Abstract
Plastid genes in higher plants are transcribed by at least two different RNA polymerases, the plastid-encoded RNA polymerase (PEP), a bacteria-like core enzyme whose subunits are encoded by plastid genes (rpoA, rpoB, rpoC1 and rpoC2), and the nuclear-encoded plastid RNA polymerase (NEP), a monomeric bacteriophage-type RNA polymerase. Both PEP and NEP enzymes are active in non-green plastids and in chloroplasts at all developmental stages. Their transcriptional activity is affected by endogenous and exogenous factors and requires a strict coordination within the plastid and with the nuclear gene expression machinery. This review focuses on the different molecular mechanisms underlying chloroplast transcription regulation and its coordination with the photosynthesis-associated nuclear genes (PhANGs) expression. Particular attention is given to the link between NEP and PEP activity and the GUN1- (Genomes Uncoupled 1) mediated chloroplast-to-nucleus retrograde communication with respect to the Δrpo adaptive response, i.e. the increased accumulation of NEP-dependent transcripts upon depletion of PEP activity, and the editing-level changes observed in NEP-dependent transcripts, including rpoB and rpoC1, in gun1 cotyledons after norflurazon or lincomycin treatment. The role of cytosolic preproteins and HSP90 chaperone as components of the GUN1-retrograde signalling pathway, when chloroplast biogenesis is inhibited in Arabidopsis cotyledons, is also discussed.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles’.
Keywords: plastid, RNA polymerases, retrograde communication, photosynthesis, cotyledons, Arabidopsis thaliana
1. Introduction
Compared to its cyanobacterial ancestor, the transcriptional apparatus of the land plant chloroplast is more complex and reflects the evolutionary integration of a prokaryotic gene expression system into a eukaryotic host cell. Unlike bacteria, chloroplasts of angiosperms and the moss Physcomitrella patens require at least two different RNA polymerases to ensure transcription of plastid genes: the plastid-encoded polymerase (PEP), a multimeric bacterial-type enzyme [1], and, in addition, the nuclear-encoded polymerase (NEP), a monomeric T3-T7 bacteriophage-type enzyme [2,3]. Correct plastid development and functionality necessitate the interplay of both PEP and NEP enzymes, together with nuclear-encoded factors, such as sigma-like factors (Sig) and PEP-associated proteins (PAPs) [4–8]. These are just a few examples of nuclear factors that exert control over the plastid gene expression (PGE), since the nuclear genome encodes most of the plastid proteins [9]. On the other hand, chloroplast biogenesis and functionality also require tight coordination of the transcription of thousands of nuclear genes with the expression of the relatively few plastid genes. This coordination is achieved through an extensive flow of information from developing plastids to the nucleus, via biogenic retrograde signalling, and from mature chloroplasts to the nucleus, via operational retrograde signalling [10–12]. The GUN1 (genomes uncoupled 1) protein, a pentatricopetide repeat protein that localizes to plastids, was proposed as the central node relaying information from multiple retrograde signalling pathways that regulate photosynthesis-associated nuclear genes (PhANGs) expression [13]. Interestingly, the involvement of GUN1 in relaying signals after norfluorazon (NF) and lincomycin (Lin) treatments [13], and after impairment of protein import [14] and PGE [15–17] raises the possibility that each of the treatments or mutant genetic backgrounds may affect a similar process, thereby using GUN1 as a common signalling component [18]. Intriguingly, GUN1 and NEP proteins have some features in common including the fact that (i) NEP, as GUN1, should be found in plastid nucleoids, since plastid DNA is exclusively located in these domains; (ii) the NEP polymerase and the GUN1 protein have never been detected in plastid proteomics analysis [6,19,20], most probably as a consequence of their very low abundance; and (iii) they are highly active during early stages of chloroplast biogenesis [2,21]. This review focuses on the recently discovered molecular mechanisms at the basis of the regulation of chloroplast transcription and its coordination with PhANGs expression during early stages of chloroplast development and upon alteration of plastid protein homeostasis in Arabidopsis cotyledons. Particular emphasis is given to the link between NEP and PEP activity and the GUN1-mediated chloroplast-to-nucleus retrograde communication in Arabidopsis cotyledons [22–24].
2. The plastid-encoded RNA polymerase
The PEP catalytic core is made of α2 (it occurs as a dimer and serves as a stabilizing agent of the PEP core), β (the catalytic subunit), β′ (unknown function) and β″ (DNA binding function) subunits, encoded by the plastid rpoA, rpoB, rpoC1 and rpoC2 genes, respectively, and inherited from the cyanobacterial ancestor [25]. Studies performed in barley, tobacco and Arabidopsis showed that rpo genes are essential for proper chloroplast biogenesis, as mutants lacking any PEP subunit exhibit an albino or yellowish phenotype [26]. As in any other bacterial RNA polymerase, the PEP enzyme needs sigma factors (σ) for promoter recognition and initiation of transcription [27]. Genes coding for plastid sigma factors are located in the nuclear genome, thus, PGE is strictly controlled by the nucleus. In Arabidopsis thaliana, six different sigma factors, SIG1–SIG6, have been identified. Their specific functions in chloroplast physiology have not been fully understood yet, however, the investigation of knockout mutants has provided the first insights [5]. The PEP enzyme is located in the nucleoids and the Rpo core subunits jointly with one of the sigma factors form the holoenzyme that, upon light exposure, recruits additional subunits driving chloroplasts biogenesis. Different nucleus-encoded proteins, unrelated to bacterial transcription factors, are consistently co-purified with PEP and collectively are named PEP-associated proteins (PAPs) [6]. PAPs are an evolutionary conquest of solely land plants, as no PAP orthologues were found in Chlamydomonas reinhardtii [6,28]. Intriguingly, functional studies on PAP knockout mutants indicate that PAP proteins are required for PEP-mediated transcription and regulation [29–36], contributing to establish a subdomain in the plastid nucleoid where PEP-mediated transcription takes place [6].
3. Origin and evolution of nuclear-encoded polymerase
In the early 1990s, researchers postulated the existence of a nuclear-encoded and plastid-located RNA polymerase, NEP. The first evidence came from the obligate parasitic angiosperm Epifagus virginiana that possesses plastids with a reduced genome (just nine genes are retained), deprived of all rpo genes [37], and from the barley mutant albostrians deficient in plastid ribosomes [38] and therefore unable to synthesize the PEP core subunits. Despite the absence of PEP, plastid transcripts were detectable both in Epifagus virginiana and in albostrians barley mutant, suggesting the existence of a nuclear-encoded plastid RNA polymerase. In the same period, a 110 kDa polypeptide was purified from spinach chloroplasts via sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The purified polypeptide actively synthesized RNA in the presence of supercoiled DNA template and nucleoside triphosphates, showing features of a single-subunit RNA polymerase of the T3-T7 bacteriophage type [39]. The plant genome era, inaugurated by the release of the Arabidopsis thaliana genome, allowed the identification of three nuclear genes, called RPOT, coding for phage-like RNA polymerases, targeted to mitochondria (RPOT1/RpoTm), chloroplasts (RPOT3/RpoTp) and to both organelles (RPOT2/RpoTmp) [40,41]. In general, the three RPOT genes are present in eudicotyledon plants, while RPOT2, encoding the dual-located RpoTmp, is absent in monocots and early-diverging angiosperms [42,43]. The basal angiosperm Nuphar advena, for instance, has two mitochondrial RNA polymerases and a proper NEP, which could be considered the phylogenetically earliest RpoTp enzyme in higher plants [44]. Unicellular green algae, such as C. reinhardtii, Ostreococcus tauri and Thalassiosira pseudonana, also have a single gene for the RpoTm and no RpoTmp enzyme [43,45]. This feature is in common with fungi and mammals, as well as the spike-moss Selaginella moellendorffii [43,46]. However, the nuclear genome of the moss P. patens bears three RPOT gene as in Arabidopsis. However, one gene codes for an exclusively mitochondrial polymerase, while the other two produce two dual-located RpoTmp enzymes [3]. In the light of these phylogenetic data, the NEP enzyme appeared to originate from duplication of the RPOT1 gene, which could have occurred multiple times in the evolution of land plants, as three copies of RPOT genes are present in mosses and dicot plants but not in spike-mosses and monocots. Alternatively, the extra dual-located enzyme could have been lost during evolution [43,44].
NEP, like PEP, is essential for chloroplast transcription. Knocking out the RPOT2 or RPOT3 genes in Arabidopsis yields plants with delayed chloroplast biogenesis, while the rpot2 rpot3 double mutant is characterized by chlorophyll deficiency and a complete arrest of growth early in development [47], indicating partially overlapping functions of RpoTp and RpoTmp in chloroplast gene transcription.
4. Nuclear-encoded polymerase and plastid-encoded polymerase and their role in plastid gene expression
Data obtained during the last two decades indicate that both PEP and NEP polymerases are active in all green and non-green tissues, but with different degrees of activity according to the developmental stages and physiological conditions (figure 1). As a matter of fact, Arabidopsis dry seeds already contain several transcripts of the plastid transcription machinery, such as the NEP RPOT2 and RPOT3, the PEP core rpoA, rpoB, rpoC1, rpoC2, and the nuclear-encoded PEP-associated sigma factors SIG2 and SIG5 [48,49]. These PEP core transcripts derive from chloroplasts present in photosynthetically active Arabidopsis embryos, before dedifferentiation to small non-green plastids, termed eoplasts, occurs during late embryo and seed maturation. In addition, during stratification (72 h at 4°C in the dark) several plastid house keeping genes, including rpoA, rpoB, rpoC1 and rpoC2, and many nuclear genes encoding SIG1–6 factors and the RpoTp, RpoTmp enzymes are actively transcribed [48,50].
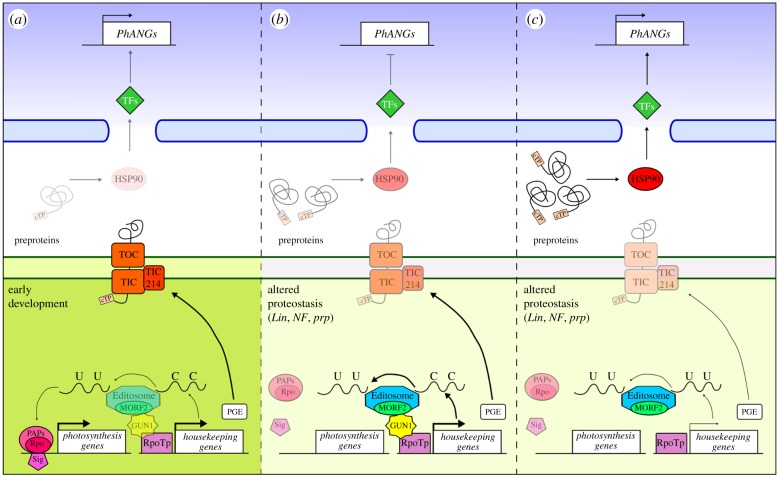
Figure 1.
Schematic overview of the GUN1-dependent coordination of plastid and nuclear transcriptional machinery in three different scenarios: early stages of chloroplast development (a), altered protein homeostasis (b) and altered protein homeostasis in the absence of GUN1 protein (c). The role of GUN1 becomes evident when chloroplast biogenesis is altered either by chemical treatments (lincomycin of norflurazon) or genetic defects that impair plastid protein homeostasis (see panels indicated as altered proteostasis) [15,16,24]. Under these conditions, GUN1 has been reported to (i) interact with NEP and favour accumulation of NEP-dependent transcripts, including ycf1 that encodes the Tic214 subunit of the protein translocon at the inner envelope membrane of chloroplasts [24]; (ii) interact with MORF2, a component of the Editosome protein complex, and edit NEP-dependent transcripts, including rpoB and rpoC1, but also rps12, rps14, ndhB and ndhD mRNAs [22]; (iii) interact with cpHSC70-1 and regulate plastid protein import (this detail is not shown) [23]. Overaccumulation of preproteins in the cytosol of gun1 cotyledon cells induces the accumulation of cytosolic HSP90 proteins that favour PhANGs transcription [23,24]. This could be obtained by either 26S proteasome-dependent degradation of negative transcriptional regulators or activation, through HSP90-promoting folding, of positive ones (transcription factors, TFs). It is conceivable that a similar mechanism could be active even at early stages of chloroplast development [21], to fine-tune the chloroplast and nuclear transcription machineries with respect to the developmental needs. However, under these conditions the accumulation of preproteins and downstream components of the signalling pathway have not been described yet. Note that the thickness and brightness of lines and shapes is directly proportional to the activity of the specific process. cTP, chloroplast transite peptide; TOC, translocon at the outer chloroplast membrane; TIC, translocon at the inner chloroplast membrane; Lin, lincomycin; NF, norflurazon; prp, plastid ribosomal protein mutants; PGE, plastid gene expression, defined as transcription and translation rate of plastid-encoded genes.
At the protein level, rpoB, RpoTp and RpoTmp are also present in dry seeds. Furthermore, a dramatic accumulation of RpoTp protein occurs shortly after imbibition, and its increased activity leads to a higher accumulation of PEP core proteins as well. Upon seed germination, NEP activities and transcripts increase and peak at 2–3 days after germination, leading to increased accumulation of NEP-dependent transcripts, followed by a severe drop as RPOT3 gene expression decreases [51,52]. Moreover, gene expression studies revealed that RPOT3 is mainly active in green photosynthetic tissues while RPOT2 plays its role in dividing and non-green cells [53].
When seed germination takes place in darkness, the eoplasts turn into etioplasts, where a soluble inactive form of the PEP holoenzyme, termed PEP-B and made of core proteins only, is present. Upon illumination, the PEP complex starts interacting with several PAP proteins and becomes part of the membrane-bound plastid transcription active chromosome (pTAC) megadalton complex. This more complex enzyme, named PEP-A, increases the PEP activity and leads to the transcription of Photosynthesis-Associated Plastid-Encoded Genes (PhAPGs) and cotyledon greening [34,54,55]. NUCLEAR CONTROL OF PEP ACTIVITY (NCP), a dual-targeted nuclear/plastidial protein, also known as MRL7-L (Mesophyll-cell RNAi Library line 7-like) [56] and SVR4-like (Suppressor of Variegation 4-like) [57] because of its essential role in chloroplast biogenesis [56,57], is required for both the nuclear and plastidial signalling steps of PhAPGs activation, promoting the assembly of the PEP complex upon illumination [58]. In particular, NCP mediates the degradation of two repressors of chloroplast biogenesis in the nucleus, PIF1 and PIF3, thus participating in the phytochrome-mediated signalling at the basis of chloroplast biogenesis. NCP also has a paralog in Arabidopsis, At4g28590 also known as ECB1 (Early Chloroplast Biogenesis 1) [59], SVR4 (Suppressor of Variegation 4) [57] and MRL7 (Mesophyll-cell RNAi Library line 7), which was renamed Regulator of Chloroplast Biogenesis (RCB) [60]. Like NCP, RCB is also a dual-targeted nuclear/plastidial protein required for the degradation of the nuclear transcriptional regulators PIF1 and PIF3 and for PEP assembly and PhAPGs expression in the plastids. These data support a model in which phytochromes control PhAPGs expression through light-dependent double nuclear and plastidial switches that are linked by evolutionarily conserved and dual-localized regulatory proteins.
Following PEP-A assembly, the enzyme takes over most of the transcription activity, including mRNAs, rRNAs and most of the tRNAs [61–65], while the NEP enzyme remains active to perform the transcription of rpoB (forming an operon with rpoC1 and rpoC2), accD (in dicots), clpP, atpB, atpI, genes coding for ribosomal proteins and a few tRNAs, and, in dicots, ycf1 and ycf2 until senescence.
5. Plastid-encoded polymerase regulation
PEP promoter recognition and transcription initiation are mediated by σ70-like factors, as occurs in prokaryotes [27]. Arabidopsis has six different sigma factors, SIG1-SIG6. All Arabidopsis sig knockout mutants fail to accumulate wild-type levels of plastid transcripts; nevertheless, only sig2 and sig6 displayed a pale-green phenotype, suggesting a more relevant role in plastid transcription and chloroplast development [66]. Furthermore, only a partial functional redundancy among Arabidopsis SIG factors during early steps of seedling development has been revealed, indicating gene-specific functions [4,5].
Unlike their bacterial counterparts, the plastid sigma factors SIG1, SIG2 and SIG6 can be modified post-translationally by phosphorylation at the N-terminal variable region in fully mature chloroplasts [4,67]. SIG2 and SIG6 phosphorylation have been related to alterations in the promoter binding efficiency of PEP [68]. Similarly, SIG1 phosphorylation is regulated in a redox-dependent manner and serves to adapt PSII/PSI stoichiometry to light changes by modulating the relative transcription of the photosynthesis reaction centre genes psbA (photosystem II, PSII) and psaA/B (photosystem I, PSI), indicating that phosphorylation is also needed to adapt the PEP-dependent transcription to the redox state of the thylakoid electron transport chain [67]. Lastly, a role for SIG6 in the operational retrograde signalling during singlet oxygen stress has been reported [69]. In particular, a genetic screen uncovered a sig6 mutant (soldat8) that was able to survive the high levels of singlet oxygen in the Arabidopsis flu mutant that has uncontrolled tetrapyrrole synthesis. This regulatory mechanism originated after the endosymbiosis event, since the N-terminal of plastid SIG factors is not conserved among bacteria and chloroplasts, and makes plastid SIG factors incompatible with the transcription machinery of bacteria [70,71].
SIG factors also contribute to the construction of PEP-A holoenzyme, together with the 12 true PAP proteins, identified through genetic and biochemical studies, and shown to be essential for chloroplast biogenesis in Arabidopsis [6]. Indeed, all pap mutants display an albinotic/chlorotic phenotype, corroborating the hypothesis that PAP proteins are fundamental for PEP activity, as Rpo-core subunits. In particular, PAPs are involved in different regulatory functions [34], including:
-
(i)
DNA/RNA metabolism-related gene expression regulation, as in the case of PAP1, 2, 3, 5, 7 and 12. For instance, PAP1 and PAP2 display pentatricopeptide repeat (PPR) motifs, known to be involved in RNA metabolism [72] and PAP3 is predicted to interact with RNAs through its S1-like domain [35].
-
(ii)
Redox-dependent gene regulation and protection against oxidative stresses, as in PAP4, 6, 9 and 10. As an example, PAP10/TrxZ physically interacts with the PLASTID REDOX INSENSITIVE 2 (PRIN2) protein, a key regulator of PEP activity, capable of transducing the redox state of thylakoid membranes into regulation of PEP-dependent transcription [73,74]. In particular, PAP10 interacts with the PRIN2 dimer and through its thioredoxin domain causes the reduction of inter-molecular disulfide bridges and the release of PRIN2 monomers, that are then able to boost the transcription of PEP-dependent genes, providing a mechanistic link between photosynthetic electron transport and activation of photosynthetic gene expression.
Chloroplast transcription has also been reported to be under the control of phytohormones during the greening of etioplasts [75] and in fully developed chloroplasts of barley (Hordeum vulgare L.). In particular, stimulatory effects of cytokinin (6-benzyladenine BA) and repressive effects of methyl jasmonate (MeJA), auxin (indole-3-acetic acid, IAA) and gibberellic acid (GA3) on chloroplast gene expression at the levels of transcription and transcript accumulation of both NEP- and PEP-transcribed genes have been reported [76,77]. More detailed information is available for abscisic acid (ABA). In particular, it has been shown that exogenously supplied ABA represses the transcription of plastid genes during greening and in fully mature chloroplasts in barley leaves, with the exception of psbA, psbD and a few other genes that remain as active as in untreated leaves [78]. Based on these findings, ABA seems to coordinate the repression of photosynthesis genes in the nuclear and chloroplast genomes while leaving active those chloroplast genes that are needed for the protection of the PSII reaction centres from damage by reactive oxygen species (ROS) [78]. This adaptive response involves the ABA-activated expression of RSH2 and RSH3 nuclear genes, coding for enzymes in the chloroplast that synthesize guanosine-3′, 5′-bisdiphosphate (ppGpp), an inhibitor of chloroplast transcription. In particular, ppGpp was shown to inhibit transcriptional activities of purified PEP preparations in vitro [79]. In addition, an assay based on the in planta incorporation of the base analogue 4-thiouridine into nascent chloroplast RNA demonstrated that the accumulation of ppGpp inhibits the transcription of PEP-dependent and, to a lesser extent, of NEP-dependent genes in developing seedlings [80], resembling the ancient bacterial stress-signalling pathway known as the stringent response (for a review see [81]). Furthermore, relatively high ABA levels increase the amount of SIG5 by activating the expression of its gene in the nucleus leading to a subpopulation of SIG5–PEP. The SIG5–PEP transcribes chloroplast genes from specific promoters, resulting in the observed escape of a few plastid genes (psbA, psbD) from the otherwise general downregulation of transcription of chloroplast genes [78].
6. Nuclear-encoded polymerase regulation
Although PEP regulation has been widely characterized and several regulatory factors identified, only a few pieces of information on NEP activity regulation are currently available. Firstly, the tRNA-Glu is able to bind and inhibit the activity of NEP in vitro, displaying specificity compared to tRNA-Val, tRNA-Gly and tRNA-Trp, used as control [51]. The tRNA-Glu is different from the other plastid-encoded tRNAs, since it is not only involved in plastid protein synthesis but is the substrate for the synthesis of 5-aminolevulinic acid, an early step of the tetrapyrrole biosynthesis pathway that leads to chlorophyll, heme and phytochrome production. In particular, the tRNA-Glu, massively transcribed by PEP during the greening process, has a pivotal role in the switch between NEP and PEP activity, by inhibiting the NEP-dependent transcription in a threshold manner, once the greening has occurred [51]. A second regulatory mechanism of NEP activity was initially observed in tobacco leaves through the impairment of the plastid rpoB gene, encoding the β-subunit of the PEP enzyme. In particular, an entire class of plastid transcripts was found to accumulate at high level upon the complete loss of PEP activity [62,82]. Such an adaptation mechanism, termed as ‘Δrpo phenotype’, was then reproduced in tobacco leaves upon depletion of the rpoA, rpoB, rpoC1 and rpoC2 genes [83], encoding the core complex of the PEP enzyme. Later, the Δrpo adaptive response was described in nearly every mutant identified as PEP regulator and impaired in PEP-dependent gene expression in Arabidopsis, including mutants with PAP gene expression defects (for further details, see table 1), together with many other mutants affected in the accumulation of key players of chloroplast protein homeostasis, such as PPR proteins involved in mRNA metabolism (CRP1 and PPR4), plastid ribosomal proteins (PRPL11 and PRPS21), chaperones (CLPR1 and cpHSP70–1), and the iron superoxide dismutase FSD2 and FSD3 (table 1). Moreover, impairment of chloroplast translation upon lincomycin treatment activates the Δrpo adaptive response in Arabidopsis seedlings [13,24]. More generally, it appears that every genetic- or chemical-induced impairment of plastid protein homeostasis leads to the upregulation of NEP-dependent transcripts, as an attempt by the PGE machinery to maintain non-photosynthetic plastid functions by expressing housekeeping genes. Strikingly, we have recently reported that GUN1 protein is able to physically interact with NEP in chloroplasts of Arabidopsis cotyledons and is required for the Δrpo adaptive response, providing an important link between plastid transcription and the GUN1-mediated plastid-to-nucleus retrograde communication [24].
Table 1.
List of genes/proteins whose impairments result in the activation of the Δrpo adaptive response, i.e. increase accumulation of NEP-dependent transcripts upon depletion of PEP activity.
| gene | accession | phenotype | molecular defect | refs |
|---|---|---|---|---|
| plastid transcription regulation | ||||
| SIG2 | AT1G08540 | pale | reduced PEP activity | [66] |
| SIG6 | AT2G36990 | pale | reduced PEP activity | [66,84] |
| DG1 | AT5G67570 | delayed greening | reduced PEP activity | [84,85] |
| PAP2/pTAC2 | AT1G74850 | pale | reduced PEP activity; RNA maturation |
[29] |
| PAP3/pTAC10 | AT3G48500 | albino | reduced PEP activity | [86] |
| PAP4/FSD3 | AT5G23310 | pale | reduced ROS scavenging; reduced PEP activity |
[87] |
| PAP5/pTAC12/HMR | AT2G34640 | pale | reduced PEP activity; altered phytochrome signalling |
[88,89] |
| PAP6/FLN1 | AT3G54090 | albino | reduced PEP activity | [89] |
| FLN2 | AT1G69200 | virescent | reduced PEP activity | [89,90] |
| PAP7/pTAC14 | AT4G20130 | albino | reduced PEP activity | [90] |
| PAP8/pTAC6 | AT1G21600 | albino | reduced PEP activity | [29] |
| PAP9/FSD2 | AT5G51100 | pale | reduced ROS scavenging; reduced PEP activity |
[87] |
| PAP10/TrxZ | AT3G06730 | albino | reduced PEP activity | [34] |
| PAP12/pTAC7 | AT5G24314 | albino | reduced PEP activity | [91] |
| pTAC5 | AT4G13670 | pale | reduced PEP activity | [92] |
| PRIN2 | AT1G10522 | albino | reduced PEP activity; impaired redox-mediated retrograde signalling |
[73,74] |
| ECB1/MRL7/SVR4/RCB | AT4G28590 | tall/albino | reduced PEP activity; altered phytochrome signalling |
[56,57,60] |
| MRL7-L/SVR4-L/NCP | AT2G31840 | tall/albino | reduced PEP activity; altered phytochrome signalling |
[57,58] |
| plastid transcripts maturation | ||||
| CRP1 | AT5G42310 | albino | impaired plastid mRNA maturation | [93] |
| PPR4 | AT5G04810 | embryo-lethal | impaired plastid mRNA maturation | [94] |
| PDM1 | AT4G18520 | albino | impaired plastid mRNA maturation | [95] |
| plastid translation | ||||
| PRPS21 | AT3G27160 | pale | reduced plastid ribosome activity | [24,96] |
| PRPL11 | AT1G32990 | pale | reduced plastid ribosome activity | [15,24,97] |
| plastid proteostasis maintenance | ||||
| HSP21 | AT4G27670 | none (22°C); albino (30°C) | reduced PEP activity under heat stress | [92] |
| cpHSC70-1 | AT4G24280 | variegated | reduced plastid import capacity; impaired protein quality control |
[24,98] |
| CLPR1 | AT1G49970 | virescent | impaired protein quality control | [99] |
7. Interplay between plastid transcription regulation and retrograde signalling
Due to the chimeric nature of PEP enzyme, made of subunits encoded by both the plastid and the nuclear genome, and the nuclear localization of NEP encoding genes, it is clear that the coordinated transcription of plastid and nuclear genes is essential for proper development of all plastid types. This coordination takes place through the nuclear control of PGE and the retrograde signalling pathways [10].
In the case of chloroplast biogenesis, multiple pieces of evidence on the coordination of plastid transcription and PhANGs expression have been reported. In particular, the status of plastid transcription and translation as a trigger for retrograde signalling has been demonstrated by repression of PhANGs expression upon chemical treatments, for instance with the plastid translation inhibitor lincomycin or by rifampicin, which selectively inhibit the PEP enzyme [12,66]. More specifically related to PEP-dependent transcription, increasing evidence indicates that several factors involved in the coordination of plastid and nuclear transcriptional machineries are characterized by the presence of both the chloroplast transit peptide (cTP) and the nuclear localization signal (NLS) in their peptide sequence, implying a plastid and nuclear dual localization [20]. These proteins have potential access to plastid and nuclear DNA and, therefore, they can be directly involved in the coordination of gene expression in both compartments. Among them are NCP and RCB, described above, and PAP1, −5, −7, −8, −9 and −12. Currently, only in the case of PAP5/pTAC12 has the dual localization been experimentally confirmed [88,100]. Indeed, PAP5 seems to be involved in the red light-mediated skotomorphogenesis-to-photomorphogenesis transition, through the physical interaction and degradation of phytochrome-interacting-factors PIF1 and PIF3, thereby affecting gene expression in the nucleus [101–103], similarly to NCP and RCB. Interestingly, genes coding for PAPs were found to be expressed in non-green tissues before the assembly of the PEP-A complex, which is induced in the light. It is, therefore, possible to speculate that the NLS-containing PAP proteins can form two different complexes, one in the nucleus and one in the chloroplast, and modulate gene expression in both compartments by interacting with different kinds of RNA polymerases [26]. In this scenario, a nuclear PAP complex could assemble in the dark at first, mediating the early steps of plastid development by interacting with RNA polymerase II, and upon illumination migrate to the chloroplast to form PEP-A, in a sort of NLS-cTP competition which could determine the PAP protein intracellular localization.
In addition to dual-located proteins, proteins specifically located in plastids are also able to coordinate plastid and nuclear gene expression. For instance, PRIN2 has been reported to be able to trigger retrograde signalling in response to light, besides its direct regulation of PEP activity, [73,74].
Furthermore, depletion of SIG2 and SIG6 in Arabidopsis allowed demonstration that the activity of both sigma factors is the source of retrograde signals that promote PhANGs expression [66]. The addition of the gun1 mutation in sig6 mutant led to a global reduction of 72 plastid transcripts, both PEP- and NEP-dependent, suggesting a possible involvement of GUN1 in the regulation of the plastid and nuclear transcriptional machineries. The physical interaction between PAP8/pTAC6 and GUN1 observed by Tadini et al. [15] points further to a direct link between PEP activity modulation and retrograde signalling.
More recently, Tadini and co-workers demonstrated that GUN1 plays a central role in the regulation of NEP activity, thus creating a direct link between retrograde signalling and PEP/NEP-dependent transcription (figure 1). In particular, GUN1 was shown to interact physically with RpoTp and to have a direct role in the upregulation of NEP-dependent transcripts upon perturbation of plastid protein homeostasis, i.e. during the Δrpo adaptive response [24]. Concomitantly, GUN1 has been reported to have a direct role in RNA editing by physically interacting with the MULTIPLE ORGANELLAR RNA EDITING FACTOR 2 (MORF2), a member of the so-called plastid RNA Editosome, which is involved in editing nearly all sites of plastid RNA [22], in agreement with previous studies where a link between plastid signalling and RNA editing was shown [18,104]. Consistent with this, gun1 mutant cotyledons have differential efficiency of RNA editing (C->U) levels of 11 sites in the plastid transcriptome, after norflurazon or lincomycin treatment, compared to wild type. Intriguingly, the target genes were NEP-dependent, including transcripts of PEP core subunits β and β’ [22]. The rpoB and rpoC1 editing sites have been observed in previous studies and lead to amino acid changes in the protein sequence [105]. However, the biological meaning of such editing events and the impact on the transcripts is not always clear. RNA editing is usually associated with the restoration of conserved codons or aims to recreate start/stop codons serving as a correction for otherwise defective transcripts [106]. In this specific case, GUN1 is responsible for three amino acid changes in rpoB sequence (S113F, S184 L and S811 L) and one in rpoC1 (S163 L) [22]. The edited residues are highly conserved in several species, suggesting that the lack of GUN1 leads to the production of altered forms of PEP core proteins. In this scenario (figure 1), GUN1 appears to be part of a protein complex that act as a positive regulator of both NEP- and PEP-dependent transcription, and that could contribute to the increase of cellular RNA amount observed during germination or even to the doubling of plant cell RNA detected within 48 h under stress conditions, i.e. in response to herbicide-induced Mg-protoporphyrin and heme accumulation or a high level of sugar treatment, as previously reported [107]. According to this model, GUN1-dependent closure of the photosynthetic apparatus, by repressing photosynthesis-associated nuclear gene expression, would contribute further to protection from oxidative stress [107]. Alternatively, the GUN1-dependent increased accumulation of NEP-dependent transcripts upon depletion of PEP activity would serve to maintain the house keeping functions of plastids, reducing to a minimum the photosynthesis-dependent oxidative damage.
Interestingly, the over-accumulation of unimported precursor proteins (preproteins) observed in the cytosol of gun1 cotyledon cells upon growth in the presence of lincomycin (figure 1) induces upregulation of cytosolic Heat Shock Protein 90 (HSP90) and in turn sustains the expression of PhANGs, indicating that this pathway might play a key role in the coordination of plastid and nuclear gene transcription [23,24]. In particular, HSP90 could mediate retrograde communication by either repressing negative regulators of transcription (for example, through delivery to the 26S proteasome for degradation) such as abscisic acid insensitive 4 (ABI4) [13], although its role in retrograde signalling has been questioned recently [108], or by activating a positive regulator of transcription (for example, through promoting folding or refolding of a transcription factor) such as Elongated Hypocotyl 5 (HY5) [109] and Golden-Like 1/2 (GLK1/2) [110], reported to be involved in retrograde communication and required for coordinated expression of key genes in chloroplast biogenesis [111,112].
8. Concluding remarks and future perspectives
The field of plastid-to-nucleus signalling has been very dynamic over the last few years, and there have been several major breakthroughs leading to a much more advanced understanding of the mechanisms involved in plastid and nuclear genome crosstalk. Three recent studies have identified GUN1-interacting proteins in Arabidopsis cotyledons, contributing to unravelling the mechanism by which the activity of plastid transcription machinery is coordinated with the nuclear gene expression apparatus [22–24]. A direct connection between GUN1 and the NEP-PEP plastid RNA polymerases has been provided by both the physical interaction of GUN1 with RpoTp and its role in favouring the NEP-dependent transcript accumulation upon alteration of plastid protein homeostasis [24], and by the editing-level changes observed in rpoB and rpoC1 transcripts, which encode subunits of plastid-encoded RNA polymerase [22]. In agreement with previous studies [66,74], it is conceivable that the alteration of the activity of the plastid RNA polymerases results in abnormal transcription of a specific set of plastid genes and, possibly, in the source of the GUN1-dependent retrograde signalling. Furthermore, the altered transcription of the NEP-dependent ycf1 gene, which encodes the Tic214 subunit of the 1 MDa TIC complex [24], together with the GUN1-cpHSC70-1 (chloroplast heat shock protein 70-1) interaction [23], have provided the connection between cytosolic folding stress and the GUN1-dependent retrograde signalling. HSP90, induced by cytosolic preproteins, has been verified to sustain the expression of nuclear photosynthesis-related genes [23], pointing to a possible connection of GUN1-retrograde signalling and the chloroplast unfolded protein response (cpUPR) [113,114].
These recent findings offer a novel point of view to elucidate the mechanisms by which the plastid transcriptional machinery and, more generally, the PGE apparatus communicates with the nucleus; however, several questions still remain. Critical future directions for the research field include efforts to understand whether cytosolic folding stress only occurs in the gun1 mutant. Also, the identity of the downstream signal components of HSP90 require further exploration. Another question is whether plastid RNA editing and cytosolic folding stress are also connected with each other. Finally, the intriguing dual localization of a few PAP proteins and their role in the coordination of plastid and nuclear transcription deserves further investigation. Nuclear proteomics analyses, and the in-depth biochemical analyses of the main players identified, could provide important clues on the molecular mechanisms at the root of plastid-nucleus transcription machinery coordination, essential during early stages of chloroplast biogenesis and upon alteration of plastid protein homeostasis.
Acknowledgements
We apologize to the authors who did not get their work discussed in this review due to space limitation.
Data accessibility
This article has no additional data.
Authors' contributions
L.T. and P.P. designed the study. N.J. and M.C. took care of table 1 and figure 1. All authors helped draft the manuscript. P.P. coordinated the study and took care of the final version of the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by Ministero dell'Istruzione dell'Università e della Ricerca – MIUR, grant no. PRIN-2017FBS8YN to P.P.
References
- 1.Lgloi GL, Kössel H. 1992. The transcriptional apparatus of chloroplasts. CRC. Crit. Rev. Plant Sci. 10, 525–558. ( 10.1080/07352689209382326) [DOI] [Google Scholar]
- 2.Börner T, Aleynikova AYU, Zubo YO, Kusnetsov VV. 2015. Chloroplast RNA polymerases: role in chloroplast biogenesis. Biochim. Biophys. Acta 1847, 761–769. ( 10.1016/j.bbabio.2015.02.004) [DOI] [PubMed] [Google Scholar]
- 3.Richter U, Kiessling J, Hedtke B, Decker E, Reski R, Börner T, Weihe A. 2002. Two RpoT genes of Physcomitrella patens encode phage-type RNA polymerases with dual targeting to mitochondria and plastids. Gene 290, 95–105. ( 10.1016/S0378-1119(02)00583-8) [DOI] [PubMed] [Google Scholar]
- 4.Schweer J, Türkeri H, Kolpack A, Link G. 2010. Role and regulation of plastid sigma factors and their functional interactors during chloroplast transcription—recent lessons from Arabidopsis thaliana. Eur. J. Cell Biol. 89, 940–946. ( 10.1016/j.ejcb.2010.06.016) [DOI] [PubMed] [Google Scholar]
- 5.Lerbs-Mache S. 2011. Function of plastid sigma factors in higher plants: regulation of gene expression or just preservation of constitutive transcription? Plant Mol. Biol. 76, 235–249. ( 10.1007/s11103-010-9714-4) [DOI] [PubMed] [Google Scholar]
- 6.Pfalz J, Pfannschmidt T. 2013. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 18, 186–194. ( 10.1016/j.tplants.2012.11.003) [DOI] [PubMed] [Google Scholar]
- 7.Yu Q-B, Huang C, Yang Z-N. 2014. Nuclear-encoded factors associated with the chloroplast transcription machinery of higher plants. Front. Plant Sci. 5, 316 ( 10.3389/fpls.2014.00316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chi W, He B, Mao J, Jiang J, Zhang L. 2015. Plastid sigma factors: their individual functions and regulation in transcription. Biochim. Biophys. Acta Bioenerg. 1847, 770–778. ( 10.1016/j.bbabio.2015.01.001) [DOI] [PubMed] [Google Scholar]
- 9.Kleine T, Maier UG, Leister D. 2009. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu. Rev. Plant Biol. 60, 115–138. ( 10.1146/annurev.arplant.043008.092119) [DOI] [PubMed] [Google Scholar]
- 10.Pogson BJ, Albrecht V. 2011. Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155, 1545–1551. ( 10.1104/pp.110.170365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodson JD, Chory J. 2008. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. ( 10.1038/nrg2348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. 2016. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. ( 10.1146/annurev-arplant-043015-111854) [DOI] [PubMed] [Google Scholar]
- 13.Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. 2007. Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. ( 10.1126/science.1140516) [DOI] [PubMed] [Google Scholar]
- 14.Kakizaki T, Matsumura H, Nakayama K, Che F-S, Terauchi R, Inaba T. 2009. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151, 1339–1353. ( 10.1104/pp.109.145987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tadini L, et al. 2016. GUN1 controls accumulation of the plastid ribosomal protein s1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 170, 1817–1830. ( 10.1104/pp.15.02033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paieri F, Tadini L, Manavski N, Kleine T, Ferrari R, Morandini P, Pesaresi P, Meurer J, Leister D. 2018. The DEAD-box RNA helicase RH50 is a 23S-4.5S rRNA maturation factor that functionally overlaps with the plastid signaling factor GUN1. Plant Physiol. 176, 634–648. ( 10.1104/pp.17.01545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P. 2016. GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front. Plant Sci. 7, 1427 ( 10.3389/fpls.2016.01427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kakizaki T, Yazu F, Nakayama K, Ito-Inaba Y, Inaba T. 2012. Plastid signalling under multiple conditions is accompanied by a common defect in RNA editing in plastids. J. Exp. Bot. 63, 251–260. ( 10.1093/jxb/err257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk KJ.. 2012. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 158, 156–189. ( 10.1104/pp.111.188474) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krupinska K, Melonek J, Krause K. 2013. New insights into plastid nucleoid structure and functionality. Planta 237, 653–664. ( 10.1007/s00425-012-1817-5) [DOI] [PubMed] [Google Scholar]
- 21.Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R. 2018. Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol. 176, 2472–2495. ( 10.1104/pp.18.00009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao X, Huang J, Chory J. 2019. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl Acad. Sci. USA 116, 10 162–10 167. ( 10.1073/pnas.1820426116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu GZ, et al. 2019. Control of retrograde signalling by protein import and cytosolic folding stress. Nat. Plants 5, 525–538. ( 10.1038/s41477-019-0415-y) [DOI] [PubMed] [Google Scholar]
- 24.Tadini L, et al. 2019. GUN1 influences the accumulation of NEP-dependent transcripts and chloroplast protein import in Arabidopsis cotyledons upon perturbation of chloroplast protein homeostasis. Plant J. 101, 1198–1220. ( 10.1111/tpj.14585) [DOI] [PubMed] [Google Scholar]
- 25.Hu J, Bogorad L. 1990. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc. Natl Acad. Sci. USA 87, 1531–1535. ( 10.1073/pnas.87.4.1531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfannschmidt T, Blanvillain R, Merendino L, Courtois F, Chevalier F, Liebers M, Grübler B, Hommel E, Lerbs-Mache S. 2015. Plastid RNA polymerases: orchestration of enzymes with different evolutionary origins controls chloroplast biogenesis during the plant life cycle. J. Exp. Bot. 66, 6957–6973. ( 10.1093/jxb/erv415) [DOI] [PubMed] [Google Scholar]
- 27.Tiller K, Link G. 1995. σ-like plastid transcription factors. Methods Mol. Biol. 37, 337–348. ( 10.1385/0-89603-288-4:337) [DOI] [PubMed] [Google Scholar]
- 28.Surzycki SJ. 1969. Genetic functions of the chloroplast of Chlamydomonas reinhardi: effect of rifampin on chloroplast DNA-dependent RNA polymerase. Proc. Natl Acad. Sci. USA 63, 1327–1334. ( 10.1073/pnas.63.4.1327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfalz J, Liere K, Kandlbinder A, Dietz K-J, Oelmüller R. 2006. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18, 176–197. ( 10.1105/tpc.105.036392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia M, Myouga F, Takechi K, Sato H, Nabeshima K, Nagata N, Takio S, Shinozaki K, Takano H. 2008. An Arabidopsis homolog of the bacterial peptidoglycan synthesis enzyme MurE has an essential role in chloroplast development. Plant J. 53, 924–934. ( 10.1111/j.1365-313X.2007.03379.x) [DOI] [PubMed] [Google Scholar]
- 31.Myouga F, et al. 2008. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell Online 20, 3148–3162. ( 10.1105/tpc.108.061341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arsova B, Hoja U, Wimmelbacher M, Greiner E, Üstün Ş, Melzer M, Petersen K, Lein W, Börnke F. 2010. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. Plant Cell 22, 1498–1515. ( 10.1105/tpc.109.071001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Z-P, Yu Q-B, Zhao T-T, Ma Q, Chen G-X, Yang Z-N. 2011. A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol. 157, 1733–1745. ( 10.1104/pp.111.184762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steiner S, Schröter Y, Pfalz J, Pfannschmidt T. 2011. Identification of essential subunits in the plastid-encoded RNA polymerase complex reveals building blocks for proper plastid development. Plant Physiol. 157, 1043–1055. ( 10.1104/pp.111.184515) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon Y, Jung HJ, Kang H, Park Y-I, Lee SH, Pai H-S. 2012. S1 domain-containing STF modulates plastid transcription and chloroplast biogenesis in Nicotiana benthamiana. New Phytol. 193, 349–363. ( 10.1111/j.1469-8137.2011.03941.x) [DOI] [PubMed] [Google Scholar]
- 36.Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T. 2012. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proc. Natl Acad. Sci. USA 109, 7541–7546. ( 10.1073/pnas.1119403109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morden CW, Wolfe KH, DePamphilis CW, Palmer JD. 1991. Plastid translation and transcription genes in a non-photosynthetic plant: intact, missing and pseudo genes. EMBO J. 10, 3281–3288. ( 10.1002/j.1460-2075.1991.tb04892.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hess WR, Prombona A, Fieder B, Subramanian AR, Börner T. 1993. Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J. 12, 563–571. ( 10.1002/j.1460-2075.1993.tb05688.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lerbs-Mache S. 1993. The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes? Proc. Natl Acad. Sci. USA 90, 5509–5513. ( 10.1073/pnas.90.12.5509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hedtke B, Börner T, Weihe A. 1997. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277, 809–811. ( 10.1126/science.277.5327.809) [DOI] [PubMed] [Google Scholar]
- 41.Hedtke B, Börner T, Weihe A. 2000. One RNA polymerase serving two genomes. EMBO Rep. 1, 435–440. ( 10.1093/embo-reports/kvd086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emanuel C, Weihe A, Graner A, Hess WR, Börner T. 2004. Chloroplast development affects expression of phage-type RNA polymerases in barley leaves. Plant J. 38, 460–472. ( 10.1111/j.0960-7412.2004.02060.x) [DOI] [PubMed] [Google Scholar]
- 43.Liere K, Weihe A, Börner T. 2011. The transcription machineries of plant mitochondria and chloroplasts: composition, function, and regulation. J. Plant Physiol. 168, 1345–1360. ( 10.1016/j.jplph.2011.01.005) [DOI] [PubMed] [Google Scholar]
- 44.Yin C, Richter U, Börner T, Weihe A. 2010. Evolution of plant phage-type RNA polymerases: the genome of the basal angiosperm Nuphar advena encodes two mitochondrial and one plastid phage-type RNA polymerases. BMC Evol. Biol. 10, 379 ( 10.1186/1471-2148-10-379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maier UG, Bozarth A, Funk HT, Zauner S, Rensing SA, Schmitz-Linneweber C, Börner T, Tillich M. 2008. Complex chloroplast RNA metabolism: just debugging the genetic programme? BMC Biol. 6, 36 ( 10.1186/1741-7007-6-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tracy RL, Stern DB. 1995. Mitochondrial transcription initiation: promoter structures and RNA polymerases. Curr. Genet. 28, 205–216. ( 10.1007/bf00309779) [DOI] [PubMed] [Google Scholar]
- 47.Hricová A, Quesada V, Micol JL. 2006. The SCABRA3 nuclear gene encodes the plastid RpoTp RNA polymerase, which is required for chloroplast biogenesis and mesophyll cell proliferation in Arabidopsis. Plant Physiol. 141, 942–956. ( 10.1104/pp.106.080069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Demarsy E, Courtois F, Azevedo J, Buhot L, Lerbs-Mache S. 2006. Building up of the plastid transcriptional machinery during germination and early plant development. Plant Physiol. 142, 993–1003. ( 10.1104/pp.106.085043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allorent G, Courtois F, Chevalier F, Lerbs-Mache S. 2013. Plastid gene expression during chloroplast differentiation and dedifferentiation into non-photosynthetic plastids during seed formation. Plant Mol. Biol. 82, 59–70. ( 10.1007/s11103-013-0037-0) [DOI] [PubMed] [Google Scholar]
- 50.Demarsy E, Buhr F, Lambert E, Lerbs-Mache S. 2012. Characterization of the plastid-specific germination and seedling establishment transcriptional programme. J. Exp. Bot. 63, 925–939. ( 10.1093/jxb/err322) [DOI] [PubMed] [Google Scholar]
- 51.Hanaoka M, Kanamaru K, Fujiwara M, Takahashi H, Tanaka K. 2005. Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep. 6, 545–550. ( 10.1038/sj.embor.7400411) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kusumi K, Chono Y, Shimada H, Gotoh E, Tsuyama M, Iba K. 2010. Chloroplast biogenesis during the early stage of leaf development in rice. Plant Biotechnol. 27, 85–90. ( 10.5511/plantbiotechnology.27.85) [DOI] [Google Scholar]
- 53.Emanuel C, von Groll U, Müller M, Börner T, Weihe A.. 2006. Development- and tissue-specific expression of the RpoT gene family of Arabidopsis encoding mitochondrial and plastid RNA polymerases. Planta 223, 998–1009. ( 10.1007/s00425-005-0159-y) [DOI] [PubMed] [Google Scholar]
- 54.Pfannschmidt T, Link G. 1994. Separation of two classes of plastid DNA-dependent RNA polymerases that are differentially expressed in mustard (Sinapis alba L.) seedlings. Plant Mol. Biol. 25, 69–81. ( 10.1007/BF00024199) [DOI] [PubMed] [Google Scholar]
- 55.Pfannschmidt T, Ogrzewalla K, Baginsky S, Sickmann A, Meyer HE, Link G. 2000. The multisubunit chloroplast RNA polymerase A from mustard (Sinapis alba L.). Integration of a prokaryotic core into a larger complex with organelle specific functions. Eur. J. Biochem. 267, 253–261. ( 10.1046/j.1432-1327.2000.00991.x) [DOI] [PubMed] [Google Scholar]
- 56.Qiao J, Ma C, Wimmelbacher M, Börnke F, Luo M. 2011. Two novel proteins, MRL7 and its paralog MRL7-l, have essential but functionally distinct roles in chloroplast development and are involved in plastid gene expression regulation in Arabidopsis. Plant Cell Physiol. 52, 1017–1030. ( 10.1093/pcp/pcr054) [DOI] [PubMed] [Google Scholar]
- 57.Powikrowska M, Khrouchtchova A, Martens HJ, Zygadlo-Nielsen A, Melonek J, Schulz A, Krupinska K, Rodermel S, Jensen PE. 2014. SVR4 (suppressor of variegation 4) and SVR4-like: two proteins with a role in proper organization of the chloroplast genetic machinery. Physiol. Plant. 150, 477–492. ( 10.1111/ppl.12108) [DOI] [PubMed] [Google Scholar]
- 58.Yang EJ, et al. 2019. NCP activates chloroplast transcription by controlling phytochrome-dependent dual nuclear and plastidial switches. Nat. Commun. 10, 1–13. ( 10.1038/s41467-019-10517-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yua Q-B, Ma Q, Kong M-M, Zhao T-T, Zhang X-L, Zhou Q, Huang C, Chong K, Yang Z-N. 2014. AtECB1/MRL7, a thioredoxin-like fold protein with disulfide reductase activity, regulates chloroplast gene expression and chloroplast biogenesis in Arabidopsis thaliana. Mol. Plant 7, 206–217. ( 10.1093/mp/sst092) [DOI] [PubMed] [Google Scholar]
- 60.Yoo CY, Pasoreck EK, Wang H, Cao J, Blaha GM, Weigel D, Chen M. 2019. Phytochrome activates the plastid-encoded RNA polymerase for chloroplast biogenesis via nucleus-to-plastid signaling. Nat. Commun. 10, 1–16. ( 10.1038/s41467-019-10518-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhelyazkova P, Sharma CM, Förstner KU, Liere K, Vogel J, Börner T. 2012. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the plastid-encoded RNA polymerase. Plant Cell 24, 123–136. ( 10.1105/tpc.111.089441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hajdukiewicz PTJ, Allison LA, Maliga P. 1997. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16, 4041–4048. ( 10.1093/emboj/16.13.4041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams-Carrier R, Zoschke R, Belcher S, Pfalz J, Barkan A. 2014. A major role for the plastid-encoded RNA polymerase complex in the expression of plastid transfer RNAs. Plant Physiol. 164, 239–248. ( 10.1104/pp.113.228726) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liere K, Kaden D, Maliga P, Börner T. 2004. Overexpression of phage-type RNA polymerase RpoTp in tobacco demonstrates its role in chloroplast transcription by recognizing a distinct promoter type. Nucleic Acids Res. 32, 1159–1165. ( 10.1093/nar/gkh285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zoschke R, Liere K, Börner T. 2007. From seedling to mature plant: Arabidopsis plastidial genome copy number, RNA accumulation and transcription are differentially regulated during leaf development. Plant J. 50, 710–722. ( 10.1111/j.1365-313X.2007.03084.x) [DOI] [PubMed] [Google Scholar]
- 66.Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. 2013. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73, 1–13. ( 10.1111/tpj.12011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimizu M, Kato H, Ogawa T, Kurachi A, Nakagawa Y, Kobayashi H. 2010. Sigma factor phosphorylation in the photosynthetic control of photosystem stoichiometry. Proc. Natl Acad. Sci. USA 107, 10 760–10 764. ( 10.1073/pnas.0911692107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Türkeri H, Schweer J, Link G. 2012. Phylogenetic and functional features of the plastid transcription kinase cpCK2 from Arabidopsis signify a role of cysteinyl SH-groups in regulatory phosphorylation of plastid sigma factors. FEBS J. 279, 395–409. ( 10.1111/j.1742-4658.2011.08433.x) [DOI] [PubMed] [Google Scholar]
- 69.Coll NS, Danon A, Meurer J, Cho WK, Apel K. 2009. Characterization of soldat8, a suppressor of singlet oxygen-induced cell death in Arabidopsis seedlings. Plant Cell Physiol. 50, 707–718. ( 10.1093/pcp/pcp036) [DOI] [PubMed] [Google Scholar]
- 70.Hakimi MA, Privat I, Valay JG, Lerbs-Mache S. 2000. Evolutionary conservation of C-terminal domains of primary sigma(70)-type transcription factors between plants and bacteria. J. Biol. Chem. 275, 9215–9221. ( 10.1074/jbc.275.13.9215) [DOI] [PubMed] [Google Scholar]
- 71.Mache R, Cottet A, Imberty A, Hakimi AM, Lerbs-Mache S. 2002. The plant sigma factors: structure and phylogenetic origin. Genome Lett. 1, 71–76. ( 10.1166/gl.2002.007) [DOI] [Google Scholar]
- 72.Schmitz-Linneweber C, Small I. 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670. ( 10.1016/j.tplants.2008.10.001) [DOI] [PubMed] [Google Scholar]
- 73.Kindgren P, Kremnev D, Blanco NE, de Dios Barajas López J, Fernández AP, Tellgren-Roth C, Small I, Strand Å. 2012. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signalling to the nucleus. Plant J. 70, 279–291. ( 10.1111/j.1365-313X.2011.04865.x) [DOI] [PubMed] [Google Scholar]
- 74.Díaz MG, Hernández-Verdeja T, Kremnev D, Crawford T, Dubreuil C, Strand Å. 2018. Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat. Commun. 9, 1–12. ( 10.1038/s41467-017-02468-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kravtsov AK, Zubo YO, Yamburenko MV, Kulaeva ON, Kusnetsov VV. 2011. Cytokinin and abscisic acid control plastid gene transcription during barley seedling de-etiolation. Plant Growth Regul. 64, 173–183. ( 10.1007/s10725-010-9553-y) [DOI] [Google Scholar]
- 76.Zubo YO, Yamburenko MV, Kusnetsov VV, Börner T. 2011. Methyl jasmonate, gibberellic acid, and auxin affect transcription and transcript accumulation of chloroplast genes in barley. J. Plant Physiol. 168, 1335–1344. ( 10.1016/j.jplph.2011.01.009) [DOI] [PubMed] [Google Scholar]
- 77.Zubo YO, et al. 2008. Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol. 148, 1082–1093. ( 10.1104/pp.108.122275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamburenko MV, Zubo YO, Börner T. 2015. Abscisic acid affects transcription of chloroplast genes via protein phosphatase 2C-dependent activation of nuclear genes: repression by guanosine-3′-5′-bisdiphosphate and activation by sigma factor 5. Plant J. 82, 1030–1041. ( 10.1111/tpj.12876) [DOI] [PubMed] [Google Scholar]
- 79.Sato M, Takahashi K, Ochiai Y, Hosada T, Ochi K, Nabeta K. 2009. Bacterial alarmone, guanosine 5′-diphosphate 3′-diphosphate (ppGpp), predominantly binds the β′ subunit of plastid-encoded plastid RNA polymerase in chloroplasts. Chembiochem 10, 1227–1233. ( 10.1002/cbic.200800737) [DOI] [PubMed] [Google Scholar]
- 80.Sugliani M, Abdelkefi H, Ke H, Bouveret E, Robaglia C, Caffarri S, Field B. 2016. An ancient bacterial signaling pathway regulates chloroplast function to influence growth and development in Arabidopsis. Plant Cell 28, 661–679. ( 10.1105/tpc.16.00045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Field B. 2018. Green magic: regulation of the chloroplast stress response by (p)ppGpp in plants and algae. J. Exp. Bot. 69, 2797–2807. ( 10.1093/jxb/erx485) [DOI] [PubMed] [Google Scholar]
- 82.Allison LA, Simon LD, Maliga P. 1996. Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J. 15, 2802–2809. ( 10.1002/j.1460-2075.1996.tb00640.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Serino G, Maliga P. 1998. RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol. 117, 1165–1170. ( 10.1104/pp.117.4.1165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chi W, Mao J, Li Q, Ji D, Zou M, Lu C, Zhang L. 2010. Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. Plant J. 64, 14–25. ( 10.1111/j.1365-313X.2010.04304.x) [DOI] [PubMed] [Google Scholar]
- 85.Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. 2008. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol. 147, 573–584. ( 10.1104/pp.108.116194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu Q-B, Zhao T-T, Ye L-S, Cheng L, Wu Y-Q, Huang C, Yang Z-N. 2018. pTAC10, an S1-domain-containing component of the transcriptionally active chromosome complex, is essential for plastid gene expression in Arabidopsis thaliana and is phosphorylated by chloroplast-targeted casein kinase II. Photosynth. Res. 137, 69–83. ( 10.1007/s11120-018-0479-y) [DOI] [PubMed] [Google Scholar]
- 87.Kuo WY, Huang CH, Liu AC, Cheng CP, Li SH, Chang WC, Weiss C, Azem A, Jinn TL. 2013. CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytol. 197, 99–110. ( 10.1111/j.1469-8137.2012.04369.x) [DOI] [PubMed] [Google Scholar]
- 88.Chen M, Galvão RM, Li M, Burger B, Bugea J, Bolado J, Chory J. 2010. Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141, 1230–1240. ( 10.1016/j.cell.2010.05.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gilkerson J, Perez-Ruiz J, Chory J, Callis J. 2012. The plastid-localized pfkB-type carbohydrate kinases FRUCTOKINASE-LIKE 1 and 2 are essential for growth and development of Arabidopsis thaliana. BMC Plant Biol. 12, 102 ( 10.1186/1471-2229-12-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang C, Yu QB, Lv RH, Yin QQ, Chen GY, Xu L, Yang ZN. 2013. The reduced plastid-encoded polymerase-dependent plastid gene expression leads to the delayed greening of the Arabidopsis fln2 mutant. PLoS ONE 8, e73092 ( 10.1371/journal.pone.0073092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu QB, Lu Y, Ma Q, Zhao TT, Huang C, Zhao HF, Zhang XL, Lv RH, Yang ZN. 2012. TAC7, an essential component of the plastid transcriptionally active chromosome complex, interacts with FLN1, TAC10, TAC12 and TAC14 to regulate chloroplast gene expression in Arabidopsis thaliana. Physiol. Plant. 148, 408–421. ( 10.1111/j.1399-3054.2012.01718.x) [DOI] [PubMed] [Google Scholar]
- 92.Zhong L, Zhou W, Wang H, Ding S, Lu Q, Wen X, Peng L, Zhang L, Lu C. 2013. Chloroplast small heat shock protein HSP21 interacts with plastid nucleoid protein pTAC5 and is essential for chloroplast development in Arabidopsis under heat stress. Plant Cell 25, 2925–2943. ( 10.1105/tpc.113.111229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferrari R, et al. 2017. CRP1 Protein: (dis)similarities between Arabidopsis thaliana and Zea mays. Front. Plant Sci. 8, 1–18. ( 10.3389/fpls.2017.00163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tadini L, et al. 2018. Trans-splicing of plastid rps12 transcripts, mediated by AtPPR4, is essential for embryo patterning in Arabidopsis thaliana. Planta 248, 257–265. ( 10.1007/s00425-018-2896-8) [DOI] [PubMed] [Google Scholar]
- 95.Wu H, Zhang L. 2010. The PPR protein PDM1 is involved in the processing of rpoA pre-mRNA in Arabidopsis thaliana. Chinese Sci. Bull. 55, 3485–3489. ( 10.1007/s11434-010-4040-4) [DOI] [Google Scholar]
- 96.Morita-Yamamuro C, Tsutsui T, Tanaka A, Yamaguchi J. 2004. Knock-out of the plastid ribosomal protein S21 causes impaired photosynthesis and sugar-response during germination and seedling development in Arabidopsis thaliana. Plant Cell Physiol. 45, 781–788. ( 10.1093/pcp/pch093) [DOI] [PubMed] [Google Scholar]
- 97.Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D. 2001. Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J. 27, 179–189. ( 10.1046/j.1365-313x.2001.01076.x) [DOI] [PubMed] [Google Scholar]
- 98.Su PH, Li HM. 2010. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell 22, 1516–1531. ( 10.1105/tpc.109.071415) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Koussevitzky S, Stanne TM, Peto CA, Giap T, Sjögren LLE, Zhao Y, Clarke AK, Chory J. 2007. An Arabidopsis thaliana virescent mutant reveals a role for ClpR1 in plastid development. Plant Mol. Biol. 63, 85–96. ( 10.1007/s11103-006-9074-2) [DOI] [PubMed] [Google Scholar]
- 100.Pfalz J, Holtzegel U, Barkan A, Weisheit W, Mittag M, Pfannschmidt T. 2015. ZmpTAC12 binds single-stranded nucleic acids and is essential for accumulation of the plastid-encoded polymerase complex in maize. New Phytol. 206, 1024–1037. ( 10.1111/nph.13248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen M, Chory J. 2011. Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21, 664–671. ( 10.1016/j.tcb.2011.07.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Galvão RM, Li M, Kothadia SM, Haskel JD, Decker PV, Van Buskirk EK, Chen M.. 2012. Photoactivated phytochromes interact with HEMERA and promote its accumulation to establish photomorphogenesis in Arabidopsis. Genes Dev. 26, 1851–1863. ( 10.1101/gad.193219.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qiu Y, et al. 2015. HEMERA couples the proteolysis and transcriptional activity of PHYTOCHROME INTERACTING FACTORs in Arabidopsis photomorphogenesis. Plant Cell 27, 1409–1427. ( 10.1105/tpc.114.136093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tseng CC, Lee CJ, Chung YT, Sung TY, Hsieh MH. 2013. Differential regulation of Arabidopsis plastid gene expression and RNA editing in non-photosynthetic tissues. Plant Mol. Biol. 82, 375–392. ( 10.1007/s11103-013-0069-5) [DOI] [PubMed] [Google Scholar]
- 105.Zeltz P, Hess WR, Neckermann K, Börner T, Kössel H. 1993. Editing of the chloroplast rpoB transcript is independent of chloroplast translation and shows different patterns in barley and maize. EMBO J. 12, 4291–4296. ( 10.1002/j.1460-2075.1993.tb06113.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stern DB, Goldschmidt-Clermont M, Hanson MR. 2010. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61, 125–155. ( 10.1146/annurev-arplant-042809-112242) [DOI] [PubMed] [Google Scholar]
- 107.Zhang Z-W, et al. 2011. Mg-protoporphyrin, haem and sugar signals double cellular total RNA against herbicide and high-light-derived oxidative stress. Plant. Cell Environ. 34, 1031–1042. ( 10.1111/j.1365-3040.2011.02302.x) [DOI] [PubMed] [Google Scholar]
- 108.Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ. 2019. Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol. 179, 18–23. ( 10.1104/pp.18.01047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ruckle ME, DeMarco SM, Larkin RM. 2007. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960. ( 10.1105/tpc.107.054312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA. 2009. GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21, 1109–1128. ( 10.1105/tpc.108.065250) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kobayashi K, et al. 2012. Regulation of root greening by light and auxin/cytokinin signaling in Arabidopsis. Plant Cell 24, 1081–1095. ( 10.1105/tpc.111.092254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kobayashi K, et al. 2013. Photosynthesis of root chloroplasts developed in Arabidopsis lines overexpressing GOLDEN2-LIKE transcription factors. Plant Cell Physiol. 54, 1365–1377. ( 10.1093/pcp/pct086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ramundo S, et al. 2014. Conditional depletion of the Chlamydomonas chloroplast ClpP protease activates nuclear genes involved in autophagy and plastid protein quality control. Plant Cell 26, 2201–2222. ( 10.1105/tpc.114.124842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dogra V, Li M, Singh S, Li M, Kim C. 2019. Oxidative post-translational modification of EXECUTER1 is required for singlet oxygen sensing in plastids. Nat. Commun. 10, 2834 ( 10.1038/s41467-019-10760-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.