SUMMARY
Genome packaging by nucleosomes is a hallmark of eukaryotes. Histones and the pathways that deposit, remove, and read histone modifications are deeply conserved. Yet, we lack information regarding chromatin landscapes in extant representatives of ancestors of the main groups of eukaryotes, and our knowledge of the evolution of chromatin-related processes is limited. We used the bryophyte Marchantia polymorpha, which diverged from vascular plants circa 400 mya, to obtain a whole chromosome genome assembly and explore the chromatin landscape and three-dimensional genome organization in an early diverging land plant lineage. Based on genomic profiles of ten chromatin marks, we conclude that the relationship between active marks and gene expression is conserved across land plants. In contrast, we observed distinctive features of transposons and other repetitive sequences in Marchantia compared with flowering plants. Silenced transposons and repeats did not accumulate around centromeres. Although a large fraction of constitutive heterochromatin was marked by H3K9 methylation as in flowering plants, a significant proportion of transposons were marked by H3K27me3, which is otherwise dedicated to the transcriptional repression of protein-coding genes in flowering plants. Chromatin compartmentalization analyses of Hi-C data revealed that repressed B compartments were densely decorated with H3K27me3 but not H3K9 or DNA methylation as reported in flowering plants. We conclude that, in early plants, H3K27me3 played an essential role in heterochromatin function, suggesting an ancestral role of this mark in transposon silencing.
In Brief
Montgomery et al. provide a chromosome-scale genome assembly of the early diverging land plant Marchantia polymorpha. Profiling of chromatin marks shows conserved roles of active marks and suggests an ancestral association between H3K27me3 and transposons that is partly retained in Marchantia and replaced by H3K9 methylation in flowering plants.
INTRODUCTION
In eukaryotes, the evolution of histones that assemble with DNA into nucleosomes generated chromatin with a more diverse composition and complex organization compared to that found in prokaryotes [1, 2]. Post-translational modifications of core histones that form nucleosomes contribute to the complexity and flexibility of chromatin [3]. The characterization of such modifications, marking transcriptionally active and inactive regions of the genome, has furthered insights into the functional organization of eukaryotic chromatin. In flowering plants, extensive meta analyses of histone modification profiles in Arabidopsis thaliana highlighted the association of H3K4me3, H3K36me3, and H3 acetylation with gene expression, while H3K27me3 marks transcriptional repression and H3K9 methylation is associated with DNA methylation (5′ methyl Cytosine) marking silenced transposons [4].
The three-dimensional (3D) organization of domains where distant regions of chromatin connect is revealed by genomic methods such as Hi-C [5] and genome architecture mapping [6]. The 3D organization of flowering plant genomes analyzed by classic cytological methods and Hi-C showed a wide variety of nuclear organization patterns [7, 8]. The diversity of chromatin organization suggests that, during land plant evolution, genome organization changed and diversified depending on genome duplications, size, and relative transposable element (TE) versus gene content. It is therefore important to extend investigations of 3D genome organization to a larger number of species representative of extant ancestral lineages to understand how genome architecture evolved in eukaryotes.
Bryophytes, composed of liverworts, mosses, and hornworts, represent ancient lineages of land plants that diverged from the vascular plant lineage over 400 Mya [9]. Analysis of the genome sequences of the liverwort Marchantia polymorpha and the moss Physcomitrella patens demonstrated that genes encoding pathways related to histone modifications are broadly conserved in land plants [10] but that heterochromatic islands of transposons and repeats alternate with genes without a clear demarcation of a region enriched in transposons around centromeres [11]. This contrasts with the vast accumulation of transposons and repeats around centromeres described in Arabidopsis and many species of flowering plants [12, 13]. Yet, the lack of Hi-C maps and the limited knowledge of chromatin modification profiles in bryophytes have limited our understanding of the ancestral functional organization of chromatin in land plants.
We obtained a new full chromosome assembly of the Marchantia polymorpha genome with updated annotations, which will be publicly accessible as reference genome version 5.1 (v5.1) for this species. Here, we present a new set of extensive profiles of key chromatin marks as well as 3D chromatin organization patterns obtained by Hi-C. Altogether, our observations lead to a model of chromatin organization in early land plants, revealing that considerable changes arose during the evolution of vascular plants.
RESULTS
A Full Chromosome Assembly of the Marchantia Genome
The previous version of the nuclear genome of Marchantia polymorpha (v3.1 from a Tak-2 backcross) comprised 2,957 scaffolds with 19,138 protein-coding genes [10]. We obtained a new set of scaffolds of the genome from the male accession Tak-1 using long-read sequencing and assembled them at a chromosomal scale using Hi-C (Figure S1). Overall, this newly assembled Tak-1 genome, referred to as Marchantia polymorpha v5.1, contains 218.7 Mb, including 215.8 Mb jointly covered by the autosomes and the male sex chromosome (chromosome V), and can be accessed at MarpolBase (http://marchantia.info/). A total of 200 Mb genomic regions showed high sequence identity (>99% identity) against the v3.1 genome. The majority of the additional 17.7 Mb was accounted for by repetitive regions (14 Mb), while the remaining 3.7 Mb showed lower similarity or no homology against the v3.1 genome. Markers associated with distinct genetic linkage groups were identified between the two accessions Tak-1 and Tak-2 (Data S1). The linkage groups and linear order of the vast majority of these genetic markers were fitted correctly with the chromosomes assembled in v5.1 (Data S1). This genetic map at low resolution validated the overall structure of the physical whole chromosome genome assembly.
The v5.1 genome harbors 19,421 predicted protein-coding loci with 24,751 transcript models including isoforms (Data S1). Among them, 24,078 transcript models were carried over from the v3.1 genome, and 673 were newly identified by de novo prediction and manual inspection. We also curated 303 new transcript models based on expression evidence from RNA sequencing (RNA-seq) and Iso sequencing (Iso-seq). The completeness of the gene set was assessed using BUSCO [14], estimating that 97.6% (296) out of 303 universal single-copy orthologs for eukaryotes were present, the same level as the v3.1 genome. We adopted a new series of unique gene identifiers following the guidelines established for the Arabidopsis genome. Examples of newly identified genes include gene clusters such as the NNP family, nitrate/nitrite transporters (Mp5g10710, Mp5g10760, Mp5g10780, Mp5g10790), metalloproteases (Mp8g14490, Mp8g14520, Mp8g14560, Mp8g14610), and DEAD-box family RNA helicases (Mp4g13200, Mp4g13270, Mp4g13330). These regions were missing or fragmented into different scaffolds in the v3.1 genome, indicating the advantage of the v5.1 assembly leveraged by long-read sequencing in reconstructing such repetitive regions. We also identified comprehensive lists tRNAs, micro-RNAs (miRNAs), transposons, and repeats (Data S2).
The male-specific sex chromosome V of Marchantia consists of two parts, YR1 and YR2, each of which has distinctive sequence content [15]. YR1 is highly enriched in repeats unique to chromosome V [15, 16]. Version 5.1 includes two novel regions of the V chromosome, a 506-kb region between Contig-A and Contig-B, and a 1.3-Mb region at the distal end of Contig-A from Contig-B. The 1.3-Mb region contains blocks of the V-specific repeats (Figure S2), most likely representing part of YR1. The extremely high repeat content still prevented this region from being properly assembled and reconstructed. Interestingly, copies of rDNA were found among the blocks of the V-specific repeats (Figure S2). Two types of rDNA were previously reported to be present in the Marchantia genome, one autosomal and the other U-chromosomal [17]. The V-chromosomal copies were more similar to the autosomal (99.64%) than to the U-chromosomal copies (97.02%). Unlike the autosomal and U-chromosomal rDNAs, the V-chromosomal rDNAs do not form a regular tandem array suggesting potential for distinct epigenetic regulation as shown for distinct rDNA clusters in Arabidopsis [18].
Telomeres, Centromeres, and Overall Nuclear Organization
Telomeres of Marchantia polymorpha are composed of tandem arrays of TTTAGGG repeats similar to that identified in Marchantia palaeceae [19]. To gauge the size of telomere tracts, we performed terminal restriction fragment analysis and observed that Marchantia telomeres are longer than in Physcomitrella and shorter than in Arabidopsis (Figure S3A). We concluded that Marchantia telomeres are comparable with those of most other plants [19-21].
In most flowering plants, centromeres are composed of specific satellite repeats interspersed with transposons and surrounded by a pericentromeric region enriched in transposons. We identified centromeric repeats composed of 162-bp satellite DNA (Figure S3B). This size is within the range found in other land plants [22] and compatible with the typical shorter length of DNA associated with centromeric nucleosomes [23]. These repeats were found close to the center of each autosome (Figure S3C). Beyond the satellite repeats, long terminal repeat (LTR) retrotransposons accumulate in centromeres and pericentromeres of flowering plants and animals [24-26]. In contrast, in Marchantia we did not find LTR transposons in proximity of the centromeres. Only the specific family LINE/RTE-X showed a sharp peak surrounding centromeres of each chromosome, indicating a high density of this family (Figure S3C) despite its modest genomic abundance (Data S2). These data indicate that Marchantia has monocentric centromeres marked by short repeats as described in the majority of land plants, but the extent of these repeats and the lack of LTR transposons do not define an extended pericentric region as observed in many flowering plants.
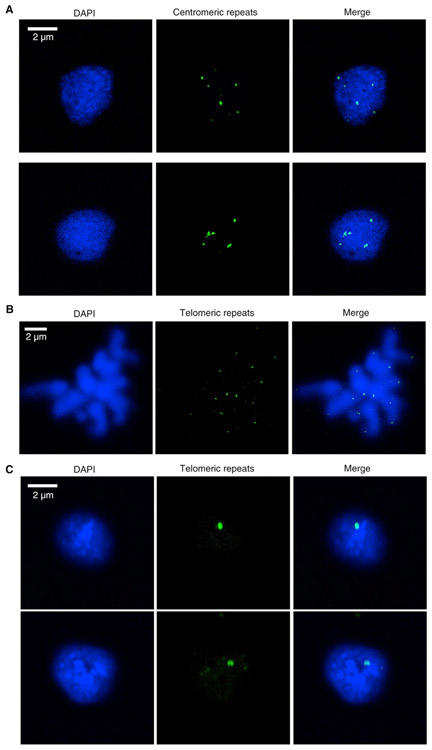
With knowledge of Marchantia centromeric and telomeric regions, we designed probes to examine their distribution in interphase nuclei in the vegetative thallus. We found up to nine dots marked by the centromeric repeat probes, which showed a dispersed localization (Figure 1A). Telomeres were located at the ends of each chromosome in metaphase (Figure 1B). In interphase, telomeres often clustered to form a single speckle (Figure 1C). A similar conformation, called a “bouquet,” has been reported in meiotic maize, wheat, and rice cells [27-29]. However, in contrast to bouquet conformation described in flowering plants, the telomere gathering in Marchantia nuclei did not display a specific association of telomeres with the nuclear periphery (Figure 1C).
Figure 1. Distribution Patterns of Centromeric Repeats and Telomeres in Marchantia.
(A) Distribution of centromeric repeats in Tak-1 nuclei isolated from vegetative thalli.
(B) Confirmation of telomere probes’ specificity by using chromosome spread. Probes labeled with digoxigenin were hybridized with Tak-1 chromosome spread.
(C) Distribution of telomeres in Tak-1 nuclei isolated from vegetative thalli.
See also Figures S3 and S4.
To examine the spatial organization of euchromatin versus heterochromatin, we immunostained Marchantia and Physcomitrella patens nuclei with antibodies against histone modifications typical of constitutive heterochromatin (H3K9me1 and H3K27me1), facultative heterochromatin (H3K27me3), and euchromatin (H3K36me3 and H3K4me3) as defined in Arabidopsis [4]. The distribution of DNA in Marchantia is more punctate, with many small foci and several larger ones (Figure S4A), in comparison to the smooth and homogeneous distribution of DNA in Physcomitrella patens (Figure S4B). In Marchantia nuclei, heterochromatic regions, denoted by denser staining, tend to overlap with H3K9me1 and H3K27me1 but also surprisingly with H3K27me3. These heterochromatic regions do not form clear compact structures comparable to chromocenters described in Arabidopsis and other flowering plants.
Organization of Chromatin Profiles
Using CUT&RUN [30, 31] in Marchantia, we obtained genomic profiles of eight histone modifications (H3K9me1, H3K27me1, H3K9ac, H3K14ac, H3K4me1, H3K36me3, H3K4me3, and H3K27me3), one histone variant (H2A.Z), and H3. This set of histone modifications together with data available for DNA methylation [32] and transcriptional activity [10] can be accessed at at MarpolBase (http://marchantia.info/). This comprehensive and integrated dataset enabled us to draw comparisons with chromatin states in Arabidopsis [4]. Biological replicates tended to cluster together in a Pearson correlation matrix (Figure S5A) and marks typically considered active (H3K9ac, H3K14ac, H3K36me3) or repressive (H3K9me1, H3K27me1) grouped among themselves (Figure S5B). Although H3K9me2 is often used to mark constitutive heterochromatin in Arabidopsis, H3K9me1 shows a similar coverage (Figure S5C) and the antibody against this mark gave more consistent results in Marchantia. Interestingly, H3K27me3 was quite distinct from other marks and correlated most strongly with H3K4me3 and H2A.Z. Accordingly, H3K27me3 peaks overlapped primarily with H3K4me3 and H2A.Z peaks (Figure S5D) but not with DNA methylation in CG, CHG, and CHH contexts [32], which was most strongly associated with H3K9me1 and H3K27me1 (Figure S5E).
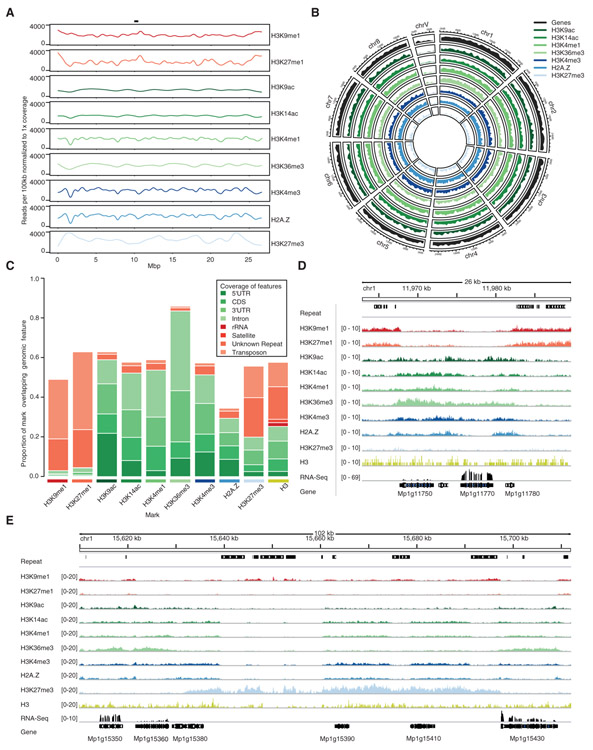
Each of the chromatin profiles was spread evenly across chromosomes (Figures 2A and 2B) following the even interspersed distribution of transposons and genes. Peaks of H3K9me1 and H3K27me1 were enriched on ribosomal RNA coding genes, satellites, repeats, and transposons (Figures 2C and 2D). In flowering plants, centromeres are surrounded by heterochromatic pericentromeric regions marked by DNA methylation, H3K9me1, H3K9me2, and H3K27me1, that target multiple families of transposons [4, 13, 24, 34]. Such accumulation was not detected around centromeres in Marchantia (Figure 2A), and we concluded that there is no detectable pericentric heterochromatin in Marchantia. Strikingly, 60% of the length of H3K27me3 peaks were found on repeats and transposons, while the remaining length was associated with genes (Figures 2C and 2E). All other chromatin modifications profiled were primarily associated with genes with a notable enrichment of H3K36me3 over the coding sequence and 3′ UTR while the 5′ UTR is relatively more enriched in H3K9ac (Figures 2C and 2D).
Figure 2. Distribution of Chromatin Marks in the Marchantia Genome.
(A) Coverage of chromatin marks across chromosome 5. Reads were normalized to 1 × coverage and binned into 100-kbp windows along the chromosome and a smoothed spline was fit to the data. Position of the putative centromere is indicated at the top.
(B) Circos plot of euchromatic marks and genes. Each band shows the density of annotated chromatin mark peaks per chromosome, relative to the greatest density per band.
(C) Distribution of chromatin marks over genomic features. The total length of chromatin mark peaks overlapping specified genomic features was divided by the total length of peaks of chromatin marks to determine each proportion. Unknown represents repeats annotated as unknown by RepeatMasker. Simple repeats are not shown as they cover less than 0.3% of chromatin mark peaks.
(D) Integrative Genomics Viewer (IGV) browser screenshot demonstrating flanking of genes by H3K9me1 and H3K27me1 marked transposons. The region shown is 26 kb in length and from the proximal arm of chromosome 1. Chromatin mark tracks are bigwig files scaled to 1 × genomic coverage in 10-bp windows. “Repeat” and “Gene” tracks are annotation files for repeats and genes, respectively. “RNA-seq” track is a bigwig of mapped RNA-seq reads from thallus tissue [33]. Scales are noted in square brackets beside each track.
(E) IGV browser screenshot demonstrating large H3K27me3 islands covering both genes and transposons. The region shown is 102 kb in length and from the distal arm of chromosome 1. Chromatin mark tracks are bigwig files scaled to 1 × genomic coverage in 10-bp windows. “Repeat” and “Gene” tracks are annotation files for repeats and genes, respectively. “RNA-seq” track is a bigwig of mapped RNA-seq reads from thallus tissue [33]. Scales are noted in square brackets beside each track.
See also Figure S5.
Histone Modifications and Gene Expression
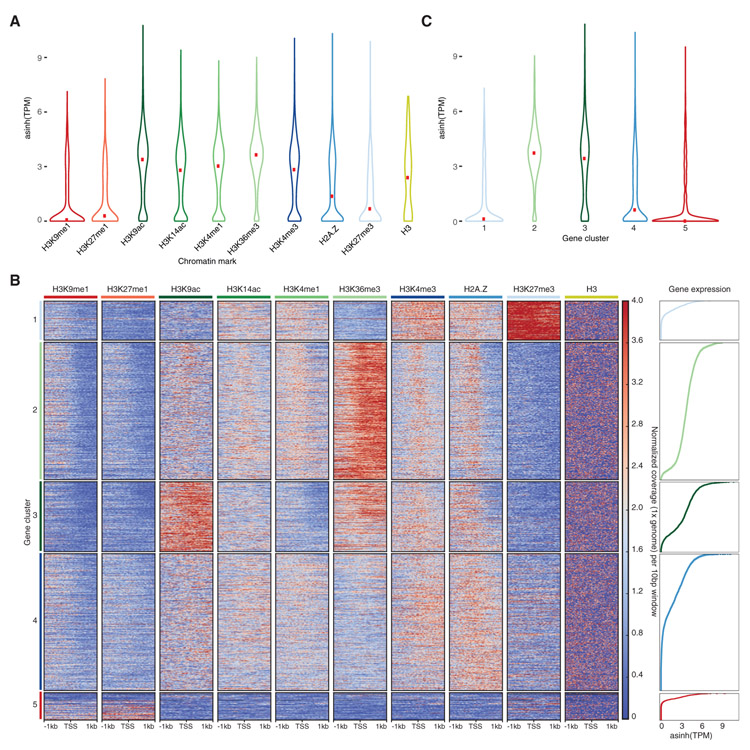
We explored preferential associations between chromatin marks and the transcriptional status of genes based on their average expression in the thallus somatic cells [10]. H3K36me3 showed the strongest association with expressed genes, which were also marked by H3K9ac, H3K14ac, and to a lesser extent by H3K4me1 and H3K4me3 (Figures 3A and S6A). In contrast, H3K9me1, H3K27me1, and H3K27me3 marked inactive genes (Figure 3A). Interestingly, H2A.Z showed a bimodal distribution of expression levels for the genes it associates with (Figure 3A), potentially linked with its correlation and overlap with H3K27me3 (Figure S5D).
Figure 3. Association of Chromatin Marks with Genes.
(A) Expression level of genes associated with profiled chromatin marks. Width is relative to the density of genes. Red dots indicate median expression values.
(B) Heatmap of k-means clustering of genes based on chromatin marks. Prevalence of each mark (columns) based on its score of normalized 1 × genomic coverage per 10 bp ± 1 kb around the transcription start site per gene, with red for enrichment and blue for depletion. Each row corresponds to one gene, with multiple genes grouped into blocks that have been defined as gene clusters 1 through 5.
(C) Expression level of genes per gene cluster. Width is relative to the density of genes. Red dots indicate median expression values.
See also Figures S5 and S6.
To untangle the relationships between chromatin profiles and genes in Marchantia, we performed k-means clustering of chromatin profiles over genes. This led to the identification of five main clusters of genes showing distinct chromatin environments (Figure 3B). Cluster 5 contained 7% of all genes and showed low levels of H3 and H3 modifications, suggesting a low nucleosome density, an inaccessibility for chromatin profiling, or difficulties in read alignment, and we did not consider this cluster further. Gene clusters 2 and 3 encompassed active genes, accounting for 33% and 17% of genes, respectively, and showed enrichment in H3K14ac, H3K4me1, and H2A.Z at the transcription start site (TSS), though this trend was less marked for cluster 3 (Figures 3B, S6A, and S6B). Genes in cluster2 and 3 shared a strong enrichment in H3K36me3 over gene bodies with additional enrichment in H3K9ac in genes of cluster3 (Figures 3B, S6A, and S6B). Inactive genes were found in clusters 1 and 4, accounted for 10% and 33% of genes, respectively, and were characterized by a prominent enrichment of H2A.Z and H3K4me3 and an absence of H3K36me3 along gene bodies (Figures 3B, 3C, S6A, and S6B). A strong enrichment of H3K27me3 distinguished genes in cluster 1 from genes in cluster4 (Figures 3B and S6A). Gene clusters were uniformly distributed across the genome, to the exception of the gene-poor sex chromosome V (Figure S6C). We observed a low density of DNA methylation in CG, CHG, and CHH contexts over genes irrespective of the nature of the dominating histone modification present (Figures S6D-S6F).
We conclude that DNA methylation on gene bodies does not correlate with chromatin states and transcriptional activity in Marchantia in contrast to Arabidopsis [35] and in agreement with a previous report [32]. In Marchantia, the enrichment in H3K36me3 over gene bodies is the best predictor of active transcription, and the combination of histone modifications that mark active genes is comparable to chromatin state 3 in Arabidopsis [4]. The TSS of active genes in Marchantia is marked by H3K4me3 and H2A.Z, similar to chromatin state 1, which marks TSS of active genes in Arabidopsis [4]. Repressed genes in Marchantia are marked with H2A.Z associated with H3K27me3 or H3K4me3 over gene bodies, similar to chromatin state 5 in Arabidopsis [4]. Altogether we conclude that the association between combination of histone modifications with gene transcriptional states in Marchantia is comparable to Arabidopsis [35], and other eukaryotes [36], although the association between H3K4me3 alongside H2A.Z on the body of inactive genes in cluster 4 appears more specific to Marchantia. The combination of H3K27me3 and H3K4me3 at some loci may reflect bivalent marks as observed in Arabidopsis [4] but might as well represent genes repressed with H3K27me3 in some cells while expressed and marked with H3K4me3 in other cells.
Heterochromatin and Transposons
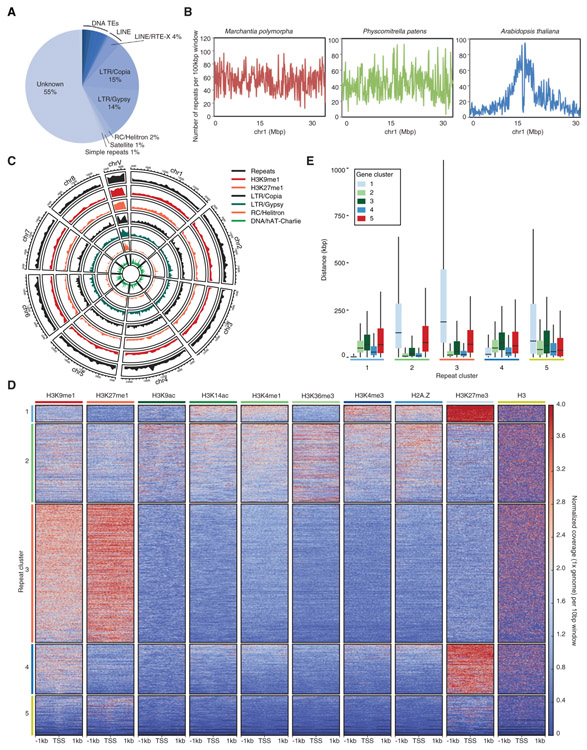
We reassessed the census of transposons and repeats in Marchantia, which comprise at least 63 Mb and represents 27% of the genome, in contrast with 56% of the Physcomitrella genome (Data S2). This lower proportion is largely attributed to the absence of the large expansion of Gypsy retrotransposons in Physcomitrella (Data S2 and [11]). In Marchantia, about two-thirds of the transposons that were ascribed to a family belonged to retrotransposons from the Copia or Gypsy families, and families of retrotransposons unique to Marchantia or Physcomitrella were identified (Figure 4A; Data S2). We also noted a comparable diversity of DNA transposons between the two species but an increased diversity of LINE families in Marchantia (Data S2), in part related to the expansion of LINE/RTE-X around centromeres (Figure S3C).
Figure 4. Association of Chromatin Marks with Transposons.
(A) Proportion of total repetitive elements belonging to major transposon superfamilies. Total counts of each transposon superfamily can be found in Data S2.
(B) Distribution of transposons across a representative chromosome from Marchantia polymorpha, Physcomitrella patens, and Arabidopsis thaliana. The number of repeats occurring in 100-kbp bins across each chromosome are shown.
(C) Circos plot of heterochromatic marks, the four most abundant transposon superfamilies in Marchantia and all repeats. Each band shows the density of annotated repetitive elements or chromatin mark peaks per chromosome, relative to the greatest density per band.
(D) Heatmap of k-means clustering of transposons based on chromatin marks. Prevalence of each mark (columns) based on its score of normalized 1 × genomic coverage per 10 bp ± 1 kb around the transcription start site per gene, with red for enrichment and blue for depletion. Each row corresponds to one transposon, with multiple transposons grouped into blocks that have been defined as repeat clusters 1 through 5.
(E) Boxplot of distances between each transposon and the nearest gene per gene cluster. Briefly each transposon is compared to all genes belonging to a gene cluster to find its nearest neighbor. Transposons are divided based on the repeat cluster they belong to. Distances are in kilobases (kbp). Colored boxes represent interquartile range, and lines represent median values. Outliers are not shown.
See also Figures S5 and S7 and Data S2.
Heterochromatic marks and transposons were distributed evenly across chromosomes (Figures 4B and 4C). We performed k-means clustering of chromatin profiles over transposons and repeats leading to the identification of five main clusters showing distinct chromatin environments (Figure 4D). Over 40% of LINE/RTE-X elements were found in cluster 5, which represented 12% of repeats and was enriched around putative centromeres (Figure S3C). These transposons appeared to be relatively depleted of all profiled chromatin marks (Figure 4D), which could reflect a low nucleosome density or their relative inaccessibility to the MNase used in CUT&RUN profiling. Cluster 3, containing 43% of repeats and transposons, was characterized by a strong enrichment of H3K9me1 and H3K27me1 (Figure 4D). Repeats from cluster 3 were much more enriched in the male sex chromosome V than on autosomes (Figures 4D andS7A). This cluster also associated with high DNA methylation levels in CG, CHG, and CHH contexts (Figures S7B-S7D), and the combination of chromatin marks in transposons and repeats from cluster 3 was comparable to chromatin states 8 and 9 in Arabidopsis [4]. 25% of repeats and transposons represented cluster 2 that was enriched in DNA transposons (Figure S7E) and showed low uniform enrichment in all marks except H3K27me3 (Figure 4D). A similar chromatin state was observed over protein coding genes from cluster 4 (Figure 3B), and these two clusters were closely associated next to each other (Figures 4E and S7F). This combination of chromatin marks associated with low expression (Figure 3C) was not reported in Arabidopsis. Contrasting with clusters 2 and 3, H3K27me3 was enriched over transposons forming clusters 1 and 4, which represented 5% and 15% of repeats, respectively (Figure 4D). The average length of elements from each cluster differed significantly with shorter transposons in cluster 1 than in cluster 4 (Figure S7G). Overall, clusters 1 and 4 marked with H3K27me3 represented circa 30% of the constitutive heterochromatin, while 54% of constitutive heterochromatin was marked jointly by H3K9me1, H3K27me1, and DNA methylation (Data S3). Repeats from cluster 4 showed higher levels of H3K9me1, whereas repeats from cluster 1 were more enriched in H3K4me3 and H2A.Z. DNA methylation levels in CG, CHG, and CHH contexts were higher in repeats from cluster 4 than from cluster 1 (Figures S7B-S7D). There was no specific association between clusters 1–3 and a single class of repeat (Figure S7E). RC/Helitron elements were mostly enriched in cluster 4, and there was preferential association of retrotransposons LTR/Copia and LTR/Gypsy with clusters 1 and 4, respectively (Figure S7E). Cluster 5 was strongly enriched in LINE/RTE-X, which surrounded centromeres (Figures S7A and S7E). We also noted that the sex chromosome V contains mostly repeats and transposons from cluster 3 (Figure S7A). These regions contrast with autosomes, where a large fraction of potentially mobile retrotransposons is marked by the repressive mark H3K27me3 (Figure S7E).
We investigated the possibility that genes and surrounding transposons and repeats share similar combinations of chromatin modifications. We measured the distance between each transposon and the nearest gene per gene cluster (Figure 4E) and vice versa (Figure S7F). Strikingly, genes from cluster 2, which are expressed at high levels, were usually surrounded by transposons and repeats strongly enriched in H3K9me1 and H3K27me1 (Figures 2D and 4E). In contrast, H3K27me3 covered inactive genes and surrounding repeats and transposons (Figures 2E, 4E, and S7F), accounting for 60% of nucleosomes that carried this mark related to the transcriptionally repressed state (Figures 2C and 3C). These account for large domains containing repressed genes and transposons covered by a high density of H3K27me3 (see an example in Figure 2E) in accord with the potential of H3K27me3 to spread [37]. We conclude that a large proportion of genes and surrounding transposons share the same chromatin state in Marchantia (Figures 4E and S7F) with the notable exception being active genes surrounded by transposons marked by H3K9me1 on autosomes and on the sex chromosome V (see the gene cluster 2 associated with the repeat cluster 3 in Figure 4E).
V Chromosome and Autosomes Have Distinct Conformations
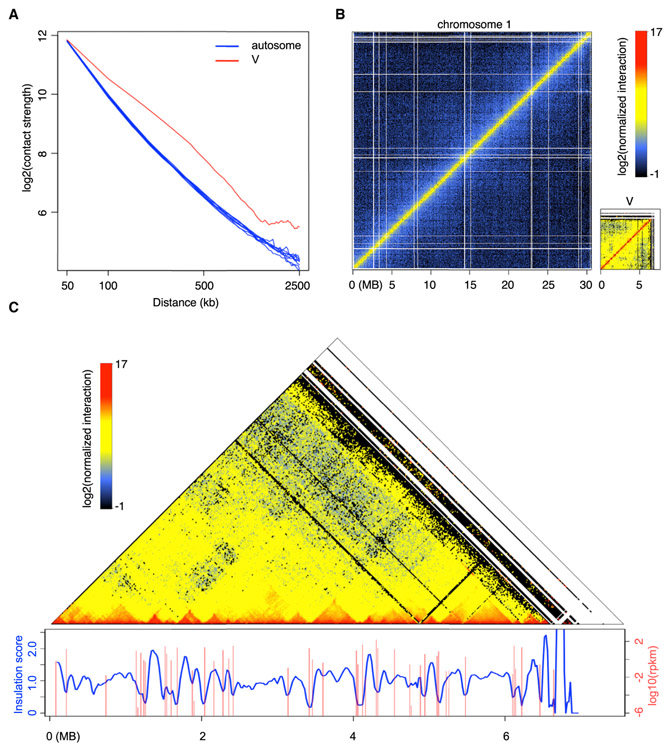
By comparing power-law decay curves of intra-chromosomal interaction strength with genomic distance in individual chromosomes, we found that the pattern of the male V chromosome was different from those of autosomes (Figures 5A and 5B). Particularly, the V chromosome Hi-C map indicated that it had stronger long-range chromatin contacts than those of autosomes, suggesting that the V chromosome was more compact. Additionally, on a chromosomal scale, the V chromosome exhibited significantly higher levels of heterochromatic marks H3K9me1 and H3K27me1 than autosomes (Figure 4C). These data indicate that the V chromosome is largely repressed and is more condensed than autosomes. Interestingly, manual inspection along the diagonal of the V chromosome Hi-C map revealed many self-interacting domains, in which chromatin contacts within one domain were stronger than those across different domains (Figure 5C). These self-interacting chromatin domains resembled topologically associated domains (TADs) discovered in mammals [39]. TADs appear as the basic structural units beyond nucleosomes, modulating higher-order chromatin organization [40]. TAD boundaries, which reflect local chromatin insulation, are enriched for insulator element binding proteins and active gene transcription [41]. Upon associating transcriptional activities at the V chromosome with the Hi-C map, we found a positive correlation in which many domain boundaries corresponded to local gene expression (Figure 5C). This suggests a tight relationship between the male sex chromosome topology and its transcriptional regulation. Previous studies reported reproductive-organ-specific expression of V chromosome-specific genes [10, 15]. In future, it would be interesting to examine possible dynamic V chromosome organization during sexual reproduction.
Figure 5. Marchantia Chromosome V Has Distinct Chromatin Packing Patterns Compared with Autosomes.
(A) Comparison of interaction decay exponents among autosomes and V chromosome. The average interaction strengths of each chromosome at various distances were calculated based on a whole-genome Hi-C map normalized at 50 kb resolution.
(B) Hi-C maps of Tak-1 chromosome 1 and chromosome V.
(C) Association between V chromosome Hi-C map (normalized at 20 kb resolution) and local gene expression. Insulation scores were calculated according to [38] with minor modifications, in which a sliding square of 100 kb × 100 kb along the matrix diagonal was used, and the ratio of observed over expected interaction strengths of this sliding square was plotted as insulation score. Genomic regions with local minima of insulation scores have strong chromatin insulation. Data of gene expression in Tak-1 thalli were from [10].
See also Figure S2.
Extensive Intra- and Inter-chromosomal Contacts of Marchantia Chromatin
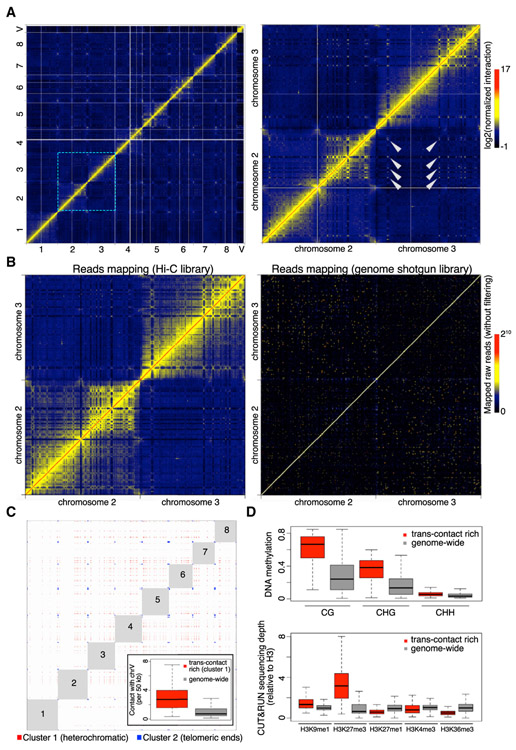
On the genome-wide Hi-C map, we found many regions showing both strong intra- and inter-chromosomal contacts (Figure 6A). A comparison between interaction matrices generated with similar amounts of mapped reads from our Hi-C and a genome shotgun library indicated that these strong long-range chromatin interaction patterns were not caused by mapping errors (Figure 6B). Depending on their interaction networks, we classified these genomic regions into two groups (Figure 6C). One group (cluster 2) comprised regions found at chromosomal ends, consistent with our fluorescence in situ hybridization (FISH) data showing telomere clustering. This appears to be a universal phenomenon across plants [42-46].
Figure 6. Marchantia Genome Shows Extensive Inter-chromosomal Interactions.
(A) Normalized Hi-C map at 50 kb resolution. The right panel shows the zoom-in image of an area containing chromosomes 2 and 3, in which selected trans-contacts among interstitial regions in different chromosomes are highlighted with arrowheads.
(B) Comparison of chromatin interaction maps (50-kb bin) generated with comparable amounts of mapped reads in Hi-C and genome shotgun libraries (110 versus 130 million), respectively. The pair-end genome shotgun library is a combination of SRA: SRR396657 and SRR396658 [10] and was mapped to the assembled TAK-1 genome as Hi-C reads. Note that the diagonal of the plot shown on the right has values larger than the maximum defined in the color bar.
(C) Genomic regions showing strong and extensive trans-interactions. Bins having at least one top 0.5% inter-chromosomal contacts in the normalized Hi-C map shown in (A) were subjected to k-means clustering based on their genome-wide inter-chromosomal contact patterns. The optimal number of clusters was determined as 3 based on the Elbow method. For the first two clusters, virtual interactions among members of each cluster are shown as red and blue dots, respectively, representing an ideal situation in which all possible contacts happen within each cluster and are visible on a Hi-C map. Numbers depict autosome names. The inset shows inter-chromosomal contacts between autosomes and the V chromosome.
(D) DNA methylation (top panel) and histone modifications (bottom panel) in genomic regions annotated as “cluster 1” in (C) and the whole genome (V chromosome not included). The DNA methylation data of Tak-1 thalli was from [32].
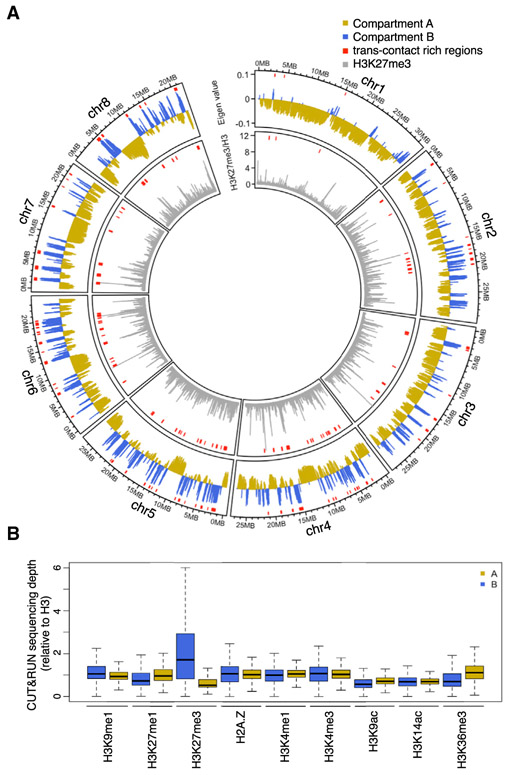
Regions in the other group (cluster 1) were interstitial in each chromosome. Members of this group showed extensive contacts with each other, which stood out as speckles on the Hi-C map (Figures 6A and 6C; Table S1). These regions were depleted of the heterochromatic mark H3K27me1 and euchromatic marks H3K4me3 and H3K36me3 and showed enrichment in DNA methylation (Figure 6D). To some extent, these results resembled those associated with a special type of region in Arabidopsis and rice genomes named IHIs/KEEs (Interactive Heterochromatic Islands or KNOT ENGAGED ELEMENTs), which are marked by H3K9 methylation and DNA methylation [47-49]. In contrast with angiosperms, high levels of H3K27me3 were the strongest marker of heterochromatic islands in Marchantia. Notably, these heterochromatic islands showed stronger interactions with the V chromosome than did the average across all autosomes (Figure 6C, inset), suggesting the existence of chromatin compartmentalization that selectively brought some repressed genomic regions into physical proximity (i.e., close to the V chromosome). Furthermore, a routine compartmentalization annotation for identifying A (active) and B (inactive) compartments [5] showed that B compartment regions were associated with trans-contact rich regions (Figure 7A). Notably, B compartments showed much higher levels of H3K27me3 and no significant association with enrichment in H3K9me1 and H3K27me1 (Figures 7A and 7B). We speculate that H3K27me3 plays an important role in shaping chromatin compartmentalization and defining heterochromatin in autosomes, while local transcriptional activities delimit TADs on the sex chromosome.
Figure 7. A/B Compartments and Their Associated Epigenetic Marks.
(A) A/B compartments and H3K27me3 chromatin marks in individual Tak-1 autosomes plotted in 50-kb windows. For each autosome, the compartment bearing the estimated centromere is labeled as “Compartment B.” Red segments shown on top of each track denote trans-contact rich regions that display strong inter-chromosomal interactions.
(B) Epigenetic features associated with A/B compartments.
DISCUSSION
In flowering plants, transposons represent 10%–90% of genomes and tend to cluster in pericentromeric heterochromatin clearly delimiting chromocenters, as shown in Arabidopsis [22, 24, 25]. In the maize genome, consisting of circa 90% of transposons and repeats, it is expected that many transposons are interspersed between genes, though they are still found in greater densities in pericentromeric heterochromatin [50, 51]. In contrast, transposons and genes are spread relatively evenly across chromosomes in the moss Physcomitrella patens [11] and the liverwort Marchantia polymorpha, although transposons and repeats represent less than 25% of the genome in this species. This even distribution is associated with the lack of chromocenters in both species, which is also observed in many other bryophytes including hornworts [52], suggesting that early land plants shared a general genome organization devoid of a linear cluster of transposons. It has been proposed that the interspersed organization of genes and transposons in Physcomitrella may be a facet of inbreeding and low recombination rates [11]. As Marchantia and many other liverworts are dioicous and reproduce by outcrossing, there are likely alternative explanations. However, the enrichment of specific classes of transposons around the centromeres of Physcomitrella and Marchantia indicates that potential mechanisms by which transposons become enriched around centromeres may have been active already in these plants.
Epigenetic and transcriptional states are key predictors of Hi-C contact maps in eukaryotes [41, 53, 54]. Similar to the observations made from Hi-C maps in other eukaryotes, the binary annotation of Marchantia autosomes based on Hi-C data largely correlates to the demarcation of active/inactive chromatin domains. On the V chromosome, DNA and H3K9 methylation are associated with transposons surrounding highly expressed genes, forming clear TADs. These associations also exist on autosomes (Figure 2D) but are relatively scarce compared with the sex chromosome V. Similar patterns are also observed in Arabidopsis chromocenters, in which the 3D folding of constitutive heterochromatin marked by DNA and H3K9 methylation is proposed to be driven by local expression levels [41]. This suggests that the function of marks typical of constitutive heterochromatin in eukaryotes [55] is conserved in Marchantia and insulates transcriptional units.
However, a major portion of the Marchantia genome exhibits low levels of DNA methylation [32], as in other bryophytes [56, 57], and we observed that a significant fraction of transposons and repeats are not marked by H3K9me1 nor H3K27me1 (Figure 4D). In Marchantia, H3K27me3 associates with the repressive B compartment and trans-contact rich regions, whereas B compartments are marked by H3K9me1 and H3K27me1 in flowering plants [58]. Remarkably, a third of constitutive heterochromatin is marked with H3K27me3. H3K27me3 is deposited by the Polycomb repressive complex 2 (PRC2) in Physcomitrella [59], and the conservation of PRC2 subunits in Marchantia [10] indicates that its function is likely conserved in bryophytes. In land plants, as in other eukaryotes, H3K27me3 is involved in maintaining repressed transcriptional states [4, 59, 60], and previous plant Hi-C studies reported that H3K27me3-marked chromatin is involved in forming long-range interactions [45, 48, 61]. Hi-C analyses in Marchantia highlight the potential dominant impact of H3K27me3 in strong intra- and inter-chromosomal contacts. The IHI/KEE-like regions marked by H3K27me3 in Marchantia (Figure 6) are likely to be distinct from heterochromatic islands marked by H3K9 methylation in flowering plants both in their genesis and association with transcriptional regulation. As in many species of eukaryotes, transposons associate primarily with H3K9me2 in flowering plants [4]. However, in Arabidopsis, a fraction of transposons are marked by H3K27me3 in reproductive tissues, which are characterized by reduced DNA methylation [62] and in mutants with reduced DNA methylation [63, 64]. In mammalian cells deprived of DNA methylation or H3K9me3, H3K27me3 also associates with transposons and represses transcription of retroelements MRVL [65, 66]. Similarly, in the ascomycete Neurospora crassa, the loss of H3K9me3 or the H3K9me3 reader Heterochromatin Protein 1 causes redistribution of H3K27me2/3 to constitutive heterochromatin [67]. These reports suggest that H3K9 methylation and the associated DNA methylation prevent association between H3K27me3 and repeats and transposons. Such an association takes place in species with low DNA methylation such as red algae [68] and diatoms [69] representing groups that diverged from the streptophyte lineage more than 900 Mya. Phylogenetic data support the emergence of PRC2 function in unicellular eukaryotes [70]. In ciliates Tetrahymena thermophila and Paramecium tetraurelia, H3K27me3 is associated with transposon repression [71, 72]. In ciliates, PRC2 deposits both H3K27me3 and H3K9me3 [71], and this activity is associated with RNAi [72]. In contrast, we observe a clear distinction between the group of transposons marked by H3K9 methylation and H3K27me3 in Marchantia, which may result from the PRC2-independent evolution of the H3K9 methylation pathway in plants [2, 73, 74]. We thus propose that PRC2 evolved as a repressor of transposons in ancestral unicellular eukaryotes. In Marchantia, the association between H3K27me3 and transposons is still extant. This might be explained by the absence of a strong feedback loop between DNA and H3K9 methylation in bryophytes [74]. It remains to be investigated whether H3K27me3 was still primarily involved in transposon silencing in charophycean algea, representative of ancestors of land plants. If that were the case, Marchantia would be an ideal model to study how and why this silencing pathway was replaced by H3K9 and DNA methylation during land plant evolution.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
All data generated in this study will be available for sharing and provided online at MarpolBase (http://marchantia.info/). Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Frédéric Berger (frederic.berger@gmi.oeaw.ac.at). Rabbit polyclonal anti-H2A.Z antibody is available upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Marchantia growth conditions
Male Takaragaike-1 (Tak-1) [75] (Marchantia polymorpha) gemmae were cultured on half-strength B5 1% (w/v) agar medium supplemented with 1% (w/v) sucrose. The light condition was set to long day (16 hr light and 8 hr dark, 3,000 lux) and the temperature was maintained at 22°C.
METHOD DETAILS
Isolation of nuclear DNA from Marchantia
Briefly, 100 g of 3-week-old thallus was rinsed with 250 mL of ice-cold ethyl ether for 3 minutes followed by washing with cold TE buffer, and homogenized with 1 L of cold MPD-based extraction buffer (1 M 2-methy-2,4-pentanediol, 10 mM PIPES-KOH, 10 mM MgCl2x6H2O, 2% polyvinylpyrrolidone (PVP), 10 mM sodium metabisulfite, 5 mM 2-mercaptoethanol, 0.5% sodium diethyldithiocarbamate, 200 mM L-lysine, and 6 mM EGTA, pH 6.0.). The slurry was filtered through a 40 μm nylon filter, and Triton X-100 was added to the flow-through to 0.5% v/v. The mixture was centrifuged at 800 x g for 20 min at 4°C, and the nuclei pellet was washed three times with MPDB buffer (0.5 M 2-methy-2,4-pentanediol, 10 mM PIPES-KOH, 10 mM MgCl2×6H2O, 0.5% Triton X-100, 10 mM sodium metabisulfite, 5 mM 2-mercaptoethanol, 200 mM L-lysine, and 6 mM EGTA, pH 6.0.). Nuclei were then lysed with 2% SDS (w/v) at 60°C for 10 min, and the released genomic DNA was extracted with phenol/chloroform/isoamyl alcohol (25:24:1) following the standard protocol. The aqueous layer was dialyzed overnight into TE buffer at 4°C. On the next day, RNase T1 and RNase A were added to the sample to a final concentration of 50 units/ml and 50 μg/ml, respectively. RNA digestion was performed at 37°C for 60 min. Subsequently, Proteinase K was added to a final concentration of 150 μg/ml, and the solution was further incubated at 37°C for 60 min. Finally, DNA was recovered by following standard phenol/chloroform/isoamyl alcohol extraction and ethanol precipitation protocols.
Hi-C library preparation and sequencing
The in situ Hi-C library preparation was performed by following a protocol established for rice seedlings [43] In total, two replicates of 3-week old Tak-1 thalli Hi-C libraries were made, and for each replicate around 0.5 g of fixed sample was homogenized for nuclei isolation. The libraries were sequenced on an Illumina HiSeq 3000 instrument with 2 × 150 bp reads.
Chromosome-scale genome assembly
PacBio reads were assembled into scaffolds with miniasm using default settings [76] except that the minimum coverage was set as -c 2. Next, Hi-C reads were mapped to these scaffolds with an iterative mapping strategy described previously [43]. Subsequently, Hi-C contacts were processed by the 3d-dna-master software to further assemble the scaffolds [77]. In brief, the whole process had two steps. First, it attempted to connect all scaffolds to build a genomic “super-scaffold.” Next, it split this “super-scaffold” into chromosomes according to the chromosome number defined by the user. For the first step, a Tak-1 “super-scaffold” was generated with following parameters: -t 1000 -s 3 -c 9 -w 25000 -n 1000 -k 5 -d 150000. Consistent with Tak-1’s karyotype, this “super-scaffold” showed 9 blocks of self-interacting domains with various sizes (Figure S1) [78]. For the second step, we split this “super-scaffold” into 9 segments (chromosomes) with the parameter set as -c 9 accordingly. Because the estimated size of the Tak-1 V chromosome (10 Mb) is much smaller than the minimum expected chromosome size to be split from the “super-scaffold” by the 3d-dna-master program, we modified two default settings to circumvent this issue [15]. We changed the resolution setting (“res”) in the “run-asm-splitter.sh” file from 100000 (default) to 50000, and the bin number setting (“m_size_threshold”) in the “recursive-chromosome-splitter.py” file from 200 (default) to 60. In this way, we modified the lower boundary of “chromosome size” that the program accepted to 3 MB (50000 kb x 60), which is smaller than that of the V chromosome. As a result, the 3d-dna-master tool generated an assembled Tak-1 reference with 9 “chromosomes” that collectively covered around 215 MB as well as 441 unplaced scaffolds adding up to 3 MB that failed to be localized to any chromosomal sequence.
Next, we manually searched for local misjoint errors by checking the diagonals of Hi-C maps at 20 kb window setting. Typically, mapping Hi-C reads to a reference containing misjoints or large-scale chromosomal rearrangements gives rise to aberrant and strong “interactions” off the diagonals in Hi-C maps. Meanwhile, these regions display depleted interactions with their neighboring chromatin (see examples in Figures S1B and S1C, left panels). Upon identifying misjoints, we rearranged the corresponding scaffolds according to the Hi-C map such that the revised scaffold ordering would generate a continuous diagonal (Figures S1B and S1C, right panels). Finally, the manually inspected and corrected chromosomes were sorted in descending order according to their size and named chromosome 1 to 8 and V.
Genome assembly polishing
The chromosome-level assembly of the Tak-1 genome was further processed with the Pilon tool for local sequence correction [79]. A subset of Illumina short reads from Tak-1 (SRA: SRR1800537), which correspond to approximately 100X genomic coverage, were preprocessed using fastp [80] with “–cut_front–cut_tail” options. They were aligned to the pre-polished Hi-C assembly using BWA v0.7.15 [81] with the MEM algorithm. The alignment result was provided to Pilon ver 1.22 to correct short indels and SNPs (–fix indels,snps). Additionally, indels and SNPs in the protein-coding regions were corrected manually based on the mapping results of RNaseq and Iso-seq.
Gap closing and additional scaffolds
Assembly gaps in the polished genome sequences were filled with the ver 3.1 sequences after checking the flanking regions and the order of protein-coding genes within and around the gap. When both of the flanking 800 bp regions of the gap matched with ver 3.1 sequences (> 99% identity) and the gene order was consistent when compared to the annotation in the ver 3.1 genome, the gap was fully patched with the ver 3.1 sequence. When only one of the flanking 800 bp regions matched the ver 3.1 sequence, the gap was partially patched with the ver 3.1 sequence containing the target genes. In total, 52 assembly gaps were fully patched and 32 were partially patched.
When gene sequences from ver 3.1 genome, whose annotation was well supported by expression evidence and/or protein homology, were not mapped to the assembled genome, genomic regions containing those ver 3.1 genes were added as unplaced scaffolds. This resulted in additional 14 scaffolds. 20 unplaced scaffolds were removed from the assembly as they were redundant or considered to be derived from chloroplast genomes. We finally obtained the genome assembly designated as ver 5.1, which consists of 9 chromosomal sequences and an additional 435 unplaced scaffolds. Genetic markers were mapped to this genome and the agreement between linkage groups and assigned chromosomes was evaluated (Data S1)
CAGE-seq, Iso-seq, and data analysis
CAGE-seq and Iso-seq were employed for improving gene annotation. For CAGE-seq analysis, total RNA was isolated with an RNeasy kit (QIAGEN) from 10 day-old Tak-1 thalli cultured from gemmae under continuous white fluorescent tube light. CAGE library construction, sequencing, and mapping onto the v5.1 genome was carried out by DNAFORM (Yokohama, Kanagawa, Japan). The mapped read distribution on the v5.1 genome was calculated by RSeQC ver.3.0.0 [82]. For Iso-seq analysis, total RNA was separately prepared by an RNeasy kit from the meristematic regions of 10 day-old thalli cultured from gemmae (vegetative tissue) and immature gametangiophores (reproductive tissue) for each of Tak-1 (male) and Tak-2 (female) plants, and then pooled to make male and female pooled samples, each of which contains RNA from two different tissues. Library construction and sequencing by PacBio Sequel (Pacific Biosciences, Menlo Park, CA, USA) were carried out by Kazusa DNA Research Institute (Kazusa, Chiba, Japan). Obtained data were processed with the IsoSeq3 pipeline of SMRT Link v6.0 (Pacific Biosciences) to generate clean sequences and they were aligned to the genome using GMAP (ver. 2018-07-04) [83].
Genome annotation
Annotation of protein-coding genes was conducted through a combination of the ver 3.1 genome and de novo prediction. A total of 24,674 predicted transcript models (including 5,387 isoforms) for the ver 3.1 genome were obtained from MarpolBase (http://marchantia.info). After excluding 134 genes putatively encoded on the female sex chromosome, they were aligned to the ver 5.1 genome sequences using BLASTN. The 23,623 transcript models (96.2%) that were aligned without insertions or deletions within coding regions were transferred from the ver 3.1 genome. Subsequently, 455 were aligned to the ver 5.1 genome with GMAP and manually modified if needed. The remaining 462 transcript models, which were not supported by expression data or protein homology, were discarded as false genes.
For de novo gene prediction, RNA-seq libraries (SRA: SRR896223-30, PRJNA251267) were mapped to the repeat-masked genome using Hi-SAT2 (ver. 2.1.0) [84]. The mapping results were used to build transcript models using Braker2 (ver. 2.0.3) [85] and StringTie (ver. 1.3.4d) [86]. Braker2 was run with the Augustus parameters pre-trained using ver. 3.1 gene models. In total, 166 and 89 transcript models were incorporated from the results of Braker2 and StringTie, respectively. Based on manual inspection using RNA-seq and Iso-seq, 418 transcript models were also added. Functional annotation for transcript modeling was performed by an RPS-BLAST search against the Eukaryotic Orthologous Groups (KOG) database [87], KEGG pathway analysis using KEGG Automatic Annotation Server (KAAS) [88], and InterProScan [89].
The completeness of the gene set was evaluated by BUSCO using 303 universal single-copy orthologous markers designed for eukaryotes (eukaryota_odb9) [14].
Repeat masking was conducted using RepeatModeler (ver 1.0.11) and RepeatMasker (ver. 4.0.7) (http://www.repeatmasker.org). A de novo repeat library was constructed using RepeatModeler, which was then subjected to RepeatMasker as a custom library to mask repetitive regions of the genome. RepeatMasker was run with ‘-s -no_low’ parameters.
The annotation of micro-RNA genes and their putative targets was based on published information [90, 91]. The mature miRNA and v5.1 mRNA profiles were used for putative target prediction by psRNATarget [92]. The degradome profile from Tak-1 thallus (SRA: SRR2179617) was used to evaluate the target prediction based on the method that was published previously [90]. Putative targets had to fit the following criteria: (1) degradome reads of the cleaved site (CS-d reads) had to be greater than or equal to 5 reads; (2) the CS-d read count was claimed significant larger than the nearby 100 bp window (±50 bp from the site) if the p value of Poisson one-tail test was less than 0.05. Details of miRNA sequences and their target gene identities can be found in Data S2.
Nuclear tRNA prediction was done with tRNAscan-SE version 2.0 using the general model parameter [93]. The data were manually curated to filter tRNA, organellar contaminations, and tRNA-like sequences. Details of each nuclear tRNA locus can be found in Data S2.
Large sequence comparison of sex chromosomes from ver. 3.1 and ver. 5.1 were aligned and visualized by D-Genies with default parameters [94].
Chromatin profiling
Marchantia Tak-1 gemmae were cultured on half-strength B5 medium under continuous light at 22°C for 14 days. Plants, excluding gemmae cups, were chopped in Galbraith buffer (45 mM MgCl2-6H2O, 30 mM Trisodium citrate, 20 mM MOPS) pH 7.0 plus 0.1% Triton X-100 with a razor blade on ice to extract nuclei. Nuclei were passed through a 40 μm filter and stained with 2 μg/mL DAPI before sorting on a BD FACSARIA III (BD Biosciences). Aliquots of 40,000 nuclei were collected in 10X binding buffer (200 mM HEPES-KOH pH 7.9, 100mM KCl, 10mM CaCl2, 10mM MnCL2, 5mM spermidine) diluted 1:10 in 1x PBS. The harvested nuclei were processed with the CUT&RUN protocol [31]. Gently resuspend Bio-Mag Plus Concanavalin A coated beads (Polysciences, Inc. #86057). Withdraw 10 x N samples μL bead slurry, and transfer to 40 x N of Binding buffer in a 2 mL Eppendorf tube. Place on a magnet stand and wash twice in 1mL 1x Binding buffer. Resuspend in 10 x N μL Binding buffer. Add bead slurry to nuclei while gently vortexing. Rotate 10 min at room temperature. Place on the magnet stand, allow to clear (~20 s->2 min) and pull off the liquid. Add 1mL Blocking buffer (1mL Wash buffer (20mM HEPES pH 7.5, 150mM NaCl, 0.5mM Spermidine, 0.1% BSA, 1cOmplete Protease Inhibitor Cocktail (Roche), 2mM EDTA) and mix either with gentle pipetting or invert ~10x. Incubate 5′ at room temperature. Place on the magnet stand and pull off the liquid. Add 1mL Wash buffer, invert ~10x. (or more Wash buffer, make sure the wash buffer has coated the whole tube). Place on the magnet stand and pull off the liquid. Resuspend in 250 μL Wash buffer. Add 2.5 μL primary antibody to Wash buffer (1:100) while gently vortexing. Incubate on rotator 2hr at 4°C. Quick spin and wash twice in 1mL Wash buffer. Pull off the liquid and resuspend each sample in 250 μL Wash buffer. Add 0.625 μL pA-MNase for a final pA-MN ase concentration of 1:400 for Batch #6. Incubate 1 hr on rotator at 4°C. Quick spin and wash twice in 1mL Wash buffer. Pull off the liquid and resuspend in 150 μL Wash buffer. Equilibrate to 0°C in in metal blocks fitted for Eppendorf tubes in ice water in cold room (5-10min). Remove a tube from 0°C, add 3 μL 100 mM CaCl2 per 150 μL while vortexing, flick quickly then return to 0°C. Stop after 30min with 150 μL 2XSTOP+ (200mM NaCl, 20mM EDTA, 4mM EGTA, 50 μg/mL RNase A, 40 μg/mL glycogen, 10pg/mL heterologous DNA (HEK293). Incubate 20’ 37°C to RNase and release CUT&RUN fragments from the insoluble nuclear chromatin. Spin 5′ 16,000 x g 4°C, and pull off supernatants to fresh tubes. To each sample add 3 μL 10% SDS (to 0.1%), and 2.5 μL Proteinase K (20 mg/ml). Mix by inversion and incubate 10 min 70°C. Add 300 μL buffered phenol-chloroform-isoamyl solution (25:24:1) and vortex. Transfer to a phase-lock tube, and spin 5 min full speed. Remove aqueous to a fresh tube containing 2 μL of 2 mg/ml glycogen. Add 750 μL 100% ethanol and mix by vortexing or tube inversion. Leave at −20°C O/N, spin 10 min full speed 4°C. Pour off the liquid and drain on a paper towel. Wash the pellet (hardly visible) in 1 mL 70% ethanol, spin briefly full speed. Carefully pour off the liquid and drain on a paper towel. Air dry. When the pellet is dry, dissolve in 50 μL nuclease-free water. Transfer to strip tubes.
Nuclei immunostaining
Marchantia Tak-1 thallus and Physcomitrella patens gametophyte were chopped in Galbraith buffer (45 mM MgCl2-6H2O, 30 mM Trisodium citrate, 20 mM MOPS) pH 7.0 plus 0.1% Triton X-100 with a razor blade on ice to extract nuclei. Nuclei were passed through a 40 μm filter and immunostained following a protocol by [95]. 16% paraformaldehyde was added to a final concentration of 4% and nuclei were incubated for 20min on ice. 2M glycine was then added to a final concentration of 125mM. 10μL of the nuclei suspension was spotted onto glass slides and dried at room temperature. Slides were then immunostained by the VBCF Histopathology as follows: Wash 5x 10min with 1xPBS + 0.1% Tween-20 (PBST). 2x 30min blocking buffer (2% BSA, 1% 1x PBS, 0.1% Tween-20). 6hr primary antibody (1:100) at room temperature. 6x 10min 1x PBST. 2hr secondary antibody (1:500) at room temperature. 8x 10min 1x PBST. Slides were dried and 200μL of 1.5μg/mL DAPI solution was added. Slides were incubated in the dark at room temperature for 20min and washed with 200μL water. Liquid was removed and slides were mounted in 10μL Vectashield + DAPI (Vector Laboratories) and sealed. Images were obtained on an LSM 780 (Zeiss) and processed using FIJI [96]. Images shown are maximum intensity projections. Contrast was enhanced for Marchantia H3K27me1 and H3K27me3 stainings and Physcomitrella H3K4me3, H3K27me1, and H3K27me3 stainings.
Chromosome spread preparation and Fluorescence in situ Hybridization (FISH)
Centromeric repeats probes were synthesized as two oligos: 5′-[DIG]TGGGCTTGTTCACGACGGCCGGGCGCACATACCTGCA AATTTTCAGCCCCAACGGAGCT[DIG]-3′ and 5′-[DIG]TTTTCAGCCCCAACGGAGCTGCTGTCAAGAAGTTGTCATTTCGAAACTTTG AGTTT[DIG]-3′ (Figure S3B), where the terminal thymidines were labeled with digoxigenin (DIG). These two oligos were mixed in a 1:1 molar ratio and used for hybridization. Telomere probes were synthesized as 5′-[DIG](TTTAGGG)7T[DIG]-3′, with their terminal thymidines labeled.
Chromosome spread preparation was performed as described [16] and placed on Superfrost Ultra Plus Adhesion Slides (ThermoFisher Scientific). For chromosome spread hybridization, 5 μl of hybridization buffer [58] containing 25 ng DIG-labeled telomere probes was used. Before applying the probes to the slides, the probes were denatured at 95°C for 5 min and cooled for 5 min on ice. For hybridization, the slides were heated at 70°C for 8 min and incubated at 37°C overnight in a humid chamber. Detection of the DIG probes was performed according to [58].
For FISH experiment with Marchantia nuclei, around 5,000 nuclei were collected with FACS as described [97] and were used for one hybridization spot (~1 cm2). After nuclei sorting, the nuclei were centrifuged for 3,000 x g at 4°C for 7 min, and the pellet was resuspended with 20 μl PBS buffer. The nuclei were incubated at 65°C for 30 min, and mixed with 5 μl 0.1 mg/ml RNase A. The mixture was transferred onto a Superfrost Ultra Plus Adhesion Slide (ThermoFisher Scientific) and incubated for 1 h at 37°C. At the end of RNase A treatment, the nuclei became attached to the glass slide. Next, the slide was washed briefly with PBS buffer and dehydrated in a graded series of alcohol solutions. All subsequent steps, including probe denaturation, hybridization, washing, and detection were performed as described for chromosome spread samples.
Centromere identification
Regions with strong Hi-C interactions among each other and occurring only once per chromosome were aligned to create dot plots using EMBOSS Dotmatcher with 10 bp windows and a threshold of 50 [98] (Figure S3D). One 165 bp repeat found in each region was identified and the centromeric FISH probes are indicated (Figure S3B).
QUANTIFICATION AND STATISTICAL ANALYSIS
Chromatin profiling analyses
CUT&RUN reads were mapped to the Tak-1 v5.1 genome presented in this paper using Bowtie2 v2.1.0 [99] and further processed using Samtools v1.3 [100] and Bedtools v2.17.0 [101]. Reads with MAPQ less than ten were removed with Samtools v1.3 and duplicates were removed with Picard v1.141 (http://broadinstitute.github.io/picard/). Inserts less than 150 bp were removed from further analyses, as these fragments are sub-nucleosomal in size and likely represent noise when profiling histones and histone modifications. Deduplicated reads from 2-4 biological replicates were merged. We called peaks for chromatin marks using HOMER v4.9 [102] and considered a gene associated with a mark if at least 50% of the gene length overlapped with peaks. We used the following settings: -style histone -size 250 -minDist 500. Bigwig files were made using deepTools v2.2.4 [103].
Pearson correlation matrices were generated using deepTools v2.5.4 [103] using multiBamSummary and plotCorrelation tools. Overlaps between features were calculated using bedtools intersect v2.27.1 [101]. Circos plots were generated using circlize [104] using bedgraphs of peaks called by HOMER. Chromosome coverage plots were generated using the smooth.spline function in R v3.4.0 (https://www.R-project.org/). IGV v2.3.97 [105] browser shot was obtained by loading bed files of peaks and bigwig files of RNA-Seq and H3 coverage data.
Clustering analyses
K-means clustering of chromatin marks was performed using deepTools v2.2.4 [103]. Matrices were computed using computeMatrix for either genes or repeats using bigwig files as input and the start of the feature as the reference point with 1 kb upstream and downstream. Heatmaps of matrices were plotted with plotHeatmap with k-means clustering. Cluster assignments can be found in Data S3.
Gene expression analyses
Gene expression data from [33] were downloaded from the SRA (samples DRR050343, DRR050344, DRR050345) and processed with RSEM v1.2.31 [106] and STAR v2.5.2a [107]. Transcript Per Million (TPM) values were averaged from three biological replicates from vegetative thalli and used for further analyses. Genes were determined to overlap with a feature of interest if at least 50% of the gene length overlapped with the feature.
DNA methylation analysis
Bisulfite sequencing data of Tak1-1 thallus was downloaded from SRA (SRA: SRP101412) and analyzed following the method described in [32]. Read mapping and the identification of methylated cytosines were performed with Bismark v0.22.1 with default settings [108]. The mean methylation percentage per gene or repeat was calculated using MethylDackel v0.4.0 (https://github.com/dpryan79/MethylDackel) from analyzed cytosines that were assigned to genes or repeats.
Hi-C map normalization
Raw Hi-C reads of the two replicates used for genome assembly were mapped to the final Tak-1 genome assembly. Read mapping and filtering were performed essentially as described [43]; at the end, about 89 million informative Hi-C reads were obtained in total (Table S2). Hi-C matrices normalization was performed as described [43] assuming equal visibility of individual genomic bins, with which a Hi-C matrix was adjusted toward having similar sum values for each row or column [109]. Normalization of the Hi-C map at 50 kb resolution was performed at the genome-wide level (i.e., all chromosomes were included), while normalization at 20 kb was done separately for each chromosome.
ChIP-Seq data analysis
Raw ChIP-Seq reads from [110] were mapped to the TAIR10 Arabidopsis thaliana genome using Bowtie2 v2.1.0 [99] and further processed using Samtools v1.3 [100] and Bedtools v2.17.0 [101]. Reads with MAPQ less than ten were removed with Samtools v1.3 and duplicates were removed with Picard v1.141 (http://broadinstitute.github.io/picard/). Broad peaks were called using MACS2 [111] using H3 as a control with the settings:–nomodel–nolambda–broad -q 0.01–broad-cutoff 0.1 -g 1.19146348e8. Overlaps between features were calculated using bedtools intersect v2.27.1 [101].
DATA AND CODE AVAILABILITY
All raw read data and assembled sequence data that support the findings of this study have been submitted to the DDBJ/ENA/NCBI public sequence databases under accession numbers SRA: PRJNA553138 and PRJDB8530.
The code supporting the current study have not been deposited in a public repository but are available from the corresponding author on request.
ADDITIONAL RESOURCES
MarpolBase genome database for Marchantia polymorpha containing a genome browser with expression and chromatin profiles, BLAST search tools and download tools for current and past genomic resources: http://marchantia.info
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-H2A.Z | This paper | N/A |
| Rabbit polyclonal anti-H3 | Abcam | Cat# ab1791; RRID:AB_302613 |
| Rabbit polyclonal anti-H3K4me1 | Abcam | Cat# ab8895; RRID:AB_306847 |
| Rabbit polyclonal anti-H3K4me3 | Abcam | Cat# ab8580; RRID:AB_306649 |
| Rabbit polyclonal anti-H3K9ac | Active Motif | Cat# 39137; RRID:AB_2561017 |
| Rabbit polyclonal anti-H3K9me1 | Abcam | Cat# ab9045; RRID:AB_306963 |
| Rabbit polyclonal anti-H3K14ac | Millipore | Cat# 07-353; RRID:AB_310545 |
| Rabbit polyclonal anti-H3K27me1 | Millipore | Cat# 17-643; RRID:AB_1587128 |
| Rabbit polyclonal anti-H3K27me3 | Millipore | Cat# 07-449; RRID:AB_310624 |
| Rabbit polyclonal anti-H3K36me3 | Abcam | Cat# ab9050; RRID:AB_306966 |
| Goat anti-rabbit IgG Alexa Fluor 488 | Abcam | Cat# ab150077; RRID:AB_2630356 |
| Goat anti-rabbit IgG Alexa Fluor 594 | Abcam | Cat# ab150080; RRID:AB_2650602 |
| Monoclonal Anti-Digoxin, Clone DI-22 | Sigma | Cat# D8156; RRID:AB_259242 |
| Goat anti-Mouse IgG Alexa Fluor 488 | Invitrogen | Cat# A-11017; RRID:AB_143160 |
| Biological Samples | ||
| HEK293 DNA | Danhua Jiang, Beijing, China | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gamborg B5 basal salt | Duchefa | Cat# G0209 |
| RNase T1 | ThermoFisher Scientific | Cat# EN0541 |
| RNase A | ThermoFisher Scientific | Cat# EN0531 |
| Proteinase K | ThermoFisher Scientific | Cat# EO0491 |
| Bio-Mag Plus Concanavalin A coated beads | Polysciences | Cat#86057 |
| cOmplete Protease Inhibitor Cocktail | Roche | Cat#11697498001 |
| pA-MNase | [30] | Henikoff lab batch #6 purified 11.01.2017 |
| Vectashield with DAPI | Vector Laboratories | Cat#H-1200 |
| Critical Commercial Assays | ||
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| Deposited Data | ||
| Marchantia polymorpha genome v3.1 | [10] | http://marchantia.info, SRA: SRR1800537 |
| Marchantia polymorpha genome v5.1 | This paper | http://marchantia.info; SRA: PRJNA553138 and PRJDB8530 |
| CAGE-seq and Iso-seq | This paper | SRA: PRJDB8530 |
| Pair-end genome shotgun library for Hi-C analysis | [10] | SRA: SRR396657 and SRR396658 |
| De novo gene prediction from RNA-seq libraries | [10] | SRA: SRR896223-30 and PRJNA251267 |
| Tak-1 bisulfite sequencing | [32] | SRA: SRR5314038 |
| Degradome for miRNA target prediction | [75] | SRA: SRR2179617 |
| Tak-1 thallus RNA-seq for expression analyses | [76] | SRA: DRR050343, DRR050344, and DRR050345 |
| Arabidopsis ChIP-Seq sequencing | [77] | SRA: SRR1005422, SRR1005423, and SRR1999291 |
| Eukaryotic Orthologous Groups (KOG) database | [78] | https://www.ncbi.nlm.nih.gov/COG/ |
| Experimental Models: Organisms/Strains | ||
| Marchantia polymorpha Tak-1 | [10] | N/A |
| Physcomitrella patens Gransden | [79] | N/A |
| Arabidopsis thaliana Col-0 | Nottingham Arabidopsis Stock Centre | N/A |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| Software and Algorithms | ||
| miniasm | [75] | https://github.com/lh3/miniasm |
| 3d-dna-master | [80] | https://github.com/theaidenlab/3d-dna/ |
| Hi-C reads processing and map normalization | [81] | https://github.com/changliu325/Arabidopsis_crwn1_chromatin/tree/master/HiC |
| fastp | [82] | https://github.com/OpenGene/fastp |
| BWA v0.7.15 | [83] | http://bio-bwa.sourceforge.net/ |
| Pilon v1.22 | [84] | https://github.com/broadinstitute/pilon |
| RSeQC v3.0.0 | [85] | http://rseqc.sourceforge.net/ |
| IsoSeq3 SMRT Link v6.0 | Pacific Biosciences | https://github.com/PacificBiosciences/IsoSeq |
| GMAP V2018-07-04 | [86] | https://omictools.com/gmap-tool |
| Hi-SAT2 v2.1.0 | [87] | https://ccb.jhu.edu/software/hisat2/index.shtml |
| Braker2 v2.0.3 | [88] | https://github.com/Gaius-Augustus/BRAKER |
| StringTie v1.3.4d | [89] | https://ccb.jhu.edu/software/stringtie/ |
| KEGG Automatic Annotation Server (KAAS) | [78] | https://www.genome.jp/kegg/kaas/ |
| InterProScan v5.33 | [90] | https://www.ebi.ac.uk/interpro/ |
| BUSCO v3.0.2 | [14] | https://busco.ezlab.org |
| RepeatModeler v1.0.11 | DFAM consortium | http://www.repeatmasker.org/RepeatModeler/ |
| RepeatMasker v4.0.7 | DFAM consortium | http://www.repeatmasker.org |
| psRNATarget | [91] | http://plantgrn.noble.org/psRNATarget/ |
| tRNAscan-SE v2.0 | [92] | http://lowelab.ucsc.edu/tRNAscan-SE/ |
| D-Genies | [93] | https://github.com/genotoul-bioinfo/dgenies |
| Bowtie2 v2.1.0 | [94] | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Samtools v1.3 | [95] | http://www.htslib.org/ |
| Bedtools v2.17.0, v2.17.1 | [96] | https://bedtools.readthedocs.io/en/latest/ |
| Picard v1.141 | Broad Institute, Boston, MA | http://broadinstitute.github.io/picard/ |
| HOMER v4.9 | [97] | http://homer.ucsd.edu/homer/ |
| deepTools v2.2.4, v2.5.4 | [98] | https://deeptools.readthedocs.io/en/develop/ |
| circlize | [99] | https://jokergoo.github.io/circlize_book/book/ |
| R v3.4.0 | R Foundation for Statistical Computing, Vienna, Austria | https://www.R-project.org/ |
| IGV v2.3.97 | [100] | https://software.broadinstitute.org/software/igv/ |
| RSEM v1.2.31 | [101] | https://github.com/deweylab/RSEM |
| STAR v2.5.2a | [102] | https://github.com/alexdobin/STAR |
| Bismark v0.22.1 | [103] | https://www.bioinformatics.babraham.ac.uk/projects/bismark/ |
| MethylDackel v0.4.0 | MPI Immunology and Epigenetics, Freiburg, Germany | https://github.com/dpryan79/MethylDackel |
| FIJI | [104] | https://fiji.sc/ |
| EMBOSS Dotmatcher | [105] | http://www.bioinformatics.nl/cgi-bin/emboss/dotmatcher |
| MACS2 | [106] | https://github.com/taoliu/MACS |
| Other | ||
| Superfrost Ultra Plus Adhesion Slides | ThermoFisher Scientific | Cat# 10417002 |
| BD FACSARIA III | BD Biosciences | https://www.bdbiosciences.com/en-us |
| CAGE library construction, sequencing and mapping | DNAFORM | https://www.dnaform.jp/en/ |
| PacBio Sequel library construction and sequencing | Kazusa DNA Research Institute | http://www.kazusa.or.jp/en/ |
Highlights.
A database combining genomic information and chromatin profiles for Marchantia
Correlations between chromatin marks and transcription are conserved in land plants
A significant portion of constitutive heterochromatin is marked by H3K27me3
Insights into the evolution of TAD organization in plants
ACKNOWLEDGMENTS
We acknowledge computing support by the High Performance and Cloud Computing Group at the Zentrum für Datenverarbeitung of the University of Tübingen, the state of Baden-Württemberg through bwHPC, and the German Research Foundation (DFG) through grant no. INST 37/935-1 FUGG. We acknowledge Ms. Fumi Hayashi and Dr. Mika Sakamoto for helping with the exhaustive manual correction of the assembly and Dr. J. Matthew Watson for proofreading the manuscript. F.B. acknowledges support from the PlantS, next-generation sequencing and histopathology facilities at the Vienna BioCenter Core Facilities (VBCF), and the BioOptics facility and Molecular Biology Services from the Institute for Molecular Pathology (IMP). C.L. and N.W. were supported by European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement no. 757600). This work was also supported by the Gregor Mendel Institute (F.B. and S.A.) and FWF grants I2163-B16, I2303-B25, P26887, and DK 1238 chromosome dynamics (S.A.M. and F.B.), NIH (R01 GM065383 to D.E.S.; R01 GM127402 to E.V.S.), Russian Science Foundation (18-74-00112 to L.V.R.), Russian Foundation for Basic Research (18-016-00146 to E.V.S.), and funds from the Russian Government Program for Competitive Growth of Kazan Federal University. We also received further support from JSPS KAKENHI grant nos. 16H06279 (Y.T., Y.N., and T.K.), 15K21758 (T.K., F.B., and Y.N.), 17H05841 (S.Y.), 25113001 (T.K.), and 25113009 (T.K.); the Project Research of the Faculty of Biology-Oriented Science and Technology, Kindai University no. 16-I-3,2017 (K.T.Y.); and the Australian Research Council, DP170100049 (J.L.B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.12.015.
REFERENCES
- 1.Talbert PB, Ahmad K, Almouzni G, Ausió J, Berger F, Bhalla PL, Bonner WM, Cande WZ, Chadwick BP, Chan SW, et al. (2012). A unified phylogeny-based nomenclature for histone variants. Epigenetics Chromatin 5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talbert PB, Meers MP, and Henikoff S (2019). Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat. Rev. Genet 20, 283–297. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T (2007). Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 4.Sequeira-Mendes J, Aragüez I, Peiró R, Mendez-Giraldez R, Zhang X, Jacobsen SE, Bastolla U, and Gutierrez C (2014). The Functional Topography of the Arabidopsis Genome Is Organized in a Reduced Number of Linear Motifs of Chromatin States. Plant Cell 26, 2351–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009). Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, et al. (2017). Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doğan ES, and Liu C (2018). Three-dimensional chromatin packing and positioning of plant genomes. Nat. Plants 4, 521–529. [DOI] [PubMed] [Google Scholar]
- 8.Sotelo-Silveira M, Chévez Montes RA, Sotelo-Silveira JR, Marsch-Martínez N, and de Folter S (2018). Entering the Next Dimension: Plant Genomes in 3D. Trends Plant Sci 23, 598–612. [DOI] [PubMed] [Google Scholar]
- 9.de Sousa F, Foster PG, Donoghue PCJ, Schneider H, and Cox CJ (2019). Nuclear protein phylogenies support the monophyly of the three bryophyte groups (Bryophyta Schimp.). New Phytol. 222, 565–575. [DOI] [PubMed] [Google Scholar]
- 10.Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, et al. (2017). Insights into Land Plant Evolution Garnered from the Marchantia polymorpha. Genome. Cell 171, 287–304. [DOI] [PubMed] [Google Scholar]
- 11.Lang D, Ullrich KK, Murat F, Fuchs J, Jenkins J, Haas FB, Piednoel M, Gundlach H, Van Bel M, Meyberg R, et al. (2018). The Physcomitrella patens chromosome-scale assembly reveals moss genome structure and evolution. Plant J. 93, 515–533. [DOI] [PubMed] [Google Scholar]
- 12.Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, et al. (2010). Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fransz P, De Jong JH, Lysak M, Castiglione MR, and Schubert I (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99, 14584–14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, and Zdobnov EM (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. [DOI] [PubMed] [Google Scholar]
- 15.Yamato KT, Ishizaki K, Fujisawa M, Okada S, Nakayama S, Fujishita M, Bando H, Yodoya K, Hayashi K, Bando T, et al. (2007). Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system. Proc. Natl. Acad. Sci. USA 104, 6472–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada S, Fujisawa M, Sone T, Nakayama S, Nishiyama R, Takenaka M, Yamaoka S, Sakaida M, Kono K, Takahama M, et al. (2000). Construction of male and female PAC genomic libraries suitable for identification of Y-chromosome-specific clones from the liverwort, Marchantia polymorpha. Plant J. 24, 421–428. [DOI] [PubMed] [Google Scholar]
- 17.Fujisawa M, Nakayama S, Nishio T, Fujishita M, Hayashi K, Ishizaki K, Kajikawa M, Yamato KT, Fukuzawa H, and Ohyama K (2003). Evolution of ribosomal DNA unit on the X chromosome independent of autosomal units in the liverwort Marchantia polymorpha. Chromosome Res. 11, 695–703. [DOI] [PubMed] [Google Scholar]
- 18.Rabanal FA, Mandáková T, Soto-Jiménez LM, Greenhalgh R, Parrott DL, Lutzmayer S, Steffen JG, Nizhynska V, Mott R, Lysak MA, et al. (2017). Epistatic and allelic interactions control expression of ribosomal RNA gene clusters in Arabidopsis thaliana. Genome Biol. 18, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki K (2004). Characterization of telomere DNA among five species of pteridophytes and bryophytes. J. Bryol 26, 175–180. [Google Scholar]
- 20.Shakirov EV, Perroud PF, Nelson AD, Cannell ME, Quatrano RS, and Shippen DE (2010). Protection of Telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell 22, 1838–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shakirov EV, and Shippen DE (2004). Length regulation and dynamics of individual telomere tracts in wild-type Arabidopsis. Plant Cell 16, 1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveira LC, and Torres GA (2018). Plant centromeres: genetics, epigenetics and evolution. Mol. Biol. Rep 45, 1491–1497. [DOI] [PubMed] [Google Scholar]
- 23.Henikoff S, and Furuyama T (2012). The unconventional structure of centromeric nucleosomes. Chromosoma 121, 341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, Birchler JA, Parrott WA, and Dawe RK (2003). A molecular view of plant centromeres. Trends Plant Sci. 8, 570–575. [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Wing RA, Bennetzen JL, and Jackson SA (2007). Plant centromere organization: a dynamic structure with conserved functions. Trends Genet. 23, 134–139. [DOI] [PubMed] [Google Scholar]
- 26.Steiner FA, and Henikoff S (2015). Diversity in the organization of centromeric chromatin. Curr. Opin. Genet. Dev. 31, 28–35. [DOI] [PubMed] [Google Scholar]
- 27.Bass HW, Riera-Lizarazu O, Ananiev EV, Bordoli SJ, Rines HW, Phillips RL, Sedat JW, Agard DA, and Cande WZ (2000). Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci 113, 1033–1042. [DOI] [PubMed] [Google Scholar]
- 28.Schwarzacher T (1997). Three stages of meiotic homologous chromosome pairing in wheat: cognition, alignment and synapsis. Sex. Plant Reprod 10, 324–331. [Google Scholar]
- 29.Zhang F, Tang D, Shen Y, Xue Z, Shi W, Ren L, Du G, Li Y, and Cheng Z (2017). The F-Box Protein ZYGO1 Mediates Bouquet Formation to Promote Homologous Pairing, Synapsis, and Recombination in Rice Meiosis. Plant Cell 29, 2597–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skene PJ, and Henikoff S (2017). An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. eLife 6 Published online January 16, 2017. 10.7554/eLife.21856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng XY, and Gehring M (2019). Low-input chromatin profiling in Arabidopsis endosperm using CUT&RUN. Plant Reprod. 32, 63–75. [DOI] [PubMed] [Google Scholar]
- 32.Schmid MW, Giraldo-Fonseca A, Rövekamp M, Smetanin D, Bowman JL, and Grossniklaus U (2018). Extensive epigenetic reprogramming during the life cycle of Marchantia polymorpha. Genome Biol. 19, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higo A, Niwa M, Yamato KT, Yamada L, Sawada H, Sakamoto T, Kurata T, Shirakawa M, Endo M, Shigenobu S, et al. (2016). Transcriptional Framework of Male Gametogenesis in the Liverwort Marchantia polymorpha L. Plant Cell Physiol. 57, 325–338. [DOI] [PubMed] [Google Scholar]
- 34.Law JA, and Jacobsen SE (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zilberman D, Gehring M, Tran RK, Ballinger T, and Henikoff S (2007). Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat. Genet 39, 61–69. [DOI] [PubMed] [Google Scholar]
- 36.Lawrence M, Daujat S, and Schneider R (2016). Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 32, 42–56. [DOI] [PubMed] [Google Scholar]
- 37.Jiang D, and Berger F (2017). DNA replication-coupled histone modification maintains Polycomb gene silencing in plants. Science 357, 1146–1149. [DOI] [PubMed] [Google Scholar]
- 38.Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, and Meyer BJ (2015). Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, and Ren B (2012). Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sexton T, and Cavalli G (2015). The role of chromosome domains in shaping the functional genome. Cell 160, 1049–1059. [DOI] [PubMed] [Google Scholar]
- 41.Rowley MJ, Nichols MH, Lyu X, Ando-Kuri M, Rivera ISM, Hermetz K, Wang P, Ruan Y, and Corces VG (2017). Evolutionarily Conserved Principles Predict 3D Chromatin Organization. Mol. Cell 67, 837–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong P, Tu X, Chu PY, Lü P, Zhu N, Grierson D, Du B, Li P, and Zhong S (2017). 3D Chromatin Architecture of Large Plant Genomes Determined by Local A/B Compartments. Mol. Plant 10, 1497–1509. [DOI] [PubMed] [Google Scholar]
- 43.Liu C, Cheng YJ, Wang JW, and Weigel D (2017). Prominent topologically associated domains differentiate global chromatin packing in rice from Arabidopsis. Nat. Plants 3, 742–748. [DOI] [PubMed] [Google Scholar]
- 44.Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, Radchuk V, Dockter C, Hedley PE, Russell J, et al. (2017). A chromosome conformation capture ordered sequence of the barley genome. Nature 544, 427–433. [DOI] [PubMed] [Google Scholar]
- 45.Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, and Weigel D (2015). Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res. 25, 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang M, Tu L, Lin M, Lin Z, Wang P, Yang Q, Ye Z, Shen C, Li J, Zhang L, et al. (2017). Asymmetric subgenome selection and cis-regulatory divergence during cotton domestication. Nat. Genet 49, 579–587. [DOI] [PubMed] [Google Scholar]
- 47.Dong Q, Li N, Li X, Yuan Z, Xie D, Wang X, Li J, Yu Y, Wang J, Ding B, et al. (2018). Genome-wide Hi-Canalysis reveals extensive hierarchical chromatin interactions in rice. Plant J. 94, 1141–1156. [DOI] [PubMed] [Google Scholar]
- 48.Feng S, Cokus SJ, Schubert V, Zhai J, Pellegrini M, and Jacobsen SE (2014). Genome-wide Hi-C analyses in wild-type and mutants reveal high-resolution chromatin interactions in Arabidopsis. Mol. Cell 55, 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grob S, Schmid MW, and Grossniklaus U (2014). Hi-C analysis in Arabidopsis identifies the KNOT, a structure with similarities to the flamenco locus of Drosophila. Mol. Cell 55, 678–693. [DOI] [PubMed] [Google Scholar]
- 50.Baucom RS, Estill JC, Chaparro C, Upshaw N, Jogi A, Deragon JM, Westerman RP, Sanmiguel PJ, and Bennetzen JL (2009). Exceptional diversity, non-random distribution, and rapid evolution of retroelements in the B73 maize genome. PLoS Genet. 5, e1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009). The B73 maize genome: complexity, diversity, and dynamics. Science 326, 1112–1115. [DOI] [PubMed] [Google Scholar]
- 52.Tatuno S (1941). Zytologische Untersuchungen Uber die Lebermoose von Japan. Journal of Science of the Hiroshima University 4, 73–188. [Google Scholar]
- 53.Di Pierro M, Cheng RR, Lieberman Aiden E, Wolynes PG, and Onuchic JN (2017). De novo prediction of human chromosome structures: Epigenetic marking patterns encode genome architecture. Proc. Natl. Acad. Sci. USA 7 114, 12126–12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qi Y, and Zhang B (2019). Predicting three-dimensional genome organization with chromatin states. PLoS Comput. Biol 15, e1007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Janssen A, Colmenares SU, and Karpen GH (2018). Heterochromatin: Guardian of the Genome. Annu. Rev. Cell Dev. Biol 34, 265–288. [DOI] [PubMed] [Google Scholar]
- 56.Takuno S, Ran JH, and Gaut BS (2016). Evolutionary patterns of genic DNA methylation vary across land plants. Nat. Plants 2, 15222. [DOI] [PubMed] [Google Scholar]
- 57.Zemach A, McDaniel IE, Silva P, and Zilberman D (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919. [DOI] [PubMed] [Google Scholar]
- 58.Bi X, Cheng YJ, Hu B, Ma X, Wu R, Wang JW, and Liu C (2017). Nonrandom domain organization of the Arabidopsis genome at the nuclear periphery. Genome Res. 27, 1162–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pereman I, Mosquna A, Katz A, Wiedemann G, Lang D, Decker EL, Tamada Y, Ishikawa T, Nishiyama T, Hasebe M, et al. (2016). The Polycomb group protein CLF emerges as a specific tri-methylase of H3K27 regulating gene expression and development in Physcomitrella patens. Biochim. Biophys. Acta 1859, 860–870. [DOI] [PubMed] [Google Scholar]
- 60.van Mierlo G, Veenstra GJC, Vermeulen M, and Marks H (2019). The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 29, 660–671. [DOI] [PubMed] [Google Scholar]
- 61.Liu C, Wang C, Wang G, Becker C, Zaidem M, and Weigel D (2016). Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res. 26, 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinhofer I, Hehenberger E, Roszak P, Hennig L, and Köhler C (2010). H3K27me3 profiling of the endosperm implies exclusion of polycomb group protein targeting by DNA methylation. PLoS Genet. 6 Published online October 7, 2010. 10.1371/journal.pgen.1001152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deleris A, Stroud H, Bernatavichute Y, Johnson E, Klein G, Schubert D, and Jacobsen SE (2012). Loss of the DNA methyltransferase MET1 Induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 8, e1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mathieu O, Probst AV, and Paszkowski J (2005). Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 24, 2783–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peters AH, Kubicek S, Mechtler K, O’Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. (2003). Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12, 1577–1589. [DOI] [PubMed] [Google Scholar]
- 66.Reddington JP, Perricone SM, Nestor CE, Reichmann J, Youngson NA, Suzuki M, Reinhardt D, Dunican DS, Prendergast JG, Mjoseng H, et al. (2013). Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol. 14, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jamieson K, Wiles ET, McNaught KJ, Sidoli S, Leggett N, Shao Y, Garcia BA, and Selker EU (2016). Loss of HP1 causes depletion of H3K27me3 from facultative heterochromatin and gain of H3K27me2 at constitutive heterochromatin. Genome Res. 26, 97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mikulski P, Komarynets O, Fachinelli F, Weber APM, and Schubert D (2017). Characterization of the Polycomb-Group Mark H3K27me3 in Unicellular Algae. Front. Plant Sci 8, 607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Veluchamy A, Rastogi A, Lin X, Lombard B, Murik O, Thomas Y, Dingli F, Rivarola M, Ott S, Liu X, et al. (2015). An integrative analysis of post-translational histone modifications in the marine diatom Phaeodactylum tricornutum. Genome Biol. 16, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shaver S, Casas-Mollano JA, Cerny RL, and Cerutti H (2010). Origin of the polycomb repressive complex 2 and gene silencing by an E(z) homolog in the unicellular alga Chlamydomonas. Epigenetics 5, 301–312. [DOI] [PubMed] [Google Scholar]
- 71.Frapporti A, Miró Pina C, Arnaiz O, Holoch D, Kawaguchi T, Humbert A, Eleftheriou E, Lombard B, Loew D, Sperling L, et al. (2019). The Polycomb protein Ezl1 mediates H3K9 and H3K27 methylation to repress transposable elements in Paramecium. Nat. Commun 10, 2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao X, Xiong J, Mao F, Sheng Y, Chen X, Feng L, Dui W, Yang W, Kapusta A, Feschotte C, et al. (2019). RNAi-dependent Polycomb repression controls transposable elements in Tetrahymena. Genes Dev. 33, 348–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krauss V (2008). Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica 133, 93–106. [DOI] [PubMed] [Google Scholar]
- 74.Schmitz RJ, Lewis ZA, and Goll MG (2019). DNA Methylation: Shared and Divergent Features across Eukaryotes. Trends Genet. 35, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ishizaki K, Chiyoda S, Yamato KT, and Kohchi T (2008). Agrobacterium-mediated transformation of the haploid liverwort Marchantia polymorpha L., an emerging model for plant biology. Plant Cell Physiol. 49, 1084–1091. [DOI] [PubMed] [Google Scholar]
- 76.Li H (2016). Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32, 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, and Aiden EL (2017). De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Okada S, Sone T, Fujisawa M, Nakayama S, Takenaka M, Ishizaki K, Kono K, Shimizu-Ueda Y, Hanajiri T, Yamato KT, et al. (2001). The Y chromosome in the liverwort Marchantia polymorpha has accumulated unique repeat sequences harboring a male-specific gene. Proc. Natl. Acad. Sci. USA 98, 9454–9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, and Earl AM (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen S, Zhou Y, Chen Y, and Gu J (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang L, Wang S, and Li W (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185. [DOI] [PubMed] [Google Scholar]
- 83.Wu TD, Reeder J, Lawrence M, Becker G, and Brauer MJ (2016). GMAP and GSNAP for Genomic Sequence Alignment: Enhancements to Speed, Accuracy, and Functionality. Methods Mol. Biol 1418, 283–334. [DOI] [PubMed] [Google Scholar]
- 84.Kim D, Langmead B, and Salzberg SL (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hoff KJ, Lomsadze A, Borodovsky M, and Stanke M (2019). Whole-Genome Annotation with BRAKER. Methods Mol. Biol 1962, 65–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, and Salzberg SL (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. (2004). A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 5, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, and Kanehisa M (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin PC, Lu CW, Shen BN, Lee GZ, Bowman JL, Arteaga-Vazquez MA, Liu LY, Hong SF, Lo CF, Su GM, et al. (2016). Identification of miRNAs and Their Targets in the Liverwort Marchantia polymorpha by Integrating RNA-Seq and Degradome Analyses. Plant Cell Physiol. 57, 339–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsuzuki M, Nishihama R, Ishizaki K, Kurihara Y, Matsui M, Bowman JL, Kohchi T, Hamada T, and Watanabe Y (2016). Profiling and Characterization of Small RNAs in the Liverwort, Marchantia polymorpha, Belonging to the First Diverged Land Plants. Plant Cell Physiol. 57, 359–372. [DOI] [PubMed] [Google Scholar]
- 92.Dai X, Zhuang Z, and Zhao PX (2018). psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 46 (W1), W49–W54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chan PP, and Lowe TM (2019). tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol. Biol 1962, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cabanettes F, and Klopp C (2018). D-GENIES: dot plot large genomes in an interactive, efficient and simple way. PeerJ 6, e4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Borg M, Buendía D, and Berger F (2019).A simple and robust protocol for immunostaining Arabidopsis pollen nuclei. Plant Reprod. 32, 39–43. [DOI] [PubMed] [Google Scholar]
- 96.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu W, Hu B, Becker C, Doğan ES, Berendzen KW, Weigel D, and Liu C (2017). Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol. 18, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, and Lopez R (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47 (W1), W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, and Durbin R; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44 (W1), W160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gu Z, Gu L, Eils R, Schlesner M, and Brors B (2014). circlize Implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. [DOI] [PubMed] [Google Scholar]
- 105.Thorvaldsdóttir H, Robinson JT, and Mesirov JP (2013). Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Krueger F, and Andrews SR (2011). Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27, 1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, and Mirny LA (2012). Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat. Methods 9, 999–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, and Jacobsen SE (2014). Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat. Struct. Mol. Biol 21, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, and Liu XS (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw read data and assembled sequence data that support the findings of this study have been submitted to the DDBJ/ENA/NCBI public sequence databases under accession numbers SRA: PRJNA553138 and PRJDB8530.
The code supporting the current study have not been deposited in a public repository but are available from the corresponding author on request.