Abstract
Chloroplast development requires communication between the progenitor plastids and the nucleus, where most of the genes encoding chloroplast proteins reside. Retrograde signals from the chloroplast to the nucleus control the expression of many of these genes, but the signalling pathway is poorly understood. Tetrapyrroles have been strongly implicated as mediators of this signal with the current hypothesis being that haem produced by the activity of ferrochelatase 1 (FC1) is required to promote nuclear gene expression. We have tested this hypothesis by overexpressing FC1 and specifically targeting it to either chloroplasts or mitochondria, two possible locations for this enzyme. Our results show that targeting of FC1 to chloroplasts results in increased expression of the nuclear-encoded chloroplast genes GUN4, CA1, HEMA1, LHCB2.1, CHLH after treatment with Norflurazon (NF) and that this increase correlates to FC1 gene expression and haem production measured by feedback inhibition of protochlorophyllide synthesis. Targeting FC1 to mitochondria did not enhance the expression of nuclear-encoded chloroplast genes after NF treatment. The overexpression of FC1 also increased nuclear gene expression in the absence of NF treatment, demonstrating that this pathway is operational in the absence of a stress treatment. Our results therefore support the hypothesis that haem synthesis is a promotive chloroplast-to-nucleus retrograde signal. However, not all FC1 overexpression lines enhanced nuclear gene expression, suggesting there is still a lot we do not understand about the role of FC1 in this signalling pathway.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles’.
Keywords: haem, plastid signalling, tetrapyrroles, mitochondria, gun phenotype
1. Introduction
Chloroplasts evolved through the integration of a free-living photosynthetic prokaryote into a non-photosynthetic eukaryote, followed by relocation of the majority of the chloroplast genome to the nucleus [1]. The chloroplast retains its own reduced genome, encoding less than 100 predicted proteins in Arabidopsis thaliana, with the remaining approximately 3000 proteins encoded in the nucleus and imported into the developing chloroplast [2]. Consequently, there is a requirement for bidirectional signalling pathways between these organelles to ensure correct provision of proteins to the chloroplast. Anterograde signalling pathways by which the nucleus controls chloroplast development are reasonably well characterized and include photoreceptor and hormone control of nuclear-encoded chloroplast proteins [1,3], some of which can control the expression of chloroplast-encoded proteins [4,5]. Signalling from the chloroplast to the nucleus during chloroplast development (termed biogenic retrograde signalling; [6]) is more poorly understood. However, treatments leading to chloroplast damage at the developmental stage result in a strong downregulation of hundreds of nuclear-encoded genes, many encoding chloroplast proteins [7,8]. In addition, the impact of the environment on photosynthesis enables chloroplasts to fulfil a sentinel function for environmental stress, and various operational retrograde signals from mature chloroplasts can regulate nuclear gene expression to acclimate to these stresses [6,9,10].
Our understanding of biogenic retrograde signalling is based on the identification of genomes uncoupled (gun) mutants in which expression of the nuclear-encoded LHCB1.2 gene is maintained after severe chloroplast damage that strongly inhibits expression of many nuclear-encoded photosynthetic genes [11]. In this case, chloroplast development was prevented by treatment with the phytoene desaturase inhibitor Norflurazon (NF), which blocks the production of photoprotective carotenoids [12,13]. Of the five originally described gun mutations, four were in genes encoding proteins required for the synthesis of tetrapyrroles. gun2 and gun3 are haem oxygenase and phytochromobilin synthase mutants, respectively, with reduced ability to convert haem to phytochromobilin [14]. The gun5 mutation is in the gene encoding the H subunit of Mg-chelatase [14] and gun4 lacks a positive regulator of Mg-chelatase [15]. Initial ideas around Mg-protoporphyrin IX (Mg-proto) functioning as a mobile retrograde signal [16] have mostly been unsupported as no correlation was observed between Mg-proto levels and Lhcb gene expression when Mg-proto levels were manipulated chemically [17] or genetically [18]. Instead, the identification of the dominant gun6 mutation that results in elevated ferrochelatase (FC) 1 activity seemed to resolve the gun mutant puzzle and led to the hypothesis that synthesis of the FC1 product, haem, was required to promote expression of nuclear-encoded photosynthetic genes [19]. As well as making sense of the impact of the gun mutations on tetrapyrrole biosynthesis, this hypothesis was consistent with an established role for haem as a signalling molecule in many systems, its relative suitability in terms of its chemistry and its known export from chloroplasts [20].
The retrograde signalling field has struggled in recent years with proposed components of the signalling pathway that have not stood up to scrutiny. Recent examples of mutants for which a reported gun phenotype has not been reproducible in other laboratories include those lacking PTM1 [21] and ABI4 [22]. However, the phenotypes of the gun mutants themselves have been observed in many laboratories over a long period, including the more recently identified gun6 mutant [21]. In the current study, we set out to test the hypothesis that FC1 overexpression results in an increase in a promotive retrograde signal, by constructing plants overexpressing FC1. In Arabidopsis (and other higher plants), there are two genes encoding ferrochelatase, FC1 and FC2. The expression profile [17,23–25] and functional analysis [26–29] of these genes are consistent with FC1 having a role in providing non-photosynthetic haem and FC2 being required for photosynthetic haem production. For example, mutants lacking FC1 show poor early development with strong alleles being embryo lethal [28,29] and reduced accumulation of extra-plastidic cytochromes [28]. By contrast, the loss of FC2 results in poor chlorophyll accumulation and reduced development of the photosynthetic apparatus [26–28]. The fc2 mutants also show reduced total haem levels. FC2 can partially compensate for the loss of FC1 if expressed from the FC1 promoter [29] and FC1 (with an FC2 transit peptide) can partially compensate for the loss of FC2 [27].
There is considerable biochemical evidence that both chloroplasts and mitochondria contain ferrochelatase activity and activity of the preceding enzyme in the pathway, protoporphyrinogen IX oxidase [30–34]. Import experiments in purified organelles also demonstrated that while FC2 was restricted to chloroplasts, FC1 was imported into both chloroplasts and mitochondria, albeit with the majority of FC1 localized in the former [35,36], and recently, haemagglutinin‐tagged FC1 was detected in mitochondrial fractions [34]. These data continue to suggest the possibility of the dual localization of FC1, although some studies do not support this (e.g. [37]). There are links between mitochondria and chloroplasts in retrograde signalling responses [38–40] and it is possible that FC1 may mediate its effect through mitochondrial localization. We have therefore expressed FC1 with its predicted transit peptide replaced with transit peptides specific for plastid (RecA) or mitochondrial import (CoxIV). The RecA and CoxIV transit peptides were selected as they have been used previously to successfully target proteins to these respective organelles [41–43]. Our results show that targeting of FC1 to plastids alone is sufficient to promote expression of nuclear-encoded photosynthetic genes, and thus, our data support the hypothesis that chloroplast-localized FC1 activity is required for retrograde signalling.
2. Material and methods
(a). Plant material and growth conditions
The gun5 [14] and gun6 [19] mutants in the Col-0 background have been described previously. For growth on plates, seeds were surface-sterilized with 70% (v/v) ethanol and 10% (v/v) bleach solutions, and plated seeds then imbibed for 3 days at 4°C in the dark. For selection of transgenics, seeds were plated onto half-strength Murashige and Skoog (MS) medium containing 1% (w/v) agar, pH 5.8, supplemented with 40 µg ml−1 hygromycin B. For the growth of transgenics to determine transgene expression levels, seeds were plated onto half-strength MS medium containing 1% (w/v) agar, pH 5.8. After imbibition, seeds were transferred to WLc (100 µmol m−2 s−1) at 23°C for 5 days. For NF screens, seeds were plated onto half-strength Linsmaier and Skoog (LS) medium containing 1% (w/v) sucrose and 1% (w/v) agar, pH 5.8 and supplemented with either 5 μM NF or 0.1% DMSO (control). After imbibition, seeds were transferred to continuous low white light (LWLc; 25 µmol m−2 s−1) at 23°C for 7 days. For growth in soil, seeds were sown directly onto compost (Levington's F2 : John Innes No. 2 : vermiculite; 1 : 1 : 1) and grown in photoperiods of 16 h white light, 8 h dark at 23°C with a relative humidity of 65%.
(b). Generation of transgenic Arabidopsis thaliana lines
The coding sequence of FC1 was fused at the 3′ end to a solubility-modified, red-shifted GFP [43], hereafter referred to as GFP. A 36 bp spacer was present between the FC1 sequence and the GFP sequence. In addition, the native transit peptide of FC1 was excluded. This was identified from predictions made using TargetP 1.1 Server [44,45], predictions of the target peptide cleavage sites based on known cleavage sequences and alignment of protein sequences to identify amino acids required for function that are conserved across other plant and cyanobacterial species. Following this analysis, the sequence encoding the first 77 amino acids of FC1 (FC1Δ1–77) was excluded. A BglII restriction site was added 5′ of the FC1Δ1–77:GFP sequence. The FC1:GFP fragment was cloned into pDONR™221 (Invitrogen, Carlsbad, CA, USA) using Gateway® technology. A transit peptide conferring localization either to plastids (RecA) or mitochondria (CoxIV) was then ligated directly upstream of the gene sequence (at the BglII site) to generate the expression cassettes. The RecA transit peptide sequence corresponded to the first 201 bp of the coding sequence of the Arabidopsis RECA gene (At1g79050; [46]), while the CoxIV transit peptide corresponded to the first 87 bp of the coding sequence of cytochrome c oxidase subunit 4 from Saccharomyces cerevisiae [47]. Control expression cassettes lacking FC1 were also created, consisting of the GFP sequence fused downstream of the RecA or CoxIV transit peptide sequences. Finally, a cassette consisting of the full-length FC1 (FL-FC1) sequence fused to GFP was created. The cassettes were recombined into the pGWB502Ω (hygR) plant expression plasmid [48] under the control of the 35S promoter from cauliflower mosaic virus, and the resulting plasmids were used to transform Agrobacterium tumefaciens GV3101. Flowering Arabidopsis Col-0 plants were transformed using the floral dip method [49], and positive transformants identified through antibiotic selection [50] were confirmed via PCR genotyping. Further details on the primers and plasmids used are given in electronic supplementary material, tables S1 and S2, respectively. Plants overexpressing FC1 targeted to both plastids and mitochondria were generated by manually crossing CoxIV:FC1:GFP lines (female) to RecA:FC1:GFP lines (male).
(c). RNA extraction, cDNA synthesis and qRT-PCR
Cotyledon tissue was homogenized in 500 µl extraction buffer (100 mM NaCl, 10 mM Tris pH 7.0, 1 mM EDTA, 1% (w/v) SDS). After the addition of 150 µl phenol (pH 4.8), samples were vortexed vigorously. After the addition of 150 μl phenol (pH 4.8), samples were vortexed vigorously. This was followed by the addition of 250 μL chloroform and the samples were again vortexed vigorously. After centrifugation (16 100g, 5 min, 4°C), the upper aqueous phase was transferred to a new tube containing 450 µl ice-cold 4 M LiCl. RNA was precipitated overnight at 4°C. After centrifugation (16 100g, 20 min, 4°C), pellets were resuspended in 300 µl DNase buffer (10 mM Tris pH 7.5, 2.5 mM MgCl2, 0.5 mM CaCl2). One microlitre DNase (Promega, Madison, WI, USA) was then added and samples incubated at 37°C for 25 min. Samples were mixed with 500 μl phenol : chloroform : isoamyl alcohol (25 : 24 : 1), pH 6.7 and vortexed vigorously. After centrifugation (16 100g, 5 min, 4°C), the aqueous upper phase was mixed with 750 μl 95% ethanol : 5% 3 M sodium acetate, pH 5.2 and RNA was precipitated at −20°C for 1 h. After centrifugation (16 100g, 20 min, 4°C), RNA pellets were air-dried and resuspended in 50 µl TE buffer (10 mM Tris pH 8.0, 1 mM EDTA).
cDNA synthesis was performed according to the manufacturer's protocols on 2 µg total RNA per sample with the nanoScript2 kit (Primerdesign, Southampton, UK), using random nonamer and oligo dT primers.
qRT-PCR was carried out on a StepOnePlus™ real-time PCR system (Applied Biosystems, Foster City, CA, USA). Each reaction contained 0.5 µl cDNA, 5 µl PrecisionPLUS SYBR green mastermix (Primerdesign) and 2.5 µl of primer mix (containing forward and reverse primers each at 2 µM), with the volume made up to 10 µl with nuclease-free water. qRT-PCR primer sequences are given in electronic supplementary material, table S3. Two technical replicates were performed for each sample/primer pair combination, and two ‘no template controls' were performed for each primer pair. qRT-PCR cycling conditions were: 95°C for 2 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min, with fluorescence determined at the end of every cycle. Melt curves (60°C to 92°C, in 0.5°C increments) were performed at the end of every run to verify amplification specificity for each primer pair. Primer efficiencies were determined using a serial dilution of Col-0 (untreated) cDNA. Relative expression values between samples were calculated using the ΔΔCt method, normalized to ACTIN DEPOLYMERISING FACTOR 2 (ADF2, At3g46000) or YELLOW-LEAF-SPECIFIC GENE 8 (YLS8, At5g08290). ADF2 and YLS8 were identified as excellent reference genes for NF screens through analysis of microarray data from Col-0 seedlings grown with/without NF [51]. Data shown were normalized to ADF2, with comparable results observed when normalized to YLS8. Full details of the qRT-PCR method to fulfil MIQE guidelines [52] are given in electronic supplementary material, datasheet S1.
(d). Chlorophyll, carotenoid and Pchlide determination
Chlorophyll and carotenoids were extracted from weighed cotyledon tissue by homogenizing in 800 µl ice-cold 80% (v/v) acetone. After centrifugation (16 100g, 5 min, 4°C), the absorbance of the supernatant was determined at A470, A647 and A663 using a U-2001 spectrophotometer (Hitachi, Tokyo, Japan). Total carotenoid and chlorophyll a and b contents were determined using previously published equations [53], and normalized to tissue weight.
Pchlide was extracted from cotyledon tissue harvested in a dark room under a dim green safe light using the method described in Terry & Kacprzak [54]. Cotyledon pairs were homogenized in ice-cold acetone : 0.1 M ammonium hydroxide (9 : 1, v : v), centrifuged (16 100g, 5 min, 4°C), and fluorescence emission spectra of the supernatants determined (excitation wavelength = 440 nm) using a F-2000 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). The height of the Pchlide peak (∼636 nm) was used to generate relative fluorescence values, which were normalized for cotyledon number.
(e). Localization of GFP by confocal imaging
Confocal imaging was used to confirm the subcellular localization of plastid- and mitochondrion-targeted FC1. Cotyledon tissue from 5 day continuous white light (WLc)-grown seedlings was mounted onto slides and the samples were flooded with the perfluorocarbon PP11. The localization of GFP was determined on a Leica TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany), using Leica Application Suite X software. GFP was imaged with an excitation wavelength of 488 nm and detection of emission between 497 and 531 nm, both using the 63× glycerol oil immersion objective lens. Chlorophyll autofluorescence was detected using 488 nm excitation and 678–695 nm emission. An HyD detector was used to image both signals, and at least six averages were taken for each acquisition.
3. Results
(a). Characterization of FC1 overexpressing lines
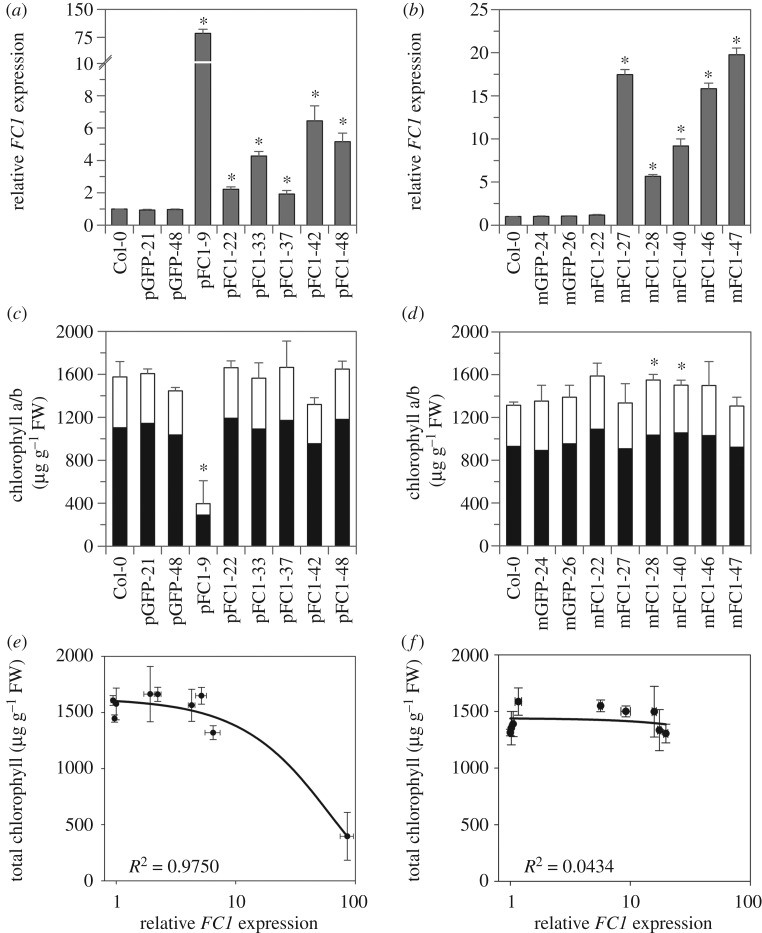
We generated transgenic lines containing either a RecA:FC1:GFP (plastid-targeted, pFC1) or a CoxIV:FC1:GFP (mitochondrion-targeted, mFC1) expression cassette driven by the constitutive CaMV 35S promoter. Selection protocols were used to identify single-insertion, homozygous transformants (T3 generation) and, subsequently, overexpressing lines were determined by measuring the FC1 expression level in cotyledon tissue from 5-day-old WLc-grown seedlings using qRT-PCR. For the pFC1 lines, expression ranged from 2- to 85-fold higher than wild-type (WT, Col-0) under these conditions (figure 1a). FC1 expression levels correlated with GFP expression levels from the same plants, while FC2 expression remained essentially at a WT level (electronic supplementary material, figure S1a). Some of these lines displayed a pale cotyledon phenotype that appeared to correlate with FC1 expression, with high overexpressors having very pale cotyledons and low overexpressors being indistinguishable from WT (electronic supplementary material, figure S2a). Control lines overexpressing only GFP targeted to plastids lacked a visible phenotype (electronic supplementary material, figures S1a and S2a). The correlation between the pale cotyledon phenotype and FC1 expression level was confirmed by analysis of the chlorophyll content of these lines when grown under the same conditions, with the highest overexpressor (pFC1–9) having significantly less total chlorophyll than WT (figure 1c,e). The pFC1–9 line also had significantly less total carotenoids than WT (electronic supplementary material, figure S2b). The next highest overexpressor (pFC1–42) also indicated reductions in chlorophyll and carotenoid content, although these were not statistically significant (figure 1c; electronic supplementary material, figure S2b). FC1 overexpressing lines using the native transit peptide have previously been reported to have a reduction in chlorophyll synthesis [19]. The chlorophyll a/b ratio of all pFC1 lines remained similar to WT (electronic supplementary material, figure S2c) and there was no significant effect of day length or light intensity on the accumulation of chlorophyll or carotenoids in these lines (electronic supplementary material, figure S3). Surprisingly, the pale phenotype of pFC1–9 was partially attenuated in mature, soil-grown plants, while the pFC1–42 line showed a paler phenotype compared to seedlings (electronic supplementary material, figure S4).
Figure 1.
The relationship between FC1 expression and chlorophyll content in the pFC1 and mFC1 transgenic lines. (a,b) FC1 expression relative to Col-0 in (a) plastid-targeted (pFC1) and (b) mitochondria-targeted (mFC1) FC1 overexpressing lines as determined by qRT-PCR. (c,d) Total chlorophyll content of the same pFC1 (c) and mFC1 (d) lines. Black bars represent chlorophyll a and white bars represent chlorophyll b. (e,f) Correlation plots between FC1 expression (log scale) and total chlorophyll content for the pFC1 (e) and mFC1 (f) lines. Seedlings were grown for 5 days in WLc for all analyses and lines overexpressing only GFP in plastids (pGFP) or mitochondria (mGFP) were included as controls. Data represent the mean + s.e.m. of three independent biological replicates and asterisks indicate a significant difference versus Col-0 (p < 0.05, Student's t-test).
For the mFC1 lines, FC1 expression in 5-day-old WLc-grown seedlings ranged from 1.2- to 20-fold higher than WT (figure 1b). GFP expression again correlated with FC1 expression, with FC2 expression fundamentally unaffected (electronic supplementary material, figure S1b). No phenotypic differences from WT were observed in these lines at any stage of growth (figure 1d,f; electronic supplementary material, figures S4–S6).
Although the paler phenotype of the two transgenic lines pFC1–9 and pFC1–42 correlated quite well with FC1 expression levels, we wanted to be certain that the observed phenotypes were not owing to the insertion site of the FC1 transgene. We therefore performed whole-genome sequencing on both lines to identify the location of the transgenes. As shown in electronic supplementary material, figure S7, the RecA-FC1-GFP transgene in pFC1–9 has interrupted the 3′ end of At1g01540 at the end of exon 6. All sequence reads indicate that insertion has occurred solely at one location in the genome and confirm our original results from antibiotic selection of T2 seed. At1g01540 is a protein related to Thylakoid-associated kinase 1, but has been determined experimentally to be a cytosolic protein [55]. A GABI-Kat mutant was reported as showing no obvious phenotype [56] and we also obtained independent T-DNA insertion lines for At1g01540 (Salk_008396, Salk_076898 and Salk_036951), but could see no visible loss of greening phenotype at the seedling stage. For pFC1–42, there was a single-insertion site in an intergenic region in chromosome 5 that lies about 600 bp upstream of the start codon of At5g67120 and about 1250 bp upstream of the start codon of At5g67130. There appear to be up to four T-DNA copies at this single-insertion site. At5g67120 and At5g67130 encode an uncharacterized RING/U-box superfamily protein predicted to be nuclear-localized and a plasma membrane-localized [57] phospholipase C-like phosphodiesterase superfamily protein with phospholipase activity [58], respectively. It is possible that the T-DNA insertion could interfere with the expression of either or both genes, but there is no evidence to suggest that this might cause the pFC1–42 phenotype.
(b). Localization of FC1-GFP proteins
To confirm the localization of the plastid and mitochondrion-targeted GFP fusion proteins, we examined 5-day-old WLc-grown seedlings using confocal imaging. GFP localization was performed on root tips and cotyledons of the highest overexpressing pFC1 and mFC1 lines. When imaging root tips, GFP-labelled structures in cells of pFC1 seedlings were significantly larger than those in mFC1 seedlings (Student's t-test p < 0.001, figure 2a,b). Moreover, the sizes of the structures in the pFC1 and mFC1 lines closely matched the known sizes of root plastids and mitochondria, respectively (pFC1 = 5.70 µm ± 0.08, mFC1 = 1.63 µm ± 0.12) [59]. In addition, the GFP-labelled structures in the mFC1 lines moved rapidly during imaging, supporting the identification of these structures as mitochondria. For lines pFC1–9 and pFC1–42, imaged cotyledons were pale with very few chlorophyll-containing cells (figure 2a). GFP was detected in plastids lacking chlorophyll, while no GFP signal was observed when chlorophyll was present. This suggests that the ability to synthesize chlorophyll is an inverse function of plastid FC1 expression such that high expression of FC1 protein necessarily limits chlorophyll accumulation. The imaging of mFC1–27 cotyledons further supported mitochondrial localization of FC1 in these lines, given the absence of overlap and difference in size between the GFP-labelled structures in this line and chloroplasts (figure 2b). As expected, a control line in which GFP was overexpressed in the absence of a transit peptide showed cytosolic localization (figure 2c).
Figure 2.
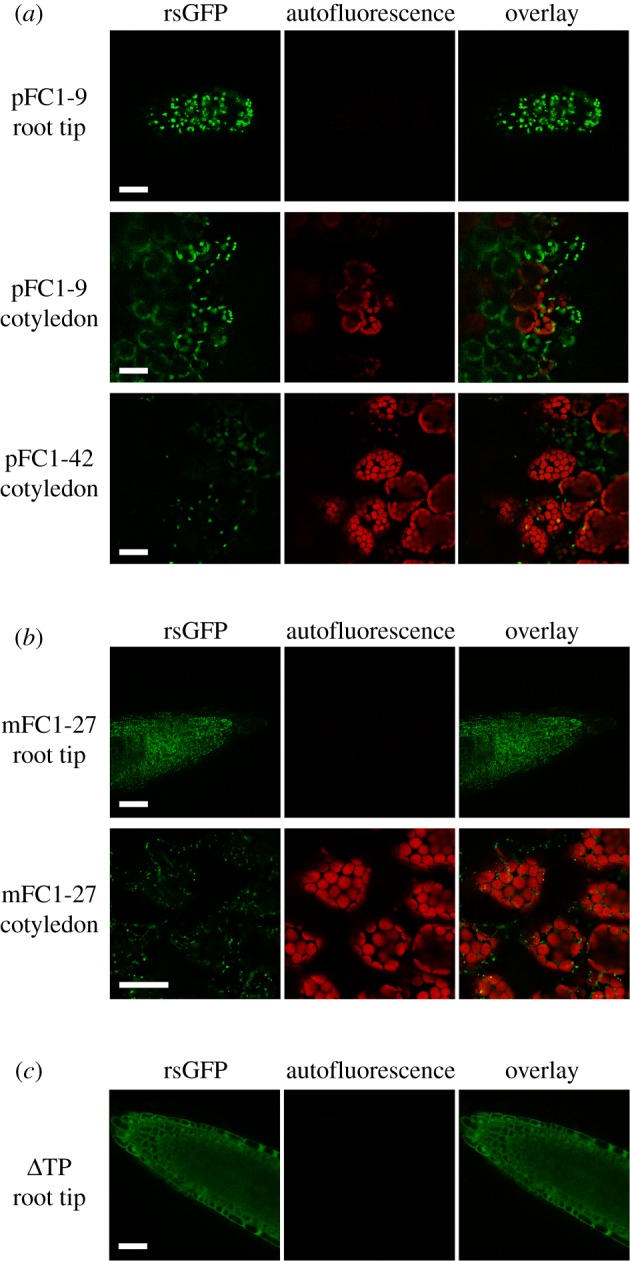
Localization of FC1 in roots and cotyledons of pFC1 and mFC1 seedlings. (a,b) Confocal microscopy was used to determine the subcellular localization of FC1:GFP fusion proteins in pFC1 (a) and mFC1 (b) lines. (c) A control line of FC1:GFP without a transit peptide (ΔTP). Scale bars, 30 µm.
(c). Retrograde signalling in FC1 overexpressing lines
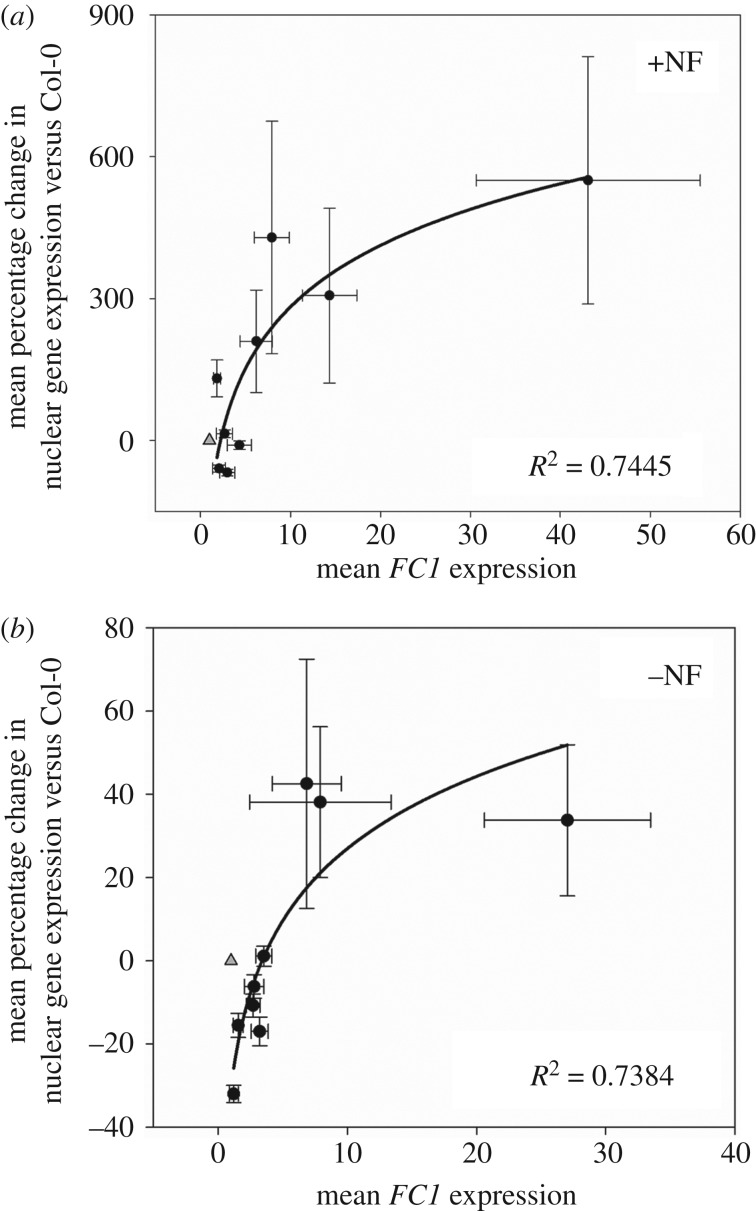
It was previously demonstrated that the overexpression of full-length FC1 with its native transit peptide rescued the expression of photosynthesis-associated nuclear genes when seedlings were grown on NF (gun phenotype) [19]. To establish whether organellar-specific overexpression of FC1 was sufficient to replicate the gun phenotype, the transgenic lines described above were grown on NF and expression of nuclear genes determined. gun5 and gun6 were included in these screens as positive controls and lines overexpressing GFP alone in either plastids (pGFP) or mitochondria (mGFP) were included as negative controls. In the presence of NF, the two highest expressors of plastid-targeted FC1 (pFC1–9 and pFC1–42) were able to significantly rescue expression of all five nuclear genes tested (GUN4, CA1, HEMA1, LHCB2.1 and CHLH), when compared with Col-0 and pGFP seedlings (figure 3a; electronic supplementary material, figure S8a). By contrast, the highest overexpressing mFC1 lines were not able to rescue the expression of any of the genes tested (figure 3b; electronic supplementary material, figure S8b). Importantly, growth on NF did not have a strong effect on the expression of FC1 in the lines tested (electronic supplementary material, figure S9) and results were independent of the reference gene used (electronic supplementary material, figure S10). Correlation plots of percentage recovery of nuclear gene expression (for all five genes pooled together) after NF treatment versus WT and FC1 expression in the presence of NF show a positive correlation for the plastid-targeted overexpressors (figure 4a), but no correlation for the mitochondrion-targeted overexpressors (electronic supplementary material, figure S11a). These results strongly support the idea that the overexpression of FC1 targeted to plastids is sufficient to rescue expression of nuclear-encoded photosynthesis genes in the presence of NF. Interestingly, both pFC1 and mFC1 lines showed a positive correlation between percentage change in nuclear gene expression and FC1 expression in the absence of NF (figure 4b; electronic supplementary material, figure S11b), although the maximum increase in expression was just 10% for mFC1 lines compared to 50% for pFC1 lines. The increase in nuclear gene expression observed in pFC1 lines demonstrates the operation of this retrograde pathway under standard plant growth conditions.
Figure 3.
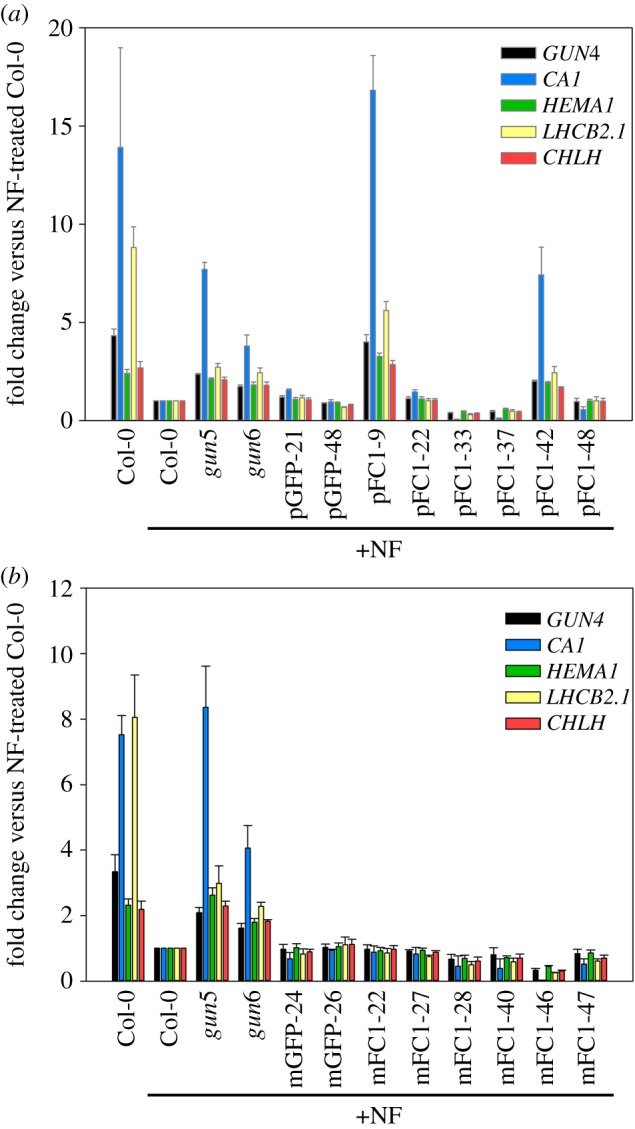
Expression of photosynthesis-associated genes on NF is enhanced in plastid FC1, but not mitochondrial FC1 overexpressors. (a,b) The expression of GUN4, CA1, HEMA1, LHCB2.1 and CHLH was determined by qRT-PCR in pFC1 (a) and mFC1 (b) seedlings grown for 7 days in LWLc on plates with NF. The control lines pGFP (a) and mGFP (b), as well as gun5 and gun6, were included. Data shown are the mean fold changes versus Col-0 on NF + s.e.m. of three independent biological replicates. The original qRT-PCR data for these graphs are given in electronic supplementary material, figure S8.
Figure 4.
Plastid-targeted FC1 expression correlates with enhanced nuclear gene expression on NF. (a,b) Correlation plots of the combined mean percentage change in expression of GUN4, CA1, HEMA1, LHCB2.1 and CHLH, versus FC1 expression for pFC1 seedlings in the presence (a) or absence (b) of NF. Data are relative to Col-0 +NF (a) or −NF (b). For both graphs, data points include gun6, the six transgenic pFC1 overexpressing lines and two F1 progenies of pFC1x mFC1 crosses. The triangles indicate WT response. SigmaPlot 13.0 was used to fit logarithmic best-fit lines and derive coefficients of determination. Data shown are the mean ± s.e.m. of three independent biological replicates.
Next, we tested whether the effect of elevated plastid FC1 expression on nuclear gene expression required photoreceptor input in order to be observed. We therefore tested the same five nuclear genes (GUN4, CA1, HEMA1, LHCB2.1 and CHLH) in seedlings grown for 4 days in the dark. In this case, we saw little difference in expression between pFC1 or mFC1 lines and WT for any genes tested (electronic supplementary material, figure S12), except for HEMA1 expression, which was slightly, yet significantly, increased in pFC1–9 in the dark compared to Col-0 (electronic supplementary material, figure S12a). An increase in HEMA1 expression in dark-grown seedlings has previously been noted for gun1 seedlings [60].
To determine if overexpression of FC1 in both organelles would modify the rescue of nuclear gene expression on NF seen in pFC1 lines, pFC1–9 (the highest expressor of plastid-targeted FC1) was independently crossed with both mFC1–27 and mFC1–47 (the two highest mitochondrion-targeted FC1 overexpressors), and the F1 generation screened on NF. The three parent lines were included in the screens for reference. F1 plants of both the mFC1–47x pFC1–9 and mFC1–27x pFC1–9 lines showed significant enhancement of gene expression after NF treatment (see CA1 and the tetrapyrrole biosynthesis genes; electronic supplementary material, figure S13a), but expression levels were reduced compared with the pFC1–9 parent line. This was most likely owing to the greatly reduced FC1 expression levels in the F1 plants compared to the parent lines (electronic supplementary material, figure S13b). The observation that F1 generation FC1 overexpressing plants can confer a gun phenotype demonstrates that this trait is semi-dominant and provides further evidence that the observed phenotype is solely the result of FC1 overexpression.
(d). Modulation of tetrapyrrole synthesis in FC1 overexpressing seedlings correlates with induction of nuclear gene expression
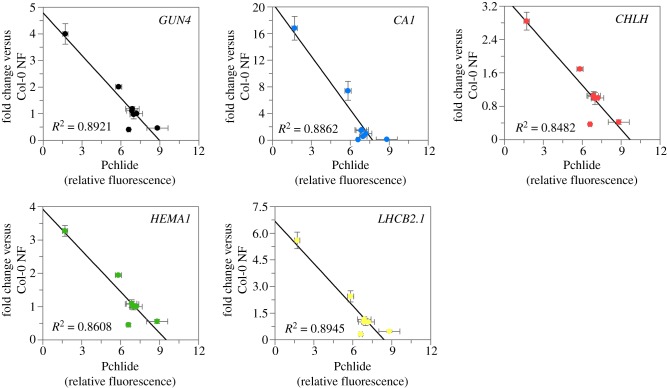
Lines overexpressing plastid-localized FC1 were able to enhance nuclear gene expression on NF, and this ability correlated with FC1 expression. To determine if this enhancement of gene expression was owing to changes in haem synthesis as proposed by the current model [19,20], we examined the impact of the overexpressing lines on tetrapyrrole synthesis and determined whether this was also correlated with nuclear gene expression. As it is difficult to measure a signalling haem pool in young seedlings, we determined the accumulation of protochlorophyllide (Pchlide) in the dark as a proxy for such a haem pool at the onset of the light treatment. It is well established that the accumulation of haem results in feedback inhibition of aminolevulinic acid (ALA) synthesis resulting in reduced Pchlide [61–64]. Previous studies have observed elevated Pchlide in fc2 mutants, but not fc1 mutants, suggesting that FC2-synthesized haem is responsible for feedback inhibition [26]. However, it has been shown that overexpression of FC1 can rescue this phenotype [27], indicating that FC1-synthesized haem can contribute to this regulatory pool. In pFC1 seedlings, Pchlide accumulation in the dark (electronic supplementary material, figure S14) showed a strong negative correlation with expression of all five nuclear genes on NF (figure 5). This correlation was not apparent for mFC1 seedlings (electronic supplementary material, figure S15). Together, these data suggest that there is an elevated regulatory haem pool in pFC1 lines that correlates well with the observed increases in nuclear gene expression in these lines. These results therefore support the hypothesis that increased FC1 activity results in the production of a promotive retrograde signal [19] and, furthermore, that activity in the plastid alone is sufficient for this response.
Figure 5.
Enhancement of nuclear gene expression on NF inversely correlates with protochlorophyllide levels in dark-grown pFC1 seedlings. Correlation plots of protochlorophyllide (Pchlide) in 4-day-old dark-grown pFC1 seedlings against fold change in expression of GUN4, CA1, HEMA1, LHCB2.1 and CHLH versus Col-0 on NF. Data represent the mean ± s.e.m. of three independent biological replicates.
4. Discussion
The interpretation of the gun mutant phenotype has been the focus of our attempts to understand chloroplast-to-nucleus retrograde signalling since these mutants were first described over 25 years ago [11]. Five of the six gun mutants isolated by the Chory laboratory had altered activities of tetrapyrrole biosynthesis-related proteins [14,15,19] and the link between tetrapyrrole synthesis and retrograde signalling has stood up to scrutiny over this period. The current hypothesis is that haem synthesized by FC1 is a promotive retrograde signal or precursor of the signal ([19]; see [20,65,66] for discussion). This hypothesis is based on the observation that both the dominant gun6 mutation that results in overexpression of FC1 and a transgenic FC1 overexpression line resulted in enhanced nuclear gene expression after NF treatment and was developed through the re-interpretation of the phenotypes of the gun2–gun5 mutants [19]. Consistent with this hypothesis, haem has a well-established role as a mobile signalling molecule in numerous biological systems [20]. Here, we have shown that overexpression of FC1 in chloroplasts results in a strong gun phenotype in two independent transgenic lines and that expression of five nuclear-encoded photosynthetic genes correlated with FC1 gene expression and the ability to feedback inhibit Pchlide synthesis. Our data therefore broadly support the hypothesis that FC1-dependent haem synthesis results in a promotive chloroplast-to-nucleus retrograde signal. Moreover, this signal is directly related to FC1 activity in the chloroplast as no evidence was observed for a gun phenotype when FC1 was targeted to mitochondria. This result is consistent with previous experiments in which overexpression of FC1 using an FC2 transit peptide could increase nuclear gene expression after NF treatment [19], although formally the localization of the FC2-targeted FC1 protein in vivo is unknown as GFP-tagged FC proteins have never been detected in mitochondria, despite the strong evidence for the presence of FC in this organelle. Although we were unable to isolate a very highly expressing mFC1 line to match the level of FC1 overexpression seen in line pFC1–9, under the conditions of the NF screen, three of the mFC1 lines had clearly higher FC1 expression than pFC1–42, a line that shows significant rescue of nuclear gene expression on NF. Our data do not therefore support a model in which a chloroplast retrograde signal could have made use of presumably pre-existing mitochondrial signals. Instead, there appears to be direct regulation of nuclear-encoded genes for chloroplast proteins during chloroplast biogenesis.
During the course of this study, we identified six lines that showed elevated expression of FC1 in cotyledon tissue under the conditions used for the retrograde signalling assays. Only two of these lines showed a gun phenotype, but we included data for all six lines as we wanted to be transparent about the issues we encountered. For example, three of the pFC1 lines (pFC1–22, pFC1–33 and pFC1–48) had similar or higher levels of FC1 expression on NF than the gun6 mutant but did not show a gun phenotype, and the pFC1–42 and pFC1–48 lines had similarly high FC1 expression but showed different gene expression responses. This discussion is complicated by the observation that FC1 expression in gun6 decreases on NF, something not observed in the overexpression lines. Only pFC1–9 and pFC1–42 showed higher FC1 expression than gun6 in the absence of NF and this may account for their ability to confer a gun phenotype while other lines were unable to. Importantly perhaps, only these two lines had expression levels that were sufficient to impact on chlorophyll accumulation. Woodson et al. [19] reported reduced chlorophyll levels in all lines that also showed a gun phenotype. We are confident that the phenotypes we observed are owing to FC1 overexpression. Genome sequencing to identify the position of each overexpression construct ruled out the likelihood of an insertional effect causing the observed phenotype and the pFC1–9 construct showed a semi-dominant phenotype following crosses with mFC1 lines. Interestingly, even the pFC1–9 and pFC1–42 lines showed slightly different phenotypes, with the former showing a stronger reduction in chlorophyll levels in seedlings and the latter having a more pronounced mature plant phenotype. This might be related to positional effects altering expression levels in different tissues. Overall, a far more detailed characterization of FC1 protein levels, localization and activity as well as haem levels for each line would be required to explain the observed phenotypic differences between the different FC1 overexpressing lines. Nevertheless, we believe that our observation that overexpression of FC1 in chloroplasts can confer a gun phenotype, which confirms and builds on the results of Woodson et al. [19], is important in helping to establish an agreed set of reliable data on the retrograde signalling response.
One interesting aspect of our data is the clear demonstration that overexpression of FC1 resulted in an increase in nuclear gene expression in the absence of NF treatment. Expression of key genes increased up to 50% in pFC1 lines. A small increase was also observed in mFC1 lines, although this was not significant for any individual line (electronic supplementary material, figure S8). One of the criticisms of the retrograde signalling field is the perceived requirement for severe treatments to observe the effects of mutations that affect signalling. Our data therefore support the idea that retrograde signalling is functioning under standard growth conditions and that the amount of signal is not necessarily limited. This result therefore supports previous data such as elevated HEMA1 expression in a gun1, gun5 double mutant during de-etiolation [60].
Finally, a central question in retrograde signalling research is whether single or multiple signals are operating during chloroplast biogenesis. The question derives from analysis of the gun1 mutation that confers elevated nuclear gene expression after treatments with either NF or the plastid translation inhibitor, lincomycin [7], which has led to the suggestion that GUN1 mediates a signal related to plastid protein synthesis. Indeed, GUN1 does seem to have a role in plastid protein homeostasis [67–69]. However, recently, other roles have also been suggested in chloroplast RNA editing [70] and import of nuclear-encoded chloroplast proteins [71]. GUN1 has also been shown to interact with tetrapyrrole biosynthesis enzymes [67] and to bind haem and a range of porphyrins and regulate FC1 enzyme activity in vitro [72]. Given the strong evidence for a tetrapyrrole signal from the haem branch of the pathway, it could be proposed that GUN1 might have a role in coordinating various chloroplast processes with production of the FC1-dependent haem signal. Certainly, an understanding of the relationship between GUN1 and FC1-mediated retrograde signalling will be crucial in determining the mechanism of this signalling pathway during chloroplast development.
Supplementary Material
Supplementary Material
Acknowledgements
Thanks to Joanne Chory and Jesse Woodson (SALK Institute, USA) for the gun5 and gun6 mutants used in this study. Thanks also to George Littlejohn (University of Exeter, UK) for the gift of PP11 and to Chiara Perico and Imogen Sparkes (University of Exeter, UK) for looking at FC1 localization in some of our transgenic lines.
Data accessibility
All datasets supporting this article have been provided as part of the electronic supplementary material.
Authors' contributions
M.T.P. performed all of the experiments and analysed data; T.G-B. contributed to making the FC1 overexpressing lines; A.S. conceived the project and analysed data. M.J.T. conceived the project and analysed data. All authors contributed to writing the article and approved the final version.
Competing interests
We have no competing interests.
Funding
M.T.P., T.G-B. and M.J.T. were supported by UK Biotechnology and Biological Sciences Research Council (BBSRC) grant no. BB/J018139/1. A.G.S. was supported by BBSRC grant no. BB/J018694/1.
References
- 1.Jarvis P, López-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 14, 787–802. ( 10.1038/nrm3702) [DOI] [PubMed] [Google Scholar]
- 2.Abdallah F, Salamini F, Leister D. 2000. A prediction of the size and evolutionary origin of the proteome of chloroplasts of Arabidopsis. Trends Plant Sci. 5, 141–142. ( 10.1016/s1360-1385(00)01574-0) [DOI] [PubMed] [Google Scholar]
- 3.Pogson BJ, Ganguly D, Albrecht-Borth V. 2015. Insights into chloroplast biogenesis and development. Biochim. Biophys. Acta 1847, 1017–1024. ( 10.1016/j.bbabio.2015.02.003) [DOI] [PubMed] [Google Scholar]
- 4.Belbin FE, Noordally ZB, Wetherill SJ, Atkins KA, Franklin KA, Dodd AN. 2017. Integration of light and circadian signals that regulate chloroplast transcription by a nuclear-encoded sigma factor. New Phytol. 213, 727–738. ( 10.1111/nph.14176) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo CY, Pasoreck EK, Wang H, Cao J, Blaha GM, Weigel D, Chen M. 2019. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun. 10, 2629 ( 10.1038/s41467-019-10518-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pogson BJ, Woo NS, Förster B, Small ID. 2008. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. ( 10.1016/j.tplants.2008.08.008) [DOI] [PubMed] [Google Scholar]
- 7.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. ( 10.1126/science.1140516) [DOI] [PubMed] [Google Scholar]
- 8.Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. 2013. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73, 1–13. ( 10.1111/tpj.12011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. 2016. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. ( 10.1146/annurev-arplant-043015-111854) [DOI] [PubMed] [Google Scholar]
- 10.de Souza A, Wang JZ, Dehesh K. 2017. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 85–108. ( 10.1146/annurev-arplant-042916-041007) [DOI] [PubMed] [Google Scholar]
- 11.Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. ( 10.1016/0092-8674(93)90459-4) [DOI] [PubMed] [Google Scholar]
- 12.Breitenbach J, Zhu C, Sandmann G. 2001. Bleaching herbicide norflurazon inhibits phytoene desaturase by competition with the cofactors. J. Agric. Food Chem. 49, 5270–5272. ( 10.1021/jf0106751) [DOI] [PubMed] [Google Scholar]
- 13.Oelmüller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. 1986. Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168, 482–492. ( 10.1007/BF00392267) [DOI] [PubMed] [Google Scholar]
- 14.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl Acad. Sci. USA 98, 2053–2058. ( 10.1073/pnas.98.4.2053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. ( 10.1126/science.1079978) [DOI] [PubMed] [Google Scholar]
- 16.Strand Å, Asami T, Alonso J, Ecker JR, Chory J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. ( 10.1038/nature01204) [DOI] [PubMed] [Google Scholar]
- 17.Moulin M, McCormac AC, Terry MJ, Smith AG. 2008. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl Acad. Sci. USA 105, 15 178–15 183. ( 10.1073/pnas.0803054105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. 2008. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl Acad. Sci. USA 105, 15 184–15 189. ( 10.1073/pnas.0803245105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodson JD, Perez-Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903. ( 10.1016/j.cub.2011.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terry MJ, Smith AG. 2013. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4, 14 ( 10.3389/fpls.2013.00014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ. 2017. Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol. 174, 21–26. ( 10.1104/pp.16.01930) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ. 2019. Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 179, 18–23. ( 10.1104/pp.18.01047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow KS, Singh DP, Walker AR, Smith AG. 1998. Two different genes encode ferrochelatase in Arabidopsis: mapping, expression and subcellular targeting of the precursor proteins. Plant J. 15, 531–541. ( 10.1046/j.1365-313X.1998.00235.x) [DOI] [PubMed] [Google Scholar]
- 24.Singh DP, Cornah JE, Hadingham S, Smith AG. 2002. Expression analysis of the two ferrochelatase genes in Arabidopsis in different tissues and under stress conditions reveals their different roles in haem biosynthesis. Plant Mol. Biol. 50, 773–788. ( 10.1023/A:1019959224271) [DOI] [PubMed] [Google Scholar]
- 25.Nagai S, Koide M, Takahashi S, Kikuta A, Aono M, Sasaki-Sekimoto Y, Ohta H, Takamiya K-I, Masuda T. 2007. Induction of isoforms of tetrapyrrole biosynthetic enzymes, AtHEMA2 and AtFC1, under stress conditions and their physiological functions in Arabidopsis. Plant Physiol. 144, 1039–1051. ( 10.1104/pp.107.100065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scharfenberg M, Mittermayr L, Von Roepenack-Lahaye E, Schlicke H, Grimm B, Leister D, Kleine T. 2015. Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 38, 280–298. ( 10.1111/pce.12248) [DOI] [PubMed] [Google Scholar]
- 27.Woodson JD, Joerns MS, Sinson AB, Gilkerson J, Salomé PA, Weigel D, Fitzpatrick JA, Chory J. 2015. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350, 450–454. ( 10.1126/science.aac7444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espinas NA, Kobayashi K, Sato Y, Mochizuki N, Takahashi K, Tanaka R, Masuda T. 2016. Allocation of heme is differentially regulated by ferrochelatase isoforms in Arabidopsis cells. Front. Plant Sci. 7, 1326 ( 10.3389/fpls.2016.01326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan T, Roling L, Meiers A, Brings L, Ortega-Rodés P, Hedtke B, Grimm B. 2019. Complementation studies of the Arabidopsis fc1 mutant substantiate essential functions of ferrochelatase 1 during embryogenesis and salt stress. Plant Cell Environ. 42, 618–632. ( 10.1111/pce.13448) [DOI] [PubMed] [Google Scholar]
- 30.Smith AG, Marsh O, Elder GH. 1993. Investigation of the subcellular location of the tetrapyrrole-biosynthesis enzyme coproporphyrinogen oxidase in higher plants. Biochem. J. 292, 503–508. ( 10.1042/bj2920503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papenbrock J, Mishra S, Mock HP, Kruse E, Schmidt EK, Petersmann A, Braun HP, Grimm B. 2001. Impaired expression of the plastidic ferrochelatase by antisense RNA synthesis leads to a necrotic phenotype of transformed tobacco plants. Plant J. 28, 41–50. ( 10.1046/j.1365-313X.2001.01126) [DOI] [PubMed] [Google Scholar]
- 32.Cornah JE, Roper JM, Pal Singh D, Smith AG. 2002. Measurement of ferrochelatase activity using a novel assay suggests that plastids are the major site of haem biosynthesis in both photosynthetic and non-photosynthetic cells of pea (Pisum sativum L). Biochem. J. 362, 423–432. ( 10.1042/bj3620423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masuda T, Suzuki T, Shimada H, Ohta H, Takamiya K. 2003. Subcellular localization of two types of ferrochelatase in cucumber. Planta 217, 602–609. ( 10.1007/s00425-003-1019-2) [DOI] [PubMed] [Google Scholar]
- 34.Hey D, Ortega-Rodes P, Fan T, Schnurrer F, Brings L, Hedtke B, Grimm B. 2016. Transgenic tobacco lines expressing sense or antisense FERROCHELATASE 1 RNA show modified ferrochelatase activity in roots and provide experimental evidence for dual localization of ferrochelatase 1. Plant Cell Physiol. 57, 2576– 2585. ( 10.1093/pcp/pcw171) [DOI] [PubMed] [Google Scholar]
- 35.Chow KS, Singh DP, Roper JM, Smith AG. 1997. A single precursor protein for ferrochelatase-I from Arabidopsis is imported in vitro into both chloroplasts and mitochondria. J. Biol. Chem. 272, 27 565–27 571. ( 10.1074/jbc.272.44.27565) [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T, Masuda T, Singh DP, Tan FC, Tsuchiya T, Shimada H, Ohta H, Smith AG, Takamiya K. 2002. Two types of ferrochelatase in photosynthetic and nonphotosynthetic tissues of cucumber: their difference in phylogeny, gene expression, and localization. J. Biol. Chem. 277, 4731–4737. ( 10.1074/jbc.M105613200) [DOI] [PubMed] [Google Scholar]
- 37.Lister R, Chew O, Rudhe C, Lee MN, Whelan J. 2001. Arabidopsis thaliana ferrochelatase-I and -II are not imported into Arabidopsis mitochondria. FEBS Lett. 506, 291–295. ( 10.1016/S0014-5793(01)02925-8) [DOI] [PubMed] [Google Scholar]
- 38.Leister D. 2005. Genomics-based dissection of the cross-talk of chloroplasts with the nucleus and mitochondria in Arabidopsis. Gene 354, 110–116. ( 10.1016/j.gene.2005.03.039) [DOI] [PubMed] [Google Scholar]
- 39.Woodson JD, Chory J. 2008. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. ( 10.1038/nrg2348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfannschmidt T. 2010. Plastidial retrograde signaling—a true ‘plastid factor’ or just metabolite signatures? Trends Plant Sci. 15, 427–435. ( 10.1016/j.tplants.2010.05.009) [DOI] [PubMed] [Google Scholar]
- 41.Köhler RH, Cao J, Zipfel WR, Webb WW, Hanson MR. 1997. Exchange of protein molecules through connections between higher plant plastids. Science 276, 2039–2042. ( 10.1126/science.276.5321.2039) [DOI] [PubMed] [Google Scholar]
- 42.Köhler RH, Zipfel WR, Webb WW, Hanson MR. 1997. The green fluorescent protein as a marker to visualize plant mitochondria in vivo. Plant J. 11, 613–621. ( 10.1046/j.1365-313X.1997.11030613.x) [DOI] [PubMed] [Google Scholar]
- 43.Akashi K, Grandjean O, Small I. 1998. Potential dual targeting of an Arabidopsis archaebacterial-like histidyl-tRNA synthetase to mitochondria and chloroplasts. FEBS Lett. 431, 39–44. ( 10.1016/S0014-5793(98)00717-0) [DOI] [PubMed] [Google Scholar]
- 44.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. ( 10.1016/j.cell.2008.06.016) [DOI] [PubMed] [Google Scholar]
- 45.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6. ( 10.1093/protein/10.1.1) [DOI] [PubMed] [Google Scholar]
- 46.Cerutti H, Osman M, Grandoni P, Jagendorf AT. 1992. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc. Natl Acad. Sci. USA 89, 8068–8072. ( 10.1073/pnas.89.17.8068) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maarse AC, Van Loon AP, Riezman H, Gregor I, Schatz G, Grivell LA. 1984. Subunit IV of yeast cytochrome c oxidase: cloning and nucleotide sequencing of the gene and partial amino acid sequencing of the mature protein. EMBO J. 3, 2831–2837. ( 10.1002/j.1460-2075.1984.tb02216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakagawa T, et al. 2007. Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotech. Biochem. 71, 2095–2100. ( 10.1271/bbb.70216) [DOI] [PubMed] [Google Scholar]
- 49.Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. ( 10.1046/j.1365-313x.1998.00343) [DOI] [PubMed] [Google Scholar]
- 50.Harrison SJ, Mott EK, Parsley K, Aspinall S, Gray JC, Cottage A. 2006. A rapid and robust method of identifying transformed Arabidopsis thaliana seedlings following floral dip transformation. Plant Methods 2, 19 ( 10.1186/1746-4811-2-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Page MT, McCormac AC, Smith AG, Terry MJ. 2017. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol. 213, 1168–1180. ( 10.1111/nph.14223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bustin SA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. ( 10.1373/clinchem.2008.112797) [DOI] [PubMed] [Google Scholar]
- 53.Lichtenthaler HK, Buschmann C. 2001. Chlorophylls and carotenoids: measurement and characterization by UV-VIS spectroscopy. Curr. Protoc. Food Anal. Chem. 1, F4.3.1–F4.3.8. ( 10.1002/0471142913.faf0403s01) [DOI] [Google Scholar]
- 54.Terry MJ, Kacprzak SM. 2019. A simple method for quantification of protochlorophyllide in etiolated Arabidopsis seedlings. In Phytochromes: methods and protocols, methods in molecular biology, vol. 2026 (ed. Hiltbrunner A.), pp. 169–177. New York, NY: Springer Nature; ( 10.1007/978-1-4939-9612-4_14) [DOI] [PubMed] [Google Scholar]
- 55.Armbruster U, et al. 2009. Chloroplast proteins without cleavable transit peptides: rare exceptions or a major constituent of the chloroplast proteome? Mol. Plant 2, 1325–1335. ( 10.1093/mp/ssp082) [DOI] [PubMed] [Google Scholar]
- 56.Bölter B, Nada A, Fulgosi H, Soll J. 2006. A chloroplastic inner envelope membrane protease is essential for plant development. FEBS Lett. 580, 789–794. ( 10.1016/j.febslet.2005.12.098) [DOI] [PubMed] [Google Scholar]
- 57.Elortza F, Mohammed S, Bunkenborg J, Foster LJ, Nühse TS, Brodbeck U, Peck SC, Jensen ON. 2006. Modification-specific proteomics of plasma membrane proteins: identification and characterization of glycosylphosphatidylinositol-anchored proteins released upon phospholipase D treatment. J. Prot. Res. 5, 935–943. ( 10.1021/pr050419u) [DOI] [PubMed] [Google Scholar]
- 58.Aryal N, Lu C. 2018. A phospholipase C-like protein from Ricinus communis increases hydroxy fatty acids accumulation in transgenic seeds of Camelina sativa. Front. Plant Sci. 9, 1576 ( 10.3389/fpls.2018.01576) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Itoh RD, Yamasaki H, Septiana A, Yoshida S, Fujiwara MT. 2010. Chemical induction of rapid and reversible plastid filamentation in Arabidopsis thaliana roots. Physiol. Plant. 139, 144–158. ( 10.1111/j.1399-3054.2010.01352) [DOI] [PubMed] [Google Scholar]
- 60.McCormac AC, Terry MJ. 2004. The nuclear genes Lhcb and HEMA1 are differentially sensitive to plastid signals and suggest distinct roles for the GUN1 and GUN5 plastid-signalling pathways during de-etiolation. Plant J. 40, 672–685. ( 10.1111/j.1365-313X.2004.02243.x) [DOI] [PubMed] [Google Scholar]
- 61.Terry MJ, Kendrick RE. 1999. Feedback inhibition of chlorophyll synthesis in the phytochrome chromophore-deficient aurea and yellow-green-2 mutants of tomato. Plant Physiol. 119, 143–152. ( 10.1104/pp.119.1.143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Terry MJ, Ryberg M, Raitt CE, Page AM. 2001. Altered etioplast development in phytochrome chromophore-deficient mutants. Planta 214, 314–325. ( 10.1007/s004250100624) [DOI] [PubMed] [Google Scholar]
- 63.Goslings D, Meskauskiene R, Kim C, Lee KP, Nater M, Apel K. 2004. Concurrent interactions of heme and FLU with glu tRNA reductase (HEMA1), the target of metabolic feedback inhibition of tetrapyrrole biosynthesis, in dark- and light-grown Arabidopsis plants. Plant J. 40, 957–967. ( 10.1111/j.1365-313X.2004.02262.x) [DOI] [PubMed] [Google Scholar]
- 64.Richter AS, Banse C, Grimm B. 2019. The GluTR-binding protein is the heme-binding factor for feedback control of glutamyl-tRNA reductase. eLife 8, e46300 ( 10.7554/eLife.46300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Terry MJ, Bampton J. 2019. The role of tetrapyrroles in chloroplast-to-nucleus retrograde signalling. Adv. Bot. Res. 91, 225–246. ( 10.1016/bs.abr.2019.05.002) [DOI] [Google Scholar]
- 66.Larkin RM. 2016. Tetrapyrrole signaling in plants. Front. Plant Sci. 7, 1586 ( 10.3389/fpls.2016.01586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tadini L, et al. 2016. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 170, 1817–1830. ( 10.1104/pp.15.02033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Llamas E, Pulido P, Rodriguez-Concepcion M. 2017. Interference with plastome gene expression and Clp protease activity in Arabidopsis triggers a chloroplast unfolded protein response to restore protein homeostasis. PLoS Genet. 13, e1007022 ( 10.1371/journal.pgen.1007022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marino G, Naranjo B, Wang J, Penzler J-F, Kleine T, Leister D. 2019. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J. 99, 521–535. ( 10.1111/tpj.14342) [DOI] [PubMed] [Google Scholar]
- 70.Zhao X, Huang J, Chory J. 2019. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signalling. Proc. Natl Acad. Sci. USA 116, 10 162–10 167. ( 10.1073/pnas.1820426116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu GZ, et al. 2019. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 5, 525–538. ( 10.1038/s41477-019-0415) [DOI] [PubMed] [Google Scholar]
- 72.Shimizu T, et al. 2019. The retrograde signalling protein GUN1 regulates tetrapyrrole biosynthesis. Proc. Natl Acad. Sci. USA 116, 24 900–24 906. ( 10.1073/pnas.1911251116) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets supporting this article have been provided as part of the electronic supplementary material.