Abstract
Background
Drug susceptibility testing for Mycobacterium tuberculosis (MTB) is difficult to perform in resource-limited settings where Acid Fast Bacilli (AFB) smears are commonly used for disease diagnosis and monitoring. We developed a simple method for extraction of MTB DNA from AFB smears for sequencing-based detection of mutations associated with resistance to all first and several second-line anti-tuberculosis drugs.
Methods
We isolated MTB DNA by boiling smear content in a Chelex solution, followed by column purification. We sequenced PCR-amplified segments of the rpoB, katG, embB, gyrA, gyrB, rpsL, and rrs genes, the inhA, eis, and pncA promoters and the entire pncA gene.
Results
We tested our assay on 1,208 clinically obtained AFB smears from Ghana (n = 379), Kenya (n = 517), Uganda (n = 262), and Zambia (n = 50). Coverage depth varied by target and slide smear grade, ranging from 300X to 12000X on average. Coverage of ≥20X was obtained for all targets in 870 (72%) slides overall. Mono-resistance (5.9%), multi-drug resistance (1.8%), and poly-resistance (2.4%) mutation profiles were detected in 10% of slides overall, and in over 32% of retreatment and follow-up cases.
Conclusion
This rapid AFB smear DNA-based method for determining drug resistance may be useful for the diagnosis and surveillance of drug-resistant tuberculosis.
Background
Globally, tuberculosis (TB) remains one of the top ten causes of death [1]. TB treatment has been complicated by a rise in drug resistance, with treatment success rates of approximately 55% for rifampicin-resistant (RR) and multidrug-resistant (MDR) TB (defined as resistance to both isoniazid (INH) and rifampicin (RIF)), and approximately 34% for extensively drug-resistant (XDR) TB (defined as MDR plus resistance to the fluoroquinolone (FQ) antibiotics and second-line injectable drugs) [1]. Commercially-available molecular assays, such as the GeneXpert® MTB/RIF (Cepheid, Sunnyvale, CA) and the GenoType MDRTBplus (Bruker-Hain Diagnostics, Nehren, Germany), have shown excellent sensitivity and specificity for detecting RIF and RIF and INH resistance respectively. However, cost and technical complexity of these tests may be prohibitive in resource-limited areas. Additionally, these assays do not identify pyrazinamide (PZA) resistance, which is strongly associated with the success or failure of World Health Organization (WHO)-recommended shortening regimens [2].
Widespread use of TB drug susceptibility testing (DST) would aid in prevention and treatment of drug-resistant TB. Universal DST for at least RIF is recommended in all TB cases [3]. DST enables expanded use of standardized short course MDR treatments, which require susceptibility testing to major components of treatment before treatment initiation [2]. Broad use of culture or molecular-based DST methods has been difficult to achieve in resource-limited settings. Global surveillance data indicates that DST for RIF was only performed for 24% of new TB cases and 70% of previously treated cases in 2017 [1]. DST for PZA has been especially difficult to perform because phenotypic testing is technically challenging and nucleic acid amplification based tests (NAAT) must be able to identify hundreds of different mutations to effectively detect most cases of PZA resistance [4]. Performing DST at centralized locations might improve access to some types of drug susceptibility results, taking advantage of the economy of scale. However, safe, inexpensive, and efficient methods to collect, store, and transport sputum samples from TB patients to these centralized facilities present some challenges.
Much of the world obtains a TB diagnosis using microscopic examination of Acid Fast Bacilli (AFB) stained sputum smears. Microscopy is also widely used for disease monitoring, even in regions that have adopted molecular testing for initial TB diagnoses [3]. AFB smears on glass slides are easily stored and shipped; they do not require temperature controlled storage and are not infectious. A number of studies have shown that DNA can be extracted from AFB smears and then used in TB DST analysis using molecular and other DNA sequencing based methods [5–12]. However, most of these studies involved small sample sizes (<100 slides) and only demonstrated the performance of their assays with one or two TB specific gene targets. Additionally, some studies used complex extraction and amplification protocols involving phenol-chloroform extractions, ethanol precipitations, and nested Polymerase Chain Reaction (PCR) amplifications requiring multiple primers for each target region, which would be impractical for widespread use.
We have developed a simple and effective method to extract Mycobacterium tuberculosis (MTB) DNA from clinically obtained AFB smears with sufficient quality for PCR amplification and next-generation DNA sequencing. We have also streamlined the process for PCR amplification of the isolated DNA for subsequent high-throughput sequencing of 17 regions of the MTB genome, enabling rapid detection of mutations associated with resistance to INH, RIF, ethambutol (EMB), PZA, FQ, and injectable aminoglycoside antibiotics. Here, we describe the method and assess the utility of our approach with 1,208 clinical AFB smears from Ghana, Kenya, Uganda, and Zambia. Since sequencing is becoming increasingly established as a definitive, reproducible, and reliable predictor of phenotypic DST, we also report the results of our new test desegregated by region and patient treatment status.
Methods
AFB direct sputum microscopy smears
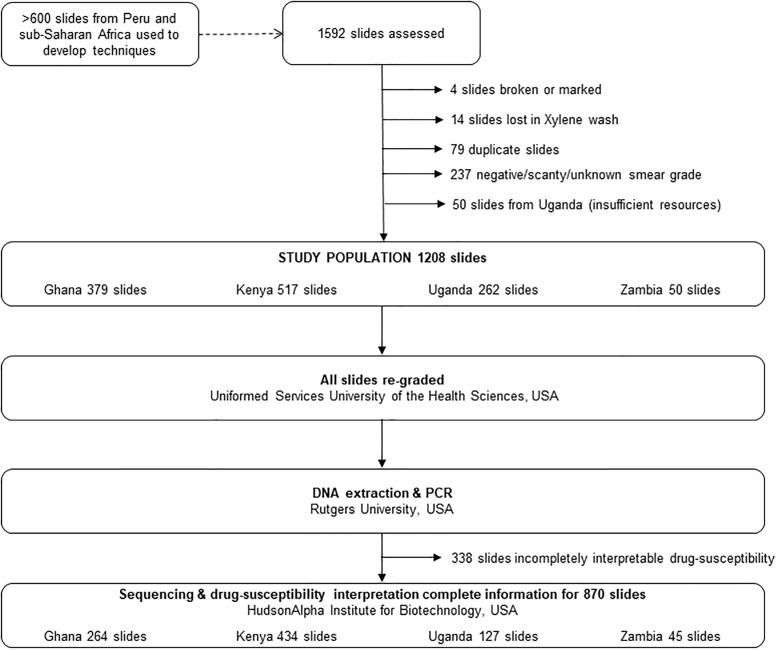
A convenience sample of clinically obtained direct AFB sputum smears (n = 2,227), collected between 2013 and 2016, was provided for evaluation with our assay. We utilized 635 of the slides to develop our assay. We processed an additional 1,208 smears (one per subject) with the final version of our assay, which is the focus of this manuscript. A summary of the study sample population and methods is presented in Fig 1. We excluded 384 smears from processing (Fig 1). We excluded scanty [1 to 9 AFB in 100 fields] smears because a preliminary evaluation of our assay showed suboptimal performance with such smears (S1 Fig). The slides included in this study consisted of 379 smears from the Ghanaian Armed Forces Health Care Beneficiary population seen at 37 Military Hospital, Accra, 517 smears from a PEPFAR population in the Western highlands of Kenya, 262 smears from the Uganda Peoples Defence Force and Ministry of Health hospitals and the Central Public Health Laboratory, Uganda, and 50 smears from Maina Soko Military Hospital serving the greater Lusaka area of Zambia.
Fig 1. Flow diagram schematic of study sample population and methods.
In each country, smears were prepared based on standard protocols recommended for clinical use by their national TB control programs. Types of stains used included Kinyoun, Auramine-Rhodamine, and Ziehl–Neelsen. All smears were centrally graded at the Uniformed Services University of the Health Sciences (USUHS) based on the International Union Against Tuberculosis and Lung Disease (IUATLD) grading system [13]; this required stripping and re-staining the Auramine-Rhodamine stained slides with Kinyoun stain (Remel, Lenexa, KS, USA). In the remainder of this manuscript, wherever a reference is made to stain type, we are specifically referring to the original stain type used on the smear.
Ethics reviews
This study was reviewed and approved as non-human subjects research by the Institutional Review Boards (IRB) at the USUHS and Rutgers New Jersey Medical School, and it was reviewed and designated as an exempt protocol by the IRBs at the Naval Health Research Center, Walter Reed Army Institute of Research, Kenya Medical Research Institute, Makerere University, 37 Military Hospital, and the University of Zambia. Therefore, informed consent was waived by all aforementioned institutions. All data was anonymized before access by this study.
Smear extraction and DNA isolation
Prior to extraction, the slides were washed with Xylene (Sigma Aldrich, Inc., Burlington, MA, USA) to remove immersion oil. For the extraction, 200 μl of Instagene Matrix (Bio-Rad Laboratories, Inc., Philadelphia, PA, USA) with 0.1% Triton X-100 (Sigma Aldrich, Inc., Burlington, MA, USA) was aliquoted into 1.5 mL Eppendorf tubes. Without disturbing the Chelex pellet, up to 100 μL of the Instagene Matrix and Triton X-100 solution was aspirated from the tube and dispensed onto the smear. The smear was then scraped off using a razor blade (Fisherbrand™ Razor Blades, Thermo Fisher Scientific, Inc., Hampton, NH, USA), aspirated off the slide, and transferred into the Eppendorf tube containing the Chelex pellet. To isolate the DNA, the extracted smear material was pulse vortexed at high speed for 30 seconds, then boiled for 20 minutes at 90°C, pulse vortexed on medium speed for 10 seconds, and centrifuged for 5 minutes at 15,000 rpm using Eppendorf Centrifuge 5424 (Eppendorf, Hamburg, Germany). The supernatant was transferred to a clean Eppendorf tube and purified using Qiagen QIAamp DNA Micro Kit (Qiagen, Inc., Germantown, MD, USA) according to manufacturer’s instructions for cleanup of genomic DNA. The purified DNA was eluted in 45 μL of AE elution buffer from the kit.
Amplification targets and primers
The drug resistance phenotypes, gene target regions, and the associated segment names and primers are shown in Table 1. These target regions were selected to cover single nucleotide polymorphisms (SNPs) which have been reported to be associated with drug resistance in previous studies [14–16]. Mutation H57D in pncA, which occurs naturally in M. bovis, was excluded from downstream SNP analyses in this study. For pncA, the entire gene, including upstream and downstream flanking regions were targeted for amplification (Table 1).
Table 1. Mycobacterium tuberculosis gene regions targeted for detection of drug resistance associated mutations and associated primers.
| Drug | Gene | Segment Name | Targeted Nucleotide Positions | Primer Direction | Primer Sequence (5’ to 3’) |
|---|---|---|---|---|---|
| Rifampicin | rpoB | rpoB | 1215 to 1383 | Forward | GATCACACCGCAGACGTTGA |
| Reverse | ACGCTCACGTGACAGACCG | ||||
| Isoniazid | inhA promoter | inhA promoter | nucleotide -160 to 44 of mabA gene | Forward | CCCAGAAAGGGATCCGTCAT |
| Reverse | GATACGAATGGGGGTTTGGC | ||||
| katG | katG | 809 to 992 | Forward | CTGTTGTCCCATTTCGTCGG | |
| Reverse | GGCGGTCACACTTTCGGTAA | ||||
| Ethambutol | embB | emb10 | 841 to 1017 | Forward | GCCGTGGTGATATTCGGCTT |
| Reverse | CAGCGCCAGCAGGTTGTAAT | ||||
| emb20 | 1156 to 1299 | Forward | GCGGCGGCCATGGTCTTG | ||
| Reverse | CAGCGCCGCCGGTGTGA | ||||
| emb40 | 1451 to 1550 | Forward | CCGCCGGCACCGTCATCCTGA | ||
| Reverse | GCCTGGCTCGGCCCGATTTTG | ||||
| Fluoroquinolones/ Moxifloxacin/ Ofloxacin | gyrA | gyrA | 159 to 395 | Forward | CCGGGTGCTCTATGCAATGT |
| Reverse | GCTTCGGTGTACCTCATCGC | ||||
| Fluoroquinolones | gyrB | gyrB | 1390 to 1639 | Forward | GAGTTGGTGCGGCGTAAGAG |
| Reverse | CCGTGATGATCGCCTGAACT | ||||
| Kanamycin | eis | eis promoter | -81 to 34 | Forward | CGTCCTCGGTCGGGCTACACAG |
| Reverse | GCATCGCGTGATCCTTTGCCAGAC | ||||
| Streptomycin | rpsL | rpsL0 | 54 to 170 | Forward | GGTCAAGACCGCGGCTCTGA |
| Reverse | AACTTCACGCGGGCAACCTTC | ||||
| rpsL1 | 183 to +53 | Forward | CGAGGTCACGGCGTACATTC | ||
| Reverse | GTAGACCGGGTCGTTGACCA | ||||
| rrs | rrs10 | 465 to 689 | Forward | TCGGATTGACGGTAGGTGGA | |
| Reverse | CATTCCACCGCTACACCAGG | ||||
| Streptomycin /Amikacin/Capreomycin/Kanamycin | rrs30 | 1368 to +59 | Forward | ATACGTTCCCGGGCCTTGTA | |
| Reverse | AGACAAGAACCCCTCACGGC | ||||
| Pyrazinamide | pncA | pncA3 | -69 to 154 | Forward | CAACAGTTCATCCCGGTTCG |
| Reverse | TCGGTATTGCCACCGATCAT | ||||
| pncA2 | 111 to 337 | Forward | CCAAGCCATTGCGTACCG | ||
| Reverse | ATCCCAGTCTGGACACGTCG | ||||
| pncA1 | 245 to 454 | Forward | CGTTCTCGTCGACTCCTTCG | ||
| Reverse | AGCGGCGGACTACCATCAC | ||||
| pncA0 | 392 to +29 | Forward | TGTGGAAGTCCTTGGTTGCC | ||
| Reverse | CCCTATATCTGTGGCTGCCG |
Polymerase Chain Reactions (PCR)
All samples were amplified in 20 μL reactions. Smears graded 1+ were amplified in uniplex reactions containing 2 μl of target DNA to increase PCR efficacy and sequencing depth for these paucibacillary samples. For smears graded 2+ or 3+ all gene target sequences were amplified in duplexed reactions using 3 or 4 μL of target DNA per reaction, except for gyrB, which was only amplified in a uniplex reaction to maintain amplification efficiency. The PCR mix consisted of the following: 2.5 mM magnesium chloride (MgCl2)), 0.25 mM deoxyribonucleotides, 5% glycerol, 1X PCR buffer without MgCl2, 1 unit Jumpstart Taq DNA polymerase, and 0.5 μM primers (all reagents from Sigma Aldrich, Inc., Burlington, MA, USA). Samples were PCR-amplified in Roche LightCycler® 480 System (Roche Molecular Systems, Inc., Branchburg, NJ, USA) and Applied Biosystems Prism 7900HT Fast Real-Time PCR System (Applied Biosystems, Foster City, USA). For duplex reactions, primers were paired as follows: rpoB-pncA3, rrs30-rpsL1, emb10-katG, emb20-eis, emb40-rpsL0, inhA-pncA2, gyrA-pncA0, and rrs10-pncA1. All PCR products for each sample were pooled post amplification. For amplification, samples were denatured at 95°C for 1 minute followed by 40 cycles of touchdown PCR with the following parameters: 1) 10 cycles with 95°C denaturation for 15 seconds, annealing for 15 seconds starting at 70°C then lowered by 1°C with each subsequent cycle, and extension at 72°C for 30 seconds; 2) 30 cycles with 95°C denaturation for 15 seconds, annealing at 60°C for 15 seconds, and extension at 72°C for 30 seconds. With each PCR batch, a negative (dH2O) control, a wild type positive control (H37Rv genomic DNA), and genomic DNA extracted from a mutant positive control drug-resistant strain (TB-TDR-0114 or TB-TDR-0115)) were included [17]. These controls were also included in the downstream sequencing and SNP detection pipeline.
Sequencing
The samples were sequenced at the HudsonAlpha GSL (Huntsville, AL, USA) using an Ilumina MiSeq™ platform (Illumina, Inc., San Diego, CA, USA) (S1 Appendix). Sequencing results have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (BioProject numbers: PRJNA608715, PRJNA608724).
Data analysis
Reads were aligned to a reference sequence from the H37Rv strain of TB using an in-house bioinformatics pipeline (S2 Appendix). For each DST target region of a given slide, results were classified as interpretable if coverage depth of 20X or more reads was obtained; a mutation frequency of 80% or greater was considered sufficient for calling SNPs. If coverage depth of a DST target region was less than 20X, the results for that target region were classified as un-interpretable. Prevalence and corresponding exact binomial confidence intervals of mutations associated with resistance to first- and second-line anti-tuberculosis drugs were estimated for all slides with interpretable results for all gene targets.
Results
Slide characteristics are summarized by country of origin in Table 2. The 1,208 processed slides consisted of 282 (23.1%) 1+ smears, 352 (29.1%) 2+ smears, and 574 (47.5%) 3+ smears. All Kenya smears and 58% of Ghana smears were from adult patients. Information on adult or child status was missing for Uganda and Zambia smears. Over 61% of the smears were classified as new TB cases. The majority of the smears (42.8%) were from Kenya (Table 2). Ziehl-Neelsen was the most frequently used stain (Table 2).
Table 2. Sample characteristics by country.
| Sample Characteristics | Ghana (%) | Kenya (%) | Uganda (%) | Zambia (%) | Total (%) |
|---|---|---|---|---|---|
| Sample Size | 379 (31.4) | 517 (42.8) | 262 (21.7) | 50 (4.1) | 1208 (100) |
| Smear Grade | |||||
| 1+ | 72 (19.0) | 174 (33.7) | 21 (8.0) | 15 (30.0) | 282 (23.3) |
| 2+ | 62 (16.4) | 141 (27.3) | 131 (50.0) | 18 (36.0) | 352 (29.1) |
| 3+ | 245 (64.6) | 202 (39.1) | 110 (42.0) | 17 (34.0) | 574 (47.5) |
| Stain Typea | |||||
| Ziehl Neelsen | 379 (100) | 0 | 49 (18.7) | 0 | 428 (35.4) |
| Kinyoun | 0 | 341 (66.0) | 0 | 50 (100) | 391 (32.4) |
| Auramine/Rhodamine | 0 | 176 (34.0) | 207 (79.0) | 0 | 383 (31.7) |
| Unknown | 0 | 0 | 6 (2.3) | 0 | 6 (0.5) |
| Treatment Status | |||||
| New case | 172 (45.4) | 352 (68.1) | 221 (84.4) | 0 | 745 (61.7) |
| Follow Up | 0 | 124 (24.0) | 35 (13.4) | 0 | 158 (13.1) |
| Retreatment | 0 | 38 (7.4) | 4 (1.5) | 0 | 43 (3.6) |
| Unknown | 207 (54.6) | 3 (0.6) | 2 (0.8) | 50 (100) | 262 (21.7) |
aStain type specified in this table is the original type of stain used on the smear at the clinical site where sample was collected.
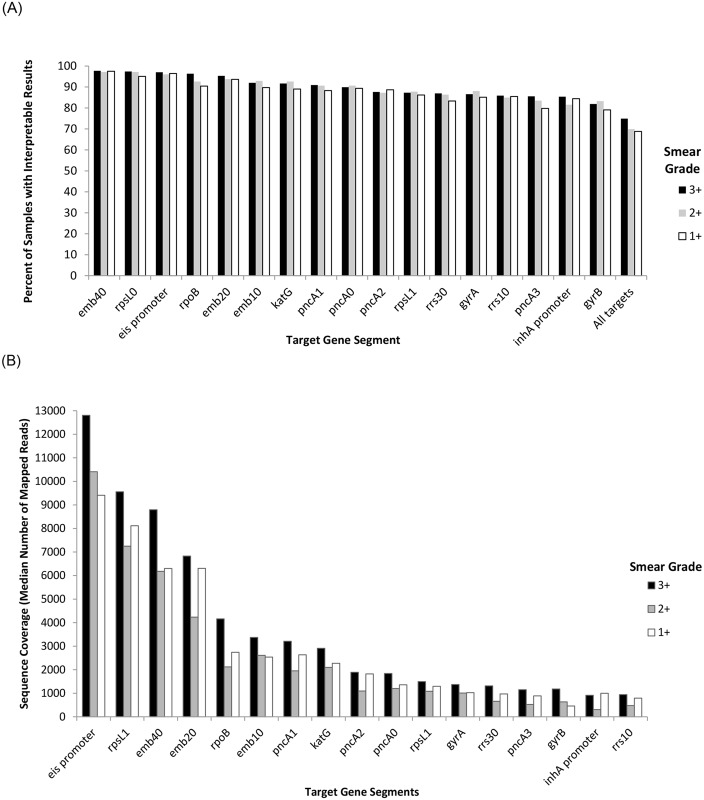
Coverage depth varied by target region. The percentage of slides per smear grade with interpretable results overall and per target gene segments is reported in Fig 2A. With the exception of gyrB (interpretable results obtained in 79% of 1+ smears), interpretable results were obtained for any given target gene segment in 80% or more of slides, regardless of smear grade (Fig 2A). Interpretable results was obtained for all targets in 194 (68.8%) 1+ smears, 246 (69.9%) 2+ smears, 430 (74.9%) 3+ smears, and 870 (72%) slides overall. The number of mapped reads varied by target gene segment and slide smear grade, ranging from approximately 300X for rrs10 to over 12000X for the eis promoter on average. Median coverage per target region, stratified by smear grade, is shown in Fig 2B. Coverage depth also varied by stain type. Interpretable results were obtained for all targeted gene segments in over 94% of Kinyoun stained smears, but only 64% of Ziehl-Neelsen stained smears, and 59% of Auramine/Rhodamine stained smears (S1 Table).
Fig 2. Percent of samples per smear grade with interpretable results and median number of mapped reads per target gene segment.
A) Percent of samples per smear grade with interpretable results (coverage depth of 20X or greater) for all and specified target gene segments; B) Sequence coverage in median number of mapped reads per target gene segment, stratified by smear grade.
A summary of all drug resistance associated mutations detected in all gene targets are presented in Table 3. RIF resistance associated mutations in rpoB were detected in 6.3% of new cases, 3.3% of follow-up cases (patients under initial treatment), 20.0% of retreatment cases (S2 Table), and 6.1% of slides overall (Table 3). The highest rate of RIF resistance associated mutations were detected in slides from Uganda (18.3%), followed by slides from Zambia (8.51%). Over 85% of RIF resistance associated mutations occurred in codon 445 (alternative numbering system: 526) of rpoB (Table 3).
Table 3. Summary of drug resistance associated mutations detected in direct AFB smears and total number of drug resistant smears from Ghana, Kenya, Uganda, and Zambia, by drug typea.
| Drug | Gene Segment | Nucleotide (Reference/Mutant) | Amino Acid Alterationb | Ghana | Kenya | Uganda | Zambia | Total |
|---|---|---|---|---|---|---|---|---|
| Rifampicin | rpoB | 1289 CTG/ CCG | L430P (L511P) | 0/345 | 0/496 | 2/246 | 0/47 | 2/1134 |
| 1295 CAA/ CCA | Q432P (Q513P) | 1/345 | 0/496 | 0/246 | 0/47 | 1/1134 | ||
| 1304 GAC/ GTC | D435V (D516V) | 0/345 | 1/496 | 0/246 | 1/47 | 2/1134 | ||
| 1333 CAC/GAC | H445D (H526D) | 1/345 | 0/496 | 41/246 | 3/47 | 45/1134 | ||
| 1333 CAC/TAC | H445Y (H526Y) | 2/345 | 10/496 | 0/246 | 0/47 | 12/1134 | ||
| 1333 CAC/AAC | H445N (H526N) | 1/345 | 0/496 | 0/246 | 0/47 | 1/1134 | ||
| 1349 TCG/ TTG | S450L (S531L) | 2/345 | 0/496 | 2/246 | 0/47 | 4/1134 | ||
| 1355 CTG/ CCG | L452P (L533P) | 2/345 | 0/496 | 0/246 | 0/47 | 2/1134 | ||
| Total Rifampicin Resistant Smears (%) | 9/345 (2.61) | 11/496 (2.22) | 45/246 (18.29) | 4/47 (8.51) | 69/1134 (6.08) | |||
| Isoniazid | inhA Promoter | -15 C/T | Promoter | 3/326 | 0/476 | 2/166 | 0/47 | 5/1015 |
| katG | 944 AGC/ACC | S315T | 24/337 | 14/501 | 16/218 | 6/47 | 60/1103 | |
| Total Isoniazid Resistant Smears (%) | 27/316 (8.54) | 14/473 (2.96) | 18/168 (10.71) | 6/47 (12.77) | 65/1004 (6.47) | |||
| Streptomycin | rpsL0 | 128 AAG/AGG | K43R | 14/365 | 13/512 | 0/242 | 0/50 | 27/1169 |
| rpsL1 | 263 AAG/AGG | K88R | 5/312 | 0/490 | 3/204 | 0/47 | 8/1053 | |
| rrs10 | 513 A/C | 0/319 | 3/480 | 0/186 | 0/48 | 3/1033 | ||
| rrs10 | 516 C/T | 0/319 | 0/480 | 2/186 | 0/48 | 2/1033 | ||
| Total Streptomycin Resistant Smears (%) | 19/292 (6.51) | 16/468 (3.42) | 5/179 (2.79) | 0/47 | 40/986 (4.06) | |||
| Ethambutol | embB10 | 916 ATG/GTG | M306V | 2/343 | 13/500 | 4/218 | 1/47 | 20/1108 |
| embB10 | 918 ATG/ATA | M306I | 1/343 | 0/500 | 2/218 | 0/47 | 3/1108 | |
| embB20 | 1190 GGC/GAC | G406D | 0/360 | 0/505 | 3/228 | 1/47 | 4/1140 | |
| Total Ethambutol Resistant Smears (%) | 3/339 (0.88) | 13/496 (2.62) | 7/218 (3.21)c | 2/47 (4.26) | 25/1100 (2.27)c | |||
| Fluoroquinolones | gyrA | 281 GAC/GGC | D94G | 2/314 | 12/486 | 0/200 | 0/47 | 14/1047 |
| Total Fluoroquinolones Resistant Smears (%)g | 2/281 (0.71) | 12/459 (2.61) | 0/180 | 0/46 | 14/966 (1.45) | |||
| Moxifloxacin/ Ofloxacin | gyrA | 281 GAC/GGC | D94G | 2/314 | 12/486 | 0/200 | 0/47 | 14/1047 |
| Total Moxifloxacin/ Ofloxacin Resistant Smears (%) | 2/314 (0.64) | 12/486 (2.47) | 0/200 | 0/47 | 14/1047 (1.34) | |||
| Pyrazinamide | pncA3 | -11 A/G | Promoter | 0/319 | 10/475 | 0/170 | 0/46 | 10/1010 |
| pncA3 | 11 TTG/TCG | L4S | 0/319 | 1/475 | 0/170 | 0/46 | 1/1010 | |
| pncA3 | 35 GAC/GCC | D12A | 0/319 | 0/475 | 1/170 | 0/46 | 1/1010 | |
| pncA3 | 37 TTC/CTC | F13L | 1/319 | 0/475 | 0/170 | 0/46 | 1/1010 | |
| pncA3 | 104 CTG/CGG | L35R | 0/319 | 0/475 | 0/170 | 2/46 | 2/1010 | |
| pncA3 | 137 GCA/GTA | A46V | 1/319 | 0/475 | 0/170 | 0/46 | 1/1010 | |
| pncA2 | 151 CAC/TAC | H51Y | 1/327 | 0/487 | 0/199 | 0/47 | 1/1060 | |
| pncA2 | 185 CCG/CAG; 185 CCG/CTG | P62Q; P62L Mixture | 1/327 | 0/487 | 0/199 | 0/47 | 1/1060 | |
| pncA2 | 188 GAC/GGC | D63G | 0/327 | 0/487 | 2/199 | 0/47 | 2/1060 | |
| pncA2 | 202 TGG/CGG | W68R | 1/327 | 0/487 | 0/199 | 0/47 | 1/1060 | |
| pncA1 | 322 GGA/AGA | G108R | 1/335 | 0/496 | 0/211 | 0/48 | 1/1090 | |
| Total Pyrazinamide Resistant Smears (%) | 6/302 (1.99) | 11/467 (2.36) | 3/160 (1.88) | 2/46 (4.35) | 22/975 (2.26) |
a A coverage depth cut-off of 20X and a frequency cut-off of 80% was used for all reported single nucleotide polymorphisms. For each target gene segment, the number of smears with SNPs out of the total number of smears with 20X or greater coverage for the given target gene segment are reported per country. For each country and in total, the percent of smears with resistance to a given drug were calculated by dividing the number of smears with at least one mutation associated with resistance to the given drug by the total number of smears in which all relevant target gene segments (listed in Table 1) were successfully screened plus any smears in which at least one associated target gene segment met the coverage cut-off and contained a drug resistance associated mutation.
b For rpoB0, the amino acid number is presented in the MTB numbering system followed by the alternative numbering system in parentheses.
c Two samples from Uganda contained a drug resistant associated mutation in both emb10 and emb20 gene segments. The numerator has been adjusted to ensure we do not double count these samples in calculating percent of etambutol resistant smears from Uganda and overall.
INH resistance associated mutations in katG and/or inhA were detected in 6% of slides overall, with the highest rate being among Zambia slides (12.8%), followed by Uganda (10.7%) (Table 3). Most (92%) of the observed INH resistance associated mutations were S315T in katG (Table 3). Overall, fluoroquinolones, moxifloxacin, and ofloxacin resistance associated mutations in gyrA were observed in over 1% of slides (Table 3). PZA resistance associated mutations in pncA were observed in over 2% of slides overall (Table 3). Majority of the distinct pncA mutations were only detected in a single slide. However, the -11 A/G mutation in the pncA promoter was observed in 2.4% of Kenya slides. Detected pncA SNPs for which there currently is little to no evidence of an association with drug resistance are reported in S3 Table. We did not detect any SNPs in our wild-type positive controls or negative controls.
Prevalence estimates and associated 95% confidence intervals for detected drug resistance mutation profiles in smears with interpretable results for all DST gene targets, stratified by country are presented in Table 4. Overall, drug resistance associated SNPs were detected in 88/870 (Estimate: 10.11%; 95% CI: 8.19%, 12.31%) of the slides with interpretable results for all DST gene targets, with mutation rates being highest in slides from Zambia (Table 4). Approximately 58% of the slides in which a drug resistance associated mutation was detected contained a SNP associated with mono-resistance to RIF, INH, or SM, with the most frequent mutation being associated with INH resistance (Table 4). A MDR mutation profile with resistance to at least RIF and INH was also observed in 8% (95% CI: 2.48%, 21.22%) of smears from Zambia and in 1.8% (95% CI: 1.05%, 2.97%) of smears overall (Table 4). No XDR mutation profiles were identified.
Table 4. Prevalence and 95% confidence intervals for detected drug resistance mutation profiles in smears with interpretable results for all target gene segments, stratified by countrya.
| Mutation Based Drug Resistance Profile | Ghana | Kenya | Uganda | Zambia | Total | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | |
| Mono-resistance | ||||||||||
| RIF only | 1/264 (0.38) | 0.01, 2.09 | 10/434 (2.30) | 1.11, 4.20 | 6/127 (4.72) | 1.75, 10.00 | 0/45 (0.00) | 0.00, 7.87 | 17/870 (1.95) | 1.14, 3.11 |
| INH only | 14/264 (5.30) | 2.93, 8.74 | 0/434 | 0.00, 0.85 | 6/127 (4.72) | 1.75, 10.00 | 2/45 (4.44) | 0.54, 15.15 | 22/870 (2.53) | 1.59, 3.80 |
| SM only | 9/264 (3.41) | 1.57, 6.37 | 2/434 (0.46) | 0.06, 1.66 | 0/127 (0.00) | 0.00, 2.86 | 0/45 (0.00) | 0.00, 7.87 | 11/870 (1.26) | 0.63, 2.25 |
| PZA only | 0/264 (0.00) | 0.00, 1.39 | 0/434 (0.00) | 0.00, 0.85 | 0/127 (0.00) | 0.00, 2.86 | 1/45 (2.22) | 0.06, 11.77 | 1/870 (0.11) | 0.00, 0.64 |
| Multi-drug Resistance | ||||||||||
| RIF and INH | 4/264 (1.52) | 0.41, 3.83 | 0/434 (0.00) | 0.00, 0.85 | 1/127 (0.79) | 0.02, 4.31 | 3/45 (6.67) | 1.40, 18.27 | 8/870 (0.92) | 0.40, 1.80 |
| RIF, INH, and one or more of the following: EMB, SM, PZA | 3/264 (1.14) | 0.23, 3.28 | 1/434 (0.23) | 0.01, 1.28 | 3/127 (2.36) | 0.49, 6.75 | 1/45 (2.22) | 0.06, 11.77 | 8/870 (0.92) | 0.40, 1.80 |
| Poly-resistance | ||||||||||
| Two or more of the following: INH, SM, EMB, PZA | 6/264 (2.27) | 0.84, 4.88 | 0/434 (0.00) | 0.00, 0.85 | 3/127 (2.36) | 0.49, 6.75 | 1/45 (2.22) | 0.06, 11.77 | 10/870 (1.15) | 0.55, 2.10 |
| FQ, MXF, and OFL | 1/264 (0.38) | 0.01, 2.09 | 0/434 (0.00) | 0.00, 0.85 | 0/127 (0.00) | 0.00, 2.86 | 0/45 (0.00) | 0.00, 7.87 | 1/870 (0.11) | 0.00, 0.64 |
| INH, EMB, SM, PZA, FQ, MXF, OFL | 0/264 (0.00) | 0.00, 1.39 | 10/434 (2.30) | 1.11, 4.20 | 0/127 (0.00) | 0.00, 2.86 | 0/45 (0.00) | 0.00, 7.87 | 10/870 (1.15) | 0.55, 2.10 |
| Total | 38/264 (14.39) | 10.39, 19.22 | 23/434 (5.30) | 3.99, 7.85 | 19/127 (14.96) | 9.25, 22.37 | 8/45 (17.78) | 8.00, 32.05 | 88/870 (10.11) | 8.19, 12.31 |
aAbbreviations: RIF = rifampicin; INH = isoniazid; SM = streptomycin; PZA = pyrazinamide; EMB = ethambutol; FQ = fluoroquinolone; MXF = moxifloxacin; OFL = ofloxacin; CI = confidence interval.
Presented in Table 5 are overall and country specific prevalence estimates and 95% confidence intervals for phenotypic interpretation of observed mutations in smears with interpretable results for all DST gene targets, stratified by case treatment status. Among smears with interpretable results for all gene targets, 7.8% of new cases, 4.1% of follow up cases, and 28.1% of retreatment cases had mutation profiles for mono-resistance, MDR, or poly-resistance.
Table 5. Overall and country specific prevalence and 95% confidence intervals for phenotypic interpretation of observed mutations in smears with interpretable results for all gene targets, by case treatment status.
| Case Treatment Status by Country | No Drug Resistance | Mono-Resistance | Multi-Drug Resistance | Poly-Resistance | Any Drug Resistant Smears | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | Prevalence (%) | 95% CI | |
| Ghana | ||||||||||
| New | 111/125 (88.80) | 81.92, 93.74 | 13/125 (10.40) | 5.65, 17.13 | 1/125 (0.80) | 0.02, 4.38 | 0/125 (0.00) | 0.00, 2.91 | 14/125 (11.20) | 6.26, 18.08 |
| Unknown | 115/139 (82.73) | 75.41, 85.61 | 11/139 (7.91) | 4.02, 13.72 | 6/139 (4.32) | 1.60, 9.16 | 7/139 (5.04) | 2.05, 10.10 | 24/139 (17.27) | 11.39, 24.59 |
| Total | 226/264 (85.61) | 80.78, 89.61 | 24/264 (9.09) | 5.91, 13.22 | 7/264 (2.65) | 1.07, 5.39 | 7/264 (2.65) | 1.07, 5.39 | 38/264 (14.39) | 10.39, 19.22 |
| Kenya | ||||||||||
| New | 287/297 (96.63) | 93.90, 98.37 | 4/297 (1.35) | 0.37, 3.41 | 1/297(0.34) | 0.01, 1.86 | 5/297 (1.68) | 0.55, 3.88 | 10/297 (3.37) | 1.63, 6.10 |
| Follow-up | 100/104 (96.15) | 90.44, 98.94 | 1/104 (0.96) | 0.02, 5.10 | 0/104 (0.00) | 0.00, 3.48 | 3/104 (2.88) | 0.60, 8.20 | 4/104 (3.85) | 1.06, 9.56 |
| Retreatment | 23/31 (74.19) | 55.39, 88.14 | 7/31 (22.58) | 9.59, 41.10 | 0/31 (0.00) | 0.00, 11.22 | 1/31 (3.23) | 0.08, 16.70 | 8/31 (25.81) | 11.86, 44.61 |
| Unknown | 1/2 (50.00) | 1.26, 98.74 | 0/2 (0.00) | 0.00, 84.19 | 0/2 (0.00) | 0.00, 84.19 | 1/2 (50.00) | 1.26, 98.74 | 1/2 (50.00) | 1.26, 98.74 |
| Total | 411/434 (94.70) | 92.15, 96.61 | 12/434 (2.76) | 1.44, 4.78 | 1/434 (0.23) | 0.01, 1.28 | 10/434 (2.30) | 1.11, 4.20 | 23/434 (5.30) | 3.39, 7.85 |
| Uganda | ||||||||||
| New | 90/107 (84.11) | 75.78, 90.46 | 12/107 (11.21) | 5.93, 18.77 | 3/107 (2.80) | 0.58, 7.98 | 2/107 (1.87) | 0.23, 6.59 | 17/107 (15.89) | 9.54, 24.22 |
| Follow-up | 17/18 (94.44) | 72.71, 99.86 | 0/18 (0.00) | 0.00, 18.53 | 1/18 (5.56) | 0.14, 27.29 | 0/18 (0.00) | 0.00, 18.53 | 1/18 (5.56) | 0.14, 27.29 |
| Retreatment | 0/1 (0.00) | 0.00, 97.50 | 0/1 (0.00) | 0.00, 97.50 | 0/1(0.00) | 0.00, 97.50 | 1/1 (100.00) | 2.50, 100.00 | 1/1 (100.00) | 2.50, 100.00 |
| Unknown | 1/1 (100) | 2.50, 100.00 | 0/1 (0.00) | 0.00, 97.50 | 0/1(0.00) | 0.00, 97.50 | 0/1 (0.00) | 0.00, 97.50 | 0/1(0.00) | 0.00, 97.50 |
| Total | 108/127 (85.04) | 77.63, 90.75 | 12/127 (9.45) | 4.98, 15.92 | 4/127 (3.15) | 0.86, 7.87 | 3/127 (2.36) | 0.49, 6.75 | 19/127 (14.96) | 9.25, 22.37 |
| Zambia | ||||||||||
| Unknown | 37/45 (82.22) | 67.95, 92.00 | 3/45 (6.67) | 1.40, 18.27 | 4/45 (8.89) | 2.48, 21.22 | 1/45 (2.22) | 0.06, 11.77 | 8/45 (17.78) | 8.00, 32.05 |
| Total | 37/45 (82.22) | 67.95, 92.00 | 3/45 (6.67) | 1.40, 18.27 | 4/45 (8.89) | 2.48, 21.22 | 1/45 (2.22) | 0.06, 11.77 | 8/45 (17.78) | 8.00, 32.05 |
| Overall | ||||||||||
| New | 488/529 (92.25) | 89.63, 94.38 | 29/529 (5.48) | 3.70, 7.78 | 5/529 (0.95) | 0.31, 2.19 | 7/529 (1.32) | 0.53, 2.71 | 41/529 (7.75) | 5.62, 10.37 |
| Follow-up | 117/122 (95.90) | 90.69, 98.66 | 1/122 (0.82) | 0.02, 4.48 | 1/122 (0.82) | 0.02, 4.48 | 3/122 (2.46) | 0.51, 7.02 | 5/122 (4.10) | 1.34, 9.31 |
| Retreatment | 23/32 (71.88) | 53.25, 86.25 | 7/32 (21.88) | 9.28, 39.97 | 0/32 (0.00) | 0.00, 10.89 | 2/32 (6.25) | 0.77, 20.81 | 9/32 (28.13) | 13.75, 46.75 |
| Unknown | 153/187 (81.82) | 75.53, 87.07 | 14/187 (7.49) | 4.15, 12.24 | 10/187 (5.35) | 2.59, 9.61 | 9/187 (4.81) | 2.22, 8.94 | 33/187 (17.65) | 12.47, 23.88 |
| Total | 782/870 (89.89) | 87.69, 91.81 | 51/870 (5.86) | 4.40, 7.64 | 16/870 (1.84) | 1.05, 2.97 | 21/870 (2.41) | 1.50, 3.67 | 88/870 (10.11) | 8.19, 12.31 |
We re-sequenced a subset of the slides using Sanger sequencing to confirm the accuracy of our MiSeq based approach. This included 16 slides with a wild type pncA gene, 61 with rpoB mutations, 18 with KatG mutations, and 27 additional slides with mutations in other gene targets. Our Sanger sequencing results matched the results of our primary study in every case except for one slide where an H445D mutation was detected in rpoB by MiSeq (coverage depth: 1700X; mutation frequency: 99%) but not Sanger sequencing (S1, S2 and S3 Data).
Discussion
We have demonstrated that DNA of sufficient quality for PCR amplification and next generation DNA sequencing can be isolated from AFB stained direct sputum smears and tested for mutations associated with resistance to all first and several second line anti-tuberculosis drugs. The DNA isolation method we have developed is simple and rapid and does not require much technical expertise. The samples used in this study were comprised of a diverse set of clinically obtained smears in terms of smear grade, AFB stain type, and geographic origin. Although the percentage of slides that met the coverage cut-off of 20X was similar for all target gene segments, the median number of reads for each target varied. This indicates that while sufficient DNA can be isolated from 1+ to 3+ smears to amplify and sequence all gene segments tested in this study, certain targets (e.g., eis promoter) are amplified more efficiently than others by the selected primer pairs. Kinyoun stained smears enabled better sequencing coverage compared to other stain types. However, the poor performance with Auramine/Rhodamine stained smears could in part be due to subsequent stripping and counterstaining of these slides with Kinyoun stain at USUHS, which may have resulted in DNA loss. Given that each country mainly used one type of stain, the variation observed in coverage depth by stain type could also be due to site-specific factors (Table 2).
Our assay demonstrated well-established associations of drug-resistance with geographical region and TB treatment status. Although the number of retreatment cases in our study was small, the rate of RIF resistance associated mutations we observed in retreatment cases as compared to new cases was similar to trends currently reported for the continent of Africa, where the rate of drug resistance mutations in retreatment cases is almost four times that observed in new TB cases [3]. The countries from which slides were analyzed in our study are on the WHO’s list of top high burden countries for TB (Kenya; Zambia), MDR-TB (Kenya), and/or TB-HIV concurrent infections (Zambia; Ghana; Uganda) [3]. Comprehensive genotypic drug resistance surveillance data for these countries is currently limited. Based on national surveillance data, less than 2% of new TB cases are drug-resistant in all countries from which slides were included in our study [3]. Although our sample size for each country was small and subject treatment status was only known for a subset of our samples, the estimated prevalence of drug resistance among new TB cases in our study was over ten times higher than reported national rates for Ghana and Uganda. Among Kenyan smears in our sample, the estimated drug resistance rate was almost three times higher than the reported national rate. It could be that a greater number of drug-resistant cases were evaluated at the facilities that we received our samples from because they are centralized laboratories. The drug resistance rate estimated among Zambian smears in our sample was similar to the national rate of 18% reported for relapse cases in Zambia [3]. However, the subject treatment status associated with the Zambian slides in our study was unknown. Similar to findings from previous phenotypic and molecular drug resistance surveillance studies [18–26], we found INH mono-resistance associated mutations to be more common in samples overall than RIF mono-resistance or RIF-INH dual resistance. Noted is that Ghana likely has both M. africanum and M. tuberculosis infection in clinically identified TB subjects, cited as up to 40% in West Africa [24]. This may be relevant to our study, as M. africanum has been reported to have less drug resistance compared to M. tuberculosis in Ghana [25]. Differentiation between M. africanum and M. tuberculosis was not possible with the target regions we sequenced.
A limitation of this study is that our specimens were obtained from central or regional reference laboratories and thus may not be generalizable to the entire population. Another limitation is that the observed drug resistance mutations could not be confirmed via phenotypic DST, given that the study was conducted retrospectively. Nevertheless, we were able to validate the in-house bioinformatics pipeline that was used to analyze our samples by reanalyzing the data for 384 slides with the previously published ASAP pipeline [26]. We observed no discordance between the variant calls made by our pipeline and the ASAP pipeline when adjusting coverage cut-off to 20X and SNP frequency to 80% (S3 Appendix). Also, limiting our screening process only to previously published drug-resistant mutations may have led to an underestimation of drug resistance in our samples.
Among the slides processed by Sanger and MiSeq sequencing, one discordant sample was observed, where rpoB mutation H445D (alternate naming system: H526D) was detected by MiSeq but not by Sanger sequencing. Given the length of the rpoB target, coverage of codon 445 was only available from the forward direction from Sanger sequencing. It is possible that the rpoB SNP was missed by Sanger sequencing in this sample. Alternatively, the portion of the discordant sample amplified by MiSeq sequencing may have been contaminated. Regardless, the overall concordance between the two approaches was very high. Although precautions were taken to prevent contamination, some samples could have been contaminated during sample processing.
Our approach has several potential advantages compared to phenotypic DSTs and to currently available rapid molecular DST tests. Our approach is free from the biohazards associated with phenotypic DST and can be performed without use of expensive biocontainment laboratories. Compared to commercially available rapid molecular tests, our use of targeted DNA sequencing enables easy expansion to detect additional resistance mutations as well as mutations to new drugs as needed. The total reagent costs for our approach, including all assay steps, was approximately $50 per slide, for slides amplified with duplex PCR, and $60 for slides amplified with uniplex PCR. However, with additional multiplexing of the PCR step and with expected decreases in next generation sequencing costs, it is likely that our approach could soon become less expensive than other DST methods. Aside from the smear scraping step, slides can be batch processed at every step of the process. Slide scraping to DNA purification can be completed for at least 12 samples in less than an hour.
In summary, we have demonstrated a simple and rapid method for determining drug resistance in a widely available sample type, AFB stained sputum microscopy smears, which is easily storable, and safely transportable without infectious risk. This approach should prove useful for diagnosing drug-resistant TB as well as for surveillance purposes.
Future research may add mycobacterial speciation targets, which may determine the frequency of positive acid-fast microscopy caused by mycobacteria other than MTB.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(ZIP)
(ZIP)
(XLSX)
Acknowledgments
We thank Dr. Peter Small for suggesting the use of AFB smears as a matrix for DNA sequencing and drug resistance detection. We thank Dr. Hassan Safi for providing TDR strain DNA for our positive controls. We thank Dr. Paridhi Sukeja and Dr. Pradeep Kumar for their assistance in slide shipping and storage. We thank Dr. James Munyao Kingoo, Kibet Shikuku, and Rosaline Bosibori for their assistance in preparing and collecting slides for this project at the Kenya Medical Research Institute/Walter Reed Project, and Dr. Eyako Wurapa, Derrick Mimbe and Allan Tindikahwa for their strong support of the study at the Walter Reed Project—Uganda. We thank Dr. Douglas Shaffer and MAJ Brett E. Swierczewski from the United States Army Medical Research Unit (USAMRU)/Walter Reed Project-Kenya and Raphael Langat from the President’s Emergency Plan for AIDS Relief (PEPFAR)/ Walter Reed Project HIV Program-Kericho, Kenya for their support and assistance. We thank the Innovation For Health And Development, Laboratory for Research and Development (IFHAD) research team for slide collection, processing and comments on this manuscript.
The sponsors had no role in study design, data collection, analysis, publication decision or manuscript preparation. The content and views expressed are those of the authors and do not reflect the official policy of the National Institutes of Health, Department of the Army/Navy/Air Force, Department of Defense, Department of Health and Human Services, or the United States Government.
Data Availability
Most of the relevant data are within the paper and its Supporting Information files. Raw sequencing data has been uploaded to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (BioProject numbers: PRJNA608715, PRJNA608724).
Funding Statement
This work was supported by a grant from the Department of Defense Armed Forces Health Surveillance Branch and the Global Emerging Infections Surveillance Section[grant number: P0028_14_HS; funding years: 2011-17] and from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [award numberRC1 AI087062] awarded to N.A. E.S.R. and C.A.E. acknowledge funding from Department For International Development (UK-AID) Civil Society Challenge Fund (DFID-CSCF)[award 419] and Wellcome Trust [award number 078340/Z/05/Z]. The funding sources mentioned above did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the ‘author contributions’ section.
References
- 1.World Health Organization. Global tuberculosis report 2018. Geneva: 2018. Licence: CC BY-NC-SA 3.0 IGO. http://apps.who.int/iris/bitstream/handle/10665/274453/9789241565646-eng.pdf?ua=1. Accessed December 1, 2018. [Google Scholar]
- 2.World Health Organization. Rapid communication: key changes to treatment of multidrug- and rifampicin-resistant tuberculosis (MDR/RR-TB). Geneva: 2018. Licence: CC BY-NC-SA 3.0 IGO. https://www.who.int/tb/publications/2018/WHO_RapidCommunicationMDRTB.pdf?ua=1. Accessed December 1, 2018. [Google Scholar]
- 3.World Health Organization. Global tuberculosis report 2017. Geneva: 2017. Licence: CC BY-NCSA 3.0 IGO. http://apps.who.int/medicinedocs/documents/s23360en/s23360en.pdf. Accessed December 1, 2018. [Google Scholar]
- 4.Whitfield MG, Soeters HM, Warren RM, York T, Sampson SL, Streicher EM, et al. A Global Perspective on Pyrazinamide Resistance: Systematic Review and Meta-Analysis. PloS one. 2015. 10(7), e0133869 10.1371/journal.pone.0133869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patnaik M, Liegmann K, Peter JB. Rapid detection of smear-negative Mycobacterium tuberculosis by PCR and sequencing for rifampicin resistance with DNA extracted directly from slides. J Clin Microbiol. 2001. 39:51–52. 10.1128/JCM.39.1.51-52.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mokrousov I, Otten T, Filipenko M, Chrapov E, Limeschenko E, Steklova L, et al. Detection of isoniazid resistant Mycobacterium tuberculosis isolates by multiplex allele specific PCR assay targeting katG codon 351 variation. J Clin Microbiol. 2002; 40: 2509–2512. 10.1128/JCM.40.7.2509-2512.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokrousov I, Otten T, Vyshnevskiy B, Narvskaya O. Allele-specific rpoB PCR assays for detection of rifampicin-resistant Mycobacterium tuberculosis in sputum smears. Antimicrob Agents Chemother. 2003; 7: 2231–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Der Zanden AG, Te Koppele-Vije EM, Vijaya Bhanu N, Van Soolingen D, Schouls LM. Use of DNA extracts from Ziehl–Neelsen-stained slides for molecular detection of rifampicin resistance and spoligotyping of Mycobacterium tuberculosis. J Clin Microbiol. 2003; 3: 1101–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suresh N, Singh UB, Gupta C, Arora J, Rana T, Samantaray JC. Rapid detection of rifampicin-resistant Mycobacterium tuberculosis directly from stained sputum smears using single-tube nested polymerase chain reaction deoxyribonucleic acid sequencing. Diagn Microbiol Infect Dis. 2007; 2: 217–222. [DOI] [PubMed] [Google Scholar]
- 10.Dubois Cauwelaert N, Ramarokoto H, Ravololonandriana P, Richard V, Rasolofo V. DNA extracted from stained sputum smears can be used in the MTBDRplus assay. J Clin Microbiol. 2011; 10: 3600–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhutia R, Narain K, Devi KR, Singh TS, Mahanta J. Direct and early detection of Mycobacterium tuberculosis complex and rifampicin resistance from sputum smears. Int J Tuberc Lung Dis. 2013; 2: 258–261. [DOI] [PubMed] [Google Scholar]
- 12.Lavania S, Anthwal D, Bhalla M, Singh N, Haldar S, Tyagi JS. Direct detection of Mycobacterium tuberculosis rifampicin resistance in bio-safe stained sputum smears. PLoS ONE. 2017. 12 (12): e0189149 10.1371/journal.pone.0189149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Union Against Tuberculosis and Lung Disease (IUATLD). Sputum examination for tuberculosis by direct microscopy in low income Countries: technical guide. 5 Paris: IUATLD; 2000. [Google Scholar]
- 14.Zaunbrecher MA, Sikes RD Jr, Metchock B, Shinnick TM, Posey JE. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences. November 2009. 106 (47) 20004–20009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salamon H, Yamaguchi KD, Cirillo DM, Miotto P, Schito M, Posey J, et al. Integration of Published Information Into a Resistance-Associated Mutation Database for Mycobacterium tuberculosis. The Journal of Infectious Diseases. 2015;211(Suppl 2):S50–S57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miotto P, Tessema B, Tagliani E, Chindelevitch L, Starks AM, Emerson C, et al. A standardised method for interpreting the association between mutations and phenotypic drug resistance in Mycobacterium tuberculosis. Eur Respir J. 2017. December 28;50(6). Print 2017 Dec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent V, Rigouts L, Nduwamahoro E, Holmes B, Cunningham J, Guillerm M, et al. The TDR Tuberculosis Strain Bank: a resource for basic science, tool development and diagnostic services. Int. J. Tuberc. Lung Dis. 2012. 16, 24–31. 10.5588/ijtld.11.0223 [DOI] [PubMed] [Google Scholar]
- 18.Addo KK, Addo SO, Mensah GI, Mosi L, Bonsu FA. Genotyping and drug susceptibility testing of mycobacterial isolates from population-based tuberculosis prevalence survey in Ghana. BMC infectious diseases. 2017. 17(1), 743 10.1186/s12879-017-2853-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otchere ID, Asante-Poku A, Osei-Wusu S, Baddoo A, Sarpong E, Ganiyu AH, et al. Detection and characterization of drug-resistant conferring genes in Mycobacterium tuberculosis complex strains: a prospective study in two distant regions of Ghana. Tuberculosis 2016;99:147e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ssengooba W, Meehan CJ, Lukoye D, Kasule GW, Musisi K, Joloba ML, et al. Whole genome sequencing to complement tuberculosis drug resistance surveys in Uganda. Infect Genet Evol. 2016;40:8–16. 10.1016/j.meegid.2016.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lukoye D, Adatu F, Musisi K, Kasule GW, Were W, Odeke R, et al. Anti-tuberculosis drug resistance among new and previously treated sputum smear-positive tuberculosis patients in Uganda: results of the first national survey. PLoS One. 2013;8(8):e70763 Published 2013 Aug 1. 10.1371/journal.pone.0070763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kigozi E, Kasule GW, Musisi K, Lukoye D, Kyobe S, Katabazi FA, et al. Prevalence and patterns of rifampicin and isoniazid resistance conferring mutations in Mycobacterium tuberculosis isolates from Uganda. PLoS One. 2018;13(5):e0198091 Published 2018 May 30. 10.1371/journal.pone.0198091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Second Zambian National Tuberculosis Drug Resistance survey—a comparison of conventional and molecular methods. Trop Med Int Health. 2015. November;20(11):1492–1500. Epub 2015 Aug 27. 10.1111/tmi.12581 [DOI] [PubMed] [Google Scholar]
- 24.World Health Organization. Global tuberculosis control: WHO report 2011. Geneva: World Health Organization; http://www.who.int/iris/handle/10665/44728. Accessed January 5, 2019. [Google Scholar]
- 25.Yeboah-Manu D, Asante-Poku A, Bodmer T, Stucki D, Koram K, Bonsu F, et al. Genotypic Diversity and Drug Susceptibility Patterns among M. tuberculosis Complex Isolates from South-Western Ghana. PLoS ONE. 2011. 6(7): e21906 10.1371/journal.pone.0021906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colman RE, Anderson J, Lemmer D, Lehmkuhl E, Georghiou SB, Heaton H, et al. Rapid Drug Susceptibility Testing of Drug-Resistant Mycobacterium tuberculosis Isolates Directly from Clinical Slides by Use of Amplicon Sequencing: a Proof-of-Concept Study. J Clin Microbiol. 2016. August;54(8):2058–67. 10.1128/JCM.00535-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(ZIP)
(ZIP)
(XLSX)
Data Availability Statement
Most of the relevant data are within the paper and its Supporting Information files. Raw sequencing data has been uploaded to the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (BioProject numbers: PRJNA608715, PRJNA608724).