We have identified a novel sleep regulatory pathway.
Abstract
Sleep is plastic and is influenced by ecological factors and environmental changes. The mechanisms underlying sleep plasticity are not well understood. We show that manipulations that impair flight in Drosophila increase sleep as a form of sleep plasticity. We disrupted flight by blocking the wing-expansion program, genetically disrupting flight, and by mechanical wing perturbations. We defined a new sleep regulatory circuit starting with specific wing sensory neurons, their target projection neurons in the ventral nerve cord, and the neurons they connect to in the central brain. In addition, we identified a critical neuropeptide (burs) and its receptor (rickets) that link wing expansion and sleep. Disrupting flight activates these sleep-promoting projection neurons, as indicated by increased cytosolic calcium levels, and stably increases the number of synapses in their axonal projections. These data reveal an unexpected role for flight in regulating sleep and provide new insight into how sensory processing controls sleep need.
INTRODUCTION
Since the discovery that sleep is an active process (1), numerous studies have identified pathways and neurotransmitter systems that promote sleep and waking in the mammalian brain (2). A key feature of these systems is that they are distributed across the neuraxis. Thus, sleep regulatory neurons are found in the preoptic area (3), the basal forebrain (4), lateral hypothalamus (5), the cortex (6), the brainstem (sublateral dorsal nucleus and parafacial zone) (7), and glia (8). In comparison to mammalian sleep, Drosophila is a relatively recent entrant to sleep research (9). However, as with the mammalian system, fly sleep and wake circuits are also distributed throughout the brain. Sleep and wake regulatory regions (and functions they regulate) include the following: mushroom body (learning and memory) (10, 11), the pars intercerebralis (PI; metabolic center) (12), the ellipsoid body (orientation, locomotion, and navigation) (13), fan-shaped body (visual learning) (14), clock neurons (15), and glia (16). The observation that sleep regulatory neurons appear to be organized in regions that subserve different functions suggests the intriguing possibility that these different sleep-promoting loci could also serve different functions and be activated under different circumstances (17).
Many of the neural circuits that regulate sleep in mammals and flies have been identified because they alter baseline sleep. However, it is increasingly clear that sleep itself is plastic and is not only influenced by ecological factors but is also responsive to environmental changes within an individual’s lifetime (18–22). These observations suggest that certain sleep regulatory centers might exist to regulate sleep and waking but only under specific circumstances. Many animals, in the wild, suppress sleep at specific times (and, on occasion, dispense with it all together) without exhibiting a sleep rebound or disrupting cognition or fitness. Examples include, but are not limited to, pectoral sandpipers (23), swallows (24), dolphins (25), elephants (26), and likely flies as well (27). In the laboratory, sleep is also responsive to environmental conditions including starvation, stress, and temperature to name a few challenges (28–30). Identifying sleep regulatory circuits that are predominantly engaged during environmental perturbations may provide new opportunities for developing therapeutics for treating a variety of sleep disorders.
Flight is critical for survival in Drosophila (31). Thus, we hypothesized that disrupting flight would present flies with major logistical challenges that would then recruit novel, downstream sleep regulatory circuits. Here, we show that manipulations that impair flight in Drosophila induce sleep plasticity as measured by an increase in sleep. We disrupted flight by blocking the wing-expansion program, genetically disrupting the ability of animals to fly, and by mechanically perturbing the wing (32, 33). We mapped the effects of wing expansion on sleep to a single pair of neurons in the subesophageal ganglion (SEG) that release the peptide hormone bursicon. In addition, by knocking down the receptor for bursicon, rickets, we identified a novel set of neurons, also within the SEG, that regulate sleep. The increase in sleep following wing cut involves some of the exact same neurons as for wing expansion. The wing cut–mediated increase in sleep is signaled by specific classes of chemosensory neurons in the fly wing that regulate sexual behavior (34). Using circuit-tracing techniques, we identified the projection neuron targets of these chemosensory neurons within the ventral nerve cord (VNC). Flight impairments activate and induce structural changes in these projection neurons to support long-lasting increases in sleep. Further, we identified postsynaptic targets of these projection neurons in the SEG and, in addition, neuromodulators of this circuit. Thus, we have described a circuit starting with peripheral sensors in the wing through two synapses to sleep regulatory neurons in the central brain. Activating components of this circuit increased sleep in flies with intact wings. These data reveal an unexpected role for flight in regulating sleep and provide new insight into how sensory processing controls sleep need.
RESULTS
Blocking wing expansion increases sleep
Newly eclosed flies emerge with unexpanded wings and, in the first 20 to 30 min after eclosion, undertake a stereotyped series of behaviors to expand their wings (33). Confining flies with unexpanded wings to a restricted space overnight greatly delays wing expansion, resulting in flies that cannot fly (Fig. 1A and fig. S1A) (35). As seen in Fig. 1B, flies placed into confinement before expansion sleep more compared to age-matched siblings that were confined for the same amount of time but immediately following wing expansion or unconfined siblings when released into recovery the following day. The increase in sleep was particularly marked during the day and associated with increased daytime sleep consolidation, as evidenced by increased sleep bout length during the day (Fig. 1, B to D). Confinement did not reduce waking activity, indicating that locomotion was not impaired (fig. S1B). Further, sleep of flies that had been confined was rapidly reversible by a mechanical stimulus (Fig. 1E) and was associated with increased arousal thresholds (Fig. 1F), indicating that confinement did not induce a behavioral malaise. To determine whether confinement increased sleep drive, we expressed the calcium-dependent nuclear import of LexA (CaLexA) system to monitor activity of the ellipsoid body R2 neurons (a known marker of sleep drive) (13). Confinement increased CaLexA signal in the R2 neurons of confined flies, suggesting that sleep drive was increased (fig. S1, C and D). Thus, confinement induces a state that meets the established criteria for sleep (9).
Fig. 1. Disrupting wing expansion increases sleep.
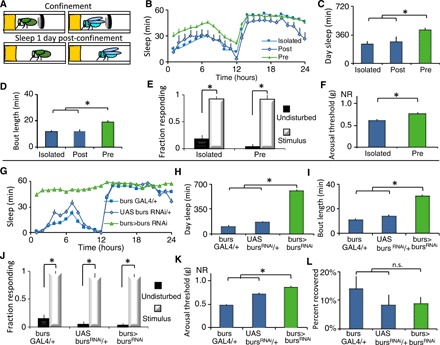
(A) Wild-type flies were confined in a restricted space before or after wing expansion following eclosion (day 0) and evaluated for sleep beginning on day 1. (B) Flies that were confined before expansion (Pre, n = 37) slept more than age-matched flies confined after expansion (Post, n = 9) and unconfined siblings (Isolated, n = 36); repeated-measures ANOVA for Time × Condition, P < 0.05. (C and D) Flies with unexpanded wings displayed increased daytime sleep and sleep bout duration compared to controls (t test, *P = 0.001). (E) Sleep in both groups was rapidly reversible in response to a mechanical stimulus at ZT15 (n = 20 to 32 flies per condition; *P < 0.01, Tukey correction). (F) Arousal thresholds were higher in flies confined before expansion than isolated controls (n = 14 flies per condition; *P < 0.01, t test). (G) burs-GAL4/+>UAS-bursRNAi/+ flies slept more than parental controls (n = 32 flies per genotype; repeated-measures ANOVA for Time × Genotype, P < 0.001). (H and I) burs-GAL4/+>UAS-bursRNAi/+ displayed increased daytime sleep and sleep bout duration compared to controls (*P < 0.01, Tukey correction). (J) Sleep was rapidly reversible in response to a mechanical stimulus for all genotypes (n = 25 to 30 flies per condition; *P < 0.01, Tukey correction). (K) Sleep in burs-GAL4/+>UAS-bursRNAi/+ flies was associated with increased arousal thresholds (n = 14 flies per condition; *P < 0.01, Tukey correction). (L) All genotypes displayed similar sleep rebound following 12 hours of sleep deprivation (n = 30 to 31 flies per condition). n.s., not significant.
Confinement results in unexpanded wings due to alterations in the release of the neurohormone bursicon (35), a cystine-knot heterodimer composed of two subunits: bursicon (burs) and partner of bursicon (pburs). Loss of function of burs or pburs completely blocks wing expansion (33). Thus, we asked whether knocking down burs or pburs would increase sleep in the absence of confinement. As seen in Fig. 1 (G to I), burs-GAL4>bursRNAi flies slept more than their parental controls, exhibiting increased daytime sleep, and sleep consolidation, without impairing locomotion (fig. S1E). burs-GAL4>bursRNAi flies were rapidly awakened by a mechanical stimulus (Fig. 1J) and exhibited elevated arousal thresholds (Fig. 1K) and a normal homeostatic response to overnight sleep deprivation (Fig. 1L). Similarly, sleep amount and consolidation were also elevated in burs-GAL4>pbursRNAi flies without impairing locomotion (fig. S1, F to I). burs-GAL4>pbursRNAi flies also met the established criteria for sleep (fig. S1, J to L). The burs-GAL4 and bursRNAi flies were outcrossed to a reference wild-type strain. Thus, RNA interference (RNAi)–mediated burs knockdown phenocopies the results using confinement. Flies do not sleep well when they are confined to a small space; however, burs-GAL4>bursRNAi flies did not lose any sleep relative to controls on the first day of adult life (fig. S1, M to O). Loss-of-function burs point mutations also increased sleep (fig. S1, P to R). RNAi knockdown of genes that coexpress with bursicon such as crustacean cardioactive peptide (CCAP), myoinhibiting peptide precursor (mip), the RNA binding protein Lark, or the histone transferase absent, small, or homeotic discs 1 (ash) did not change sleep or affect wing expansion (fig. S1, S and T) (36, 37), supporting the specificity of the burs knockdown experiments above. Confinement and loss of burs function both perturb wing expansion and increase sleep.
Two burs+ neurons modulate sleep
burs is transiently expressed in a small subset of neurons in the fly central nervous system (CNS), 2 neurons in the SEG (“Bseg”) and 12 to 14 neurons in the abdominal ganglion (“Bag”) (burs-GAL4>GFP; Fig. 2A). Thus, we conducted a series of experiments to determine whether wing expansion and sleep could be dissociated functionally, temporally, or spatially. First, burs+ neurons were chronically inhibited by expressing the inward rectifying potassium channel UAS-Kir2.1. burs-GAL4>UAS-Kir2.1 disrupted wing expansion and increased both sleep and sleep consolidation during the day (Fig. 2, B to D). The sleep episodes displayed the defining behavioral hallmarks of sleep without inhibiting locomotion (fig. S2, A to D). Second, we constitutively activated burs+ neurons by expressing the bacterial sodium channel UAS-NaChBac with burs-GAL4. This manipulation also disrupted wing expansion and increased sleep while meeting the behavioral criteria for sleep and without inhibiting locomotion (fig. S2, E to K). Similar results were obtained when the larger group of CCAP+ neurons (that includes burs+ neurons) was activated (fig. S2, L to S). Activation of CCAP neurons was shown to deplete bursicon levels in central processes, suggesting a possible mechanism by which activation and inhibition yield the same phenotype. Third, we used the TARGET system to determine whether burs GAL4 activity that supports sleep and wing expansion were temporally dissociable (fig. S2, T and U). Consistent with burs expression peaking at eclosion, these experiments indicated that burs neuron activity was required in pharate adults/early adult life for wing expansion and sleep. Last, we used split-GAL4 lines to specifically inactivate subsets of burs GAL4-expressing neurons. Inactivation of the Bseg (Bseg>UAS-Kir2.1) had a partially penetrant effect on wing expansion; the flies with wing defects increased sleep (Fig. 2, E to G, and fig. S3, A to D). In contrast, inactivating the Bag did not affect wing expansion but had a modest effect on sleep (Fig. 2, H to J, and fig. S3E). These data suggest that signals from the Bag might influence the Bseg or that the Bag neurons affect sleep independently from their impact on flight; these possibilities will be evaluated in future studies. The involvement of neurons in the SEG was confirmed using the Flipase-induced intersectional GAL80/GAL4 repression (FINGR) method to disrupt subsets of CCAP+ neurons (fig. S3, F to L). Collectively, these results suggest that modulating the activity of just two burs+ neurons in a restricted time window perturbs wing expansion and increases sleep.
Fig. 2. The excitability of burs neurons regulates wing expansion and sleep.
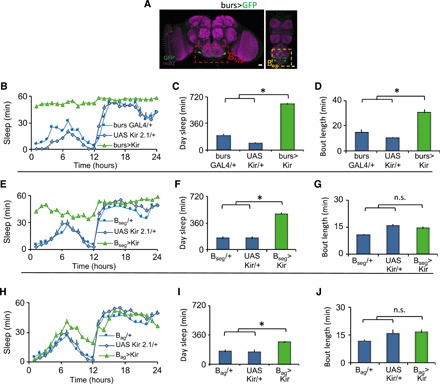
(A) burs-GAL4/+>UAS-GFP/+ labels two neurons in the SEG (Bseg) and 12 to 14 neurons in the abdominal ganglion (Bag). Scale bar, 20 μm. (B) burs-GAL4/+>UAS-Kir2.1/+ flies slept more than parental controls (n = 20 to 27 flies per genotype; repeated-measures ANOVA for Time × Genotype, P < 0.001). (C and D) burs-GAL4/+>UAS-Kir2.1/+ flies displayed increased daytime sleep and sleep bout duration compared to controls (*P < 0.01, Tukey correction). (E) Bseg>UAS-Kir2.1 flies with unexpanded wings slept more than parental controls (n = 16 to 32 flies per genotype; repeated-measures ANOVA for Time × Genotype, P < 0.001). (F and G) Bseg>UAS-Kir2.1 had more daytime sleep, but sleep consolidation was not altered (*P < 0.01, Tukey correction). (H) Bag>UAS-Kir2.1 flies had normal wings and modestly increased sleep (n = 18 to 30 flies per genotype; repeated-measures ANOVA for Time × Genotype, P < 0.001). (I and J) Bag>UAS-Kir2.1 flies modestly increased daytime sleep without altering sleep consolidation (*P < 0.01, Tukey correction).
Defining the role of the bursicon receptor rk
The burs receptor is rickets (rk). rk is a leucine-rich repeat containing G protein–coupled receptor (GPCR) that signals through adenyl cyclase and protein kinase A (PKA). The strong effects we observed with manipulating burs function led us to explore the effects of manipulating rk signaling using rk-GAL4 (Fig. 3A). We first knocked down rk with RNAi in rk+ neurons. This manipulation had a partially penetrant effect on wing expansion. rk-GAL4>rkRNAi flies with wing expansion defects exhibited increases in daytime sleep amount and consolidation without impairing locomotion (Fig. 3, B to D, and fig. S4, A to D). rk-GAL4>rkRNAi flies with expanded wings had a small increase in daytime sleep (fig. S4, A to D). Point mutants in rk also increased sleep (fig. S4, E to G). We next blocked PKA signaling in rk+ neurons by expressing a dominant-negative PKA (PKA-DN) with rk-GAL4. This manipulation blocked wing expansion and increased sleep amount and consolidation compared to parental controls, without impairing locomotion (Fig. 3, E to G, and fig. S4H). Similar results were obtained when cyclic adenosine 3′,5′-monophosphate (cAMP) levels were reduced in rk+ neurons by overexpressing the cAMP phosphodiesterase dunce (fig. S4, I to K). When is rk required for wing expansion and sleep? To address this question, we transiently inactivated rk+ neurons by expressing the temperature-sensitive dynamin shibire (Shits) with rk-GAL4. Transient inactivation for 1.5 hours after eclosion (and before wing expansion) was sufficient to block wing expansion and increase sleep (fig. S4, L to R). Collectively, these data indicate that loss of rk function phenocopies the effects seen with loss of burs function. We next examined the effects of activation of rk+ neurons. Chronic activation of rk+ neurons with NaChBac blocked wing expansion and increased sleep (fig. S5, A to D). burs labels 14 to 16 cells that die by apoptosis in the first 48 hours after eclosion (33, 38). rk-GAL4, in contrast, labels a large number of cells, which persist throughout adult life. Can rk+ neurons regulate sleep in older flies? To address this, we activated rk+ neurons by expressing the heat-sensitive transient receptor potential 1 (UAS-TrpA1) channel with rk-GAL4 in 4- to 5-day-old flies. Transient TRPA1-mediated activation of rk+ neurons increased sleep (fig. S5, E to G), indicating that rk+ neurons are sleep promoting. Precisely how rk+ neurons can increase sleep in response to such a diverse number of genetic manipulations is unclear and is the subject of ongoing investigations.
Fig. 3. rickets neurons regulate wing expansion and sleep.
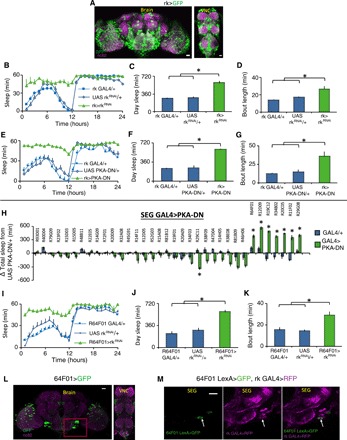
(A) rk-GAL4/+>UAS-GFP/+ labels a large number of cells in the fly CNS. (B and E) Sleep was increased in both rk-GAL4/+>UAS-rkRNAi/+ and rk-GAL4/+>UAS-PKADN/+ flies compared to parental controls (n = 16 to 32 flies per genotype; repeated-measures ANOVA for Time × Genotype, P < 0.001). rk-GAL4/+>UAS-rkRNAi/+ and rk-GAL4/+>UAS-PKADN/+ displayed increased daytime sleep (C and F) and sleep bout duration (D and G) compared to controls (*P < 0.01, Tukey correction). (H) Screen for SEG GAL4 drivers that increase sleep when expressing UAS-PKADN; sleep is expressed as change in sleep in minutes relative to the UAS-PKADN/+ controls (*P < 0.01, Tukey correction). The names of the GAL4 lines tested are listed above. (I) 64F01-GAL4/+>UAS-rkRNAi flies with unexpanded wings slept more than parental controls (n = 16 to 32 flies per genotype; repeated-measures ANOVA for Time × Genotype, P < 0.001). (J and K) 64F01-GAL4/+>UAS-rkRNAi flies displayed increased daytime sleep and sleep bout duration compared to controls (*P < 0.01, Tukey correction). (L) R64F01-GAL4/+>UAS-GFP/+ labels a sparse population of cells in the CNS, including the SEG (red box). (M) R64F01LexA/+>LexAopGFP/+; rk-GAL4/+>UAS-RFP/+ (red fluorescent protein) overlap in one cell in the SEG (white arrow). (L) Maximal intensity z projections counterstained with nc82 (magenta). (M) Single confocal slices. Scale bar, 20 μm.
Where is rk required? We focused on two candidate regions—the PI, a known sleep regulatory center (12) (where we see rk-GAL4 expression), and the SEG (as our results above implicated the Bseg). We anatomically selected a panel of GAL4 lines that label subsets of PI and SEG cells to drive the expression of PKA-DN. Blocking PKA in the PI did not increase sleep (fig. S5H). In contrast, eight GAL4 lines (selected from the large Rubin collection that express in the SEG) increase sleep when expressing PKA-DN (Fig. 3H). Seven of these eight lines blocked wing expansion when crossed to PKA-DN. One line, R20C05GAL4, generated flies with normal wings that could not fly and increased sleep when crossed to UAS PKA-DN. To verify specificity for rk, we knocked down rk with RNAi using the primary screen hits (fig. S6A). Seven of the eight lines increased sleep and blocked wing expansion in this secondary screen. We focused on one of the hits—R64F01-GAL4. This line is derived from enhancer elements of the CCAP receptor (CCAPR) gene, and because burs and CCAP are coexpressed, we reasoned that R64F01-GAL4 might express in a subset of rk+ neurons. We first evaluated this possibility with functional experiments. Expressing rkRNAi (Fig. 3, I to K, and fig. S6, B to E) or Gβ13FRNAi (a known component of rk signaling; fig. S6, F to H) with R64F01-GAL4 blocked wing expansion and increased sleep. In addition, expression of UAS-NaChBac (fig. S6, I to L) or UAS-TrpA1 (fig. S6, M to O) with R64F01-GAL4 increased sleep, similar phenotypes to those obtained with rk-GAL4. R64F01-GAL4 displays a restricted expression in the fly CNS, enriched in the SEG (Fig. 3L). Although R64F01LexA does not fully recapitulate the R64F01-GAL4 expression pattern, expressing LexAop-GFP using R64F01-LexA and UAS-RFP using rk-GAL4 identifies at least one common neuron in the SEG (Fig. 3M).
Last, to identify a minimal subset of R64F01 neurons that mediate the effects on wing expansion and sleep, we used an intersectional approach where we combined GAL80s with R64F01-GAL4>rkRNAi. dvGlut GAL80;R64F01-GAL4>rkRNAi flies had normal (expanded) wings and unchanged sleep (fig. S7, A to D), suggesting that it is the glutamatergic R64F01-GAL4 neurons that are the critical subset for wing expansion and sleep. Most R64F01-GAL4 neurons appear to be glutamatergic (fig. S7E).
Disrupting wings increases sleep
Wing damage occurs in adults to negatively affect flight (39). To determine whether the adaptive inactivity role for sleep would be observed in adults, we cut wings of flies on the first day of adult life after they had expanded their wings and examined sleep 2 days later. Flies with wings cut increased both sleep and sleep consolidation during the day (Fig. 4, A to C) and night (fig. S7, F and G) compared to their siblings with intact wings; locomotion was not impaired (fig. S7H). Sleep of flies with cut wings was rapidly reversible (fig. S7I) and associated with increased daytime arousal thresholds (fig. S7J). Robust increases in sleep were observed following wing cut in wild-type and per01 mutant flies maintained in constant darkness (fig. S7K). Thus, wing cut–induced increase in sleep does not require a functional circadian system. Further, a careful examination of the time course of wing cut–induced sleep revealed that wing cut increased sleep immediately following wing cut (fig. S7L), and sleep remained elevated in flies with cut wings for the 4 days that sleep was recorded (fig. S7, M to O). To determine whether flies with cut wings were under higher sleep drive, we again turned to the CaLexA system to monitor activity of the sleep-drive responsive ellipsoid body R2 neurons (13). Wing cut increased CaLexA signal in the R2 neurons, suggesting that sleep drive was increased (Fig. 4, D and E). The wing cut–mediated sleep increase was not a response to wing damage/injury, as mutations in immune response genes did not impair the ability of wing cut to induce sleep (fig. S7P). Further, increases in sleep were also observed when wings were glued (fig. S7, Q to T). Last, we evaluated a number of genetic manipulations that impair flight. Expressing the cell death activator reaper (UAS-rpr) in the wing disc (fig. S7, U to W)—or mutations in wingless (wg), protein kinase c (pkcΔ), and the commonly used CyO marker (fig. S8, A and B)—all impair flight (32) and increase sleep. Because many of these mutations are likely to have pleiotropic effects, we focused on wing cut.
Fig. 4. Cutting wings increases sleep.
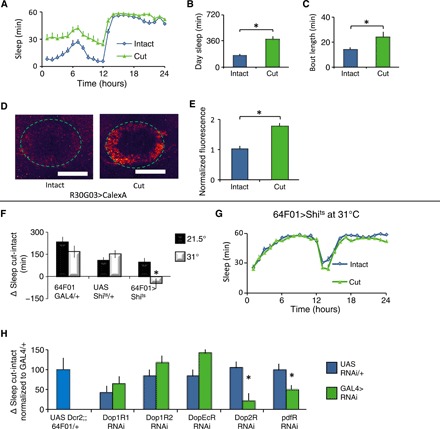
(A) Flies slept more the second day following wing cut than age-matched controls (n = 32 flies per condition; repeated-measures ANOVA for Time × Condition, P < 0.001). (B and C) Flies with cut wings displayed increased daytime sleep and sleep bout duration compared to controls (t test, *P < 0.001). (D) Representative images of R30G03-GAL4/+>CalexA/+ with intact and cut wings. (E) Quantification of CaLexA. Signal was higher in brains of flies with cut wings compared to controls (n = 8 to 9 flies per condition; *P < 0.001, t test). (F) Silencing R64F01-GAL4/+>UAS-shits1/+ neurons by raising the temperature from 21.5° to 31°C blocked the increase in sleep following wing cut. Sleep is expressed as a change in sleep in cut versus intact siblings (n = 16 flies per genotype per condition; *P < 0.01, Tukey correction). (G) Sleep in minutes per hour for R64F01-GAL4/+>UAS-shits1/+ flies shown in (F). (H) R64F01-GAL4/+>UAS-Dop2RRNAi/+ and R64F01-GAL4/+>UAS-PdfrRNAi/+ flies displayed an attenuated change in sleep following wing cut; R64F01-GAL4/+>UAS-Dop1R1RNAi/+, R64F01-GAL4/+>UAS-Dop1R2RNAi/+, and R64F01-GAL4/+>UAS-DopEcrRNAi/+ were not different from controls (n = 16 to 32 flies per genotype; *P < 0.05, Tukey correction).
Disrupting wings in adult flies increases sleep. Transient activation of wing-expansion circuits in adults also increases sleep (see above). Together, these results suggest that wing cut may recruit wing expansion circuits to increase sleep. To test this hypothesis, we expressed a thermosensitive mutant form of dynamin, Shibire (UAS-shits1), to block clathrin-mediated endocytosis of neurotransmitter release only at nonpermissive temperatures (31°C). As seen in Fig. 4 (F and G) at 31°C, wing cut did not result in an increase in sleep in 64F01-GAL4>UAS-shits1 flies compared to either siblings maintained at 21.5°C or parental controls. Although bursicon neurons undergo apoptosis (33, 38), both dopamine and Pigment-dispersing factor (Pdf) are known to regulate flight and sleep in adulthood (15, 40). Knocking down the D2 dopamine receptor (Dop2R), the Pdf receptor (Pdfr), and their downstream signaling components in R64F01 neurons using RNAi mitigated the increased sleep following wing cut (Fig. 4H and fig. S8, C to E). Although we have only used one RNAi line to evaluate Dop2R and Pdfr, these are commonly used RNAi lines that also produce robust phenotypes in our laboratory. Nonetheless, the role of Dop2R and Pdfr is the topic of active investigation in the laboratory and will be discussed in future work. Knocking down the other known Drosophila dopamine receptors did not appear to influence the extent of wing cut–induced sleep, although the precise role of the Dop1R1 receptor remains ambiguous and will be revisited later (Fig. 4H). Further, dopaminergic neural processes were detected in close proximity to the R64F01-GAL4 neurons in the SEG (fig. S8F), suggesting that the SEG is the relevant site of dopaminergic modulation. Although it is possible that distinct subsets of neurons mediate wing expansion and the response to wing cut, we feel that this scenario is unlikely because the wing cut response was normal in dvGlutGAL80; 64F01-GAL4>Dop2RRNAi flies (fig. S8G). Impairing flight by cutting wings increases sleep, requires R64F01-GAL4 neurons, and is not immune-mediated.
A neural pathway for wing cut–induced sleep
We hypothesized that a neural pathway from the wing conveys information about wing integrity to the brain to modulate sleep. To test this hypothesis, we inactivated subsets of wing chemosensory and mechanosensory neurons using UAS-Kir2.1. Expressing UAS-Kir2.1 with either Ir52aGAL4 or Ir76b-GAL4 attenuated but did not block the increase in sleep following wing cut (Fig. 5A). These data suggest that the combined input from Ir52a-GAL4 and Ir76b-GAL4 are partially redundant such that the smaller increase in sleep may result from losing input from the nonsilenced set of neurons during wing cut. To test this hypothesis, we expressed UAS Kir2.1 with both Ir52a-GAL4 and Ir76b-GAL4 simultaneously. As seen in Fig. 5A, no statistical increase in sleep was observed in Ir76b-GAL4/+; Ir52a-GAL4/+>UAS-Kir2.1 flies following wing cut (Fig. 5A). Baseline sleep was increased in Ir76b-GAL4/+; Ir52a-GAL4/+>UAS-Kir2.1 flies, while no change in sleep was observed in Ir76b-GAL4/+>UAS-Kir2.1 or Ir52a-GAL4/+>UAS-Kir2.1 flies (fig. S9, A to I). Ir52a GAL4 is strongly expressed in putative chemosensory neurons along the wing margin that project into the wing neuromere of the VNC, is weakly expressed in leg sensory neurons that also project axons into the VNC, but not detected in the brain (Fig. 5B) (41). Ir76b GAL4 expression is similar to Ir52a GAL4 and is also expressed in other classes of sensory neurons (fig. S9J). We focused on Ir52a GAL4, as its expression pattern was more restricted.
Fig. 5. Circuit for wing cut–induced sleep.
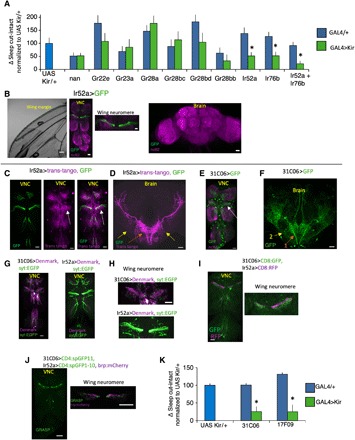
(A) Change in sleep (cut-intact) in flies expressing UAS-Kir2.1 in GAL4 lines associated with wing chemo- and mechanosensation normalized to the UAS-Kir2.1 parental control. Ir52a-GAL4/+>UAS-Kir2.1/+ and Ir76b-GAL4/+>UAS-Kir2.1/+ flies displayed reduced sleep in response to wing cut relative to controls (n = 20 to 45 flies per condition; *P < 0.05, Tukey correction). Ir76GAL4/+; Ir52a-GAL4/+>UAS-Kir2.1/+ flies did not increase sleep in response to wing cut. (B) Ir52a-GAL4/+>UAS-GFP/+ labels subsets of wing neurons (left) that project into the wing neuromere of the VNC (middle). Weak expression was also detected in nerves from leg neurons that project into the VNC (middle). No expression was detected in the brain (right). (C) Ir52a-GAL4/+>Trans-tango/+ (magenta) detects neurites in close proximity to the projections of Ir52a-GAL4 axons (green) in the VNC, with prominent labeling in the wing neuromere (white arrow), and two projection neuron axon tracts that exit the VNC and project to the lateral protocerebrum. (D) Ir52a-GAL4/+>Trans-tango/+ labels VNC neurons that project axons out of the VNC into the brain in two tracts with arborizations in the SEG and the VLP (orange and yellow arrows). (E) In the VNC, 31C06-GAL4/+>UAS-GFP labels neurites that resemble the Ir52a>trans-tango pattern in (C) with strong labeling in the wing neuromere (white arrow). (F) In the brain, 31C06-GAL4/+>UAS-GFP/+ labels neurons that project in patterns similar to the Ir52a>trans tango–labeled axons (orange and yellow arrows, “1” and “2”). (G and H) 31C06-GAL4/+>UAS-Denmark,UAS syt_EGFP/+ and Ir52a-GAL4/+>UAS-Denmark,UAS syt:EGFP/+ staining patterns. UAS-Denmark (magenta) labels dendrites; syt:EGFP (green) labels presynaptic sites. (I) 31C06LexA/+>LexAop CD8:GFP/+ and Ir52a-GAL4/+>UAS CD8:RFP/+ expression patterns reveal that 31C06 dendrites (GFP, green) are in close proximity to Ir52a-GAL4 axons (RFP, red), particularly in the wing neuromere (right). (J) Strong GRASP signal was detected between 31C06LexA dendrites and Ir52a-GAL4 axons in the VNC (left). GRASP signal was in close proximity to Ir52a-GAL4 presynaptic sites (right, brp:mcherry in magenta). (K) 31C06-GAL4/+>UAS-Kir2.1 and 17F09GAL4/+>UAS-Kir2.1 blocked the increase in sleep following wing cut compared to parental controls (n = 27 to 46 flies per condition; *P < 0.01, Tukey correction). (B to J) Maximum intensity confocal projections. Wing neuromere image to the right in (J) is a single confocal slice. Scale bar, 20 μm.
What circuits are downstream of Ir52a+ neurons? Second-order neurons to wing sensory neurons are hitherto unknown. To identify postsynaptic partners of Ir52a-GAL4 neurons, we used the trans-tango system, which labels neurons one synapse from a given presynaptic neuron. Ir52a-GAL4>trans-tango labeled two classes of projection neurons with neurites in close proximity to Ir52a sensory axons in the VNC, particularly in the wing neuromere (Fig. 5C). These axon tracts exit the VNC, arborize in the SEG, and terminate in the ventrolateral protocerebrum (VLP) in the brain (Fig. 5D). In analogy to olfactory projection neurons, we call these tracts the medial and lateral VNC-VLP tract. Ir76b-GAL4>trans-tango labeled a broader neural population including a tract that resembled the medial VNC-VLP tract above (fig. S9, K to M). From a visual screen of images of GAL4 lines, we identified one line, 31C06 GAL4, whose expression pattern resembles the Ir52a>trans-tango pattern (Fig. 5, E and F, and fig. S9N). 31C06-GAL4 projection neurons are likely postsynaptic to Ir52a-GAL4 sensory neurons. 31C06-GAL4 neurites are largely dendritic, and Ir52a-GAL4 neurons are largely axonal in the VNC (Fig. 5, G and H). The processes of 31C06LexA projection neurons and Ir52a-GAL4 sensory neurons are in close proximity (Fig. 5I) and make physical contacts that appear to be synaptic [as evidenced by GFP Reconstitution Across Synaptic Partners (GRASP) signal; Fig. 5J]. Last, a recently developed tool to identify enhancers that overlap with a given line (42) identified a second GAL4 line, 17F09 GAL4, that overlaps with 31C06 GAL4 in projection neurons of the medial VNC-VLP tract. Inactivation of VNC-VLP projection neurons with 31C06 GAL4 or 17F09 GAL4 abrogated the wing-cut response (Fig. 5K).
Flight impairments activate and induce plasticity in projection neurons
These results describe a neural pathway that connects the wings to higher brain centers and is required for wing cut–induced sleep. Further, they suggest the possibility that wing cut or flight impairment more generally directly modulates these projection neurons to increase sleep. We found that activation of 31C06-GAL4 neurons with dTRPA1 increased sleep (fig. S10, A to C) and that wing cut elevated CaLexA signal in 31C06-GAL4 VNC projection neurons, indicating that wing cut activates this pathway (Fig. 6, A to C). What are the mechanisms that support the changes in activity? We hypothesized that flight impairments would induce plastic changes in the number of synapses in 31C06-GAL4 projection neurons to stably modulate sleep. To test this possibility, we used the Synaptic Tagging with Recombination (STaR) system that labels active zones in neurons of interest via recombinase-based tagging of the active zone protein Bruchpilot (BRP; Fig. 6, D to G). We observed more BRP puncta and increased BRP intensity per punctum, along the medial and lateral VNC-VLP tracts referenced above, of 31C06-GAL4>STaR flies with curly or cut wings relative to controls with normal wings (Fig. 6, H to K). In addition, we also observed increased BRP intensity per punctum in the terminal arborizations of the 31C06-GAL4 projection neurons (fig. S10, D and E). Wing cut–induced sleep increases are apparent in the first day following wing surgery (fig. S7L). Consistent with these observations, we found that wing cut–induced structural changes (i.e., more BRP puncta and increased BRP intensity per punctum) were also observed 24 hours following wing cut (fig. S10, F and G). Last, 31C06LexA projection neurons are presynaptic to the 64F01-GAL4 wing-expansion/wing-cut neurons—their processes are in close proximity (Fig. 6L), exhibit complimentary axonal and dendritic profiles (fig. S10, F and G), and make physical contacts that appear to be synaptic (Fig. 6, M and N). 31C06LexA projection neurons are functionally coupled to 64F01-GAL4 neurons, as adenosine triphosphate (ATP)–mediated stimulation of the P2X2 cation channel in 31C06LexA projection neurons induced changes in calcium levels in 64F01-GAL4 neurons (fig. S10, J to L). Thus, while 31C06 neurons project to several brain areas that may influence sleep, 64F01-GAL4 neurons likely represent one sleep-promoting output of 31C06 neurons.
Fig. 6. Wing cut induces structural plasticity in 31C06-GAL4 projection neurons.
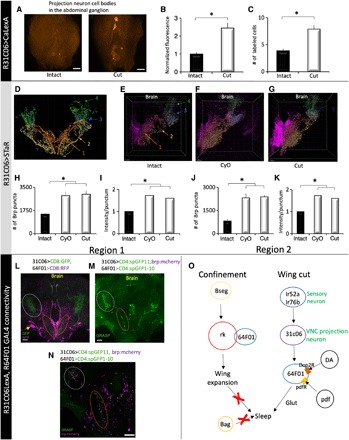
(A) Pseudo-colored representative images of 31C06-GAL4/+>CalexA expression in the abdominal ganglion. Wing cut increases the number (B) and intensity (C) of labeled cells compared to intact siblings (n = 8 to 10 flies per condition; *P < 0.01 and *P < 0.01, t test, respectively). (D) Schematic of projections from R31C06-GAL4. Region 1 (orange-red) captures the medial projection, region 2 (gold) reflects the lateral projection, and regions 3 and 4 reflect the lateral and dorsal VLP arborizations of the two projections (see Materials and Methods for details). (E to G) Representative images of BRP puncta in intact (E) and cut (G) R31C06-GAL4/+>STaR flies as well as in R31C06-GAL4/+>CyO/+;STaR flies (F). (H and I) In region 1, the number and intensity of BRP puncta were increased in cut R31C06-GAL4/+>STaR and R31C06-GAL4/+>CyO/+;STaR flies compared to intact controls. (J and K) The number and intensity of BRP puncta were also increased in region 2 in cut R31C06-GAL4/+>STaR and R31C06-GAL4/+>CyO/+;STaR flies compared to intact controls (n = 7 per group; *P < 0.002, Tukey correction). (L) R64F01-GAL4/+>UAS-RFP/+ (magenta) and 31C06LexA+>LexAop-GFP/+ (green) neurites are in close proximity in the SEG (orange ellipse) and the VLP (gray circle). (M) GRASP signal (green) was detected in these regions (orange and gray circles). (N) GRASP signal was adjacent to 31C06LexA presynaptic sites (brp:mcherry, magenta) in SEG and the VLP. (O) Schematic of identified flight and sleep-regulating circuitry. DA, dopamine; pdf, pigment-dispersing factor; Glut, glutamate. (A, L, and M) Maximal intensity confocal projections. (N) Single confocal slice of brain in (M). (E to G) Snapshots from Imaris software. Scale bars, 20 μm.
DISCUSSION
Sleep and waking are known to be responsive to internal and external cues (e.g., stress, starvation, light, temperature, and immune system) (29, 43–46). Research in this area has primarily focused on how various cues affect the central brain circuits that directly regulate sleep and waking; the peripheral inputs to these central circuits have received much less attention (47). With this in mind, we hypothesized that novel sleep regulatory centers might be identified by determining how peripheral signals modulate sleep. To test this hypothesis, we disrupted wing functioning using independent strategies: confinement, wing cut, and wing glue (32, 33). Using this approach, we identified specific sensory neurons, their target projection neurons in the VNC, and a previously unknown set of neurons in the central brain that regulate sleep. In addition, we have identified a critical neuropeptide (burs) and its receptor (rickets) that link wing expansion and sleep. Thus, these data trace a novel and unexpected sleep-promoting pathway from peripheral inputs to central brain neurons and describe an environmental manipulation that activates this circuit (Fig. 6O). Our data emphasize that sensory processing does much more than simply allow an individual to recognize a chemical for the sole purpose of avoiding it or being attracted to it. That is, sensory cues seem to provide critical information to circuits regulating cognitive behaviors.
Blocking wing expansion increases sleep
The molecular mechanisms, neuronal circuitry, and temporal dynamics controlling wing expansion are extremely well characterized (33). Thus, we asked whether blocking wing expansion using established genetic tools would alter sleep. To our surprise, we found that every environmental or genetic manipulation that disrupted wing expansion led to an increase in sleep. Initially, we used an environmental perturbation, confinement, to block wing expansion and thereby increase sleep (35). The episodes of sleep met the historical criteria for sleep, including increased arousal thresholds and rapid reversibility; locomotion was not impaired by any manipulation investigated. The increase in sleep was unlikely due to the stress of confinement because siblings that had been confined for a similar amount of time after wing expansion did not increase sleep. Moreover, several independent genetic manipulations that block wing expansion, without inducing stress, including knocking down bursicon or pburs, burs mutants, and altering the excitability of bursicon neurons all increased sleep. Last, we found that the increase in sleep was primarily mediated by a pair of bursicon-expressing neurons in the SEG.
The receptor for bursicon, rickets, is known to play a prominent role in wing expansion (33). Knocking down rickets using RNAi or altering the activity of rk-GAL4 neurons increased sleep, consistent with the observations reported for bursicon. Unfortunately, the precise sites of requirement of rickets within the central brain are not well understood. To address this problem, we mapped the neurons responsible for wing expansion and increased sleep by knocking down rickets or altering signaling pathways used by rickets (e.g., PKA-DN). Given the role of bursicon in the SEG, we focused our attention on subsets of neurons that primarily express in the SEG. These data revealed candidate GAL4 lines that express in nonoverlapping subsets of cells in the SEG that are capable of modulating both phenotypes. R64F01-GAL4–expressing neurons are of particular interest. That is, bursicon is coexpressed with CCAP, and the enhancer elements within R64F01-GAL4 are derived from the CCAPR promoter. Thus, while we primarily focused on R64F01-GAL4, rk appears to function in multiple distributed sites. How these other neuronal groups modulate sleep is a topic of active investigation.
Although our data have identified a novel sleep regulatory set of neurons in Drosophila, the extent to which the role of burs/rk signaling will apply to the regulation of mammalian sleep is unknown. There is no known homolog of bursicon in humans. rickets, on the other hand, is part of a larger family of leucine-rich repeat glucohormone receptors. The closest human homologs of rickets are LGR4-6 (48). The LGR4-6s are orphan receptors, which might suggest that the human genome encodes an as yet unidentified human analog of burs. The expression of LGR6 was found to be modulated by conditions of partial sleep restriction in humans (49). Thus, burs/rk signaling may have conserved roles in sleep regulation.
Cutting wings increases sleep
rk-GAL4–expressing neurons are able to regulate sleep in adults (fig. S5, E to G). This result suggests that these neurons maintain the capacity to modulate sleep, although bursicon neurons die by apoptosis in the first 48 hours of adult life (38). Thus, we hypothesize that subsets of rk-GAL4 neurons might be reactivated in adult flies to modulate sleep under some circumstances. One way rk-GAL4 neurons could be reactivated may be through wing damage. Wing damage and flight impairments might be fairly common. A recent study found that male flies frequently inflicted sufficient wing damage during aggressive bouts to disrupt flight (39). To test this hypothesis, we disrupted wings by cutting or gluing them. These complimentary studies both increased sleep. The wings remained intact and undamaged following treatment with glue indicating that the increase in sleep was not due to neurodegeneration or activation of the immune system. Additional experiments testing the involvement of the immune system revealed that flies mutant for immune response genes still increased sleep following wing cut (fig. S7P). It is important to note that wing cut did not disrupt locomotor activity and that the subsequent increase in sleep was characterized by elevated arousal thresholds and was rapidly reversible. Furthermore, wing cut increased sleep drive as evidenced by increased CaLexA signal in the R2 ellipsoid body ring neurons, a well-characterized marker of sleep drive (13). Last, wing cut–induced sleep did not require a functional circadian system (fig. S7K). Thus, mechanically disrupting wings in adults phenocopies wing-expansion deficits. We find that, like its role in wing expansion, R64F01-GAL4–expressing neurons also play an important role in regulating sleep following wing cut.
Specific wing chemosensory neurons mediate wing cut–induced sleep
There are ~400 neurons distributed along the anterior margin of the Drosophila wing that are organized into mechanosensory and chemosensory sensilla. To determine which neurons are capable of modulating sleep, we expressed UAS-Kir2.1 in subsets of these neurons throughout development. We hypothesized that constitutively silenced neurons would not change their output upon wing cut and thus would not increase sleep. We find that two GAL4 lines that express in sensory neurons, Ir52a GAL4 and Ir76b GAL4, mitigated the increase in sleep following wing cut (Fig. 5A). Unexpectedly, silencing neither Ir52a-GAL4 nor Ir76b-GAL4–expressing neurons alone completely abrogated the increase in sleep following wing damage. We interpret this result as indicating that Ir52a-GAL4 and Ir76b-GAL4 are expressed in partially nonoverlapping cells, and their effect on sleep is partially redundant. As a consequence, the nonsilenced neurons would be able to signal a change in wing integrity when the other neurons are silenced. This interpretation is supported by the observation that Ir52a and Ir76b mediate functionally distinct gustatory responses (34, 41). Inhibiting both Ir52a-GAL4 and Ir76b GAL4 neurons together abrogated the increase in sleep following wing cut. Baseline sleep was also increased when Ir52a-GAL4 and Ir76b-GAL4 neurons were simultaneously silenced in the absence of wing cut. A recent study has shown that hydrophobic molecules can be transferred from one fly to the wing margin of another to influence male and female sexual behavior via Ir52a (34). These data support a role for Ir52a in sleep regulation given the interaction between sleep and sexual behavior (21). Nonetheless, our data suggest that independent sets of wing sensory neurons convey information on wing integrity and modulate sleep.
Specific projection neurons mediate wing cut–induced sleep
Because the downstream neurons of wing sensory neurons have not been well characterized, we used the circuit-tracing techniques, trans-tango and GRASP, to identify candidate circuits downstream of Ir52a neurons. We identified two specific classes of projection neurons that connect the wing neuromere in the VNC to the SEG and VLP along a medial and lateral tract. The path of the medial wing projection neuron tract appears to follow a similar path to other taste projection neurons (50). Moreover, our expression results are also consistent with a recent report that used functional circuit mapping to describe neurons downstream from Ir52a neurons (34). However, in contrast to He et al., we also identified GAL4 lines that display similar expression patterns and allow us genetic access to these critical neurons. Silencing R31C06-GAL4 neurons with UAS-Kir2.1 blocked the increase in sleep following wing cut. Furthermore, we demonstrate that wing cut activates 31C06-GAL4 neurons, as evidenced by increased CaLexA signal (Fig. 6, A to C). The increased activity is associated with an increase in BRP puncta (Fig. 6, D to K). The number and distribution of BRP levels have been associated with basal activity as well as activity-dependent plastic changes. Other manipulations that increase sleep drive, such as sleep deprivation and social enrichment, also lead to increases in synapses (13, 18). It has been suggested that structural changes such as these provide a mechanism to ensure persistence of sleep drive (13).
Wing projection neurons connect to wing-expansion neurons
Our data identify nonoverlapping subsets of cells in the SEG, including R6401GAL4-expressing neurons, that are capable of modulating both sleep and wing-expansion phenotypes. We also identify projection neurons, e.g., R31C06-GAL4 neurons, that project to the SEG that mediate the effects of wing cut on sleep. We show, using GRASP and functional calcium imaging, that R31C06-GAL4 projection neurons connect to glutamatergic R64F01-GAL4 neurons (Fig. 6, L to N, and fig. S10, H to L). This interconnected circuitry between wing expansion and wing cut may help explain why flies with unexpanded wings continue to sleep more, even after bursicon neurons die on day 2 of adult life. We speculate that the change in sleep in flies with unexpanded wings might use the wing-cut pathway after bursicon neurons have been eliminated. This possibility will be explored in future studies.
It is important to note that sleep and wake regulatory neurons that project into the SEG have been previously described (12). However, their targets in the SEG have not yet been isolated. Further, glutamatergic wake and sleep-promoting neurons have been suggested in flies and mammals, although, in many cases, their precise identity is not known (51). Glutamatergic neurons have been proposed as one of the targets of vestibular sleep-promoting neurons (52), suggesting that peripheral inputs might converge onto glutamatergic central brain sleep-promoting neurons. We find that dopaminergic modulation of R64F01 GAL4 neurons mediated by the D2R dopamine receptor is critical for wing cut–induced sleep. A role for the D2R receptor in regulating sleep and wake was recently reported; our data localize the effects of D2R to a novel set of neurons (53). Collectively, our characterization of the neurobiological mechanisms in R64F01 GAL4 neurons thus extends our understanding of sleep regulatory mechanisms in important ways.
Impairing flight alters behavior
Classic studies from McEwen (54) and Benzer (55) examined the effects of disrupting wings on flies’ phototactic behavior. These pioneering investigations disrupted flight by clipping wings or through mutations that disrupted wing development (e.g., vestigial). Both classes of flight-disrupting manipulations greatly impaired phototaxis. These observations were confirmed and extended in a recent elegant study (32) that examined a number of flight-disrupting manipulations and found that they impaired normal phototactic behavior. Thus, previous manuscripts indicate that disrupting the wing can have substantial impact on motivated behavior. Our data support their hypotheses and extend the results to an additional behavior, sleep.
Sleep as a form of adaptive inactivity
The increases in sleep we observe when flight is impaired could be viewed as an adaptive response, enabling flies to modify their behavioral repertoire to meet new challenges (32). An influential theory of sleep function posits that ecological factors that place animals in harm’s way increase sleep as a state of adaptive inactivity (56). An unstated assumption of the adaptive inactivity hypothesis is that inactivity must be regulated and must be under the influence of natural selection. Unexpectedly, neither the underlying circuitry nor the molecular mechanisms regulating sleep during dangerous or life-threatening conditions are known (57). We propose that the increase in sleep that occurs when flight is disrupted may be an example of adaptive inactivity. By identifying the underlying circuitry and neuromodulators, our data provide support for a key prediction of this idea and extend its applicability. We report that more than eight manipulations that impair flight also increase sleep. These manipulations include (i) disruptions in burs/rk signaling, (ii) expressing rpr in the wing disc, (iii) flies with structural wing abnormalities (CyO flies), and (iv) flies lacking one wing (wg1 mutants). In addition, we show that flies with intact wings that cannot fly show increases in sleep including (v) R20C05>UAS- PKA-DN (Fig. 3H) and (vi) mutants for pkcΔ. Last, sleep is increased following mechanical wing disruption (vii) by wing cut or (viii) with wings glued. Together, these data suggest that an alternative strategy for identifying novel sleep regulatory pathways is to examine animals during a variety of species-specific challenges rather than focusing only on sleep during optimal laboratory conditions. Identifying sleep regulatory circuits that are predominantly engaged during environmental perturbations may provide new opportunities for developing therapeutics for treating a variety of sleep disorders.
MATERIALS AND METHODS
Flies
Flies were cultured at 25°C with ~50% relative humidity and kept on a standard yeast, corn syrup, and agar diet while being maintained on a 12-hour light:12-hour dark cycle. Female flies were used as subjects in all experiments. Crosses with UAS rk RNAi were set up at 29°C as per established protocols.
Fly strains
burs GAL4, rk GAL4 (rk-pan GAL4), Bseg GAL4 (ETVP16AD-99 ∩ burs Gal4DBDU6A1), Bag GAL4 (ETVP16AD-N9A88A ∩ burs Gal4DBDU6A1), and UAS dnc were gifts of B. White (National Institute of Mental Health). bursZ1091, bursZ5569, and bw; st flies were gifts of C. Zuker (Columbia). UAS PKA-DN (homozygous viable second chromosome insert), a constitutively active regulatory PKA subunit (UAS R*), was a gift of D. Kalderon (Columbia). Sifa GAL4, kurs58 GAL4, and DH44VT GAL4 were gifts of A. Sehgal (University of Pennsylvania). 9-30 GAL4 and 12-230 GAL4 were gifts of F. Wolf (UC Merced). UAS dTRPA1 (homozygous viable second chromosome insert) was a gift of P. Garrity (Brandeis). UAS Kir2.1EGFP (homozygous viable third chromosome insert) was a gift of R. Baines (Manchester). UAS rk RNAi (8930-R1) was obtained from NIG-FLY (National Institute of Genetics, Mishima, Japan). W4 (CCAP-GAL4, UAS-GluedDN, tubP>stop>GAL80, and UAS mCD8GFP) flies and the enhancer-trap flipase (Et-flp) lines used in fig. S3 were gifts of B. Zhang (University of Missouri). UAS Shits (pJFRC100-20XUAS-TTS-Shibire-ts1-p10 in attp2) was a gift of G. Rubin (Janelia Farms Research Campus). GRASP reagents—UAS CD4:SpGFP1–10, LexAop CD4:SpGFP11, UAS brp:mcherry (third chromosome insert), and lexaop brp:mcherry (third chromosome insert)—were gifts of C.-H. Lee (Academia Sinica, Taiwan). LexAop P2X2 was a gift of O. Shafer (U. Michigan).
All other GAL4 and LexA lines were obtained from the Bloomington Drosophila Stock Center. The following lines were also obtained from the Bloomington Drosophila Stock Center:
rk1, rk4, wg1, pkcΔe04408, relE20, imd1, UAS NaChBac (UAS NaChBacEGFP4), 20XUAS-IVS-mCD8GFP (in attP2), 10XUAS-IVS-mCD8::RFP (attP18), 13XLexAop2-mCD8::GFP (attP8), UAS rpr (UAS-rpr.C14 on second chromosome), dvGlut GAL80 (VGlutMI04979-T3XG80.2), trans-tango (UAS myrGFP.QUAS mtdTomatoHA; trans tango), UAS Denmark, UAS syt.EGFP (on second chromosome), CaLexA (LexAop-CD8::GFP-2A-CD8::GFP; LexAop-CD2::GFP; UASmLexA-VP16-NFAT/TM6B), STaR [UAS FLP, brp(FRT.stop)V5-2A-LexA-VP16 in VK00033], UAS burs RNAiJ02260, UAS pburs RNAiHMC04211, UAS CCAP RNAiHMJ23953, UAS CCAPR RNAiJF01338, UAS ash1 RNAiHMS00582, UAS lark RNAiJF02783, UAS Mip RNAiHMS02244, UAS Gβ13F RNAiHMS01455, UAS Gβ76c RNAiJF03127, UAS Gγ30a RNAiHMS01455, UAS Gαi RNAiHMS01273, UAS Gαo RNAiHMS01129, UAS Gαq RNAiHMJ30300, UAS plc21c RNAiHMS00600, UAS Itpr RNAiHMC03351, UAS Stim RNAi HMC03651, UAS Irk1 RNAi HMS02480, UAS Irk2 RNAiHMS02379, UAS Irk3 RNAiJF02262, UAS Dop1R1 RNAi HM04077, UAS Dop1R2 RNAi HMC06293, UAS Dop2R RNAiHMC02988, UAS DopEcR RNAiJF03415, and UAS pdfR RNAiHMS01815.
Genetics
burs GAL4, rk GAL4, rk1, rk4, UAS burs RNAiJ02260, UAS pburs RNAiHMC04211, UAS NaChBac, UAS Kir2.1, ETVP16AD-99, burs Gal4DBDU6A1, and ETVP16AD-N9A88A were all outcrossed to a reference yw line for five generations. Using balancer chromosomes, a yw; CyO/Sco line was generated where the first, third, and Y chromosomes were identical to a reference yw strain.
Behavioral analysis
Sleep
Sleep was assessed as previously described (58). Briefly, individual virgin female flies were placed into 65-mm tubes, and their locomotor activity was continuously measured using the Drosophila Activity Monitoring (DAM) system (TriKinetics, Waltham, MA). Locomotor activity was binned in 1-min intervals; sleep defined as periods of inactivity of 5 min or more was computed using custom Excel scripts. In sleep plots, sleep in minutes per hour is displayed as a function of zeitgeber time (ZT). ZT0 represents the beginning of the fly’s subjective day (lights on), and ZT12 represents the transition from lights on to lights off.
Sleep homeostasis
Four- to 7-day-old female flies were placed into 65-mm tubes in DAM monitors, and sleep was recorded for 2 days to establish a baseline. Flies were then sleep-deprived for 12 hours during the dark phase (ZT12 to ZT0) using the sleep nullifying apparatus (SNAP) with procedures previously described (59). For each individual fly, the difference between the sleep between the sleep time on the recovery day and baseline was calculated as the sleep gained/lost. Sleep rebound was calculated as the ratio of this sleep gained/lost to the sleep lost during baseline, expressed as a percentage.
Reversibility
Female flies were placed into 65-mm tubes in DAM monitors. A mechanical stimulus was delivered for 10 min at ZT15. Only flies that had been inactive for at least 5 min preceding the stimulus were considered for analysis. The fraction of flies aroused by this stimulus was computed for flies subjected to this stimulus and undisturbed controls.
Arousal thresholds
Arousal thresholds were calculated using the Drosophila Arousal Tracking (DART) system as previously described (60). Female flies were housed individually in 80-mm-long glass tubes, and their activity was monitored using video tracking. Fourteen flies were used per genotype. Flies so housed were probed hourly for 24 hours, with a train of vibrational stimuli of increasing strength from 0 to 1.2 g. Each stimulus consisted of five pulses of 200 ms and was delivered in 0.24-g increments 15 s apart. The arousal threshold for each fly was calculated as the weakest vibration intensity (g) required to elicit a response (walking at least half the length of the glass tube) in quiescent flies that had been inactive for least the preceding minute. The average arousal threshold across the day was then calculated for each strain.
Confinement
Individual female flies were collected before expansion and confined overnight in a 4.9 × 10−2 cm3 space in the 65-mm tubes used in standard DAM system sleep recording as previously described (35). Following confinement overnight, flies were placed in 65-mm tubes for sleep recording.
Wing cut
Female flies were collected on the day they eclosed, and both wings were cut at their base to remove the wings in their entirety; wings were cut under CO2 anesthesia after wings had expanded. Flies with cut wings, and their siblings with intact wings that had been subject to the same anesthesia protocol, were then placed in 65-mm glass tubes in DAM monitors. Sleep data are reported for second day after wing cut, i.e., for 2-day-old flies.
Wing glue
Wings of 3-day-old female flies were glued with a small amount of ultraviolet (UV)–activated glue (Bondic, ON) under CO2 anesthesia and short (~5 s) UV light exposure. As a control, a small amount of glue was applied to the abdomen of siblings. Unglued flies were also subject to the same anesthesia and UV curing light exposure. All flies were then housed in 65-mm glass tubes and placed in DAM monitors.
Immunohistochemistry
Whole flies were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) in phosphate-buffered saline (PBS; Sigma-Aldrich) + 0.3% Triton X-100 (PBST). Following fixation, the CNS was dissected in PBS, washed in PBST, and incubated in blocking solution (PBST + 5% normal goat serum) at 4°C overnight. The following day, brains and VNCs were incubated in primary antibodies (diluted in blocking solution) for 2 days at 4°C. Primary antibodies (and dilutions used) were as follows: chicken anti-GFP (green fluorescent protein) (1:1000; Abcam), mouse monoclonal anti-GFP (1:100; Sigma-Aldrich), rabbit anti-Dsred (1:250; Takara Bio), rat anti-HA (hemagglutinin) (1:500; Sigma-Aldrich), mouse anti-TH (tyrosine hydroxylase) (1:500; Immunostar), monoclonal antibody nc82 (1:400; Developmental Studies Hybridoma Bank), and mouse anti-V5 (1:400; Invitrogen). Following incubation in primary antibodies, brains were washed and incubated overnight in secondary antibody solution. Secondary antibodies used included goat anti-chicken Alexa Fluor 488 (1:400; Invitrogen), goat anti-mouse Alexa Fluor 488 (1:400; Invitrogen), goat anti-rabbit Alexa Fluor 568 (1:300), goat anti-rabbit Alexa Fluor 633 (1:300; Invitrogen), goat anti-rat Alexa Fluor 568 (1:200; Invitrogen), goat anti-mouse Alexa Fluor 568 (1:200; Invitrogen), and goat anti-mouse Alexa Fluor 633 (1:200; Invitrogen). Following incubation in secondary antibodies, brains were washed in PBST and mounted in Vectashield (Vector Laboratories). Images were obtained using an Olympus FV1200 laser scanning confocal microscope, using one of a UAPO 20× air, 40× water immersion, 63× water immersion, or with a Zeiss LSM 880 confocal microscope with a 40× oil objective. Confocal z stacks were acquired with sequential scanning frame by frame, to prevent bleedthrough across channels, and at a depth of 1.0 or 0.5 μm. Images were processed using Fiji or Imaris (Bitplane) software. Unless otherwise specified, 5- to 7-day-old female flies were used for immunostaining experiments.
CaLexA measurements
CaLexA was expressed in R2 ellipsoid body neurons with R30G03 GAL4 or in VNC projection neurons with 31C06 GAL4. Three-day-old CaLexA-expressing female flies with cut wings and their siblings with intact wings were fixed at ZT0 to ZT1. Two-day-old CaLexA-expressing confined female flies and their siblings with intact wings were fixed at ZT0 to ZT1. CaLexA-driven GFP signal in brains and VNCs was enhanced by staining with chicken anti-GFP. Images were acquired with a 40× water immersion objective at 1024 × 1024 pixels on an Olympus FV1200 confocal microscope. Cut and intact groups were imaged using the same settings. Images were analyzed using Fiji/ImageJ. To measure fluorescent intensities, the sum of all pixels of a stack in a region of interest (ROI) was calculated. ROI intensities were corrected for background by measuring and subtracting background fluorescent intensity from a region adjacent to the ROI.
BRP measurements
STaR was expressed with 31C06 Gal4 to label BRP puncta in VNC projection neurons. Three-day-old 31c06>STaR female flies with cut, curly, and intact wings were fixed at ZT0 to ZT1. BRP puncta were visualized by immunostaining with an anti-V5 antibody. For the experiments in fig. S10 (F and G), 1-day-old 31c06>STaR female flies were fixed at ZT0 to ZT1. Images were acquired with a 60× water immersion objective at 1024 × 1024 pixels on an Olympus FV1200 confocal microscope. All groups were imaged with the same settings. 31C06 GAL4 labeled neurons with dendritic arborizations in the VNC, including in the wing neuromere, and axonal projections that exited the VNC forming two tracts along the SEG: a medial tract and a lateral tract. This nomenclature also reflected the position of these tracts in the VNC brain connective. Both tracts were terminated in the VLP where they made extensive arborizations. Although the entire CNS was dissected and immunostained, we focused on the region of the brain containing the projections of the 31C06 wing VLP projection neurons from the ventral SEG to the VLP. Anatomical experiments suggested that the 31C06 projection neurons are largely axonal in this region. Images were processed with Imaris software (Bitplane), which allows visualization and quantification of data in three dimensions. Images were analyzed and quantified while being blinded to condition. Confocal z stacks were carefully segmented by manual annotation of a ~150-μm z stack into four regions: region1 corresponding to the medial tract from the VNC to the VLP, region 2 to the lateral tract, region 3 to the lateral (and anterior) VLP arborizations, and region 4 to the dorsal (and posterior) arborizations of the two tracts in the VLP. Background intensity was automatically calculated for each region. Thresholding criteria for identifying BRP puncta were automatically generated and occasionally manually adjusted. The number of BRP puncta and average staining intensity for each punctum was automatically generated.
Live brain imaging
Flies were chilled for approximately 5 min before pinning them onto a sylgaard dissection dish. Brains were dissected in calcium-free HL3 and then transferred onto a polylysine-treated dish (35 × 10 mm Falcon polystyrene) containing 3.6 ml of 1.5 mM calcium HL3. Images were captured using an Olympus BX61 microscope, and x-, y-, and z-stage movements were set via SlideBook 5.0 (Intelligent Imaging Innovations), which controlled Prior H105Plan Power Stage through Prior ProScan II. GCaMP fluorescence images were acquired at 1 Hz. Following 1 min of baseline measurements, 0.4 ml of 50 mM ATP was perfused onto the dish to activate the P2X2 receptor, yielding an effective concentration of 5 mM ATP.
Statistical analysis
Data are presented as mean values accompanied by the SEM. Statistical analyses were carried out in Systat software. Statistical comparisons were done with a Student’s t test for comparisons between two groups, or analysis of variance (ANOVA) followed by Tukey’s post hoc comparisons for tests involving multiple comparisons. Unless otherwise specified, the most commonly used statistical analysis was a one-way ANOVA for genotype/condition.
Supplementary Material
Acknowledgments
We thank L. Salkoff for crucial input; T. Y. Lin and C. H. Lee for sharing reagents; H. Dierick, S. Dissel, M. Thimgan, and B. White for helpful discussions and comments on the manuscript; D. Oakley and M. Shih for technical input on image acquisition and analysis; and L. Cao, Z. Koch, and D. Chan for technical assistance. Funding: This work was supported by NIH grants 5R01NS051305-14 and 5R01NS076980-08 to P.J.S. The confocal facility is supported by NIH shared instrument grant S1OD21629-01A1. Author contributions: K.M., B.Z., and P.J.S. conceived the project, designed and performed experiments, interpreted the data, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/19/eaaz2166/DC1
REFERENCES AND NOTES
- 1.Moruzzi G., Magoun H. W., Brain stem reticular formation and activation of the EEG. Electroencephalogr. Clin. Neurophysiol. 1, 455–473 (1949). [PubMed] [Google Scholar]
- 2.Scammell T. E., Arrigoni E., Lipton J. O., Neural circuitry of wakefulness and sleep. Neuron 93, 747–765 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sherin J. E., Shiromani P. J., McCarley R. W., Saper C. B., Activation of ventrolateral preoptic neurons during sleep. Science 271, 216–219 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Xu M., Chung S., Zhang S., Zhong P., Ma C., Chang W.-C., Weissbourd B., Sakai N., Luo L., Nishino S., Dan Y., Basal forebrain circuit for sleep-wake control. Nat. Neurosci. 18, 1641–1647 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Konadhode R. R., Pelluru D., Blanco-Centurion C., Zayachkivsky A., Liu M., Uhde T., Glen Jr W. B., van den Pol A. N., Mulholland P. J., Shiromani P. J., Optogenetic stimulation of MCH neurons increases sleep. J. Neurosci. 33, 10257–10263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morairty S. R., Dittrich L., Pasumarthi R. K., Valladao D., Heiss J. E., Gerashchenko D., Kilduff T. S., A role for cortical nNOS/NK1 neurons in coupling homeostatic sleep drive to EEG slow wave activity. Proc. Natl. Acad. Sci. U.S.A. 110, 20272–20277 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anaclet C., Ferrari L., Arrigoni E., Bass C. E., Saper C. B., Lu J., Fuller P. M., The GABAergic parafacial zone is a medullary slow wave sleep-promoting center. Nat. Neurosci. 17, 1217–1224 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halassa M. M., Florian C., Fellin T., Munoz J. R., Lee S.-Y., Abel T., Haydon P. G., Frank M. G., Astrocytic modulation of sleep homeostasis and cognitive consequences of sleep loss. Neuron 61, 213–219 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shaw P. J., Cirelli C., Greenspan R. J., Tononi G., Correlates of sleep and waking in Drosophila melanogaster. Science 287, 1834–1837 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Joiner W. J., Crocker A., White B. H., Sehgal A., Sleep in Drosophila is regulated by adult mushroom bodies. Nature 441, 757–760 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Pitman J. L., McGill J. J., Keegan K. P., Allada R., A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature 441, 753–756 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Foltenyi K., Greenspan R. J., Newport J. W., Activation of EGFR and ERK by rhomboid signaling regulates the consolidation and maintenance of sleep in Drosophila. Nat. Neurosci. 10, 1160–1167 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Liu S., Liu Q., Tabuchi M., Wu M. N., Sleep drive is encoded by neural plastic changes in a dedicated circuit. Cell 165, 1347–1360 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Donlea J. M., Thimgan M. S., Suzuki Y., Gottschalk L., Shaw P. J., Inducing sleep by remote control facilitates memory consolidation in Drosophila. Science 332, 1571–1576 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheeba V., Fogle K. J., Kaneko M., Rashid S., Chou Y.-T., Sharma V. K., Holmes T. C., Large ventral lateral neurons modulate arousal and sleep in Drosophila. Curr. Biol. 18, 1537–1545 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerstner J. R., Vanderheyden W. M., Shaw P. J., Landry C. F., Yin J. C. P., Fatty-acid binding proteins modulate sleep and enhance long-term memory consolidation in Drosophila. PLOS ONE 6, e15890 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D., Dan Y., A motor theory of sleep-wake control: Arousal-action circuit. Annu. Rev. Neurosci. 42, 27–46 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Donlea J. M., Ramanan N., Shaw P. J., Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Joiner W. J., Unraveling the evolutionary determinants of sleep. Curr. Biol. 26, R1073–R1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keene A. C., Duboué E. R., McDonald D. M., Dus M., Suh G. S. B., Waddell S., Blau J., Clock and cycle limit starvation-induced sleep loss in Drosophila. Curr. Biol. 20, 1209–1215 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machado D. R., Afonso D. J., Kenny A. R., Öztürk-Çolak A., Moscato E. H., Mainwaring B., Kayser M., Koh K., Identification of octopaminergic neurons that modulate sleep suppression by male sex drive. eLife 6, e23130 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thimgan M. S., Suzuki Y., Seugnet L., Gottschalk L., Shaw P. J., The perilipin homologue, lipid storage droplet 2, regulates sleep homeostasis and prevents learning impairments following sleep loss. PLOS Biol. 8, e1000466 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesku J. A., Rattenborg N. C., Valcu M., Vyssotski A. L., Kuhn S., Kuemmeth F., Heidrich W., Kempenaers B., Adaptive sleep loss in polygynous pectoral sandpipers. Science 337, 1654–1658 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Rattenborg N. C., Mandt B. H., Obermeyer W. H., Winsauer P. J., Huber R., Wikelski M., Benca R. M., Migratory sleeplessness in the white-crowned sparrow (Zonotrichia leucophrys gambelii). PLOS Biol. 2, e212 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lyamin O., Pryaslova J., Lance V., Siegel J., Animal behaviour: Continuous activity in cetaceans after birth. Nature 435, 1177 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gravett N., Bhagwandin A., Sutcliffe R., Landen K., Chase M. J., Lyamin O. I., Siegel J. M., Manger P. R., Inactivity/sleep in two wild free-roaming African elephant matriarchs - Does large body size make elephants the shortest mammalian sleepers? PLOS ONE 12, e0171903 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanin S., Bhutani S., Montelli S., Menegazzi P., Green E. W., Pegoraro M., Sandrelli F., Costa R., Kyriacou C. P., Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature 484, 371–375 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Arrigoni E., Chee M. J. S., Fuller P. M., To eat or to sleep: That is a lateral hypothalamic question. Neuropharmacology 154, 34–49 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Harding E. C., Franks N. P., Wisden W., The temperature dependence of sleep. Front. Neurosci. 13, 336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szymusiak R., Chapter 20-Body temperature and sleep. Handb. Clin. Neurol. 156, 341–351 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Giraldo Y. M., Leitch K. J., Ros I. G., Warren T. L., Weir P. T., Dickinson M. H., Sun navigation requires compass neurons in Drosophila. Curr. Biol. 28, 2845–2852.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorostiza E. A., Colomb J., Brembs B., A decision underlies phototaxis in an insect. Open Biol. 6, 160229 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White B. H., Ewer J., Neural and hormonal control of postecdysial behaviors in insects. Annu. Rev. Entomol. 59, 363–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z., Luo Y., Shang X., Sun J. S., Carlson J. R., Chemosensory sensilla of the Drosophila wing express a candidate ionotropic pheromone receptor. PLOS Biol. 17, e2006619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peabody N. C., Pohl J. B., Diao F., Vreede A. P., Sandstrom D. J., Wang H., Zelensky P. K., White B. H., Characterization of the decision network for wing expansion in Drosophila using targeted expression of the TRPM8 channel. J. Neurosci. 29, 3343–3353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim Y.-J., Zitnan D., Cho K.-H., Schooley D. A., Mizoguchi A., Adams M. E., Central peptidergic ensembles associated with organization of an innate behavior. Proc. Natl. Acad. Sci. U.S.A. 103, 14211–14216 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNeil G. P., Zhang X., Genova G., Jackson F. R., A molecular rhythm mediating circadian clock output in Drosophila. Neuron 20, 297–303 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Gatto C. L., Broadie K., Fragile X mental retardation protein is required for programmed cell death and clearance of developmentally-transient peptidergic neurons. Dev. Biol. 356, 291–307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis S. M., Thomas A. L., Liu L., Campbell I. M., Dierick H. A., Isolation of aggressive behavior mutants in Drosophila using a screen for wing damage. Genetics 208, 273–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andretic R., van Swinderen B., Greenspan R. J., Dopaminergic modulation of arousal in Drosophila. Curr. Biol. 15, 1165–1175 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Koh T.-W., He Z., Gorur-Shandilya S., Menuz K., Larter N. K., Stewart S., Carlson J. R., The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83, 850–865 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.H. Otsuna, M. Ito, T. Kawase, Color depth MIP mask search: A new tool to expedite Split-GAL4 creation. bioRxiv 318006 [Preprint]. 9 May 2018. 10.1101/318006. [DOI]
- 43.Borbély A. A., Sleep in the rat during food deprivation and subsequent restitution of food. Brain Res. 124, 457–471 (1977). [DOI] [PubMed] [Google Scholar]
- 44.de la Iglesia H. O., Fernández-Duque E., Golombek D. A., Lanza N., Duffy J. F., Czeisler C. A., Valeggia C. R., Access to electric light is associated with shorter sleep duration in a traditionally hunter-gatherer community. J. Biol. Rhythms 30, 342–350 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Imeri L., Opp M. R., How (and why) the immune system makes us sleep. Nat. Rev. Neurosci. 10, 199–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kroeger D., Absi G., Gagliardi C., Bandaru S. S., Madara J. C., Ferrari L. L., Arrigoni E., Münzberg H., Scammell T. E., Saper C. B., Vetrivelan R., Galanin neurons in the ventrolateral preoptic area promote sleep and heat loss in mice. Nat. Commun. 9, 4129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harding E. C., Yu X., Miao A., Andrews N., Ma Y., Ye Z., Lignos L., Miracca G., Ba W., Yustos R., Vyssotski A. L., Wisden W., Franks N. P., A neuronal hub binding sleep initiation and body cooling in response to a warm external stimulus. Curr. Biol. 28, 2263–2273.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Loy T., Vandersmissen H. P., Van Hiel M. B., Poels J., Verlinden H., Badisco L., Vassart G., Broeck J. V., Comparative genomics of leucine-rich repeats containing G protein-coupled receptors and their ligands. Gen. Comp. Endocrinol. 155, 14–21 (2008). [DOI] [PubMed] [Google Scholar]
- 49.Aho V., Ollila H. M., Rantanen V., Kronholm E., Surakka I., van Leeuwen W. M. A., Lehto M., Matikainen S., Ripatti S., Härmä M., Sallinen M., Salomaa V., Jauhiainen M., Alenius H., Paunio T., Porkka-Heiskanen T., Partial sleep restriction activates immune response-related gene expression pathways: Experimental and epidemiological studies in humans. PLOS ONE 8, e77184 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim H., Kirkhart C., Scott K., Long-range projection neurons in the taste circuit of Drosophila. eLife 6, e23386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Allada R., Cirelli C., Sehgal A., Molecular Mechanisms of Sleep Homeostasis in Flies and Mammals. Cold Spring Harb. Perspect. Biol. 9, a027730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kompotis K., Hubbard J., Emmenegger Y., Perrault A., Mühlethaler M., Schwartz S., Bayer L., Franken P., Rocking promotes sleep in mice through rhythmic stimulation of the vestibular system. Curr. Biol. 29, 392–401.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Deng B., Li Q., Liu X., Cao Y., Li B., Qian Y., Xu R., Mao R., Zhou E., Zhang W., Huang J., Rao Y., Chemoconnectomics: Mapping chemical transmission in Drosophila. Neuron 101, 876–893.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 54.McEwen R. S., The reactions to light and to gravity in Drosophila and its mutants. J. Exp. Zool. 25, 49–106 (1918). [Google Scholar]
- 55.Benzer S., Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl. Acad. Sci. U.S.A. 58, 1112–1119 (1967). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siegel J. M., Sleep viewed as a state of adaptive inactivity. Nat. Rev. Neurosci. 10, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allada R., Siegel J. M., Unearthing the phylogenetic roots of sleep. Curr. Biol. 18, R670–R679 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shaw P. J., Tononi G., Greenspan R. J., Robinson D. F., Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature 417, 287–291 (2002). [DOI] [PubMed] [Google Scholar]
- 59.Seugnet L., Suzuki Y., Vine L., Gottschalk L., Shaw P. J., D1 receptor activation in the mushroom bodies rescues sleep-loss-induced learning impairments in Drosophila. Curr. Biol. 18, 1110–1117 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Alphen B., Yap M. H. W., Kirszenblat L., Kottler B., van Swinderen B., A dynamic deep sleep stage in Drosophila. J. Neurosci. 33, 6917–6927 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/19/eaaz2166/DC1


