A biomimetic peptide recognizes and traps bacteria in vivo as human defensin-6 for inhibiting the invasion of host cells.
Abstract
Using broad-spectrum antibiotics for microbial infection may cause flora disequilibrium, drug-resistance, etc., seriously threatening human health. Here, we design a human defensin-6 mimic peptide (HDMP) that inhibits bacterial invasion in vivo through mimicking the mechanisms of human defensin-6 with high efficiency and precision. The HDMP with ligand and self-assembling peptide sequence recognizes bacteria through ligand-receptor interactions and subsequently traps bacteria by an in situ adaptive self-assembly process and resulting nanofibrous networks; these trapped bacteria are unable to invade host cells. In four animal infection models, the infection rate was markedly decreased. Notably, administration of HDMP (5 mg/kg) nanoparticles increased the survival rate of mice with methicillin-resistant S. aureus bacteremia by as much as 100%, even more than that of vancomycin treatment (5 mg/kg, 83.3%)–treated group, the golden standard of antibiotics. This biomimetic peptide shows great potential as a precise and highly efficient antimicrobial agent.
INTRODUCTION
Microbial infections are severely harmful to human health even if there are many antibiotics, antibacterial peptides, and so on (1–3). The development of broad microbial killing agents leads to flora disequilibrium and drug resistance, resulting in secondary infections and antibiotics failure (4, 5). The innate immune system of human beings maintaining microbiota homeostasis of intestine inspired us to develop new strategies for the specific therapy of microbial infections. Recently, it has been found that the antimicrobial peptide human defensin-6 (HD6), expressed and secreted by Paneth cells, can recognize the invading microbes and effectively inhibit infection when encountering invading microbes (6–8). The HD6 does not kill microbes directly, but self-assembles into entangled fibrous networks, traps microbial pathogens, and blocks the invasion (9). The HD6 performs an “encircle without attacking” natural supramolecular strategy, which is different from general human defensin attacking microbes directly. The finding of an antibacterial mechanism of HD6 provides a biomimetic model as a new antimicrobial strategy (Fig. 1). From the point of view of materials science, the peptide can self-assemble into fibrous networks with stabilized β-sheet structures and may mimic the natural HD6, achieving the ligand-receptor interaction–induced self-assembly process for bacteria recognizing and trapping. It is easy to in situ construct fibrous networks by self-assembling peptides to trap bacteria in vitro (10, 11). However, it needs to overcome huge barriers such as transportation of self-assembling peptides to the bacteria invasion region, realizing in situ construction of self-assembling fibrous networks for bacteria trapping in vivo. The dynamic process of protein/peptide self-assembly in vivo (12) further inspired us to design a programmable self-assembling HD6 mimic peptide (HDMP) for inhibition of bacteria invasion in vivo (Fig. 1).
Fig. 1. Antimicrobial mechanism of natural HD6 and artificial HDMP.
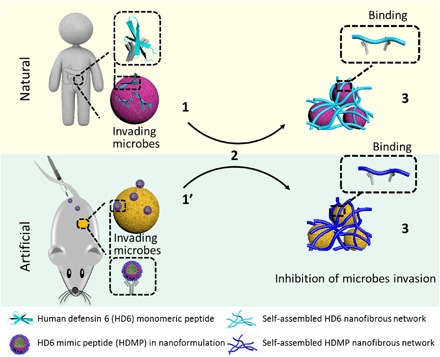
The HDMP mimics the HD6 that (2) in situ self-assembles on microbial surfaces by ligand-receptor interactions, resulting in (3) self-assembled trapping fibrous networks for the inhibition of microbial invasion. Different from the HD6 that is (1) in situ secreted from Paneth cells, the HDMP is (1′) transported into the targeted region through intravenous administration as nanoparticle (NP) formulation, overcoming the barrier of self-assembling peptides to application in vivo.
The HDMP bis-pyrene-KLVFF-RLYLRIGRR consists of three modules (Fig. 2A). (i) A ligand peptide sequence RLYLRIGRR can bind to lipoteichoic acid (LTA), a unique component of Gram-positive bacteria, and impart the recognition functionality of HDMP, which is superior to the HD6 without specific recognition capability (13). (ii) The KLVFF sequence is a peptide skeleton for β-sheet fibrous structures, mimicking the β-sheet structured HD6 networks (14). (iii) Aromatic bis-pyrenes (BP) enable the synthesis of HDMP in particulate form, providing the potential for transport of HDMP through intravenous administration. Moreover, the fluorescence arising from the aggregation-induced emission (AIE) of BP can be used to monitor the biodistribution of HDMP in vivo (15). Mechanistically, HDMP first self-assembles into nanoparticle formulation and then specifically bound to the cell wall of Staphylococcus aureus and transformed into nanofibers (NFs) to form fibrous networks, triggered by ligand-receptor interactions and stabilized by hydrogen-bonding and π-π interactions (16). As a result, bacteria are trapped by fibrous networks, thus inhibiting bacterial invasion. The combination of the three components enables the programmable self-assembly process, precisely mimicking the process of HD6, and safely and effectively inhibits infection by Gram-positive bacteria in vivo.
Fig. 2. Preparation of HDMP NPs and its transformation into NFs.
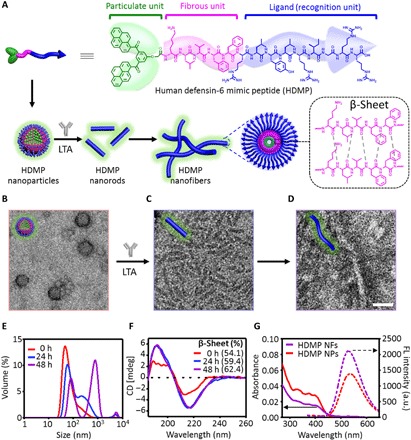
(A) Molecular structure of HDMP and schematic illustration of HDMP assembly into NPs, transforming into nanorods and nanofibers (NFs) upon the incubation of lipoteichoic acid (LTA). Transmission electron microscope (TEM) image of HDMP NPs (B), HDMP nanorods (C), and NFs (D) triggered by LTA at 24 and 48 hours, respectively. (E) Dynamic light scattering (DLS) and (F) circular dichroism (CD) of HDMP assembly from NPs into NFs in the presence of LTA in aqueous solutions. (G) Ultraviolet-visible (UV-vis) and fluorescence (FL) spectra of HDMP NPs and HDMP NFs, respectively. The TEM images are representative of three independent experiments. Scale bar, 50 nm. a.u., arbitrary units.
RESULTS
Preparation of HDMP and its particle formulation
The HDMP was synthesized by a solid-phase synthesis technique, and the molecular structure was determined by matrix-assisted laser desorption/ionization–time-of-flight–mass spectrometry (MALDI-TOF-MS) (fig. S1, A and B). HDMP could form self-assembled NPs when HDMP in dimethyl sulfoxide (DMSO) was injected into water with the final concentration of 30 μM (Vwater/VDMSO = 99/1) (Fig. 2A) (17). The transmission electron microscope (TEM) images revealed that HDMP NPs was 35.2 ± 7.1 nm in diameter (Fig. 2B), which were confirmed by the dynamic light scattering (DLS) measurements (41.9 ± 9.1 nm) (Fig. 2E). The HDMP self-assembly into NPs was mainly driven by the hydrophobic and π-π interactions of BP, which could be confirmed by the UV-vis absorption and fluorescence spectra. The absorption peaks of HDMP monomers around 300 to 400 nm decreased, and the bathochromic absorption peak increased (fig. S1C), indicating the aggregation of BP units. The peptide sequence was also aggregated, which was proven by the broadened absorption at 277 nm. Meanwhile, as shown in fig. S1D, as the water ratio in the mixed solution increased, the fluorescence intensity at 520 nm increased gradually, indicating the aggregation of AIE-active BP (18, 19).
HDMP NPs recognize LTA and transform into fibers in solution
To study the specificity of RLYLRIGRR (ligand peptide), HDMP NPs were incubated with the LTA, a receptor molecule in Gram-positive bacterial cell membrane. As shown in Fig. 2C, the HDMP NPs transformed into nanorods with a length of 72.3 ± 23.9 (Φ = 9.8 ± 0.9 nm) at 24 hours, followed by further self-assembly into NFs with a length of 202.4 ± 55.9 (Φ = 11.8 ± 1.3 nm) at 48 hours after incubation (Fig. 2, D and E). However, HDMP NPs without LTA incubation were still NPs with an average diameter of 48.0 ± 18.4 nm from TEM and 55.9 ± 20.2 nm from DLS (fig. S2A), without morphological transformation within 2 days. Furthermore, the lipopolysaccharides (LPS), a feature component on Gram-negative bacteria surfaces, were used to incubate with HDMP NPs. The TEM (fig. S2B) images revealed that HDMP NPs generally retained their particulate morphology (44.8 ± 12.5 nm) for 2 days. These results validated the specificity of RLYLRIGRR to LTA, and HDMP NPs could transform the morphology from NPs to NFs through binding LTA, not LPS. To mimic self-assembling β-sheet structured fibrous networks of the natural HD6, KLVFF, a β-sheet structured sequence, was introduced into HDMP. In addition, the control peptide with weak β-sheet structured sequence (C-HDMP), BP-KAAGG-RLYLRIGRR (fig. S2C), and C-HDMP NPs were prepared (fig. S3). The TEM (fig. S4) and DLS (fig. S5) measurements showed that C-HDMP NPs did not undergo morphological transformation with/without LTA incubation, indicating the importance of KLVFF fibrillation sequence with the strong hydrogen-bonding and hydrophobic interactions.
During the structural transformation from HDMP NPs to NFs, circular dichroism (CD) spectroscopy may give insight into the secondary structure information of HDMP. As shown in Fig. 2F, the percentage of β-sheet in LTA-incubated HDMP NPs increased from 54.1 to 62.4% in 2 days with morphological transformation. The 8.3% increase in β-sheet indicated more highly ordered packing structures of peptides in NFs than that in HDMP NPs. So did BP, which was validated by the decrease in absorption at 300 to 400 nm and the increase in emission at 520 nm (Fig. 2G). The percentage of β-sheet in HDMP NPs in the absence of LTA remained almost constant (about 48%) in 2 days (fig. S6A). In the presence of LPS, HDMP NPs exhibited an almost constant percentage of β-sheet (about 50%) in 2 days (fig. S6B). These results suggested that the specific interaction between LTA and RLYLRIGRR disrupted the dynamic equilibrium of HDMP NPs, and the HDMP in situ self-assembled into thermodynamically stable HDMP NFs. The control C-HDMP peptide with the sequence of KAAGG showed 25.3% (fig. S6, C and D) β-sheet structure upon the formation of C-HDMP NPs and was almost unchanged in 2 days (28.1%), as were the C-HDMP NPs incubating with LTA (from 26.1% at 0 hours to 29.6% at 48 hours). The high percentage of β-sheet structure in HDMP nanostructures validated the importance of β-sheet structured KLVFF for formation and stabilization of NFs.
HDMP NPs recognize and trap S. aureus and inhibit its invasion in vitro
The recognition ability of HDMP NPs to bacteria was first investigated by confocal laser scanning microscope (CLSM) observation (Fig. 3A). Gram-negative Escherichia coli and Gram-positive S. aureus were incubated with HDMP NPs and C-HDMP NPs (30 μM) in 1.5-ml centrifuge tubes for 4 hours, respectively. HDMP NPs treated with S. aureus showed strong fluorescence signals, which could completely cover the surface of S. aureus. C-HDMP NPs could light up the S. aureus somehow with much lower fluorescence intensity. However, the E. coli treated with HDMP NPs and C-HDMP NPs showed negligible fluorescence (fig. S7A). These results indicated that the RLYLRIGRR sequence could recognize Gram-positive bacteria, not Gram-negative bacteria. Furthermore, HDMP NPs even recognized S. aureus from the mixture of both E. coli and S. aureus (Fig. 3, C and D). HDMP NP–incubated S. aureus showed a clear layer out of the S. aureus cell wall, indicating in situ self-assembled HDMP NFs on the surface of S. aureus. Scattered NPs were found on the surface of C-HDMP NPs treated with S. aureus (Fig. 3F), suggesting the attachment of C-HDMP NPs on S. aureus. Obviously, much more HDMP attached on surfaces of S. aureus than C-HDMP due to the in situ self-assembly from HDMP NPs into NFs, which could explain much higher fluorescence intensity of S. aureus treated with HDMP NPs than that of S. aureus treated with C-HDMP NPs. There were almost no differences between HDMP NP–incubated (Fig. 3G) and C-HDMP NP–incubated (Fig. 3H) E. coli and blank E. coli, revealing no NP attachment on E. coli, leading to non-fluorescent E. coli. These results validated the specific recognition capability of ligand peptide in HDMP to S. aureus, not E. coli. The ligand peptide and the β-sheet structured KLVFF enabled the HDMP transformation from NPs into NFs upon ligand-receptor interaction–induced self-assembly on surfaces of S. aureus.
Fig. 3. HDMP NPs recognize and transform into NFs on S. aureus in vitro.
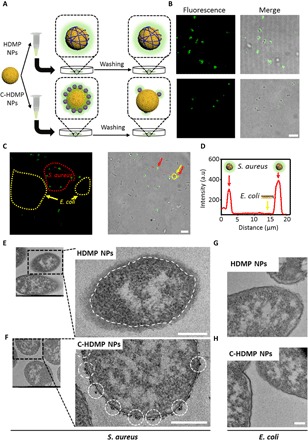
(A) Schematic illustration of HDMP NP– and C-HDMP NP–treated bacteria procedure. The bacteria were first treated with HDMP (30 μM) or C-HDMP (30 μM) NPs for 4 hours, followed by washing with phosphate-buffered saline (PBS) for confocal imaging. (B) Confocal microscopy showing specific recognition of HDMP NPs to S. aureus (top) and low targeting of C-HDMP to S. aureus (bottom). Scale bar, 5 μm. (C) Confocal imaging of S. aureus and E. coli mixture treated by the HDMP NPs (30 μM), showing the specific recognition of HDMP NPs to S. aureus, not E. coli. The red and yellow arrows indicate S. aureus and E. coli, respectively. Scale bar, 10 μm. (D) The normalized intensity profile for regions of interest across the red line in (C). (E to H) TEM images of S. aureus and E. coli treated with HDMP NPs (30 μM) and C-HDMP NPs (30 μM), exhibiting transformed HDMP fibrous networks out of the S. aureus wall (white dashed line) (E) and remained C-HDMP NPs (white dashed circles) on the S. aureus wall (F); the E. coli only without HDMP (G) and C-HDMP NPs (H). Scale bar, 200 nm. The confocal images and TEM images are representative of three independent experiments.
The HDMP fibrous networks were expected to form and trap S. aureus. The scanning electron microscope (SEM) showed that HDMP NP–incubated S. aureus was covered and trapped by fibrous networks (Fig. 4A). The phenomena were similar to in situ self-assembled fibrous networks of natural HD6-trapping bacteria (8). However, the C-HDMP NP–treated S. aureus did not form fibrous networks and trap S. aureus (fig. S7B), similar to the bare S. aureus (Fig. 4B).The E. coli was also tested with HDMP and C-HDMP NPs, and there were no fibrous networks entangled with E. coli for both HDMP and C-HDMP NPs (fig. S7C).
Fig. 4. HDMP NPs transform into fibrous networks for trapping S. aureus and inhibiting its invasion in vitro.
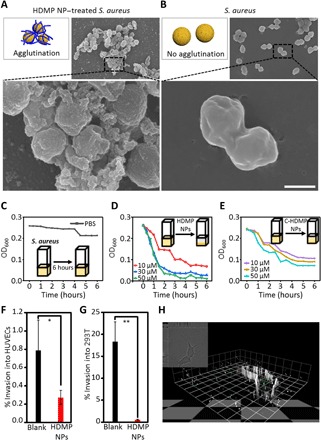
Schematic illustrations and SEM images of S. aureus treated with (A) HDMP NPs (30 μM) and bare S. aureus (B), indicating that the HDMP NPs transformed into fibrous networks trapping the S. aureus. The SEM images are representative of three independent experiments. Scale bar, 1 μm. (C to E). The time-dependent S. aureus agglutination assay, showing that HDMP NPs could trap bacteria effectively. (C) S. aureus suspension without treatment in 6 hours. (D) S. aureus suspension treated with HDMP NPs in 6 hours and (E) S. aureus suspension treated with C-HDMP NPs in 6 hours. (F and G) The invasion inhibition of bacteria to cells, showing the highly efficient inhibition rate of HDMP NPs. Invasion of human umbilical vein endothelial cells (HUVECs) (F) and human embryonic kidney 293 (293T) cells (G) by S. aureus [106 colony-forming units (CFU)/ml] with/without HDMP NPs (10 μM). (H) Three-dimensional confocal image of invasion of 293T cells by S. aureus. The confocal image is representative of three experiments. Scale bar, 1 unit = 9.5 μm. The * represents significant difference between blank and HDMP NP–treated groups (*P < 0.05, **P < 0.01).
The trapping of S. aureus by entangled HDMP NFs could lead to bacterial agglutination, which was evaluated by the measurement of turbidity of bacterial solution treated with HDMP NPs and C-HDMP NPs as controls. As shown in Fig. 4C, the turbidity of S. aureus without treatment was almost kept constant in 6 hours. However, the turbidity of S. aureus in the cuvette was substantially decreased when incubated with HDMP NPs (Fig. 4D). In particular, the S. aureus solution was substantially clear after 3 hours in 50 μM HDMP. C-HDMP NPs could somewhat reduce the turbidity of bacteria, but not as much as HDMP NPs (Fig. 4E). The C-HDMP NPs that interacted with S. aureus in NP formulation could not coagulate S. aureus effectively. These results revealed that HDMP NPs had the ability to recognize and trap S. aureus through in situ self-assembled fibrous networks, similar to HD6.
S. aureus is an invasive Gram-positive bacterium, which can bind to and invade host cells, causing skin and soft tissue infection and endovascular inflammation. The self-assembly on S. aureus of HDMP NPs was expected to inhibit the invasion to host cells and was validated by the invading experiment between S. aureus and two cell lines, human umbilical vein endothelial cells (HUVECs) and human embryonic kidney 293 (293T) cells. The results revealed that the percentage of intrusion for 293T cells by S. aureus dropped from about 18.37% to 0.58% once S. aureus was pretreated with HDMP NPs (31.7 times, 10 μM) (Fig. 4F). The percentage of intrusion of S. aureus for HUVECs decreased from 0.79 to 0.27% after pre-incubation with the HDMP NPs (10 μM) (Fig. 4G). Meanwhile, free S. aureus stained with green fluorescence invaded into 293T cells (Fig. 4H). In contrast, it was almost impossible to observe the HDMP NP–treated S. aureus in 293T cells, which was trapped by in situ self-assembled fibrous networks. The cytotoxicity of HDMP NPs and C-HDMP NPs for bacteria was respectively carried out (fig. S8). The HDMP NPs and C-HDMP NPs were not toxic to bacteria when the concentration was less than 50 μM. Therefore, these experimental results indicated that HDMP NPs could trap the S. aureus and inhibit the invasion significantly, not killing bacteria directly. For the in vitro cellular experiment, HDMP showed similar inhibition of bacterial invasion (31.7 times, 10 μM) with the natural HD6 (100 times, 10 μM) (9).
HDMP NPs recognize and transform into NFs on S. aureus in vivo
To study transformation of HDMP NPs on S. aureus in vivo, a mouse model inoculated with S. aureus was established. S. aureus (50 μl, 108 CFU) was injected into the right leg muscle of mice, and the same amount of 0.9% saline was injected into the left leg muscle of mice. HDMP NPs were injected through the tail vein, and the fluorescence signal was observed by an in vivo fluorescence imaging system (Fig. 5, A and B) (20). It can be visually seen that the fluorescence signal at the muscle inoculated with S. aureus on the right side was much stronger than that on the left side inoculated with 0.9% saline, indicating that HDMP NPs could recognize S. aureus in vivo. The above mice were dissected, and the leg muscle tissue was taken to prepare frozen tissue sections and observed by CLSM (Fig. 5C). The green fluorescence of BP in HDMP and the red fluorescence of propidium iodide staining dead bacteria were merged, indicating that HDMP NPs had excellent binding ability to S. aureus in vivo. Meanwhile, the HDMP NPs could accumulate in a region of bacteria in the right leg muscle through an active targeting mechanism, which was beneficial for in situ self-assembly to construct fibrous networks. From the bio-TEM of muscle tissue (Fig. 5D), it was observed that HDMP NPs transformed into NFs surrounding the surface of S. aureus. However, the C-HDMP NP–treated group showed that NPs surrounded S. aureus. These TEM images were similar to those treated with HDMP NPs and C-HDMP NPs in vitro (Fig. 3E), respectively. Therefore, we concluded that HDMP NPs could self-assemble on the surface of S. aureus, transforming into NFs in vivo.
Fig. 5. HDMP NPs recognize and transform into NFs on S. aureus in vivo.
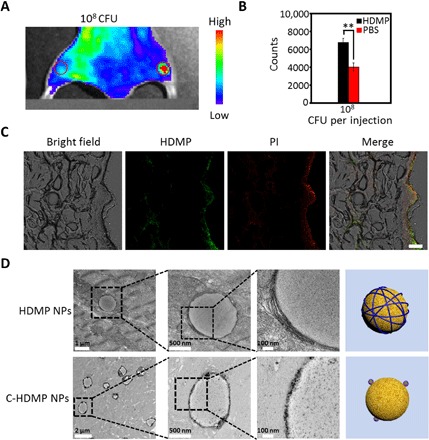
(A) In vivo fluorescence images of leg muscle inoculated with 108 CFU bacteria (right) and PBS (left), followed by HDMP NP intravenous administration. (B) The quantification of (A). The * represents significant difference between the PBS group and the experiment group (**P < 0.01). (C) Ex vivo confocal images of infected muscle tissue treated with HDMP NPs, showing specific recognition of HDMP to bacteria in vivo. Green fluorescence is HDMP. Red fluorescence is propidium iodide. Scale bar, 40 μm. (D) The TEM images of muscle tissue slices inoculated with S. aureus and treated with HDMP and C-HDMP NPs, showing transformed HDMP NFs and maintained C-HDMP NPs on bacterial surfaces, respectively. The TEM images are representative of three independent experiments.
HDMP NPs fight against S. aureus in vivo
Before the evaluation of the inhibition effect of HDMP NPs on bacteria invasion in vivo, cell viability (fig. S8) and hemolysis test (fig. S9) were evaluated, showing good biocompatibility of HDMP NPs for the in vivo experiment (21). There were some prevalence of S. aureus in soft-tissue infections and severe sepsis (21). Therefore, four in vivo infection models based on mouse abscess and bacteremia were set up and tested. First, 12 mice were randomly divided into two groups and were inoculated with S. aureus alone and HDMP NP–incubated S. aureus on the right leg muscle of the mouse, respectively (Fig. 6A). There was no sign of infection in the HDMP NP–incubated S. aureus group 48 hours after injection. However, there were three mice exhibiting obvious infection (abscess) in the S. aureus group. These results showed that HDMP NPs could notably restrict S. aureus-induced muscle lesions in vivo. The muscle tissue was further analyzed by the hematoxylin and eosin (H&E) staining. As shown in Fig. 6B, the typical leg muscle tissue in the HDMP NP–incubated S. aureus group exhibited the same normal tissue morphology as the PBS group, indicating that the mice in the HDMP NP–incubated S. aureus group were not infected. In contrast, there were many inflammatory cells in the muscle tissue of the S. aureus group.
Fig. 6. In vivo antibacterial effect of HDMP NPs.
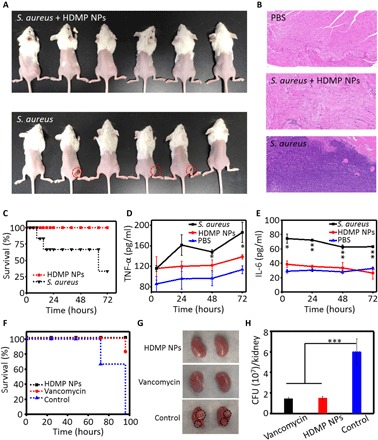
(A) Images of S. aureus inoculated in the right leg muscle in mice in the presence and absence of HDMP NPs (n = 6). (B) The representative hematoxylin and eosin (H&E) staining images of the leg muscle tissue of mice, indicating that the HDMP NP–treated S. aureus did not induce the bacterial infection. (C) Survival curve of the bacteremia mice infected by S. aureus treated with/without HDMP NPs (n = 6), showing the highly efficient antibacterial ability of HDMP NPs. Amount of (D) tumor necrosis factor–α (TNF-α) and (E) interleukin-6 (IL-6) in mouse sera in the bacteremia model during the experiment. The * represents significant difference between the control group and the experiment group (*P < 0.05, **P < 0.01). (F) Survival curve of the bacteremia mice infected by methicillin-resistant S. aureus (MRSA) treated with HDMP NPs, compared with vancomycin (n = 6). (G) Representative images of the MRSA-infected kidney (96 hours, untreated) and MRSA-infected kidney after 1-hour infection treated with vancomycin (5 mg/kg i.v.) or HDMP NPs (5 mg/kg i.v.) (96 hours). The white areas are indicated by black dashed circles. (H) CFU levels at 96 hours in kidney of mice intravenously infected with MRSA with treatment of vancomycin (5 mg/kg), HDMP NPs (5 mg/kg), or PBS 1 hour after infection. n = 3 per treatment group. The * represents significant difference between the control group and the experiment group (***P < 0.001). Photo credit: Yu Fan, NCNST.
Another similar experiment was carried out with the same S. aureus (50 μl, 108 CFU) inoculation on the right leg muscles of all 12 mice, 6 of which were intravenously injected with HDMP NPs from the tail vein. After 48 hours, only one mouse was infected (showing signs of swelling) in the HDMP NP–treated group. However, there were four mice that showed signs of swelling in the right leg muscle, indicating infection (fig. S10a). These results revealed that HDMP NP treatment could effectively inhibit the invasion of the S. aureus into muscle.
To confirm the efficacy of the biomimetic in situ self-assembly strategy based on the HDMP NPs, we investigated the therapeutic effects of HDMP NPs on the bacteremia mouse model. Mice were divided into three groups, and each group involved six mice. Two groups (12 mice) were injected with S. aureus (108 CFU) suspended in 50 μl of 0.9% saline through the tail vein for bacteremia model groups and 50 μl of 0.9% saline for one control group. One bacteremia group was treated with HDMP (200 μM, 150 μl, 3.7 mg/kg) NPs, and the other group was treated with PBS. As shown in Fig. 6C, in the bacteremia model, there was significantly reduced mortality of mice injected with HDMP NPs, and all the six mice were alive in 72 hours. However, four of six mice were dead in the PBS-treated bacteremia group. Furthermore, the amount of the inflammatory cytokine tumor necrosis factor–α (TNF-α) (Fig. 6D) and interleukin-6 (IL-6) (Fig. 6E) was significantly reduced in blood samples of mice, indicating that sepsis was markedly inhibited through intravenous injection of HDMP NPs. Our HDMP NPs increased survival rate by 66.7% in the S. aureus–infected bacteremia mouse model.
Last, we used HDMP NPs as a novel, encircle-without-attacking natural supramolecular agent for treating the methicillin-resistant S. aureus (MRSA)–infected bacteremia mouse model, and compared its effects with that of vancomycin. Mice were divided into three groups (n = 6 per group), which were injected through the tail vein with MRSA (5 × 107 CFU) suspended in 50 μl of 0.9% saline for bacteremia model groups. The three groups were treated with HDMP (200 μl, 5 mg/kg) NPs, vancomycin (200 μl, 5 mg/kg), and PBS (200 μl) 1 hour later (21). As shown in Fig. 6F, all the mice in the PBS-treated control group were dead in 96 hours. However, only one mouse in the vancomycin-treated group died, and all the mice in HDMP NP–treated group were alive in 96 hours. Mice that received HDMP NPs or vancomycin after treatment had no significant pathological changes, such as white areas in the kidney surfaces (Fig. 6G) (22). To quantitatively evaluate the antibacterial activity of HDMP, the bacteria in the kidney (Fig. 6H) and liver (fig. S10B) 96 hours after injection were cultured and counted. There was a marked reduction of CFU in kidney and liver tissues of the HDMP NP–treated group compared with untreated controls, similar to vancomycin treatment. These results suggested that the biomimetic HDMP NPs displayed a similar efficacy to the vancomycin, the golden antibiotics.
The biodistribution and the side effects of HDMP NPs were characterized. The mice were treated with HDMP NPs through intravenous injection and euthanized to obtain the main organs 24 hours after injection. The ex vivo fluorescence imaging of main organs showed that the HDMP NPs and C-HDMP NPs were mainly metabolized by the liver (fig. S10, C and D). The main organs were further characterized by H&E staining. As shown in fig. S10e, the HDMP NPs showed negligible toxicity to the main organs.
DISCUSSION
The antibacterial strategy of direct killing is highly efficient, but may induce drug resistance, flora disequilibrium, etc. HD6 traps bacteria with self-assembled nanofibrous networks for inhibition of bacteria invasion, providing a new concept of anti-bacterial therapy. In vivo antibacterial investigation of HD6 in a transgenic mouse model has provided insight into its antibacterial mechanism, that is, recognition and capture of bacteria. However, it is difficult to apply a transgenic strategy in clinical antibacterial therapy. In this study, we design an HDMP, which utilizes in situ ligand-receptor–induced self-assembly for recognizing and trapping bacteria in vivo. Our HDMP NPs can be administered intravenously and show high efficiency of antibacterial activity, indicating high applicability. The specificity of HDMP is superior to that of the natural HD6 due to the specificity of its ligand peptide, RLYLRIGRR, for Gram-positive bacteria. The difficulty of transporting raw self-assembling peptide materials to the area of bacterial invasion is overcome by intravenous administration of pre-assembled nanostructures (HDMP NPs). The ligand peptide enabled the strong accumulation of pre-assembled HDMP nanostructures and induced the subsequent self-assembly process for trapping the bacteria and inhibiting bacterial invasion. Meanwhile, the in situ self-assembly process on bacteria increases the accumulation of HDMP NPs. In all, the programmable self-assembly process from NPs (for transport via intravenous administration) to NFs (in situ trapping of bacteria) is realized by the combination of three components, inhibiting the infection of S. aureus in vivo.
The HDMP NPs show comparable inhibition (31.7 times reduction by 10 μM) of bacterial invasion with HD6 (100 times reduction by 10 μM) at the cellular level. In the MRSA bacteremia mouse model, which is reaching epidemic proportions, causing morbidity, mortality, and chronic disease, the mice treated with HDMP (5 mg/kg) NPs showed 100% survival rate, even higher than that of the vancomycin (5 mg/kg, 83.3%)–treated mice. This strategy provides a new in vivo process biomimetic concept to prevent bacteria invasion, not for Gram-negative bacteria or cells, probably solving the current awkward situation of antibiotics.
The biomimetic materials once used for biomedical applications become more complicated and sophisticated. It is difficult to simply mimic the ingredient and structure of the natural materials for their properties and functions. The precise control of the biological process, such as the asymmetric ion transport process (23) and protein-folding process modulated by chaperone (24), should be paid much more attention.
MATERIALS AND METHODS
Materials
All reagents and solvents for organic synthesis were purchased from commercially available sources and used without further purification unless otherwise stated. The BP-COOH was synthesized as per the previous report by our group. Dichloromethane and N,N-dimethylformamide (DMF) were treated by molecular sieve. Methanol, O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU), 1-hydroxybenzotriazole (Hobt) piperidine, 4-methylmorpholine (NMM), and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich Chemical Co. All Fmoc amino acids and Wang resins were obtained from GL Biochem (Shanghai) Ltd. Cell counting kit-8 assay was obtained from Beyotime Institute of Biotechnology, China. Mouse TNF-α enzyme-linked immunosorbent assay (ELISA) kit (SEKM-0034-48T) and Mouse IL-6 ELISA kit (SEKM-0007-48T) were obtained from Solarbio, China. The cell lines RAW 264.7, L929, HUVEC, and 293T were purchased from the Cell Culture Center of the Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China). Cell culture medium and fetal bovine serum were purchased from Wisent Inc. (Multicell, Wisent Inc., St. Bruno, Quebec, Canada). 0.25% Trypsin-EDTA and antibiotic solution (penicillin and streptomycin) were purchased from Invitrogen (Invitrogen, Carlsbad, CA).
The preparation of HD6 mimic peptide HDMP and control molecule C-HDMP
The molecules HDMP and C-HDMP were synthesized by standard solid-phase peptide synthesis techniques using Fmoc-coupling chemistry. His-resin (loading, 0.32 mM/g) or Lys-resin (loading, 0.50 mM/g) was used as phase supports. Piperidine (20% v/v) in anhydrous DMF was used as deprotection agents to the Fmoc group. The complete deprotection was confirmed by using the ninhydrin test (ninhydrin, phenol, VC 1:1:1 v/v). Then, the activated amino acids (160 mM) by HBTU (160 mM) and Hobt was added into NMM (5%) in anhydrous DMF. This process was repeated until the last amino acid (BP-COOH) was coupled. The final production was obtained through cleaving from the resin by the mixture of TFA (95%, volume percentage), triisopropyl silane (2.5%, volume percentage), and water (2.5%, volume percentage) for 2.5 hours in an ice bath. The resin and the lysate were suction-filtered to obtain a filtrate, and then the filtrate was blown to 1/10 of the total volume with nitrogen. The remaining filtrate was added to ice diethyl ether to give a precipitated solid. The obtained solid was washed three times with ice diethyl ether and dried in a vacuum oven to give the final product. The molecular structures of peptides were confirmed by MALDI-TOF-MS (Bruker Daltonics).
Preparation and characterization of HDMP NPs and C-HDMP NPs
The HDMP and C-HDMP were dissolved in DMSO to form a solution (3 mM). The 20 μl of HDMP solution was further diluted with DMSO (880, 680, 480, 280, and 80 μl) and mixed with deionized water (100, 300, 500, 700, and 900 μl), respectively. The UV-vis and fluorescence spectra of the resulting solution were measured to monitor the formation process of HDMP NPs and C-HDMP NPs.
HDMP NPs and C-HDMP NPs recognize bacteria by CLSM
S. aureus and E. coli were incubated with HDMP NPs and C-HDMP NPs (30 μM) in 500 μl of 0.9% saline for 4 hours, respectively. Subsequently, after washing bacteria once with 0.9% saline, it was suspended in 500 μl of 0.9% saline, and the bacteria solution (10 μl) was dropped on the center of the confocal dish and covered with a cover glass for CLSM measurements (Ultra-VIEW Vox, PerkinElmer).
HDMP NPs were used to incubate with the mixture of two bacteria (S. aureus and E. coli) in 500 μl of 0.9% saline for 4 hours. After washing with 0.9% saline, the bacteria solution in 0.9% saline (10 μl) was dropped on a confocal dish, covered with a cover glass for CLSM measurements (Ultra-VIEW Vox, PerkinElmer).
HDMP NPs and C-HDMP NPs in situ self-assembly on bacteria by TEM
The HDMP and C-HDMP NP solutions were mixed with S. aureus and E. coli, respectively, and incubated for 4 hours in a constant-temperature incubator, followed by centrifugation at 10,000 rpm for 5 min. The precipitated bacteria were washed three times with 0.9% saline. The obtained bacteria were used for TEM measurements by a bio-TEM (HT7700 HITACHI TEM).
HDMP NPs and C-HDMP NPs in situ self-assembly and trapping bacteria by SEM
SEM was used to detect in situ assembly on bacterial surfaces. First, S. aureus and E. coli were incubated on silicon wafer in 0.9% saline, which was placed in 24-well plates overnight. The next day, the 0.9% saline was aspirated, and HDMP NPs and C-HDMP NPs (30 μM, 1 ml) were added and incubated for 4 hours. After removal of excess solution, the bacteria were solidified with glutaraldehyde (4%) overnight and then coated with gold for 2 min for SEM observation (Hitachi-SU8220).
Entanglement and sedimentation of bacteria by HDMP NPs and C-HDMP NPs
S. aureus from frozen stock bacteria was grown overnight with shaking (37°C) in 5 ml of tryptone soy broth to saturation. The bacterial suspension (840 μl) was mixed with 6.16 ml of 0.9% saline to obtain an OD600 (optical density at 600 nm) value of 0.25. The suspension was centrifuged and resuspended in 1 ml of 0.9% saline containing 10, 30, and 50 μM HDMP NPs or C-HDMP NPs and then transferred to a sterile cuvette. The OD600 value was measured at a determined time point until 6 hours.
HDMP NPs and C-HDMP NPs inhibit the invasion of bacteria
HUVECs and 293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were cultured at 37°C and 5% CO2. The cultured 293T cells were counted to prepare 6 ml of suspension (about 104 cells per 1 ml). The concentration of HUVECs and 293T cells was quantified by using a manual hemocytometer and adjusted to 105 cells/ml. A 1-ml aliquot of cells was then added to 24-well tissue culture plates and incubated for 24 hours.
S. aureus from frozen stock bacteria was grown overnight with shaking (37°C) with 5 ml of TBS to saturation. Ten microliters of the bacterial suspension was taken out and added to 2 ml of 0.9% saline to obtain a suspension with an OD600 value of 0.1, representing 108 CFU/ml of bacteria. Then, the bacterial suspension was diluted to obtain 12 ml of 106 CFU/ml of bacterial suspension. The bacterial suspension was centrifuged (3500 rpm, 10 min) to remove the supernatant. HDMP NPs (30 μM) were prepared in 6 ml of serum- and antibiotic-free DMEM and added to bacteria sediment, incubating for 4 hours in a 37°C incubator.
Then, the cells were washed once with PBS and infected with 1 ml of HDMP NP–treated S. aureus [multiplicity of infection (MOI) = 10]. MOI is a ratio of the number of bacterial cells to cells when infection is initiated. The medium was removed after 1.5 hours and washed twice with PBS. The cells were subsequently incubated in 200 μl of serum-free DMEM containing kanamycin (100 μg/ml) for 1.5 hours at 37°C to kill any remaining extracellular bacteria. The medium was removed after 1.5 hours and washed twice with PBS. The cells were then incubated in 200 μl of sterile-filtered 1% Triton X-100 in PBS for 10 min at room temperature.
The solution in each well was treated with sodium phosphate, diluted 10-, 100-, and 1000-fold, respectively, and then 500 μl of the solution was spin-coated on LB agar plates and incubated at 37°C for 20 hours, followed by counting colonies on the plate by the colony-counting method for determination of the invasion rate. The result averages and SDs were reported on the basis of three independent experiments for each cell line.
Hemolytic properties of HDMP NPs and C-HDMP NPs
The blood of mice was taken into a centrifuge tube, and the blood was agitated with a glass rod to remove fibrinogen. About 10 times of the 0.9% sodium chloride solution was added, shaken well, and centrifuged at 1000 to 1500 rpm for 15 min, removing the supernatant. Next, the precipitated red blood cells were washed three times with the 0.9% sodium chloride solution, until the supernatant is no longer red. The centrifuged red blood cells were centrifuged as a 20% (v/v) red blood cell suspension with the 0.9% sodium chloride solution. A 2% (v/v) red blood cell suspension was prepared by mixing 20% (v/v) red blood cell suspension with HDMP NPs and C-HDMP NPs (10, 30, 50, and 100 μM). The mixture was immediately incubated in an incubator at 37°C. Hemolysis reaction was observed in 1 hour.
In situ self-assembly of HDMP NPs and C-HDMP NPs on bacteria in vivo
Female BALB/c mice between 4 and 6 weeks were inoculated with S. aureus (108 CFU) suspended in 50 μl of 0.9% saline in the right leg and 50 μl of 0.9% saline in the left leg. After 30 min, HDMP NPs (150 μl, 200 μM) were injected through the tail vein and observed by in vivo fluorescence imaging technique for 4 hours. The quantified fluorescence results were obtained from a histogram of photon counts. After that, the mice were euthanized and two leg muscles were dissected. After washing with 0.9% saline, they were divided into two parts, one part was fixed with 0.4% paraformaldehyde tissue fixative for 12 hours for bio-TEM, and the other part was embedded with embedding agent and frozen (−80°C) and sliced by a cryostat for CLSM observation.
HDMP NP treatment of a mouse model of muscle infection
Animal experiments were carried out complying with National Institutes of Health guidelines for the Care and Use of Laboratory Animals, and the study protocol was approved by the Institutional Animal Care and Use Committee of National Center for Nanoscience and Technology, China. Twelve female BALB/c mice between 4 and 6 weeks were randomly divided into two groups with six mice in each group. HDMP NPs (30 μM) and 1 ml of the 0.9% saline solution were incubated with S. aureus (108 CFU) for 4 hours. The bacterial suspension was then centrifuged, and after removing the supernatant, the bacteria were suspended in 50 μl of 0.9% saline and inoculated into the right leg muscle of six mice. Six mice were inoculated with 50 μl of 0.9% saline of untreated S. aureus (108 CFU) in the right leg muscle. Both groups were observed for 48 hours to evaluate the infection. The infection was further confirmed by H&E stain of mouse muscle tissue. Similarly, the two groups of mice were inoculated with 50 μl of 0.9% saline of S. aureus (108 CFU) in the right leg muscle. One group was further treated with 200 μl of HDMP NPs (30 μM) 30 min later through intravenous administration. The other group was intravenously injected with the same amount of PBS. Both groups were observed to evaluate the infection at 48 hours.
HDMP NP treatment of a mouse model of bacteremia
Female BALB/c mice between 4 and 6 weeks were divided into three groups, and each group involved six mice. S. aureus (108 CFU) suspended in 50 μl of 0.9% saline was injected through the tail vein in two groups and 50 μl of 0.9% saline in the control group. One was intravenously injected with HDMP (150 μl, 200 μM) NPs 0.5 hours later. The other was intravenously injected with 150 μl of PBS. Then, the health of mice was monitored every 3 hours for the first 24 hours, and then every 8 hours for up to 72 hours. The mice were euthanized immediately if they showed signs of inability to eat and drink, respiratory distress, or mobility loss. Meanwhile, serum samples were harvested from test animals. Serum samples at different time points were used to detect TNF-α and IL-6. Detailed steps were referred to the ELISA kit.
HDMP NP treatment of a mouse model of bacteremia with MRSA
Female BALB/c mice between 4 and 6 weeks were divided into three groups, and each group involved six mice. MRSA (5 × 107 CFU) suspended in 50 μl of 0.9% saline was injected through the tail vein in three groups. The HDMP group was intravenously injected with HDMP (200 μl, 200 μM, 5 mg/kg) NPs 1 hour later. The vancomycin group was intravenously injected with vancomycin (200 μl, 337 μM, 5 mg/kg). The control group was intravenously injected with 200 μl of PBS. Mice were monitored and euthanized at various time points according to experimental design. The livers and kidneys were obtained and homogenized. For determination of CFU, the tissue homogenates were mixed, followed by treatment with sodium phosphate for dilution of 10-, 100-, and 1000-fold, respectively. Then, 500 μl of tissue homogenates was spin-coated on LB agar plates and incubated at 37°C for 20 hours. Last, the bacterial colonies were counted.
Statistical analysis
All data are reported as mean ± SD. The in vitro experiments were performed in three independent experiments with at least three technical replicates. The in vivo experiments were performed with three to six mice for each group. Statistical analysis of the samples was performed using Student’s t test, and a P value of <0.05 was considered significant.
Supplementary Material
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (51890891, 51890894, 51573031, 51573032, and 51725302), the Science Fund for Creative Research Groups of the National Natural Science Foundation of China (11621505), the CAS Interdisciplinary Innovation Team, the Jilin Province Key Laboratory of Organic Functional Molecular Design and Synthesis (130028911), and The Fundamental Research Funds for the Central Universities, South-Central University for Nationalities (CZD19014). Author contributions: L.W. conceived the idea and developed the project. Y.F. and X.-D.L. conducted all experiments and analyzed the data. X.-X.H., P.-P.H., and J.-Q.F. assisted with the chemical synthesis, data analysis, and cell culture studies. K.Z. performed bio-TEM. P.-P.Y. and L.J. assisted with the animal studies. H.-Y.Z., W.T., and Z.-M.C. assisted with artwork. H.W. and X.D.L. assisted with the design and supervision of the research. L.W., Y.F., H.W., and X.-D.L. co-wrote the paper and all authors commented on the manuscript. Competing interests: The authors filed patents pertaining to the results presented in the paper. L.W., H.W., and Y.F. are the co-inventors of a pending patent on the biomimetic antibacterial strategy. The remaining authors declare no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/19/eaaz4767/DC1
REFERENCES AND NOTES
- 1.Baym M., Stone L. K., Kishony R., Multidrug evolutionary strategies to reverse antibiotic resistance. Science 351, aad3292 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Didelot X., Walker A. S., Peto T. E., Crook D. W., Wilson D. J., Within-host evolution of bacterial pathogens. Nat. Rev. Microbiol. 14, 150–162 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laxminarayan R., Matsoso P., Pant S., Brower C., Røttingen J. A., Klugman K., Davies S., Access to effective antimicrobials: A worldwide challenge. Lancet 387, 168–175 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Coates A., Hu Y., Bax R., Page C., The future challenges facing the development of new antimicrobial drugs. Nat. Rev. Drug Discov. 1, 895–910 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Brauner A., Fridman O., Gefen O., Balaban N. Q., Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat. Rev. Microbiol. 14, 320–330 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Akira S., Uematsu S., Takeuchi O., Pathogen recognition and innate immunity. Cell 124, 783–801 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Ayabe T., Satchell D. P., Wilson C. L., Parks W. C., Selsted M. E., Ouellette A. J., Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2, 113–118 (2000). [DOI] [PubMed] [Google Scholar]
- 8.Chu H., Pazgier M., Jung G., Nuccio S.-P., Castillo P. A., de Jong M. F., Winter M. G., Winter S. E., Wehkamp J., Shen B., Salzman N. H., Underwood M. A., Tsolis R. M., Young G. M., Lu W., Lehrer R. I., Bäumler A. J., Bevins C. L., Human α-defensin 6 promotes mucosal innate immunity through self-assembled peptide nanonets. Science 337, 477–481 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chairatana P., Nolan E. M., Molecular basis for self-assembly of a human host-defense peptide that entraps bacterial pathogens. J. Am. Chem. Soc. 136, 13267–13276 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chairatana P., Chiang I. L., Nolan E. M., Human α-defensin 6 self-assembly prevents adhesion and suppresses virulence traits of Candida albicans. Biochemistry 56, 1033–1041 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Chairatana P., Nolan E. M., Human α-defensin 6: A small peptide that self-assembles and protects the host by entangling microbes. Acc. Chem. Res. 50, 960–967 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hynes R. O., The extracellular matrix: Not just pretty fibrils. Science 326, 1216–1219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saido-Sakanaka H., Ishibashi J., Momotani E., Amano F., Yamakawa M., In vitro and in vivo activity of antimicrobial peptides synthesized based on the insect defensin. Peptides 25, 19–27 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Kumar D. K. V., Choi S. H., Washicosky K. J., Eimer W. A., Tucker S., Ghofrani J., Lefkowitz A., McColl G., Goldstein L. E., Tanzi R. E., Moir R. D., Amyloid-β peptide protects against microbial infection in mouse and worm models of Alzheimer’s disease. Sci. Transl. Med. 340, 340ra372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L., Li W., Lu J., Zhao Y.-X., Fan G., Zhang J.-P., Wang H., Supramolecular nano-aggregates based on Bis(Pyrene) derivatives for lysosome-targeted cell imaging. J. Phys. Chem. C 117, 26811–26820 (2013). [Google Scholar]
- 16.He P.-P., Li X.-D., Wang L., Wang H., Bispyrene-based self-assembled nanomaterials: In vivo self-assembly, transformation, and biomedical effects. Acc. Chem. Res. 52, 367–378 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Le P., Kunold E., Macsics R., Rox K., Jennings M. C., Ugur I., Reinecke M., Chaves-Moreno D., Hackl M. W., Fetzer C., Mandl F. A. M., Lehmann J., Korotkov V. S., Hacker S. M., Kuster B., Antes I., Pieper D. H., Rohde M., Wuest W. M., Medina E., Sieber S. A., Repurposing human kinase inhibitors to create an antibiotic active against drug-resistant Staphylococcus aureus, persisters and biofilms. Nat. Chem. 12, 145–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X.-X., He P.-P., Qi G.-B., Gao Y.-J., Lin Y.-X., Yang C., Yang P.-P., Hao H., Wang L., Wang H., Transformable nanomaterials as an artificial extracellular matrix for inhibiting tumor invasion and metastasis. ACS Nano 11, 4086–4096 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Yang P.-P., Luo Q., Qi G.-B., Gao Y.-J., Li B.-N., Zhang J.-P., Wang L., Wang H., Host materials transformable in tumor microenvironment for homing theranostics. Adv. Mater. 29, 1605869 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Li L.-L., Ma H.-L., Qi G.-B., Zhang D., Yu F., Hu Z., Wang H., Pathological-condition-driven construction of supramolecular nanoassemblies for bacterial infection detection. Adv. Mater. 28, 254–262 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Wang R., Braughton K. R., Kretschmer D., Bach T.-H. L., Queck S. Y., Li M., Kennedy A. D., Dorward D. W., Klebanoff S. J., Peschel A., DeLeo F. R., Otto M., Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13, 1510–1514 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Surewaard B. G., Deniset J. F., Zemp F. J., Amrein M., Otto M., Conly J., Omri A., Yates R. M., Kubes P., Identification and treatment of the Staphylococcus aureus reservoir in vivo. J. Exp. Med. 213, 1141–1151 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo W., Tian Y., Jiang L., Asymmetric ion transport through ion-channel-mimetic solid-state nanopores. Acc. Chem. Res. 46, 2834–2846 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Ma F.-H., Li C., Liu Y., Shi L., Mimicking molecular chaperones to regulate protein folding. Adv. Mater. 32, 1805945 (2020). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/19/eaaz4767/DC1


