Abstract
In recent years, it has become evident that plants perceive, integrate and communicate abiotic stress signals through chloroplasts. During the process of acclimation plastid-derived, retrograde signals control nuclear gene expression in response to developmental and environmental cues leading to complex genetic and metabolic reprogramming to preserve cellular homeostasis under challenging environmental conditions. Upon stress-induced dysfunction of chloroplasts, GENOMES UNCOUPLED (GUN) proteins participate in the repression of PHOTOSYNTHESIS-ASSOCIATED NUCLEAR GENES (PHANGs). Here, we show that the retrograde signal emitted by, or communicated through, GUN-proteins is also essential to induce the accumulation of photoprotective anthocyanin pigments when chloroplast development is attenuated. Comparative whole transcriptome sequencing and genetic analysis reveal GUN1 and GUN5-dependent signals as a source for the regulation of genes involved in anthocyanin biosynthesis. The signal transduction cascade includes well-known transcription factors for the control of anthocyanin biosynthesis, which are deregulated in gun mutants. We propose that regulation of PHANGs and genes contributing to anthocyanin biosynthesis are two, albeit oppositely, co-regulated processes during plastid biogenesis.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles’.
Keywords: retrograde communication, genomes uncoupled, anthocyanin, flavonoid
1. Background
Owing to their sessile lifestyle, plants had to evolve strategies to cope instantaneously with biotic and abiotic stress such as pathogens, changing light intensities, temperature and nutrient availability. On a molecular level, the stress response includes a complex genetic and metabolic reprogramming to preserve cellular homeostasis and to allow growth under challenging conditions. The processes of acclimation to a changing environment are active from the beginning of germination through adolescence. In recent years, it became evident that abiotic stress is perceived, integrated and communicated through chloroplasts, the energy facility of photoautotrophic organisms. Stress responses which are initiated by chloroplast-derived signals feed so-called retrograde signalling pathways that aim at regulating the expression of nucleus-encoded genes. Although they cannot always be strictly separated, retrograde signals can be distinguished between those important during early developmental stages when the proplastid-to-chloroplast transition occurs (biogenic control) and signals from fully developed chloroplasts which are part of the so-called ‘operational control’ [1]. The latter group of signals, encompasses metabolites of biochemical pathways such as the isoprenoid biosynthesis [2–4], nucleotide metabolism [5,6], tetrapyrroles [7,8], the redox-status of plastid components [9–11] as well as reactive oxygen species (ROS) emerging from photosynthesis [12–16], intermediates, end and breakdown products of carotenoid biosynthesis [17,18] and fatty acids (derivatives). Plastid gene expression also contributes to the regulation of nuclear-encoded genes [11,19–22]. The retrograde signalling pathways preserve cellular homeostasis mainly through transcriptional and post-transcriptional regulation of (stress) specific genes in the nucleus [23–26] but also through, for example, degradation of ROS-damaged plastids [27] or adjustment of morphological traits [28]. One of the retrograde signalling pathways acting during plastid biogenesis depends on the plastid-localized GENOMES UNCOUPLED (GUN) proteins, but the identity of the precise signals and function of the contributing components and mechanisms are still limited. In the course of plastid development, nucleus-encoded PHOTOSYNTHESIS-ASSOCIATED NUCLEAR GENES (PHANGs), such as genes encoding components of the photosynthetic electron transfer chain (LIGHT HARVESTING COMPLEX (LHCs) or PLASTOCYANIN (PC)), are highly expressed. When plastid biogenesis is disturbed, PHANGs are repressed [29–31]. GUN1, a plastid-localized pentatricopeptide repeat protein identified as one component in the retrograde communication pathway to regulate PHANG expression, was isolated almost three decades ago [30]. Extensive genetic and biochemical analysis revealed contributions of GUN1 to various aspects of plastid physiology and biochemistry such as plastid gene expression, RNA editing, proteostasis and protein import [32–42]. In addition to GUN1, genetic perturbation of Mg-chelatase (MgCh), a key enzyme of plastid-localized chlorophyll biosynthesis, leads to uncoupling of PHANG expression from the developmental state of plastids. Mutants of GUN4 (a positive regulator of MgCh), GUN5 (the catalytic subunit of MgCh) [43,44] and, although to a lesser extend compared to gun5, also of the I and D subunit of MgCh show a gun phenotype, i.e. reduced repression of PHANGs when chloroplast development is perturbed in Arabidopsis and barley [45,46]. Alteration of haeme metabolism in gun2 (HEME OXYGENASE), gun3 (PHYTOCHROMOBILLIN SYNTHASE), gun6 (FERROCHELATASE I overexpressor line) and potentially also in gun4 and gun5 placed haeme, rather than other tetrapyrrolic intermediates [47,48], in the centre of current models for retrograde signal(s) emerging from tetrapyrrole biosynthesis [7,37,49]. In addition, CRYPTOCHROME 1 (CRY1) and downstream signalling components such as ELONGATED HYPOCOTYL 5 (HY5) [31] or GOLDEN2-LIKE 1 and 2 [50–52] play an important role for the retrograde control of nuclear gene expression.
One of the major traits of plants suffering from adverse environmental conditions, like high light or temperature shifts, is the accumulation of coloured anthocyanin pigments in above-ground tissues. Although dispensable for growth under optimal conditions, these secondary metabolites were shown to protect plants from excessive amounts of light [53–55]. Anthocyanins are end products of a combined pathway of cytosolic phenylpropanoid and flavonoid biosynthesis, which produces a great diversity of polyphenolic plant secondary metabolites [56]. After the conversion of plastid-derived phenylalanine to p-coumaroyl CoA through PHENYLALANINE AMMONIA LYASE (PAL), CINNAMIC ACID-4-HYDROXYLASE (C4H) and 4-COUMAROYL COA LIGASE (4CL), CHALCONE SYNTHASE (CHS) catalyses the initial step of flavonoid biosynthesis (figure 1a). Subsequent reactions provide the precursor(s) for various flavonoid derivatives [57]. The main route for the synthesis of anthocyanins branching from core flavonoid/anthocyanin biosynthesis (FAB) is initiated by DIHYDROFLAVONOL 4-REDUCTASE (DFR) and LEUCOANTHOCYANIDIN DIOXYGENASE (LDOX) [58–60]. The FAB pathway is divided into two major parts which consist of enzymes encoded by ‘early biosynthetic genes' (EBGs), like CHS, and the ‘late biosynthetic genes’ (LBGs), such as DFR and LDOX. Diminished pathway activity in FAB mutants often coincides with the lack of oxidized tannins in the seed coat and a transparent testa (tt) (seed coat) phenotype [58]. In leaves, enzymes and regulators of FAB are targets for post-translational protein modifications [61–63]. EBGs are controlled by a set of partially redundant transcription factors (TF) MYB11, MYB12 and MYB111 [64,65]. By contrast, LBG transcription is mainly regulated by an MBW-complex consisting of MYB, bHLH and WD40 TF. Together with one WD40 variant (TRANSPARENT TESTA GLABRA 1), MYB75 encoding PRODUCTION OF ANTHOCYANIN PIGMENT 1 (PAP1) and the bHLH TFs TT8, GLABRA3 (GL3) and ENHANCER GL3 (EGL3) these major factors concertedly regulate the composition and activity of the MBW-complex [66–68]. The exchange of PAP1 by MYBL2 inactivates the complex [69–72]. EBGs, LBGs and TFs have been shown to be under the control of light signalling pathways involving ultraviolet-, blue and red-light photoreceptors and the downstream component HY5 [73–77]. The role of chloroplast-derived signals (including phytohormones and ROS) and their downstream components on the regulation of the cytosolic FAB pathway have been previously discussed [16,55,78–80]. However, the identity of signal molecules and the underlying mechanisms by which, in particular, plastid biogenesis contributes to the regulation of FAB are still scarce.
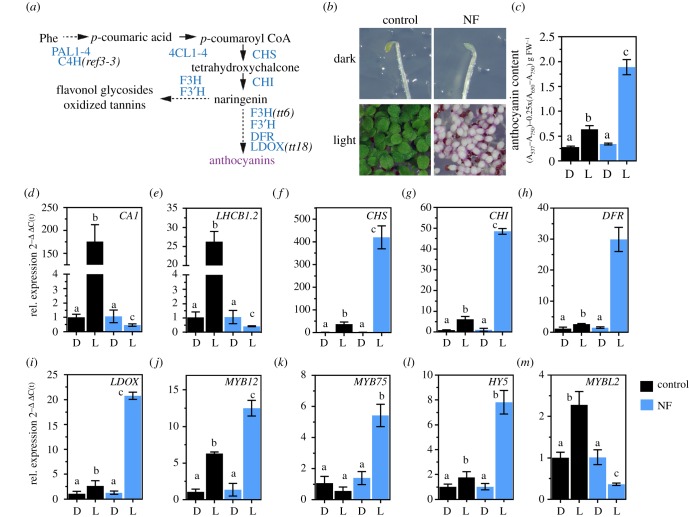
Figure 1.
Induction of flavonoid/anthocyanin biosynthesis (FAB) upon chloroplast dysfunction. (a) Schematic overview of the FAB pathway leading to the formation of polyphenolic compounds. PAL1-4, PHENYLALANINE AMMONIA LYASE; C4H, CINNAMIC ACID 4-HYDROXYLASE; 4CL1-4, 4-COUMAROYL COA LIGASE 1-4; CHS, CHALCONE SYNTHASE; CHI, CHALCONE ISOMERASE; F3H, FLAVANONE 3-HYDROXYLASE; F3'H, FLAVONOID 3′-HYDROXYLASE; DFR, DIHYDROFLAVONOL-4-REDUCTASE; LDOX, LEUCOANTHOCYANIDIN DIOXYGENASE. (b) Representative photograph of etiolated (dark) and continuous light-grown Arabidopsis thaliana Col-0 seedlings germinated in the absence (control) or presence of norflurazon (NF). (c) Quantification of anthocyanins in seedlings shown in (b). Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05). FW, fresh weight. Black (control) and blue (NF). (d–m) qRT-PCR analysis of gene expression for genes encoding PHANGs (CARBONIC ANHYDRASE1 and LIGHT HARVESTING COMPLEX B1.2), enzymes involved in FAB and transcription factors (MYB12, MYB75, HY5, MYBL2). Seedlings were grown as described in (b). D, dark; L, light. Gene expression was calculated relative to the WT etiolated in the absence of NF and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05).
2. Results
(a). Induction of anthocyanin biosynthesis upon block of chloroplast development
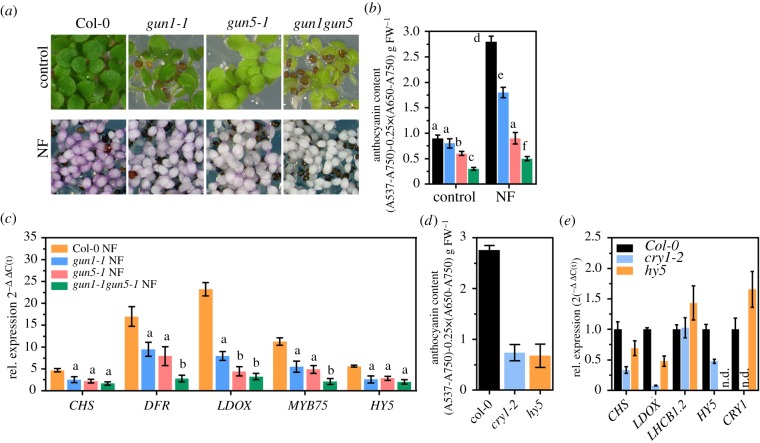
Norflurazon (NF) is an inhibitor of plastid-localized carotenoid biosynthesis which is routinely applied to germinating seedlings to suppress chloroplast development from undifferentiated pro-plastids [30,81]. When germinated in the presence of NF, plant leaves lack carotenoids and do not accumulate chlorophyll and chlorophyll-binding proteins resulting in the development of white cotyledons (figure 1b). Also, and in contrast to control plants grown without NF, light-grown, NF-treated Arabidopsis thaliana wild-type plants (WT, Col-0) showed a purple coloration, which was attributable to the accumulation of anthocyanins (figure 1b,c). While etiolated plants showed the same basal level of anthocyanins in the presence and absence of NF, anthocyanin accumulation was stimulated by three to fourfold in light, when chloroplast development was perturbed (figure 1c). At the same time, two light-inducible representative PHANGs (CA1 and LHCB1.2), were repressed in the light, but not in the dark-grown NF-treated plants (figure 1d,e). Visible accumulation of anthocyanins in the NF-treated tissues resulted from strong induction of EBG and LBG expression. In contrast to the untreated control, transcripts coding for CHS, CHI, DFR and LDOX were enriched by 5–10-fold when plants germinated in the presence of NF (figure 1f–i). Higher expression of FAB was explained by induction of three major TFs (MYB12, MYB75 (PAP1) and HY5) as well as a reduced transcription of the MYBL2 repressor in the light (figure 1j–m).
(b). Involvement of the GUN-dependent signalling pathway
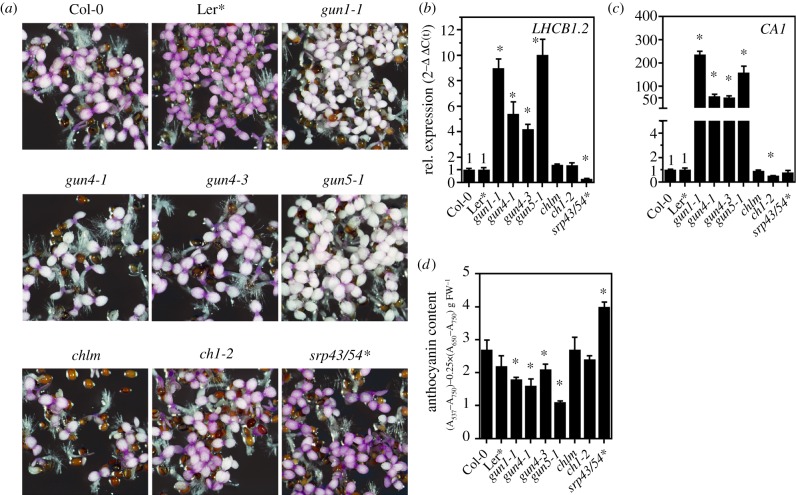
Because PHANGs were shown to be under GUN-dependent retrograde control when chloroplast development is altered, we tested if the same plastid-to-nucleus communication is responsible for the induction of anthocyanin biosynthesis. For this purpose, previously published leaky gun1-1 (electronic supplementary material, figure S4), gun4-1, gun4-3 and gun5-1 were compared with NF-treated WT plants, other mutants of the tetrapyrrole biosynthesis pathway (e.g. chlm (encoding MgP methyltransferase), ch1-2 (chlorophyll a oxygenase)) and for components of the thylakoid protein integration pathway (cpsrp43/54, see the electronic supplementary material, table SII). After growth on NF, cotyledons of all genotypes—except the four gun mutants—showed a strong purple coloration (figure 2a). The gun mutants defective in retrograde signalling, on the other hand, showed less purple coloration and accumulated only 40–70% of the anthocyanin content detected in the WT (figure 2d). Metabolic profiling confirmed the reduced accumulation of the major anthocyanins but not of the flavonoid/anthocyanin precursor phenylalanine in gun1, gun4 and gun5 mutants germinated in the presence of NF (electronic supplementary material, figure S1). As expected, the expression of two representative PHANGs (CA1 and LHCB1.2) was repressed to the WT level in chlm, ch1-2 and srp43/54, but were less downregulated in gun1-1, gun4-1, gun4-3 and gun5-1 mutants (figure 2a,b). We also examined the role of GUN-dependent signalling during acclimation to environmental stress and treated soil-grown WT, gun and non-gun mutant genotypes with high light (HL) for a period of 24 h and determined anthocyanin and chlorophyll (Chl) content (electronic supplementary material, figure S2). While the gun1-1 mutant showed WT-like Chl content, mutant genotypes affected in Chl synthesis (gun4, gun5, chl27 (MgPMME oxidative cyclase), ch1- 2) accumulated 50−70% of the Chl amounts detected in the WT plants at the beginning of the HL shift (electronic supplementary material, figure S2A). During the 24 h of HL treatment, the Chl content decreased by approximately 30% in the WT, and a similar trend was observed for the mutants. HL exposure induced WT-like accumulation of anthocyanins in gun1-1 mutant plants with a gradual increase of anthocyanin pigment content (electronic supplementary material, figure S2B). By contrast, gun4-1, gun4-3, gun5-1 as well as the non-gun genotypes (chl27, ch1-2) showed diminished accumulation of anthocyanins during and at the end, respectively, of the HL treatment (electronic supplementary material, figure S2B). This finding indicates that de-regulation of FAB is a gun specific property during early plastid biogenesis only.
Figure 2.
Induction of FAB depends on genomes uncoupled retrograde signalling. (a) Representative photograph of 5-day-old Arabidopsis thaliana genotypes grown in continuous light in the presence of norflurazon. For the double mutant of cpsrp43/54, the Landsberg errecta ecotype was used as control (*). (b,c) qRT-PCR analysis of gene expression for genes encoding PHANGs (CARBONIC ANHYDRASE1 and LIGHT HARVESTING COMPLEX B1.2). Gene expression was calculated relative to the NF-treated WT as control and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 3). Asterisks indicate statistical significance relative to the WT controls determined by Student's t-test (p < 0.05). (d) Quantification of anthocyanins in seedlings shown in (a). Data are given as mean ± s.d. (n = 3). Asterisks indicate statistical significance relative to the WT controls determined by Student's t-test (p < 0.05). FW, fresh weight.
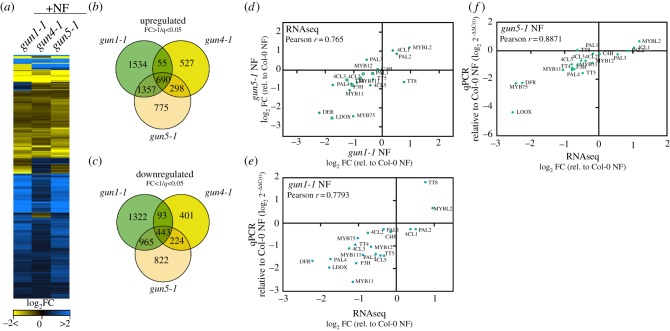
To analyse the anthocyanin deficiency in gun mutants after NF treatment further, we analysed global transcriptome profiles of NF-treated WT, gun1-1, gun4-1 and gun5-1 using RNA-sequencing. Heat map representation of the approximately 9300 significantly deregulated genes (SDGs) in at least one of the gun mutants (0 < log2 fold change > 0, adjusted p-value 0.05, electronic supplementary material, table SI) revealed a strikingly similar transcriptome response of the NF-treated gun mutants compared to the NF-treated WT (figure 3a). Because gun4-1 showed only a weak gun phenotype in comparison to, for example, gun5 (figure 2b,c) the overlap of SDG between gun1 and gun5 was always bigger than the overlap of gun4-1 with the other genotypes (figure 3b,c). Out of the 5056 upregulated transcripts, 690 were significantly deregulated in all gun mutants (figure 3b). Within this cluster, PHANGs were identified (electronic supplementary material, table SI). By contrast, the cluster of commonly downregulated transcripts (443 in all gun, figure 3c) included genes of the phenylpropanoid/FAB explaining the anthocyanin deficiency of the gun mutants. We, therefore, analysed both, the upregulated and downregulated clusters for transcripts whose gene products are involved in or related to FAB. Because gun1-1 and gun5-1 showed an almost similar transcriptome response, we focused on gun1-1 and gun5-1 in further experiments. Expression of mRNAs coding for enzymes involved in anthocyanin biosynthesis (PAL4, 4CL2,3,5, TT4, TT5, F3H, DFR and LDOX) as well as transcripts of major TFs such as MYB11, MYB111, MYB75 were found to be downregulated with the same tendency in gun1-1 and gun5-1 (figure 3d). On the other hand, 4CL1, PAL2 and the transcriptional repressor MYBL2 were stronger expressed in gun1-1 and gun5-1 compared to the NF-treated WT, while the bHLH TF TT8 was repressed in gun5-1, but induced in gun1-1 (figure 3d). We confirmed the differential expression of transcripts identified by RNAseq with quantitative real-time polymerase chain reaction (qRT-PCR) analysis using cDNA obtained from independently grown seedlings and compared the expression of the FAB genes in NF-treated gun relative to NF-treated WT (figure 3e,f). A strong positive correlation between expression values of RNAseq and qRT-PCR confirmed the de-regulation of FAB gene expression in gun mutants when plastid development is suppressed. However, it is important to mention that the expression of LBGs (DFR, LDOX) was always stronger affected than EBGs (CHS, CHI).
Figure 3.
RNA-seq analysis of transcript changes in norflurazon (NF)-treated WT Col-0, gun1-1, gun4-1 and gun5-1 mutants. (a) Heat map representation of significantly deregulated genes (SDGs) for each genotype relative to the WT control (q < 0.05, 0 < log2(fold change) > 0). The RNA-seq experiment was performed, and the results analysed as described in the Materials and methods section. (b,c) Venn diagrams representing the degree of overlap between upregulated (B, fold change > 1) and downregulated (C, fold change < 1) SDGs in the gun mutants. SDGs relative to the WT control were selected based on q < 0.05 (p-value adjusted for the false discovery rate). (d) Comparison of FAB transcript abundance in the gun1-1 and gun5-1 transcriptome datasets. Closed circles denote statistical significance relative to the NF-treated WT (q < 0.05). Abbreviations for genes are explained in figure 1 and in the main text. (f,e) Confirmation of the RNA-seq result by qRT-PCR analysis. Expression of selected transcripts was analysed with cDNA obtained from independent growth of seedlings and was compared to the expression values obtained from the RNA-seq experiments. Gene expression in NF-treated gun mutants was calculated relative to the NF-treated WT and ACTIN2 as reference (ΔΔC(t)method). (d,e) Pearson r > 0.75 indicates a positive correlation between the different datasets and analysis.
(c). Anthocyanins do not control PHANG expression
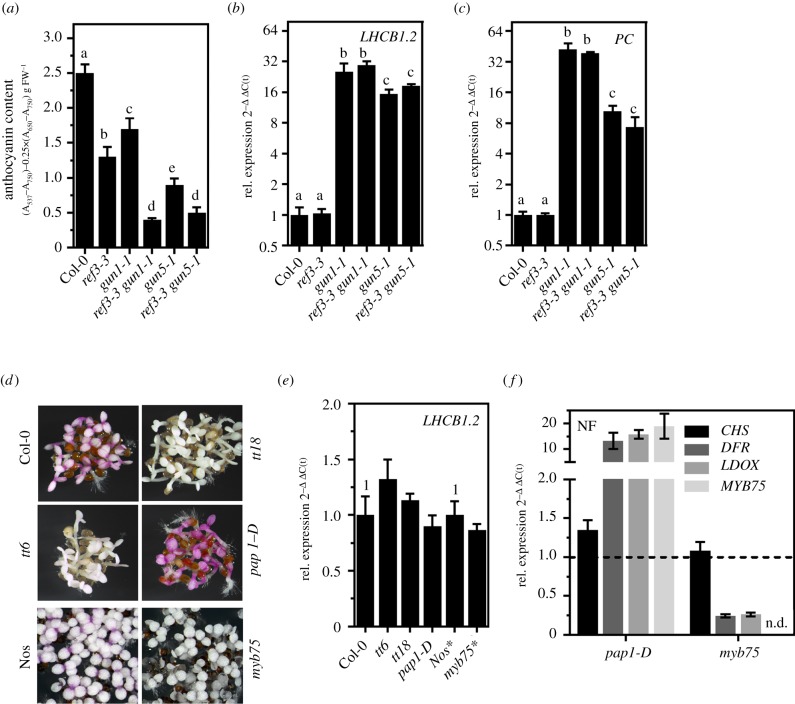
The strong negative correlation between PHANG and FAB expression, i.e. gun phenotype and anthocyanin accumulation, prompted us to hypothesize that anthocyanins might be involved in the GUN-dependent signalling pathway responsible for the repression of PHANGs. Therefore, a mutant of the early steps of the phenylpropanoid biosynthesis preceding FAB was analysed. As expected, the ref3-3 mutant, expressing a point-mutated C4H gene (figure 1a; electronic supplementary material, figure S3A/B), showed reduced accumulation of anthocyanins compared to WT when germinated in the presence of NF (figure 4a). Although anthocyanin accumulation was even more impaired in ref3-3 compared to gun1, PHANG expression was suppressed to WT levels after NF treatment (figure 4b,c), indicating that anthocyanin levels do not determine the transcription rate of PHANGs. This finding was further supported by the fact that diminished anthocyanin accumulation in ref3-3 gun1-1 and ref3-3 gun5-1 compared to the single mutants did not lead to enhanced PHANG expression (figure 4a–c). Likewise, knockout of the genes encoding the FAB enzymes flavanone 3-hydroxylase (F3H), and LDOX designated tt6 and tt18, respectively (electronic supplementary material, figure S3C/D) prevented the accumulation of anthocyanins after NF treatment but not the repression of LHCB1.2 (figure 4d,e). Additionally, overexpression (pap1-D) and knockout of MYB75 (myb75) positively and negatively affected anthocyanin biosynthesis upon NF treatment, respectively, but did not trigger a gun phenotype (figure 4d–f). The lack of a gun phenotype of mutants defective in FAB is in agreement with gun mutant screens which yielded no FAB mutant. In summary, anthocyanins are excluded to control PHANG expression, but the results reveal FAB genes as an additional target of the GUN-dependent retrograde signalling pathway when chloroplast development is perturbed.
Figure 4.
Anthocyanins do not control PHANG expression. (a) Quantification of anthocyanins in seedlings grown for 5 d in continuous light in the presence of NF. WT, gun1-1, gun5-1, ref3-3 (point mutant for C4H) and double mutants were analysed. Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05). FW, fresh weight. (b,c) qRT-PCR analysis of gene expression for genes encoding PHANGs (CARBONIC ANHYDRASE1 and PLASTOCYANIN). Data are given as mean ± s.d. (n = 3). Letters indicate statistical groups determined by Student's t-test (p < 0.05). (d) Representative photograph of 5 d-old WT (Col-0 and Nos), transparent testa (tt) 6, tt18, the dominant pap1-D overexpression line and myb75 mutant grown in continuous light in the presence of norflurazon. (e) qRT-PCR analysis of gene expression for LIGHT HARVESTING COMPLEX B1.2 in genotypes shown in (d). (f) qRT-PCR analysis of CHS, DFR, LDOX and MYB75 expression in pap1-D and myb75 mutant grown in the presence of NF. For the analysis shown in (b,c,e,f) the expression was calculated relative to the NF-treated WT and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 3). The Nossen ecotype was used as control for the myb75 mutant (*).
(d). Activation of anthocyanin biosynthesis depends on GUN1 and GUN5
To obtain detailed insights on how the GUN1 and GUN5 specific signals influence FAB, we compared light-exposed gun1-1, gun5-1, and gun1-1 gun5-1 with the WT in the absence or presence of NF (figure 5). While gun1-1 showed WT-like anthocyanin levels in the control condition without NF, gun5-1 accumulated less anthocyanins, and anthocyanin deficiency was even more pronounced in gun1-1 gun5-1 plants (figure 5a,b). Stimulated expression of FAB genes (figure 1) led to 2-3-fold higher anthocyanin content in the treated WT compared to the control without NF. By contrast, NF-treated gun1-1 and gun5-1 accumulated only 60% and 30%, respectively, of the WT anthocyanin content. To test if knock out of GUN1 exacerbates anthocyanin deficiency in comparison to leaky gun1-1 mutant and gun5-1, we additionally analysed a loss-of-function allele of GUN1 (gun1-102, [38]). While knock out of GUN1 enhanced uncoupling of PHANG expression (electronic supplementary material, figure S4C), the phenotype (electronic supplementary material, figure S4A) and anthocyanin content (electronic supplementary material, figure S4B) of gun1-102 was indistinguishable from that of gun1-1 after NF-treatment. In agreement, expression of FAB pathway genes was reduced to the same extent in both GUN1 alleles (electronic supplementary material, figure S4D). Stimulation of FAB was hardly detectable in NF-treated gun1-1 gun5-1 compared to the control condition, and anthocyanin level did not even reach the level of the untreated WT (figure 5b). In stark contrast to induced FAB genes in the NF-treated WT (figure 1 and figure 5c), NF-treated gun mutants showed diminished induction of FAB genes compared to the control condition (figure 5c). Furthermore, an additive effect of GUN1 and GUN5 mutation was found in gun1-1 gun5-1 relative to the single mutants on the expression, for example, of MYB75 and DFR (figure 5c). Consequently, gun1-1 gun5-1 accumulated fewer anthocyanin pigments than the single mutants (figure 5b). The blue light photoreceptor CRY1 and the downstream acting TF HY5 are essential for light induction of FAB. As reported previously, cry1-2 and hy5 mutants showed a modified expression of CHS and LDOX and, consequently, reduced anthocyanin content compared to WT seedlings when treated with NF (figure 5d,e). Reduced FAB gene expression was more pronounced in cry1-2 seedlings and resembled the FAB transcript level of NF-treated gun mutants. In summary, the results suggest that FAB genes were downregulated by two independent GUN signals and proper activation of FAB depended on a concerted action of both plastid signals and light signalling pathways.
Figure 5.
Induction of FAB involves GUN1, GUN5, and components of light signalling. (a) Representative photograph of 5 d-old Col-0, gun1-1, gun5-1 and gun1-1 gun5-1 double mutant grown in continuous light in the absence (control) or presence of norflurazon (NF). (b) Quantification of anthocyanins in the seedlings shown in (a). Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05). FW, fresh weight. (c) qRT-PCR analysis of gene expression for gene products involved in FAB and transcription factors. Gene expression was calculated relative to the control without NF for each genotype and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 4). Letters indicate statistical significance relative to the NF-treated WT determined by Student's t-test (p < 0.05). (d) Quantification of anthocyanins in Col-0, cry1-2 and hy5 mutants grown in continuous light in the presence of NF. Data are given as mean ± s.d. (n = 4). FW, fresh weight. (e) qRT-PCR analysis of gene expression for genes encoding CHS, LDOX, LHCB1.2, HY5 and CRY1. Gene expression was calculated relative to the NF-treated WT and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 3). n.d., not detectable.
(e). Evidence for a function of MYBL2
Regulation of gene expression for FAB is mediated through the activity of the MBW-complex, which includes the essential MYB component, such as MYB75 (PAP1). When MYBL2 replaces PAP1, the activity of the MBW-complex is diminished. In the presence of NF, an MYBL2 knockout line (mybl2-2) accumulated more anthocyanins compared to the WT (figure 6a), which was explained by transcriptional induction of MYB75 (figure 6c), DFR and LDOX (figure 6d). This result confirmed the important function of MYBL2 in counteracting FAB gene expression and anthocyanin biosynthesis, respectively, in NF-treated Arabidopsis seedlings [69,71]. Interestingly, MYBL2 was expressed to a higher level in the gun mutants compared to WT (figures 3 and 6c). We, therefore, analysed crosses of mybl2-2 with either gun1-1 or gun5-1 for the potential impact of MYBL2 on the downregulated FAB expression (figure 6c). MYBL2 deficiency did not interfere with the gun phenotype observed in the single mutants (figure 6b). Surprisingly, and in stark contrast to mybl2-2, MYB75 transcript levels did not differ between gun single and mybl2-2 gun1-1 or mybl2-2 gun5-1 (figure 6c). DFR and LDOX expression were recovered to a mybl2-2 level in mybl2-2 gun1 but only to WT-like level in mybl2-2 gun5-1 (figure 6d). As a result, stimulated expression of FABs in mybl2 gun double mutants gave rise to higher anthocyanin contents, which increased compared to the gun single mutants but were still lower compared to mybl2-2 (figure 6a).
Figure 6.
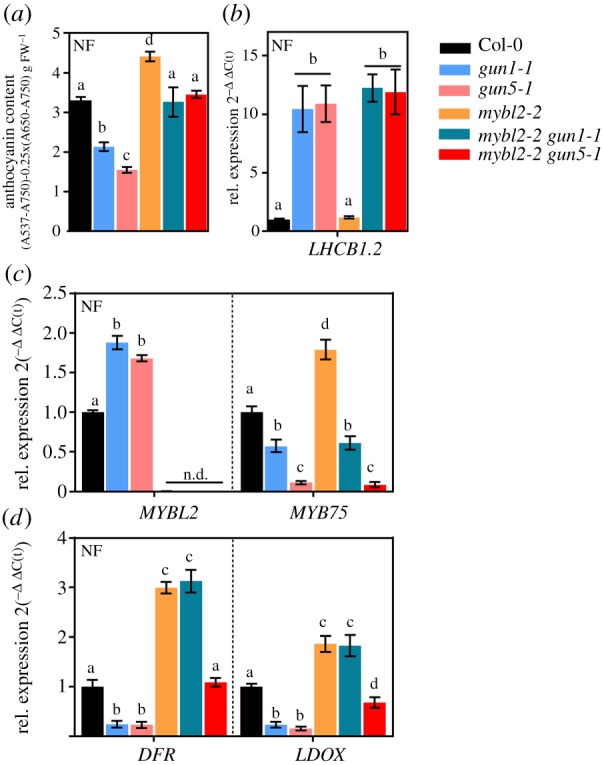
The impact of MYBL2 on FAB in gun mutants. (a) Quantification of anthocyanins in seedlings grown for 5d in continuous light in the presence of NF. WT, gun1-1, gun5-1, mybl2-2 and double mutants of gun and mybl2-2 were analysed. Data are given as mean ± SD (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05). FW, fresh weight. (b–d) qRT-PCR analysis of gene expression for LHCB1.2 (b), MYBL2 and MYB75 (c) and DFR and LDOX (d) in NF-treated seedlings. Gene expression was calculated relative to the NF-treated WT and ACTIN2 as reference (ΔΔC(t)method). Data are given as mean ± s.d. (n = 4). Letters indicate statistical groups determined by Student's t-test (p < 0.05). n.d., not detectable.
3. Discussion
(a). Plastid signals control flavonoid/anthocyanin biosynthesis
Stimulation of FAB and accumulation of end-products of the pathway, like anthocyanins, has been reported for plants suffering from nutrient starvation, changes in light quality and quantity as well as growth temperature (e.g. Kovinich, Kayanja [82]). Essential factors for the transcriptional and post-translational regulation of FAB, such as photoreceptors [75,76] and components of hormone signalling [72,80] have been described. Previous analysis also revealed a function of GUN1 in the regulation of FAB when chloroplast biogenesis is inhibited [16,36,41,83]. However, the knowledge about molecular mechanisms and factors of (gun-dependent) plastid signalling functioning in the regulation of FAB and, in particular, the importance of the tetrapyrrole-dependent signalling pathway is still incomplete. NF is commonly used to prevent the development of chloroplasts from proplastids. By blocking phytoene desaturase, NF diminishes the accumulation of carotenoids, which are essential factors for the stabilization, accumulation and function of the light-absorbing photosystems. When germinated in the presence of NF in the light, plants develop white cotyledons lacking functional chloroplasts (figure 1 and [30], Nott et al. [81]). As a result of the inhibition of plastid biogenesis, transcriptional induction of FAB pathway genes and strong accumulation of anthocyanins was observed (figure 1). Expression of genes encoding positive transcriptional regulators of EBGs (MYB12) and LBGs (MYB75) and of MYB75 itself (i.e. HY5, Shin et al. [74]) was induced upon NF treatment. By contrast, a negative regulator, MYBL2, which concurrently interacts with MYB75 (PAP1) for binding to the MBW-complex [71], was downregulated in NF-treated light-grown seedlings, allowing high expression of FAB pathway genes and anthocyanin accumulation. Diminished expression of MYBL2 probably results from the stimulated expression of HY5 [70,71,84]. Reduced accumulation of anthocyanins in myb75 knockout mutants and elevated contents in a dominant PAP1 (MYB75) overexpression line (figure 4), as well as over-accumulation of anthocyanins in a NF-treated knockout mutant line for the negative regulator MYBL2 (figure 6) also verified their function in regulating the composition and activity of the MBW-complex for the accumulation of anthocyanins when plastid development is suppressed. Stimulated mRNA transcription and activity of CHS, a key enzyme in the FAB pathway, was also observed in mustard seedlings treated with NF and the plastid specific translation inhibitor chloramphenicol, respectively [85]. Also, perturbed plastid development stimulated the strong accumulation of anthocyanins in young seedlings [41,83,86]. In summary, the results emphasize the important role of plastid-derived communication in regulating the expression and activity of the FAB pathway and synthesis of anthocyanins during plastid biogenesis.
(b). Regulation of flavonoid/anthocyanin biosynthesis by two GUN-dependent signals
Simultaneously to the induction of FAB genes, the block in chloroplast development led to suppressed PHANG expression in WT seedlings (figure 1). The repression of PHANGs is known to be mediated, at least partially, by the GUN-dependent retrograde signalling pathway and, consequently, gun mutants accumulated more CA1 and LHCB1.2 mRNAs upon perturbed plastid development (figure 2). At the same time, the modified function of GUN1, GUN4 and GUN5 prevented WT-like accumulation of anthocyanins (figure 2). Anthocyanin deficiency of gun mutants was found to be a result of modulated FAB gene expression (figures 3 and 5), rather than a shortfall in the supply of phenylalanine, the precursor of all phenylpropanoids and their derivatives (electronic supplementary material, figure S1). Even more important, although strongly induced in NF-treated WT seedlings, FAB genes were markedly less induced in gun mutants compared to the control without NF (figure 5). This result is indicative for either a negative signal emitted from gun plastids or a stimulating action of a FAB repressor (e.g. a TF) in gun mutants. Indeed, FAB is negatively affected by MYBL2, which interferes with the composition of the MBW TF complex [69,71]. MYBL2 was significantly upregulated in gun1, and gun5 (figure 6) and knockout of MYBL2 rescued the modified LBG expression of gun1 but only partially that of gun5. These results indicated that MYBL2 contributes to GUN1-dependent control of FAB but plays a minor role in GUN5-mediated signalling. Hence, FAB/FAB regulation might also involve another dominant repressor (or the lack of an activator) in gun5.
The altered accumulation of anthocyanins in gun1-1 seedlings observed here is in agreement with previous studies revealing anthocyanin deficiency of gun1 mutants when plastid development is inhibited [36,41,83]. The recently demonstrated impairment of plastid protein import in gun1 seedlings upon block of chloroplast biogenesis, helped, in addition to other studies, to distinguish GUN1-dependent from the tetrapyrrole biosynthesis-dependent signalling (i.e. GUN4/5) and verifies the synergistic effects of two independent signals emerging from GUN1 and tetrapyrrole biosynthesis, respectively [34,87]. In this context, double mutation of GUN1 and GUN5 exacerbated the anthocyanin deficiency of the single mutants (figure 5). Interestingly, the NF-treated gun1 gun5 mutant accumulated similar amounts of anthocyanins as etiolated WT seedlings and the anthocyanin content did not even reach that of the light-grown WT in the absence of NF. Therefore, it could be assumed that, during the process of plastid biogenesis, regulation of FAB genes exclusively depends on plastid-derived, GUN-mediated signals. The blue light receptor CRY1 and the TF HY5 were shown to be positive regulators of (abiotic stress-induced) anthocyanin biosynthesis [74,75,88,89] and knockout of both factors also led to anthocyanin deficiency after NF-treatment (figure 5 and Ruckle & Larkin [83]). Because anthocyanin content of cry1-2 and hy5 resembled that of gun1 gun5, it is reasonable to speculate that GUN-dependent signalling acts in concert with components of light signalling pathways to regulate FAB [31,83]. Indeed, the two FAB representatives CHS and LDOX were downregulated in hy5 and to a stronger extent in cry1-2 compared to WT, where the expression of these genes was comparable to that in gun1 and gun5. On the other hand, while CRY1 expression was slightly induced in gun1 and gun5 compared to the NF-treated WT, expression of HY5, which is induced in NF-treated WT (figure 1) and in a mutant with defects in plastid protein import [51], was reduced in gun1 and gun5 (figure 5; electronic supplementary material, table SI). Reduced expression of HY5 correlated with modified expression of FAB genes in gun mutants. Furthermore, accelerated proteasomal degradation of HY5 in cry1-2 [90,91] can, at least partially, explain the deregulation of FAB expression in the photoreceptor mutant. Although differently interpreted at that time, previous work already indicated that GUN1-dependent signalling requires CRY1 and HY5 for WT-like accumulation of anthocyanins when plastid development is blocked by lincomycin in blue light [83]. Therefore, a yet unknown connection of GUN1 and tetrapyrrole specific retrograde signals and the action of CRY1 and HY5 regulating the expression of FAB genes is proposed.
We cannot rule out that GUNs are also involved in a putative retrograde signalling cascade regulating FAB genes in high light (i.e. during operational control, Pogson et al. [1]), but our results rather suggest a correlation between the chlorophyll content and the ability to accumulate anthocyanins in various genotypes (electronic supplementary material, figure S2). While CRY1 and HY5 are essential, lack of GUN1 did not interfere with the accumulation of anthocyanins when plants germinated in high light [83]. In conclusion, photosynthesis or a connected process might serve as a signal for or is involved in FAB induction under increased growth light intensities, which is in agreement with previous assumptions [55,78,79]. It is also not excluded that alteration of plastid tetrapyrrole biosynthesis interferes with light signalling pathways needed to perceive and integrate ultraviolet, blue or red light stimuli in order to induce anthocyanin biosynthesis.
(c). Connection of flavonoid/anthocyanin biosynthesis and PHANG
Although reduced expression of PHANGs correlated with a stimulated transcription of FAB genes in NF-treated WT (figure 1) and gun mutants show higher expression of PHANGs but reduced induction of FAB genes after NF treatment (figures 2 and 3), anthocyanins are probably not involved in the retrograde signalling-mediated control of PHANG expression. Firstly, lack of anthocyanins in FAB pathway mutants (figure 4) [92], as well as cry1-2 and hy5 (figure 5), did not prevent repression of PHANGs and, secondly, stronger accumulation of anthocyanins in the dominant pap1-D mutant resulted in WT-like repression of PHANGs when chloroplast development was perturbed (figure 4). In addition, even diminished accumulation of anthocyanins in ref3-3 gun double mutants compared to the single mutants, did not further uncouple PHANG expression from the developmental state of the plastids (figure 4). Thus, we propose that PHANGs and FABs are inversely co-regulated sets of nuclear-encoded genes whose regulation depends on GUN-proteins during biogenesis of plastids. The proposed co-regulation depends on TFs recognizing common regulatory motives in the promoter of their target genes. Indeed, promotor regions of classical targets of GUN-dependent signalling contain the G-box motif (CACGTA) needed for light-induced regulation of PHANGs [93,94]. Previous reports also revealed that FAB genes, including CHS, DFR, LDOX and MYB75, are controlled through TFs binding to G-Box and ACE elements. One of the most important TFs for light-dependent stimulation of gene products involved in FAB is HY5 binding to the regulatory elements [74,95–97] supporting our and the results of other studies (figure 5; Ruckle & Larkin [83]). HY5 has also been shown to act as a negative factor in GUN-dependent regulation of PHANGs when plastid development is arrested [31,98]. Based on the previous findings, the NF-induced expression in WT (figure 1) and reduced expression of HY5 in gun mutants after NF-treatment (figure 5), the assumed co-regulation of PHANGs and FABs by a GUN signalling could be achieved through HY5.
In summary, our data reveal and emphasize the important function of GUN-dependent retrograde communication for the induction of FAB gene expression and accumulation of protective anthocyanin pigments during plastid biogenesis, one of the most sensitive processes in the course of plant development and establishment of photoautotrophy. Repression of PHANGs and induction of FABs through the same signalling pathway allows proper acclimation when plants suffer from adverse environmental conditions. Future research has to reveal how the import of plastid proteins (GUN1 specific), tetrapyrrole biosynthesis as well as known and unknown factors of light signalling cascades are connected to the regulation of secondary metabolism during plastid development.
4. Methods
(a). Plant material and growth conditions
Unless otherwise stated, Arabidopsis seeds (genotypes are listed in the electronic supplementary material, table SII) were surface-sterilized and plated on half-strength Murashige-Skoog (MS) medium (4.4 g l−1 MS medium, 0.5 g l−1 2-(N-morpholino)ethanesulfonic acid, 1% agar, pH 5.7). After stratification for 2 days at 4°C, plates were exposed to continuous light (100 µmol photons m−2 s−1) at 20–22°C. For etiolation experiments, plates were illuminated for 2 h immediately after stratification and then transferred to darkness. In each case, samples were harvested 5 days later. High light experiments were performed with soil-grown plants. After two weeks of growth at short day (10 h light) plants were transferred to continuous light for 2 days. Plants were then treated with high light (350 µmol photons m−2 s−1) for the indicated time points. Mutation, the presence of T-DNA or knockout of genes in the mutants was confirmed with PCR-based methods using genomic DNA or cDNA and primers listed in the electronic supplementary material, table SIII.
(b). Treatments
All treatments of seedlings were carried out by diluting the appropriate compounds in the growth medium. NF was added (from a 5 mM stock solution in 100% acetone; Sigma-Aldrich) to a final concentration (f.c.) of 5 µM.
(c). RNA isolation, cDNA synthesis and quantitative real-time polymerase chain reaction analysis
For qPCR analysis at least four samples, each consisting of about 50 pooled seedlings, were harvested, frozen in liquid nitrogen and crushed in a steel-ball mill. Samples were stored at −80°C until further use. RNA was isolated from crushed plant material using the citric acid method [99]. Quality and quantity of RNA were checked on 1.2% TBE agarose gels. After DNaseI treatment (Thermo Scientific) aliquots of RNA (1–2 µg) were transcribed into cDNA using Moloney Murine Leukemia Virus (M-MuLV) reverse transcriptase (Thermo Scientific) and oligo dT(18) primer. All steps were performed essentially according to the manufacturer's protocol. qPCR analysis was carried out in a CFX96-C1000 96-well plate thermocycler (Bio-Rad) using SYBR green dye (Bio-Tool). The primers used are listed in the electronic supplementary material, table SIII. Calculation of gene expression was performed with the Bio-Rad CFX-Manager Software 1.6 using the ΔΔC(t)method [100] and ACTIN2 as reference.
(d). RNA sequencing
RNA sequencing (RNA-seq) analyses were performed with total RNA from 5-day-old seedlings grown in the presence of 5 µM NF. RNA was extracted as described above. After DNase treatment, RNA was purified using phenol/chloroform/isoamyl alcohol precipitation protocol. After washing with 70% ethanol (v/v), RNA was dried and resuspended in RNase-free water. RNA-seq was performed by Novogene (China). mRNA was enriched using oligo(dT) beads. mRNA was fragmented randomly by adding fragmentation buffer, cDNA was then synthesized using the fragmented mRNA as template and random hexamer primers. After ligation and sequencing-adaptor ligation, the double-stranded cDNA library was subjected to size selection and PCR enrichment. Paired-end read libraries were sequenced on the Illumina Platform HiSeq4000. The RNA-seq experiment was performed once with three biological replicates, each consisting of a pool of about 50 seedlings.
(e). RNA sequencing data analysis
Base-calling was performed using bcl2fastq (v. 2.16.0.10). Differential expression analysis was carried out using a local installation of the GALAXY platform [101]. Sequenced reads were processed with Trimmomatic (v. 0.36.2) [102] to eliminate adapters and low-quality sequences. If at least one of the reads of a pair had a length of less than 15 bp after trimming, both reads were discarded. Remaining reads were mapped to the Arabidopsis genome (TAIR 10) using the TopHat2 (v. 2.1.1) gapped-read mapper [103]. After mapping, we used Cufflinks (v. 2.2.1.2) to assemble our transcriptome using the Araport11 transcriptome annotation as reference for each library independently. The assemblies were merged using Cuffmerge (v. 2.2.1.2). Differential expression analysis was performed using Cuffdiff (v. 2.2.1.5) [103]. Tools were run with default parameters, except that the maximum intron length was set to 3000 bp. Venn diagrams were constructed using the VENNY 2.1 web interface (http://bioinfogp.cnb.csic.es/tools/venny/). Heat maps were built using Perseus software [104]. Gene ontology term (GoT) enrichment analysis was performed with the GoTermFinder tool [105].
(f). Quantification of anthocyanins
Anthocyanins were quantified according to the method described previously [65]. Briefly, seedlings were harvested, frozen in liquid nitrogen and homogenized using a ball-mill. Then, the frozen powder was resuspended in 1 ml of extraction buffer (18% 1-propanol and 1% HCl in water) and incubated for at least 2 h at room temperature in darkness. After centrifugation for 15 min (maximum speed, 4°C) absorbance (A) of the supernatant at 537 nm, 650 nm and 750 nm were determined. Anthocyanin content was calculated using the following formula: (A537-A750)-0.25×(A650-A750)/g fresh weight or dry weight, respectively.
(g). Metabolite profiling
Profiling of anthocyanins and flavonoids was carried out exactly as described previously [106].
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We acknowledge the gifts of mutant seeds by Wout Boerjan and Ruben Vanholme (University Gent, Belgium), Steven J. Rothstein (University of Guelph, Canada), Clint Chapple (Purdue University, West Lafayette, Ind., USA) and Tatjana Kleine and Dario Leister (Ludwig-Maximilians-Universität Munich). We also thank José M. Muino (Humboldt University Berlin) for the support with the RNA-seq analysis and Leonardo Perez de Souza (Max Planck Institute for Molecular Plant Physiology, Potsdam-Golm) for supportive analysis of GC/LC-MS data.
Data accessibility
RNA-seq data acquired during this study are publicly available from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the accession no. GSE104868.
Authors' contributions
A.S.R. conceived and designed the study, performed the experiments and wrote the article. T.T. performed mass spectrometric analysis. A.S.R., T.T., A.R.F. and B.G. revised the manuscript. All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
The authors declare no competing interests.
Funding
The work was financed by a grant to B.G. (Subproject C04 of TRR 175 ‘The Green Hub’, DFG, Germany) and to A.R.F. (subproject B04 of TRR 175 ‘The Green Hub’, DFG, Germany). Work in the laboratory of A.S.R. is supported by a startup grant from the DFG funded TRR 175 ‘The Green Hub’.
References
- 1.Pogson BJ, Woo NS, Forster B, Small ID. 2008. Plastid signaling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. ( 10.1016/j.tplants.2008.08.008) [DOI] [PubMed] [Google Scholar]
- 2.Xiao Y, et al. 2012. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149, 1525–1535. ( 10.1016/j.cell.2012.04.038) [DOI] [PubMed] [Google Scholar]
- 3.Benn G, Bjornson M, Ke H, De Souza A, Balmond EI, Shaw JT, Dehesh K.. 2016. Plastidial metabolite MEcPP induces a transcriptionally centered stress-response hub via the transcription factor CAMTA3. Proc. Natl Acad. Sci. USA 113, 8855–8860. ( 10.1073/pnas.1602582113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang J, et al. 2018. Interplay of the two ancient metabolites auxin and MEcPP regulates adaptive growth. Nat. Commun. 9, 2262 ( 10.1038/s41467-018-04708-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Estavillo GM, et al. 2011. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012. ( 10.1105/tpc.111.091033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang X, Zhao G, Zhang S, Li Y, Gu H, Li Y, Zhao Q, Qi Y. 2019. Chloroplast-to-nucleus signaling regulates MicroRNA biogenesis in Arabidopsis. Dev. Cell 48, 371–382. ( 10.1016/j.devcel.2018.11.046) [DOI] [PubMed] [Google Scholar]
- 7.Woodson JD, Perez-Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903. ( 10.1016/j.cub.2011.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duanmu D, et al. 2013. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl Acad. Sci. USA 110, 3621–3626. ( 10.1073/pnas.1222375110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dietzel L, et al. 2015. Identification of early nuclear target genes of plastidial redox signals that trigger the long-term response of Arabidopsis to light quality shifts. Mol. Plant 8, 1237–1252. ( 10.1016/j.molp.2015.03.004) [DOI] [PubMed] [Google Scholar]
- 10.Dietz KJ, Turkan I, Krieger-Liszkay A. 2016. Redox- and reactive oxygen species-dependent signaling into and out of the photosynthesizing chloroplast. Plant Physiol. 171, 1541–1550. ( 10.1104/pp.16.00375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz MG, Hernandez-Verdeja T, Kremnev D, Crawford T, Dubreuil C, Strand A. 2018. Redox regulation of PEP activity during seedling establishment in Arabidopsis thaliana. Nat. Commun. 9, 50 ( 10.1038/s41467-017-02468-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee KP, Kim C, Landgraf F, Apel K. 2007. EXECUTER1- and EXECUTER2-dependent transfer of stress-related signals from the plastid to the nucleus of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 104, 10 270–10 275. ( 10.1073/pnas.0702061104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Page MT, McCormac AC, Smith AG, Terry MJ. 2016. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol. 213, 1168–1180. ( 10.1111/nph.14223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim C, Meskauskiene R, Zhang S, Lee KP, Lakshmanan Ashok M, Blajecka K, Herrfurth C, Feussner I, Apel K. 2012. Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039. ( 10.1105/tpc.112.100479) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Exposito-Rodriguez M, Laissue PP, Yvon-Durocher G, Smirnoff N, Mullineaux P. M. 2017. Photosynthesis-dependent H2O2 transfer from chloroplasts to nuclei provides a high-light signaling mechanism. Nat. Commun. 8, 49 ( 10.1038/s41467-017-00074-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willems P, Mhamdi A, Stael S, Storme V, Kerchev P, Noctor G, Gevaert K, Van Breusegem F.. 2016. The ROS wheel: refining ROS transcriptional footprints. Plant Physiol. 171, 1720–1733. ( 10.1104/pp.16.00420) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avendano-Vazquez AO, et al. 2014. An uncharacterized apocarotenoid-derived signal generated in ζ-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26, 2524–2537. ( 10.1105/tpc.114.123349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylides C, Havaux M. 2012. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl Acad. Sci. USA 109, 5535–5540. ( 10.1073/pnas.1115982109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grubler B, et al. 2017. Light and plastid signals regulate different sets of genes in the albino mutant Pap7–1. Plant Physiol. 175, 1203–1219. ( 10.1104/pp.17.00982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kindgren P, Kremnev D, Blanco NE, de Dios Barajas Lopez J, Fernandez AP, Tellgren-Roth C, Kleine T, Small I, Strand A. 2012. The plastid redox insensitive 2 mutant of Arabidopsis is impaired in PEP activity and high light-dependent plastid redox signaling to the nucleus. Plant J. 70, 279–291. ( 10.1111/j.1365-313X.2011.04865.x) [DOI] [PubMed] [Google Scholar]
- 21.Kleine T, Voigt C, Leister D. 2009. Plastid signaling to the nucleus: messengers still lost in the mists? Trends Genet. 25, 185–192. ( 10.1016/j.tig.2009.02.004) [DOI] [PubMed] [Google Scholar]
- 22.Hernandez-Verdeja T, Strand A. 2018. Retrograde signals navigate the path to chloroplast development. Plant Physiol. 176, 967–976. ( 10.1104/pp.17.01299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Souza A, Wang JZ, Dehesh K.. 2017. Retrograde signals: integrators of interorganellar communication and orchestrators of plant development. Annu. Rev. Plant Biol. 68, 85–108. ( 10.1146/annurev-arplant-042916-041007) [DOI] [PubMed] [Google Scholar]
- 24.Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ. 2015. Learning the languages of the chloroplast: retrograde signaling and beyond. Annu. Rev. Plant Biol. 67, 25–53. ( 10.1146/annurev-arplant-043015-111854) [DOI] [PubMed] [Google Scholar]
- 25.Kleine T, Leister D. 2016. Retrograde signaling: organelles go networking. Biochim. Biophys. Acta 1857, 1313–1325. ( 10.1016/j.bbabio.2016.03.017) [DOI] [PubMed] [Google Scholar]
- 26.Petrillo E, et al. 2014. A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344, 427–430. ( 10.1126/science.1250322) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodson JD, Joens MS, Sinson AB, Gilkerson J, Salome PA, Weigel D, Fitzpatrick JA, Chory J. 2015. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 350, 450–454. ( 10.1126/science.aac7444) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Alcaide M, Llamas E, Gomez-Cadenas A, Nagatani A, Martinez-Garcia J. F, Rodriguez-Concepcion M. 2019. Chloroplasts modulate elongation responses to canopy shade by retrograde pathways involving hy5 and abscisic acid. Plant Cell 31, 384–398. ( 10.1105/tpc.18.00617) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hess WR, Muller A, Nagy F, Borner T. 1994. Ribosome-deficient plastids affect transcription of light-induced nuclear genes: genetic evidence for a plastid-derived signal. Mol. Gen. Genet. 242, 305–312. ( 10.1007/BF00280420) [DOI] [PubMed] [Google Scholar]
- 30.Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. ( 10.1016/0092-8674(93)90459-4) [DOI] [PubMed] [Google Scholar]
- 31.Ruckle ME, DeMarco SM, Larkin RM. 2007. Plastid signals remodel light signaling networks and are essential for efficient chloroplast biogenesis in Arabidopsis. Plant Cell 19, 3944–3960. ( 10.1105/tpc.107.054312) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao X, Huang J, Chory J. 2019. GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc. Natl Acad. Sci. USA 116, 201820426 ( 10.1073/pnas.1820426116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu GZ, Meyer EH, Wu S, Bock R. 2019. Extensive post-transcriptional regulation of nuclear gene expression by plastid retrograde signals. Plant Physiol. 180, 421 ( 10.1104/pp.19.00421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu GZ, et al. 2019. Control of retrograde signaling by protein import and cytosolic folding stress. Nat Plants 5, 525–538. ( 10.1038/s41477-019-0415-y) [DOI] [PubMed] [Google Scholar]
- 35.Woodson JD, Perez-Ruiz JM, Schmitz RJ, Ecker JR, Chory J. 2012. Sigma factor-mediated plastid retrograde signals control nuclear gene expression. Plant J. 73, 1–13. ( 10.1111/tpj.12011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voigt C, Oster U, Bornke F, Jahns P, Dietz KJ, Leister D, Kleine T. 2010. In-depth analysis of the distinctive effects of norflurazon implies that tetrapyrrole biosynthesis, organellar gene expression and ABA cooperate in the GUN-type of plastid signaling. Physiol. Plant. 138, 503–519. ( 10.1111/j.1399-3054.2009.01343.x) [DOI] [PubMed] [Google Scholar]
- 37.Terry MJ, Smith AG. 2013. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4, 14 ( 10.3389/fpls.2013.00014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tadini L, et al. 2016. GUN1 controls accumulation of the plastid ribosomal protein S1 at the protein level and interacts with proteins involved in plastid protein homeostasis. Plant Physiol. 170, 1817–1830. ( 10.1104/pp.15.02033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pesaresi P, Kim C. 2019. Current understanding of GUN1: a key mediator involved in biogenic retrograde signaling. Plant Cell Rep. 38, 819–823. ( 10.1007/s00299-019-02383-4) [DOI] [PubMed] [Google Scholar]
- 40.Marino G, Naranjo B, Wang J, Penzler JF, Kleine T, Leister D. 2019. Relationship of GUN1 to FUG1 in chloroplast protein homeostasis. Plant J. 99, 521–555. ( 10.1111/tpj.14342) [DOI] [PubMed] [Google Scholar]
- 41.Cottage A, Mott EK, Kempster JA, Gray J. C. 2010. The Arabidopsis plastid-signaling mutant gun1 (genomes uncoupled1) shows altered sensitivity to sucrose and abscisic acid and alterations in early seedling development. J. Exp. Bot. 61, 3773–3786. ( 10.1093/jxb/erq186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colombo M, Tadini L, Peracchio C, Ferrari R, Pesaresi P. 2016. GUN1, a jack-of-all-trades in chloroplast protein homeostasis and signaling. Front. Plant Sci. 7, 1427 ( 10.3389/fpls.2016.01427) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peter E, Grimm B. 2009. GUN4 is required for posttranslational control of plant tetrapyrrole biosynthesis. Mol. Plant 2, 1198–1210. ( 10.1093/mp/ssp072) [DOI] [PubMed] [Google Scholar]
- 44.Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. ( 10.1126/science.1079978) [DOI] [PubMed] [Google Scholar]
- 45.Huang YS, Li HM. 2009. Arabidopsis CHLI2 can substitute for CHLI1. Plant Physiol. 150, 636–645. ( 10.1104/pp.109.135368) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gadjieva R, Axelsson E, Olsson U, Hansson M. 2005. Analysis of gun phenotype in barley magnesium chelatase and Mg-protoporphyrin IX monomethyl ester cyclase mutants. Plant Physiol. Biochem. 43, 901–908. ( 10.1016/j.plaphy.2005.08.003) [DOI] [PubMed] [Google Scholar]
- 47.Mochizuki N, Tanaka R, Tanaka A, Masuda T, Nagatani A. 2008. The steady-state level of Mg-protoporphyrin IX is not a determinant of plastid-to-nucleus signaling in Arabidopsis. Proc. Natl Acad. Sci. USA 105, 15 184–15 189. ( 10.1073/pnas.0803245105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moulin M, McCormac AC, Terry MJ, Smith AG. 2008. Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl Acad. Sci. USA 105, 15 178–15 183. ( 10.1073/pnas.0803054105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larkin R. M. 2016. Tetrapyrrole signaling in plants. Front. Plant Sci. 7, 1586 ( 10.3389/fpls.2016.01586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leister D, Kleine T. 2016. Definition of a core module for the nuclear retrograde response to altered organellar gene expression identifies GLK overexpressors as gun mutants. Physiol. Plant. 157, 297–309. ( 10.1111/ppl.12431) [DOI] [PubMed] [Google Scholar]
- 51.Kakizaki T, Matsumura H, Nakayama K, Che FS, Terauchi R, Inaba T. 2009. Coordination of plastid protein import and nuclear gene expression by plastid-to-nucleus retrograde signaling. Plant Physiol. 151, 1339–1353. ( 10.1104/pp.109.145987) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ. 2019. Plastid-to-nucleus retrograde signaling during chloroplast biogenesis does not require ABI4. Plant Physiol. 179, 18–23. ( 10.1104/pp.18.01047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gould KS, Jay-Allemand C, Logan BA, Baissac Y, Bidel LPR. 2018. When are foliar anthocyanins useful to plants? Re-evaluation of the photoprotection hypothesis using Arabidopsis thaliana mutants that differ in anthocyanin accumulation. Environ. Exp. Bot. 154, 11–22. ( 10.1016/j.envexpbot.2018.02.006) [DOI] [Google Scholar]
- 54.Gould KS. 2004. Nature's Swiss army knife: the diverse protective roles of anthocyanins in leaves. J. Biomed. Biotechnol. 2004, 314–320. ( 10.1155/S1110724304406147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Z, Mahmood K, Rothstein SJ. 2017. ROS Induces anthocyanin production via late biosynthetic genes and anthocyanin deficiency confers the hypersensitivity to ROS-generating stresses in Arabidopsis. Plant Cell Physiol. 58, 1364–1377. ( 10.1093/pcp/pcx073) [DOI] [PubMed] [Google Scholar]
- 56.Vogt T. 2010. Phenylpropanoid biosynthesis. Mol. Plant 3, 2–20. ( 10.1093/mp/ssp106) [DOI] [PubMed] [Google Scholar]
- 57.Fraser CM, Chapple C. 2011. The phenylpropanoid pathway in Arabidopsis. Arabidopsis Book/Am. Soc. Plant Biol. 9, e0152 ( 10.1199/tab.0152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Appelhagen I, Thiedig K, Nordholt N, Schmidt N, Huep G, Sagasser M, Weisshaar B. 2014. Update on transparent testa mutants from Arabidopsis thaliana: characterisation of new alleles from an isogenic collection. Planta 240, 955–970. ( 10.1007/s00425-014-2088-0) [DOI] [PubMed] [Google Scholar]
- 59.Passeri V, Koes R, Quattrocchio F. M. 2016. New challenges for the design of high value plant products: stabilization of anthocyanins in plant vacuoles. Front. Plant Sci. 7, 153 ( 10.3389/fpls.2016.00153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaakola L. 2013. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 18, 477–483. ( 10.1016/j.tplants.2013.06.003) [DOI] [PubMed] [Google Scholar]
- 61.Maier A, et al. 2013. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 74, 638–651. ( 10.1111/tpj.12153) [DOI] [PubMed] [Google Scholar]
- 62.Zhang X, Abrahan C, Colquhoun T. A, Liu C. J. 2017. A proteolytic regulator controlling chalcone synthase stability and flavonoid biosynthesis in Arabidopsis. Plant Cell 29, 1157–1174. ( 10.1105/tpc.16.00855) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang X, Gou M, Liu CJ. 2013. Arabidopsis Kelch repeat F-box proteins regulate phenylpropanoid biosynthesis via controlling the turnover of phenylalanine ammonia-lyase. Plant Cell 25, 4994–5010. ( 10.1105/tpc.113.119644) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. 2007. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. ( 10.1111/j.1365-313X.2007.03078.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lotkowska ME, Tohge T, Fernie AR, Xue GP, Balazadeh S, Mueller-Roeber B. 2015. The Arabidopsis transcription factor MYB112 promotes anthocyanin formation during salinity and under high light stress. Plant Physiol. 169, 1862–1880. ( 10.1104/pp.15.00605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shi MZ, Xie DY. 2014. Biosynthesis and metabolic engineering of anthocyanins in Arabidopsis thaliana. Recent Pat. Biotechnol. 8, 47–60. ( 10.2174/1872208307666131218123538) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lloyd A, Brockman A, Aguirre L, Campbell A, Bean A, Cantero A, Gonzalez A. 2017. Advances in the MYB-bHLH-WD Repeat (MBW) pigment regulatory model: addition of a wrky factor and co-option of an anthocyanin myb for betalain regulation. Plant Cell Physiol. 58, 1431–1441. ( 10.1093/pcp/pcx075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang F, Gonzalez A, Zhao M, Payne C. T, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869. ( 10.1242/dev.00681) [DOI] [PubMed] [Google Scholar]
- 69.Dubos C, et al. 2008. MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55, 940–953. ( 10.1111/j.1365-313X.2008.03564.x) [DOI] [PubMed] [Google Scholar]
- 70.Nguyen NH, Jeong CY, Kang GH, Yoo SD, Hong S. W, Lee H. 2015. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 84, 1192–1205. ( 10.1111/tpj.13077) [DOI] [PubMed] [Google Scholar]
- 71.Wang Y, Wang Y, Song Z, Zhang H. 2016. Repression of MYBL2 by both microRNA858a and HY5 leads to the activation of anthocyanin biosynthetic pathway in Arabidopsis. Mol. Plant 9, 1395–1405. ( 10.1016/j.molp.2016.07.003) [DOI] [PubMed] [Google Scholar]
- 72.Xie Y, Tan H, Ma Z, Huang J. 2016. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol. Plant 9, 711–721. ( 10.1016/j.molp.2016.01.014) [DOI] [PubMed] [Google Scholar]
- 73.Stracke R, Favory JJ, Gruber H, Bartelniewoehner L, Bartels S, Binkert M, Funk M, Weisshaar B, Ulm R. 2010. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 33, 88–103. ( 10.1111/j.1365-3040.2009.02061.x) [DOI] [PubMed] [Google Scholar]
- 74.Shin DH, Choi M, Kim K, Bang G, Cho M, Choi SB, Choi G, Park YI. 2013. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. ( 10.1016/j.febslet.2013.03.037) [DOI] [PubMed] [Google Scholar]
- 75.Gao J, Wang X, Zhang M, Bian M, Deng W, Zuo Z, Yang Z, Zhong D, Lin C. 2015. Trp triad-dependent rapid photoreduction is not required for the function of Arabidopsis CRY1. Proc. Natl Acad. Sci. USA 112, 9135–9140. ( 10.1073/pnas.1504404112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Warnasooriya SN, Porter KJ, Montgomery BL. 2011. Tissue- and isoform-specific phytochrome regulation of light-dependent anthocyanin accumulation in Arabidopsis thaliana. Plant Signal Behav. 6, 624–631. ( 10.4161/psb.6.5.15084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heijde M, et al. 2013. Constitutively active UVR8 photoreceptor variant in Arabidopsis. Proc. Natl Acad. Sci. USA 110, 20 326–20 331. ( 10.1073/pnas.1314336110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Das PK, Shin DH, Choi SB, Yoo SD, Choi G, Park YI. 2012. Cytokinins enhance sugar-induced anthocyanin biosynthesis in Arabidopsis. Mol. Cells 34, 93–101. ( 10.1007/s10059-012-0114-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Das PK, Geul B, Choi SB, Yoo S. D, Park Y. I. 2011. Photosynthesis-dependent anthocyanin pigmentation in Arabidopsis. Plant Signal Behav. 6, 23–25. ( 10.4161/psb.6.1.14082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Qi T, et al. 2011. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23, 1795–1814. ( 10.1105/tpc.111.083261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nott A, Jung HS, Koussevitzky S, Chory J. 2006. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759. ( 10.1146/annurev.arplant.57.032905.105310) [DOI] [PubMed] [Google Scholar]
- 82.Kovinich N, Kayanja G, Chanoca A, Otegui M. S, Grotewold E. 2015. Abiotic stresses induce different localizations of anthocyanins in Arabidopsis. Plant Signal Behav. 10, e1027850 ( 10.1080/15592324.2015.1027850) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruckle ME, Larkin RM. 2009. Plastid signals that affect photomorphogenesis in Arabidopsis thaliana are dependent on GENOMES UNCOUPLED 1 and cryptochrome 1. New Phytol. 182, 367–379. ( 10.1111/j.1469-8137.2008.02729.x) [DOI] [PubMed] [Google Scholar]
- 84.Kim S, Hwang G, Lee S, Zhu JY, Paik I, Nguyen TT, Kim J, Oh E. 2017. High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 8, 1787 ( 10.3389/fpls.2017.01787) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Oelmuller R, Levitan I, Bergfeld R, Rajasekhar V. K, Mohr H. 1986. Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168, 482–492. ( 10.1007/Bf00392267) [DOI] [PubMed] [Google Scholar]
- 86.Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjogren LL, Clarke AK, Straus M, Apel K. 2011. ‘happy on norflurazon’ (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J. 65, 690–702. ( 10.1111/j.1365-313X.2010.04454.x) [DOI] [PubMed] [Google Scholar]
- 87.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl Acad. Sci USA 98, 2053–2058. ( 10.1073/pnas.98.4.2053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Zheng S, Liu Z, Wang L, Bi Y. 2011. Both HY5 and HYH are necessary regulators for low temperature-induced anthocyanin accumulation in Arabidopsis seedlings. J. Plant Physiol. 168, 367–374. ( 10.1016/j.jplph.2010.07.025) [DOI] [PubMed] [Google Scholar]
- 89.Liu CC, Chi C, Jin LJ, Zhu J, Yu J. Q, Zhou Y. H. 2018. The bZip transcription factor HY5 mediates CRY1a-induced anthocyanin biosynthesis in tomato. Plant Cell Environ. 41, 1762–1775. ( 10.1111/pce.13171) [DOI] [PubMed] [Google Scholar]
- 90.Osterlund MT, Hardtke CS, Wei N, Deng X. W. 2000. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. ( 10.1038/35013076) [DOI] [PubMed] [Google Scholar]
- 91.Jia KP, Luo Q, He SB, Lu XD, Yang HQ. 2014. Strigolactone-regulated hypocotyl elongation is dependent on cryptochrome and phytochrome signaling pathways in Arabidopsis. Mol. Plant 7, 528–540. ( 10.1093/mp/sst093) [DOI] [PubMed] [Google Scholar]
- 92.Xu D, Dhiman R, Garibay A, Mock H-P, Leister D, Kleine T. 2020. Cellulose defects in the Arabidopsis secondary cell wall promote early chloroplast development. plantJ. 101, 156–170. [DOI] [PubMed] [Google Scholar]
- 93.Strand A, Asami T, Alonso J, Ecker JR, Chory J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. ( 10.1038/nature01204) [DOI] [PubMed] [Google Scholar]
- 94.Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. 1998. Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10, 673–683. ( 10.1105/tpc.10.5.673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kleine T, Kindgren P, Benedict C, Hendrickson L, Strand A. 2007. Genome-wide gene expression analysis reveals a critical role for CRYPTOCHROME1 in the response of Arabidopsis to high irradiance. Plant Physiol. 144, 1391–1406. ( 10.1104/pp.107.098293) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gangappa S. N, Botto J. F. 2016. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 9, 1353–1365. ( 10.1016/j.molp.2016.07.002) [DOI] [PubMed] [Google Scholar]
- 97.Dao TT, Linthorst HJ, Verpoorte R. 2011. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 10, 397–412. ( 10.1007/s11101-011-9211-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kindgren P, Noren L, Lopez Jde D, Shaikhali J, Strand A. 2012. Interplay between Heat Shock Protein 90 and HY5 controls PhANG expression in response to the GUN5 plastid signal. Mol. Plant 5, 901–913. ( 10.1093/mp/ssr112) [DOI] [PubMed] [Google Scholar]
- 99.Oñate-Sánchez L, Vicente-Carbajosa J. 2008. DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res. Notes 1, 93 ( 10.1186/1756-0500-1-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 ( 10.1093/nar/29.9.e45) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gruning BA, et al. 2017. The RNA workbench: best practices for RNA and high-throughput sequencing bioinformatics in Galaxy. Nucleic Acids Res. 45, W560–W566. ( 10.1093/nar/gkx409) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Trapnell C, et al. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. ( 10.1038/nprot.2012.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, Mann M, Cox J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. ( 10.1038/nmeth.3901) [DOI] [PubMed] [Google Scholar]
- 105.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. 2004. GO::TermFinder–open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics 20, 3710–3715. ( 10.1093/bioinformatics/bth456) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Alseekh S, et al. 2015. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27, 485–512. ( 10.1105/tpc.114.132266) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data acquired during this study are publicly available from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) under the accession no. GSE104868.