Abstract
The early steps in germination and development of angiosperm seedlings often occur in the dark, inducing a special developmental programme called skoto-morphogenesis. Under these conditions photosynthesis cannot work and all energetic requirements must be fulfilled by mitochondrial metabolization of storage energies. Here, we report the physiological impact of mitochondrial dysfunctions on the skoto-morphogenic programme by using the Arabidopsis rpoTmp mutant. This mutant is defective in the T7-phage-type organellar RNA polymerase shared by plastids and mitochondria. Lack of this enzyme causes a mitochondrial dysfunction resulting in a strongly reduced mitochondrial respiratory chain and a compensatory upregulation of the alternative-oxidase (AOX)-dependent respiration. Surprisingly, the mutant exhibits a triple-response-like phenotype with a twisted apical hook and a shortened hypocotyl. Highly similar phenotypes were detected in other respiration mutants (rug3 and atphb3) and in WT seedlings treated with the respiration inhibitor KCN. Further genetic and molecular data suggest that the observed skoto-morphogenic alterations are specifically dependent on the activity of the AOX1a enzyme. Microarray analyses indicated that a retrograde signal from mitochondria activates the ANAC017-dependent pathway which controls the activation of AOX1A transcription. In sum, our analysis identifies AOX as a functional link that couples the formation of a triple-response-like phenotype to mitochondrial dysfunction.
This article is part of the theme issue ‘Retrograde signalling from endosymbiotic organelles'.
Keywords: Arabidopsis, mitochondrial respiration, alternative oxidase, retrograde signal, organellar RPOTmp RNA polymerase, skoto-morphogenesis
1. Introduction
Mitochondria are essential cell organelles that convert organic matter into chemical energy in the form of ATP and reducing power through the process of respiration [1]. To this end, in seeds of oilseed plants such as Arabidopsis, the triacylglycerol lipids present in the oil bodies are oxidatively degraded into citrate, which is fed into the tricarboxylic acid (TCA) cycle within the mitochondrial matrix [2]. Here, the carbon skeleton is completely oxidized and CO2, ATP and reducing equivalents are released by the cycle. The reducing power then is used to drive electron transport through the mitochondrial KCN-sensitive electron transport chain (ETC) within the inner mitochondrial membrane towards oxygen as end acceptor. The ETC consists of three large multi-subunit protein complexes (complexes I, III and IV) that are connected by mobile electron carriers. The electron transport through this chain is coupled to a proton-translocation across the mitochondrial membranes, which is finally used by the ATP synthase to build ATP. The oxidized forms of the reducing equivalents produced by the ETC are then available again for the TCA cycle as electron sink. In this way, the TCA cycle and ETC are functionally coupled and any restriction in metabolic flux in one of them affects also the other [1].
Plant mitochondria display an extraordinary plasticity in their respiratory pathways as they possess additional oxidoreductases that can release excess levels of reducing power by bypassing parts of the ETC without concomitant ATP production [1,3]. At the ETC input site electrons can be donated to the alternative type II NADH dehydrogenases which bypass complex I. A second shunt is found at the end of the ETC, where electrons can be transferred to oxygen by the alternative oxidase (AOX), which bypasses complexes III and IV. The role of both the alternative NADH dehydrogenases and AOX is still a matter of debate, but they are proposed to avoid generation of dangerous reactive oxygen species (ROS) and to stabilize the NAD+/NADH balance by preventing a metabolic feedback inhibition of the TCA cycle [1]. In addition, these enzymes are also suggested to serve as safety valves for photosynthesis under various stress conditions. Experimental evidences support the function of AOX as a releasing enzyme of excess reducing power transferred from the chloroplast under high light [3]. The additional enzymes also provide high flexibility to the ETC of plant mitochondria, allowing genetic studies. Mutants in nuclear genes that are necessary for the proper expression of mitochondrial-encoded proteins are defined as surrogate mutants [4]. The surrogate mutants rpoTmp and rug3 present a defective expression of ETC complex subunits but they are viable in that they maintain residual complexes. Even if they display delayed growth phenotypes, they still generate rosettes that are comparable in size to wild-type (WT) and produce seeds [5–7]. These mutants accumulate high levels of AOX protein. In rpoTmp plants, the AOX pathway has been shown to be activated [8].
Recent studies have shown that stress induced by block of mitochondrial translation or respiration as well as by environmental constraints activates retrograde signalling pathways from mitochondria towards the nucleus [9,10]. One of the target genes is AOX1a (ALTERNATIVE OXIDASE 1a), which is now commonly used as a marker to investigate mitochondrial retrograde signalling [7,11,12]. Also, recently identified regulators of AOX1A expression (e.g. RAO1/CDKE1 and RAO2/ANAC017) seem to respond to mitochondrial as well as to chloroplast retrograde signals [13,14]. Furthermore, chloroplast redox signals from photosynthetic electron transport impact the expression of mitochondria-located genes [15]. These last studies investigating the signalling functions of the two energy-converting organelles have all been performed on plants grown in the light, where both chloroplasts and mitochondria are active. Because of the multiple connections between these two organelles, a clear separation of mitochondrial and chloroplast retrograde signals, therefore, remains difficult and partial overlaps and interactions occur in green tissues [14,16]. How a mitochondrial dysfunction would affect plant growth and development when chloroplasts and energy supply from photosynthesis are not available, i.e. during germination in the dark, is, however, not well investigated. Angiosperms such as Arabidopsis undergo a special developmental programme when they germinate and grow in the dark, skoto-morphogenesis [17]. When a germinating seed is buried by humus or soil the seedling re-allocates all storage energy into shoot elongation, while the cotyledons remain small, being directed downwards and forming an apical hook that protects the sensitive apical meristem. In this phase, the seedling relies completely on its storage energies and their metabolization by mitochondria. As soon as the cotyledons breach the surface and perceive light, photo-morphogenesis is initiated and chloroplasts are formed [18]. Mutants with mitochondrial dysfunction, therefore, should be expected to show major defects in the performance of skoto-morphogenesis.
Arabidopsis rpoTmp mutants are defective in a phage-type single-subunit RNA polymerase that is bi-localized to chloroplasts and mitochondria [19,20]. This RNA polymerase is required for the proper expression of the organelle genomes; however, it was shown that its impact in chloroplasts is restricted to the recognition of the PC promoter in front of the 16S rRNA operon [21], while it displays a major deficiency in the transcription of several mitochondrial genes, including those for complexes I and IV [5,22]. The mutant exhibits strongly reduced complex I and complex IV accumulation, but displays only a mild phenotype in the light, with delayed growth and curly leaves in the fully developed rosette [5].
In this article, we analyse the impact of restrictions of respiration on early seedling development in the dark using the respiration deficient rpoTmp mutant from Arabidopsis. We found that limitations of the ETC lead to drastic changes in the early developmental programme of etiolated seedlings and that this reprogramming of skoto-morphogenesis is triggered by the AOX1a enzyme.
2. Material and methods
(a). Plant material
Arabidopsis thaliana WT (ecotype Columbia) and mutant lines (rpoTmp-2 SALK_132842 and rpoTmp-3 SALK_086115 [5,21], rug3 SALK_092071 [6], atphb3 SALK_020707 [23], rpoTmp SALK_132842/anac017 (anac017, rao2-1) [7,24], aox1A SALK_084897 [8] and rpoTmp SALK_132842/aox1A SALK_084897 [8]) were used in this study. Seeds were surface-sterilized and spread on Murashige and Skoog (MS) agar plates supplemented with sucrose (1%). Activated charcoal (powder, SIGMA-ALDRICH, 0.08%) was added to the plant culture medium in order to improve image quality of etiolated plants by increasing background contrast. Seeds were stratified for 72 h (at 4°C in darkness), exposed to light (100 µE white light) for 6 h and then grown at 23°C in darkness. Plants were harvested after 3 days. For microarray, qRT-PCR and western-immuno blot analyses the etiolated plants were harvested in the dark under a green safe-light, immediately frozen in liquid nitrogen, ground in a mortar and subjected to further techniques as described below. The germination percentage was variable according to the seed lot and the drug treatment. For chemical treatments, MS agar plates were supplemented with 1-aminocyclopropane-carboxylic acid (ACC, Sigma-Aldrich A3903, 20 µM), KCN (1 mM) or salicylhydroxamic acid (SHAM, Sigma-Aldrich S7504, 1 mM in dimethyl sulfoxide (DMSO)). In the case of KCN or SHAM treatments, seeds were first spread on black nitrocellulose membrane filters (Whatman 10409770, 0.45 µm) on MS agar plates, stratified for 72 h and light-exposed for 6 h, and then transferred (on filter) to KCN/SHAM-containing MS agar plates for growth in the dark. DMSO-containing MS agar plates were used as mock-control for the SHAM treatment.
(b). Measurements of phenotypic parameters
Phenotypic appearance of plantlets was analysed with a dissection microscope (Olympus SZX12) using the ACT-1C for DXM1200C software. Digital images of individual plants were taken and the angles formed in the apical hook between hypocotyl and cotyledons were subsequently determined using the ImageJ program. In the absence of any curvature (cotyledons straight and along the same axis as the hypocotyl but oppositely oriented from the roots) the angle was considered as 0°. In a hook where cotyledons were aligned with the same axis as the hypocotyl, but oriented towards the roots, the angle was considered as 180°. Any hook angle larger than 180° was considered as twisted. The length of the hypocotyls was measured by determining the shoot length between the origins of roots and cotyledons.
(c). Statistical analyses of phenotypic parameter measurements
All statistical tests were performed using the computing environment R [25], with a significance level of 0.05. Details about the statistical analyses are found in the electronic supplementary material, Methods.
(d). Microarray analysis
Total RNA purification and microarray analysis were performed as indicated in [26]. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) with on-column DNase I (Quiagen) treatment. Affymetrix whole transcriptome microarray analysis was performed on three biological replicates by the commercial service Kompetenz-Zentrum für Fluoreszente Bioanalytik (KFB) (Regensburg, Germany). cDNAs were prepared using the Ambion® Whole Transcriptome (WT) Expression Kit and fragmented and labelled using the Affymetrix GeneChip® WT Terminal Labeling Kit. Expression analyses were performed using the GeneChip® Arabidopsis Gene 1.0 ST Array. Since the Ambion® WT Expression Kit uses a mixture of oligo-dT and random hexamer primers for the generation of the first-strand cDNA, the resulting hybridization signals reflect the organellar transcript accumulation as well as that from the nucleus.
(e). Analysis of microarray data
Analysis of microarray data was performed as indicated in [26]. The cell files containing the scanned chips received from KFB were analysed with RobiNA [27] with the background correction method RMA (robust multi-array average expression measure) [28], p-value correction method Benjamini–Hochberg [29] and the analysis strategy Limma for the quality control [30]. For the main analysis, the normalization method RMA with p-value correction method of Benjamini–Hochberg and a multiple testing strategy ‘nestedF’ was chosen. In the analysis using Excel (MS-Office), data with the description ‘Multiple Hits’ were ignored and only results with one-to-one correspondence to a given gene identity were employed for further analysis.
(f). RT-qPCR analysis of RNA levels
Ground plant material was resuspended in three volumes of solution A (10 mM Tris-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 1% SDS) and two volumes of phenol/chloroform/isoamyl alcohol (25 : 24 : 1, v/v/v). After centrifugation, RNAs in the aqueous phase were extracted twice more with an equal volume of phenol–chloroform (1 : 1) and finally once with chloroform. After overnight precipitation in 2 M LiCl at 4°C, RNAs were then precipitated in ethanol, washed in 70% ethanol and resuspended in water. DNAse (300 ng; DNAse Max Kit, Quiagen)-treated RNAs were retro-transcribed using random hexa nucleotides and the Super-Script II enzyme (Invitrogen) according to the manufacturer's protocol. The PCR reaction was performed in a Biorad CFX384TM Real-Time System PCR machine in the presence of forward and reverse gene-specific primers (0.5 µM; electronic supplementary material, table S1) using the SYBR® Premix Ex Taq™ (Tli RNaseH Plus) from Takara Bio. Data were analysed using the CFX Manager Software. The expression levels of the transcript of interest were normalized with the levels of ACTIN 2-8 expression, used as reference gene. The mean values of biological and/or technical replicates were plotted, together with the error bars corresponding to standard errors.
(g). Protein immuno-western analysis
Total protein extracts were prepared by re-suspending 100 mg of ground plant material in 100 µl of protein lysis buffer (50 mM Tris pH 8.0, 2% SDS, 10 mM EDTA, and protease inhibitors (Roche (04693159001) cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail)) and incubating at room temperature for 30 min. Extracts were cleared by centrifugation for 30 min at 12000g at 4°C and dilutions of the supernatant were quantified with the Bradford method. Samples were then denatured for 5 min at 95°C in 4× reducing electrophoresis sample buffer. Increasing amounts of WT and mutant protein extracts (10, 20, 40 µg) were fractionated on a 12% gel by SDS-PAGE, electro-blotted on nitrocellulose or PVDF membranes and immuno-decorated with specific polyclonal rabbit antisera against mitochondrial protein NAD9 [31], mitochondrial AOX1/2 plant alternative oxidase 1 and 2 (Agrisera, AS04 054), plastidial RBCL (Agrisera, AS03 037) and vacuole V-ATPase (Agrisera, AS07 213). The blots were developed with Clarity Western ECL substrate (Bio-Rad, Hercules, USA). Images of the blots were obtained using a CCD imager (Chemidoc MP, Bio-Rad) and the Image Lab program (Bio-Rad, Hercules, USA). Image analysis for AOX protein quantification was performed using the Image Lab program and normalized with the levels of V-ATPase.
(h). In planta O2 consumption measurements
Etiolated plantlets were harvested from Petri dishes and immediately soaked in the dark with 1 ml of oxygenated phosphate buffer (10 mM sodium phosphate buffer pH 7.2, 10 mM KCl, 10 mM glucose) in the measurement cell of a Clark oxygen electrode (Hansatech). For electron transport inhibition experiments, KCN (0.1 M stock, inhibitor of KCN-sensitive chain at the level of complex IV) and SHAM (0.1 M stock in DMSO, inhibitor of the AOX-dependent pathway) were added directly into the electrode chamber to a final concentration of 1 mM once the recorded signal reached a constant regime. The capacity (or maximum activity) of the AOX-dependent pathway (corresponding to KCN-insensitive pathway) and of the COX-dependent pathway (corresponding to KCN-sensitive pathway) was calculated as ratio of O2 consumption rate upon addition of KCN or SHAM and total O2 consumption rate, respectively. The contribution of extra mitochondrial O2 consumption to mitochondrial respiration was evaluated at the end of each measurement after addition of both KCN and SHAM, and systematically subtracted from O2 consumption rates.
3. Results
(a). rpotmp mutants display a triple-response-like phenotype
To investigate the impact of mitochondrial dysfunction on skoto-morphogenesis we compared 3-day-old dark-grown (etiolated) WT with rpoTmp mutant plants grown under identical conditions (figure 1a). WT seedlings displayed a typical skoto-morphogenesis with elongated hypocotyl, apical hook and yellow cotyledons. By contrast, seedlings of two allelic rpoTmp mutant lines (rpoTmp-2 SALK_132842 and rpoTmp-3 SALK_086115) developed a significant exaggeration of apical hook curvature (twist) and a marked shortening of the hypocotyls. The phenotype of both alleles was identical, indicating that indeed the genetic defect in rpoTmp causes the massive change in the seedling developmental programme. The observed mutant phenotype was reminiscent of morphological changes reported for etiolated seedlings exposed to high levels of ethylene, commonly known as triple-response [32,33]. To investigate this in more detail, we compared the rpoTmp phenotype with that of WT plants grown in the presence of 20 µM ACC, a biosynthetic precursor of ethylene known to induce the triple-response [34] (figure 1a). We observed comparable values for the increased apical hook curvature (figure 1b) and the hypocotyl shortening (figure 1c) in the two plant samples. Only the increase of the hypocotyl diameter in response to ACC treatment was not observable in rpoTmp (figure 1a), suggesting that the genetic defect in the RPOTmp locus causes a strong but not perfect phenocopy of the ACC treatment. Therefore, we termed the observed phenotypic response a triple-response-like phenotype.
Figure 1.
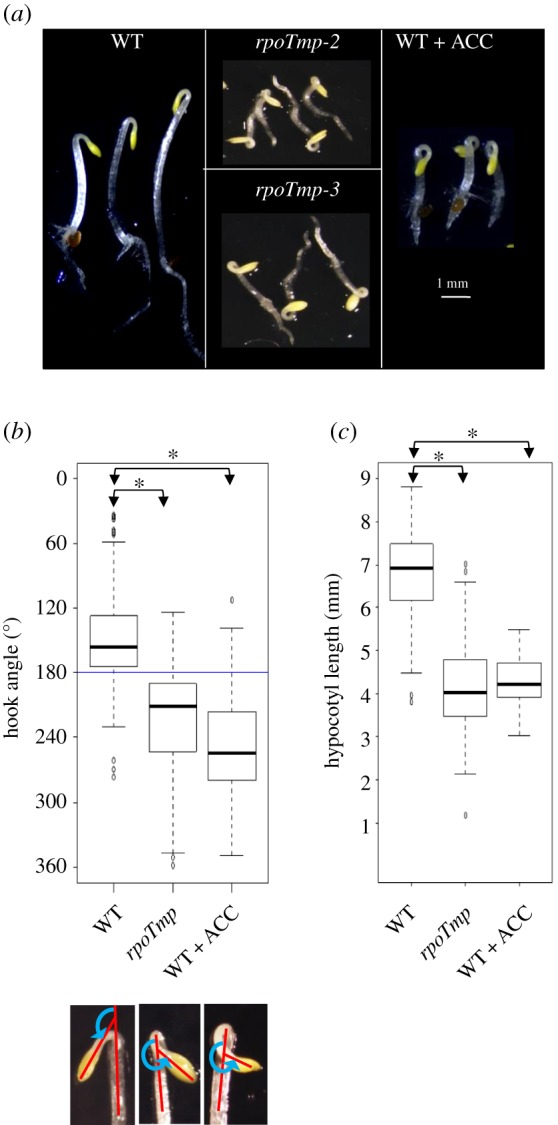
Etiolated rpoTmp mutant plantlets exhibit a triple-response-like phenotype. (a) Images of etiolated rpoTmp mutant (two mutant alleles, rpoTmp-2 SALK_132842 and rpoTmp-3 SALK_086115) and WT plants (grown in the presence or in the absence of 20 µM ACC) were taken under a dissection microscope. Scale bar corresponds to 1 mm. (b) Top panel: median values of apical hook angle measurements are box-plotted for WT plants grown in the absence or in the presence of 20 µM ACC and for rpoTmp plants. The number of pooled individuals (N) that were measured in seven independent hook angle analyses corresponds to 304 for WT plants; N (measured in three independent analyses) corresponds to 130 for WT + ACC plants; N (measured in five independent analyses) corresponds to 211 for rpoTmp plants. Data are box-plotted using versions of the first and third quartiles [25] with median values (horizontal lines inside the boxes). The external horizontal lines show the largest and smallest observations that fall within a distance of 1.5 times the box size from the nearest hinge. Additional points are extreme value observations. The asterisks (*) indicate significant difference between two samples. Bottom panel: principal of angle determination. Magnified images of the hook from WT, rpoTmp and ACC-treated WT. The apical hook curvature was measured as the angle (blue line) that is formed by the two straight lines (red) passing through the hypocotyl and the cotyledon axes. Any hook angle larger than 180° (blue horizontal line) was considered as twisted. Images were analysed with the ImageJ program. (c) Median values of hypocotyl length measurements are box-plotted for WT plants grown in the absence or in the presence of 20 µM ACC and for rpoTmp plants. N (measured in two independent analyses) corresponded to 92 for WT plants, to 75 for ACC-treated WT plants, and to 98 for rpoTmp plants. Data are box-plotted as in figure 1b. (Online version in colour.)
(b). The triple-response-like phenotype is dependent on an active AOX1a enzyme
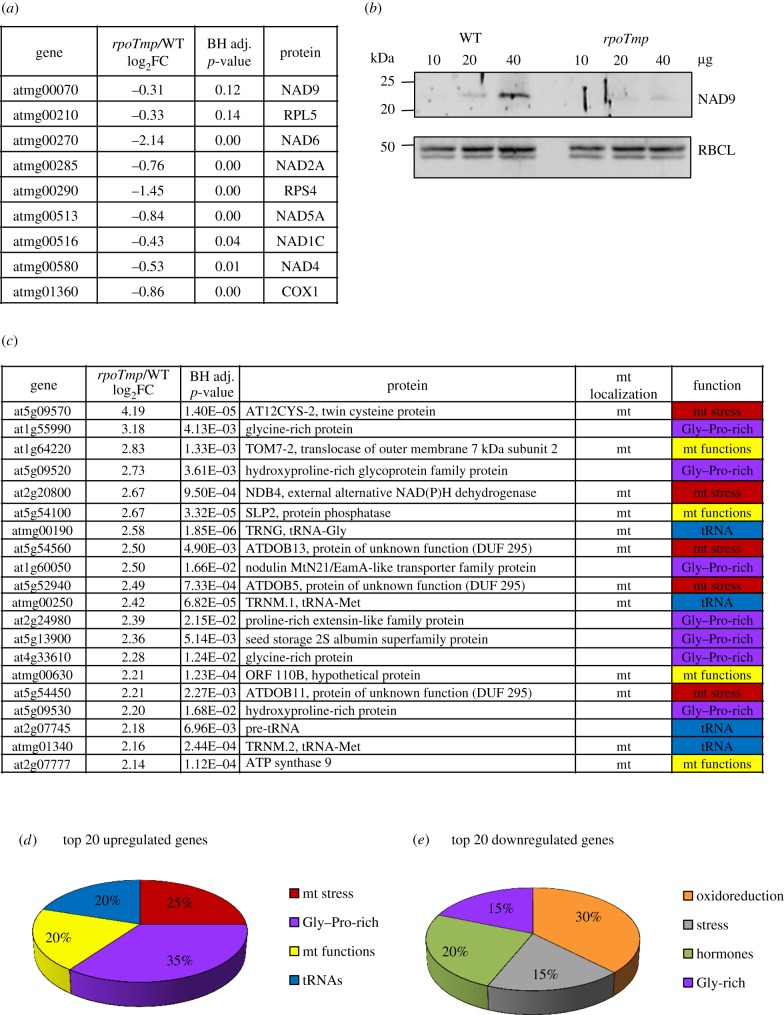
In order to find a potential link between the genetic inactivation of the RPOTmp gene and the morphological changes, we performed a transcriptomic profiling of etiolated mutant seedlings in comparison with WT (electronic supplementary material, table S2) and compared these data with previous expression analyses performed in light-grown plants. An overview of the gene expression profiles in etiolated plants in relation to the corresponding cellular metabolic pathways using the MapMan tool indicated that the general impact of the mutation in RPOTmp was relatively weak (electronic supplementary material, figure S1). Only a small number of genes displayed a strong expression change of more than log2 fold ≥2 (29 genes) or ≤−2 (60 genes, electronic supplementary material, table S2). These data suggest that the observed triple-response-like phenotype is a specific effect rather than the result of pleiotropic disturbance in the mutant. By analysing the mitochondrial transcriptome, we found that from the 20 mitochondrial genes that were presenting the highest degree of transcriptional variations in previous analyses in light-grown rpoTmp adult plants [7], 17 were also differentially expressed in etiolated rpoTmp seedlings and, here, in the same direction of expression change, even though to a lower extent (electronic supplementary material, table S3). More particularly, the subset of mitochondrial genes encoding ETC complex subunits that was previously described as RPOTmp-dependent [5] was also downregulated in etiolated rpoTmp seedlings (figure 2a). Analysis of the accumulation of the mitochondrial-encoded NAD9 protein as marker for complex I accumulation indicated that these gene expression defects were also reflected at the protein level (figure 2b). While clearly present in WT, NAD9 was almost undetectable in rpoTmp mutant plants in contrast to the control RBCL, the plastid-encoded large subunit of ribulose-1,5-bisphosphate-carboxylase/oxygenase. This indicated strong deficiency of complex I in rpoTmp plants in the dark also, which was accompanied by a decrease in the capacity of the KCN-sensitive pathway in rpoTmp seedlings (electronic supplementary material, figure S2). By analysing the transcriptomic profile in more detail, we found that more than half of the 20 most upregulated genes encode proteins that are localized in the mitochondria (figure 2c). Functional sub-grouping of these genes identified four groups: one class containing genes corresponding to markers of mitochondrial stress and targets of the retrograde-mitochondrial pathway, such as AT12CYS and its co-expression partner NDB4 [11] and three DUF295 Organellar B genes [35]; one tRNA class including one nucleus- and three mitochondria-encoded tRNAs; one class corresponding to general mitochondrial functions, including TOM7-2 (coding for a member of the family of the TOM7 translocases of outer membrane), SLP2 phosphatase [36], ATP synthase or an undefined gene; and a final group including genes coding for proline- and glycine-rich proteins, often involved in stress response (figure 2c,d). Functional sub-grouping of the 20 most downregulated genes also identified a group of genes coding for glycine-rich proteins and/or factors involved in response to stress. An additional group of genes was also defined corresponding to the response to hormonal stimuli, and a final class of genes related to reduction/oxidation activities (figure 2e; electronic supplementary material, table S4).
Figure 2.
Relative gene expression profiles of rpoTmp mutants versus WT etiolated plants. (a) The relative expression values for the RPOTmp-dependent subset of mitochondrial genes are indicated in the middle columns together with the corresponding p-values, which are derived from a t-test adjusted for false discovery rate (FDR) using the Benjamini–Hochberg (BH) procedure. Gene identities are indicated in the left, and protein identities in the right columns. Given values represent log2-fold changes (FC). (b) Analysis of accumulation of NAD9 protein used as marker for complex I accumulation. Protein extracts (10, 20 and 40 µg) from etiolated WT and rpoTmp plants were fractionated and immunoblotted with specific antisera against the mitochondrial proteins NAD9 (mitochondrial-encoded) and the plastidial protein RBCL (plastidial-encoded). (c) Relative expression values for the 20 most upregulated genes in rpoTmp plants. Details as in (a). Mitochondrial (mt) localization and protein function are also indicated on the right side of the table. ‘mt stress’ stands for ‘response to mitochondrial stress'. (d) Functional sub-grouping of the 20 most upregulated genes listed in (c). The four groups identified are colour-coded as in (c). For details see text. (e) Functional sub-grouping of the 20 most downregulated genes (electronic supplementary material, table S4). The four groups identified are colour-coded as in (c). The terms ‘hormones’ and ‘stress’ for protein functions stand for ‘response to hormonal stimuli’ and ‘response to stress’, respectively.
We have compared our transcriptomic data (threshold 1.5) concerning dark-grown rpoTmp seedlings with previous analyses performed by Van Aken and colleagues with light-grown adult plants [7]. We found 33 up DEGs (differentially expressed genes) and 40 down DEGs in common (electronic supplementary material, table S5). Of these genes, four are among the top 20 up DEGs in both studies (highlighted in yellow and by an asterisk in electronic supplementary material, table S5), eight are in the list of our top 20 up DEGs and seven are among our top 20 down DEGs (highlighted in yellow). These data emphasize the strong similarity in the gene-expression profiles of rpoTmp plants that differ for developmental stages and growth conditions. Remarkably, among the common up DEGs, 20 genes encode mitochondrial localized proteins and at least nine genes are induced by mitochondrial dysfunction and transcriptionally regulated by mitochondrial retrograde signalling, e.g. AT12CYS2 [11], NDB4 [11], five different DUF295 Organellar genes [35] and UPOX, which is an hallmark of oxidative stress [37]. All these data pointed to the presence of a strong mitochondrial stress in rpoTmp plants and prompted us to explore the possibility that defects in mitochondrial respiration are at the origin of the observed skoto-morphogenetic alteration. In order to test this hypothesis by an independent experimental line we chose to grow etiolated WT plants on a medium containing 1 mM KCN, an inhibitor of complex IV in the ETC. Here, a statistically significant increment in apical hook bending could be detected (figure 3a; electronic supplementary material, figure S3). This indicates that limitation of KCN-sensitive ETC results in the same phenotype as in the rpoTmp mutant, suggesting that respiration restriction is indeed at the origin of the phenotypic deviations. To obtain independent proof for this, we observed the phenotype of dark-grown rug3 and atphb3 Arabidopsis mutant seedlings (figure 3b). The rug3 mutant is disturbed in complex I biogenesis [6] and the atphb3 mutant is defective in the mitochondrial membrane protein PROHIBITIN 3, that was suggested to be a structural support for membrane protein complexes, such as the ETC [23]. Both mutants possess a fully functional RPOTmp enzyme, but as in rpoTmp mutants or KCN-treated WT we found a twisted apical hook. This supports our hypothesis of ETC limitation as cause of the reprogramming of skoto-morphogenesis and excludes also any side-effect from the defective RPOTmp enzyme in the plastid.
Figure 3.
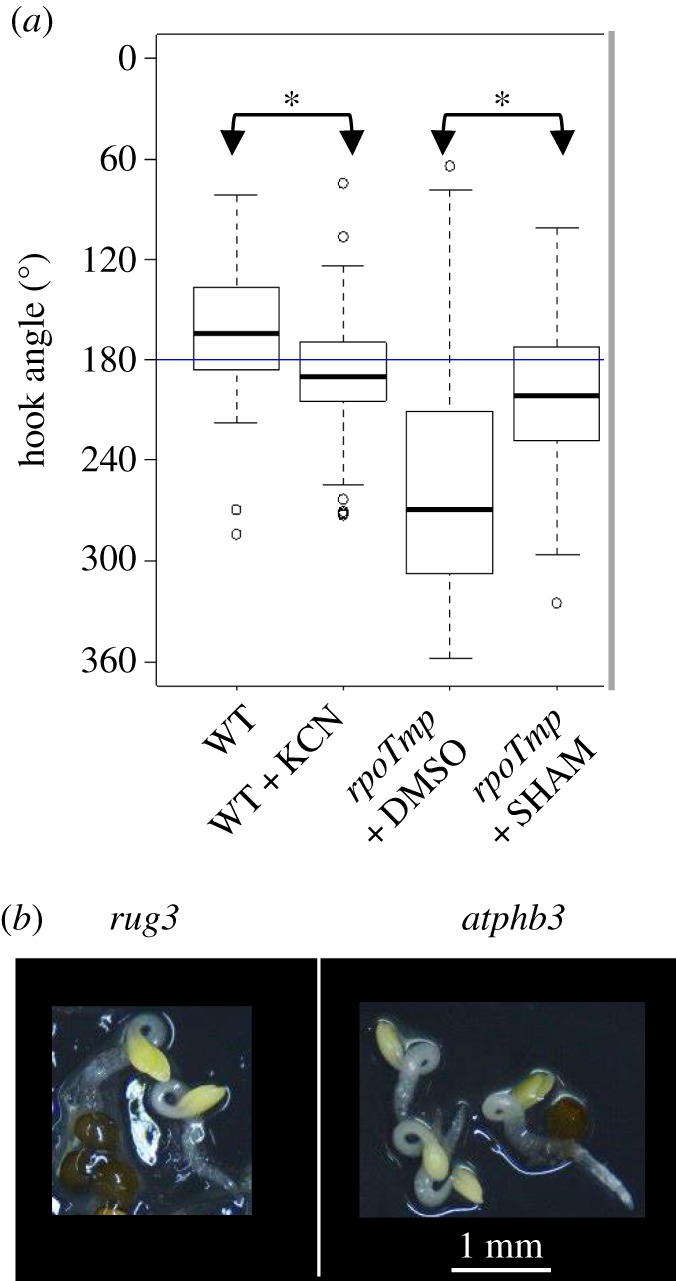
Plants that are defective in respiration present a triple-response-like phenotype. (a) Median values of apical hook angle measurements are box-plotted for WT plants that were grown in the presence of 1 mM KCN or H20 as mock-control and mutant rpoTmp plants that were grown in the presence of 1 mM SHAM or DMSO as mock-control. N (measured in four independent analyses) corresponds to 178 for H2O-treated WT plants and to 128 for KCN-treated WT plants. N (measured in two independent analyses) corresponds to 148 for DMSO-treated rpoTmp plants and to 85 for SHAM-treated rpoTmp plants. Details of box-plot representation are explained in figure 1b. (b) Images of etiolated mutant rug3 and atphb3 plants were taken under a dissection microscope. Scale bar corresponds to 1 mm. (Online version in colour.)
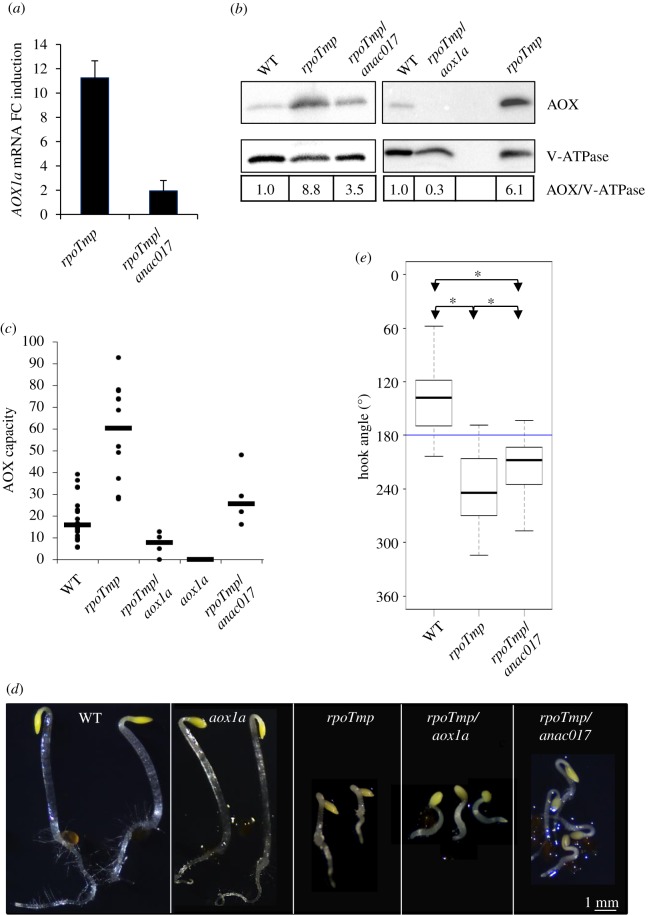
Upregulation of the AOX pathway has already been reported in light-grown rpoTmp plants [8]. In etiolated rpoTmp plants AOX1a expression was also strongly activated at both transcript and protein levels (figure 4a,b). Together with the AOX1a gene, the levels of two other markers of the cellular response to oxidative stress, AT12CYS [11,38] and UPOX [37,38], were also highly induced in rpoTmp plants (electronic supplementary material, figure S4a). Transcriptional activation of AOX1a and partially of AT12CYS mRNAs was impaired by the inactivation of the mitochondrial retrograde master regulator ANAC017 [7] (figure 4a; electronic supplementary material, figure S4b). The activation of the AOX1a gene expression corresponded to an increase in the AOX-dependent respiration capacity that is KCN-insensitive (figure 4c; electronic supplementary material, figure S5). To investigate whether or not the AOX-dependent pathway is involved in the triple-response-like phenotype we grew rpoTmp plantlets in the presence of 1 mM SHAM, a chemical inhibitor of the AOX-dependent pathway. Under this condition a statistically significant decrease of the twisting of the apical hook angle could be detected (figure 3a; electronic supplementary material, figure S3 right panel).
Figure 4.
The triple-response-like phenotype in rpoTmp mutant plants requires AOX1a. (a) Linear fold change (FC) induction of AOX1a mRNAs in mutant rpoTmp and rpoTmp/anac017 plants versus WT determined by RT-qPCR. The AOX1a expression levels were normalized with the mean of ACTIN2-8 expression, used as reference gene. The mean values of two biological replicates are plotted. Error bars correspond to standard errors. (b) Protein extracts from etiolated WT, rpoTmp, rpoTmp/anac017 plants (left panel) and WT, rpoTmp/aox1A, rpoTmp plants (right panel) were fractionated and immunoblotted with specific antisera against the mitochondrial AOX isoforms and the vacuolar protein V-ATPase as loading control. Linear fold change variation of AOX protein levels in mutant rpoTmp plants versus WT using V-ATPase for normalization is shown in the bottom panel. (c) Capacity of the AOX-dependent pathway in etiolated mutant plants. AOX capacity was calculated upon addition of 1 mM KCN (final concentration) into the measurement cell. The figure presents the ratio (as a percentage) of the KCN-insensitive O2 consumption rate to the total consumption rate. Median values (thick horizontal lines) of N independent measurements (filled circles) are scatter-plotted for WT (N = 19), rpoTmp (N = 12), rpoTmp/aox1a (N = 5), aox1a (N = 4) and rpoTmp/anac017 plants (N = 4). (d) Images of etiolated plants—WT, aox1a, rpoTmp, rpoTmp/aox1a, rpoTmp/anac017—were taken under a dissection microscope. Scale bar corresponds to 1 mm. (e) Median values of apical hook angle measurements are box-plotted for WT, mutant rpoTmp and rpoTmp/anac017 plants. Values were measured in two independent analyses. N = 53 for WT plants; N = 57 for rpoTmp plants; N = 57 for rpoTmp/anac017 plants. Details of box-plot representation are as given for figure 1b. (Online version in colour.)
To further prove independently the involvement of the AOX pathway, we aimed to characterize the rpoTmp/aox1a double mutant plants, in which the AOX1a isoform is lacking, in the dark. The AOX enzyme is known to be expressed in various isoforms that potentially could replace each other. In western-blot analyses of rpoTmp/aox1a protein extracts using an antiserum detecting the five AOX isoforms no signal was detected (figure 4b, right panel), indicating that AOX1a is the main AOX isoform in the rpoTmp genetic background in the dark. Analysis of rpoTmp/aox1a mutant seedlings, thus, should provide conclusive evidence about the involvement of the AOX1a enzyme in the phenotypic appearance of rpoTmp mutants. A strong decrease in the AOX capacity was observed in rpoTmp/aox1a double mutants when compared with rpoTmp plants (figure 4c; electronic supplementary material, figure S5). Importantly, we observed a suppression of the twisting of the apical hook in rpoTmp/aox1a mutants (figure 4d; electronic supplementary material, figure S6). However, the hypocotyl elongation was not restored, suggesting that it is regulated by a different pathway.
(c). The triple-response-like phenotype requires a retrograde signal from mitochondria
While the genetic defect in rpoTmp mutant lines causes a dysfunction in mitochondria, the upregulated AOX1a enzyme is encoded in the nucleus. The upregulation for involvement in the observed developmental change, therefore, requires a mito-nuclear retrograde signal. Recently, the endoplasmic reticulum-bound transcription factor ANAC017 was shown to be responsible for the retrograde activation of the AOX1a gene [7,24]. We thus proposed that the double mutant rpoTmp/anac017, which keeps the basal AOX1a expression but lacks the capacity to upregulate it to its full extent, should also show a reduction of the triple-response-like phenotype. Indeed, we found intermediate levels of AOX proteins and AOX capacity in the double mutant rpoTmp/anac017 plants when compared with rpoTmp and rpoTmp/aox1a mutant plants (figure 4b,c; electronic supplementary material, figure S5). As expected, we detected a partial suppression of the twisting of the apical hook in the rpoTmp/anac017 double mutant when compared with rpoTmp plants (figure 4d,e; electronic supplementary material, figure S6) indicating that the ANAC017-dependent pathway is required for the induction of the triple-response-like phenotype.
In sum, the results with the rpoTmp/aox1a and rpoTmp/anac017 double mutants provide compelling evidence that the hook exaggeration in rpoTmp requires AOX1a. A strong decrease in the AOX capacity was also observed in aox1a single mutants (figure 4c; electronic supplementary material, figure S5), but these plants displayed a normal skoto-morphogenesis without any aberrations (figure 4d). We concluded that a lack of AOX1a becomes critical for skoto-morphogenesis only under conditions when the ETC is limited.
4. Discussion
In the light, Arabidopsis rpoTmp mutants display defects in ETC but only mild phenotypes with slightly retarded growth and few macroscopic changes such as curled leaves [5,21]. In the dark, we expected stronger impact on the phenotype as no energy compensation from photosynthesis can occur. Here, we report that the expression of the mitochondrial subset of genes that were previously identified as RPOTmp-dependent [5] was highly impacted in rpoTmp plants in the dark also (figure 2a,b; electronic supplementary material, table S3). In addition, the strong activation of stress marker genes indicates the presence of a severe mitochondrial stress in etiolated mutant plants (figures 2c,d and 4a; electronic supplementary material, figure S4a). Comparison of our transcriptomic data obtained from dark-grown seedlings with previous analyses done on light-grown adult plants [7] identified high similarities at the level of the response to mitochondrial dysfunction (electronic supplementary material, table S5). These data underline the robustness of the plant stress reaction to the genetic defect in the RPOTMP gene locus, which is largely independent of the developmental stage and the light regime. However, to our surprise, we did not observe any retardation in development of the dark-grown seedlings (as observed in the light), but instead we found a strong modification of the skoto-morphogenic programme (figure 1).
We could confirm that limitation of the ETC leads to shortened hypocotyls and twisted apical hooks during the development of the seedlings by both chemical means (in KCN-treated plants, figure 3a; electronic supplementary material, figure S3 left panel) and independent genetic tools (in rug3 and atphb3 mutant plants, figure 3b). This phenotype is highly reminiscent of the triple-response of seedlings exposed to ethylene (or precursors of ethylene such as ACC) (figure 1). The third parameter, a widened hypocotyl, could not be significantly determined. Therefore, we termed the observed phenotypic response a triple-response-like phenotype. In the rpoTmp/aox1a double mutant the twisting of the hook was prevented; however, the hypocotyl shortening was maintained (figure 4d). These observations suggest that the three phenotypic traits may be controlled by different specific pathways and/or different hormone thresholds.
A recent study reported that limitation of mitochondrial translation causes proteotoxic stress and activates an unfolded protein response in mitochondria by MAP kinase cascades, which leads to shortened hypocotyls [10]. Partial inhibition of mitochondrial translation, however, leads also to a general downregulation of mitochondrial respiration and, concomitantly, respiration stress does occur. This stress was apparently not as strong as in our system since an exaggerated hook was not observed in that study. Interestingly, an upregulation of AOX1a could be observed also in that study, but its potential involvement in the signalling of the proteotoxic stress was not investigated.
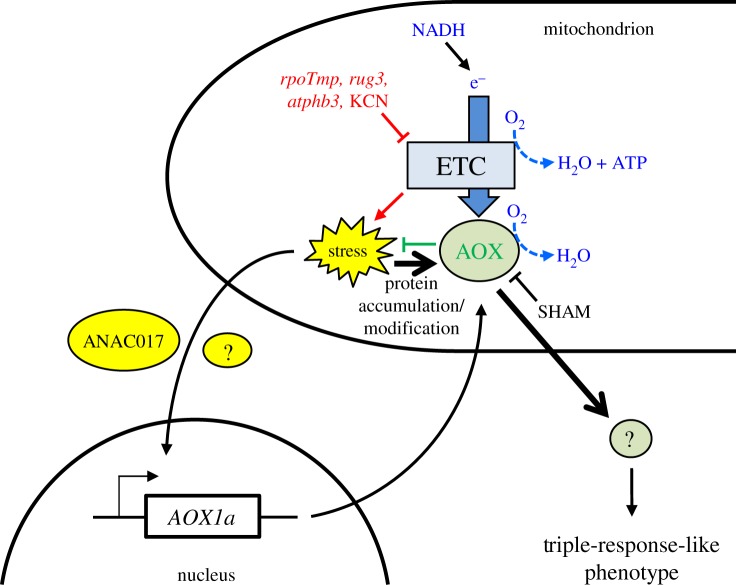
Here, we show that limitation of the ETC leads to drastic changes in skoto-morphogenesis and that the increased capacity of the AOX pathway is the critical parameter that triggers the hook exaggeration (figure 5). Both pharmaceutical interference of AOX enzyme activity (by SHAM) or genetic impairment of the AOX1a isoform resulted in a significant suppression of the twisting phenotype in rpoTmp plants (figures 3a and 4d; electronic supplementary material, figures S3 (right panel) and S6). Our western-analyses indicate also, that AOX1a appears to be the sole isoform being expressed in the dark (figure 4b). Recently, it was reported that the AOX1a enzyme expression is induced at the transcriptional level by the action of the ANAC017-dependent retrograde pathway in response to mitochondrial stress [7,24]. In our study the genetic impairment of the ANAC017-dependent transcriptional activation of the AOX1a gene (figure 4a) affects moderately both the increase of the accumulation (figure 4b) and the enzymatic capacity of AOX proteins (figure 4c) and the induction of the twisting phenotype (figure 4d,e; electronic supplementary material, figure S6) in rpoTmp seedlings. These data highlight the dependence of the triple-response-like phenotype on the AOX1a protein levels and the mitochondrial retrograde pathway. It is important to note that in the rpotmp/anac017 double mutant seedlings, the basal AOX1a expression is not reduced (as in an aox1a knockout line). The plants just lack the ability to upregulate AOX1a expression to its full extent. As such the phenotypical effect is expected to be less pronounced than in rpotmp/aox1a mutants (figure 4d; electronic supplementary material, figure S6). This observation suggests that transcriptional activation of AOX1a by the ANAC017-dependent retrograde pathway does not have an exclusive role in the dark and likely additional translational or post-translational regulation of the AOX1a enzyme might also be implemented. Based on in vitro experiments, it was proposed that AOX activity might be regulated at the post-translational level, at the redox state and by the interaction with TCA cycle intermediates, leading to a very tight and rapid metabolic control of the mitochondrial respiration process [39,40].
Figure 5.
Scheme of the proposed functional link between the respiratory chain capacities (KCN-sensitive and AOX-dependent) and the skoto-morphogenic programme. In mitochondria, the KCN-sensitive transport of electrons (ETC) from NADH is used for O2 reduction and ATP production. Only a limited fraction of electrons is diverted to the AOX enzyme for reduction of O2 without concomitant ATP production. In etiolated rpoTmp, rug3, atphb3 or KCN-treated WT plants, the KCN-sensitive electron flux is strongly decreased, inducing a strong dysfunctional stress (indicated in red). This stress activates the canonical (ANAC017-dependent) mitochondrial retrograde pathway which activates AOX1a expression. It may be supported by yet unknown factors (represented by a yellow oval with question mark). Additional AOX regulation by dysfunctional stress at the level of protein accumulation and modification might be possible (thick black arrow). As ultimate effect the capacity of the AOX-dependent chain is upregulated and more electrons can be diverted by this pathway, releasing the stress (indicated in green). The dysfunctional stress in combination with the highly increased AOX capacity, however, sends a signal via an unknown transmitter (represented by a grey circle with question mark) that triggers the formation of the triple-like response (or parts of it). (Online version in colour.)
We conclude that the limitation of respiration, therefore, induces the triple-like response by promoting the abundance and capacity of the AOX1a enzyme (figure 5). Our study strongly suggests that the AOX1a enzyme is not just a biomarker for stress [3,41] that accumulates as a side effect, but it represents a key component of the stress reaction network that triggers subsequent cellular responses to mitochondrial dysfunction. How precisely the AOX1a enzyme induces the reprogramming remains to be elucidated, but one can imagine that especially under oxygen limitation high capacities of AOX1a activity could reduce alternative substrates, generating products that may serve as signalling molecules, such as nitric oxide [42].
A physiological role for the triple-response was obscure for a long time, until recently a study uncovered that the triple-response can be triggered by the soil overlay of buried seeds [43]. The triple morphogenic change provides higher resistance to soil friction and allows the seedling to breach the surface, while protecting the apical meristem. A recent study investigated the influence of hypoxic conditions on skoto-morphogenesis and uncovered that a low oxygen tension applied after the apical hook maintenance phase prevented the opening of the hook in subsequent light conditions [44]. The study identified that seedlings are able to sense the oxygen tension via the N-end rule pathway-controlled stability of ERFVII transcription factors. However, the hypoxic condition in that study was imposed on the seedlings after the apical hook was formed and, thus, represents a phase later than that in our study. An attractive hypothesis is that dense soil overlay may lead to hypoxic conditions that in turn induce respiration stress (owing to limitation of the final electron acceptor of the KCN-sensitive ETC, the oxygen) and a subsequent triple-response-like phenotype. How soil and O2 sensing mechanisms relate to our proposed model will be the subject of future research.
Our proposed model (figure 5) is based on respiration deficiency generated in the laboratory by genetic or pharmaceutical blocking. However, it would fit also with other natural scenarios in which the final electron acceptor for KCN-sensitive electron transport (i.e. oxygen) becomes limiting, for instance in response to water flooding, drought or acute ozone exposure [9]. In addition, rapid responses to touch and wounding stress involve changes in the expression of genes for mitochondria-localized proteins that are responsible for the availability of the substrates required for mitochondrial energy metabolism [14]. It will be of great interest to investigate whether the proposed model also applies in these conditions and which developmental effects could be triggered.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We thank Robert Blanvillain and Stéphane Ravanel from LPCV (Grenoble, France), and Etienne Delannoy and Emmanuelle Issakidis-Bourguet from IPS2 (Orsay, France) for helpful discussions, and Juliette Jouhet (LPCV, Grenoble, France) and Géraldine Bonnard (IBMP, Strasburg, France) for sharing NAD9 antisera. We thank Olivier Van Aken (Lund University, Sweden) for sharing rpoTmp/anac017 and atphb3 seeds, Renate Scheibe (Universität Osnabrück, Germany) for aox1a seeds and Kristina Kühn (Universität Halle, Germany) for rpoTmp/aox1a and rug3 seeds. RNA sample processing and Affymetrix microarray hybridization were carried out at the genomics core facility: Center of Excellence for Fluorescent Bioanalytics (KFB, University of Regensburg, Germany).
Data accessibility
Microarray data are available at the Gene Expression Omnibus (GEO) under accession number GSE144694.
Authors' contributions
L.M. and T.P. designed research, L.M., F.Co., B.G., V.S. and F.Ch. performed experiments, O.B. performed statistical analyses, C.L. contributed analytical tools. L.M., F.Co., S.L.-M. and T.P. analysed data, L.M. and T.P. wrote the manuscript with the help of all co-authors. All authors read and approved the manuscript.
Competing interests
The authors declare no conflict of interest.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft (PF323-5-2 to T.P.) and the DFG research group FOR 804, the Centre National de la Recherche Scientifique (PEPS to T.P.), the French Ministry of Education and the Grenoble Alliance for Integrated Structural Cell Biology (LabEx GRAL, ANR-10-LABX-49-01). IPS2 benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS).
References
- 1.Millar AH, Whelan J, Soole KL, Day DA. 2011. Organization and regulation of mitochondrial respiration in plants. Annu. Rev. Plant Biol. 62, 79–104. ( 10.1146/annurev-arplant-042110-103857) [DOI] [PubMed] [Google Scholar]
- 2.Pracharoenwattana I, Cornah JE, Smith SM. 2005. Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17, 2037–2048. ( 10.1105/tpc.105.031856) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vanlerberghe G. 2013. Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int. J. Mol. Sci. 14, 6805–6847. ( 10.3390/ijms14046805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colas des Francs-Small C, Small I.. 2014. Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100, 234–242. ( 10.1016/j.biochi.2013.08.019) [DOI] [PubMed] [Google Scholar]
- 5.Kühn K, et al. 2009. Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21, 2762–2779. ( 10.1105/tpc.109.068536) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kühn K, et al. 2011. The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J. 67, 1067–1080. ( 10.1111/j.1365-313X.2011.04658.x) [DOI] [PubMed] [Google Scholar]
- 7.Van Aken O, Ford E, Lister R, Huang S, Millar AH.. 2016. Retrograde signalling caused by heritable mitochondrial dysfunction is partially mediated by ANAC017 and improves plant performance. Plant J. 88, 542–558. ( 10.1111/tpj.13276) [DOI] [PubMed] [Google Scholar]
- 8.Kühn K, et al. 2015. Decreasing electron flux through the cytochrome and/or alternative respiratory pathways triggers common and distinct cellular responses dependent on growth conditions. Plant Physiol. 167, 228–250. ( 10.1104/pp.114.249946) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner S, Van Aken O, Elsässer M, Schwarzländer M.. 2018. Mitochondrial energy signaling and its role in the low-oxygen stress response of plants. Plant Physiol. 176, 1156–1170. ( 10.1104/pp.17.01387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Auwerx J. 2017. Systems phytohormone responses to mitochondrial proteotoxic stress. Mol. Cell 68, 540–551. ( 10.1016/j.molcel.2017.10.006) [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, et al. 2016. Inactivation of mitochondrial complex I induces the expression of a twin cysteine protein that targets and affects cytosolic, chloroplastidic and mitochondrial function. Mol. Plant 9, 696–710. ( 10.1016/j.molp.2016.01.009) [DOI] [PubMed] [Google Scholar]
- 12.Schwarzländer M, König A-C, Sweetlove LJ, Finkemeier I. 2012. The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J. Exp. Bot. 63, 1735–1750. ( 10.1093/jxb/err374) [DOI] [PubMed] [Google Scholar]
- 13.Blanco NE, Guinea-Díaz M, Whelan J, Strand Å. 2014. Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Phil. Trans. R. Soc. B 369, 20130231 ( 10.1098/rstb.2013.0231) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Aken O, De Clercq I, Ivanova A, Law SR, Van Breusegem F, Millar AH, Whelan J.. 2016. Mitochondrial and chloroplast stress responses are modulated in distinct touch and chemical inhibition phases. Plant Physiol. 171, 2150–2165. ( 10.1104/pp.16.00273) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietzel L, et al. 2015. Identification of early nuclear target genes of plastidial redox signals that trigger the long-term response of Arabidopsis to light quality shifts. Mol. Plant 8, 1237–1252. ( 10.1016/j.molp.2015.03.004) [DOI] [PubMed] [Google Scholar]
- 16.Van Aken O, Pogson BJ.. 2017. Convergence of mitochondrial and chloroplastic ANAC017/PAP-dependent retrograde signalling pathways and suppression of programmed cell death. Cell Death Differ. 24, 955–960. ( 10.1038/cdd.2017.68) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Solymosi K, Schoefs B. 2010. Etioplast and etio-chloroplast formation under natural conditions: the dark side of chlorophyll biosynthesis in angiosperms. Photosynth. Res. 105, 143–166. ( 10.1007/s11120-010-9568-2) [DOI] [PubMed] [Google Scholar]
- 18.Arsovski AA, Galstyan A, Guseman JM, Nemhauser JL. 2012. Photomorphogenesis. Arabidopsis Book 10, e0147 ( 10.1199/tab.0147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hedtke B, Börner T, Weihe A. 1997. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277, 809–811. ( 10.1126/science.277.5327.809) [DOI] [PubMed] [Google Scholar]
- 20.Hedtke B, Börner T, Weihe A. 2000. One RNA polymerase serving two genomes. EMBO Rep. 1, 435–440. ( 10.1093/embo-reports/kvd086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courtois F, Merendino L, Demarsy E, Mache R, Lerbs-Mache S. 2007. Phage-type RNA polymerase RPOTmp transcribes the rrn operon from the PC promoter at early developmental stages in Arabidopsis. Plant Physiol. 145, 712–721. ( 10.1104/pp.107.103846) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tarasenko VI, Katyshev AI, Yakovleva TV, Garnik EY, Chernikova VV, Konstantinov YM, Koulintchenko MV. 2016. RPOTmp, an Arabidopsis RNA polymerase with dual targeting, plays an important role in mitochondria, but not in chloroplasts. J. Exp. Bot. 67, 5657–5669. ( 10.1093/jxb/erw327) [DOI] [PubMed] [Google Scholar]
- 23.Van Aken O, et al. 2007. Mitochondrial type-I prohibitins of Arabidopsis thaliana are required for supporting proficient meristem development. Plant J. 52, 850–864. ( 10.1111/j.1365-313X.2007.03276.x) [DOI] [PubMed] [Google Scholar]
- 24.Ng S, et al. 2013. A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. ( 10.1105/tpc.113.113985) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org/. [Google Scholar]
- 26.Grübler B, et al. 2017. Light and plastid signals regulate different sets of genes in the albino mutant pap7-1. Plant Physiol. 175, 1203–1219. ( 10.1104/pp.17.00982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohse M, Bolger AM, Nagel A, Fernie AR, Lunn JE, Stitt M, Usadel B. 2012. RobiNA: a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 40, W622–W627. ( 10.1093/nar/gks540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4, 249–264. ( 10.1093/biostatistics/4.2.249) [DOI] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300. ( 10.2307/2346101) [DOI] [Google Scholar]
- 30.Ritchie ME, et al. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 ( 10.1093/nar/gkv007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamattina L, Gonzalez D, Gualberto J, Grienenberger JM. 1993. Higher plant mitochondria encode an homologue of the nuclear-encoded 30-kDa subunit of bovine mitochondrial complex I. Eur. J. Biochem. 217, 831–838. ( 10.1111/j.1432-1033.1993.tb18311.x) [DOI] [PubMed] [Google Scholar]
- 32.Guzmán P, Ecker JR. 1990. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 2, 513–523. ( 10.1105/tpc.2.6.513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neljubow D. 1901. Uber die horizontale Nutation der Stengel von Pisum Sativum und einiger anderen Planzen [Concerning the horizontal mutation of the stem of Pisum sativum and some other plants]. Beih. Bot. Zbl. 10, 128–139. [In German.] [Google Scholar]
- 34.Larsen PB, Chang C. 2001. The Arabidopsis eer1 mutant has enhanced ethylene responses in the hypocotyl and stem. Plant Physiol. 125, 1061–1073. ( 10.1104/pp.125.2.1061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lama S, Broda M, Abbas Z, Vaneechoutte D, Belt K, Säll T, Vandepoele K, Van Aken O.. 2019. Neofunctionalization of mitochondrial proteins and incorporation into signaling networks in plants. Mol. Biol. Evol. 36, 974–989. ( 10.1093/molbev/msz031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhrig RG, Labandera A-M, Tang L-Y, Sieben NA, Goudreault M, Yeung E, Gingras A-C, Samuel MA, Moorhead GBG. 2017. Activation of mitochondrial protein phosphatase SLP2 by MIA40 regulates seed germination. Plant Physiol. 173, 956–969. ( 10.1104/pp.16.01641) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ho LHM, Giraud E, Uggalla V, Lister R, Clifton R, Glen A, Thirkettle-Watts D, Van Aken O, Whelan J. 2008. Identification of regulatory pathways controlling gene expression of stress-responsive mitochondrial proteins in Arabidopsis. Plant Physiol. 147, 1858 ( 10.1104/PP.108.121384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Aken O, Whelan J.. 2012. Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front. Plant Sci. 3, 281 ( 10.3389/fpls.2012.00281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selinski J, Scheibe R, Day DA, Whelan J. 2018. Alternative oxidase is positive for plant performance. Trends Plant Sci. 23, 588–597. ( 10.1016/j.tplants.2018.03.012) [DOI] [PubMed] [Google Scholar]
- 40.Del-Saz NF, Ribas-Carbo M, McDonald AE, Lambers H, Fernie AR, Florez-Sarasa I. 2018. An in vivo perspective of the role(s) of the alternative oxidase pathway. Trends Plant Sci. 23, 206–219. ( 10.1016/j.tplants.2017.11.006) [DOI] [PubMed] [Google Scholar]
- 41.Clifton R, Lister R, Parker KL, Sappl PG, Elhafez D, Millar AH, Day DA, Whelan J. 2005. Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol. Biol. 58, 193–212. ( 10.1007/s11103-005-5514-7) [DOI] [PubMed] [Google Scholar]
- 42.Vishwakarma A, Kumari A, Mur LAJ, Gupta KJ. 2018. A discrete role for alternative oxidase under hypoxia to increase nitric oxide and drive energy production. Free Radic. Biol. Med. 122, 40–51. ( 10.1016/j.freeradbiomed.2018.03.045) [DOI] [PubMed] [Google Scholar]
- 43.Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW. 2014. Ethylene-orchestrated circuitry coordinates a seedling's response to soil cover and etiolated growth. Proc. Natl Acad. Sci. USA 111, 3913–3920. ( 10.1073/pnas.1402491111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abbas M, et al. 2015. Oxygen sensing coordinates photomorphogenesis to facilitate seedling survival. Curr. Biol. 25, 1483–1488. ( 10.1016/j.cub.2015.03.060) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data are available at the Gene Expression Omnibus (GEO) under accession number GSE144694.