Abstract
Anxiety disorders (ANX), namely generalized anxiety, panic disorder, and phobias, are common, etiologically complex syndromes that show increasing prevalence and comorbidity throughout adolescence and beyond. Few genome-wide association studies (GWAS) examining ANX risk have been published and almost exclusively in individuals of European ancestry. In this study, we phenotyped participants from the Army Study To Assess Risk and Resilience in Servicemembers (STARRS) to approximate DSM-based ANX diagnoses. We factor-analyzed those to create a single dimensional anxiety score for each subject. GWAS were conducted using that score within each of three ancestral groups (EUR, AFR, LAT) and then meta-analyzed across ancestries (NTotal=16,510). We sought to (i) replicate prior ANX GWAS findings in ANGST; (ii) determine whether results extended to other ancestry groups; and (iii) meta-analyze with ANGST for increased power to identify novel susceptibility loci. No reliable genome-wide significant SNP associations were detected in STARRS. However, SNPs within the CAMKMT gene located in region 2p21 associated with shared ANX risk in ANGST were replicated in EUR soldiers but not other ancestry groups. Combining EUR STARRS and ANGST (N=28,950) yielded a more robust 2p21 association signal (p=9.08×10−11). Gene-based analyses supported three genes within 2p21 and LBX1 on chromosome 10. More powerful ANX genetic studies will be required to identify further loci.
Keywords: anxiety, anxiety disorder, panic disorder, genetics, genome-wide association study
INTRODUCTION
Anxiety disorders (ANX), namely generalized anxiety disorder, panic disorder, social phobia, agoraphobia, and specific phobias, are common, etiologically complex syndromes. These disorders often begin in childhood and variably increase in prevalence throughout adolescence and early adulthood (Beesdo, Knappe, & Pine, 2009) with an overall lifetime ANX risk of 25%. Twin studies suggest that genetic variation accounts for 30–50% of the variance in ANX risk (Hettema, Neale, & Kendler, 2001). As with other psychiatric syndromes, linkage and candidate gene association studies have failed to identify robust ANX susceptibility loci (Meier & Deckert, 2019; Shimada-Sugimoto, Otowa, & Hettema, 2015). Few genome-wide association studies (GWAS) examining ANX risk have been published thus far. The first replicated finding emerging from ANX GWAS involved an association of two single nucleotide polymorphisms (SNPs) in the transmembrane protein 16B (TMEM16B) gene with panic disorder using a three-stage association design including 909 cases and 915 controls of European ancestry (EUR) (Erhardt et al., 2011); that finding was later supported in independent samples (Erhardt et al., 2012). More recently, a large discovery GWAS examining shared genetic risk across the five primary ANX was conducted in the Anxiety NeuroGenetics STudy (ANGST) Consortium (Otowa et al., 2016) described in detail below.
ANX exhibit high comorbidity with each other (Merikangas & Swanson, 2010) and substantially increase risk for subsequent mood disorders (Horn & Wuyek, 2010) making the study of their individual pathoetiologies particularly challenging. Twin studies indicate that this broad comorbidity is partially due to shared genetic and environmental risk factors (Hettema, Prescott, Myers, Neale, & Kendler, 2005) suggesting the importance of genetically analyzing ANX in a coordinated manner. We have previously applied multivariate phenotypic approaches to estimate the covariance structure of syndromal ANX to identify and extract a common factor score representing an overall liability dimension for use as the outcome phenotype in genetic association studies (Hettema et al., 2011). Identifying shared (‘transdiagnostic”) dimensional phenotypes is consistent with the efforts of the NIMH’s Research Domain Criteria (RDoC) initiative to optimally understand complex pathophysiological mechanisms (Insel et al., 2010). Most recently, we used this approach to conduct the ANGST GWAS meta-analysis in over 18,000 EUR subjects from seven international sites. Using a factor score phenotype that captured ANX covariance, we reported a genome-wide significant locus within a large linkage disequilibrium (LD) block on chromosome 2p21 encompassing three brain-expressed genes. The most significant p-value (p=2.86×10−9) came from SNP rs1067327 within the CAMKMT gene encoding the calmodulin-lysine N-methyltransferase. Independent replication of this finding has not yet been reported, as no other GWAS has attempted to create a similar, factor-derived dimensional score representing shared ANX risk. Furthermore, since ANGST was limited to participants of EUR ancestry, generalizability of that finding to other ancestry groups is unknown.
The current study attempts to further characterize and extend this finding in several ways using the Army Study To Assess Risk and Resilience in Servicemembers (STARRS) (Ursano et al., 2014). As described below, STARRS includes phenotypic and genotypic data from more than 17,000 US soldiers including those of EUR, African (AFR), and Latino (LAT) ancestry. Our first aim was to determine whether the ANGST 2p21 association signal for shared ANX risk replicated in the EUR component of this independent sample of young adult soldiers. We also examined genome-wide overlap of ANX association signals between the STARRS and ANGST datasets using polygenic approaches. Next, we explored whether findings emerging from these analyses generalize to AFR and LAT samples. Finally, we conducted a meta-analysis of the STARRS and ANGST GWAS data in order to maximize statistical power for the detection of novel susceptibility loci. In additional analyses, we estimated the SNP-based genetic correlation between ANX and related internalizing phenotypes using data from other GWAS to test the hypothesis generated by twin studies that they substantially share molecular genetic risk (Hettema, Neale, Myers, Prescott, & Kendler, 2006; Middeldorp, Cath, van Dyck, & Boomsma, 2005). We also investigated the biological implications of our findings using gene-based and functional pathway analyses.
METHODS
Information about specific methods pertinent to this study are included below. Additional detailed information about the design and conduct of STARRS is available in a separate report (Ursano et al., 2014). The recruitment, consent, human subject, and data protection procedures were approved by all collaborating organizations.
Subjects
The analyses presented here involved two large study components of STARRS: New Soldier Study (NSS). New soldiers enrolled in the NSS at the beginning of their basic training which took place between April 2011 and November 2012 at one of three Army installations. Soldiers completed a computerized self-administered questionnaire (SAQ) and 83.2% gave blood samples for DNA. Genotyping was conducted in samples from the first half of the cohort (NSS1; N=7,999) and a smaller subset of the second half (NSS2; N=2,835). Pre/Post Deployment Study (PPDS). US Army soldiers from three Brigade Combat Teams participated in the PPDS (N=7,927 genotyped) that began in early 2012. The data included in this report were collected at baseline, 4–6 weeks prior to deployment to Afghanistan. Among all the samples included in these analyses, 65% were EUR, 20% were LAT, and 15% were AFR. Their mean age was 21 years, and 83% were male.
Measures
The SAQ included a computerized version of the Composite International Diagnostic Interview screening scales (CIDI-SC; (R. C. Kessler et al., 2013)). Among the ANX, CIDI-SC scales provided proxies for diagnostic level phenotypes for lifetime generalized anxiety disorder and panic disorder. Detailed criteria for phobias were not assessed in the SAQ; however, screening stem items and other clinical information were included for social phobia, agoraphobia, and specific phobia. We used these data to create phobic-like phenotypes - see Supplementary File 1 for the scoring algorithm. To maximize phenotypic consistency with ANGST, we created triordinal versions (“case”, “subsyndromal” or “control”) of each of the five ANX phenotypes for input to factor analyses in all subjects with available SAQ data (N=47,065). This provides greater information than purely binary diagnostic categories that are limited by dependence on a clinically-chosen threshold to separate affected from unaffected individuals that likely does not optimally reflect differences in underlying genetic liability. Besides attempting to identify subjects meeting full symptomatic criteria for these disorders (“cases”, score=2), we sought to roughly differentiate subjects who were highly symptomatic but not meeting full criteria (“sub-syndromal”, score=1) versus those with few or no symptoms (“unaffected controls”, score=0). This sub-syndromal classification was operationalized for each of the five ANX by either (i) keeping full symptomatic criteria but removing the diagnostic requirements of distress/impairment or (ii) reducing symptomatic severity or duration.
These five triordinal phenotypes were entered into a factor analysis in OpenMx (M. C. Neale et al., 2016) that confirmed the previously observed unitary anxiety factor structure as a better fit to the data than a more complex two-factor model. The same solution was obtained in each of the ancestry groups. The loadings of this factor on each of the five ANX outcomes approximated those seen in the individual ANGST samples. Furthermore, neither the overall structure nor individual factor loadings significantly differed by ethnicity or from those observed in ANGST (Supplementary File 1). These results were verified using multi-group measurement invariance testing like that previously conducted in ANGST (Lee et al., 2016). We imposed a single factor confirmatory model on these data and estimated a dimensional factor score for each subject for input to the GWAS.
Genotyping and imputation
Detailed information on genotyping, imputation, population assignment, and principal component analysis for population stratification adjustment are included in our previous report (Stein et al., 2016). Briefly, whole blood samples were shipped to Rutgers University Cell & DNA Repository where they were frozen for later DNA extraction using standard methods. NSS1 and PPDS samples were genotyped using the Illumina OmniExpress+Exome array with additional custom content (NSNP=967,537). NSS2 samples were genotyped on the Illumina PsychChip (NSNP=571,054; 477,757 SNPs overlap with OmniExpress+Exome array).
Relatedness was tested with PLINK v1.90 (Chang et al., 2015; S. Purcell et al., 2007), and pairs of subjects with π>0.2 were identified, randomly retaining one member of each relative pair. We used a two-step pre-phasing/imputation approach for genotype imputation with reference to the 1000 Genomes Project multi-ethnic panel (August 2012 phase 1 integrated release; 2,186 phased haplotypes with 40,318,245 variants). We removed SNPs not present in the 1000 Genomes Project reference panel, had non-matching alleles to 1000 Genome Project reference, or had ambiguous, unresolvable alleles (AT/GC SNPs with minor allele frequency [MAF]>0.1). For Illumina OmniExpress array 664,457 SNPs and for Illumina PsychChip 360,704 SNPs entered the imputation procedure.
Ancestry Assignment and Population Stratification Adjustment
Given the ancestral heterogeneity of the STARRS subjects, samples were assigned into major population groups (EUR, AFR, LAT, Asian). To avoid long-range LD structure from interfering with the principal components analysis, we excluded SNPs in the MHC region (Chr 6:25–35Mb) and Chr 8 inversion (Chr 8:7–13Mb). Principal components within each population group were then obtained for further population stratification adjustment. Details of these procedures are described in an earlier STARRS publication (Stein et al., 2016).
Genomic and Sample Quality Control (QC)
We performed the following sequential QC procedure on the genotype data. We kept autosomal SNPs with missing rate < 0.05; kept samples with individual-wise missing rate < 0.02; then kept SNPs with missing rate < 0.02. After QC, we merged our study samples with HapMap3 samples. We kept SNPs with minor allele frequency (MAF) > 0.05 and performed LD pruning at R2 > 0.02. We excluded SNPs in the MHC region (chr6:25–35Mb) and Chr 8 inversion (chr8:7–13Mb). We performed additional quality control on the association test results by filtering out SNPs with imputation quality score (INFO) < 0.6 or significant Hardy-Weinberg equilibrium tests (p-value < 10–6). Final sample numbers included in these analyses are displayed in Table 1.
Table 1.
Comparison of the regression coefficients for the three STARRS ancestry groups with the ANGST results for the most significant SNP in ANGST (rs1067327) and in the combined meta-analysis (rs1067394).
| rs1067327 | rs1067394 | ||||
|---|---|---|---|---|---|
| Sample | N | Beta | p-value | Beta | p-value |
| ANGST | 18,186 | 0.0283 | 2.86e-09 | 0.0283 | 2.48e-09 |
| NSS1 (EUR) | 4591 | 0.0523 | 0.003 | 0.0529 | 0.003 |
| NSS2 (EUR) | 1630 | 0.0329 | 0.320 | 0.0337 | 0.308 |
| PPDS (EUR) | 4543 | 0.0023 | 0.897 | 0.0022 | 0.901 |
| Meta (EUR) | 10,764 | 0.0278 | 0.01828 | 0.0281 | 0.01644 |
| NSS1 (AFR) | 1319 | 0.0023 | 0.937 | 0.0013 | 0.967 |
| NSS2 (AFR) | 360 | 0.0273 | 0.662 | 0.0282 | 0.648 |
| PPDS (AFR) | 803 | 0.0045 | 0.910 | 0.0043 | 0.914 |
| Meta (AFR) | 2482 | 0.0062 | 0.7814 | 0.0058 | 0.7965 |
| NSS1 (LAT) | 1395 | −0.0377 | 0.192 | −0.0351 | 0.231 |
| NSS2 (LAT) | 452 | 0.0261 | 0.649 | 0.0373 | 0.514 |
| PPDS (LAT) | 1417 | −0.0022 | 0.940 | 0.0001 | 0.997 |
| Meta (LAT) | 3264 | −0.0147 | 0.4416 | −0.0109 | 0.5715 |
| Meta (STARRS All) | 16,510 | 0.0144 | 0.1143 | 0.0157 | 0.08694 |
| Meta (ANGST + EUR) | 28,950 | 0.0282 | 2.15e-10 | 0.0283 | 9.08e-11 |
| Meta (All) | 34,696 | 0.0253 | 2.62e-09 | 0.0257 | 8.35e-10 |
NSS1 and NSS2: New Soldier Study cohorts 1 and 2, respectively.
PPDS: Pre/Post Deployment Study
Statistical Analyses
GWAS. We used PLINK v1.90 to perform linear regression of the quantitative factor score against imputed SNP dosage for an additive genetic model in each of the three studies (NSS1, NSS2, and PPDS) separately within each of the three ancestral groups. Sex, age and the first 10 within-ancestry principal components were included as covariates. Since ANX and major depression share a substantial proportion of genetic risk, we removed subjects from the analyses (N=352) that had reported a lifetime history of pure major depression (i.e., not comorbid with ANX) so that genetic risk for depression in subjects with low anxiety factor scores would not confound the associations.
Meta-analysis
Inverse-variance weighted, fixed-effects meta-analyses were conducted using PLINK v1.90, first across samples within each ethnic group and then across all groups. A p-value < 5×10−8 was used as the threshold for genome-wide significance. We compared this to a novel, trans-ethnic meta-regression approach that models allelic effects as a function of genetic variation as implemented in MR-MEGA (Magi et al., 2017). To increase detection power, MR-MEGA combines Bayesian inference with population genetics information to account for heterogeneity in allelic effects correlated with ancestry which cannot be accommodated through traditional fixed-effects meta-analysis. Results for EUR subjects were then meta-analyzed with the ANGST summary statistics available at https://www.med.unc.edu/pgc/results-and-download).
SNP-based Heritability
We estimated SNP-based heritability using LD score regression (LDSC v1.0.0, (Bulik-Sullivan et al., 2015)) for the larger EUR ancestry cohort since the other groups were too small to obtain meaningful results. We note that this cannot be readily performed across ancestries due to the differences in LD structure.
Genetic Correlations
LDSC v1.0.0 was used to estimate genome-wide SNP-based genetic correlations between the current results and a representative group of psychological/psychiatric traits including ANGST data. We did not correct for multiple testing, as many of these phenotypes are correlated amongst themselves, and these analyses were primarily conducted as an overall check of the validity of the genetic signals rather than attempting to accurately estimate the correlations. Given the limited sample size, we did not attempt to perform this with all available traits on, say, LD Hub (http://ldsc.broadinstitute.org/), as that would involve correction for hundreds of tests that would not be meaningful. Rather, we selected internalizing traits most related to ANX such as neuroticism and depression from the Psychiatric Genomics Consortium downloads site (https://www.med.unc.edu/pgc/results-and-download) and compared these with less relevant ones like schizophrenia.
The second aim extends our findings to other ancestral groups by testing for association in AFR and LAT subgroups in two ways. First, we selected the most genome-wide significant SNPs from the chromosome 2 associated region in ANGST. Second, we conducted pilot genome-wide trans-ethnic meta-analyses. However, we are aware that smaller samples like the current AFR and LAT cohorts have insufficient power to reliably detect genetic signals. This limits the ability to evaluate genetic overlap versus heterogeneity of association across ethnicities as attempted in MR-MEGA. Nonetheless, some information about the concordance of association signals can be derived using the sign test for direction of SNP effects across groups (S. M. Purcell et al., 2009). To evaluate this, we computed the sign test across the three ethnicities for SNPs with trend-level associated p-values<10−5 from MR-MEGA. However, including all SNPS would be anticonservative, as SNPs in this rather large genomic region are in LD and, thus, their tests are correlated. To eliminate most of the LD between SNPs in computing the sign test, we (i) randomly chose 100 sets containing only one SNP from any group of SNPs lying within 250 Kbp of each other and (ii) averaged the p-values among those obtained from these sets.
Gene-based and pathway analyses
For most complex traits, genetic risks are the result of the joint effect of multiple SNPs in genes and multiple genes located in causal pathways. Consequently, pooling information across SNPs in a gene and genes in a pathway is likely to improve signal detection (B. M. Neale & Sham, 2004). Given that gene expression is the critical causal mechanism linking variant to phenotype (Emilsson et al., 2008), pooling of signals should be informed by this biologically mediating factor. Gene expression-based methods, also denoted as transcriptomics, exist for gene-level inference, i.e. JEPEGMIX2 and TWAS (Chatzinakos et al., 2018; Gamazon et al., 2015; Gusev et al., 2016). These gene level methods use eQTL SNPs to impute the association statistics between gene expression and the trait. However, due to the extreme computational burden of computing LD between gene transcriptomic statistics, there are no pure transcriptomic tools for assessing the association between a trait and the expression of all genes in a biological pathway. However, recently our group developed JEPEGMIX2-P, a pathway-level method for transcriptomic analysis that includes a larger reference panel (https://www.biorxiv.org/content/early/2018/07/20/373050). It uses a novel algorithm of linear complexity for LD computation for building a Mahalanobis pathway statistic. We applied JEPEGMIX2 and JEPEGMIX2-P transcriptomic tools to the meta-analytic data sets for EUR, AFR and LAT. For a comprehensive treatment, we also applied MAGMA software that makes inferences using all SNPs in a gene agnostic to gene expression (de Leeuw, Mooij, Heskes, & Posthuma, 2015). In addition to applying these tools to data within STARRS, we conducted gene-based association testing in the combined STARRS EUR and ANGST datasets to maximize power.
RESULTS
GWAS
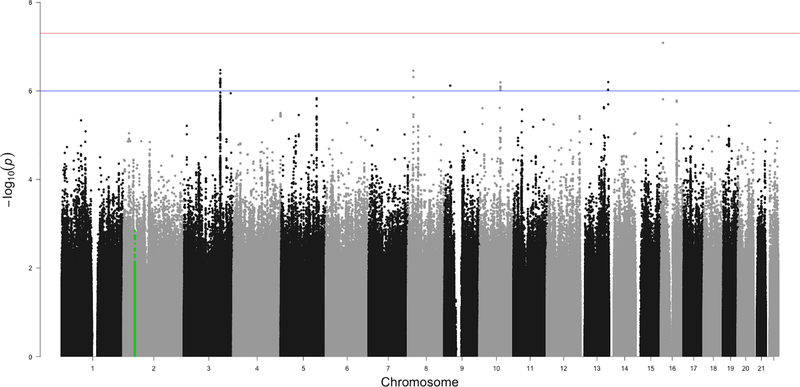
Nine separate GWAS (three ancestral groups by three studies) were conducted using PLINK v1.90 with linear regression of the quantitative factor score regressed against imputed SNP dosage for approximately ten million common SNPs after applying post-imputation QC, with sex, age and the first 10 within-ancestry principal components included as covariates. The genomic inflation factor λ ranged from 0.996 to 1.01 for all analyses suggesting little effect of population stratification consistent with q-q plots (Supplemental Figure S2). GWAS conducted within each study by ancestry group, across studies by ancestry group, and across all groups produced no genome-wide significant associations with the factor score. One exception is the EUR PPDS group for which a cluster of SNPs on chromosome 2 in the MYO1H gene had association p-values around 1E10−9; this association did not replicate in the other groups nor survive meta-analysis. The Manhattan plot for the STARRS trans-ethnic meta-analysis conducted in METAL is presented in Figure 1 (N=16,510). Loci containing SNPs with suggestive association (p<1×10−6) are listed in Supplementary Table S5.
Figure 1:
Manhattan plot of the STARRS GWAS results combined across all subjects and ethnicities (N=16,510).
Note: Manhattan plot of the STARRS genome-wide association study meta-analysis of NSS1, NSS2 and PPDS cohorts in European, African American, and Latino ancestry groups. The x-axis is chromosomal position and the y-axis is the –log10 of the p-value for the association derived by linear regression. The blue line denotes the suggestive significance threshold (1 × 10−6) and the red line denotes genome-wide significance (5 × 10−8). SNPs highlighted in green represent the region of chromosome 2 previously associated with the latent anxiety dimension in ANGST.
The findings from MR-MEGA that included data from EUR, AFR, and LAT participants essentially mirror those from the METAL meta-analysis. That is, we obtained only trend-level association signals that do not reach genome-wide significance. Supplementary Table S6 lists the magnitude and direction of SNP associations across the three ancestry groups estimated for SNPS in regions with trend-level associations using MR-MEGA. The SNPs display little evidence of heterogeneity across ethnicities with signs that are predominantly concordant, and formal tests of heterogeneity were non-significant. Bolded locations are the same SNPs that METAL identified. The p-value for the overall sign test across all these loci was significant (p=0.024).
The prior ANGST GWAS conducted in EUR data identified an association between SNPs in the CAMKMT gene on chromosome 2 (ch2:44,589,147–44,683,729) and a similarly constructed anxiety factor score. Within this genomic region, 69 of the 387 SNPs (17.8%) were nominally significant in the STARRS EUR cohort (Supplementary File 2).
Meta-analysis
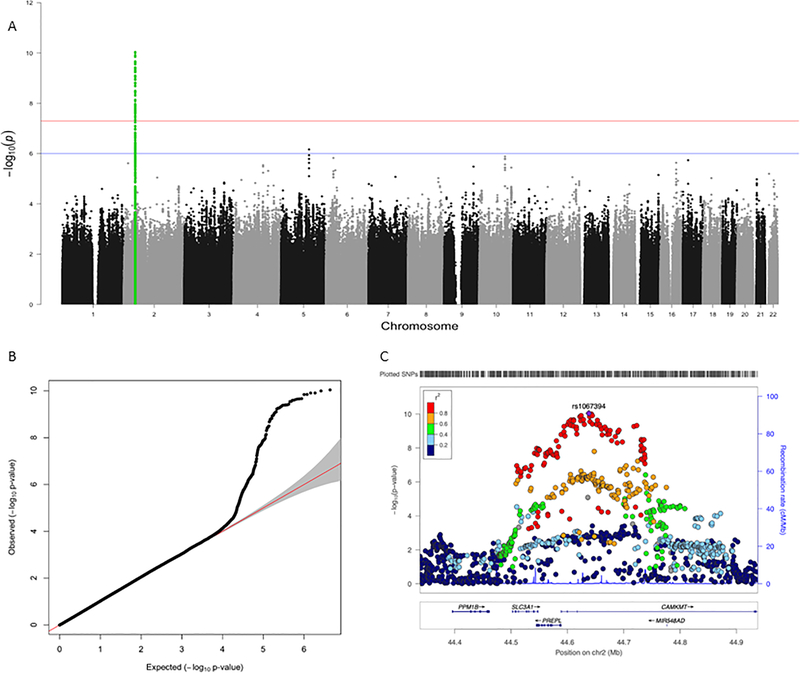
We conducted two SNP-based meta-analyses with the ANGST data to examine consistency with current results. First, only EUR data from STARRS were included to minimize heterogeneity (combined N=28,963). Figure 2a depicts the Manhattan plot and 2b the QQ plot. In Figure 2a, the genomic region on chromosome 2 from the ANGST discovery GWAS (green) is the most significantly associated region in the meta-analysis. The locus zoom plot for that region is shown in Figure 2c with lead SNP rs1067394. Next, all the STARRS data were included (combined N=34,709). The results of this trans-ancestry meta-analysis resemble the EUR only meta-analysis, but most associations are less significant.
Figure 2:
Results for the combined meta-analysis of the European ancestry subjects from STARRS+ANGST (N=28,963).
Notes: Panel A presents the Manhattan plot. The x-axis is chromosomal position and the y-axis is the –log10 of the p-value for the association derived by linear regression. The blue line denotes the suggestive significance threshold (1 × 10−6), and the red line denotes genome-wide significance (5 × 10−8). SNPs highlighted in green represent the region of chromosome 2 that was previously associated with the latent anxiety dimension in ANGST. Panel B presents the QQ plot of the associations for the meta-analysis. Panel C displays the locus zoom plot for the associated region.
Table 1 presents the beta coefficients and p-values in each sample for the two SNPs with the strongest association in each analysis (rs1067327 and rs1067394 respectively. These genomewide significant SNPs in ANGST were nominally significant in the EUR STARRS meta-analysis (bolded), providing an independent replication. Notably, for each SNP the effect sizes are remarkably similar to the ANGST results. P-values were considerably smaller in the combined of ANGST+STARRS meta-analysis, indicating increased precision. The associations are not evident in the AFR and LAT subsamples, suggesting there may be population-variable effects on ANX risk for this locus.
SNP-based Heritability and Genetic Correlations
We applied LDSC to obtain a SNP-based heritability estimate of 0.073 (p=0.007) in the EUR participants. This was essentially identical to that obtained in ANGST (0.072). We also used LDSC to estimate genetic correlations between the STARRS EUR ANX association signal and selected related traits. As indicated in Table 2, the STARRS anxiety dimension is genetically correlated with other relevant internalizing phenotypes with the highest correlation appropriately for the ANGST factor score. As expected, the estimates were positive with all except extraversion and subjective well-being. As predicted by genetic epidemiological studies, genetic correlations with less related phenotypes like schizophrenia were much smaller. However, none of the estimates were statistically different from zero.
Table 2.
SNP-based genetic correlations (Rg) estimated by LDSC between the STARRS anxiety factor results (European sample, N=10,764) and related constructs from other samples.
| PHENOTYPE | Rg | Standard Error | P-Value | N |
|---|---|---|---|---|
| ANGST Anxiety Factor Score (Otowa et al., 2016) | 0.891 | 0.659 | 0.176 | 18,186 |
| Obsessive-Compulsive Disorder ((OCGAS), 2018) | 0.292 | 0.347 | 0.400 | 9,607 |
| Major Depressive Disorder (Wray et al., 2018) | 0.507 | 0.336 | 0.132 | 173,005 |
| Depressive Symptoms (Okbay et al., 2016) | 0.718 | 0.461 | 0.119 | 161,460 |
| Neuroticism (GPC) (de Moor et al., 2015) | 0.851 | 0.622 | 0.172 | 63,661 |
| Neuroticism (SSGAC) (Okbay et al., 2016) | 0.653 | 0.422 | 0.122 | 170,911 |
| Schizophrenia (Consortium, 2014) | 0.115 | 0.16 | 0.719 | 150,064 |
| Extraversion (GPC) (van den Berg et al., 2015) | −0.206 | 0.014 | 0.516 | 63,030 |
| Loneliness (Gao et al., 2017) | 0.332 | 0.581 | 0.567 | 7,556 |
| Subjective Well Being (SSGAC) (Okbay et al., 2016) | −0.922 | 0.707 | 0.192 | 298,420 |
Gene-based and pathway analyses
No gene-based or pathway analyses yielded significant findings for any STARRS cohorts alone. However, significant results did emerge when we applied MAGMA and JEPEGMIX2 in the combined STARRS EUR+ANGST meta-analysis. MAGMA gene-level analyses (Supplementary Table S7) detected two regions harboring significant signals. The first includes CAMKMT plus two other genes (PREPL and SLC3A1) in the same LD block on chromosome 2; these were also identified in the prior ANGST gene-based analysis. The second is LBX1 on chromosome 10 encoding a homeobox transcription factor. JEPEGMIX2 also detected two highly significant transcriptomic signals, the first in thyroid tissue for pseudogene RP11–815N9.2 on chromosome 4 and the second in pituitary tissue for PREPL (Supplementary Table S8). However, neither MAGMA nor JEPEGMIX2 identified any significantly enriched pathways.
DISCUSSION
In this study, we performed GWAS for a dimensional anxiety factor score in the Army STARRS study population (N=16,510). This factor score, representing the largest proportion of variance shared between five ANX, was estimated by statistically modeling the covariance structure of triordinal ANX proxy phenotypes created from the survey data. This procedure attempted to maximally approximate a similar phenotype used in prior GWAS in ANGST (Otowa et al., 2016). Its validity was verified by measurement invariance analyses across samples. GWAS were conducted first separately by cohort (NSS1, NSS2, PPDS) and ancestral background (EUR, AFR, LAT), then these were combined via meta-analyses. The study was conducted to address three overarching aims, the results of which we discuss in turn.
First, we successfully replicated the ANGST 2p21 association signal for shared ANX risk in the EUR component of the current study (N=10,764). Table 1 displays the results for the two SNPs possessing the most significant p-values in ANGST (rs1067327) and in the current meta-analysis (rs1067394). Both SNPs are intronic to CAMKMT and are in very high LD with each other (r2=0.99), essentially representing the same association. Their direction and effect sizes in EUR were highly consistent with those from ANGST, resulting in stronger association signals (p-values 2.15E-10 and 9.08E-11, respectively) when data were meta-analyzed from the two studies. CAMKMT is an evolutionarily conserved class I protein methyltransferase that acts in the formation of trimethyllysine in calmodulin which is involved in calcium-dependent signaling (Magnani, Dirk, Trievel, & Houtz, 2010). A recent study delineating the function of this gene reported that mutant mice had deficits in motor learning, complex coordination, and learning of aversive stimuli (Haziza et al., 2015).
No robust genome-wide significant associations were identified in analyses conducted within STARRS alone (Figure 1). This is not surprising given the relatively limited sample sizes in each ancestry component for GWAS. However, LDSC results indicated substantial genetic correlation between STARRS EUR ancestry data and ANGST (Rg=0.891) although not statistically significant due to limited power. It was also variably correlated with related internalizing phenotypes like depression and neuroticism consistent with predictions from prior twin studies and ANGST data. These results provide genome-wide support for the association signals present in these data that complements the 2p21 region-specific findings.
We attempted to extend our findings to other ancestry participants by testing for association in the AFR and LAT subgroups. Consistency of sign and magnitude of association were observed for the selected chromosome 2 SNPs within the AFR but not the LAT cohorts (Table 1). Overall, no genome-wide significant associations were identified when meta-analyzing across ancestries. Nonetheless, promising trend-level signals within peaks of multiple associated SNPs were seen for several regions in the combined, trans-ethnic meta-analyses (Supplementary Tables S5 and S6). Furthermore, SNPs in these regions showed overall statistically significant concordance of direction of effects across ancestries, supporting the need for future, more powerful trans-ethnic studies.
Given the overall consistency between the association signals present in STARRS EUR subjects and those from ANGST, we combined these data via meta-analysis to maximize power. Apart from the consistent signals in the 2p21 region, no additional genome-wide susceptibility variants were identified in the SNP-based meta-analysis. This is somewhat surprising given the combined EUR effective sample size of almost 29K subjects, about 50% larger than ANGST alone. It might be that association effect sizes for the next tier of anxiety susceptibility genes are smaller than for loci identified in ANGST, requiring substantially larger samples to detect them.
Gene-level analyses uncovered signals only for the combined EUR sample of STARRS and ANGST. The lack of findings for the remaining analyses is likely due to small sample sizes, the possibility of ancestry-specific effects, and a clinically-defined psychiatric phenotype with moderate heritability and complex biology. For the combined EUR samples, we found consistent evidence supporting a role for the chromosome 2 region containing SLC3A1, PREPL and CAMKMT previously identified in ANGST. The first two genes are involved in two contiguous gene-deletion syndromes, the hypotonia-cystinuria syndrome and the more severe 2p21 deletion syndrome (Martens, Jaeken, Matthijs, & Creemers, 2008). PREPL was also identified in the transcriptomic based analysis in pituitary tissue. As discussed in detail in the ANGST study, in silico analyses suggested rs698775 in PREPL as the most likely functional candidate in this region with a cis-regulatory effect specific to that gene. Two more genes were tentatively identified by these gene-based analyses. LBX1 on chromosome 10 encodes a highly conserved homeobox transcription factor required for the development of GABAergic interneurons. It is expressed in skeletal muscle, cerebellum, and spinal cord and involved in idiopathic scoliosis. RP11–815N9.2 is an unusual pseudogene region of unknown function on chromosome 4. These latter two findings require replication in independent studies.
We note that two other anxiety disorder GWAS have been published since these analyses were performed. The first, conducted in 12,655 individuals with registry-derived ICD-10 anxiety and/or stress-related disorders and 19,225 unscreened controls from the Danish Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) cohort, reported one genome-wide significant association with variants in the PDE4B gene (Meier et al., 2019). The second, conducted in 25,453 cases with self-reported any lifetime anxiety disorder and 58,113 controls from the UK Biobank cohort, identified five novel genome-wide significant loci including variants in the gene for NTRK2, a receptor for brain-derived neurotropic factor (Purves et al., 2019). Both utilized case-control phenotypes rather than factor-analytic dimensional scores, so direct comparisons with the current study may not apply.
Results from the current study should be considered in the context of several limitations. First, sample sizes for each ancestry group were modest by current psychiatric GWAS standards; this is particularly true for the AFR and LAT groups. Nor did we include Asians due to their even smaller representation in these cohorts. Second, comprehensive diagnostic data were only available for generalized anxiety and panic disorders in the SAQ; phobia phenotypes were approximated using a truncated set of fear-related items. While prior twin studies support combining fear and phobia phenotypes, it is unclear what effect this has on our outcomes. Encouragingly, the latent factor structure using these phenotypes in STARRS was essentially identical to that obtained in ANGST. Third, although our findings increase confidence that the 2p21 region contains an ANX susceptibility locus, it is not clear which gene in this region is most etiologically relevant. With such strong LD across the region (Figure 2C), further work will be needed to fine map the causal variant(s). Fourth, the STARRS sample primarily consists of male soldiers, precluding the ability to test for sex-specific effects. Although the lifetime prevalence of anxiety disorders in women is nearly twice that in men (Kessler et al., 1994), twin studies of anxiety disorders did not detect differences in their genetic risk factors (Hettema et al., 2005).
In summary, this study successfully replicated one of the few previously reported genome-wide associated loci for ANX. Our attempt to extend this finding to other ancestry groups met with limited success likely due to reduced power and genomic heterogeneity in the non-EUR groups. Failure to identify additional associated SNPs in our combined meta-analysis of nearly 29K EUR subjects implies the need for even larger sample sizes as was the case for other high-prevalence, moderately heritable psychiatric disorders like major depression (Wray et al., 2018). Given the unknown molecular genetic architecture of anxiety compared with other psychiatric disorders, it is unclear at what sample size threshold one achieves sufficient power to detect substantially more risk variants. However, one might get a sense from the iPSYCH and UK Biobank results described above. The iPSYCH sample included 4584 participants with any lifetime anxiety disorder amongst the 12,655 cases. This is similar to that of the alternate case-control analysis in ANGST with 7016 anxiety cases, both reporting only one genome-wide significant association. UK Biobank found five associated loci with 25,453 anxiety disorder cases, about four to five times those in iPSYCH or ANGST. Speculation suggests that the next GWAS with more anxiety cases might detect linearly proportionate numbers of significant loci (Visscher, Brown, McCarthy, & Yang, 2012). The newly-formed anxiety disorders working group of the Psychiatric Genomics Consortium (PGC-ANX) is gathering data from other international groups to add to these cohorts and achieve substantially larger total sample sizes.
Supplementary Material
ACKNOWLEDGEMENTS
Army STARRS was sponsored by the Department of the Army and funded under cooperative agreement number U01MH087981 (2009–2015) with the U.S. Department of Health and Human Services, National Institutes of Health, National Institute of Mental Health (NIH/NIMH). Subsequently, STARRS-LS was sponsored and funded by the Department of Defense (USUHS grant number HU0001-15-2-0004). The contents are solely the responsibility of the authors and do not necessarily represent the views of the Department of Health and Human Services, NIMH, or the Departments of the Army or the Department of Defense. Dr. Smoller is Tepper Family MGH Research Scholar and is supported by the Demarest Lloyd, Jr, Foundation and NIH grant K24MH094614. Drs. Hettema, Verhulst, and Bacanu were supported by NIH R01 MH113665.
Grant funding: NIH R01MH113665, U01MH087981
Footnotes
CONFLICTS OF INTEREST: NONE
The Army STARRS Team consists of:
Co-Principal Investigators: Robert J. Ursano, MD (Uniformed Services University of the Health Sciences) and Murray B. Stein, MD, MPH (University of California San Diego and VA San Diego Healthcare System)
Site Principal Investigators: Steven Heeringa, PhD (University of Michigan), James Wagner, PhD (University of Michigan) and Ronald C. Kessler, PhD (Harvard Medical School)
Army liaison/consultant: Kenneth Cox, MD, MPH (US Army Public Health Center)
Other team members: Pablo A. Aliaga, MS (Uniformed Services University of the Health Sciences); COL David M. Benedek, MD (Uniformed Services University of the Health Sciences); Tianxi Cai, ScD (Harvard School of Public Health); Laura Campbell-Sills, PhD (University of California San Diego); Chia-Yen Chen, ScD (Harvard Medical School); Karmel Choi, PhD (Harvard Medical School); Carol S. Fullerton, PhD (Uniformed Services University of the Health Sciences); Nancy Gebler, MA (University of Michigan); Joel Gelernter, MD (Yale University); Robert K. Gifford, PhD (Uniformed Services University of the Health Sciences); Feng He, MS (University of California San Diego); Meredith House, BA (University of Michigan); Paul E. Hurwitz, MPH (Uniformed Services University of the Health Sciences); Sonia Jain, PhD (University of California San Diego); Tzu-Cheg Kao, PhD (Uniformed Services University of the Health Sciences); Lisa Lewandowski-Romps, PhD (University of Michigan); Holly Herberman Mash, PhD (Uniformed Services University of the Health Sciences); Adam X. Maihofer (University of California San Diego); James A. Naifeh, PhD (Uniformed Services University of the Health Sciences); Tsz Hin Hinz Ng, MPH (Uniformed Services University of the Health Sciences); Caroline M. Nievergelt, PhD (University of California San Diego); Matthew K. Nock, PhD (Harvard University); Stephan Ripke, MD (Harvard Medical School); Nancy A. Sampson, BA (Harvard Medical School); Ronen Segman, MD (Hadassah University Hospital, Israel); Jordan W. Smoller, MD, ScD (Harvard Medical School); Xiaoying Sun, MS (University of California San Diego); Erin Ware PhD (University of Michigan); LTC Gary H. Wynn, MD (Uniformed Services University of the Health Sciences); Alan M. Zaslavsky, PhD (Harvard Medical School); and Lei Zhang, MD (Uniformed Services University of the Health Sciences).
DISCLOSURES
Dr. Stein has in the past three years been a consultant for Actelion, Alkermes, Aptinyx, Bionomics, Dart Neuroscience, Healthcare Management Technologies, Janssen, Neurocrine Biosciences, Oxeia Biopharmaceuticals, Pfizer, and Resilience Therapeutics. Dr. Stein has stock options in Oxeia Biopharmaceticals. Dr. Smoller is an unpaid member of the Scientific Advisory Board of PsyBrain Inc. and the Bipolar/Depression Research Community Advisory Panel of 23andMe. In the past 3 years, Dr. Kessler received support for his epidemiological studies from Sanofi Aventis; was a consultant for Johnson & Johnson Wellness and Prevention, Sage Pharmaceuticals, Shire, Takeda; and served on an advisory board for the Johnson & Johnson Services Inc. Lake Nona Life Project. Kessler is a co-owner of DataStat, Inc., a market research firm that carries out healthcare research. The other authors report nothing to disclose.
References
- (OCGAS), I. O. C. D. F. G. C. I.-G. a. O. C. G. A. S. (2018). Revealing the complex genetic architecture of obsessive-compulsive disorder using meta-analysis. Mol Psychiatry, 23(5), 1181–1188. doi: 10.1038/mp.2017.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beesdo K, Knappe S, & Pine DS (2009). Anxiety and anxiety disorders in children and adolescents: developmental issues and implications for DSM-V. Psychiatr Clin North Am, 32(3), 483–524. doi: 10.1016/j.psc.2009.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Patterson N, … Neale BM (2015). LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet, 47(3), 291–295. doi:ng.3211 [pii]; 10.1038/ng.3211 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, & Lee JJ (2015). Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience, 4, 7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatzinakos C, Lee D, Webb BT, Vladimirov VI, Kendler KS, & Bacanu SA (2018). JEPEGMIX2: improved gene-level joint analysis of eQTLs in cosmopolitan cohorts. Bioinformatics, 34(2), 286–288. doi: 10.1093/bioinformatics/btx509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium, S. W. G. o. t. P. G. (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature, 511(7510), 421–427. doi:nature13595 [pii]; 10.1038/nature13595 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leeuw CA, Mooij JM, Heskes T, & Posthuma D (2015). MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol, 11(4), e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MH, van den Berg SM, Verweij KJ, Krueger RF, Luciano M, Arias VA, … Boomsma DI (2015). Meta-analysis of Genome-wide Association Studies for Neuroticism, and the Polygenic Association With Major Depressive Disorder. JAMA Psychiatry, 72(7), 642–650. doi:2294268 [pii]; 10.1001/jamapsychiatry.2015.0554 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, … Stefansson K (2008). Genetics of gene expression and its effect on disease. Nature, 452(7186), 423–428. doi: 10.1038/nature06758 [DOI] [PubMed] [Google Scholar]
- Erhardt A, Akula N, Schumacher J, Czamara D, Karbalai N, Muller-Myhsok B, … Binder EB (2012). Replication and meta-analysis of TMEM132D gene variants in panic disorder. Transl. Psychiatry, 2, e156. doi:tp201285 [pii]; 10.1038/tp.2012.85 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt A, Czibere L, Roeske D, Lucae S, Unschuld PG, Ripke S, … Binder EB (2011). TMEM132D, a new candidate for anxiety phenotypes: evidence from human and mouse studies. Mol Psychiatry, 16(6), 647–663. doi: 10.1038/mp.2010.41 [DOI] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, … Im HK (2015). A gene-based association method for mapping traits using reference transcriptome data. Nat Genet, 47(9), 1091–1098. doi: 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Davis LK, Hart AB, Sanchez-Roige S, Han L, Cacioppo JT, & Palmer AA (2017). Genome-Wide Association Study of Loneliness Demonstrates a Role for Common Variation. Neuropsychopharmacology, 42(4), 811–821. doi: 10.1038/npp.2016.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, … Pasaniuc B (2016). Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet, 48(3), 245–252. doi: 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haziza S, Magnani R, Lan D, Keinan O, Saada A, Hershkovitz E, … Parvari R (2015). Calmodulin Methyltransferase Is Required for Growth, Muscle Strength, Somatosensory Development and Brain Function. PLoS Genet, 11(8), e1005388. doi: 10.1371/journal.pgen.1005388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema JM, Neale MC, & Kendler KS (2001). A review and meta-analysis of the genetic epidemiology of anxiety disorders. Am. J. Psychiatry, 158(10), 1568–1578. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Neale MC, Myers JM, Prescott CA, & Kendler KS (2006). A population-based twin study of the relationship between neuroticism and internalizing disorders. Am. J. Psychiatry, 163(5), 857–864. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Prescott CA, Myers JM, Neale MC, & Kendler KS (2005). The structure of genetic and environmental risk factors for anxiety disorders in men and women. Arch. Gen. Psychiatry, 62(2), 182–189. [DOI] [PubMed] [Google Scholar]
- Hettema JM, Webb BT, Guo AY, Zhao Z, Maher BS, Chen X, … van den Oord EJ (2011). Prioritization and association analysis of murine-derived candidate genes in anxiety-spectrum disorders. Biol. Psychiatry, 70(9), 888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, & Wuyek LA (2010). Anxiety disorders as a risk factor for subsequent depression. Int J Psychiatry Clin Pract, 14(4), 244–247. doi: 10.3109/13651501.2010.487979 [DOI] [PubMed] [Google Scholar]
- Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, … Wang P (2010). Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am. J. Psychiatry, 167(7), 748–751. doi:167/7/748 [pii]; 10.1176/appi.ajp.2010.09091379 [doi] [DOI] [PubMed] [Google Scholar]
- Kessler RC, Calabrese JR, Farley PA, Gruber MJ, Jewell MA, Katon W, … Wittchen HU (2013). Composite International Diagnostic Interview screening scales for DSM-IV anxiety and mood disorders. Psychol Med, 43(8), 1625–1637. doi: 10.1017/s0033291712002334 [DOI] [PubMed] [Google Scholar]
- Kessler RC, McGonagle KA, Zhao S, Nelson CB, Hughes M, Eshleman S, … Kendler KS (1994). Lifetime and 12-month prevalence of DSM-III-R psychiatric disorders in the United States. Results from the National Comorbidity Survey. Arch Gen Psychiatry, 51(1), 8–19. [DOI] [PubMed] [Google Scholar]
- Lee M, Aggen SH, Otowa T, Castelao E, Preisig M, Grabe HJ, … Hettema JM (2016). Assessment and characterization of phenotypic heterogeneity of anxiety disorders across five large cohorts. Int. J. Methods Psychiatr. Res doi: 10.1002/mpr.1519 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magi R, Horikoshi M, Sofer T, Mahajan A, Kitajima H, Franceschini N, … Morris AP (2017). Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum Mol Genet, 26(18), 3639–3650. doi: 10.1093/hmg/ddx280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani R, Dirk LM, Trievel RC, & Houtz RL (2010). Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat Commun, 1, 43. doi: 10.1038/ncomms1044 [DOI] [PubMed] [Google Scholar]
- Martens K, Jaeken J, Matthijs G, & Creemers JW (2008). Multi-system disorder syndromes associated with cystinuria type I. Curr Mol Med, 8(6), 544–550. [DOI] [PubMed] [Google Scholar]
- Meier SM, & Deckert J (2019). Genetics of Anxiety Disorders. Curr Psychiatry Rep, 21(3), 16. doi: 10.1007/s11920-019-1002-7 [DOI] [PubMed] [Google Scholar]
- Meier SM, Trontti K, Purves KL, Als TD, Grove J, Laine M, … Mors O (2019). Genetic Variants Associated With Anxiety and Stress-Related Disorders: A Genome-Wide Association Study and Mouse-Model Study. JAMA Psychiatry. doi: 10.1001/jamapsychiatry.2019.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merikangas KR, & Swanson SA (2010). Comorbidity in anxiety disorders. Curr Top Behav Neurosci, 2, 37–59. [PubMed] [Google Scholar]
- Middeldorp CM, Cath DC, van Dyck R, & Boomsma D (2005). The co-morbidity of anxiety and depression in the perspective of genetic epidemiology. A review of twin and family studies. Psychological Medicine, 35(May), 611–624. [DOI] [PubMed] [Google Scholar]
- Neale BM, & Sham PC (2004). The future of association studies: gene-based analysis and replication. Am. J Hum. Genet, 75(3), 353–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kirkpatrick RM, … Boker SM (2016). OpenMx 2.0: Extended Structural Equation and Statistical Modeling. Psychometrika, 81(2), 535–549. doi: 10.1007/s11336-014-9435-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, … Cesarini D (2016). Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet, 48(6), 624–633. doi: 10.1038/ng.3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG, … Hettema JM (2016). Meta-analysis of genome-wide association studies of anxiety disorders. Mol. Psychiatry. doi:mp2015197 [pii]; 10.1038/mp.2015.197 [doi] [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, … Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet, 81(3), 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, & Sklar P (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature, 460(7256), 748–752. doi: 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, … Eley TC (2019). A major role for common genetic variation in anxiety disorders. Mol Psychiatry. doi: 10.1038/s41380-019-0559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada-Sugimoto M, Otowa T, & Hettema JM (2015). Genetics of anxiety disorders: Genetic epidemiological and molecular studies in humans. Psychiatry Clin. Neurosci, 69(7), 388–401. doi: 10.1111/pcn.12291 [doi] [DOI] [PubMed] [Google Scholar]
- Stein MB, Chen CY, Ursano RJ, Cai T, Gelernter J, Heeringa SG, … Smoller JW (2016). Genome-wide Association Studies of Posttraumatic Stress Disorder in 2 Cohorts of US Army Soldiers. JAMA Psychiatry, 73(7), 695–704. doi:2521460 [pii]; 10.1001/jamapsychiatry.2016.0350 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursano RJ, Colpe LJ, Heeringa SG, Kessler RC, Schoenbaum M, & Stein MB (2014). The Army study to assess risk and resilience in servicemembers (Army STARRS). Psychiatry, 77(2), 107–119. doi: 10.1521/psyc.2014.77.2.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg SM, de Moor MH, Verweij KJ, Krueger RF, Luciano M, Arias VA, … Boomsma DI (2015). Meta-analysis of Genome-Wide Association Studies for Extraversion: Findings from the Genetics of Personality Consortium. Behav. Genet. doi: 10.1007/s10519-015-9735-5 [doi];10.1007/s10519–015-9735–5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, & Yang J (2012). Five years of GWAS discovery. Am. J. Hum. Genet, 90(1), 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, … Sullivan PF (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet, 50(5), 668–681. doi: 10.1038/s41588-018-0090-3 [doi];10.1038/s41588–018-0090–3 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.