Abstract
STUDY QUESTION
Are reproductive characteristics associated with genome-wide DNA methylation and epigenetic age?
SUMMARY ANSWER
Our data suggest that increasing parity is associated with differences in blood DNA methylation and small increases in epigenetic age.
WHAT IS KNOWN ALREADY
A study of 397 young Filipino women (ages 20–22) observed increasing epigenetic age with an increasing number of pregnancies.
STUDY DESIGN, SIZE, DURATION
We used data from 2356 non-Hispanic white women (ages 35–74) enrolled in the Sister Study cohort.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Data on reproductive history were ascertained via questionnaire. Of the 2356 women, 1897 (81%) reported at least one live birth. Among parous women, 487 (26%) women reported ever experiencing a pregnancy complication. Three epigenetic clocks (i.e. Hannum, Horvath and Levine) and genome-wide methylation were measured in DNA from whole blood using Illumina’s HumanMethylation450 BeadChip. We estimated association β-values and 95% CIs using linear regression.
MAIN RESULTS AND THE ROLE OF CHANCE
All three epigenetic clocks showed weak associations between number of births and epigenetic age (per live birth; Hannum: β = 0.16, 95% CI = 0.02, 0.29, P = 0.03; Horvath: β = 0.12, 95% CI = −0.04, 0.27, P = 0.14; Levine: β = 0.27, 95% CI = 0.08, 0.45, P = 0.01); however, additional adjustment for current BMI attenuated the associations. Among parous women, a history of abnormal glucose tolerance during pregnancy was associated with increased epigenetic age by the Hannum clock (β = 0.96; 95% CI = 0.10, 1.81; P = 0.03) and Levine clocks (β = 1.69; 95% CI = 0.54, 2.84; P < 0.01). In epigenome-wide analysis, increasing parity was associated with methylation differences at 17 CpG sites (Bonferroni corrected P≤ 1.0 × 10-7).
LIMITATIONS, REASONS FOR CAUTION
We relied on retrospective recall to ascertain reproductive history and pregnancy complications.
WIDER IMPLICATIONS OF THE FINDINGS
Our findings suggest that parity is associated with small increases in epigenetic age and with DNA methylation at multiple sites in the genome.
STUDY FUNDING/COMPETING INTEREST(S)
This research was supported by the Intramural Research program of the NIH, National Institute of Environmental Health Sciences (Z01-ES049033, Z01-ES049032 and Z01-ES044055). None of the authors have a conflict of interest.
TRIAL REGISTRATION NUMBER
Not applicable.
Keywords: parity, pregnancy complications, epigenetic clock, DNA methylation, biological age
Introduction
To meet the demands of the developing fetus, a woman experiences various costs of reproduction, including extensive physiological changes to endocrine, cardiovascular, immune and metabolic functions, as well as long-term increases in oxidative stress (Lain and Catalano, 2007; Melchiorre et al., 2012; Somerset et al., 2004; Soma-Pillay et al., 2016; Ziomkiewicz et al., 2016). These costs may diminish somatic maintenance systems, thereby accelerating senescence and biological aging. This hypothesis finds some support in recent studies that have found an association between reproduction and telomere shortening (Kresovich et al., 2018; Ryan et al., 2018).
Epigenetic clocks are a new class of biological age estimator that may be more sensitive than telomeres in quantifying the effects of reproduction on biological age. To date, three epigenetic clocks have been developed. Briefly, the Hannum and Horvath clocks were developed using DNA methylation to predict chronological age (Hannum et al., 2013; Horvath, 2013). To estimate DNA methylation age (DNAm age), the developers selected the most informative set of cytosine–guanine dinucleotide (CpG) sites using machine learning algorithms. Differences between DNAm age and chronological age are termed epigenetic ‘age acceleration’, which can be either positive (when DNAm is greater than chronological age) or negative (Chen et al., 2016). The third clock, by Levine et al. (2018), differs from the Hannum and Horvath clocks because, rather than being designed to predict chronological age, it was designed to predict ‘PhenoAge’. PhenoAge is a biological age estimator based on a combination of chronological age and selected blood parameters related to inflammation, metabolic and immune function (albumin, creatinine, glucose, C-reactive protein, lymphocyte percent, mean cell volume, red cell distribution width, alkaline phosphatase and white blood cell count). Levine et al. (2018) then used elastic net regularization to identify a set of CpG sites that could predict PhenoAge. As Levine’s clock indirectly incorporates these clinical biomarkers, it was proposed as a more sensitive marker of mortality risk. Although reproductive history has not been examined in relation to the Hannum and Levine clocks, an initial study using Horvath’s clock suggests an association between increasing gravidity and epigenetic age acceleration (Ryan et al. 2018).
While epigenetic clocks utilize small, a priori sets of CpGs, methylation at other CpGs throughout the genome may also be influenced by reproduction. Compared to nulligravid women, there is some evidence that women in early pregnancy have differential leukocyte methylation, most often lower methylation (White et al., 2012). These methylation changes may be stable during pregnancy and return to pre-pregnancy levels postpartum (Chen et al., 2017; White et al., 2012). However, some changes may be enduring; parous women and women with earlier ages at first birth have been reported to have higher global DNA methylation in adulthood (Terry et al., 2008). Whether reproduction has long-term effects on methylation of specific genes remains unknown.
The primary goal of this study is to determine whether reproductive history is associated with biological age. Using an epigenome-wide approach, we also explore broader associations between reproduction and DNA methylation.
Materials and Methods
Study population
The Sister Study is a prospective cohort study of 50 884 women living in the USA and Puerto Rico between 2003 and 2009 (Sandler et al., 2017). To be eligible, women had to be between 35 and 75 years old, could not have had breast cancer themselves and must have had a biological sister who was diagnosed with breast cancer. At enrollment, women donated a fasting blood sample and completed a computer-assisted telephone interview. The interview included information on demographics, physical characteristics, lifestyle factors (e.g. current and past alcohol and tobacco use, current perceived stress) and reproductive history. In July 2014, a case-cohort sample of women was selected for DNA methylation analysis to determine associations with breast cancer risk. This sample was limited to non-Hispanic white women with an available baseline blood sample. It included 2878 women: 1542 women were selected because they developed breast cancer during the years after blood draw (average of 6 years to diagnosis) and 1336 women were randomly selected from the full cohort (91 of whom subsequently developed incident breast cancer, Data Release 6.0). Informed consent and blood samples were obtained during a home visit. The study was approved by the institutional review boards of the National Institute of Environmental Health Sciences and the Copernicus Group and conforms to the Declaration of Helsinki for Medical Research involving Human Subjects.
Reproductive history
At enrollment, women self-reported detailed information regarding their reproductive history. Participants were asked about the total number of pregnancies, pregnancy outcomes (i.e. live birth, still birth and miscarriage) and their age at first birth. Among the women who reported any live births, self-reported data were collected on whether they ever experienced complications, which included abnormal bleeding during pregnancy, gestational hypertension, preeclampsia/eclampsia or abnormal glucose levels. For the latter, women were asked the following: ‘Did you have pregnancy-related diabetes, an abnormal glucose tolerance test, or were you told that you were borderline during this pregnancy?’
DNA methylation processing and DNAm age calculation
Details on the DNA methylation processing procedures have been reported (O'Brien et al., 2018). Briefly, genomic DNA was extracted from aliquots of whole blood samples in the NIEHS Molecular Genetics Core Facility using an automated system (Autopure LS, Gentra Systems) or at BioServe Biotechnologies LTD (Beltsville, MD) using DNAQuik. From each sample, 1 μg of DNA was bisulfite-converted in 96-well plates using the EZ DNA Methylation Kit (Zymo Research, Orange County, CA). Methylation analysis was carried out at the NIH Center for Inherited Disease Research at Johns Hopkins University (Baltimore, MD). Samples were tested for complete bisulfite conversion, and converted DNA was analysed using Illumina’s Infinium HumanMethylation450 BeadChip following the manufacturer’s protocol. High-throughput robotics was used to minimize batch effects.
For each CpG site, we calculated a β-value, representing percent methylation, based on each individual’s proportion of unmethylated (U) and methylated (M) sites at a given locus: β = M/(U + M + 100). Methylation data preprocessing and quality control was completed using the ENmix R software package (Xu et al., 2016). This included the following steps: reducing background noise with the ENmix method, correcting fluorescent dye-bias using the RELIC method (Xu et al., 2017), quantile normalization to make overall array fluorescence intensity distribution comparable between arrays and reducing Infinium I and II probe design bias using the Regression on Correlated Probes (RCP) method (Niu et al., 2016). Of the 2878 methylation samples, 102 were excluded due to data quality issues. We restricted the analysis to autosomal CpGs. The Sister Study data can be requested via https://sisterstudy.niehs.nih.gov/English/coll-data.htm. Using the processed DNAm data, DNAm age was calculated for the three epigenetic clocks using CpG weights provided by the clock developers (Hannum et al., 2013; Horvath, 2013; Levine et al., 2018).
Statistical analysis
Epigenetic age acceleration, for each of the three clocks, was calculated using linear regression models by regressing DNAm age on chronological age and predicting the residuals. This approach generates age acceleration metrics that are independent of chronological age and can be either positive (represented as older DNAm age than chronological age) or negative. To avoid disproportional effects of outliers, we excluded participants if the absolute value of any of their age acceleration estimates was greater than four standard deviations from the mean (n = 10) (Kresovich et al., 2019; White et al., 2019).
In the age acceleration analyses, we used linear regression models to estimate association betas values and 95% CIs, treating the reproductive characteristics as the independent variable and age acceleration as the dependent variable. We first examined age acceleration ordinally (per live birth) and categorically (women with only 1, 2, 3 or 4+ live births, compared to nulliparous). In analyses of age at first birth, parous women were categorized as < 20, 20–24, 25–29, 30–34 and 35+ years where 25–29 years was treated as the referent category. Associations with pregnancy complications, among parous women, were tested comparing ‘never’ versus ‘ever’ experiencing the complication.
For the genome-wide DNA methylation analysis, we used linear regression models with robust standard errors. Percent DNA methylation β-values were logit-transformed to M-values to avoid issues with statistical heteroscedasticity (Du et al., 2010). We adjusted for a priori covariates, estimated blood cell proportion and technical variation, including batch effects (Houseman et al., 2012). To correct for multiple testing, we considered Bonferroni-corrected P-values and determined statistical significance at P ≤ 1.0 × 10-7. We further examined whether significant CpGs overlapped with those used to calculate the epigenetic age measures.
Model covariates for both the age acceleration and genome-wide analyses were determined using a directed acyclic graph. All models were adjusted for early adulthood characteristics (i.e. relative weight during teens compared to peers (reported as ‘lighter’, ‘about the same’ or ‘heavier’) as well as smoking (pack years) and alcohol use (drinks/week) before the age of 30). Among parous women, in models exploring associations with age at first birth, we adjusted for the previously listed covariates and total number of births. To test whether the age acceleration associations were potentially mediated by characteristics of later adulthood, we examined if the associations changed after adjusting for current BMI (kg/m2), current alcohol intake (drinks/week) and smoking (pack years) levels, highest educational attainment (high school or less, attended college or advanced degree) and current perceived stress (0–16 point scale). Participants were excluded if they were missing parity or covariate information (parity, n = 1; early adulthood smoking, n = 7; early adulthood alcohol use, n = 401; or relative teenage weight, n = 1) or had extreme age acceleration estimates (n = 10). We conducted a complete case analysis with the final sample size of 2356. All analyses were conducted using Stata (version 14.2, College Station, TX) and R.
Results
In our sample population, 2046 of 2356 women (87%) reported a previous pregnancy, of whom 1897 (93%) reported at least one live birth. The mean age of at first birth was 25.1 (SD = 5.2), and the mean number of births was 1.9 (SD = 1.3). Parous women were significantly older at blood draw, used birth control for a shorter duration, drank less alcohol, smoked less, were less likely to have attained an advanced degree and were more likely to perceive themselves as weighing the same or less than their peers during their teens compared to nulliparous women (Table I). Among women who reported at least one live birth, the most common self-reported complication was abnormal bleeding during pregnancy (n = 210, 11%). As expected, DNAm age estimates by all three epigenetic clocks were strongly correlated with chronological age (Fig. 1, top row). The three epigenetic age acceleration metrics all had approximate means of zero (Fig. 1, bottom row), and these were not associated with chronological age (Supplemental Fig. S1).
Table I.
Participant characteristics by parity (n= 2356).
| Characteristic | Nulliparous | Parous |
|---|---|---|
| n = 459 | n = 1,897 | |
| Mean (SD) | ||
| Age (at blood draw, yrs.) | 53.7 (9) | 57.0 (9) |
| Perceived stress (0-16 scale) | 2.7 (3) | 2.5 (3) |
| Menarche age (yrs.) | 12.6 (1) | 12.7 (1) |
| Hormonal birth control use (yrs.) | 7.0 (7) | 6.3 (6) |
| Alcohol history (drinks/wk.) | ||
| <30 yr. old | 3.7 (4) | 2.4 (4) |
| Last 12 months | 4.0 (5) | 3.4 (5) |
| Tobacco smoking (pack yrs.) | ||
| <30 yr. old | 4.3 (6) | 3.3 (5) |
| Lifetime | 8.8 (14) | 7.2 (13) |
| N (%) | ||
| Educational attainment | ||
| High school or less | 37 (8) | 308 (16) |
| Attended college | 249 (54) | 1,141 (60) |
| Advanced degree | 173 (38) | 448 (24) |
| Weight in teens (relative to peers) | ||
| Lighter | 125 (27) | 662 (35) |
| About same | 230 (50) | 916 (48) |
| Heavier | 104 (23) | 319 (17) |
| Body Mass Index (current, kg/m2) | ||
| Underweight/Normal (≤ 24.9) | 190 (41) | 758 (40) |
| Overweight (25-30) | 152 (33) | 619 (33) |
| Obese (30+) | 117 (26) | 518 (27) |
| Missing | 2 | |
| Complications | ||
| Abnormal bleeding | ----- | 210 (11) |
| Gestational hypertension | ----- | 98 (5) |
| Preeclampsia/Eclampsia | ----- | 143 (8) |
| Abnormal glucose tolerance1 | ----- | 113 (6) |
| Number of births | ||
| Zero | 459 (100) | ----- |
| One | ----- | 347 (18) |
| Two | ----- | 878 (46) |
| Three | ----- | 439 (23) |
| Four or more | ----- | 233 (12) |
1Includes gestational diabetes
Note: 6 women missing perceived stress; 2 women missing age at menarche; 13 missing birth control duration; 2 missing alcohol intakes in the last year; 2 missing lifetime smoking behavior.
Figure 1.
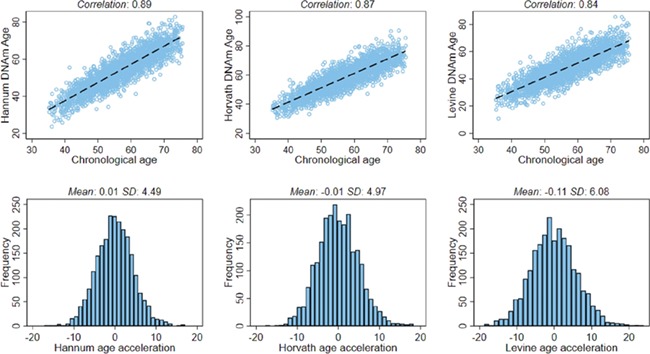
Scatter plots between chronological age and DNAm age estimated using the three epigenetic clocks and distributions of the three age acceleration metrics. The scatter plots (top row) show the Pearson correlations. The distributions of the age acceleration metrics (bottom row) include the mean and SD.
In the minimally adjusted model, after accounting for weight (when the subject was in their teenage years, relative to peers) and early adulthood alcohol and smoking levels, we observed small increases in epigenetic age acceleration per live birth for all three metrics (Hannum: β = 0.16, 95% CI = 0.02, 0.29, P= 0.03; Horvath: β = 0.12, 95% CI = −0.04, 0.27, P = 0.14; Levine: β = 0.27, 95% CI = 0.08, 0.45, P = 0.01) (Fig. 2, top row). After additional adjustment for current BMI and other potential mediators, the associations estimates were partly attenuated, particularly for the Levine clock (per live birth; Hannum: β = 0.12, 95% CI = −0.02, 0.26, P = 0.09; Horvath: β = 0.10, 95% CI= −0.06, 0.25, P = 0.23; Levine: β = 0.19, 95% CI= −0.00, 0.37, P = 0.05). When we examined associations by number of births, although the Levine clock showed an increasing trend with each additional birth, the Hannum and Horvath age acceleration increases were predominantly observed among women with four or more live births (Fig. 2, bottom row). Because of the case-cohort study design a high proportion of women in our study sample went on to be diagnosed with breast cancer. Additional adjustment for future breast cancer diagnosis did not appreciably change the association estimates (data not shown). Age at first birth was inversely associated with the age acceleration estimates from all three clocks (Hannum P-trend = 0.03; Horvath P-trend = 0.08; Levine P-trend = 0.15) (Supplemental Fig. S2).
Figure 2.
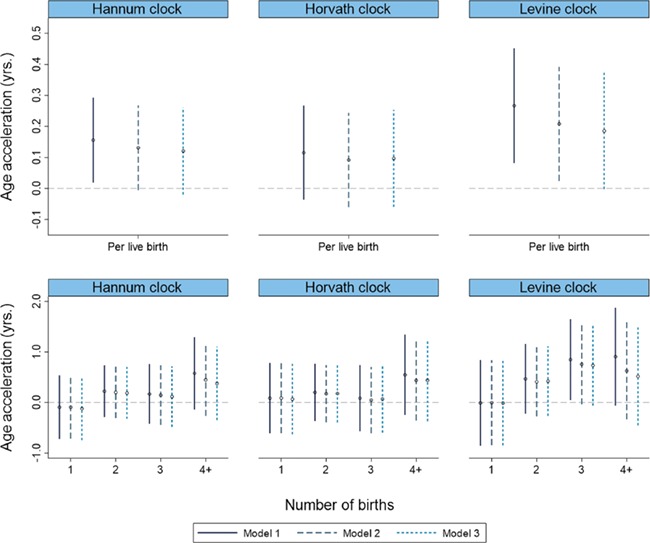
Epigenetic age acceleration associations with parity using three separate models. (Top row: treated ordinally. Bottom row: live birth categories, relative to nulliparous women) Model 1 adjusts for weight in teens (relative to peers) and early adulthood alcohol and smoking levels (n = 2356); model 2 additionally adjusts for current BMI (n = 2354); model 3 additionally adjusts for educational attainment, current alcohol use and smoking and perceived stress (n = 2344).
Because women who experience pregnancy complications have higher risks of long-term adverse health events, we tested whether pregnancy complications were associated with age acceleration. Among parous women, we did not find strong associations with abnormal bleeding, gestational hypertension or preeclampsia/eclampsia. However, women who self-reported an abnormal glucose test or gestational diabetes had higher age acceleration with both the Hannum (β = 0.96; 95% CI = 0.10, 1.81; P = 0.03) and Levine age acceleration metrics (β= 1.69; 95% CI = 0.54, 2.84; P < 0.01) (Table II).
Table II.
Complications during pregnancies and age acceleration (n =1,897).
| Hannum | Horvath | Levine | |||||
|---|---|---|---|---|---|---|---|
| N (%) | β (95% CI) | p-value | β (95% CI) | p-value | β (95% CI) | p-value | |
| Pregnancy complication | |||||||
| Abnormal bleeding | 210 (11) | -0.05 (-0.70, 0.60) | 0.88 | -0.13 (0.85, 0.59) | 0.73 | 0.58 (-0.29, 1.45) | 0.19 |
| Gestational hypertension | 98 (5) | 0.63 (-0.28, 1.55) | 0.18 | 0.43 (-0.59, 1.45) | 0.41 | 0.70 (-0.53, 1.94) | 0.26 |
| Preeclampsia/Eclampsia | 143 (8) | 0.14 (-0.63, 0.90) | 0.73 | -0.22 (-1.07, 0.64) | 0.62 | 0.55 (-0.48, 1.58) | 0.30 |
| Abnormal glucose tolerance1 | 113 (6) | 0.96 (0.10, 1.81) | 0.03 | 0.43 (-0.53, 1.38) | 0.38 | 1.69 (0.54, 2.84) | < 0.01 |
1Includes gestational diabetes
Models adjusted for relative weight in teens (lighter, about same, heavier) and early adulthood smoking (pack yrs.) and alcohol use (drinks/wk.).
Using a genome-wide approach, we found 17 CpGs that were differentially methylated and statistically significant after Bonferroni correction (P ≤ 1.0 × 10-7) (Fig. 3). At 13 (76%) of the significant sites, increasing parity was associated with higher methylation (Table III). Parity had the strongest association with methylation in the promoter region of the NGF gene (cg22354782, per live birth: β = 0.04, P = 6.2 × 10-9). We also observed higher methylation in the gene bodies of PCDHAC2 (cg18991549: β = 0.02, P = 7.1 × 10-9) and SP8 (cg01003961: β = 0.04, P = 1.1 × 10-8). None of the differentially methylated sites found in genome-wide analysis were components of any of the three epigenetic clocks.
Figure 3.
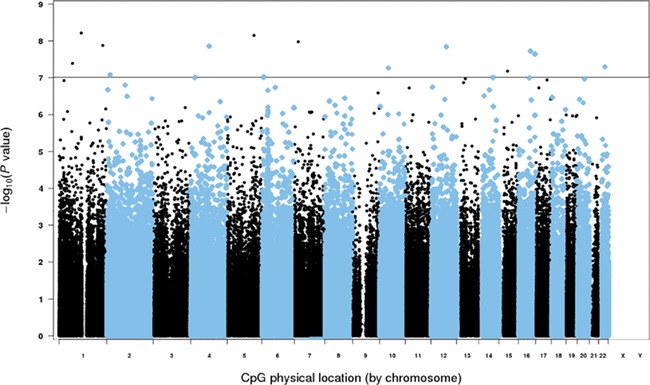
Manhattan plot for the epigenome-wide association of parity. The data account for relative teenage weight compared to peers (lighter, about same, heavier) and early adulthood smoking (pack yrs.) and alcohol use (drinks/wk.). There were 17 significant CpG sites, using a Bonferroni-corrected P ≤ 1.0 × 10-7 (solid black line).
Table III.
Genome-wide CpG sites associated with increasing parity at Bonferroni significance (P ≤ 1.0 × 10-7).
| Marker | Model coef. | Gene | Transcript | P-value |
|---|---|---|---|---|
| cg22354782 | 0.035153 | NGF | NM_002506 | 6.2×10-09 |
| cg18991549 | 0.024485 | PCDHAC2 | NM_031883 | 7.1×10-09 |
| cg01003961 | 0.042774 | SP8 | NM_182700 | 1.1×10-08 |
| cg08299045 | -0.018750 | MRPL55 | NM_181465 | 1.3×10-08 |
| cg11723848 | 0.026436 | UNC5C | NM_003728 | 1.4×10-08 |
| cg13320585 | -0.018310 | MIR1252 | NR_031700 | 1.5×10-08 |
| cg02172312 | 0.025002 | CDH8 | NM_001796 | 1.9×10-08 |
| cg26650638 | 0.030679 | LOC146513 | NR_038438 | 2.3×10-08 |
| cg00989492 | 0.025676 | N.A. | N.A. | 4.1×10-08 |
| cg24556441 | 0.035817 | SYN3 | NM_001135774 | 5.1×10-08 |
| cg09373037 | 0.024558 | SYT15 | NM_181519 | 5.4×10-08 |
| cg01138448 | -0.017970 | TRIM69 | NM_182985 | 6.6×10-08 |
| cg07098902 | 0.023914 | VSNL1 | NM_003385 | 8.3×10-08 |
| cg14909730 | 0.027713 | TFAP2A | NM_003220 | 9.5×10-08 |
| cg00688962 | 0.032648 | KCNIP4 | NM_147182 | 9.9×10-08 |
| cg13829550 | 0.025788 | SYNDIG1L | NM_001105579 | 1.0×10-07 |
| cg07839457 | -0.035600 | N.A. | N.A. | 1.0×10-07 |
Abbreviations: not applicable, N.A.
Discussion
Using three different epigenetic clocks to estimate biological age, we find evidence that reproduction is weakly associated with epigenetic age acceleration. In addition, the Hannum and Levine clocks were associated with a self-reported history of abnormal glucose tolerance. Women who ever reported an abnormal glucose tolerance test or gestational diabetes had a 1- to 2-year increase in biological age. We also observed an inverse association between age acceleration and age at first birth, such that later age at first birth was associated with decreased epigenetic age acceleration. Finally, epigenome-wide analysis of 413 652 CpGs identified 17 sites associated with increasing parity, most notably in the promoter region of NGF, a growth stimulating protein. Together, our findings suggest that reproductive history is associated with modifications of blood DNA methylation and may marginally increase biological age.
Epigenetic clocks were originally developed as predictors of chronological age and may be associated with age-related disease and mortality risk (Chen et al., 2016; Marioni et al., 2015; Christiansen et al., 2016; Perna et al., 2016). Recent studies have suggested that age acceleration may be influenced by lifestyle factors (Irvin et al., 2018; Quach et al., 2017; Levine et al., 2018). Importantly, the epigenetic clocks were developed using different procedures; the Hannum and Horvath clocks were developed solely to predict chronological age. In contrast, the Levine clock was designed to predict a previously proposed biological age estimator, PhenoAge. The stronger association estimates we observe with Levine’s clock may reflect the incorporation of information related to inflammation, immune and metabolic function in the PhenoAge estimate as these factors may be influenced by reproduction.
Pregnancy is associated with extensive physiological changes. While some changes may revert to pre-pregnancy levels postpartum, lasting changes in metabolic and cardiovascular functions have been identified and may increase the risk for adverse health events, particularly for women reporting more than three live births (Kritz-Silverstein et al., 1989; Gunderson et al., 2008; Gunderson et al., 2009; Lawlor et al., 2003). Using all three epigenetic clocks, we show that increasing parity is associated with nominal increases in age acceleration; our results are consistent with an earlier report that used Horvath’s clock (Ryan et al., 2018). Notably, we observed partial attenuation after adjustment for BMI at blood draw, particularly for Levine’s clock. The partial mediation of the reproduction and age acceleration associations by current measures of body composition fits with prior studies that show the association between increasing parity and coronary heart disease and metabolic syndrome is partly, but not entirely, attenuated after adjustment for obesity (Lawlor et al., 2003; Gunderson et al., 2009). These findings suggest returning to a healthy weight after pregnancy may help mitigate some of the impact on biological aging.
To our knowledge, this is the first report of an association between age acceleration and a history of abnormal glucose tolerance. Pregnancy may represent a medical stress test for maternal health and complications during pregnancy are reported to have lasting health consequences (Williams, 2003). For instance, women who are diagnosed with gestational diabetes or hypertension experience higher rates of type-2 diabetes and cardiovascular events as well as increased mortality risk (Lee et al., 2015; Lo et al., 2013; Kaufmann et al., 1995; Sattar and Greer, 2002; Theilen et al., 2016). We found that women who reported having abnormal glucose tolerance during pregnancy had greater age acceleration using two of the three clocks, with Levine’s clock showing the strongest association. As Levine’s clock was developed to incorporate information related to serum glucose, we suggest biological age measured using the Levine clock may be sensitive to a history of metabolic dysfunction.
We also find evidence that reproductive history is associated with long-term changes in blood DNA methylation at individual CpG sites. There were 17 CpGs identified after Bonferroni correction, of which a majority showed higher methylation; this observation parallels a previous finding that, compared to nulliparous women, parous women have higher levels of global methylation (Terry et al., 2008). Our strongest finding for a single CpG was for higher methylation at cg22354782, located in the promoter region of NGF. This gene encodes a growth factor that has been implicated in cell growth and differentiation (Aloe et al., 2016). Although there is limited evidence linking this gene to reproduction, a previous report among women diagnosed with multiple sclerosis found increasing parity was associated with differential expression of UNC5C, SYN3 and KCNIP4 (Mehta et al., 2019). We observed differential methylation at CpGs within these genes. Our findings offer additional evidence that specific locations in the genome may undergo persistent changes resulting from reproduction.
Although we found evidence that reproductive history has lasting influences on DNA methylation, this study had limitations. First, it is difficult to determine whether the associations with epigenetic age acceleration are due to parity or the downstream consequences of having children (e.g. reduced sleep, increased stress, etc.). After adjusting for potential mediators, including current BMI, educational attainment, alcohol and smoking behaviors and current perceived stress, associations were only partly attenuated suggesting some effect may be due to parity itself. However, it is notable that the association estimates were small, suggesting a minimal effect of reproduction on estimates of biological age. Our study also depends on epigenetic age acceleration metrics measured in blood. Although the Hannum and Levine clocks were developed for use in blood, the Horvath clock was designed to be used across tissues and associations may differ by tissue type. For instance, obesity has been observed to increase epigenetic aging in liver tissue, but not in blood (Horvath et al., 2014). We had detailed information regarding behaviors in early adulthood that may influence both reproductive history and DNA methylation, thereby limiting the likelihood our results are the consequence of confounding. This study is the first to identify lasting gene-specific methylation changes related to parity and is the largest study of reproductive history and DNA methylation to date.
In summary, we found evidence that reproductive history is associated with differences in blood DNA methylation decades after childbirth. We also find that age acceleration was marginally increased for women with a greater number of live births and was higher for women who experienced abnormal glucose tolerance during pregnancy. These findings suggest that a woman’s reproductive history is associated with persistent epigenetic changes that may in turn be associated with future health.
Supplementary Material
Acknowledgements
We would like to thank the participants of the Sister Study. We would also like to thank Kaitlyn Gam and Thanh Hoang for providing the internal review and Jenna Waggoner for the data check.
Authors’ roles
D.P.S. and J.A.T. conceived, designed and acquired the data for the Sister Study, and J.K.K. contributed to the conception and design of the current study. J.K.K., Q.E.H. and Z.X. conducted the statistical analysis. J.K.K. drafted the manuscript. All authors (J.K.K., Q.E.H., Z.X., H.B.N., D.P.S. and J.A.T.) provided input on the interpretation of the data and manuscript revisions critical for important intellectual content. All authors (J.K.K., Q.E.H., Z.X., H.B.N., D.P.S. and J.A.T.) approved the final manuscript version for publication.
Funding
Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (Z01-ES049033, Z01-ES049032 and Z01-ES044055).
Conflict of interest
None of the authors have any conflicts of interest.
References
- Aloe L, Rocco ML, Balzamino BO, Micera A. Nerve growth factor: role in growth, differentiation and controlling cancer cell development. J Exp Clin Cancer Res 2016;35:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Mukherjee N, Janjanam VD, Arshad SH, Kurukulaaratchy RJ, Holloway JW, Zhang H, Karmaus W. Consistency and variability of DNA methylation in women during puberty, young adulthood, and pregnancy. Genet Epigenet 2017;9:1179237X17721540. doi: 10.1177/1179237X17721540. eCollection 2017.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K. DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell 2016;15:149–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010;11:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson EP, Chiang V, Lewis CE, Catov J, Quesenberry CP, Sidney S, Wei GS, Ness R. Long-term blood pressure changes measured from before to after pregnancy relative to nonparous women. Obstet Gynecol 2008;112:1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson EP, Jacobs DR, Chiang V, Lewis CE, Tsai A, Quesenberry CP, Sidney S. Childbearing is associated with higher incidence of the metabolic syndrome among women of reproductive age controlling for measurements before pregnancy: the CARDIA study. Am J Obstet Gynecol 2009;201:177.e1–177.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 2013;49:359–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. DNA methylation age of human tissues and cell types. Genome Biol 2013;14:R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Erhart W, Brosch M, Ammerpohl O, von W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A 2014;111:15538–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 2012;13:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin MR, Aslibekyan S, Do A, Zhi D, Hidalgo B, Claas SA, Srinivasasainagendra V, Horvath S, Tiwari HK, Absher DM et al. Metabolic and inflammatory biomarkers are associated with epigenetic aging acceleration estimates in the GOLDN study. Clin Epigenetics 2018;10:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann RC, Schleyhahn FT, Huffman DG, Amankwah KS. Gestational diabetes diagnostic criteria: long-term maternal follow-up. Am J Obstet Gynecol 1995;172:621–625. [DOI] [PubMed] [Google Scholar]
- Kresovich JK, Parks CG, Sandler DP, Taylor JA. Reproductive history and blood cell telomere length. Aging (Albany NY) 2018;10:2383–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kresovich JK, Xu Z, O'Brien KM, Weinberg CR, Sandler DP, Taylor JA. Methylation-based biological age and breast cancer risk. J Natl Cancer Inst 2019; pii: djz020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kritz-Silverstein D, Barrett-Connor E, Wingard DL. The effect of parity on the later development of non-insulin-dependent diabetes mellitus or impaired glucose tolerance. N Engl J Med 1989;321:1214–1219. [DOI] [PubMed] [Google Scholar]
- Lain KY, Catalano PM. Metabolic changes in pregnancy. Clin Obstet Gynecol 2007;50:938–948. [DOI] [PubMed] [Google Scholar]
- British Women's Heart and Health Study, British Regional Heart Study . Is the association between parity and coronary heart disease due to biological effects of pregnancy or adverse lifestyle risk factors associated with child-rearing? Findings from the British Women's Heart and Health Study and the British Regional Heart Study. Circulation 2003;107:1260–1264. [DOI] [PubMed] [Google Scholar]
- Lee KK, Raja EA, Lee AJ, Bhattacharya S, Norman JE, Reynolds RM. Maternal obesity during pregnancy associates with premature mortality and major cardiovascular events in later life. Hypertension 2015;66:938–944. [DOI] [PubMed] [Google Scholar]
- Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 2018;10:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JO, Mission JF, Caughey AB. Hypertensive disease of pregnancy and maternal mortality. Curr Opin Obstet Gynecol 2013;25:124–132. [DOI] [PubMed] [Google Scholar]
- Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 2015;16:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Wani S, Wallace L, Henders AK, Wray NR, McCombe PA. Cumulative influence of parity-related genomic changes in multiple sclerosis. J Neuroimmunol 2019;328:38–49. [DOI] [PubMed] [Google Scholar]
- Melchiorre K, Sharma R, Thilaganathan B. Cardiac structure and function in normal pregnancy. Curr Opin Obstet Gynecol 2012;24:413–421. [DOI] [PubMed] [Google Scholar]
- Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina methylation BeadChip. Bioinformatics 2016;32:2659–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien KM, Sandler DP, Xu Z, Kinyamu HK, Taylor JA, Weinberg CR. Vitamin D, DNA methylation, and breast cancer. Breast Cancer Res 2018;20:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics 2016;8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, Ritz B, Bandinelli S, Neuhouser ML, Beasley JM et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 2017;9:419–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CP, Hayes MG, Lee NR, McDade TW, Jones MJ, Kobor MS, Kuzawa CW, Eisenberg DTA. Reproduction predicts shorter telomeres and epigenetic age acceleration among young adult women. Sci Rep 2018;8:11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler DP, Hodgson ME, Deming-Halverson SL, Juras PS, D'Aloisio AA, Suarez LM, Kleeberger CA, Shore DL, DeRoo LA, Taylor JA et al. The sister study cohort: baseline methods and participant characteristics. Environ Health Perspect 2017;125:127003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattar N, Greer IA. Pregnancy complications and maternal cardiovascular risk: opportunities for intervention and screening? BMJ 2002;325:157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma-Pillay P, Nelson-Piercy C, Tolppanen H, Mebazaa A. Physiological changes in pregnancy. Cardiovasc J Afr 2016;27:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerset DA, Zheng Y, Kilby MD, Sansom DM, Drayson MT. Normal human pregnancy is associated with an elevation in the immune suppressive CD25+ CD4+ regulatory T-cell subset. Immunology 2004;112:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry MB, Ferris JS, Pilsner R, Flom JD, Tehranifar P, Santella RM, Gamble MV, Susser E. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev 2008;17:2306–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theilen LH, Fraser A, Hollingshaus MS, Schliep KC, Varner MW, Smith KR, Esplin MS. All-cause and cause-specific mortality after hypertensive disease of pregnancy. Obstet Gynecol 2016;128:238–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AJ, Kresovich JK, Xu Z, Sandler DP, Taylor JA. Shift work, DNA methylation and epigenetic age. Int J Epidemiol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White WM, Brost BC, Sun Z, Rose C, Craici I, Wagner SJ, Turner S, Garovic VD. Normal early pregnancy: a transient state of epigenetic change favoring hypomethylation. Epigenetics 2012;7:729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D. Pregnancy: a stress test for life. Curr Opin Obstet Gynecol 2003;15:465–471. [DOI] [PubMed] [Google Scholar]
- Xu Z, Langie SA, De P, Taylor JA, Niu L. RELIC: a novel dye-bias correction method for Illumina Methylation BeadChip. BMC Genomics 2017;18:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Niu L, Li L, Taylor JA. ENmix: a novel background correction method for Illumina HumanMethylation450 BeadChip. Nucleic Acids Res 2016;44:e20 doi: 10.1093/nar/gkv907. Epub 2015 Sep 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziomkiewicz A, Sancilio A, Galbarczyk A, Klimek M, Jasienska G, Bribiescas RG. Evidence for the cost of reproduction in humans: high lifetime reproductive effort is associated with greater oxidative stress in post-menopausal women. PLoS One 2016;11: e0145753. doi: 10.1371/journal.pone.0145753. eCollection 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


