Abstract
In plants, tetrapyrrole biosynthesis occurs in chloroplasts, the reactions being catalysed by stromal and membrane-bound enzymes. The tetrapyrrole moiety is a backbone for chlorophylls and cofactors such as sirohaems, haems and phytochromobilins. Owing to this diversity, the potential cytotoxicity of some precursors and the associated synthesis costs, a tight control exists to adjust the demand and the fluxes for each molecule. After synthesis, haems and phytochromobilins are incorporated into proteins found in other subcellular compartments. However, there is only very limited information about the chaperones and membrane transporters involved in the trafficking of these molecules. After summarizing evidence indicating that glutathione transferases (GST) may be part of the transport and/or degradation processes of porphyrin derivatives, we provide experimental data indicating that tau glutathione transferases (GSTU) bind protoporphyrin IX and haem moieties and use structural modelling to identify possible residues responsible for their binding in the active site hydrophobic pocket. Finally, we discuss the possible roles associated with the binding, catalytic transformation (i.e. glutathione conjugation) and/or transport of tetrapyrroles by GSTUs, considering their subcellular localization and capacity to interact with ABC transporters.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles’.
Keywords: glutathione transferase, protoporphyrin, haem, ligandin, glutathione conjugation, tetrapyrrole
1. Introduction
Tetrapyrrole-containing molecules (e.g. chlorophylls, haems, sirohaems and bilins) have critical roles in plants. Indeed, these molecules or cofactors are found attached to proteins involved in photosynthesis and respiration but also in signalling, for instance, during light perception and photomorphogenesis [1]. For this latter aspect, phytochromes, which sense red/far-red light, bind an open-chain tetrapyrrole (bilin), named phytochromobilin in higher plants. Compared to non-photosynthetic organisms, plants have thus a larger diversity of tetrapyrrole-containing molecules. They have also the peculiarity that tetrapyrrole synthesis occurs in plastids (figure 1), whereas the corresponding pathway in non-photosynthetic eukaryotes takes place in mitochondria, with some steps in the cytosol [2]. Hence, given the specific redox reactions occurring in plastids, this compartmentation implies the existence of very specific and tightly regulated mechanisms. For instance, several enzymes are subject to post-translational redox regulatory mechanisms, notably controlled by the thioredoxin reducing systems [2,3]. Also, the presence of light may be detrimental in some conditions as porphyrin derivatives such as protoporphyrin IX (PPIX), protochlorophyllide, free chlorophyll molecules themselves or to a lesser extent their degradation products are harmful photosensitizers.
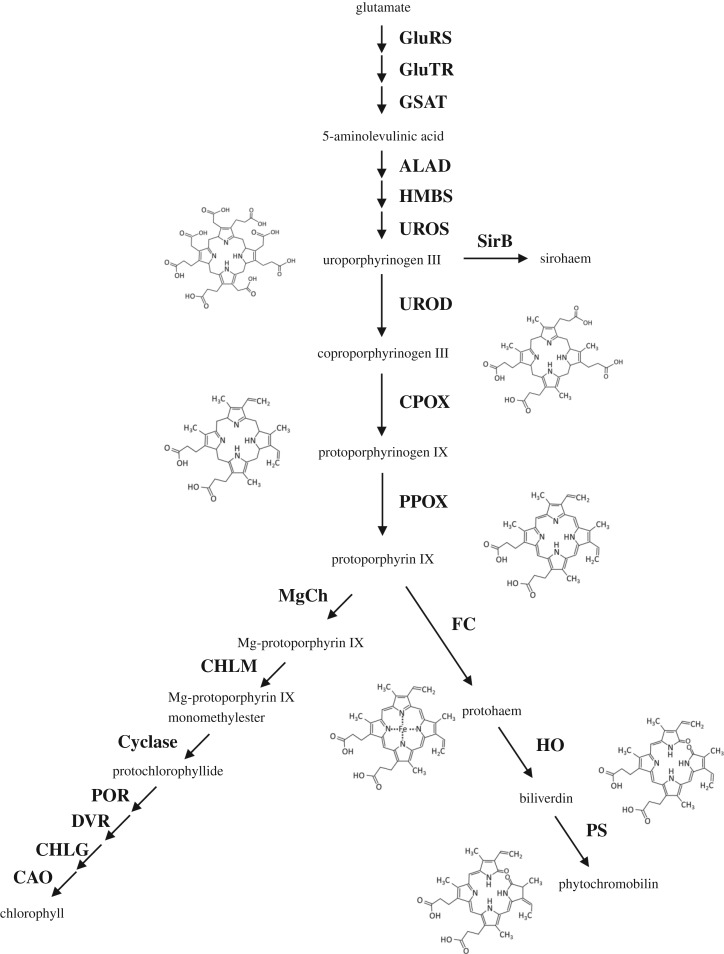
Figure 1.
Key steps of the tetrapyrrole biosynthesis pathway in plastids including the structures of some representative moieties. Enzymatic steps in this biosynthetic pathway, starting from glutamate, are represented by arrows with the names of the enzymes indicated in bold. Only the names and structures of tetrapyrroles mentioned in the text are shown. Abbreviations of the enzyme names are as follows: glutamyl-tRNA synthase (GluRS), glutamyl-tRNA reductase (GluTR), glutamate-1-semialdehyde aminotransferase (GSAT), 5-aminolevulinate dehydratase (ALAD), porphobilinogen deaminase (HMBS), uroporphyrinogen III synthase (UROS), uroporphyrinogen III decarboxylase (UROD), coproporphyrinogen oxidase (CPOX), protoporphyrinogen oxidase (PPOX), sirohydrochlorin ferrochelatase (SirB), ferrochelatase (FC), haem oxygenase (HO), phytochromobilin synthase (PS), Mg-protoporphyrin IX chelatase (MgCh), Mg-protoporphyrin IX methyltransferase (CHLM), Mg-Protoporphyrin IX monomethylester cyclase (Cyclase), protochlorophyllide oxidoreductase (POR), C8-vinyl-reductase (DVR), chlorophyll a synthase (CHLG), chlorophyll a oxygenase (CAO).
Whereas there is no need to export sirohaem (a prosthetic group only found in the chloroplastic nitrite and sulfite reductases), phytochromobilin has to be inserted into apo-phytochromes synthesized in the cytosol, possibly in an autocatalytic reaction. Also, chlorophyll catabolites, referred to as phyllobilins, are exported from the chloroplasts notably during stress response, fruit ripening or senescence. These linear tetrapyrroles are exported from plastids, further modified outside, i.e. at the endoplasmic reticulum (ER) or in the cytosol, before reaching the vacuoles through ATP-dependent transport [4]. Moreover, chlorophyll intermediates and haem moieties were proposed to act in the plastid-to-nucleus retrograde signalling, controlling the expression of nuclear-encoded photosynthetic genes [5,6]. The haem trafficking is even more complex as, in addition to a function in retrograde signalling, haems are found in a number of proteins outside chloroplasts, including the cytosol but also other organelles, notably mitochondria and peroxisomes. This implies the existence of haem transport systems across membranes for intracellular, inter-organellar movements but also possibly for intercellular exchanges as documented in non-photosynthetic organisms. Unlike the process of haem synthesis and degradation, which is relatively well documented, most actors involved in haem transport and mobilization await identification, but several lines of evidence suggest a role for glutathione transferases (GSTs).
2. Synthesis, transport and degradation of haems
Haems are protein cofactors that are essential for several key biological processes, including oxygen transport as part of haemoglobins, but also of the respiratory and photosynthetic electron transfer chains being present in cytochromes and in reaction centres/complexes. They are also crucial for detoxification processes, being present in ascorbate peroxidases and catalases and in cytochrome P450 monooxygenases. The latter proteins are required for xenobiotic detoxification and/or secondary metabolism. After synthesis, haems must be transferred to their final destination and client proteins. This implies the existence of a labile cellular haem pool consisting of haems associated with chaperones. Using a recently developed genetically encoded fluorescent haem sensor, it was deduced that the labile haem pool in the cytosol or mitochondria from yeast is in the nanomolar range [7]. However, having free/labile haems may be deleterious to macromolecules and the cells have to strictly regulate both synthesis and degradation of haems, but also their transport and delivery to client proteins.
In plants, the synthesis starts in the chloroplast with glutamate that is converted through nine successive steps into PPIX (figure 1). These steps are common with the chlorophyll synthesis pathway and PPIX is the branch point where haem and chlorophyll syntheses bifurcate. The insertion of Mg2+ by the Mg-chelatase will generate the chlorophyll precursor Mg-protoporphyrin IX, whereas the insertion of Fe by ferrochelatases will generate the protohaem giving rise to the formation of a, b- and c-type haems. The haem oxygenases (HOs) catalyse the oxidative degradation of haems into carbon monoxide (CO), biliverdin (BV) and Fe2+, which has to be recycled. Terrestrial plants use BV for the synthesis of phytochromobilin in one reaction step catalysed by the phytochromobilin synthase HY2, whereas in animals, BV is converted into bilirubin by biliverdin reductase [8]. In Arabidopsis, there are four genes coding for HOs (HO1/HY1, HO2, HO3 and HO4) and a single gene coding for phytochromobilin synthase, all proteins being located in chloroplasts [8–10]. The activity of both enzyme families is dependent on ferredoxins in plants [8,11]. The absence of HO outside plastids raises the question of whether and how haems present in non-chloroplastic haemoproteins are recycled or degraded. Among the four HOs, HO1 has a predominant role over HO3 and HO4, whereas HO2 has no HO activity, i.e. it does not bind or degrade haem [9,12]. This is consistent with the absence of a conserved histidine residue that binds the central Fe atom of haem in other HOs. However, HO2 binds PPIX quite strongly, maybe explaining the existence of a phenotype which also suggested a defect in phytochromobilin synthesis [9,13].
Two pools of haems are generally considered in cells, the haems tightly bound within haemoproteins versus labile haems required for signalling and transfer into proteins. This theoretically requires soluble haem chaperones for holding and delivering haems within and towards the different subcellular compartments where haemoproteins are found, starting from the chloroplastic site of synthesis. This also implies the existence of several haem membrane transporters for inter-organellar exchanges. A good candidate protein for intracellular haem trafficking should bind haem with a good but not too high affinity to deliver haems successfully to haem transporters or target proteins. In animal systems, several proteins have been proposed to serve in haem trafficking, including the haem-binding proteins (HBPs) [14], some fatty acid-binding proteins [15], the glycolytic protein glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [16] and also glutathione S-transferases (see §3). In fact, the in-cell haem-binding capacity of such candidate haem chaperones has not been often demonstrated, but it was recently reported that human GAPDH binds haem in cells and delivers it to downstream targets such as the inducible nitric-oxide synthase (iNOS) [16]. There are two cytosolic GAPDH in Arabidopsis thaliana (GAPC1 and GAPC2). At this time, it is not known whether they could also play a similar role, but they do possess the histidine residue, which was shown to be crucial in the human orthologue for haem binding and exchange. Five HBPs have been so far characterized in A. thaliana, including two in the chloroplasts (HBP3 and 5) and three in the cytosol [17,18]. The cytosolic HBP1 and HBP2 from A. thaliana were shown to bind PPIX and haem [17], with HBP1, HBP2 and HBP5 but not HBP3 being able to bind to a haemin-agarose affinity column, possibly indicating different functions for HBP3 and HBP5 in chloroplasts [18]. In addition, Arabidopsis HBP5, but not other HBPs, was found to interact with HY1, suggesting that it might be specifically involved in the delivery of haem to HY1 [18].
In fact, a strategy or adaptation to avoid implicating too many intermediates in haem trafficking is via the formation of complexes, notably those involving ferrochelatases. There are two genes (FC1 and FC2) in A. thaliana, which have different functions in accordance with their different expression patterns and the phenotypic analysis of plant mutants, but both are important for chloroplast development. The FC1 may more specifically produce the haem portion used as a retrograde signal to coordinate the expression of specific genes required during chloroplast development [19]. The FC2 may be more specifically important for the maturation of photosynthetic cytochromes [20]. Both FC1 and FC2 are important for stress responses [21,22]. These plant ferrochelatases may be located in both the thylakoid and chloroplast envelope membranes [23]. This might allow the release of haems within the chloroplasts without relying on a chaperone or the export of haems to the cytosolic side, either alone or by making direct interactions with some haem transporter(s), the identity of which remains, however, enigmatic so far. In animals, several transporters including ATP-binding cassette (ABC) transporters have been proposed to serve for the transport of haems or haem intermediates such as coproporphyrinogen III between the cytosol and mitochondria (reviewed in [24]), suggesting that plant ABC transporter orthologues may perform similar functions in the chloroplast envelope and elsewhere. One possible candidate for the translocation of haems and other tetrapyrrole intermediates across organelle membranes is the translocator protein (TSPO). In Arabidopsis, there is a single gene coding for this haem- and PPIX-binding, stress-responsive membrane protein [25], which is virtually found in all subcellular compartments (mitochondria, plastids, nuclear fractions, ER and Golgi vesicles) where haem exchanges occur [26]. AtTSPO is regulated at the transcriptional level in tetrapyrrole biosynthetic mutants and in response to stresses, notably salt stress [26].
In conclusion, further work on all these potential haem chaperones and transporters is needed in plants as in other organisms to delineate a more complete model of haem trafficking and signalling.
3. The glutathione transferases: versatile, promiscuous proteins possessing both catalytic and ligandin functions
Glutathione (GSH) S-transferases represent a widespread and diversified protein family found in almost all organisms. In non-photosynthetic organisms, the number of GSTs ranges from 2 in Plasmodium falciparum to 44 in the fungus Postia placenta [27]. The number of isoforms has greatly expanded in photosynthetic organisms, resulting in the existence of 14 GST classes and up to 110 GST genes in Eucalyptus grandis [28,29]. Overall, there are over 30 different classes with only a few classes shared by all organisms (electronic supplementary material, table S1). GSTs catalyse two opposite reactions. Those having a conserved serine (Ser-GSTs) or tyrosine (Tyr-GSTs) perform GSH-conjugation reactions on electrophilic molecules, whereas those having a conserved catalytic cysteine (Cys-GSTs) catalyse deglutathionylation reactions, i.e. the removal of glutathione from small molecules [29,30]. For these reactions, the (co)-substrates (i.e. either glutathionylated substrates or both GSH and the electrophilic substrates) bind to the active site, which is traditionally defined as formed by a G-site for glutathione binding and by an H-site for hydrophobic substrate binding. The role of GSTs catalysing GSH-conjugation reactions has been well documented in the context of herbicide detoxification by crops, as part of the xenobiotic detoxification system [31]. In this process, GSTs conjugate GSH onto electrophilic molecules often pre-activated by reaction with cytochromes P450, before ABC transporters target the GSH-conjugated molecules to the vacuole or to the extracellular compartment. In some other circumstances, GSTs bind various types of molecules without catalysing GSH conjugation. Virtually all types of GSTs possess this so-called ligandin property. The ligands can be either accommodated in the active site or bound to ligandin (L) sites, which are usually situated at the dimer interface in the case of dimeric GSTs [32]. Thus, GSTs act as carrier/transport proteins serving for the storage or trafficking of various sorts of molecules, often towards ABC transporters. Another function of the ligandin activity of GSTs may be to provide a labile pool of molecules, in particular endogenous specialized metabolites.
Hence, GSTs are able to bind a plethora of structurally unrelated compounds. While the in vitro biochemical characterization of GSTs relies on a battery of model compounds or inhibitors, the physiological substrates or ligands often remain enigmatic. Concerning GSH-conjugated molecules, only very few natural reaction products of GSTs have been identified, possibly because (i) they are unstable and undergo reversible glutathionylation, (ii) the conjugation products of GSTs are very rapidly processed to derived metabolites, or (iii) the use of inactive proteins is required for freezing interactions which is often not achieved [33]. In plants, it was described that GSTF6 participates in the synthesis of camalexin by catalysing the conjugation of GSH onto the indole-3-acetonitrile precursor [34] and GSTU13 in the metabolism of indole glucosinolates [35]. Concerning the non-catalytic, ligandin functions of GSTs, i.e. the transport or sequestration of specialized metabolites or some of their intermediates, one would reasonably assume that interactions are more stable and should allow for the isolation of these ligands. Still, fishing experiments performed in bacterial or plant cells are not so trivial and only a few GST ligands are known, considering the high number of isoforms in plants. The best documented example of a non-catalytic ligandin property of GSTs relates to the accumulation of anthocyanins in vacuoles. In A. thaliana, GSTF12, also referred to as TRANSPARENT TESTA 19 (TT19), plays a key role in the control of anthocyanin and proanthocyanidin vacuolar accumulation [36], ensuring the transport (without GSH conjugation) of these cytosolic flavonoids to a tonoplastic ABC transporter named TT12 [37]. Other studies have aimed at isolating and identifying substrates/ligands, mostly for phi and tau GSTs, either from plants or eventually Escherichia coli and using both in vitro and in vivo approaches (recently summarized in [29]). To cite a few examples, both GSTLs and GSTFs bind flavonoids [30,38–42], GSTFs and GSTUs bind glutathionylated conjugates of oxylipins [38], with GSTUs also binding stilbenes (trans-resveratrol) [39], fatty acid derivatives [40] and porphyrin derivatives [38,41–43]. Together, this indicates that GSTs are generally promiscuous proteins having a rather broad ligand/substrate spectrum, although oriented towards heterocyclic molecules.
4. Evidence for the connection between glutathione transferases and haem metabolism in both non-plant and plant cells
In the 1970s and 1980s, several studies reported a tight connection between GSTs and haem detoxification and/or transport, notably in mammals, with numerous publications describing interactions of GSTs of the alpha, pi and mu classes with steroids, bile acids, haem and its degradation product bilirubin [44–47]. Later on, studies using GSTs from natural sources or using recombinant systems have analysed the capacity of these proteins to bind haematin, haemin or bilirubin but also some precursors, such as PPIX and coproporphyrin, essentially using inhibition kinetics and intrinsic fluorescence quenching (table 1). Whereas some effects/interactions have been observed for most tested proteins, there is no unifying conclusion as competitive, non-competitive and uncompetitive inhibitions have been observed depending on the proteins considered. This suggested that some GSTs bind porphyrins in their active sites, whereas some others bind it elsewhere, very likely in their ligandin sites. When analysed, only one binding site was usually described. Inhibition studies using the cytosolic GST from Plasmodium falciparum revealed an uncompetitive inhibition mechanism towards GSH. It suggested also that haem preferentially binds to a preformed enzyme–GSH complex and that a high-affinity binding site with a dissociation constant (Kd) value of 30 nM was present [48,49,55]. To the contrary, a study performed with a placenta pi GST suggested the existence of two binding sites with affinities of 20 and 400 nM, which are modulated by GSH [53]. While there is no exhaustive comparative study, this is clearly complicated by the existence of as many as 30 different GST classes (all organisms considered) exhibiting different catalytic and structural properties (electronic supplementary material, table S1) [56]. In most studies, Kd constants, inhibitor binding affinity (Ki) constants or half maximal inhibitory concentration (IC50) values ranging from the nanomolar to the micromolar range have been measured (table 1). Although we consider that values above 10–15 µM obtained in these in vitro studies are less relevant, many factors, such as the presence of GSH or the nature of the bound porphyrins, might modulate the affinities of GSTs for porphyrins in cellulo. On a physiological note, the binding of porphyrins was often perceived as a detoxification mechanism, removing or recycling degradation products, but a haem chaperone function has been suggested for a cytosolic liver GST from rat as early as in the 1980s because it facilitated the haem reconstitution into an apocytochrome b5 from a mitochondrial fraction [57].
Table 1.
Summary table presenting affinity constants obtained from inhibition studies performed using GSTs from various sources and several tetrapyrroles. GSH, glutathione; CDNB, 1-chloro-2,4-dinitrobenzene; ANS, 8-anilino-1-naphthalenesulfonic acid.
| organisms | GST names | tested ligands/inhibitors | affinity constants | type of inhibition | approaches | references |
|---|---|---|---|---|---|---|
| Plasmodium falciparum | PfGST | haemin |
Ki 6.5 µM, IC50 4 µM |
uncompetitive (GSH) | inhibition kinetics | [48] |
| protoporphyrin IX |
Ki 10 µM, IC50 11 µM |
mixed-type (GSH) | inhibition kinetics | [48] | ||
| ferriprotoporphyrin IX |
Kd 0.03 µM (+1 mM GSH) |
uncompetitive | intrinsic fluorescence quenching | [49] | ||
| ferriprotoporphyrin IX | Ki 3 µM | uncompetitive | inhibition kinetics | [49] | ||
| haemin | Ki 0.4 µM | non-competitive (GSH) | inhibition kinetics | [50] | ||
| Ki 0.6 µM | competitive (CDNB) | inhibition kinetics | [50] | |||
| protoporphyrin IX | Ki 26 µM | non-competitive (GSH) | inhibition kinetics | [50] | ||
| Ki 29 µM | non-competitive (CDNB) | inhibition kinetics | [50] | |||
| Exp1 (MAPEG-like) | haematin | Ki 0.17 µM | competitive (CDNB) | inhibition kinetics | [51] | |
| haematin | KM 0.25 µM | GSH conjugation | mass spectrometry | [51] | ||
| human | mPGES-2 | haem | Kd 0.54 µM | — | intrinsic fluorescence quenching | [52] |
| mouse | GSTM1 | haemin | Ki 1.5 µM | non-competitive (GSH) | inhibition kinetics | [50] |
| Ki 0.6 µM | non-competitive (CDNB) | inhibition kinetics | [50] | |||
| protoporphyrin IX | Ki 7.3 µM | non-competitive (GSH) | inhibition kinetics | [50] | ||
| Ki 35 µM | non-competitive (CDNB) | inhibition kinetics | [50] | |||
| human | GSTP1 | haemin | Ki 2.5 µM | competitive (GSH) | inhibition kinetics | [50] |
| Ki 3.8 µM | competitive (CDNB) | inhibition kinetics | [50] | |||
| protoporphyrin IX | Ki > 50 µM | competitive (GSH) | inhibition kinetics | [50] | ||
| Ki > 100 µM | competitive (CDNB) | inhibition kinetics | [50] | |||
| human | placenta GSTP1 | haemin | Kd 20 and 400 nM | intrinsic fluorescence quenching | [53] | |
| human | haemin | Ki 4 µM | non-competitive (ClPh(NO2)2) | inhibition kinetics | [53] | |
| Taenia solium | Ts26GST | haematin |
Ki 0.3 µM Kd 0.7 µM |
non-competitive (CDNB) | inhibition kinetics, intrinsic fluorescence quenching (or with ANS) | [54] |
| mesoporphyrin |
Ki 0.5 µM Kd 1.1 µM |
non-competitive (CDNB) | inhibition kinetics, intrinsic fluorescence quenching (or with ANS) | [54] | ||
| protoporphyrin |
Ki 4 µM Kd 2.7 µM |
non-competitive (CDNB) | inhibition kinetics, intrinsic fluorescence quenching (or with ANS) | [54] | ||
| coproporphyrin |
Ki 1 µM Kd 2.6 µM |
non-competitive (CDNB) | inhibition kinetics, intrinsic fluorescence quenching (or with ANS) | [54] | ||
| Zea mays | I–I | protoporphyrin IX | IC50 1–5 µM | non-competitive (CDNB), competitive (GSH) |
inhibition kinetics | [42] |
| coproporphyrin | Kd 0.89 µM | non-competitive (CDNB), competitive (GSH) | porphyrin fluorescence emission changes | [42] | ||
| I–II | protoporphyrin IX | inhibition kinetics | [42] | |||
| II–II | protoporphyrin IX | IC50 10–25 µM | non-competitive (CDNB), competitive (GSH) | inhibition kinetics | [42] | |
| coproporphyrin | Kd 0.51 µM | porphyrin fluorescence emission changes | [42] | |||
| III–III | protoporphyrin IX | IC50 5–10 µM | inhibition kinetics | [42] | ||
| coproporphyrin | Kd 0.27 µM | porphyrin fluorescence emission changes | [42] | |||
| mesoporphyrin | Kd 1.93 µM | porphyrin fluorescence emission changes | [42] |
During their life cycle, haematophagous insects or parasites that affect humans encounter massive amounts of haems, which act as prooxidant molecules once liberated from the hydrolysis of host haemoglobin. In the kissing bug Rhodnius prolixus, a vector of Chagas disease, haem degradation proceeds via the formation of thioether bonds between a cysteine and the vinyl side chains of biliverdin [58]. Whereas only species with one or two cysteinyl-glycine residues bound to biliverdin have been detected by mass spectrometry, it is very likely that these dipeptides derive from glutathione molecules from which the glutamate was removed. Whether this step of haem digestion is spontaneous or catalysed by some GSTs present notably in insect heart will have to be explored. In parasites such as P. falciparum, an alternative detoxification strategy to haem polymerization is to degrade it despite the absence of an HO activity. In fact, GSH spontaneously disrupts haems at neutral pH in the presence of oxygen [59]. The presence of iron seems mandatory as there is no reaction with Zn-protoporphyrin or an iron-free protoporphyrin. A role for GSH and GSTs in haem degradation in this organism is supported by the observation that GSH levels and GST activity are significantly increased in parasites resistant to the anti-malarial drug chloroquine, that promotes intracellular haemin accumulation [60]. There are two GSTs in P. falciparum. The soluble PfGST is uncompetitively inhibited by haemin (Ki around 6.5 µM), indicating that free haemin can be bound by the enzyme [48]. The essential microsomal MAPEG-type GST, EXP1, has proven to degrade haematin efficiently in the presence of GSH [51], suggesting a physiological role in coping with the cytotoxic effects of haems. The affinity of EXP1 for haem is likely much better than that of the soluble PfGST as the GST activity of EXP1 is competitively inhibited by haematin with a Ki of 170 nM [51]. Interestingly, this activity towards haematin was not observed for a human microsomal GST (MGST1). On another note, a recombinant form of the soluble domain of the human mitochondrial prostaglandin E synthase (mPGES2) was purified with a bound haem and displays a Kd of 0.54 µM [52]. A GSH molecule also binds to the active site and forms an S–Fe coordination bond with the haem iron atom.
The first report of an interaction of tetrapyrroles with GSTs from photosynthetic organisms was published in 1988. The GST activity of a crude extract of etiolated oat seedlings was inhibited by chlorophyllin and haemin and to a lesser extent by bilirubin and biliverdin [61] and this was later confirmed by using a purified GST heterodimer [62]. In 2003, it was reported that maize GST isoforms, comprising GSTF and GSTU homodimers or GSTF/U heterodimers, were able to bind PPIX but also other porphyrin precursors such as coproporphyrin, uroporphyrin and Mg-protoporphyrin with Kd values in the low micromolar range (table 1) [42]. Haemin had no significant inhibitory effect. As there was no evidence for the formation of glutathionylated products, it was suggested that porphyrin binding relies on the ligandin properties of GSTs [42,43]. In fact, these GSTs were reported as preventing the non-enzymatic autoxidation of protoporphyrinogen and reducing the oxidative degradation of haemin. The observed competitive and non-competitive inhibitions with GSH and CDNB, respectively, suggested that porphyrins were bound to the G-site. In following studies, it was noted that the overexpression in E. coli of two maize GSTUs, ZmGSTU1 and ZmGSTU2 to a lesser extent, but not of ZmGSTU3, ZmGSTF1 and ZmGSTF3, perturbed tetrapyrrole metabolism causing a reduction in haem B levels and an accumulation of porphyrin precursors (uroporphyrin, pentacarboxyl porphyrin, coproporphyrin and glutathionylated harderoporphyrin) [41]. Similarly, the expression of AtGSTU7 and AtGSTU19 in E. coli allowed their purification with protoporphyrin- and harderoporphyrin–glutathione conjugates, respectively [38]. Expression of strep-tagged proteins in planta (Arabidopsis or tobacco) neither perturbed the tetrapyrrole metabolism nor allowed for fishing of porphyrin molecules, likely because of their cytosolic localization [38,41]. However, expression of a ZmGSTU1–ZmGSTU2 chimeric protein in tobacco chloroplasts resulted in the accumulation of harderoporphyrin(ogen)–glutathione conjugates. In in vitro assays, both ZmGSTU1 and ZmGSTU2 catalysed the GSH conjugation of reduced protoporphyrinogen, used as a substrate instead of the related harderoporphyrin(ogen) [41]. However, they could not conjugate GSH on an oxidized PPIX form. This suggests that GSH conjugation occurs only on reduced porphyrins and might explain why it was not observed in former studies [42]. However, the enzymes were rapidly inactivated presumably because the GSH-conjugated products formed a stable complex with them. As porphyrin synthesis in chloroplasts proceeds via reduced porphyrinogen intermediates, it may be that some GSTUs or other GSTs present in this compartment catalyse their conjugation to GSH. Altogether, although the biological significance of these interactions is not yet clear, these observations suggest a role of GSTU in porphyrin binding, transport and/or scavenging in plants.
5. Further observations that tau glutathione transferases have the capacity to bind haem and protoporphyrin IX
(a). Arabidopsis thaliana GSTU8 binds a haem b upon expression in Escherichia coli
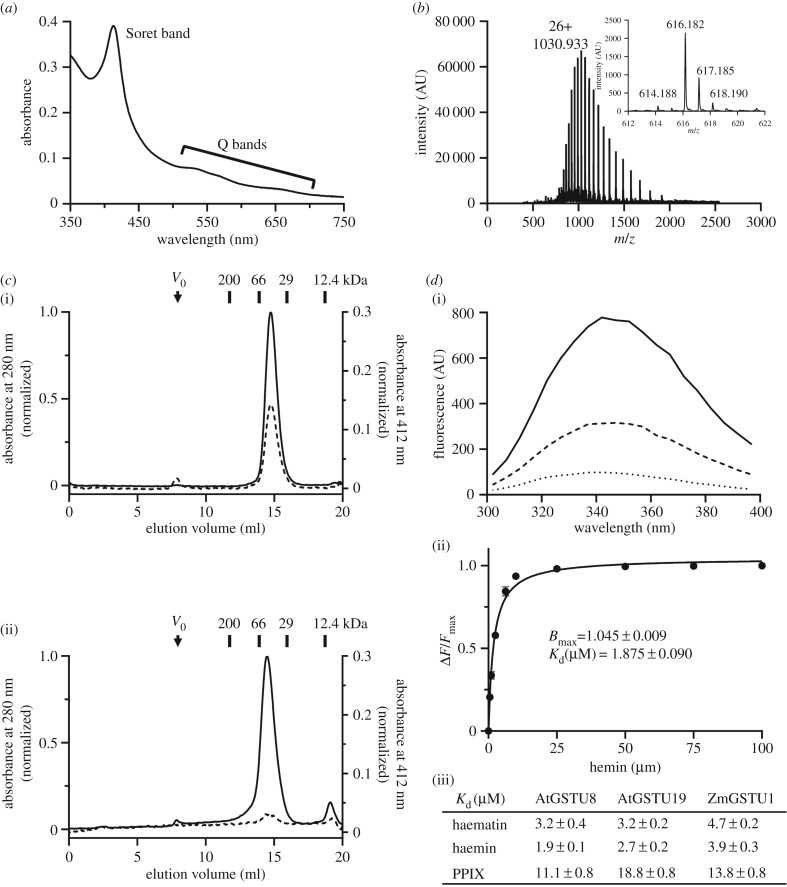
A C-terminal His6-tagged version of AtGSTU8 was expressed in E. coli and purified by affinity chromatography. Immediately upon loading the bacterial lysate, a pink coloration was visible on the resin. The UV–visible absorption spectrum of the purified protein exhibited a particular absorption band at 410 nm (Soret band) and two smaller absorption bands around 550 and 650 nm (Q bands), which are characteristic of tetrapyrrole-containing molecules (figure 2a). To determine the nature of the metabolite co-purifying with the protein, mass spectrometry analyses in denaturing conditions were carried out. In addition to the AtGSTU8 polypeptide detected at the expected theoretical molecular mass of 26 777.2 Da, an isotopic massif with an m/z of 616.182 Da was also detected, which corresponded to the mass of a haem b as found in myoglobin for instance (figure 2b). In order to test whether AtGSTU8 is able to bind PPIX, we have incubated the apo-protein, that was obtained by expressing it in minimal medium and extensive dialysis, with an excess of PPIX. Analytical gel filtration experiments performed using both the apo-protein and the PPIX-incubated protein showed the presence of a major peak eluted at around 14 ml, a volume corresponding to an apparent molecular mass of ca44 kDa that we interpreted as dimers (figure 2c). The fact that AtGSTU8 and PPIX co-eluted at the same volume indicated that PPIX was bound to the protein. Then, we sought to determine the dissociation constant (Kd) of AtGSTU8 towards PPIX but also haematin and haemin (iron-hydroxylated and iron-chloride forms of haem b, respectively) using tryptophan fluorescence quenching titrations. We included two additional proteins, the GSTU1 from maize and GSTU19 from A. thaliana, for which an interaction with porphyrin derivatives was already reported [38,41]. The fluorescence of these proteins was quenched by adding increasing concentrations of porphyrin derivatives as shown for haematin and AtGSTU8 (figure 2d). By fitting the different curves obtained to the equation of a hyperbola, we determined the maximum number of binding sites (Bmax) and Kd values (figure 2d). The Bmax values were comprised between 1.05 and 1.1 site per protein meaning that there should be only one binding site. Using AtGSTU8, the dissociation constants measured for haematin and haemin were around 2–3 µM, whereas a value of 11 µM was found for PPIX (figure 2d). Values in the same range have been measured for ZmGSTU1 and AtGSTU19. The fact that the dissociation constants and thus affinities are better with Fe-bound haems as compared with an Fe-free protoporphyrin suggests that the presence of the Fe atom has some influence on ligand binding. Altogether, these results allowed us to conclude that AtGSTU8 was co-purified from E. coli cells with a haem b and has an affinity that would be favourable for haems exchange reactions.
Figure 2.
Arabidopsis GSTU8 binds a haem b. (a) Absorbance spectrum of as-purified AtGSTU8 showing an absorption band centred at 410 nm (Soret band) and several weaker absorption bands around 550 and 650 nm (Q bands). (b) Electrospray ionization (ESI) mass-to-charge ratio (m/z) spectra of AtGSTU8 and its co-purified metabolite (inset) obtained using a Bruker microTOF-Q spectrometer in denaturing conditions. From the multiply charged ion spectra initially obtained, we focused on the peak with 26 charges on the ion. The deconvolution of the ESI spectra of denatured AtGSTU8 indicated that the ion with an m/z of 1030.933 atomic mass units (amu) corresponded to a molecular mass of 26 777.2 Da. A molecule with a molecular mass of 616.182 Da corresponding to a haem b was also detected (inset). The intensity of the signal is represented as arbitrary units (AU). (c) Analytical gel filtration chromatogram of AtGSTU8. One hundred micrograms of AtGSTU8 at 3 µM were incubated in the absence (i) or presence (ii) of PPIX (100 µM) and then loaded on analytical gel filtration (Sephadex S200 10/300 connected to an AKTA purifier system) chromatography. Protein absorption was followed at 280 nm (solid line), and PPIX absorption was followed at 412 nm (dashed line). It is noteworthy that most unbound PPIX was retained on a filter that was added before the gel filtration column. Vo means void volume. The calibration curve was obtained with the indicated standards. (d) Quenching of tryptophan intrinsic fluorescence (λexc = 290 nm) of AtGSTU8 (solid line) by PPIX (2.5 µM dashed line or 6.25 µM dotted line) (i). Data were fitted to an equation describing binding to a single binding site (ii). Results are means ± s.d. (n = 3). Bmax value represents the maximum number of binding site. The apparent dissociation constant (Kd) values of AtGSTU8, AtGSTU19 and ZmGSTU1 for haematin, haemin and PPIX have been calculated from tryptophan fluorescence quenching experiments (iii).
Using a culture medium supplemented with FeCl3 and aminolevulinic acid, we then sought to determine whether preparations containing more holoproteins can be obtained. Moreover, to assess how specific is this observation, we additionally expressed in E. coli: AtGSTU16 that belongs to the third major clade of A. thaliana GSTUs (electronic supplementary material, figure S1), AtGSTU22 that is close to AtGSTU19 and AtGAPC1 based on the assumption that it could bind similar prosthetic groups (see above). After purification using a single affinity chromatography step and dialysis, a UV–visible absorption spectrum was recorded for each protein (electronic supplementary material, figure S2) and the presence of porphyrin moieties analysed by LC-MS analysis (electronic supplementary material, figure S3). The results are summarized in table 2. No sign for the presence of porphyrin(s) was visible for AtGAPC1 in these conditions. Based on the sample coloration observed during IMAC and the ratio of absorbance at 280 and 412 nm, we concluded that all expressed GSTUs except AtGSTU22 were able to bind porphyrins. Accordingly, we could detect the presence of haem b in AtGSTU8 and AtGSTU16 and of harderoporphyrin and/or glutathionylated harderoporphyrin in AtGSTU19 and ZmGSTU1, as described previously [41]. All these data point to the existence of a certain specificity among GSTUs, with one isoform being apparently unable to bind porphyrins, and some other binding different types (at least preferentially) of porphyrins (haem b versus harderoporphyrin). In the absence of structural information, structural modelling and sequence comparisons have been initiated to understand these differences.
Table 2.
Ligands bound to E. coli expressed recombinant GSTUs. All these proteins were expressed in E. coli using a medium supplemented with FeCl3 and aminolevulinic acid, purified in a single IMAC step and dialysed before recording a UV–visible absorption spectrum (electronic supplementary material, figure S2) and performing an LC-MS analysis (electronic supplementary material, figure S3), as described in the electronic supplementary material. The A280 nm/A412 nm column corresponds to the ratio between the recorded absorbance values at these specific wavelengths. The identification of haem b in AtGSTUs was confirmed by analysing a myoglobin sample in the same conditions. The assignment of the compounds found to co-purify with AtGSTU19 and ZmGSTU1 as harderoporphyrin derivatives is based on previous results obtained with ZmGSTU1 [41].
| protein | A280 nm/A412 nm | ligand(s) detected | m/z |
|---|---|---|---|
| AtGSTU8 | 10 | haem b | 616.1780 |
| AtGSTU16 | 9 | haem b | 616.1865 |
| AtGSTU19 | 7 | harderoporphyrin | 609.2541 |
| AtGSTU22 | 29 | none | |
| ZmGSTU1 | 5 | harderoporphyrin; GS-harderoporphyrin | 609.2816; 916.3718 |
(b). Modelling of protoporphyrin/haem binding by tau glutathione transferases
Most GSTs are homodimeric assemblies constituted by two domains at the monomer scale (figure 3). The N-terminal domain exhibits the TRX-fold (β1α1β2α2β3β4α3), whereas the C-terminal domain is mainly helical (α4α5α6α7α8α9). Concerning the tau class, 10 different GSTU crystal structures (not considering the structures corresponding to the same protein bound with different ligands) have been elucidated so far [29]. Overall, GSTUs exhibit a V-shaped, open dimer configuration with only 2200 A2 of accessible surface area buried at the dimer interface, a value lower than most other GST classes. Notably, conserved salt bridges are found around the twofold axis at the solvent-exposed side of the dimer interface.
Figure 3.
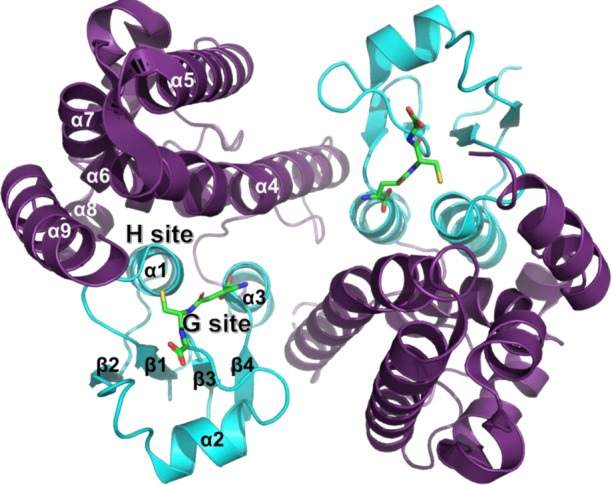
Crystal structure of OsGSTU1 dimer in complex with glutathione (PDB 1OYJ). Oryza sativa GSTU1 crystal structure [63] in complex with glutathione (green sticks). The N-terminal domain is coloured cyan and the C-terminal domain is coloured violet. Secondary structures and binding sites (for glutathione and for hydrophobic molecules) are labelled.
Structural features of GSTUs include specificities for the active site region. As for most other canonical GSTs, the GSTU catalytic region includes a glutathione binding site (G-site) at the N-terminal domain and a hydrophobic site (H-site) made of residues from both the N-terminal and the C-terminal domains. GSTUs exhibit a conserved serine residue at the N-terminal end of helix α1. This residue interacts with the Sγ atom of glutathione through hydrogen bonding. Other polar contacts with the tripeptide include residues K42 (loop β2–α2), I56 (loop α2–β3), E68 and S69 (helix α3) (PDB 1OYJ, OsGSTU1 numbering). Concerning the H-site, studies have pointed to residues located in the vicinity of the conserved serine, in loop β1–α1, helix α4, helix α6 and helix α9 [64–68]. A third site called the ligandin site was reported for Glycine max GSTU4-4 which binds one (4-nitrophenyl)methanethiol molecule in a hydrophobic region made of residues from helix α1, strand β2 and helix α8 [66].
In 2008, Dixon et al. [41] identified two regions in which mutations potentially affected protoporphyrin binding using chimeric maize enzymes. These regions, located in the active site, were (i) the C-terminal part of helix α4 with an aromatic residue (Y112 in ZmGSTU1) putatively involved in ligandin activity and (ii) the loop β2–α2 that notably bears the K42 residue of the G-site. Furthermore, molecular modelling studies suggested hydrophobic contributions of residues located in helix α6 and α9.
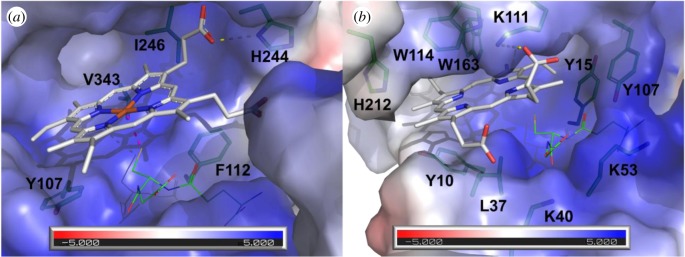
Our biochemical data indicate that AtGSTU8, AtGSTU16 and AtGSTU19 bind PPIX and/or haem. In the absence of structural information for these proteins, we used molecular docking to predict whether and how PPIX is accommodated in the active site of the recently solved AtGSTU23-GSH structure (PDB 6EP7) [69]. This isoform shares 68% sequence identity and high conservation of the active site residues with AtGSTU19. The best position predicted by Vina (affinity of −8.8 kcal mol) suggests PPIX binding at the H-site, in close vicinity of the G-site (figure 4). A hydrophobic cleft is provided by the side chains of the aromatic residues Y10 and Y15 (loop β1–α1), Y107 and W114 (helix α4), W163 (helix α6) and H212 (helix α9), whereas an electropositive entry (K40 and K53 from helix α2 region, K111 from helix α4) putatively stabilizes the negatively charged carboxylic groups of PPIX. Such an exposed and amphiphilic site was reported in the crystal structure of the protoporphyrin-binding protein GUN4 (PDB 4XKB) [71] with a putative role in transport. Interestingly, W114 was previously found as interacting with a glycerol molecule bound at the H-site of AtGSTU23-GSH (PDB 6EP7), whereas the side chain of Y107 showed flexibility upon GSH binding in the same Protein Data Bank entry [69]. Three residues, namely K40, Y107 and W114 (AtGSTU23 numbering for clarity), were previously identified from the docking of PPIX in the OsGSTU1 active site [41]. Thus, our docking study suggests the binding of PPIX mainly at the H-site and very close to the G-site, which is consistent with the substrate competition observed during enzymatic measurements reported in many studies.
Figure 4.
Comparison of the active sites from mPGES2 bound to haem and AtGSTU23 docked with protoporphyrin IX. (a) Crystal structure of human mPGES2 bound to GSH and haem (PDB code 2PBJ). (b) Docking of PPIX onto AtGSTU23 crystal structure (PDB code 6EP7). Molecular surfaces coloured according to the electrostatic potential were calculated with APBS software [70] and are shown in transparency. PPIX and haem are represented as white sticks. Residues that line the active sites are shown as green sticks. Putative H-bonds are shown as yellow dashed lines. Coordination bond between haem and GSH is shown as pink dashed line. GSH is shown as green lines.
We compared the PPIX-bound AtGSTU23 model with the sole structure of a GST in complex with a porphyrin ligand, i.e. the one of human mPGES2 where a haem molecule in interaction with GSH is bound in the active site [52]. One coordination bond is found between the haem Fe atom and GSH Sγ atom. Additional contacts include one H-bond with the side chain of H244 and hydrophobic contributions of Y107, F112, V243 and I246. Although the C-terminal domains of AtGSTU23 and mPGES2 show significant structural differences, they both bear an H-site, particularly large and solvent-exposed with a hydrophobic bottom and electropositive residues that line the entry of the pocket. Furthermore, conformations of the PPIX ring in both structures are similar, which indicates that PPIX binding in the AtGSTU23 active site is sterically possible. However, our structural analysis failed to identify a conserved residue that could assure coordination of the metal atom in the case of metalloporphyrins such as haems (usually a Cys, His, Asn or Gln residue). Whether a contribution of glutathione in the stabilization or GSH conjugation of tetrapyrroles exists in AtGSTU23, through an Fe–S coordination bond or via a covalent bonding at the vinyl groups, remains to be determined. Interestingly, previous crystallographic results obtained on fungal GST omega (which are close structural homologues of plant GSTUs) showed that the binding of both GSH and one polyphenol at the active site occurs without reaction between both molecules [72], suggesting that co-transport is also possible.
The putative residues identified via the docking study were mapped onto a multiple alignment performed with the sequences of all A. thaliana GSTUs and of the PPIX-binding ZmGSTU1 (electronic supplementary material, figure S4). Two residues (K40 and K52) that contribute to the electropositive environment are conserved in nearly all isoforms because they are also part of the highly conserved G-site. A contribution of aromatic or hydrophobic residues from helix α9 was suggested in the present work (H212) as well as a previous docking study (F213 and M217 in ZmGSTU1) [41]. The observed general trend for this helix is to bear hydrophobic residues at these positions. Importantly, it was previously shown that the conformation of helix α9 varies from one GSTU isoform to another, which putatively confers different binding properties to GSTUs [67]. However, such variations are not predictable on the sole sequence information. In one group of Arabidopsis isoforms (from AtGSTU19 to AtGSTU28), as well as in ZmGSTU1, aromatic residues are conserved at the positions of residues Y107, W114 and W163, suggesting that their H-site is adapted for tetrapyrrole binding. However, the fact that AtGSTU8 does not have these specific residues indicates differences in PPIX-binding mode from one isoform to another, as well as the existence of other structural determinants.
6. Hypotheses about the role(s) of glutathione transferases in relation to tetrapyrrole metabolism
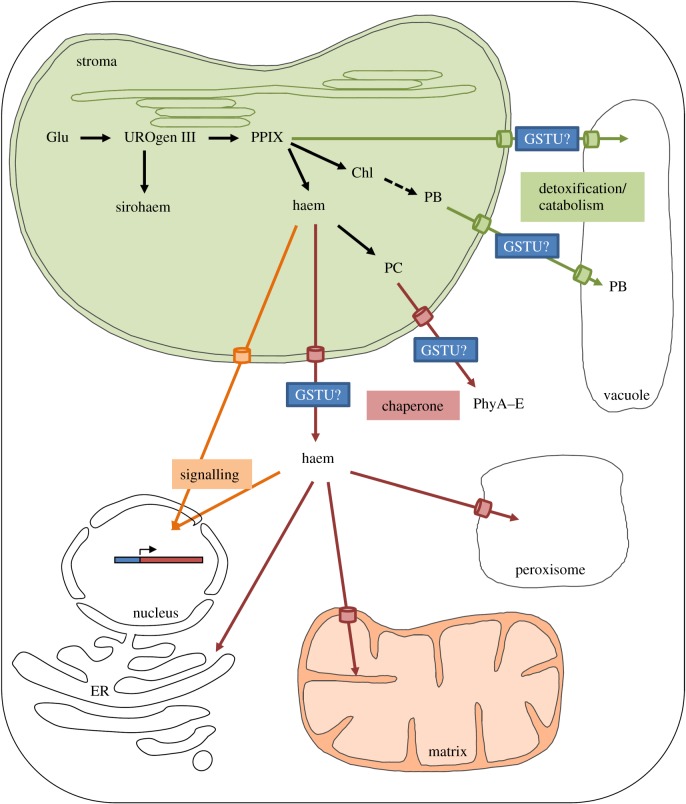
As already mentioned, the GST family has expanded in plants with members of the overrepresented GSTU class (28 members in A. thaliana) having been reported as binding porphyrin moieties, but the potential associated physiological role(s) have never been discussed in detail [29]. The absence of genetic evidence may be due to redundancies existing among these multiple isoforms. Except for A. thaliana GSTU12, which has an N-terminal extension containing a nuclear localization signal (KKRKK) (electronic supplementary material, figure S4) and is therefore found in the nucleus [73], GSTUs are mostly predicted as cytosolic proteins. However, both GSTU19 and GSTU20 have been found in chloroplast proteomes (chloroKB database), suggesting either their presence in the stroma or at least an external association with the envelope [74]. Moreover, GSTU2 from Vitis vinifera was detected at the plasma membrane and its interaction with the plasma membrane-bound HIR protein serves for the export of trans-resveratrol [39]. Hence, the proposed roles for GSTUs have to be discussed taking into account these localizations and their documented functional connexion with ABC transporters, notably of the MRP/ABCC family. It is noteworthy that other GSTs may play additional roles in specific compartments because GSTs have usually quite similar ligandin or substrate-binding sites that should also allow the accommodation of tetrapyrroles. In such a coupled chaperone-transport system and considering that GSTUs represent general stress-response factors, are quite abundant and are able to bind and/or to conjugate porphyrin moieties to GSH, they could exert several possible roles that are discussed in the following sections (figure 5).
Figure 5.
Hypothetical model for the involvement of GSTUs in the intracellular trafficking of tetrapyrroles. Unlike sirohaems and chlorophylls that remain in the chloroplastic site of synthesis, other tetrapyrroles translocated from the chloroplasts may react with GSTUs for their detoxification (phyllobilins (PB) and chlorophyll derivatives (Mg-PPIX, Pchlide)), for their trafficking to other subcellular compartments (phytochromobilins (PC) and haems) or for signalling functions (haems). Required transporter proteins are represented, but their identities are so far unknown. Other abbreviations are as follows: Glu, glutamate; UROgen III, uroporphyrinogen III; PPIX, protoporphyrin IX; Chl, chlorophylls; PhyA–E, phytochromes A–E.
(a). Functions in detoxification
GSTUs have well-documented roles in xenobiotic detoxification or sequestration of specialized metabolites and their expression is often strongly regulated during stress conditions [29]. For instance, the TT19/TT12 (GST/ABC transporter) couple is involved in anthocyanin vacuolar sequestration in A. thaliana [36]. A similar system may operate in V. vinifera where ABCC1 ensures the co-transport of glycosylated anthocyanidin and GSH, confirming that GSH conjugation is not an essential prerequisite for anthocyanin transport [75]. In other respects, these ABCC/MRP transporters are defined as GS-X pumps, being responsible for a large number of transport processes, including the transport of glutathione and glucuronide conjugates as well as bile acids in humans. For instance, human MRP1 and MRP2 act synergistically with GSTP1-1 in the detoxification of cytotoxic or genotoxic agents [76,77]. Some tonoplastic A. thaliana orthologues (MRP2/ABCC2 and MRP3/ABCC3) transport glutathione conjugates, notably glutathionylated herbicides, but also chlorophyll catabolites, notably a non-fluorescent chlorophyll catabolite (NCC) when expressed in yeast [78,79]. Thus, it is tempting to hypothesize that GSTUs, in association with yet to be identified chloroplastic envelope and tonoplastic transporters, are required as a soluble transport system for the detoxification or vacuolar sequestration of phyllobilins for their subsequent recycling (figure 5).
Another possible role of GSTUs associated with porphyrin binding is to detoxify and thus prevent the action of some reactive porphyrins. First, although tetrapyrrole synthesis and degradation are usually carefully adjusted to the cellular requirements and the potentially toxic intermediates of tetrapyrrole synthesis are usually maintained at low levels in chloroplasts, it may be that some GSTs, either uncharacterized chloroplastic GSTUs or other GSTs, prevent their reactivity via their ligandin function. On the other hand, the concentration threshold of some photosensitizing porphyrins (PPIX and its Mg2+ derivatives, including protochlorophyllide) under environmental constraints may overload the antioxidant systems that normally maintain these metabolites in their reduced, non-toxic forms. In such a situation, these reactive porphyrins are released from the chloroplasts and GSTUs may be part of a protective mechanism preventing subsequent oxidative damage by binding or targeting these molecules to the vacuoles via enzymatic (GSH conjugation) or non-enzymatic (ligandin) reactions.
(b). Functions as haem/phytochromobilin chaperones
The current knowledge about the mechanisms of intracellular phytochromobilin and haem trafficking in plants remains very limited, as in most eukaryotes. This contrasts with the more extensively characterized bacterial haem transport systems [80]. Conceptually, these tetrapyrrole entities, which are synthesized in plastids, have to be translocated outside this organelle and distributed to cytosolic phytochromes (PHYA–E) in the case of phytochromobilin or to a large number of proteins found in diverse compartments, e.g. mitochondrial matrix, mitochondrial inter-membrane space, nucleus, ER surface, peroxisomes and cytosol in the case of haems (figure 5). As free haem is hydrophobic and cytotoxic, specific molecules and pathways must ensure the efficient targeting of these cofactors to final client haemoproteins. Thinking more specifically about the evolutionary conservation of the haem chaperones and trafficking pathways in eukaryotes, it is expected that the actors are somehow conserved. Indeed, most chaperones and transporters proposed as being involved (HBPs, GAPDH, GSTs, MRP/ABC transporters) are ubiquitous. In plants, considering their capacity to bind haems and to interact with ABC transporters, members of the large GSTU class might well be involved in the cytosolic trafficking of haems, but also phytochromobilins, to final client proteins. Given their abundance, they may serve as reservoir or transient haem storage pools that can become rapidly available in specific physiological circumstances. An extension of this chaperone function would be an involvement in haem signalling functions and notably in the plastid-to-nucleus retrograde signalling in plants (discussed below).
(c). Functions in plastid-to-nucleus retrograde signalling
The exchange of signals between cellular compartments allows the coordination of development and differentiation, the modulation of metabolic pathways and triggers responses to environmental conditions [5]. For changes occurring in plastids, communication with the nucleus is essential and so-called plastid-to-nucleus retrograde signals operate. A difference is made between a biogenic control that consists of signals associated with chloroplast biogenesis and an operational control that consists of signals originating from developed chloroplasts in mature organs in response to environmental changes. This retrograde signalling is a complex and multi-layered process involving a number of components, i.e. metabolites, reactive oxygen species, proteins or tetrapyrroles, that ultimately regulate nuclear gene expression, notably photosynthesis-associated nuclear genes [5]. The mutant screens developed to understand plastid-to-nucleus signalling mechanisms in Arabidopsis notably made use of norflurazon, a herbicide, inhibitor of carotenoid biosynthesis, that blocks chloroplast biogenesis [81]. This led to the isolation of so-called genomes uncoupled (gun) mutants in which the expression of the Lhcb nuclear gene is maintained. Most proteins encoded by the defective genes in the selected loss-of-function mutants are involved in tetrapyrrole metabolism. The gun2 mutant is modified for the HO1/HY1 gene, gun3 for HY2 and gun5 for the gene encoding the H subunit of the Mg chelatase (CHLH) [81,82]. The gun4 mutant is affected for a regulator of Mg chelatase activity [83]. An additional gain-of-function mutant overexpressing FC1 (gun6-1D) also exhibited a gun phenotype [19]. Different forms of tetrapyrroles (Mg-PPIX, bilin, haem) have been described as signalling molecules in either terrestrial plants or algae, but a role of Mg-PPIX has then been refuted (reviewed in [84,85]). Recent investigations in Chlamydomonas using the hmox1 mutant for the HO pointed to a possible role of bilin as a biogenic retrograde signal that would be essential for photoacclimation and functional chloroplast maintenance during dark-to-light transition. Two mechanisms may operate: a bilin-based, blue-light-sensing system and a bilin-based retrograde signalling pathway [86,87]. The literature in the field is extremely abundant and we refer the readers to recent reviews for more details [5,84,85].
In brief, the current model, at least for the biogenic control, is that there is a positive haem-related signal that is exported from chloroplasts reflecting their developmental status [84]. Among the two ferrochelatases present in A. thaliana, only FC1 seems implicated in this signalling pathway [19]. Because FC1 is anchored to the chloroplast envelope and free haem is toxic, we speculate that other proteins participate in the signalling process, i.e. transporter(s) and cytoplasmic and/or nuclear factors/chaperones. In fact, the human mitochondrial ferrochelatase interacts with ABC transporters [88]. Hence, a similar trafficking towards an ABC transporter present in the chloroplast envelope is possible but remains to be characterized. There is also an urgent need to identify whether and which protein(s) would serve as soluble haem or haem-derivative receptors in plastids, cytosol or nucleus for further trafficking. As these factors may be similar to those involved in the trafficking of haems towards haemoproteins (haem chaperone function), proteins belonging to the HBP or GST families are obvious candidates (figure 5). In addition to this positive haem-dependent signal, other signals are produced by chloroplasts in response to environmental variations, and it is proposed that singlet oxygen (1O2), which can be produced from Mg-porphyrins under specific conditions, initiates a second inhibitory light-dependent signal, repressing both photosynthesis and tetrapyrrole synthesis genes [89]. As for the haem-dependent signal, it is not clear whether 1O2 itself or a derived molecule comes into play. Indeed, 1O2 will react in particular with carotenoids present in PSII, forming degradation products such as β-cyclocitral (β-CC), and with lipids leading to their peroxidation that can further generate toxic reactive carbonyl species. Both types of molecules have signalling functions and a recent study highlighted that β-CC induced the expression of genes encoding proteins implicated in the xenobiotic detoxification response, including many GSTs [90]. Hence, GSTs may also participate in this signalling pathway, independently of their interaction with tetrapyrroles, owing to the binding or GSH conjugation of β-CC or other carotenoid derivatives, to their GSH-dependent peroxidase activity towards peroxidized lipids or to their capacity to conjugate GSH onto reactive carbonyl species to prevent their reactivity.
7. Conclusion
Among eukaryotes, plants are unique in having the whole tetrapyrrole synthesis pathway in chloroplasts. Some of the synthesized tetrapyrrole-containing cofactors are required outside this compartment, but information about their trafficking (binding, transport) is almost nonexistent. By providing experimental data and structural models indicating that GSTUs bind PPIX and haem moieties in their substrate-binding sites, we have speculated about the possible roles played by GSTUs particularly as haem chaperones and as actors of the plastid-to-nucleus retrograde signalling, the topic of this special theme issue. There is now an urgent need to accumulate more biochemical and structural data on diverse GST isoforms, to identify transporters for haems, phytochromobilins or other important intermediates and their connection with GSTs, if any. Even more essential will be to obtain genetic and physiological evidence for the contribution of GSTUs in tetrapyrrole metabolism, despite the drawback of having a multigene family.
Supplementary Material
Acknowledgements
We thank Fabien Lachaud and François Dupire from the mass spectrometry facility platform.
Data accessibility
Data will be publicly available. The material and methods used are described in the electronic supplementary files.
Authors' contributions
E.S.-G. and J.-M.G. performed the experiments designed together with A.H. and N.R. M.S. carried out the structural modelling analyses. All authors participated in the writing of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
A.H., J.-M.G. and N.R. are supported by the French National Research Agency (grant nos ANR-11-LABX-0002-01, ANR-17-CE20-0008-01). M.S. salary was funded by a grant from Lorraine Université Excellence.
References
- 1.Tanaka R, Tanaka A. 2007. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 58, 321–346. ( 10.1146/annurev.arplant.57.032905.105448) [DOI] [PubMed] [Google Scholar]
- 2.Brzezowski P, Richter AS, Grimm B. 2015. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochim. Biophys. Acta 1847, 968–985. ( 10.1016/j.bbabio.2015.05.007) [DOI] [PubMed] [Google Scholar]
- 3.Richter AS, Grimm B. 2013. Thiol-based redox control of enzymes involved in the tetrapyrrole biosynthesis pathway in plants. Front. Plant Sci. 4, 371 ( 10.3389/fpls.2013.00371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuai B, Chen J, Hörtensteiner S. 2018. The biochemistry and molecular biology of chlorophyll breakdown. J. Exp. Bot. 69, 751–767. ( 10.1093/jxb/erx322) [DOI] [PubMed] [Google Scholar]
- 5.Kleine T, Leister D. 2016. Retrograde signaling: organelles go networking. Biochim. Biophys. Acta 1857, 1313–1325. ( 10.1016/j.bbabio.2016.03.017) [DOI] [PubMed] [Google Scholar]
- 6.Strand A, Asami T, Alonso J, Ecker JR, Chory J. 2003. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421, 79–83. ( 10.1038/nature01204) [DOI] [PubMed] [Google Scholar]
- 7.Hanna DA, Harvey RM, Martinez-Guzman O, Yuan X, Chandrasekharan B, Raju G, Outten FW, Hamza I, Reddi AR. 2016. Heme dynamics and trafficking factors revealed by genetically encoded fluorescent heme sensors. Proc. Natl Acad. Sci. USA 113, 7539–7544. ( 10.1073/pnas.1523802113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohchi T, Mukougawa K, Frankenberg N, Masuda M, Yokota A, Lagarias JC. 2001. The Arabidopsis HY2 gene encodes phytochromobilin synthase, a ferredoxin-dependent biliverdin reductase. Plant Cell 13, 425–436. ( 10.1105/tpc.13.2.425) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gisk B, Yasui Y, Kohchi T, Frankenberg-Dinkel N. 2010. Characterization of the haem oxygenase protein family in Arabidopsis thaliana reveals a diversity of functions. Biochem. J. 425, 425–434. ( 10.1042/BJ20090775) [DOI] [PubMed] [Google Scholar]
- 10.Davis SJ, Kurepa J, Vierstra RD. 1999. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc. Natl Acad. Sci. USA 96, 6541–6546. ( 10.1073/pnas.96.11.6541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muramoto T, Tsurui N, Terry MJ, Yokota A, Kohchi T. 2002. Expression and biochemical properties of a ferredoxin-dependent heme oxygenase required for phytochrome chromophore synthesis. Plant Physiol. 130, 1958–1966. ( 10.1104/pp.008128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emborg TJ, Walker JM, Noh B, Vierstra RD. 2006. Multiple heme oxygenase family members contribute to the biosynthesis of the phytochrome chromophore in Arabidopsis. Plant Physiol. 140, 856–868. ( 10.1104/pp.105.074211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis SJ, Bhoo SH, Durski AM, Walker JM, Vierstra RD. 2001. The heme-oxygenase family required for phytochrome chromophore biosynthesis is necessary for proper photomorphogenesis in higher plants. Plant Physiol. 126, 656–669. ( 10.1104/pp.126.2.656) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taketani S, Adachi Y, Kohno H, Ikehara S, Tokunaga R, Ishii T. 1998. Molecular characterization of a newly identified heme-binding protein induced during differentiation of urine erythroleukemia cells. J. Biol. Chem. 273, 31 388–31 394. ( 10.1074/jbc.273.47.31388) [DOI] [PubMed] [Google Scholar]
- 15.Vincent SH, Muller-Eberhard U. 1985. A protein of the Z class of liver cytosolic proteins in the rat that preferentially binds heme. J. Biol. Chem. 260, 14 521–14 528. [PubMed] [Google Scholar]
- 16.Sweeny EA, et al. 2018. Glyceraldehyde-3-phosphate dehydrogenase is a chaperone that allocates labile heme in cells. J. Biol. Chem. 293, 14 557–14 568. ( 10.1074/jbc.RA118.004169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi S, Ogawa T, Inoue K, Masuda T. 2008. Characterization of cytosolic tetrapyrrole-binding proteins in Arabidopsis thaliana. Photochem. Photobiol. Sci. 7, 1216–1224. ( 10.1039/b802588f) [DOI] [PubMed] [Google Scholar]
- 18.Lee H-J, Mochizuki N, Masuda T, Buckhout TJ. 2012. Disrupting the bimolecular binding of the haem-binding protein 5 (AtHBP5) to haem oxygenase 1 (HY1) leads to oxidative stress in Arabidopsis. J. Exp. Bot. 63, 5967–5978. ( 10.1093/jxb/ers242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Woodson JD, Perez-Ruiz JM, Chory J. 2011. Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903. ( 10.1016/j.cub.2011.04.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Espinas NA, Kobayashi K, Sato Y, Mochizuki N, Takahashi K, Tanaka R, Masuda T. 2016. Allocation of heme is differentially regulated by ferrochelatase isoforms in Arabidopsis cells. Front. Plant Sci. 7, 1326 ( 10.3389/fpls.2016.01326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scharfenberg M, Mittermayr L, Von Roepenack-Lahaye E, Schlicke H, Grimm B, Leister D, Kleine T.. 2015. Functional characterization of the two ferrochelatases in Arabidopsis thaliana. Plant Cell Environ. 38, 280–298. ( 10.1111/pce.12248) [DOI] [PubMed] [Google Scholar]
- 22.Fan T, Roling L, Meiers A, Brings L, Ortega-Rodés P, Hedtke B, Grimm B. 2019. Complementation studies of the Arabidopsis fc1 mutant substantiate essential functions of ferrochelatase 1 during embryogenesis and salt stress. Plant Cell Environ. 42, 618–632. ( 10.1111/pce.13448) [DOI] [PubMed] [Google Scholar]
- 23.Roper JM, Smith AG. 1997. Molecular localisation of ferrochelatase in higher plant chloroplasts. Eur. J. Biochem. 246, 32–37. ( 10.1111/j.1432-1033.1997.t01-1-00032.x) [DOI] [PubMed] [Google Scholar]
- 24.Severance S, Hamza I. 2009. Trafficking of heme and porphyrins in Metazoa. Chem. Rev. 109, 4596–4616. ( 10.1021/cr9001116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhee C, Zapotoczny G, Masquelier D, Ghislain M, Batoko H. 2011. The Arabidopsis multistress regulator TSPO is a heme binding membrane protein and a potential scavenger of porphyrins via an autophagy-dependent degradation mechanism. Plant Cell 23, 785–805. ( 10.1105/tpc.110.081570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balsemão-Pires E, Jaillais Y, Olson BJSC, Andrade LR, Umen JG, Chory J, Sachetto-Martins G. 2011. The Arabidopsis translocator protein (AtTSPO) is regulated at multiple levels in response to salt stress and perturbations in tetrapyrrole metabolism. BMC Plant Biol. 11, 108 ( 10.1186/1471-2229-11-108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morel M, Meux E, Mathieu Y, Thuillier A, Chibani K, Harvengt L, Jacquot J-P, Gelhaye E. 2013. Xenomic networks variability and adaptation traits in wood decaying fungi. Microb. Biotechnol. 6, 248–263. ( 10.1111/1751-7915.12015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plomion C, et al. 2018. Oak genome reveals facets of long lifespan. Nat. Plants 4, 440–452. ( 10.1038/s41477-018-0172-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sylvestre-Gonon E, Law SR, Schwartz M, Robe K, Keech O, Didierjean C, Dubos C, Rouhier N, Hecker A. 2019. Functional, structural and biochemical features of plant serinyl-glutathione transferases. Front. Plant Sci. 10, 608 ( 10.3389/fpls.2019.00608) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lallement P-A, Brouwer B, Keech O, Hecker A, Rouhier N. 2014. The still mysterious roles of cysteine-containing glutathione transferases in plants. Front. Pharmacol. 5, 192 ( 10.3389/fphar.2014.00192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cummins I, Dixon DP, Freitag-Pohl S, Skipsey M, Edwards R. 2011. Multiple roles for plant glutathione transferases in xenobiotic detoxification. Drug Metab. Rev. 43, 266–280. ( 10.3109/03602532.2011.552910) [DOI] [PubMed] [Google Scholar]
- 32.Ahmad L, Rylott EL, Bruce NC, Edwards R, Grogan G. 2017. Structural evidence for Arabidopsis glutathione transferase AtGSTF2 functioning as a transporter of small organic ligands. FEBS OpenBio 7, 122–132. ( 10.1002/2211-5463.12168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dixon DP, Skipsey M, Edwards R. 2010. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry 71, 338–350. ( 10.1016/j.phytochem.2009.12.012) [DOI] [PubMed] [Google Scholar]
- 34.Su T, Xu J, Li Y, Lei L, Zhao L, Yang H, Feng J, Liu G, Ren D. 2011. Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23, 364–380. ( 10.1105/tpc.110.079145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piślewska-Bednarek M, et al. 2018. Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol. 176, 538–551. ( 10.1104/pp.17.01455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitamura S, Shikazono N, Tanaka A. 2004. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 37, 104–114. ( 10.1046/j.1365-313X.2003.01943.x) [DOI] [PubMed] [Google Scholar]
- 37.Sun Y, Li H, Huang J-R. 2012. Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol. Plant 5, 387–400. ( 10.1093/mp/ssr110) [DOI] [PubMed] [Google Scholar]
- 38.Dixon DP, Edwards R. 2018. Protein-ligand fishing in planta for biologically active natural products using glutathione transferases. Front. Plant Sci. 9, 1659 ( 10.3389/fpls.2018.01659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Márquez A, Martínez-Esteso MJ, Vilella-Antón MT, Sellés-Marchart S, Morante-Carriel JA, Hurtado E, Palazon J, Bru-Martínez R. 2017. A tau class glutathione-S-transferase is involved in trans-resveratrol transport out of grapevine cells. Front. Plant Sci. 8, 1457 ( 10.3389/fpls.2017.01457) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon DP, Edwards R. 2009. Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J. Biol. Chem. 284, 21 249–21 256. ( 10.1074/jbc.M109.020107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dixon DP, Lapthorn A, Madesis P, Mudd EA, Day A, Edwards R. 2008. Binding and glutathione conjugation of porphyrinogens by plant glutathione transferases. J. Biol. Chem. 283, 20 268–20 276. ( 10.1074/jbc.M802026200) [DOI] [PubMed] [Google Scholar]
- 42.Lederer B, Böger P. 2003. Binding and protection of porphyrins by glutathione S-transferases of Zea mays L. Biochim. Biophys. Acta 1621, 226–233. ( 10.1016/S0304-4165(03)00073-4) [DOI] [PubMed] [Google Scholar]
- 43.Lederer B, Böger P. 2005. A ligand function of glutathione S-transferase. Z. Naturforsch. C J. Biosci. 60, 166–171. [DOI] [PubMed] [Google Scholar]
- 44.Ketley JN, Habig WH, Jakoby WB. 1975. Binding of nonsubstrate ligands to the glutathione S-transferases. J. Biol. Chem. 250, 8670–8673. [PubMed] [Google Scholar]
- 45.Boyer TD. 1986. Covalent labeling of the nonsubstrate ligand-binding site of glutathione S-transferases with bilirubin-Woodward's reagent K. J. Biol. Chem. 261, 5363–5367. [PubMed] [Google Scholar]
- 46.Boyer TD, Vessey DA. 1987. Inhibition of human cationic glutathione S-transferase by nonsubstrate ligands. Hepatology 7, 843–848. ( 10.1002/hep.1840070509) [DOI] [PubMed] [Google Scholar]
- 47.Mannervik B, Alin P, Guthenberg C, Jensson H, Tahir MK, Warholm M, Jörnvall H. 1985. Identification of three classes of cytosolic glutathione transferase common to several mammalian species: correlation between structural data and enzymatic properties. Proc. Natl Acad. Sci. USA 82, 7202–7206. ( 10.1073/pnas.82.21.7202) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harwaldt P, Rahlfs S, Becker K. 2002. Glutathione S-transferase of the malarial parasite Plasmodium falciparum: characterization of a potential drug target. Biol. Chem. 383, 821–830. ( 10.1515/BC.2002.086) [DOI] [PubMed] [Google Scholar]
- 49.Hiller N, Fritz-Wolf K, Deponte M, Wende W, Zimmermann H, Becker K. 2006. Plasmodium falciparum glutathione S-transferase—structural and mechanistic studies on ligand binding and enzyme inhibition. Protein Sci. 15, 281–289. ( 10.1110/ps.051891106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Al-Qattan MN, Mordi MN, Mansor SM. 2016. Assembly of ligands interaction models for glutathione-S-transferases from Plasmodium falciparum, human and mouse using enzyme kinetics and molecular docking. Comput. Biol. Chem. 64, 237–249. ( 10.1016/j.compbiolchem.2016.07.007) [DOI] [PubMed] [Google Scholar]
- 51.Lisewski AM, et al. 2014. Supergenomic network compression and the discovery of EXP1 as a glutathione transferase inhibited by artesunate. Cell 158, 916–928. ( 10.1016/j.cell.2014.07.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada T, Takusagawa F. 2007. PGH2 degradation pathway catalyzed by GSH−heme complex bound microsomal prostaglandin E2 synthase type 2: the first example of a dual-function enzyme. Biochemistry 46, 8414–8424. ( 10.1021/bi700605m) [DOI] [PubMed] [Google Scholar]
- 53.Caccuri AM, Aceto A, Piemonte F, Di Ilio C, Rosato N, Federici G.. 1990. Interaction of haemin with placental glutathione transferase. Eur. J. Biochem. 189, 493–497. ( 10.1111/j.1432-1033.1990.tb15514.x) [DOI] [PubMed] [Google Scholar]
- 54.Plancarte A, Romero JR, Nava G, Reyes H, Hernández M. 2014. Evaluation of the non-catalytic binding function of Ts26GST a glutathione transferase isoform of Taenia solium. Exp. Parasitol. 138, 63–70. ( 10.1016/j.exppara.2014.02.004) [DOI] [PubMed] [Google Scholar]
- 55.Liebau E, et al. 2005. Cooperativity and pseudo-cooperativity in the glutathione S-transferase from Plasmodium falciparum. J. Biol. Chem. 280, 26 121–26 128. ( 10.1074/jbc.M503889200) [DOI] [PubMed] [Google Scholar]
- 56.Oakley A. 2011. Glutathione transferases: a structural perspective. Drug Metab. Rev. 43, 138–151. ( 10.3109/03602532.2011.558093) [DOI] [PubMed] [Google Scholar]
- 57.Senjo M, Ishibashi T, Imai Y. 1985. Purification and characterization of cytosolic liver protein facilitating heme transport into apocytochrome b5 from mitochondria. Evidence for identifying the heme transfer protein as belonging to a group of glutathione S-transferases. J. Biol. Chem. 260, 9191–9196. [PubMed] [Google Scholar]
- 58.Paiva-Silva GO, Cruz-Oliveira C, Nakayasu ES, Maya-Monteiro CM, Dunkov BC, Masuda H, Almeida IC, Oliveira PL. 2006. A heme-degradation pathway in a blood-sucking insect. Proc. Natl Acad. Sci. USA 103, 8030–8035. ( 10.1073/pnas.0602224103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Atamna H, Ginsburg H. 1995. Heme degradation in the presence of glutathione. A proposed mechanism to account for the high levels of non-heme iron found in the membranes of hemoglobinopathic red blood cells. J. Biol. Chem. 270, 24 876–24 883. ( 10.1074/jbc.270.42.24876) [DOI] [PubMed] [Google Scholar]
- 60.Dubois VL, Platel DF, Pauly G, Tribouley-Duret J. 1995. Plasmodium berghei: implication of intracellular glutathione and its related enzyme in chloroquine resistance in vivo. Exp. Parasitol. 81, 117–124. ( 10.1006/expr.1995.1099) [DOI] [PubMed] [Google Scholar]
- 61.Singh BR, Shaw RW. 1988. Selective inhibition of oat glutathione-S-transferase activity by tetrapyrroles. FEBS Lett. 234, 379–382. ( 10.1016/0014-5793(88)80120-0) [DOI] [Google Scholar]
- 62.Zachariah VT, Walsh-Sayles N, Singh BR. 2000. Isolation, purification, and characterization of glutathione S-transferase from oat (Avena sativa) seedlings. J. Protein Chem. 19, 425–430. ( 10.1023/a:1026513528654) [DOI] [PubMed] [Google Scholar]
- 63.Dixon DP, McEwen AG, Lapthorn AJ, Edwards R. 2003. Forced evolution of a herbicide detoxifying glutathione transferase. J. Biol. Chem. 278, 23 930–23 935. ( 10.1074/jbc.M303620200) [DOI] [PubMed] [Google Scholar]
- 64.Thom R, Cummins I, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ. 2002. Structure of a tau class glutathione S-transferase from wheat active in herbicide detoxification. Biochemistry 41, 7008–7020. ( 10.1021/bi015964x) [DOI] [PubMed] [Google Scholar]
- 65.Valenzuela-Chavira I, et al. 2017. Insights into ligand binding to a glutathione S-transferase from mango: structure, thermodynamics and kinetics. Biochimie 135, 35–45. ( 10.1016/j.biochi.2017.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Axarli I, Dhavala P, Papageorgiou AC, Labrou NE. 2009. Crystallographic and functional characterization of the fluorodifen-inducible glutathione transferase from Glycine max reveals an active site topography suited for diphenylether herbicides and a novel L-site. J. Mol. Biol. 385, 984–1002. ( 10.1016/j.jmb.2008.10.084) [DOI] [PubMed] [Google Scholar]
- 67.Skopelitou K, Muleta AW, Papageorgiou AC, Chronopoulou E, Labrou NE. 2015. Catalytic features and crystal structure of a tau class glutathione transferase from Glycine max specifically upregulated in response to soybean mosaic virus infections. Biochim. Biophys. Acta Proteins Proteom. 1854, 166–177. ( 10.1016/j.bbapap.2014.11.008) [DOI] [PubMed] [Google Scholar]
- 68.Yang Q, Han X-M, Gu J-K, Liu Y-J, Yang M-J, Zeng Q-Y. 2019. Functional and structural profiles of GST gene family from three Populus species reveal the sequence-function decoupling of orthologous genes. New Phytol. 221, 1060–1073. ( 10.1111/nph.15430) [DOI] [PubMed] [Google Scholar]
- 69.Tossounian M-A, et al. 2018. Disulfide bond formation protects Arabidopsis thaliana glutathione transferase tau 23 from oxidative damage. Biochim. Biophys. Acta Gen. Subjects 1862, 775–789. ( 10.1016/j.bbagen.2017.10.007) [DOI] [PubMed] [Google Scholar]
- 70.Jurrus E, et al. 2018. Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128. ( 10.1002/pro.3280) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen X, Pu H, Wang X, Long W, Lin R, Liu L. 2015. Crystal structures of GUN4 in complex with porphyrins. Mol. Plant 8, 1125–1127. ( 10.1016/j.molp.2015.04.013) [DOI] [PubMed] [Google Scholar]
- 72.Schwartz M, et al. 2018. Molecular recognition of wood polyphenols by phase II detoxification enzymes of the white rot Trametes versicolor. Sci. Rep. 8, 8472 ( 10.1038/s41598-018-26601-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dixon DP, Hawkins T, Hussey PJ, Edwards R. 2009. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 60, 1207–1218. ( 10.1093/jxb/ern365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gloaguen P, et al. 2017. ChloroKB: a web application for the integration of knowledge related to chloroplast metabolic network. Plant Physiol. 174, 922–934. ( 10.1104/pp.17.00242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Francisco RM, et al. 2013. ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. ( 10.1105/tpc.112.102152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Paumi CM, Ledford BG, Smitherman PK, Townsend AJ, Morrow CS. 2001. Role of multidrug resistance protein 1 (MRP1) and glutathione S-transferase A1-1 in alkylating agent resistance. Kinetics of glutathione conjugate formation and efflux govern differential cellular sensitivity to chlorambucil versus melphalan toxicity. J. Biol. Chem. 276, 7952–7956. ( 10.1074/jbc.M009400200) [DOI] [PubMed] [Google Scholar]
- 77.Morrow CS, Smitherman PK, Townsend AJ. 2000. Role of multidrug-resistance protein 2 in glutathione S-transferase P1-1-mediated resistance to 4-nitroquinoline 1-oxide toxicities in HepG2 cells. Mol. Carcinog. 29, 170–178. () [DOI] [PubMed] [Google Scholar]
- 78.Tommasini R, Vogt E, Fromenteau M, Hörtensteiner S, Matile P, Amrhein N, Martinoia E. 1998. An ABC-transporter of Arabidopsis thaliana has both glutathione-conjugate and chlorophyll catabolite transport activity. Plant J. 13, 773–780. ( 10.1046/j.1365-313X.1998.00076.x) [DOI] [PubMed] [Google Scholar]
- 79.Lu YP, Li ZS, Drozdowicz YM, Hortensteiner S, Martinoia E, Rea PA. 1998. AtMRP2, an Arabidopsis ATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: functional comparisons with Atmrp1. Plant Cell 10, 267–282. ( 10.1105/tpc.10.2.267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang W, Wilks A. 2017. Extracellular heme uptake and the challenge of bacterial cell membranes. Annu. Rev. Biochem. 86, 799–823. ( 10.1146/annurev-biochem-060815-014214) [DOI] [PubMed] [Google Scholar]
- 81.Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. ( 10.1016/0092-8674(93)90459-4) [DOI] [PubMed] [Google Scholar]
- 82.Mochizuki N, Brusslan JA, Larkin R, Nagatani A, Chory J. 2001. Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl Acad. Sci. USA 98, 2053–2058. ( 10.1073/pnas.98.4.2053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larkin RM, Alonso JM, Ecker JR, Chory J. 2003. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. ( 10.1126/science.1079978) [DOI] [PubMed] [Google Scholar]
- 84.Terry MJ, Smith AG. 2013. A model for tetrapyrrole synthesis as the primary mechanism for plastid-to-nucleus signaling during chloroplast biogenesis. Front. Plant Sci. 4, 14 ( 10.3389/fpls.2013.00014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Larkin RM. 2016. Tetrapyrrole signaling in plants. Front. Plant Sci. 7, 1586 ( 10.3389/fpls.2016.01586) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Duanmu D, et al. 2013. Retrograde bilin signaling enables Chlamydomonas greening and phototrophic survival. Proc. Natl Acad. Sci. USA 110, 3621–3626. ( 10.1073/pnas.1222375110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wittkopp TM, et al. 2017. Bilin-dependent photoacclimation in Chlamydomonas reinhardtii. Plant Cell 29, 2711–2726. ( 10.1105/tpc.17.00149) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maio N, Kim KS, Holmes-Hampton G, Singh A, Rouault TA. 2019. Dimeric ferrochelatase bridges ABCB7 and ABCB10 homodimers in an architecturally defined molecular complex required for heme biosynthesis. Haematologica 104, 1756–1767. ( 10.3324/haematol.2018.214320) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Page MT, McCormac AC, Smith AG, Terry MJ. 2017. Singlet oxygen initiates a plastid signal controlling photosynthetic gene expression. New Phytol. 213, 1168–1180. ( 10.1111/nph.14223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.D'Alessandro S, Ksas B, Havaux M. 2018. Decoding β-cyclocitral-mediated retrograde signaling reveals the role of a detoxification response in plant tolerance to photooxidative stress. Plant Cell 30, 2495–2511. ( 10.1105/tpc.18.00578) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be publicly available. The material and methods used are described in the electronic supplementary files.