinical Practice Guideline for the Management of Asymptomatic Bacteriuria: 2019 Update by the Infectious Diseases Society of America. Clin Infect Dis, 2019. 68(10): p. 1611–1615. 10.1093/cid/ciz021 [DOI] [PubMed] [Google Scholar]
- 7.Krieger J.N., Urinary tract infections: what's new? J Urol, 2002. 168(6): p. 2351–8. [DOI] [PubMed] [Google Scholar]
- 8.Bonnet R., Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother, 2004. 48(1): p. 1–14. 10.1128/AAC.48.1.1-14.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford P.A., Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev, 2001. 14(4): p. 933–51, table of contents. 10.1128/CMR.14.4.933-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emmerson A.M. and Jones A.M., The quinolones: decades of development and use. J Antimicrob Chemother, 2003. 51 Suppl 1: p. 13–20. [DOI] [PubMed] [Google Scholar]
- 11.Khawcharoenporn T., et al. , High rates of quinolone resistance among urinary tract infections in the ED. Am J Emerg Med, 2012. 30(1): p. 68–74. 10.1016/j.ajem.2010.09.030 [DOI] [PubMed] [Google Scholar]
- 12.Martínez-Martínez L., Pascual A., and Jacoby G.A., Quinolone resistance from a transferable plasmid. Lancet, 1998. 351(9105): p. 797–9. 10.1016/S0140-6736(97)07322-4 [DOI] [PubMed] [Google Scholar]
- 13.Strahilevitz J., et al. , Plasmid-mediated quinolone resistance: a multifaceted threat. Clin Microbiol Rev, 2009. 22(4): p. 664–89. 10.1128/CMR.00016-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordmann P. and Poirel L., Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J Antimicrob Chemother, 2005. 56(3): p. 463–9. 10.1093/jac/dki245 [DOI] [PubMed] [Google Scholar]
- 15.Alsan M., et al. , Poverty and Community-Acquired Antimicrobial Resistance with Extended-Spectrum β-Lactamase-Producing Organisms, Hyderabad, India. Emerg Infect Dis, 2018. 24(8): p. 1490–1496. 10.3201/eid2408.171030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Development, U.A.f.I., Antimicrobial resistance module for population-based surveys. 2008.
- 17.Espinar M.J., et al. , Extended-spectrum β-lactamases of Escherichia coli and Klebsiella pneumoniae screened by the VITEK 2 system. J Med Microbiol, 2011. 60(Pt 6): p. 756–760. 10.1099/jmm.0.024075-0 [DOI] [PubMed] [Google Scholar]
- 18.Ahmed O., Asghar A., and Elhassan M., Comparison of three DNA extraction methods for polymerase chain reaction (PCR) analysis of bacterial genomic DNA. Afr J Microbiol Res, 2014. 8(6): p. 598–602. [Google Scholar]
- 19.Lavilla S., et al. , Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother, 2008. 61(2): p. 291–5. 10.1093/jac/dkm448 [DOI] [PubMed] [Google Scholar]
- 20.Pitout J.D., et al. , Development and clinical validation of a molecular diagnostic assay to detect CTX-M-type beta-lactamases in Enterobacteriaceae. Clin Microbiol Infect, 2007. 13(3): p. 291–7. 10.1111/j.1469-0691.2006.01645.x [DOI] [PubMed] [Google Scholar]
- 21.Pathak A., et al. , Frequency and factors associated with carriage of multi-drug resistant commensal Escherichia coli among women attending antenatal clinics in central India. BMC Infect Dis, 2013. 13: p. 199 10.1186/1471-2334-13-199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout J.D. and Laupland K.B., Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis, 2008. 8(3): p. 159–66. 10.1016/S1473-3099(08)70041-0 [DOI] [PubMed] [Google Scholar]
- 23.Sujatha R. and Nawani M., Prevalence of asymptomatic bacteriuria and its antibacterial susceptibility pattern among pregnant women attending the antenatal clinic at kanpur, India. J Clin Diagn Res, 2014. 8(4): p. DC01-3. 10.7860/JCDR/2014/6599.4205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandel L.R., et al. , Prevalance of pregnancy associated asymptomatic bacteriuria: a study done in a tertiary care hospital. J Obstet Gynaecol India, 2012. 62(5): p. 511–4. 10.1007/s13224-011-0071-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma J., Sharma M., and Ray P., Detection of TEM & SHV genes in Escherichia coli & Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res, 2010. 132: p. 332–6. [PubMed] [Google Scholar]
- 26.Kaur M. and Aggarwal A., Occurrence of the CTX-M, SHV and the TEM Genes Among the Extended Spectrum β-Lactamase Producing Isolates of Enterobacteriaceae in a Tertiary Care Hospital of North India. J Clin Diagn Res, 2013. 7(4): p. 642–5. 10.7860/JCDR/2013/5081.2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manoharan A., et al. , Correlation of TEM, SHV and CTX-M extended-spectrum beta lactamases among Enterobacteriaceae with their in vitro antimicrobial susceptibility. Indian J Med Microbiol, 2011. 29(2): p. 161–4. 10.4103/0255-0857.81799 [DOI] [PubMed] [Google Scholar]
- 28.Gautam V., et al. , Molecular characterization of extended-spectrum β-lactamases among clinical isolates of. Indian J Med Res, 2019. 149(2): p. 208–215. 10.4103/ijmr.IJMR_172_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Basu S. and Mukherjee M., Incidence and risk of co-transmission of plasmid-mediated quinolone resistance and extended-spectrum β-lactamase genes in fluoroquinolone-resistant uropathogenic Escherichia coli: a first study from Kolkata, India. J Glob Antimicrob Resist, 2018. 14: p. 217–223. 10.1016/j.jgar.2018.03.009 [DOI] [PubMed] [Google Scholar]
- 30.Briales A., et al. , Prevalence of plasmid-mediated quinolone resistance determinants qnr and aac (6')-Ib-cr in Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Int J Antimicrob Agents, 2012. 39(5): p. 431–4. 10.1016/j.ijantimicag.2011.12.009 [DOI] [PubMed] [Google Scholar]
- 31.Robicsek A., et al. , qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob Agents Chemother, 2006. 50(8): p. 2872–4. 10.1128/AAC.01647-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X., et al. , Prevalence of qnr, aac (6')-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother, 2012. 56(6): p. 3423–7. 10.1128/AAC.06191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong J.Y., et al. , Detection of qnr in clinical isolates of Escherichia coli from Korea. Antimicrob Agents Chemother, 2005. 49(6): p. 2522–4. 10.1128/AAC.49.6.2522-2524.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel L., Leviandier C., and Nordmann P., Prevalence and genetic analysis of plasmid-mediated quinolone resistance determinants QnrA and QnrS in Enterobacteriaceae isolates from a French university hospital. Antimicrob Agents Chemother, 2006. 50(12): p. 3992–7. 10.1128/AAC.00597-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minarini L.A., et al. , Plasmid-mediated quinolone resistance determinants among enterobacterial isolates from outpatients in Brazil. J Antimicrob Chemother, 2008. 62(3): p. 474–8. 10.1093/jac/dkn237 [DOI] [PubMed] [Google Scholar]
- 36.Yugendran T. and Harish B.N., High incidence of plasmid-mediated quinolone resistance genes among ciprofloxacin-resistant clinical isolates of Enterobacteriaceae at a tertiary care hospital in Puducherry, India. PeerJ, 2016. 4: p. e1995 10.7717/peerj.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duerink D.O., et al. , Determinants of carriage of resistant Escherichia coli in the Indonesian population inside and outside hospitals. J Antimicrob Chemother, 2007. 60(2): p. 377–84. 10.1093/jac/dkm197 [DOI] [PubMed] [Google Scholar]
- 38.Pathak A., et al. , Factors associated with carriage of multi-resistant commensal Escherichia coli among postmenopausal women in Ujjain, India. Scand J Infect Dis, 2012. 44(12): p. 973–7. 10.3109/00365548.2012.697635 [DOI] [PubMed] [Google Scholar]
- 39.Butcher C.R., et al. , Risk Factors Associated with Community-Acquired Urinary Tract Infections Caused by Extended-Spectrum β-Lactamase-Producing Escherichia coli: a Systematic Review Current Epidemiology Reports, 2019. 6: p. 300–309. [Google Scholar]
- 40.Kraupner N., et al. , Selective concentration for ciprofloxacin resistance in Escherichia coli grown in complex aquatic bacterial biofilms. Environ Int, 2018. 116: p. 255–268. 10.1016/j.envint.2018.04.029 [DOI] [PubMed] [Google Scholar]
- 41.Bengtsson-Palme J., Kristiansson E., and Larsson D.G.J., Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev, 2018. 42(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutgersson C., et al. , Fluoroquinolones and qnr genes in sediment, water, soil, and human fecal flora in an environment polluted by manufacturing discharges. Environ Sci Technol, 2014. 48(14): p. 7825–32. 10.1021/es501452a [DOI] [PubMed] [Google Scholar]
- 43.Larsson D.G., de Pedro C., and Paxeus N., Effluent from drug manufactures contains extremely high levels of pharmaceuticals. J Hazard Mater, 2007. 148(3): p. 751–5. 10.1016/j.jhazmat.2007.07.008 [DOI] [PubMed] [Google Scholar]
- 44.Fick J., et al. , Contamination of surface, ground, and drinking water from pharmaceutical production. Environ Toxicol Chem, 2009. 28(12): p. 2522–7. 10.1897/09-073.1 [DOI] [PubMed] [Google Scholar]
- 45.Kristiansson E., et al. , Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One, 2011. 6(2): p. e17038 10.1371/journal.pone.0017038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bengtsson-Palme J., et al. , Shotgun metagenomics reveals a wide array of antibiotic resistance genes and mobile elements in a polluted lake in India. Front Microbiol, 2014. 5: p. 648 10.3389/fmicb.2014.00648 [DOI] [PMC free article] [PubMed] [Google Scholar]