Abstract
The major histocompatibility complex (MHC) is a core part of the adaptive immune system. As in other vertebrate taxa, it may also affect human chemical communication via odour-based mate preferences, with greater attraction towards MHC-dissimilar partners. However, despite some well-known findings, the available evidence is equivocal and made complicated by varied approaches to quantifying human mate choice. To address this, we here conduct comprehensive meta-analyses focusing on studies assessing: (i) genomic mate selection, (ii) relationship satisfaction, (iii) odour preference, and (iv) all studies combined. Analysis of genomic studies reveals no association between MHC-dissimilarity and mate choice in actual couples; however, MHC effects appear to be independent of the genomic background. The effect of MHC-dissimilarity on relationship satisfaction was not significant, and we found evidence for publication bias in studies on this area. There was also no significant association between MHC-dissimilarity and odour preferences. Finally, combining effect sizes from all genomic, relationship satisfaction, odour preference and previous mate choice studies into an overall estimate showed no overall significant effect of MHC-similarity on human mate selection. Based on these findings, we make a set of recommendations for future studies, focusing both on aspects that should be implemented immediately and those that lurk on the far horizon. We need larger samples with greater geographical and cultural diversity that control for genome-wide similarity. We also need more focus on mechanisms of MHC-associated odour preferences and on MHC-associated pregnancy loss.
This article is part of the Theo Murphy meeting issue ‘Olfactory communication in humans’.
Keywords: HLA, complementary genes, inbreeding, sexual selection, attractiveness, body odour
1. Introduction
(a). Major histocompatability complex function in the immune system
The genes of the major histocompatibility complex (MHC) are found in all jawed vertebrates and comprise the heart of the adaptive immune system, originating over 500 Ma [1]. In humans, they are called human leucocyte antigen (HLA) genes and occur on the short arm of chromosome 6, spanning 3.6 megabase (Mb) pairs [2]. The MHC gene family is divided into three classes, with the first two being primarily involved in immune system functioning (class III genes code for proteins with other functions and will not be discussed here further). The products of classical MHC class I genes (HLA-A, -B and -C) are glycoproteins which are expressed on virtually all nucleated cells. In addition, class I genes include non-classical MHC class Ib genes (HLA-E, -F and -G), which show considerably less polymorphism. There is a growing body of evidence showing that HLA-E and -G (and to a lesser extent -F) play a key role in immunotolerance of the foetus by the maternal immune system in general and uterine natural killer (NK) cells in particular [3]. The proteins derived from intracellular pathogens are degraded in the cytosol into peptide fragments which are subsequently bound by the MHC class I molecules and presented at the cell surface. The complex of the MHC molecule and the peptide are recognized by cytotoxic T-cells which destroy the infected cell. By contrast, MHC class II (HLA-DR, -DQ and -DP) molecules are expressed only by antigen-presenting cells, such as macrophages and dendritic cells. Extracellular pathogens are phagocytozed by the antigen-presenting cell, degraded in the lysosome and derived peptide fragments are presented at the cell surface, where they are recognized by T-helper cells, stimulating further immune responses such as B cell activation or inflammation [4].
The MHC includes the most polymorphic genes in the human genome, HLA-B being the most polymorphic with hundreds of known alleles [5]. Individual alleles code for proteins that vary in the binding groove, which allows them to differ in the range of peptides they bind and transport across the cellular membrane. Because both alleles at an MHC locus are expressed, heterozygous individuals may show a selective advantage. There is robust evidence that most MHC loci are under selection that maintains allelic diversity in the population (i.e. balancing selection) [6]. The frequency of individual MHC alleles varies highly across human populations, probably depending on the presence of diverse infections, and the epidemic and demographic history of a given population [7]. Indeed, extensive research has shown associations between individual alleles and susceptibility to infectious diseases such as HIV, tuberculosis, leprosy and malaria [8–11].
(b). Major histocompatibility complex and mate choice
Because resistance to infection has direct evolutionary consequences for humans [12] as for other species, patterns of human mate choice may be influenced by MHC genotype of potential mates. Across vertebrates, individuals tend to prefer MHC-dissimilar mates [13], increasing the likelihood of eventual offspring being MHC-heterozygous and thus more resistant to a wider pathogen spectrum [14]. The first studies that tested preferences for MHC-dissimilar mates, in mice, revealed that preferences are mediated by odour cues [15]. Similar odour-mediated mating preferences have since been demonstrated in other vertebrate taxa [16], although a recent meta-analysis showed that the preference for MHC-dissimilar individuals is relatively weak [17].
MHC-associated mate preferences have also been tested in humans. An initial study reported preferences for male odours of MHC-dissimilar individuals in naturally cycling women and an opposite effect in hormonal contraceptive (HC) users [18]. Since then, other studies have tested both odour and facial preferences as well as MHC-similarity in actual couples. The results are contradictory in all three domains (for a review, see [19]).
Recently, Winternitz et al. [20] conducted a meta-analysis to quantify the overall effects of MHC-heterozygosity and MHC-dissimilarity and to explore potential moderating variables, such as HC use. The analysis was based on two types of data: (i) results of experimental studies testing MHC-associated body odour and facial preferences, and (ii) genetic data from real couples, testing whether they are more dissimilar than expected by chance. The results showed a systematic, although moderate, preference for heterozygous individuals in both odour and facial tests, which was stronger in women than men. However, the results concerning MHC-dissimilarity showed no overall consistent effect. While this might be a consequence of conflating studies which show opposite patterns (e.g. by combining effects in HC users and non-users), follow-up analyses testing moderating effects including HC use, stimulus type, and rater sex, also showed no significant effect. In fact, studies on actual mate choice (as compared to mate preference) showed a significant positive effect of MHC-similarity. Further analysis revealed that samples from genetically heterogeneous populations show higher positive MHC assortment compared to those from genetically homogenous populations, probably owing to strong preference for ethnic homogamy. In other words, individuals in ethnically heterogeneous populations tend to pair with partners of the same ethnicity, and as ethnicity also affects MHC variation, the observed pattern can initially provide an impression of preference for MHC-similarity.
Based on the results of this meta-analysis, one might conclude that variation in MHC-similarity does not contribute to human mate choice and that researchers should move on to other topics. However, we think that such a conclusion is premature, as there remain several unresolved questions that deserve further investigation before any definitive conclusion can be made. Furthermore, since the Winternitz et al. meta-analysis, eight new studies have been published, several with considerably higher statistical power than those preceding.
(c). Current study
The main aims of the current paper are twofold. First, we conducted four different meta-analyses primarily focusing on aspects which were not targeted by the Winternitz et al. study [20]. Second, based on these results, we aim to identify outstanding questions and unresolved issues in order to provide specific guidelines for future studies in MHC-associated mate choice. We focus on MHC-similarity, rather than diversity, because only one of the eight new studies investigated diversity preferences.
Our meta-analyses were divided in the following ways. (1) Genomic studies. Genomic data are increasingly being used to test for MHC-mediated mate preferences and mate choice, having three major benefits over traditional genotyping studies: (i) they can control for population stratification using major dimensions of genetic variation (e.g. principal components [21]); (ii) they can test for social and ethnic constraints in mate choice by comparing background genomic similarity between real mates to that of permuted pairs; and (iii) they can test if the MHC region is being specifically targeted by mate choice by comparing similarity at the MHC region with similarity at genomic regions of similar size and with similar recombination rates. (2) Relationship satisfaction studies. Studies have investigated whether MHC-similarity is associated with several aspects of relationship satisfaction, including sexual satisfaction, in-pair attraction and overall partnership satisfaction. It was previously predicted that MHC-similarity may specifically influence the sexual satisfaction between individuals in long-term relationships [22], but this has not previously been tested meta-analytically. We, therefore, primarily focused on sexual satisfaction. (3) Odour preference studies. These are the most common form of MHC-mate preference studies, and it is beneficial to assess the current state of knowledge and to form recommendations for future work. We include four new studies (12 effect sizes) to the six previous studies (11 effect sizes) analysed by Winternitz et al. (4) Mate selection. This analysis combined effects from all studies involved in ‘genomic’, ‘relationship satisfaction’ and ‘odour preference’ meta-analyses, and included nine previous studies analysed by Winternitz et al., to provide an overall estimate of MHC effects in human mate selection, including mate choice and mate preferences (for geographical distribution of the studied populations; figure 1). One exception is that effects from facial preference studies were excluded because it is not clear how MHC-dissimilarity may be perceived through visual traits, and thus, the direction which MHC-linked preferences should take is also unclear [19]. In addition, no newly added studies investigated facial preferences, while four new studies have contributed genomic effects, two have contributed relationship satisfaction effects and four have contributed odour preference effects (with some new studies contributing effects to multiple meta-analyses). Another exception is that we did not include effects from HC users from odour preference studies because we did not want to conflate effects predicted to have opposite patterns. We do note, however, that excluding HC users means keeping HC non-users but also men and samples with unknown HC status, mostly couples. The variables measured in these studies include genomic-based and HLA-typing based MHC-similarity effects between mates, genomic-based MHC-similarity effects between preferred and non-preferred potential mates, MHC-similarity effects for odour preferences, and MHC-similarity effects for sexual satisfaction in relationships.
Figure 1.
Map of the number of individuals investigated in the current study for MHC-linked mate selection by geographical regions of the world. Region codes are as follows: Australia (AUS), Europe (EUR), North Africa (NAF), North America (NAM), North East Asia (NEA), Oceania (OCE), South Central America (SAM), South Asia (SAS), South East Asia (SEA), Sub-Saharan Africa (SSA) and Western Asia (WAS). (Online version in colour.)
2. Materials and methods
(a). Literature search
Dataset compilation methods are described in detail in [5]. Briefly, studies were compiled from the reviews by Havlíček & Roberts [19] and Winternitz et al. [20]. Additional studies were identified from 2017 to April 2019 via Web of Science and Google Scholar using the topic ‘MHC’, ‘Major Histocompatibility Complex’, ‘HLA’, ‘Human Leukocyte Antigen’, and ‘mate choice’ or ‘mate selection’ or ‘mate preference’ and searching within results for ‘human’. Studies were retained if they tested for human mate choice or mating preferences for MHC-similarity. Studies were included if MHC genotypes (or their approximations via single nucleotide polymorphisms, e.g. HapMap data) were obtained for the individuals tested. Studies were excluded if they did not explicitly test for MHC influence on mating preferences (e.g. [23]). The study by Khankhanian et al. [24] was excluded because the sample population consisted of couples with a child affected by multiple sclerosis (MS). As MS is a complex genetic disease with strong associations with MHC class II genes, the sample population has higher frequencies of specific MHC risk alleles and is not a fair representation of the general population. Three other studies were excluded because summary statistics from pairwise tests were unavailable [25], because the study only presented a minority of genes showing extreme similarity or dissimilarity and not test statistics for the full HLA region [26], and because the study did not test if ‘male ornaments’ were correlated with MHC-dissimilarity [23].
To focus on modern developments and provide recommendations to promote progress in the field, we confined our analyses to four sets of studies related to MHC-similarity: (i) studies using genomic datasets to test for mate selection, (ii) studies testing for relationship satisfaction, (iii) studies testing for odour preferences, and (iv) all studies combined (excluding facial preference studies and effect sizes from HC users). Lists of full references and explanation for exclusions are provided in the electronic supplementary material.
(b). Data extraction and effect size calculation
We chose r effect size (correlation coefficients) as the measure of associations between MHC-dissimilarity and strength of mating preference/outcome. Studies have mostly measured dissimilarity as categories of allele-sharing (e.g. none and ≥1). Other test statistics were converted to r effect sizes following Nakagawa & Cuthill [27]. When studies provided multiple effect sizes that we could not independently evaluate with moderator variables (e.g. results from multiple loci), we calculated weighted means by first converting measures to r and then weighting them by the underlying sample sizes. We accounted for non-independence of multiple effect sizes extracted from the same study by including study as a random effect in our statistical models. The number of raters was recorded to test for potential effects of sample size on the resulting effect size. The number of individuals rated (number of independent repeats) in the study was recorded to calculate the variance in effect size (variance = 1/(n study rated − 3)). When weighted effect size means were calculated, we also recorded the mean number of individuals rated and used this estimate to calculate the variance of the weighted mean. Raw data and converted effect sizes were checked by independent extraction (by J.H.) and any inconsistency was discussed (between J.W. and J.H.) until a consensus was reached. We converted effect sizes into Fisher's Z (Zr) to stabilize variance across effect sizes, and Zr and its variance (defined above) were used for meta-analyses. The final dataset consisted of 17 effect sizes from six studies for genomic mate selection, nine effect sizes from three studies for relationship satisfaction, 23 effect sizes from 10 studies for odour preference and 55 effect sizes from 26 studies for mate selection (excluding HC users). Effect sizes from relationship satisfaction studies were taken from analyses based on sexual satisfaction only (e.g. not from analyses based on overall relationship satisfaction). The full dataset and effect size extractions and conversions are provided in the electronic supplementary material.
Previous work has shown that biological and methodological differences between studies can affect MHC-linked mating patterns in human populations [19,20]. We accounted for these potential sources of heterogeneity by considering moderator variables that could help explain within- and between-study variance in effect sizes. The following data were extracted from each study as methodological predictors: (i) study ID and (ii) year of publication, for publication bias testing, (iii) choice cue used for mating preference (i.e. genomic, relationship satisfaction and odour preference), (iv) the number of individual raters (n of rater), and (v) HC use (female HC users, female non-HC users, unknown and males). Effect sizes from HC using women were present only in the odour preference dataset, so we ran odour preference models including and excluding HC-use effect sizes (n = 4) (both sets of results provided in all tables). Biological predictors included (vi) MHC class (class I, class II or both), (vii) number of MHC loci investigated, (viii) unit of investigation (i.e. male or female raters, or couples), (ix) population, and (x) geographical region of heritage. The geographical population was determined by the ancestral population listed in the study, and if the population was mixed or not explicitly stated, the geographical location was listed as the geographical population (e.g. mixed US populations were labelled as North American). Eleven geographical populations (figure 1) were based on www.allelefrequencies.net classifications and Immunogenetics Data Analysis Working Group recommendations. Region codes are as follows: Australia (AUS), Europe (EUR), North Africa (NAF), North America (NAM), North East Asia (NEA), Oceania (OCE), South Central America (SAM), South Asia (SAS), South East Asia (SEA), Sub-Saharan Africa (SSA) and Western Asia (WAS).
An additional set of methodological moderators was recorded for genomic mate selection studies. (xi) The effect size for the extremeness of MHC relatedness compared to genome-wide relatedness within spouses. This moderator allowed us to test if MHC effect sizes—derived from the extremeness of MHC relatedness within spouses versus permuted spouses, with no insight from the rest of the genome—are related to how extreme MHC relatedness is compared to the rest of the genome within spouses. A null or negative relationship would indicate that observed MHC effect sizes are confounded by socio-ethnic processes. For instance, a negative relationship, where spouses are more MHC-similar compared to random pairs but show more extreme MHC-dissimilarity compared to background regions, could imply that population stratification is required for MHC-mediated mate choice. A null relationship, where MHC relatedness is no different from genome relatedness between spouses for varying values of MHC relatedness compared to permuted spouses, would mean that couples could be using the MHC as an indicator of genome-wide relatedness, and not choosing the MHC specifically. By contrast, a positive relationship would indicate that observed MHC effect sizes are a good indicator of the extremeness of the MHC compared to the genome within couples. (xii) Genome-wide (background) similarity effect sizes to test if MHC effects are related to genome-wide effects. A positive relationship would indicate mate choice is not MHC-specific, a negative relationship would indicate that preferences for MHC-dissimilarity increase as background relatedness increases, and no relationship would indicate MHC effects are independent of genome-wide effects. (xiii) The number of permutations used to create the null distribution for model-free approaches was recorded to test if increasing the number of permutations reduced the strength of MHC effect sizes (e.g. [28]). Other methodological moderators included (xiv) Phi-hat cryptic relatedness cut-off to test if the relatedness threshold in genomic studies influenced MHC effect sizes and (xv) the span of the MHC region (Mb) under investigation, as including an extended region beyond the classical 3.6 Mb of the MHC region has been criticized [29].
(c). Statistical analyses
(i). Meta-analytic procedures
Meta-analyses were conducted with mixed effects models using the R package metafor [30]. Study ID was included as a random effect to control for non-independence owing to some studies contributing more than one effect size. Individual numbers were given to effect sizes within the datasets for genomic studies, relationship satisfaction and odour preference and included as a random effect using the rma.mv() function to account for potential heterogeneity in the true effects (e.g. random-effects model).
To examine the impact of moderator variables (listed above) on study effect sizes, we constructed a series of meta-regression models. We conducted univariate fixed-effect mixed models to estimate the mean effect size for each moderator separately (we avoided complex models with multiple predictors given the limited sample size). Models with categorical moderators were run without the intercept to test each trait against no effect. All effect sizes are reported as Fisher's normalized correlation coefficients (Zr) with 95% confidence intervals (CIs). In the ecological literature, r ≈ 0.1 (Zr ≈ 0.10) is generally considered a small effect, r ≈ 0.3 (Zr ≈ 0.31) a medium effect and r ≈ 0.5 (Zr ≈ 0.55) a strong effect [31,32].
(ii). Publication bias and sensitivity analysis
We tested for publication bias using three different approaches. First, we tested for funnel plot asymmetry using a modified version of Egger's regression [33] for random-effects meta-analytic models including study ID with the rma.mv() function and the standard error (square root of the variance) as a moderator in the metafor R package [30]. For the mate choice model, we included ‘Region code’ as a covariate, as this was shown to reduce a large portion of heterogeneity present (73.92% down to 64.1% I2) and explained 30.1% of the residual variance (R2). Second, we tested for temporal-bias in publication results (e.g. if non-significant studies are suppressed immediately after the first significant publication) by including the centered publication year of the study as a moderator in the meta-analytic model. Third, we used the number of independent observations (number of raters) as a moderator in the meta-regression to test if sample size (and power) is significantly related to MHC effect size. Diminishing effect sizes with increasing sample sizes would be an indication of publication bias suppressing small studies showing non-significant effects.
To assess the impact of publication bias and test the robustness of our results, we used the non-parametric trim and fill method [34,35] in the metafor R package. This method adjusts the mixed-model intercept for potentially missing studies. We conducted these tests for each dataset using meta-random effects models without including study ID as a random effect, which we believe is justified because this term accounted for almost no variance and most models showed very low heterogeneity (table 1). The one exception was the model for mate choice, and for this test, we used the meta-analytic residuals which consisted of within-study effects and sample errors (what was left after taking the mean and between-study effects from the effect sizes).
Table 1.
Heterogeneity estimates for a set of random-effect meta-analytical models for human MHC-similarity. (The number of levels refers to the number of studies. Q is Cochran's Q to test for heterogeneity. tau2 is the estimated between-study variance. The p-value indicates if the heterogeneity present is significant. The heterogeneity (I2) value is the per cent of variability between studies (i.e. variance in effect sizes not owing to sampling error). I2 = 25%, 50% and 75% are considered as low, moderate and high heterogeneity, respectively [36].)
| dataset | n | random effects | number of levels | Q | tau2 | d.f. | p-value | I2 (%) |
|---|---|---|---|---|---|---|---|---|
| genomic studies | 17 | study ID | 6 | 8.888 | 0.000 | 16 | 0.918 | 3.17 |
| relationship satisfaction | 9 | study ID | 3 | 13.579 | 0.006 | 8 | 0.093 | 46.46 |
| odour preference | 23 | study ID | 10 | 16.877 | 0.000 | 22 | 0.770 | 0.00 |
| odour preference (no HC) | 19 | study ID | 10 | 10.719 | 0.000 | 18 | 0.906 | 0.00 |
| mate selection | 55 | study ID | 26 | 259.865 | 0.015 | 54 | <0.001 | 73.92 |
We also tested the robustness of our models to outliers and influential data points as part of our sensitivity analyses. This was done by examining studentized residuals and hat matrix values for our mean meta-analytic models. The studentized residuals, or externally standardized residuals, follow a standard normal distribution. A large standardized residual for a study, therefore, may suggest that the study does not fit the assumed model and may be an outlier. Points below –2 or above 2 could be considered outliers. The hat matrix values provide the use of a data point. Points farther away from the predicted values (e.g. those pulling the regression line away from a better fit) will have more leverage. A hat value larger than 3 (number of moderators/number of data points) could be considered an influential point [30].
All statistical analyses were carried out in the R environment (v.3.6.0) [37], and all R code is provided in the electronic supplementary material, appendix. The R packages we used were metafor [30], ggplot2 [38], ggpubr [39], ggstance [40], erer [41], stringr [42], maps [43], rgdal [44], truncnorm [45] and wesanderson [46].
3. Results
(a). Genomic studies
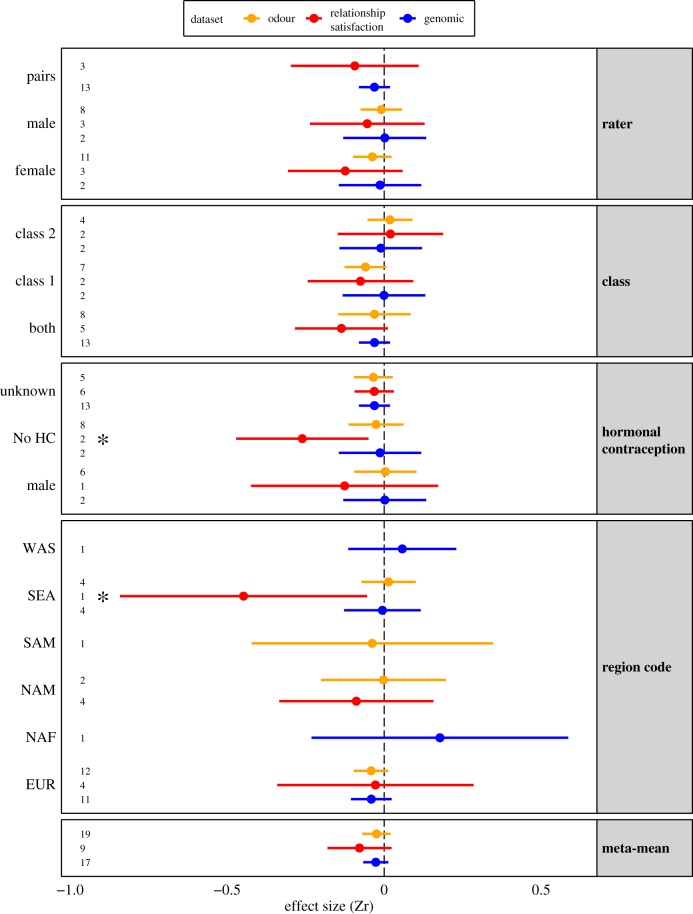
The mean effect size calculated over all genomic studies indicated no significant correlation between MHC-similarity and mate choice (intercept-only mean Zr (95% CI) = −0.027 (−0.067 to 0.013), n = 17, p = 0.191; figure 2). Very low heterogeneity (I2) was found in effect sizes (3.2%; table 1), indicating that confidence intervals for all effect sizes mostly overlapped and total variation was mostly attributable to variation within studies (electronic supplementary material, figure S1). We ran univariate meta-regression models to identify moderators that explained patterns in effect sizes of genomic MHC-linked mate choice. The only significant moderator was the effect for the extremeness of MHC relatedness compared to genome-wide relatedness within spouses (MHC | background, electronic supplementary material, table S1; figure 3a). The significant positive relationship (n = 12, β = 0.778, p = 0.023, R2 = 19.0%) indicates that observed MHC effect sizes (Zr) are related to the extremeness of the MHC compared to genome-wide similarity between spouses (figure 3b). In other words, MHC effects are relatively independent of socio-demographic processes that would affect spouses genome-wide. Additional support that MHC effects are independent of socio-demographic processes comes from the non-significant regression between observed MHC-similarity effect sizes for spouses and background genomic similarity for spouses (figure 3c; n = 12, β = 0.276, p = 0.405, R2 = 0.0%). While the relationship is positive, it is not significant. The non-significant relationship implies that MHC-dissimilarity between spouses cannot be explained by socio-demographic processes because such effects would affect the whole genome and the two effect sizes would be correlated.
Figure 2.
Forest plot of categorical moderators and meta-mean effect sizes from random-effects models run separately for genomic, relationship satisfaction and odour preference datasets. The model results can be found in the electronic supplementary material, tables S1–3, respectively. The overall meta-means were not significant (genomic studies: n = 17, Zr = −0.027 (−0.067 to 0.013), p = 0.191; relationship satisfaction: n = 9, Zr = −0.078 (−0.180 to 0.023), p = 0.131; odour preference (without pill effect sizes): n = 19, Zr = −0.024 (−0.069 to 0.021), p = 0.289). Numbers indicate the number of effect sizes, and stars indicate significant effects. (Online version in colour.)
Figure 3.
Meta-regression plots for the genomic dataset. (a) Model predictions for the relationship between the effect size that the MHC is extreme in comparison to the rest of the genome within couples and MHC effect size (Zr) between true couples compared to permuted couples. The significant positive relationship (n = 12, β = 0.778, p = 0.023) indicates that observed MHC effect sizes are relatively independent of socio-demographic processes that would affect spouses genome-wide. (b) An illustration of how effect sizes ‘MHC versus genomic similarity effect size (r)’ were calculated. The red, orange and yellow dots represent the mean relatedness for MHC between couples, and the correlation effect sizes ‘r’ are above. The density plot represents mean relatedness coefficients for genomic windows of varying recombination rates. The more extreme the MHC relatedness is compared to genomic relatedness, the further correlation coefficient is from zero. (c) Model predictions for the relationship between background genomic similarity effect size between true couples compared to permuted couples and MHC effect size (Zr) between true couples compared to permuted couples. The regression is not significant (n = 12, β = 0.276, p = 0.405), implying that MHC-dissimilarity between spouses cannot be explained by socio-demographic processes because such effects would affect the whole genome and the two effect sizes would be correlated. Coloured lines represent model predictions and grey regions represent 95% CIs. The size of points is proportional to their weight (inverse SE). (Online version in colour.)
(b). Relationship satisfaction
The mean effect size calculated over all studies indicated no significant correlation between MHC-similarity and relationship satisfaction measured as sexual satisfaction among couples (intercept-only mean Zr (95% CI) = −0.078 (−0.180 to 0.023), n = 9, p = 0.131; figure 2). The total heterogeneity (I2) in effect sizes was moderate (46.5%) and non-significant (p = 0.093; table 1). Univariate meta-regression models identified four significant and one borderline significant moderators of MHC similarity-linked relationship satisfaction effect sizes (electronic supplementary material, table S2). ‘Year’ and ‘number of raters’ will be detailed below in the section ‘Publication bias and sensitivity analysis’. Other borderline significant moderators included ‘HC use’ (n = 9, p = 0.057, R2 = 85.0%), with the level ‘no HC’ having a significant negative effect size estimate (Zr (CI) = −0.261 (−0.472 to −0.050), n = 2, p = 0.015) indicating that normally cycling women with higher levels of MHC-similarity with their partners experienced lower in-pair attraction. No significant effects were observed for men or for women or pairs with unknown HC-use status (electronic supplementary material, table S2). Lower in-pair attraction was most pronounced in partners with Asian and South-East Asian ancestry (Asian population and SEA region code Zr (CI) = −0.448 (−0.841 to −0.054), n = 1, p = 0.026; figure 2).
(c). Odour preference
The mean effect size for all effects (n = 23) and for effects excluding HC users (n = 19) both indicated no significant correlation between MHC-similarity and odour preferences (all intercept-only mean Zr (95% CI) = −0.020 (−0.064 to 0.023), p = 0.360; excluding HC users −0.024 (−0.069 to 0.021), p = 0.289; electronic supplementary material, tables S3 and S4, respectively; figure 2). The total heterogeneity (I2) in effect sizes for all data and data excluding HC users was non-existent (table 1). This indicates that confidence intervals for all effect sizes overlapped and total variation was attributable to variation within studies (electronic supplementary material, figure S1). The moderator ‘population’ was significant in the dataset excluding HC users, with Swiss individuals showing significantly reduced preference for body odours from donors with higher levels of MHC-similarity (Switzerland population Zr (CI) = −0.309 (−0.615 to −0.003), n = 3, p = 0.048).
(d). Mate selection
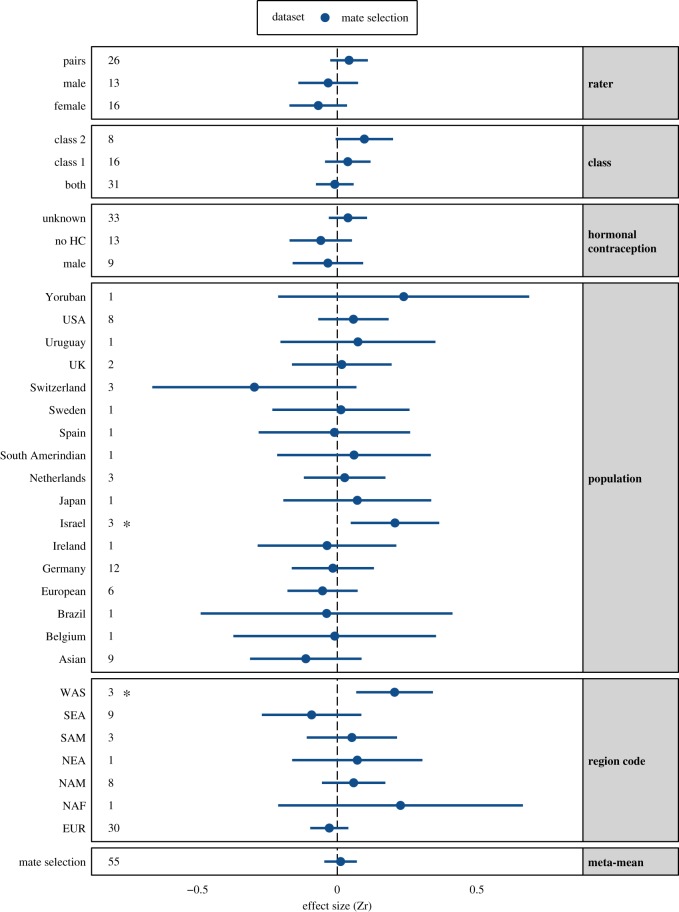
The mean effect size for all effects (n = 55) indicated no significant correlation between MHC-similarity and mate choice (intercept-only mean Zr (95% CI) = 0.012 (−0.046 to 0.070), p = 0.685; electronic supplementary material, table S5; figure 4). Mate selection studies showed significant heterogeneity (73.92%, I2, p < 0.001). Univariate meta-regression models identified three significant moderators of MHC similarity-linked mate selection effect sizes (electronic supplementary material, table S5). ‘Number of raters’ will be detailed below in the section ‘Publication bias and sensitivity analysis’. The moderator ‘population’ did not explain a significant amount of heterogeneity (n = 55, p = 0.522, R2 = 2.7%), but the population ‘Israeli’ was significant, with Israeli individuals showing significant preference for mates with higher levels of MHC-similarity (Zr (CI) = 0.207 (0.048 to 0.365), n = 3, p = 0.011). This preference for MHC-similar mates was repeated for the significant moderator ‘region code’ (n = 55, p = 0.031, R2 = 30.14%) for the Western Asian geographical region, to which Israel belongs (WAS region code Zr (CI) = 0.205 (0.068 to 0.343), n = 3, p = 0.003).
Figure 4.
Forest plot of categorical moderators and meta-mean effect sizes from random-effects models of mate selection dataset. The model results can be found in the electronic supplementary material, table S5. The overall meta-mean was not significant (mate selection: n = 55, Zr = 0.012 (−0.046 to 0.070), p = 0.685). Numbers indicate the number of effect sizes and stars indicate significant effects.
(e). Publication bias and sensitivity analysis
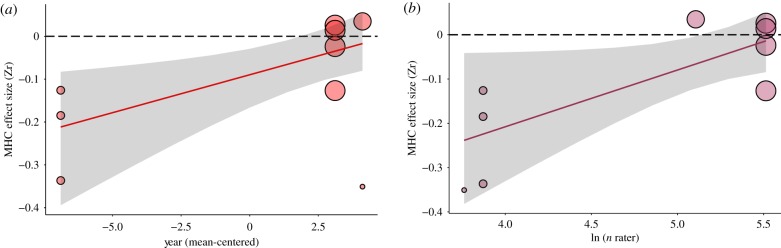
We found evidence of publication bias for the relationship satisfaction dataset from all three methods of bias testing. Egger's regression test for funnel plot asymmetry was significant (p = 0.008; electronic supplementary material, table S6 and figure S2), indicating that stronger MHC-dissimilarity effects on relationship satisfaction had a larger standard error. The meta-regression ‘year’ for temporal-bias was also borderline significant (β = 0.018, n = 9, p = 0.051, R2 = 89.5%; figure 5a), indicating that more recent studies show a reduced MHC-dissimilarity effect size. The third test for publication bias showed a significant positive relationship between natural log number of raters and MHC effect size (Zr), suggesting that greater power diminishes the effect of MHC-dissimilarity on relationship satisfaction (β = 0.128, n = 9, p = 0.012, R2 = 84.8%; figure 5b).
Figure 5.
Meta-regression plots for relationship satisfaction dataset indicate the evidence of publication bias. (a) Publication bias as a temporal-bias where more recent studies (with larger sample sizes) show a borderline significantly reduced dissimilarity effect size (n = 9, β = 0.018, p = 0.051). (b) The significant positive relationship between log number of raters and MHC effect size (Zr) suggests that greater power diminishes the effect of MHC-dissimilarity on relationship satisfaction (n = 9, β = 0.128, p = 0.012). Coloured lines represent model predictions and grey regions represent 95% CIs. The size of points is proportional to their weight (inverse SE). (Online version in colour.)
Egger's modified regression test for mate selection studies including region code as a covariate was significant (p = 0.010; electronic supplementary material, table S6 and figure S2), suggesting that publication bias, or residual heterogeneity not accounted for by region code in the studies, was present and creating asymmetry in the funnel plots. Besides relationship satisfaction studies, other studies did not show evidence of publication bias (electronic supplementary material, table S6). Sensitivity analyses using the trim and fill test found that our datasets were missing studies, but all but one of these tests were not significant and did not change the interpretation of meta-mean effect estimates for any dataset (table 2). The mate selection model was the exception, which went from a non-significant meta-mean intercept estimate of 0.012 (−0.046 to 0.070) to a positive estimate with a confidence interval that did not cross zero (0.065 (0.007 to 0.123); table 2; electronic supplementary material, figure S3) with the addition of the intercept estimate from the test on meta-analytic residuals. In other words, including 17 missing (positive) data points to all mate choice studies shifted the interpretation from no MHC-linked mate choice to MHC similarity-linked mate choice. It should be noted that the trim and fill method assumes that funnel plot asymmetry is only owing to publication bias, and so may be misleading when substantial between-study heterogeneity is present which can also induce funnel plot asymmetry [47].
Table 2.
Sensitivity analysis using the trim and fill test. Original intercept estimate is from mixed effects models with study ID as a random effect. The H0 is the hypothesis that the meta-mean effect size does not differ from zero. K, the sample size including missing studies.
| Trim and fill test | K | Estimated number of missing studies | SE | Original intercept estimate (95% CI) | Intercept estimate with points added (95% CI) | Test of H0 p-value |
|---|---|---|---|---|---|---|
| Genomic studies | 20 | 3 | 2.799 | −0.027 (−0.067 to 0.013) | −0.018 (−0.055 to 0.018) | 0.317 |
| Relationship satisfaction | 12 | 3 | 2.010 | −0.073 (−0.172 to 0.026) | −0.025 (−0.110 to 0.060) | 0.567 |
| Odour preference | 26 | 3 | 3.199 | −0.020 (−0.064 to 0.023) | −0.026 (−0.069 to 0.017) | 0.234 |
| Odour preference (no HC) | 19 | 0 | 2.375 | −0.024 (−0.069 to 0.021) | −0.024 (−0.069 to 0.021) | 0.289 |
| Mate selection (no HC) | 72 | 17 | 4.751 | 0.012 (−0.046 to 0.070) | 0.065 (0.007 to 0.123) | NA |
In addition to the trim and fill method, we checked for outliers and influential data points using studentized residuals and hat matrix values, respectively. In our genomic studies meta-mean model, the data point from Qiao et al. [48] was identified as influential, but removing it did not change the qualitative result of a non-significant meta-mean (Zr (CI) = −0.029 (−0.085 to 0.027), n = 16, p = 0.316). For our mate selection model, we identified three data points as influential (from [48–50]) and two as outliers (from [51]). However, their removal did not affect the overall interpretation and mate selection for MHC-dissimilarity remained non-significant (Zr (CI) = −0.023 (−0.059 to 0.013), n = 50, p = 0.218).
4. Discussion
(a). Meta-analyses
A recent meta-analysis on MHC-associated mate choice concluded that there is a consistent preference for MHC-heterozygous individuals [20]. By contrast, there was no systematic preference for MHC-dissimilarity. Here, we provide the results of further meta-analyses primarily focusing on genomic studies and relationship satisfaction, together with updated meta-analyses on odour preferences and human mate selection studies. Overall, the genomic studies show no significant association between MHC-similarity and mate choice in actual couples nor in mate preferences. However, we also found that the effect of MHC-similarity is independent of the genomic background. The overall effect of MHC-similarity on sexual satisfaction was not significant, but we found a negative association between MHC-similarity and sexual satisfaction in non-HC using women. Nevertheless, several lines of evidence for publication bias in studies investigating MHC-similarity and sexual satisfaction suggest that these results should be interpreted with caution. Furthermore, we found no significant effect of MHC-similarity on odour preferences among currently available studies. Finally, combining each of the effect sizes analysed above with previously extracted effect sizes for mate choice among couples into an overall estimate, showed no overall significant effect of MHC-similarity on human mate selection.
(b). Near horizons: issues arising from the meta-analyses
Our meta-analyses raise a number of pressing outstanding issues that should, and can be, addressed in future studies. Perhaps the strongest conclusion one can draw from the available data is that our knowledge is patchy across different populations. Even a brief inspection of figure 1 shows that most studies are based on populations of European ancestry; there is a notable absence or near-absence of data from two of the largest populations, China and India, from smaller populations in Oceania and Sub-Saharan Africa, and from small-scale societies. Why is this important? First, individual populations vary considerably in cultural norms regarding the level of consanguinity [52]. In addition, while all populations show some amount of admixture, this tends to be higher in large-scale populations such as those from Western European or Eastern Asian complex societies [53]. Owing to high MHC polymorphism, mating with almost any unrelated individual would probably lead to a sufficient level of dissimilarity. It is thus possible that humans, as in other species [54–56], tend to avoid individuals with high MHC-similarity, but show no systematic preference beyond a certain threshold (see [28] for a similar suggestion). However, large-scale populations are a relative novelty in human evolutionary history [57]; it is therefore of key importance to focus on small-scale societies with comparatively higher levels of inbreeding, which better reflect likely population structure during most of human evolution. To our knowledge, the only available study from small-scale societies comes from South Amerindian couples [58], which showed they were not significantly MHC dissimilar compared to random pairing. In that study, however, MHC typing was of relatively low sensitivity (serotyping of HLA-A and -B loci to the level of two-digit allele groups, no class II loci were recorded), sample size was too small to detect selection below a selection coefficient s = 0.45, and there is cultural promotion of cross-cousin marriages in some tribes [58].
Most previous studies have specifically targeted the MHC region, assuming that their findings are a consequence of selective processes in that region. While this is a reasonable assumption in view of MHC polymorphism and allele-specific associations with some diseases [8–11], apparent MHC-similar mate selection might be an epiphenomenon of more general population stratification (e.g. positive assortment [59]). In support of this, a recent meta-analysis found that MHC-similarity in couples was observed in ethnically heterogeneous, but not homogeneous, populations [20]. However, our new analysis of studies that control for genomic similarity shows that MHC-dissimilarity among couples is independent of genome-wide similarity (although the association is positive). In addition, the positive relationship detected between MHC effects (spouses versus permuted pairs) and the extremeness of the MHC within spouses indicates that observed MHC effects are relatively independent of socio-demographic processes that would affect spouses genome-wide. For example, if spouses were highly dissimilar at the MHC compared to randomly assigned mates, but had levels of MHC-similarity in line with the rest of the genome, we may conclude that the MHC does not play an independent role in mate choice and mate choice may be for inbreeding avoidance. But this was not what we observed.
The overall effect of MHC-associated mate selection was not significant but was restricted to some populations. In other words, we may observe MHC-associated preferences in some populations but not in others. For example, we found that Israeli individuals showed a significant preference for mates with higher levels of MHC-similarity. Dandine-Roulland et al. [29] contributed one of the three effect sizes to this result, and using principal component analysis detected genetic stratification, with clusters of samples lying between European and Middle Eastern populations. The two other effect sizes contributed by Israeli et al. [51] came from unmarried couples to determine paternity status and from married couples undergoing infertility treatment. The study did not specifically detail testing for population stratification, and it is likely that a random sample of the population would capture multiple ethnic groups, as Dandine-Roulland et al. [29] demonstrated. Thus, MHC-similarity preferences most likely reflect social homogamy in a genetically heterogeneous population. The Swiss individuals' significant preference for MHC-dissimilar odours was observed in the dataset without HC-using individual effects and included both female and male odour preferences. These MHC-dissimilar preferences might be related to relatively low levels of genetic variation and were specifically present in German-speaking cantons, perhaps as a consequence of geographical isolation in Alpine valleys [60]. By contrast, studies based on other European populations (such as in neighbouring Germany) did not report MHC-dissimilar preferences, emphasizing the need for investigations across diverse populations which differ in levels of genetic variation. For example, cultural practices vary related to body care. If body odour is a primary source of information about one's MHC profile, then practices such as armpit hair shaving and use of extrinsic fragrances or deodorants may impact perceptibility of MHC-associated odours. Although there is conjecture that fragrance selection may be linked to a wearer's own MHC [61,62], perhaps as a mechanism to complement body odour rather than cover it [63], we do not yet know how such cultural effects influence odour perceptibility and MHC-associated preference. Further, in cultures which idealize an ‘odourless human body’, it is considered inappropriate to overtly smell other people; under such circumstances, the effect of MHC-associated preferences might go unrealized. Clearly, our understanding of the interplay between cultural and biological evolution is far from complete, and MHC-associated mate choice is no exception.
Many cultures also practice various types of positive assortment such as ethnic, socio-economic, religious, and caste-based endogamy. Even within a single culture, mate choice is a multidimensional process based on a set of preferences for various traits which might not be linked to MHC, such as physical appearance, socio-economic status, personality, attitudes, age and many others [64,65]. Each of these may be prioritized over genotypic factors [66], including MHC. Furthermore, if positive assortment occurs for any trait with a genetic component, even subtle assortment on such traits might interfere with MHC-associated preferences.
Beyond actual mate choice, it remains possible that MHC-associated preferences exert effects on the quality of resulting relationships. Indeed, in a study of 48 couples, Garver-Apgar et al. [22] found that more MHC-similar couples report relatively lower sexual satisfaction. Subsequent investigations have recorded considerably larger sample sets [67,68]. Here, we quantitatively assessed these studies for a possible link between MHC-similarity and sexual satisfaction. The overall effect was not significant. However, in the subset of women not using HC, there was a negative association between MHC-similarity and sexual satisfaction: couples sharing fewer HLA alleles experienced greater sexual satisfaction. This pattern of results is consistent with the studies by Wedekind et al. [18] who found odour preferences for MHC-dissimilarity only in women not using HC, and by Roberts et al. [69,70] who report higher sexual satisfaction in women who did not use HC when they met their current partner. Nevertheless, the robustness of the HC-associated preferences was neither confirmed by a previous meta-analysis [20] nor in our updated analysis. There is another reason why the link between MHC-similarity and sexual satisfaction should be interpreted with extreme caution. The meta-analysis on relationship satisfaction found three different types of evidence for publication bias. First, there was a significant asymmetry in a funnel plot suggesting missing studies with a negative outcome, particularly those with small effect sizes. Second, there was a temporal effect suggesting the unequal distribution of the effect sizes over time; specifically, the initial study [22] found a considerably stronger effect than subsequent studies. Finally, studies with larger samples (i.e. having a higher power to detect possible effects) show significantly smaller effect sizes.
(c). Far horizons on major histocompatibility complex-associated mate choice
Beyond those issues raised above, we believe there are two further matters that require significant attention in the longer-term. The first of these concerns the generation of MHC-associated odours. Understanding this may be of fundamental interest in itself, but a clearer picture of the underlying mechanisms may also clarify how some cultural and contextual factors (e.g. fragrance use) affect odour variability. Several hypotheses have been proposed relating to interactions between MHC molecules and skin microflora, which produces volatile compounds that can subsequently be perceived. However, most evidence supports an idea that body odour is affected by antigen peptides bound by specific MHC molecules. It was first shown in mice that these peptides can be perceived by the vomeronasal organ [71]; however, subsequent research shows that the main olfactory system can perceive MHC peptide ligands via the olfactory epithelium [72]. MHC peptide ligands can be detected in mouse urine, although at very low concentration [73]. Evidence extends beyond mice, as sticklebacks prefer water enriched with MHC-dissimilar peptides [74]. So far, only one study addressed this mechanism in humans [75]. Two commercially available peptides were added to body odour samples, and neurophysiological responses were recorded using functional magnetic resonance imaging while participants attempted to recognize their own odour. The results showed a higher preference for odour samples enriched with peptides corresponding to the MHC of the smeller and activity in brain areas related to self-recognition. However, it is not clear whether the self-recognition paradigm can be simply generalized to mate preferences. More importantly, the study was criticized for not providing an explanation for the transduction mechanism, as peptide molecules are involatile and considerably larger than molecules usually perceived by smell [73,76,77]. Furthermore, it is also not clear whether the MHC-associated peptides are commonly present in human axillae or more generally on human skin.
A second area which requires more attention is the nature of potential selective benefits arising from MHC-associated mate choice. While it is usually assumed that MHC-preferences are a consequence of infection-driven selection, it might be alternatively (or additionally) driven by the probability of successful pregnancy. A foetus expresses paternal alloantigens which must be tolerated by the maternal immune system. It has been proposed that MHC allele-sharing between father and mother may lead to insufficient stimulation of the maternal immune system by paternal antigens—a factor that was expected to be important for maternal tolerance and inflammatory immune response—and thus decrease the chance of successful implantation [78]. Several studies suggest that MHC allele-sharing is associated with recurrent pregnancy loss (RPL) [79,80], with a recent meta-analysis indicating that HLA-B and -DR are especially important [81]. However, these results should be viewed with caution because many studies used serological genotyping resolving only to allele groups, which may miss related alleles that are functionally different [82]. More critically, classical MHC class I and II proteins (except for HLA-C) are not expressed on the trophoblast, a part of conceptus which subsequently develops into the embryonic part of the placenta and is in direct contact with the maternal immune system. Researchers have, therefore, recently focused on classical HLA-C and non-classical MHC class Ib, which are expressed on the trophoblast. In contrast to previous studies, it was reported that a mismatch, i.e. not sharing, at HLA-C*07 between mother and father was related to a higher risk of RPL [83]. These authors also observed a higher incidence of HLA-C antibodies in RPL patients than in the controls. There is a growing body of evidence showing that HLA-E, -G, and to some extent also HLA-F, all play a key role in immunotolerance of the foetus by the maternal immune system in general and uterine NK cells in particular (for a review, see [3]). Some studies report higher RPL in women with the HLA-E*101 allele [84], although others find no difference in HLA-E polymorphism between controls and couples with RPL [85,86]. Most studies on non-classical MHC Ib polymorphism and its role in pregnancy disorders focused on HLA-G polymorphism. For example, it was reported that a 14 bp insertion HLA-G allele is associated with a smaller placenta and higher probability of RPL [87], although this may be restricted to cases with three and more abortions [88]. In summary, there appears to be some evidence that couples sharing alleles at HLA-B and -DR loci are at higher risk of reproductive failure. Although these genes are not expressed on the trophoblast, this might arise through linkage disequilibrium with other functionally important MHC genes. Moreover, there is inconsistency across studies in both the association between HLA-G and -E polymorphism and reproductive failures, perhaps partly owing to factors such as variation in the diagnosis of the RPL. More importantly, most existing studies on MHC polymorphism and reproductive problems focused solely on RPL, but MHC polymorphism might affect pregnancy success much earlier as HLA-C and -G expression can be detected even before implantation [89,90]. Because a vast majority of unsuccessful early pregnancies are not detected, this may, in turn, bias the results of studies that rely solely on RPL (i.e. recognizable spontaneous miscarriage).
(d). Suggestions for future studies
Above, we have discussed in detail the current state of knowledge on MHC-associated mate choice in light of results from our meta-analysis, that should inform approaches in the immediate future. We also commented on two important wider and relatively unexplored perspectives that lurk on the far horizon of this area of inquiry. In light of these, we here outline some recommendations for future work that we hope will help to ultimately clarify the extent to which MHC influences human mating. The suggestions (i–iii) highlight methodological issues, (iv–vi) focus on population- and culture-related questions, and (vii–x) stress several associated issues such as developmental and mechanistic questions.
-
(i)
Researchers should always perform a priori power analysis to obtain a sufficient sample size (see also [91]). Power analysis is becoming a standard procedure in other fields of behavioural research, but it is particularly needed here owing to both extreme variability in MHC genes and what appear to be, at best, small effect sizes.
-
(ii)
To provide more complex insights, future studies should control for genome-wide similarity. The same applies to studies on MHC-heterozygosity. Genomic studies further allow assessment of the overall level of inbreeding in the given population. This is an important issue as MHC-associated mate choice might play a role only in relatively inbred populations.
-
(iii)
Researchers should test for specificity of the MHC region. As was discussed above, without controlling for genome-wide level of similarity/heterozygosity, we cannot decide whether the observed effects are specific to the MHC region or whether we are dealing with more general phenomena.
-
(iv)
We urgently need more studies on populations of non-European descent, and particularly those with a relatively high level of inbreeding (e.g. from small-scale societies).
-
(v)
We need more cross-cultural comparisons assessing how shared cultural practices affect MHC-associated preferences. These include marriage practices such as various forms of endogamy.
-
(vi)
In any study, researchers should obtain and clearly document detailed information about interindividual differences in cultural practices of the studied population, as some practices may interfere with MHC-associated effects. These include HC use and personal hygiene practices such as fragrance use (see also [92] for a similar proposal).
-
(vii)
We need to distinguish between a threshold-based avoidance of very similar individuals and a fluid preference for the most dissimilar individuals.
-
(viii)
Currently, there is not, to our knowledge, a single study focusing on the development of MHC-associated preferences. Therefore, we do not know when in ontogeny preferences might form and how family structure affects the development of these preferences. Rodent studies show that cross-fostering tends to reverse MHC-associated preferences [93], thus similar phenomena might be expected in humans. For instance, studies with adoptive families might be particularly informative.
-
(ix)
We need studies testing possible mechanisms of MHC-associated preferences. These include bioassay studies testing the presence and abundance of the MHC peptide ligands. Similarly, studies testing effect of the MHC peptide ligands in the context of mate choice are of primary importance.
-
(x)
Finally, we should link research on MHC-associated mate choice and research on MHC-associated pregnancy loss. The two areas have to date been studied separately; however, they may jointly provide key insights into this complex area of human reproduction. Such research may also examine links between pregnancy loss and infertility with the prevalence of cultural practices (e.g. fragrance use) that may have disrupted MHC-associated mate preferences at the beginning of the relationship.
5. Concluding remarks
We finish this paper almost a quarter of a century after the initial discovery of MHC-associated odour preferences in humans by Wedekind et al. [18]. We must humbly admit that our knowledge remains far from complete. Sadly enough, we still cannot even conclude whether MHC-associated preferences affect real-life mate choice and if so, under what circumstances. Interestingly, many of the issues that we raise are ones currently being discussed in the behavioural sciences and psychology in particular (see also [91]). For one, we base most of our knowledge on studies from Western populations and often too readily generalize them to all human beings [94]. In addition, some research areas may suffer from various types of publication bias [95]. Finally, exciting initial discoveries which become textbook staple examples are sometimes difficult to replicate, a case in point here being the effect of HC. Such conclusions may give a dark impression to some, but we see the future quite optimistically. We hope that renewed efforts, addressing some of the key issues we raise here, will bring more realistic views about the MHC-associated mate choice in the coming years.
Supplementary Material
Acknowledgements
We would like to thank Jindra Havlíčková and two anonymous reviewers for valuable comments on the previous version of the paper.
Data accessibility
The datasets and R code accompanying this article are deposited on figshare (https://doi.org/10.6084/m9.fighshare.8869505).
Authors' contributions
All authors contributed to the conception and design of the study. J.W. performed the data analysis, and J.H. contributed to the data interpretation. J.H. and J.W. drafted the article, and S.C.R. provided critical revision. All authors approved the final version of the article.
Competing interests
We declare we have no competing interests.
Funding
J.H. is supported by the Czech Science Foundation grant (no. 18-15168S), and by the Charles University Research Centre (UNCE) program UNCE/HUM/025 (204056). J.W. is supported by the German Research Foundation (DFG) as part of the SFB TRR 212 (316099922).
References
- 1.Flajnik MF, Kasahara M. 2001. Comparative genomics of the MHC: glimpses into the evolution of the adaptive immune system. Immunity 15, 351–362. ( 10.1016/s1074-7613(01)00198-4) [DOI] [PubMed] [Google Scholar]
- 2.Horton R, et al. 2004. Gene map of the extended human MHC. Nat. Rev. Genet. 5, 889–899. ( 10.1038/nrg1489) [DOI] [PubMed] [Google Scholar]
- 3.Persson G, Melsted WN, Nilsson LL, Hviid TVF. 2017. HLA class Ib in pregnancy and pregnancy-related disorders. Immunogenetics 69, 581–595. ( 10.1007/s00251-017-0988-4) [DOI] [PubMed] [Google Scholar]
- 4.Klein J, Saito A. 2000. The HLA system. N. Engl. J. Med. 343, 702–709. ( 10.1056/NEJM200009073431006) [DOI] [PubMed] [Google Scholar]
- 5.Winternitz J, Abbate J. 2015. Examining the evidence for major histocompatibility complex-dependent mate selection in humans and nonhuman primates. Res. Rep. Biol. 6, 73–88. ( 10.2147/RRB.S58514) [DOI] [Google Scholar]
- 6.Sanchez-Mazas A, Mack SJ, Single RM, Tsai Y, Lancaster AK, Solberg OD, Thomson G. 2008. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69, 443–464. ( 10.1016/j.humimm.2008.05.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prugnolle F, Manica A, Charpentier M, Guégan JF, Guernier V, Balloux F. 2005. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 15, 1022–1027. ( 10.1016/j.cub.2005.04.050) [DOI] [PubMed] [Google Scholar]
- 8.Hill AV, et al. 1991. Common West African HLA antigens are associated with protection from severe malaria. Nature 352, 595– 600 ( 10.1038/352595a0) [DOI] [PubMed] [Google Scholar]
- 9.Trachtenberg E, et al. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9, 928–935. ( 10.1038/nm893) [DOI] [PubMed] [Google Scholar]
- 10.Sveinbjornsson G, et al. 2016. HLA class II sequence variants influence tuberculosis risk in populations of European ancestry. Nat. Genet. 48, 318–322. ( 10.1038/ng.3498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause-Kyora B, et al. 2018. Ancient DNA study reveals HLA susceptibility locus for leprosy in medieval Europeans. Nat. Commun. 9, 1569 ( 10.1038/s41467-018-03857-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fumagalli M, Sironi M, Pozzoli U, Ferrer-Admettla A, Pattini L, Nielsen R. 2011. Signatures of environmental genetic adaptation pinpoint pathogens as the main selective pressure through human evolution. PLoS Genet. 7, e1002355 ( 10.1371/journal.pgen.1002355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Apanius V, Penn D, Slev PR, Ruff LR, Potts WK. 1997. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 37, 75–120. ( 10.1615/CritRevImmunol.v37.i2-6.10) [DOI] [PubMed] [Google Scholar]
- 14.Penn DJ, Damjanovich K, Potts WK. 2002. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proc. Natl Acad. Sci. USA 99, 11 260–11 264. ( 10.1073/pnas.162006499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamazaki K, Boyse EA, Miké V, Thaler HT, Mathieson BJ, Abbott J, Boyse J, Zayas ZA, Thomas L. 1976. Control of mating preferences in mice by genes in the major histocompatibility complex. J. Exp. Med. 144, 1324–1335. ( 10.1084/JEM.144.5.1324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernatchez L, Landry C. 2003. MHC studies in nonmodel vertebrates: what have we learned about natural selection in 15 years? J. Evol. Biol. 16, 363–377. ( 10.1046/j.1420-9101.2003.00531.x) [DOI] [PubMed] [Google Scholar]
- 17.Kamiya T, O'Dwyer K, Westerdahl H, Senior A, Nakagawa S. 2014. A quantitative review of MHC-based mating preference: the role of diversity and dissimilarity. Mol. Ecol. 23, 5151–5163. ( 10.1111/mec.12934) [DOI] [PubMed] [Google Scholar]
- 18.Wedekind C, Seebeck T, Bettens F, Paepke AJ. 1995. MHC-dependent mate preference in humans. Proc. R. Soc. Lond. B 260, 245–249. ( 10.1098/rspb.1995.0087) [DOI] [PubMed] [Google Scholar]
- 19.Havlíček J, Roberts SC. 2009. MHC-correlated mate choice in humans: a review. Psychoneuroendocrinology 34, 497–512. ( 10.1016/j.psyneuen.2008.10.007) [DOI] [PubMed] [Google Scholar]
- 20.Winternitz J, Havlíček J, Garamszegi LZ, Huchard E, Abbate JL. 2017. Patterns of MHC-dependent mate selection in humans and nonhuman primates: a meta-analysis. Mol. Ecol. 26, 668–688. ( 10.1111/mec.13920) [DOI] [PubMed] [Google Scholar]
- 21.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909. ( 10.1038/ng1847) [DOI] [PubMed] [Google Scholar]
- 22.Garver-Apgar CE, Gangestad SW, Thornhill R, Miller RD, Olp JJ. 2006. Major histocompatibility complex alleles, sexual responsivity, and unfaithfulness in romantic couples. Psychol. Sci. 17, 830–835. ( 10.1111/j.1467-9280.2006.01789.x) [DOI] [PubMed] [Google Scholar]
- 23.Zaidi AA, White JD, Mattern BC, Liebowitz CR, Puts DA, Claes P, Shriver MD. 2019. Facial masculinity does not appear to be a condition-dependent male ornament and does not reflect MHC heterozygosity in humans. Proc. Natl Acad. Sci. USA 116, 1633–1638. ( 10.1073/pnas.1808659116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khankhanian P, Gourraud P-A, Caillier SJ, Santaniello A, Hauser SL, Baranzini SE, Oksenberg JR. 2010. Genetic variation in the odorant receptors family 13 and the MHC loci influence mate selection in a multiple sclerosis dataset. BMC Genomics 11, 626 ( 10.1186/1471-2164-11-626) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giphart MJ, D'Amaro J. 1983. HLA and reproduction? J. Immunogenet. 10, 25–29. ( 10.1111/j.1744-313X.1983.tb01013.x) [DOI] [PubMed] [Google Scholar]
- 26.Laurent R, Chaix R. 2012. MHC-dependent mate choice in humans: why genomic patterns from the HapMap European American dataset support the hypothesis. BioEssays 34, 267–271. ( 10.1002/bies.201100150) [DOI] [PubMed] [Google Scholar]
- 27.Nakagawa S, Cuthill IC. 2007. Effect size, confidence interval and statistical significance: a practical guide for biologists. Biol. Rev. 82, 591–605. ( 10.1111/j.1469-185X.2007.00027.x) [DOI] [PubMed] [Google Scholar]
- 28.Derti A, Cenik C, Kraft P, Roth FP. 2010. Absence of evidence for MHC-dependent mate selection within HapMap populations. PLoS Genet. 6, e1000925 ( 10.1371/journal.pgen.1000925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dandine-Roulland C, Laurent R, Dall'Ara I, Toupance B, Chaix R. 2019. Genomic evidence for MHC disassortative mating in humans. Proc. R. Soc. B 286, 20182664 ( 10.1098/rspb.2018.2664) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Viechtbauer W. 2015. Conducting meta-analyses in R with the metafor package. J. Stat. Softw. 36, 1–48. ( 10.18637/jss.v036.i03) [DOI] [Google Scholar]
- 31.Cohen J. 1988 Statistical power analysis for the behavioral sciences, 2nd edn Hillsdale, NJ: L. Erlbaum. [Google Scholar]
- 32.Moller AP, Jennions MD, Møller A. 2002. How much variance can be explained by ecologists and evolutionary biologists? Oecologia 132, 492–500. ( 10.1007/s00442-002-0952-2) [DOI] [PubMed] [Google Scholar]
- 33.Egger M, Davey Smith G, Schneider M, Minder C. 1997. Bias in meta-analysis detected by a simple, graphical test. Brit. Med. J. 315, 629–634. ( 10.1136/bmj.315.7109.629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duval S, Tweedie R. 2000. A nonparametric ‘trim and fill’ method of accounting for publication bias in meta-analysis. J. Am. Stat. Assoc. 95, 89–98. ( 10.1080/01621459.2000.10473905) [DOI] [Google Scholar]
- 35.Duval S, Tweedie R. 2000. Trim and fill: a simple funnel-plot-based method. Biometrics 56, 455–463. ( 10.1111/j.0006-341X.2000.00455.x) [DOI] [PubMed] [Google Scholar]
- 36.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. 2003. Measuring inconsistency in meta-analyses. Brit. Med. J. 327, 557–560. ( 10.1136/bmj.327.7414.557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.R Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. See https://www.R-project.org/.
- 38.Wickham H. 2016. ggplot2: elegant graphics for data analysis. New York, NY: Springer. [Google Scholar]
- 39.Kassambara A. 2018. ggpubr: ‘ggplot2’ based publication ready plots. R package, version 0.2. See https://CRAN.R-project.org/package=ggpubr. [DOI] [PubMed]
- 40.Henry L, Wickham H, Chang W. 2018. ggstance: horizontal ‘ggplot2’ components. R package, version 0.3.1 See https://CRAN.R-project.org/package=ggstance.
- 41.Sun C.2016. erer: empirical research in economics with R. R package, version 2.5. See https://CRAN.R-project.org/package=erer .
- 42.Wickham H. 2019. stringr: simple, consistent wrappers for common string operations. R package, version 1.4.0. See https://CRAN.R-project.org/package=stringr.
- 43.Becker RA, Wilks A, Brownrigg R, Minka T, Deckmyn A. 2018. maps: draw geographical maps. R package, version 3.3.0. See https://CRAN.R-project.org/package=maps.
- 44.Bivand R, Keitt T, Rowlingson B. 2019. rgdal: bindings for the ‘geospatial’ data abstraction library. R package, version 1.4-3. See https://CRAN.R-project.org/package=rgdal.
- 45.Mersmann O, Trautmann H, Steuer D, Bornkamp B. 2018. truncnorm: truncated normal distribution. R package, version 1.0-8. See https://CRAN.R-project.org/package=truncnorm.
- 46.Ram K, Wickham H. 2018. wesanderson: a Wes Anderson palette generator. R package, version 0.3.6. See https://CRAN.R-project.org/package=wesanderson.
- 47.Peters JL, Sutton AJ, Jones DR, Abrams KR, Rushton L. 2007. Performance of the trim and fill method in the presence of publication bias and between-study heterogenity. Stat. Med. 26, 4544–4562. ( 10.1002/sim.2889) [DOI] [PubMed] [Google Scholar]
- 48.Qiao Z, Powell JE, Evans DM. 2018. MHC-dependent mate selection within 872 spousal pairs of European ancestry from the Health and Retirement Study. Genes (Basel) 9, 53. ( 10.3390/genes9010053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nordlander C, Hammarström L, Lindblom B, Smith CI. 1983. No role of HLA in mate selection. Immunogenetics 18, 429–431. ( 10.1007/BF00372474) [DOI] [PubMed] [Google Scholar]
- 50.Rosenberg LT, Cooperman D, Payne R. 1983. HLA and mate selection. Immunogenetics 17, 89–93. ( 10.1007/BF00364292) [DOI] [PubMed] [Google Scholar]
- 51.Israeli M, Kristt D, Nardi Y, Klein T. 2014. Genetic considerations in human sex-mate selection: partners share human leukocyte antigen but not short-tandem-repeat identity markers. Am. J. Reprod. Immunol. 71, 467–471. ( 10.1111/aji.12213) [DOI] [PubMed] [Google Scholar]
- 52.Bittles AH, Black ML. 2010. Consanguinity, human evolution, and complex diseases. Proc. Natl Acad. Sci. USA 107, 1779–1786. ( 10.1073/pnas.0906079106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elhaik E, et al. 2014. Geographic population structure analysis of worldwide human populations infers their biogeographical origins. Nat. Commun. 5, 1–13. ( 10.1038/ncomms4513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts SC, Gosling LM. 2003. Genetic similarity and quality interact in mate choice decisions by female mice. Nat. Genet. 35, 103–106. ( 10.1038/ng1231) [DOI] [PubMed] [Google Scholar]
- 55.Reichard M, Spence R, Bryjová A, Bryja J, Smith C. 2012. Female rose bitterling prefer MHC-dissimilar males: experimental evidence. PLoS ONE 7, e40780 ( 10.1371/journal.pone.0040780) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olsson M, Madsen T, Nordby J, Wapstra E, Ujvari B, Wittsell H. 2003. Major histocompatibility complex and mate choice in sand lizards. Proc. R. Soc. Lond. B 270, 254–256. ( 10.1098/rsbl.2003.0079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hellenthal G, Falush D, Myers S, Busby GBJ, Band G, Wilson JF, Capelli C. 2014. A genetic atlas of human admixture history. Science 747, 747–751. ( 10.1126/science.1243518) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hedrick PW, Black FL. 1997. HLA and mate selection: no evidence in South Amerindians. Am. J. Hum. Genet. 61, 505–511. ( 10.1086/515519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Štěrbová Z, Valentová JV. 2012. Influence of homogamy, complementarity, and sexual imprinting on mate choice. Anthropologie L/1, 47–59. [Google Scholar]
- 60.Barrai I, Scapoli C, Beretta M, Nesti C, Mamolini E. 1996. Isonymy and the genetic structure of Switzerland. I. The distributions of surnames. Ann. Hum. Biol. 23, 431–455. ( 10.1080/03014469600004672) [DOI] [PubMed] [Google Scholar]
- 61.Milinski M, Wedekind C. 2001. Evidence for MHC-correlated perfume preferences in humans. Behav. Ecol. 12, 140–149. ( 10.1093/beheco/12.2.140) [DOI] [Google Scholar]
- 62.Hämmerli A, Schweisgut C, Kaegi M, Kacgi M. 2012. Population genetic segmentation of MHC-correlated perfume preferences. Int. J. Cosmet. Sci. 34, 161–168. ( 10.1111/j.1468-2494.2011.00696.x) [DOI] [PubMed] [Google Scholar]
- 63.Lenochová P, Vohnoutová P, Roberts SC, Oberzaucher E, Grammer K, Havlíček J. 2012. Psychology of fragrance use: perception of individual odor and perfume blends reveals a mechanism for idiosyncratic fragrance choice. PLoS ONE 7, e33810 ( 10.1371/journal.pone.0033810) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fletcher GJO, Simpson JA, Thomas G, Giles L. 1999. Ideals in intimate relationships. J. Pers. Soc. Psychol. 76, 72 ( 10.1037/0022-3514.76.1.72) [DOI] [PubMed] [Google Scholar]
- 65.Csajbók Z, Berkics M. 2017. Factor, factor, on the whole, who's the best fitting of all?: factors of mate preferences in a large sample. Pers. Individ. Dif. 114, 92–102. ( 10.1016/j.paid.2017.03.044) [DOI] [Google Scholar]
- 66.Zietsch BP, Verweij KJH, Heath AC, Martin NG. 2011. Variation in human mate choice: simultaneously investigating heritability, parental influence, sexual imprinting, and assortative mating. Am. Nat. 177, 605–616. ( 10.1086/659629) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kromer J, Hummel T, Pietrowski D, Giani AS, Sauter J, Ehninger G, Schmidt AH, Croy I. 2016. Influence of HLA on human partnership and sexual satisfaction. Sci. Rep. 6, 6–11. ( 10.1038/srep32550) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saphire-Bernstein S, Larson CM, Gildersleeve KA, Fales MR, Pillsworth EG, Haselton MG. 2017. Genetic compatibility in long-term intimate relationships: partner similarity at major histocompatibility complex (MHC) genes may reduce in-pair attraction. Evol. Hum. Behav. 38, 190–196. ( 10.1016/j.evolhumbehav.2016.09.003) [DOI] [Google Scholar]
- 69.Roberts SC, Klapilová K, Little AC, Burriss RP, Jones BC, DeBruine LM, Petrie M, Havlíček J. 2012. Relationship satisfaction and outcome in women who meet their partner while using oral contraception. Proc. R. Soc. B 279, 1430–1436. ( 10.1098/rspb.2011.1647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roberts SC, Little AC, Burriss RP, Cobey KD, Klapilová K, Havlíček J, Jones BC, DeBruine L, Petrie M. 2014. Partner choice, relationship satisfaction, and oral contraception: the congruency hypothesis. Psychol. Sci. 25, 1497–1503. ( 10.1177/0956797614532295) [DOI] [PubMed] [Google Scholar]
- 71.Leinders-Zufall T, et al. 2004. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 306, 1033–1037. ( 10.1126/science.1102818) [DOI] [PubMed] [Google Scholar]
- 72.Spehr M, Kelliher KR, Li XH, Boehm T, Leinders-Zufall T, Zufall F. 2006. Essential role of the main olfactory system in social recognition of major histocompatibility complex peptide ligands. J. Neurosci. 26, 1961–1970. ( 10.1523/JNEUROSCI.4939-05.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sturm T, Leinders-zufall T, Mac B, Zufall F, Overath P, Rammensee H. 2013. Mouse urinary peptides provide a molecular basis for genotype discrimination by nasal sensory neurons. Nat. Commun. 4, 1616 ( 10.1038/ncomms2610) [DOI] [PubMed] [Google Scholar]
- 74.Milinski M, Griffiths S, Wegner KM, Reusch TBH, Haas-Assenbaum A, Boehm T. 2005. Mate choice decisions of stickleback females predictably modified by MHC peptide ligands. Proc. Natl Acad. Sci. USA 102, 4414–4418. ( 10.1073/pnas.0408264102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Milinski M, Croy I, Hummel T, Boehm T. 2013. Major histocompatibility complex peptide ligands as olfactory cues in human body odour assessment. Proc. R. Soc. B 280, 20122889 ( 10.1098/rspb.2012.2889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Natsch A, Emter R. 2020. The specific biochemistry of human axilla odour formation viewed in an evolutionary context. Phil. Trans. R. Soc. B 375, 20190269 ( 10.1098/rstb.2019.0269) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Natsch A. 2013. A human chemosensory modality to detect peptides in the nose? Proc. R Soc. B 280, 20131678 ( 10.1098/rspb.2013.1678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas ML, Harger JH, Wagener DE, Rabin BS, Gill TJ. 1985. HLA sharing and spontaneous abortion in humans. Am. J. Obstet. Gynecol. 151, 1053–1058. ( 10.1016/0002-9378(85)90379-5) [DOI] [PubMed] [Google Scholar]
- 79.Ober C. 1999. Studies of HLA, fertility and mate choice in a human isolate. Hum. Reprod. Update 5, 103–107. ( 10.1093/humupd/5.2.103) [DOI] [PubMed] [Google Scholar]
- 80.Kishore R, Agarwal S, Halder A, Das V, Shukla BRK, Agarwal S. 1996. HLA sharing, anti-paternal cytotoxic antibodies and MLR blocking factors in women with recurrent spontaneous abortion. J. Obstet. Gynaecol. Res. 22, 177–183. ( 10.1111/j.1447-0756.1996.tb00962.x) [DOI] [PubMed] [Google Scholar]
- 81.Meuleman T, Lashley LELO, Dekkers OM, Van Lith JMM, Claas FHJ, Bloemenkamp KWM. 2015. HLA associations and HLA sharing in recurrent miscarriage: a systematic review and meta-analysis. Hum. Immunol. 76, 362–373. ( 10.1016/j.humimm.2015.02.004) [DOI] [PubMed] [Google Scholar]
- 82.Rull K, Nagirnaja L, Laan M. 2012. Genetics of recurrent miscarriage: challenges, current knowledge, future directions. Front. Genet. 3, 1–13. ( 10.3389/fgene.2012.00034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meuleman T, Haasnoot GW, Van Lith JMM, Bloemenkamp VKWM, Claas FHJ. 2018. Paternal HLA-C is a risk factor in unexplained recurrent miscarriage. Am. J. Reprod. Immunol. 79, e12797 ( 10.1111/aji.12797) [DOI] [PubMed] [Google Scholar]
- 84.Tripathi P, Naik S, Agrawal S. 2006. HLA-E and immunobiology of pregnancy. Tissue Antigens 67, 207–213. ( 10.1111/j.1399-0039.2005.00550.x) [DOI] [PubMed] [Google Scholar]
- 85.Kanai T, et al. 2001. Polymorphism of human leukocyte antigen-E gene in the Japanese population with or without. Am. J. Reprod. Immunol. 45, 168–173. ( 10.1111/j.8755-8920.2001.450308.x) [DOI] [PubMed] [Google Scholar]
- 86.Pfeiffer KA, Fimmers R, Engels G, Van Der Ven H, Van Der Ven K. 2001. The HLA-G genotype is potentially associated with idiopathic recurrent spontaneous abortion. Mol. Hum. Reprod. 7, 373–378. ( 10.1093/molehr/7.4.373) [DOI] [PubMed] [Google Scholar]
- 87.Hviid TV, Hylenius S, Hoegh AM, Kruse C, Christiansen OB. 2002. HLA-G polymorphisms in couples with recurrent spontaneous abortions. Tissue Antigens 60, 122–132. ( 10.1034/j.1399-0039.2002.600202.x) [DOI] [PubMed] [Google Scholar]
- 88.Fan W, Li S, Huang Z, Chen Q. 2014. Relationship between HLA-G polymorphism and susceptibility to recurrent miscarriage: a meta-analysis of non-family-based studies. J. Assist. Reprod. Genet. 31, 173–184. ( 10.1007/s10815-013-0155-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jurisicova A, Casper RF, Maclusky NJ, Millst GB, Librach CL. 1996. HLA-G expression during preimplantation human embryo development. Proc. Natl Acad. Sci. USA 93, 161–165. ( 10.1073/pnas.93.1.161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Q, Zhuang G, Zhou C, Li T, Li J, Xu Y, Gu X, Li Y. 2009. Expression of certain HLA-I types in cleavage-stage embryos. Reprod. Biomed. Online 18, 244–250. ( 10.1016/S1472-6483(10)60262-3) [DOI] [PubMed] [Google Scholar]
- 91.Wyatt TD. 2020. Reproducible research into human chemical communication by cues and pheromones: learning from psychology's renaissance. Phil. Trans. R Soc. B 375, 20190262 ( 10.1098/rstb.2019.0262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ferdenzi C, Richard Ortegón S, Delplanque S, Baldovini N, Bensafi M. 2020. Interdisciplinary challenges for elucidating human olfactory attractiveness. Phil. Trans. R. Soc. B 375, 20190268 ( 10.1098/rstb.2019.0268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Penn D, Potts W. 1998. MHC-disassortative mating preferences reversed by cross-fostering. Proc. R. Soc. Lond. B 265, 1299–1306. ( 10.1098/rspb.1998.0433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Henrich J, Heine SJ, Norenzayan A. 2010. Most people are not WEIRD. Nature 466, 29 ( 10.1038/466029a) [DOI] [PubMed] [Google Scholar]
- 95.Dwan K, Gamble C, Williamson PR, Kirkham JJ. 2013. Systematic review of the empirical evidence of study publication bias and outcome reporting bias—an updated review. PLoS ONE 8, e66844 ( 10.1371/journal.pone.0066844) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and R code accompanying this article are deposited on figshare (https://doi.org/10.6084/m9.fighshare.8869505).