Abstract
Intravital imaging, the direct visualization of cells and tissues within a living animal, is a technique that has been employed for the better part of a century. The advent of confocal and multiphoton microscopy has dramatically improved the power of intravital imaging, making it possible to obtain optical sections of tissues non-destructively. This review discusses the various techniques used for intravital imaging, describes how intravital imaging provides information about cellular and tissue dynamics not possible to be garnered by other techniques, and details several ways in which intravital imaging is making a direct impact on the clinical care of patients.
Keywords: multiphoton, intravital, imaging windows, clinical applications
For the last three decades, high-resolution multiphoton and confocal intravital imaging (IVI) has become a quintessential tool in the armamentarium of biological and biomedical research. Intravital imaging provides researchers the ability to visualize and capture, in vivo and with single cell resolution, dynamic biological processes in multiple organ systems.(1) Though intravital imaging is most commonly performed in mice and rodents (due to their small size, genetic similarity to humans,(2) and their ability to be experimentally manipulated to model numerous diseases(3)), successful application of the technique has been accomplished in a variety of species(4) including humans.(5)
In this review, we discuss the use and application of IVI in biomedical research with the aim of giving readers a greater understanding of (1) the various techniques used for intravital imaging and (2) how these techniques are capable of giving insights into the dynamics of cells and tissues that were not possible to be gained with other techniques. Finally, we describe the role of IVI in translational research and its direct impact in clinical care and medical treatments.
We begin with a brief description of the advantages of confocal multiphoton and intravital microscopy and then turn to discuss a variety of protocols for accessing different tissue types. We proceed in order of increasing sophistication, from the simplest transdermal and ocular imaging, through externalization of tissues and terminal surgeries, and finally on to survival surgeries with implantable imaging windows. We next discuss some of the new biological insights that have been garnered from intravital imaging studies of cellular dynamics in the fields of immunology, metabolomics, and cancer metastasis. Finally, we conclude with a description of how intravital imaging is being brought directly into the clinic through the development of new instrumentation and clinical protocols.
Intravital Imaging Visualizes in vivo Cellular Dynamics and Interactions, Nondestructively
Intravital imaging is the process of imaging cells and tissues of a living animal. Though able to be accomplished with numerous imaging technologies (e.g. PET, MRI, CT, etc.), we here focus on those techniques capable of resolving the individual cells of imaged samples (i.e. multiphoton, confocal, and wide-field fluorescence). In particular, we limit our discussion to those microscopy techniques capable of optical sectioning, that is, those able to illuminate, or collect light from, a single slice of an otherwise intact tissue (multiphoton and confocal imaging).
By limiting the generation or detection of signal to a single geometrical plane within the tissue, optical sectioning generates images of the cells within this plane that are strikingly similar to those generated by standard histopathology techniques (Fig. 1). However, these images are acquired nondestructively and not subject to the same artifacts (e.g. tissue dehydration, shrinkage, tearing, etc.) as tissues that are fixed, chemically processed, and mechanically cut into thin sections. (6,7) Further, the inherent registration of optical sections makes generating three-dimensional (3D) reconstructions (computationally combining the image data from a stack of images) simple.(8)
Figure 1.
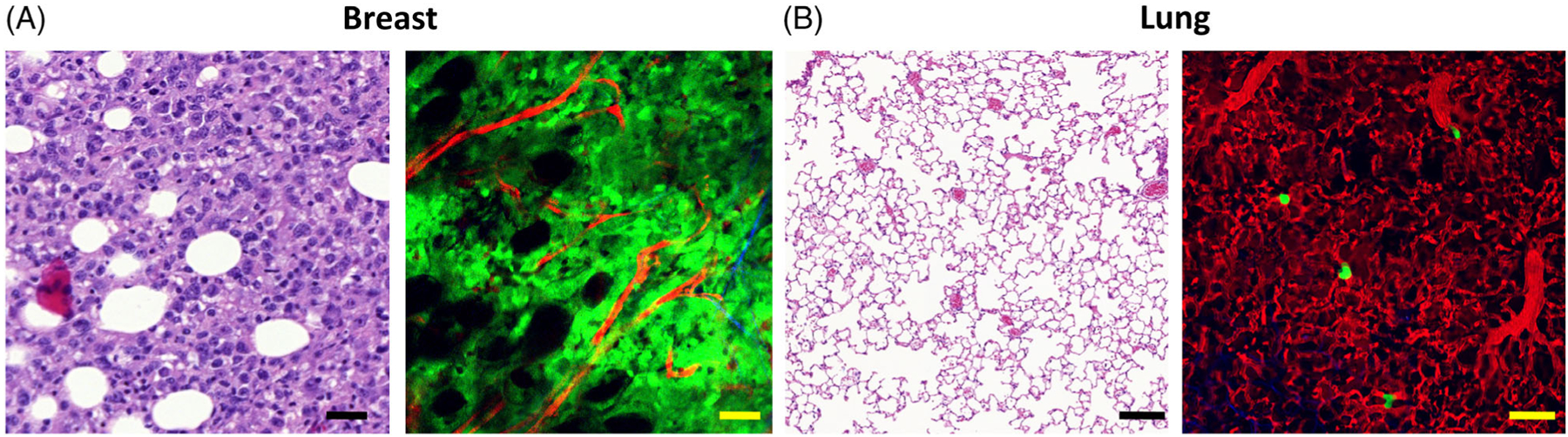
Comparison between intravital imaging and fixed tissue histology. The optical sectioning capabilities of multiphoton and confocal microscopes generate images of living tissues that are similar to those obtained by mechanical sectioning and staining of fixed tissues. However, as the tissues may be imaged live, cellular dynamics and cell–cell interactions can be visualized in real time. A, Mechanical and optical sections of breast carcinoma tissue. Left: H&E, white circles = adipocytes. Bar = 20 μm. Right: Multiphoton, green = tumor cells, red = vasculature, black circles = adipocytes. Bar = 20 μm. B, Lung tissue. Left: H&E. Bar = 20 μm. Right: Red = vasculature, green = disseminated tumor cells. Bar = 20 μm.
To accomplish optical sectioning, confocal microscopy (Fig. 2) focuses light to a single point within a sample and then limits the reflected or generated fluorescence light, which returns from the sample by employing a pinhole aperture before the detector. This pinhole only allows light originating within the focal point of the illumination to reach the detector and blocks all light emanating from regions above, below, and lateral to this point.(9)
Figure 2.
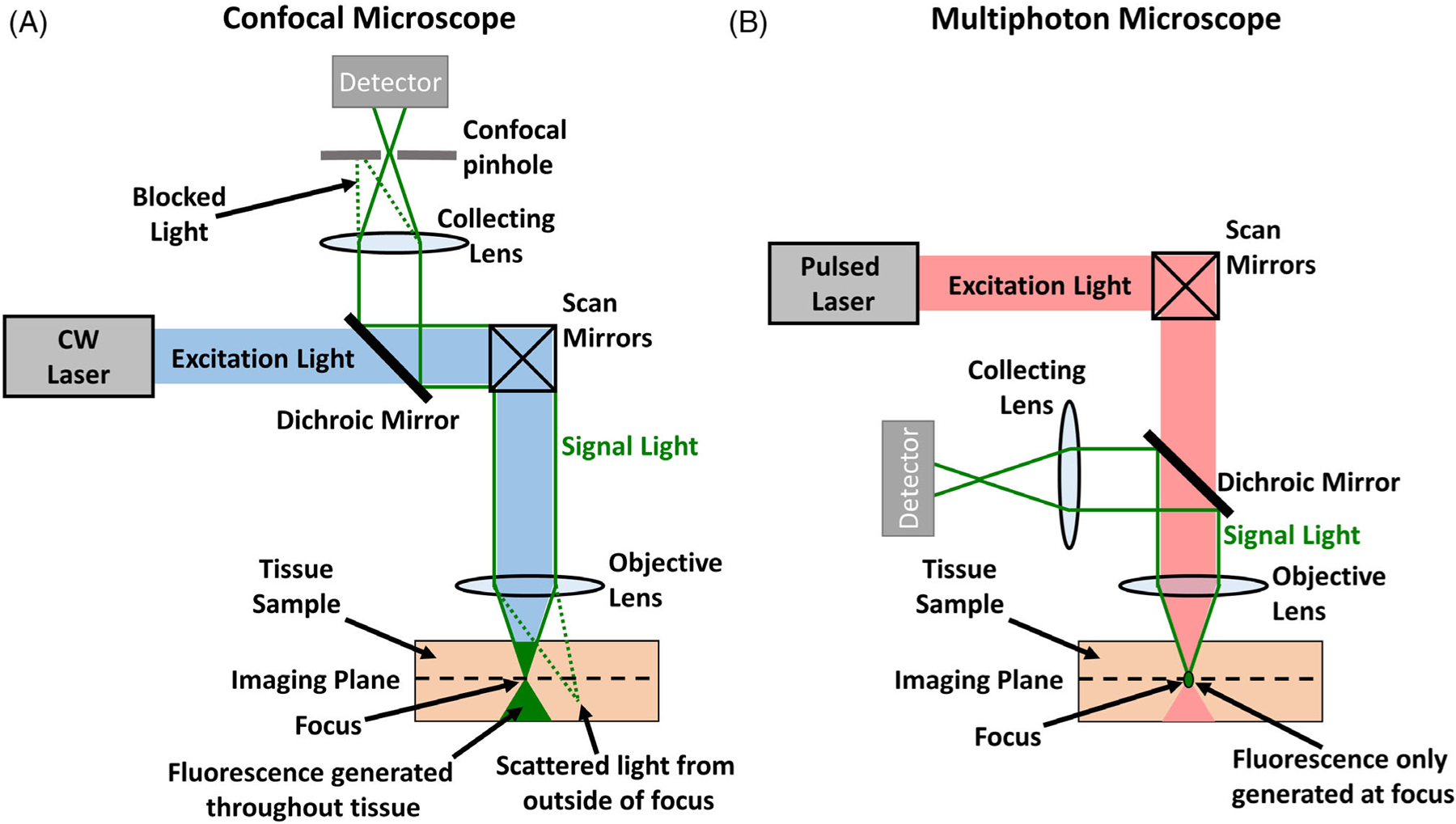
Confocal and multiphoton microscopes capture images from a single plane within solid tissue. A, Optical layout of a confocal microscope. Though the continuous wave (CW) laser generates signal throughout the tissue sample, out of focus light is eliminated by focusing returning signal light through a pinhole with a collecting lens. Light originating from within the focal point of the objective lens passes through the pinhole and can reach the detector. Light originating outside of the focus is blocked by the pinhole. B, Optical layout of a multiphoton microscope. The use of a femtosecond pulsed laser allows multiphoton excitation of the sample and signal light is only generated at the exact focus. In this case, a pinhole is not required and all signal can be collected on the detector. In both confocal and multiphoton microscopy, signal from the entire imaging plane is generated by scanning the focal point over the imaging plane with the scan mirrors.
Multiphoton microscopy (a fluorescence technique, Fig. 2) in comparison achieves optical sectioning also by focusing its illumination light to a single point, but then limiting absorption outside of this focal point. This is made possible by utilizing a nonlinear optical phenomenon based on the near simultaneous absorption (usually within a few femtoseconds) of two or more photons whose energies sum up to the excitation energy gap of the molecules to be excited. In terms of wavelength, this is most commonly accomplished by utilizing a femtosecond pulsed laser with a wavelength that is twice that ordinarily needed to excite the molecules. Thus, for molecules, which excite within the visible spectrum, illumination wavelengths are required to lie in the near infrared or infrared.(10,11)
In general, the multiphoton microscope offers significant advantages over confocal including: reduced photobleaching, increased penetration depth, more robust and stable optical alignment, and increased sensitivity to weak fluorophores.(12) However, the significantly higher cost of multiphoton microscopes (mostly due to the cost of the femtosecond pulsed laser) make them less commonly available than confocal microscopes. In particular, multiphoton microscopy allows roughly five times the imaging depth of confocal(13) but this depth is highly dependent upon the optical properties of the tissue to be imaged. Imaging is deepest in homogeneous tissue such as the brain where depths of up to 1.2 mm have been achieved(14) but is approximately several hundred microns in various other organs.(10,15) In some tissues such as the pancreas or the living lung, imaging depths are even shallower due to the high degree of scatter from, and absorption of, red blood cells.
Transdermal and Ocular Imaging
Given the relatively limited penetration depth of the technique (compared to MRI or PET), a tissue of interest must be exposed for multiphoton or confocal-based intravital imaging. This can be accomplished in a variety of ways. The simplest, and least invasive of these techniques, is utilized when the cells of interest lie within a depth that is readily accessible by the microscope. This is the case for those cells at the back of the eye, such as the retina(16) and the trabecular meshwork (17) (accessed through the cornea using long working distance objective lenses), or those that either exist within, or are injected into, the skin (Fig. 3). Generally, imaging in the skin is limited to the epidermal layers. However, the dorsal skin area of the mouse ear is thin enough for both the epidermis and dermis to be accessed by multiphoton microscopy.(18)
Figure 3.
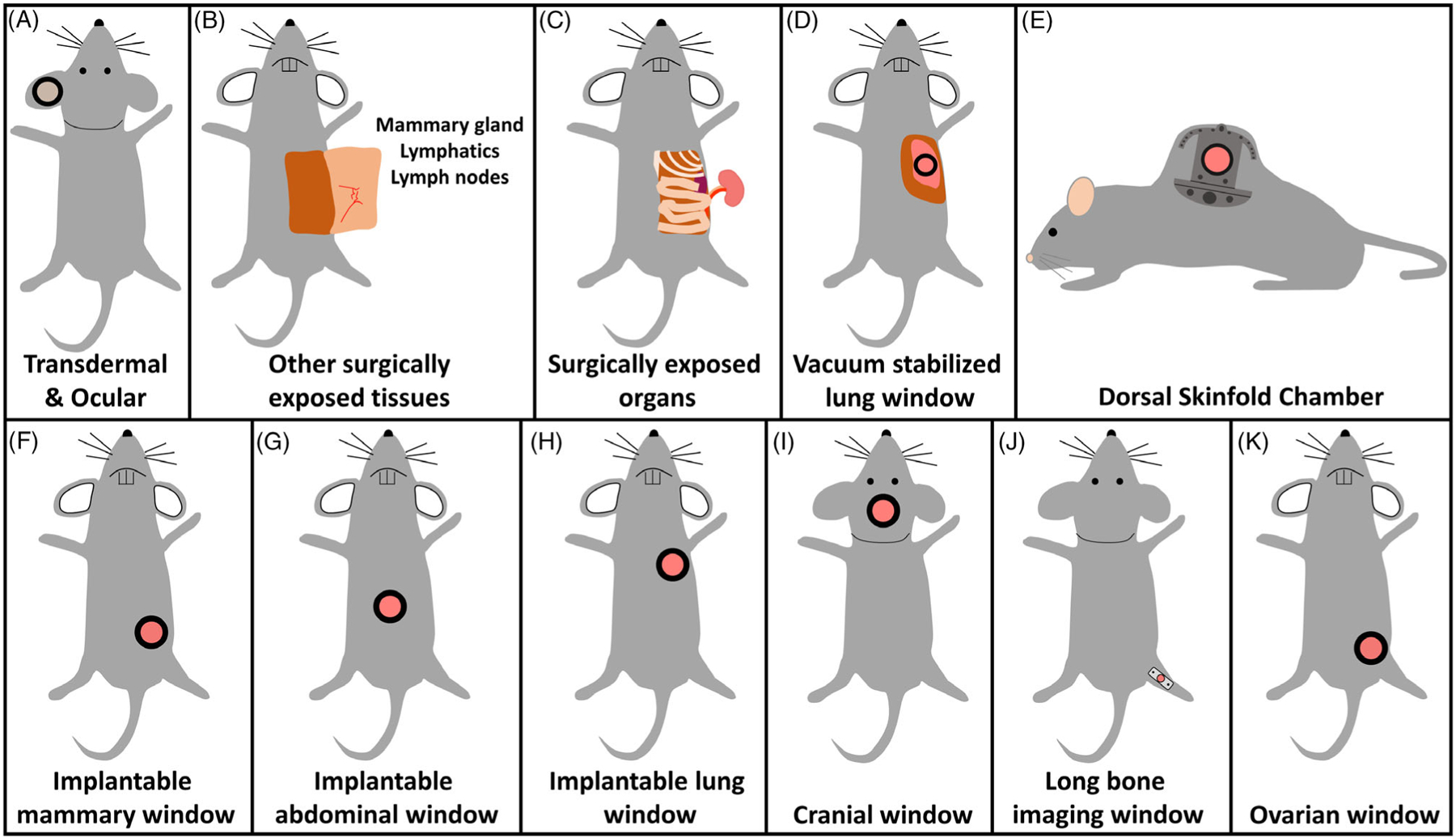
A myriad of techniques is available for accessing tissues with intravital imaging. A, The least invasive methods are transdermal and ocular imaging, which requires no surgical manipulation of the mouse. B-D, Access to deeper tissues requires surgical exposure. Due to their invasive nature, these protocols are almost always terminal. E-K, In recent years a wide number of survival surgical protocols have been developed to access a variety of tissues, including visceral and vital organs, throughout the mouse. These protocols maintain the vitality of the mouse so as to enable repeated imaging sessions.
Transdermal imaging requires no preparation other than affixing a coverslip to the tissue (if a coverslip is required by the objective lens) and immobilizing the region to be imaged. Structures such as the different layers of the skin,(19,20) hair follicles,(18) and blood vessels; individual cells such as platelets and Langerhans cells(21,22); and large proteins and molecules such as thrombin,(23,24) can all be directly visualized over time.
Further, the progression and behavior of diseases such as cancer can be investigated either in mice—using cancer cells directly injected into the epidermis(25) or chemically induced on the tongue(26)—or directly in patients in the clinic.(27) The noninvasive nature of this imaging means that cancer progression and the effect of treatments on individual cells can be tracked over multiple days.(28) Relocalization of the same fields of view can be accomplished by an electromechanical coordinate positioning system that utilizes easily identifiable micro-tattooing marks on the skin as references point to allow relocalization of the imaged areas.(28)
Externalization of Tissues
In contrast to transdermal imaging, other tissues, which lie below the skin, require surgery to expose. Surgical engineering is a field which has typically utilized the expertise of engineers to develop novel instruments, materials, and techniques for the operating room.(29,30) However, lately surgical engineering has been employed in the reverse direction: bringing the skills of the surgeon into the imaging lab.(31,32) This has led to the development of protocols that combine engineering designs with novel surgical techniques, all aimed at increasing the level of sophistication of surgical tissue exposure, while simultaneously reducing the difficulty and necessity for specialized surgical skills for successful application. These protocols have enabled imaging of more tissue over longer periods of time and in more locations than previously possible.(31)
There now exist a number of surgical techniques, of varying degrees of sophistication, to accomplish intravital imaging in many different mouse organs and tissues.(33) These procedures can be classified as terminal (Fig. 3) or survival surgeries (Fig. 3), depending on the invasiveness of the operation and ability of the animal to recover after one imaging session.
Terminal Surgeries
The simplest of the terminal procedures is the skin flap (Fig. 3). This method involves exposure of the tissue of interest by means of a simple incision through the overlying tissue and externalization of the tissue to be imaged. The technique has been used to image mammary tumors,(13) inguinal lymph nodes,(25,34) the cremaster muscle,(35) and salivary gland.(26) Although the skin flap is a terminal surgical procedure (due to its invasiveness and poor recovery), extended imaging sessions are possible with one group having imaged mammary tumors for up to 40 h.(36) The skin flap technique has been used to capture time-lapse movies of processes such as gene expression, drug infiltration, cancer cell death,(37) and stromal and tumor cell migration in the mammary gland.(38–40)
Another use of the skin flap is to visualize the dynamics of cells (e.g. cellular migration and translocation) within intact lymph nodes such as the inguinal(34) and popliteal nodes.(41) These techniques have successfully exposed the tissues, while still maintaining the integrity of the delicate nodes and lymphatic vessels, and have measured parameters such as the speed of migration of lymphocytes within B cell follicles.
For tissues deeper than subcutaneous, externalization protocols have enabled imaging of internal organs such as the kidney,(42,43) liver,(44–46) and the mesentery(47) (Fig. 3). This is accomplished with either a vertical left flank incision, giving access to the kidney, spleen, and pancreas, or a horizontal incision just below the level of the xyphoid process for accessing the liver.(48) Stable IVI using kidney externalization has been utilized to look at cell vitality, apoptosis, fluid transport, receptor-mediated endocytosis, blood flow, and leukocyte trafficking.(43)
In the liver, real-time images were captured during chemotherapy treatment to visualize the dynamic interaction between metastatic tumor cells and host stromal cells as they respond to chemotherapy.(44) In the intestinal tract, investigators have been able to probe the lumen, serosal Peyer’s patches, and the villi of the murine intestinal tract to study several pathological conditions such as inflammatory bowel disease, celiac disease, and colorectal cancer.(49)
Given the high-resolution images that multiphoton and confocal microscopy generate, it would seem impossible to image perpetually moving organs such as the lung and the heart. However, several techniques have been developed to either stabilize these tissues or to take advantage of their pre-dictable cyclic motion. The application of vacuum suction to immobilize lung tissue dates back to 1939(50) but has recently been miniaturized for use with the murine lung(51) (Fig. 3), and further applied to the murine heart.(52) Besides vacuum stabilization, motion artifacts can additionally be minimized utilizing high-speed gated imaging (where images are acquired at frame rates much faster than the motion of the tissue) and triggering acquisition in synchrony with the cyclic motion of the tissue.(52,53) Regularization of the breathing rate by the use of mechanical ventilation (a necessity since these techniques breach the thoracic cavity) aids in this process.
Survival Surgeries
As mentioned, the high level of invasiveness and poor recovery of skin flap and externalization surgeries make them terminal. Thus, with very few exceptions,(54) serial imaging is not possible with these techniques. To address this, a number of protocols have been developed aimed at preserving the vitality of the animal and minimizing the impact on the imaged tissue (Fig. 3–3).
Skinfold chamber
One of the oldest and most widely used of these protocols is the skinfold chamber (Fig. 3): a technique which was originally developed for the rabbit ear(55) and then subsequently adapted for use in the dorsal skin of mice.(56) The most commonly used design of the dorsal skinfold chamber consists of two pieces, a rigid backing designed to keep the skin straight and taut, and a front cover piece, which contains an aperture that accepts a cover glass, most often with a retaining ring, which allows the glass to be removed and replaced repeatedly. Implantation involves forming a skinfold from the depilated dorsal skin of the mouse, suturing in place the ridged backing and the front piece, and then surgically removing the skin from the region under the aperture. Finally, the exposed epidermis is covered over by the placement of a cover glass. After implantation, direct and repeated observation of the epidermis under the cover glass is possible over a period of weeks. In addition, since the coverslip may be removed, foreign materials, cells, or tissues may be introduced into the chamber and their impact upon the epidermis visualized over time. Studies using these windows have focused on angiogenesis,(57) biocompatibility of materials,(58–60) wound healing,(61) and tumor cell growth and invasion.(62)
It is important to note that an important, often over-looked, limitation of the skinfold chamber for cancer studies is that very few cancers are naturally found within the epidermis. This is problematic because ectopically grown cancer cells have been observed to show gene expression patterns that are different from those grown orthotopically.(63)
Mammary imaging window
The mammary imaging window (MIW, Fig. 3) was developed to eliminate the limitations of the terminal skin flap and dorsal skin fold chamber techniques and allow serial visualization the mammary gland and orthotopic mammary tumors at high-resolution and over a period of time spanning days to weeks.(64) In this protocol, a small incision is made through skin and a window frame containing a cover glass is sutured in place to reseal the incision. This window, combined with the photoconvertable protein Dendra2(65), has allowed fate mapping and tracking of tumor cells from primary tumors to secondary sites such as the lung.(66,67)
Lymph node imaging window
To address the limitations of the skin flap procedure, a chronic inguinal lymph node window (CLNW) was developed to provide serial access to this important immunological tissue. The CLNW surgical protocol closely follows the surgical procedure for either the mammary imaging window or the dorsal skin fold window and involves localization of the murine lymph node with a tracer dye (such as Evan’s Blue) followed by removal of the skin and subcutaneous fat (which are bluntly dissected), thus exposing the lymph node. After implantation of the window, placement of a glass coverslip re-seals the surgical site. This procedure is well tolerated without any significant physiologic changes in the mouse 14 days after CLNW implantation.(68)
Abdominal imaging window
To access tissues deeper than the mammary gland, the abdominal imaging window (AIW, Fig. 3) can be used to visualize distinct biological processes in the visceral organs such as the spleen, kidney, small intestine, pancreas, and liver. (69) Utilizing a similar design and implantation protocol to the mammary imaging window, the AIW is implanted by making a small incision through both the skin and the abdominal wall and then affixing the window frame in place with a purse string suture. Different visceral organs can be targeted for imaging by repositioning them to the center of the window’s aperture and affixing them in place with adhesive.
Originally used to visualize, at a subcellular resolution, the behavior of metastatic colon carcinoma cells during out-growth from single cells to micro-metastases in the liver, the window has become a commonly employed tool. It has also been used to observe tumor cell fragmentation, condensation and swelling, and intracellular vacuoles of metastatic colorectal tumor cells after chemotherapy treatment of 5-FU or CPT-11.(44)
Lung imaging window
While the vacuum-stabilized lung window has allowed researchers the ability to image the arrival, proliferation, and motility of metastatic circulating tumor cells in the lung with subcellular resolution,(70) ultimately, the window involves a terminal procedure and does not allow for serial imaging of the murine lung. To address this need, a permanent window for high-resolution imaging of the lung (WHRIL, Fig. 3) was developed.(32) The implantation protocol for this window involves a small incision (~5 mm) through the skin, the underlying muscle, and the ribcage, exposing the lung tissue. This is followed by insertion of the window, which, through a combination of sutures and adhesive, reseals the thoracic cavity and allows the mouse to breathe independently. Thus, the WHRIL allows for serial imaging for up to 2 weeks without adverse effects to mouse. Utilization of the aforementioned coordinate transformation technique(28) allows return to selected areas of interest, which can then be imaged repeatedly over time.
Cranial imaging windows
Imaging of the cortex of the murine brain can be accomplished with two different cranial imaging windows (CIWs, Fig. 3). The first is known as a thinned-skull CIW, where the thickness of the skull is reduced by grinding the bone away down the dura matter with a dental drill. The thinned bone is then protected by adherence of a cover glass to the exposed bone using either cyanoacrylate or dental adhesive. This procedure, which remains stable for months, reduces light scattering from irregularities on the bone surface, inhibits bone regrowth, and allows deep imaging into the brain tissue. (71–73) The second method is the chronic CIW. In this procedure, the cortex is exposed through a craniotomy, which is then resealed using a cover glass.(74) While both methods have gained adoption in the field, some concern about the chronic CIW method has been raised as it has been linked with a higher spinal turnover and significant glial cell activation for up to one month postoperation.(75) Importantly, these windows have been used in both anesthestized and awake mice.(76–78) Finally, protocols have been developed to allow the removal of the coverglass, which dramatically extends the usable duration of the window and allows the implantation of cells, or even micro-prisms, after the initial surgery.(78)
Cerebellar windows
Closely related to the cranial imaging windows is the cerebellar imaging window.(79) While the CIW gives access to the cortex, a common site of growth of glioblastoma and breast cancer metastasis, the cerebellar window is positioned over the brainstem and can reveal the dynamics and tumor-stromal interactions that occur with medulloblastoma. Prior to this window, imaging-based investigations of medulloblastoma were performed with comparatively low-resolution MRI and focused upon measuring tumor volume.(80) With this window, direct high-resolution imaging of changes in tumor vasculature over time demonstrated the supportive role of placental growth factor in the maintenance of medulloblastoma.(81)
Spinal cord imaging
The spinal cord is a unique organ within the body and is of particular interest to researchers investigating the mechanisms underlying spinal cord injury and repair. However, the spinal cord is also a particularly challenging organ to image; its proximity to the heart and lungs make it very susceptible to motion artifacts. Early studies performed either single imaging sessions, or repeatedly exposed the cord with a surgery per imaging session.(82–85) In response, chronic imaging windows were developed that allow repeated access to this tissue for intravital imaging for periods up to months.(86–88)
Long bone imaging window
Given that the bone marrow is one of the major sites of cancer metastasis, some groups have attempted to utilize the thinned CIW to look at the calvarium.(89) Unfortunately, this region of the bone marrow, while easily accessible, is not a common site of bone metastasis. Thus, a novel technique was recently developed to achieve long-term imaging in the long bone using multi-photon imaging through a gradient refractive index (GRIN) microendoscopic lens permanently implanted into the bone marrow of the murine femur (Fig. 3). This approach, known as LIMB (longitudinal intravital imaging of the bone marrow) allowed researchers, for the first time, to observe, at a subcellular level, vascular remodeling, lymphocyte maturation, and hematopoietic cell-stromal cell interactions in the diaphyseal and metaphyseal regions of the femur.(90) The LIMB could offer valuable insights into cancer cell and immune interactions within the bone marrow.
Ovarian imaging window
Finally, the ovary, an important reproductive organ, has also been accessed with stable, long-term imaging windows (Fig. 3). The ovarian imaging window was developed to study the maturation of the ovarian follicle in response to gonadotropin analogues, as well as tumor invasion into the ovary.(91)
Intravital Imaging Leads to New Biological Insights
All of the techniques outlined above have provided biomedical researchers in a number of fields with the unparalleled ability to collect real-time in-vivo information about the dynamic processes that cells and tissues undergo in their native environs.
Homing of stem cells and leukocytes
By removing the spatial and temporal sampling restrictions imposed by fixed tissue or low-resolution imaging techniques (e.g. bioluminescence), intravital imaging through a cranial imaging window allowed direct visualization of the arrival and engraftment of circulating cells into the calvarial bone marrow. By tracking fluorescently labeled cells over extended periods of time, Sipkins et al. found that the bone marrow contains unique and spatially limited anatomic regions of vasculature, which serve as attraction points for both circulating leukemia cells and for healthy hematopoietic stem/progenitor cells.(92) Combining these imaging studies with in vivo immunolabeling of cell surface antigens, the group was able to determine that the locations of cell arrest corresponded to hot spots of stromal-cell-derived factor 1 (SDF-1) overexpression, thus establishing the molecular basis for cellular arrest in these locations as these cells express CXCR4, the receptor for SDF-1.
Blood glucose metabolism
IVI has also been used to understand the regulation of blood flow in pancreatic islets, the mechanism of which has been poorly understood due to the difficulty of accessing the living pancreas. Blood flow to the pancreatic islet is vital in its main function to control blood glucose concentration. Using an in vivo platform in non-human primates, Diez et al. transplanted islets autologously into the anterior chamber of the eye and monitored them non-invasively and longitudinally at single-cell resolution. Their investigation found that blood flow to islets is dynamic and is controlled by a vasoconstriction of the capillaries, potentially under the control of autonomic innervation.(93) This study also demonstrated no changes in velocity with glucose or the GLP-1 analog, liraglutide; in contrast to the findings of previous studies in the rodent pancreatic model.(94)
Breast cancer cell dissemination
Intravital imaging of autochthonous models of cancer revealed that perivascular macrophages interact with tumor cells and endothelial cells to create dynamic, transient openings in neoangiogenic vessels through which high-molecular weight molecules, and most importantly, motile tumor cells can pass to access the blood vasculature and spread hematogenously. (95) These doorways, termed the Tumor microenvironment of Metastasis (TMEM), have been shown to correlate with distant metastasis, and have been clinically validated as a biomarker for distant metastasis in breast cancer patients.(96)
Early steps of the metastatic cascade in secondary sites
Immediately after dissemination from primary tumors, cancer cells arrive to capillary beds within secondary sites and progress through several steps, ultimately leading to the formation of macro-metastatic lesions. However, the exact nature and sequencing of these steps were controversial until their direct visualization with intravital imaging. Two studies, one in the brain(97) using a cranial imaging window, and one in the lung(32) using a newly developed implantable lung-imaging window, were able to surveil the respective tissues over extended periods of time and track disseminated cells as they arrived to the tissue vasculature, extravasated into the parenchyma, and reinitiated growth. Further, visualization of the formation of functional TMEM structures in the lung indicates that re-dissemination from metastatic lesions may be an important source of circulating tumor cells, even after resection of a primary tumor.
These are just a few of the many examples of how intravital imaging is capable of providing insights that were not possible to be attained using static or low-resolution methods.
Intravital Imaging as a Clinical Tool
Intravital imaging is a powerful tool that has not only provided new insights in translational research, but has also become an important clinical tool in its own right with several groups pushing the application and adoption of IVI directly into the clinical setting. These efforts have focused upon using in vivo imaging for (1) diagnosis and surveillance of pathologic pre-cancerous and cancerous lesions (comparable to a traditional tissue biopsy), (2) as a guide for directing standard biopsies in sensitive tissues, and (3) as a rapid pathology tool during Mohs surgery.
The standard of care for diagnosis of cancerous lesions involves removal of the tissue through a biopsy followed by preparation of the tissue for histological analysis. This preparation involves numerous chemical processing steps starting with fixation: chemical cross-linking of proteins to preserve the tissue. Next the tissue may either be rapidly frozen in liquid nitrogen and embedded in optimum cutting temperature (OCT) compound(98) or dehydrated and embedded in paraffin wax.(99,100) Thin sectioning on a cryotome or micro-tome, respectively, produces mechanical sections, which are then stained with a combination of dyes and/or chromogen-labeled antibodies. Each of these steps takes a considerable amount of time and is susceptible to artifacts such as folds, tears, and retractions which could potentially alter the final tissue assessment.
In response to these issues, researchers have pushed forward the adoption and use of reflectance confocal microscopy (RCM) as a non-invasive imaging technique in place of biopsy and mechanical sectioning for the diagnosis of cancerous lesions. Used for the morphological assessment of the dermis and the dermal-epidermal junction (DEJ), it has been shown to be a highly effective tool in diagnosing skin cancer with high levels of specificity and sensitivity.(101)
Both reflection and fluorescence confocal microscopy have also been utilized for the analysis of tissues deeper than the DEJ by utilizing endoscopes instead of standard objective lenses. Known as confocal laser endomicroscopy (CLE), this technique was developed to obtain “optical biopsies,” high-resolution histologic analyses of targeted tissues, in vivo and in real time, during endoscopy.(102) Currently, there are two FDA-approved CLE platforms available for clinical use; the integrated endoscope confocal microscope (iCLE) and the probe-based confocal laser endomicroscope (pCLE).(103) iCLE utilizes a fiber-optic cable to convey blue laser light to a miniaturized confocal microscope that is integrated into the 12-mm-diameter tip of an endoscope, allowing the capture of videoendoscopic and endomicroscopic images, which are then displayed simultaneously on two monitors. Alternatively, pCLE utilizes a standard endoscope connected by a fiber-optic probe bundle to a standard confocal microscope situated outside the patient.(103) Both the iCLE and pCLE platforms allow for collection of an optical section without interference from out-of-plane light.(104) An increase in contrast can also be attained through the use of exogenous contrast agents that can be applied topically or intravenously.(104)
Clinically, pCLE has been used to obtain targeted biopsies and has been shown to reduce the number of physical biopsies needed and increase the accuracy of the procedure, even with endoscopists who have limited experience with the tool.(105) The technology has played a critical role in the diagnosis and surveillance of malignant transformation in Barrett’s esophagus where it has been found to be a reliable modality to identify different epithelial cell types at the squamocolumnar junction. Several studies have clinically validated the use of this technique and demonstrated high interobserver agreement as well as accuracy in pathological diagnosis (e.g. high-grade dysplasia of the esophagus and adenocarcinoma in Barrett’s esophagus(106,107)). Fluorescein-aided pCLE was also shown to be a reliable method with extremely high levels of sensitivity and specificity for diagnosis of Barrett mucosa and low-or high-grade dysplasia, using traditional histology as the gold standard.(108)
Beyond the esophagus, pCLE has been used for accurate detection of other diseases such as gastric carcinoma,(109–111) helicobacter pylorus infection,(112) celiac disease,(113) and inflammatory bowel diseases (IBD) such as colitis and Crohn’s disease.(108,114) In IBD, pCLE has been shown to improve clinical outcome during surveillance of patients by detecting colitis and Crohn’s disease-related mucosal inflammation. (108,114) pCLE has also been demonstrated to markedly improve identification of mucosal alterations in ulcerative colitis where a 4.75-fold higher neoplasia detection rate (over conventional colonoscopy) has been attained through the use of methylene blue staining of the tissue (chromoendoscopy).(115) In non-IBD patients, pCLE has been found to differentiate neoplastic from non-neoplastic colonic mucosa and to have a higher sensitivity and similar specificity to histopathology when classifying colorectal polyps (66–91% and 77–83%, respectively).(116,117) CLE has been used for diagnosis of other solid organ diseases such as the liver by identifying the tissue microarchitecture associated with liver disease(118) and increasing the sensitivity of detection of biliary neoplasia by differentiating benign from malignant biliary strictures.(119)
Finally, needle-based confocal laser endomicroscopy (nCLE) has recently been utilized during endoscopic ultrasound-guided fine-needle aspiration (EUS-FNA) of the pancreas. This new technology is enabling real-time imaging of pancreatic cysts to determine the need for possible surgical resection of pre-malignant cystic neoplasms.(120)
In addition to direct in vivo imaging, confocal microscopy has been making a significant clinical impact as a rapid pathology tool during Mohs surgery. Mohs surgery is the standard of care for non-melanoma skin cancers (e.g. basal cell carcinoma and squamous cell carcinoma) and allows surgeons to obtain clear tumor margins, while maximizing normal tissue preservation. Preservation of normal tissue is especially important when operating on cosmetically sensitive areas such as the face, where excessive tissue removal leads to disfigurement. In the standard surgical procedure, precise tumor clearance during Mohs surgery is assured by sequentially excising thin layers and analyzing the surface of the excised tissue for the presence of residual cancer with histopathology. The use of histopathology for analysis means that the initial operation is followed by a time-consuming tissue preparation process (freezing, sectioning, and staining) and analysis by a pathologist. If the excised tissue margin is deter-mined to contain residual cancer, then surgery is resumed and the process repeated until margins are deemed clear of residual cancer. During this time, the patient must wait with an open wound throughout the whole process, which may take from one to several hours. Beyond the inconvenience of an extended surgery time, the analysis is also susceptible to complications by other factors including embedding the tissue in the wrong orientation and various sectioning artifacts (e.g. as folds, tears, retractions), which could alter the final margin assessment.(5,121)
In response to these difficulties, researchers have designed microscopes specifically for use in patient care that are capable of confocal mosaicing microscopy (CMM). CMM enables rapid imaging of large areas of freshly excised tissue, at single cell resolution, and without the processing that is necessary for conventional histology. This dramatically reduces the cycle time for Mohs surgery, allows surgery to proceed to completion without interruption, and eliminates tissue processing artifacts. CMM has now been clinically validated and is currently embedded in clinical service at several institutions as an adjunct for Mohs surgeons to determine the correct depth of surgery in real-time.(122) The success of CMM in Mohs surgery has further fueled interest in applying this technique to other surgical procedures such as on the breast, lung, kidney, and liver.(123)
CONCLUSIONS
While space limitations prevent an exhaustive review of every intravital imaging technique in the literature, in this review, we have endeavored to describe many of the numerous protocols that have been developed over the years and that have made intravital imaging a versatile and invaluable tool for biomedical research.
While IVI cannot replace other techniques such as fixed tissue, FACS, or in vitro assays, it does provide a powerful complementary role, overcoming many of limitations of these end-point assays. The ability of IVI to non-destructively visualize cells and tissues, with single cell resolution, and over an exceedingly wide range of temporal and spatial scales, means that biological mechanisms can be tested while connections with the rest of the organism (e.g. circulatory system, endocrine system, etc.) are maintained.
Further, advances in commercial microscopes and developments in methodology over the past two decades have simplified IVI protocols and enabled access to a wide range of tissues throughout the body.(31) This has dramatically reduced barriers to entry for laboratories interested in under-taking IVI experiments.
Finally, IVI is becoming an important tool not only for researchers, but also for patient care with new microscopes and techniques being employed directly in the clinical care of patients. We expect that the coming decade will prove to be exceptionally fruitful as the techniques outlined in this review are utilized to definitively answer biological questions, resolve outstanding controversies, and improve health care for patients.
ACKNOWLEDGMENTS
This work was supported by the NIH grants CA216248, 1S10OD019961, and Montefiore’s Ruth L. Kirschstein T32 Training Grant of Surgeons for the Study of the Tumor Microenvironment (CA200561) and the Gruss-Lipper Biophotonics Center and its Integrated Imaging Program.
Grant sponsor: National Cancer Institute, Grant numberCA200561, Grant numberCA216248; Grant sponsor: NIH Office of the Director, Grant number1S10OD019961
Footnotes
CONFLICTS OF INTEREST
There are no conflicts of interest to report.
REFERENCES
- 1.Pittet MJ, Weissleder R. Intravital imaging. Cell 2011;147:983–991. 10.1016/j.cell.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masedunskas A, Porat-Shliom N, Tora M, Milberg O, Weigert R. Intravital microscopy for imaging subcellular structures in live mice expressing fluorescent proteins. J Vis Exp 2013;79:e50558 10.3791/50558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandamme TF. Use of rodents as models of human diseases. J Pharm Bioallied Sci 2014;6:2–9. 10.4103/0975-7406.124301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ericsson AC, Crim MJ, Franklin CL. A brief history of animal modeling. Mo Med 2013;110:201–205. [PMC free article] [PubMed] [Google Scholar]
- 5.Malvehy J, Pellacani G. Dermoscopy, confocal microscopy and other non-invasive tools for the diagnosis of non-melanoma skin cancers and other skin conditions. Acta Derm Venereol 2017;218:22–30. 10.2340/00015555-2720. [DOI] [PubMed] [Google Scholar]
- 6.Chandler DE, Roberson RW. Bioimaging: Current concepts in light and electron microscopy. Sudbury, Massachusetts: Jones & Bartlett Publishers, 2009. [Google Scholar]
- 7.Chatterjee S. Artefacts in histopathology. J Oral Maxillofac Pathol 2014;18: S111–S116. 10.4103/0973-029X.141346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trusk TC. 3D Reconstruction of Confocal Image Data In: Gray JW, Price RL, editors Basic Confocal Microscopy. 2nd ed. Cham, Switzerland: Springer Nature; 2018;p. 279–307. [Google Scholar]
- 9.Minsky M. Memoir on inventing the confocal scanning microscope. Scanning 1988;10:128–138. [Google Scholar]
- 10.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science 1990;248:73–76. [DOI] [PubMed] [Google Scholar]
- 11.Sheppard CJ, Kompfner R. Resonant scanning optical microscope. Appl Opt 1978; 17:2879–2882. 10.1364/AO.17.002879. [DOI] [PubMed] [Google Scholar]
- 12.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: Multiphoton microscopy in the biosciences. Nat Biotechnol 2003;21:1369–1377. 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 13.Wyckoff J, Gligorijevic B, Entenberg D, Segall J, Condeelis J. High-resolution multiphoton imaging of tumors in vivo. Cold Spring Harb Protoc 2011;2011: 1167–1184. 10.1101/pdb.top065904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouzounov DG et al. In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nat Methods 2017;14:388–390. 10.1038/nmeth.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Helmchen F, Denk W. New developments in multiphoton microscopy. Curr Opin Neurobiol 2002;12:593–601. [DOI] [PubMed] [Google Scholar]
- 16.Alt C, Runnels JM, Mortensen LJ, Zaher W, Lin CP. In vivo imaging of microglia turnover in the mouse retina after ionizing radiation and dexamethasone treatment. Invest Ophthalmol Vis Sci 2014;55:5314–5319. 10.1167/iovs.14-14254. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez JM Jr, Hamm-Alvarez S, Tan JC. Analyzing live cellularity in the human trabecular meshwork. Invest Ophthalmol Vis Sci 2013;54:1039–1047. 10.1167/iovs.12-10479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pineda CM et al. Intravital imaging of hair follicle regeneration in the mouse. Nat Protoc 2015;10:1116–1130. 10.1038/nprot.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.New K et al. In vivo imaging of human teeth and skin using real-time confocal microscopy. Scanning 1991;13:369–372. [Google Scholar]
- 20.Rajadhyaksha M, Grossman M, Esterowitz D, Webb RH, Anderson RR. In vivo confocal scanning laser microscopy of human skin: Melanin provides strong contrast. J Invest Dermatol 1995;104:946–952. [DOI] [PubMed] [Google Scholar]
- 21.Vishwanath M et al. Development of intravital intermittent confocal imaging system for studying Langerhans cell turnover. J Invest Dermatol 2006;126:2452–2457. 10.1038/sj.jid.5700448. [DOI] [PubMed] [Google Scholar]
- 22.Kissenpfennig A, Malissen B. Langerhans cells—revisiting the paradigm using genetically engineered mice. Trends Immunol 2006;27:132–139. 10.1016/j.it.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Goh CC et al. Real-time imaging of dendritic cell responses to sterile tissue injury. J Invest Dermatol 2015;135:1181–1184. 10.1038/jid.2014.506. [DOI] [PubMed] [Google Scholar]
- 24.Honkura N et al. Intravital imaging-based analysis tools for vessel identification and assessment of concurrent dynamic vascular events. Nat Commun 2018;9:2746 10.1038/s41467-018-04929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Entenberg D et al. Multimodal microscopy of immune cells and melanoma for longitudinal studies. Proc. SPIE 2006;6081:62–73. [Google Scholar]
- 26.Amornphimoltham P, Thompson J, Melis N, Weigert R. Non-invasive intravital imaging of head and neck squamous cell carcinomas in live mice. Methods 2017; 128:3–11. 10.1016/j.ymeth.2017.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimitrow E et al. Sensitivity and specificity of multiphoton laser tomography for in vivo and ex vivo diagnosis of malignant melanoma. J Invest Dermatol 2009;129: 1752–1758. 10.1038/jid.2008.439. [DOI] [PubMed] [Google Scholar]
- 28.Dunphy MP, Entenberg D, Toledo-Crow R, Larson SM. In vivo microcartography and subcellular imaging of tumor angiogenesis: A novel platform for translational angiogenesis research. Microvasc Res 2009;78:51–56. 10.1016/j.mvr.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kishikova L, Norris JM, Smith MD. Engineering the future of surgery: The place of the surgical engineering faculty. Surgery 2013;153:135 10.1016/j.surg.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 30.Riskin DJ, Longaker MT, Gertner M, Krummel TM. Innovation in surgery: A historical perspective. Ann Surg 2006;244:686–693. 10.1097/01.sla.0000242706.91771.ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Entenberg D et al. Time-lapsed, large-volume, high-resolution intravital imaging for tissue-wide analysis of single cell dynamics. Methods 2017;128:65–77. 10.1016/j.ymeth.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Entenberg D et al. A permanent window for the murine lung enables high-resolution imaging of cancer metastasis. Nature Methods 2018;15:73–80. 10.1038/nmeth.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alieva M, Ritsma L, Giedt RJ, Weissleder R, van Rheenen J. Imaging windows for long-term intravital imaging: General overview and technical insights. Dermatol Int 2014;3:e29917 10.4161/intv.29917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MJ, Wei SH, Parker I, Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science 2002;296:1869–1873. 10.1126/science.1070051. [DOI] [PubMed] [Google Scholar]
- 35.Morris VL, MacDonald IC, Koop S, Schmidt EE, Chambers AF, Groom AC. Early interactions of cancer cells with the microvasculature in mouse liver and muscle during hematogenous metastasis: Videomicroscopic analysis. Clin Exp Metastasis 1993;11:377–390. [DOI] [PubMed] [Google Scholar]
- 36.Nakasone ES et al. Imaging tumor-stroma interactions during chemotherapy reveals contributions of the microenvironment to resistance. Cancer cell 2012;21: 488–503. 10.1016/j.ccr.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egeblad M et al. Visualizing stromal cell dynamics in different tumor microenvi-ronments by spinning disk confocal microscopy. Disease Models Mech 2008;1: 155–167. 10.1242/dmm.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ewald AJ et al. Mammary collective cell migration involves transient loss of epithelial features and individual cell migration within the epithelium. J Cell Sci 2012; 125:2638–2654. 10.1242/jcs.096875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farina KL, Wyckoff JB, Rivera J, Lee H, Segall JE, Condeelis JS, Jones JG. Cell motility of tumor cells visualized in living intact primary tumors using green fluorescent protein. Cancer Res 1998;58:2528–2532. [PubMed] [Google Scholar]
- 40.Entenberg D et al. in Curr Protoc Cell Biol Vol. Chapter 19 Unit19 17 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liou HL, Myers JT, Barkauskas DS, Huang AY. Intravital imaging of the mouse popliteal lymph node. J Visualized Experiments: JoVE 2012:e3720 10.3791/3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hato T, Winfree S, Dagher PC. Intravital imaging of the kidney. Methods 2017; 128:33–39. 10.1016/j.ymeth.2017.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dunn KW, Sutton TA, Sandoval RM. Live-animal imaging of renal function by multiphoton microscopy. Curr Protoc Cytom 2007;41:12.9.1–12.9.18. 10.1002/0471142956.cy1209s41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka K et al. In vivo real-time imaging of chemotherapy response on the liver metastatic tumor microenvironment using multiphoton microscopy. Oncol Rep 2012;28:1822–1830. 10.3892/or.2012.1983. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka K et al. Intravital dual-colored visualization of colorectal liver metastasis in living mice using two photon laser scanning microscopy. Microsc Res Tech 2012; 75:307–315. 10.1002/jemt.21059. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka K et al. In vivo time-course imaging of tumor angiogenesis in colorectal liver metastases in the same living mice using two-photon laser scanning microscopy. J Oncol 2012;2012:265487 10.1155/2012/265487(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh T, Yaegashi K, Kosaka T, Kinoshita T, Morimoto T. A closed chamber for intravital microscopy of the rat mesentery under controlled oxygen tension and temperature. Jpn J Physiol 1993;43:847–854. [DOI] [PubMed] [Google Scholar]
- 48.Rhodes GJ. Surgical preparation of rats and mice for intravital microscopic imaging of abdominal organs. Methods 2017;128:129–138. 10.1016/j.ymeth.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolesnikov M, Farache J, Shakhar G. Intravital two-photon imaging of the gastrointestinal tract. J Immunol Methods 2015;421:73–80. 10.1016/j.jim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Terry RJ. A thoracic window for observation of the lung in a living animal. Science 1939;90:43–44. 10.1126/science.90.2324.43. [DOI] [PubMed] [Google Scholar]
- 51.Looney MR et al. Stabilized imaging of immune surveillance in the mouse lung. Nat Methods 2011;8:91–96. 10.1038/nmeth.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vinegoni C, Lee S, Gorbatov R, Weissleder R. Motion compensation using a suctioning stabilizer for intravital microscopy. Dermatol Int 2012;1:115–121. 10.4161/intv.23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Presson RG Jr et al. Two-photon imaging within the murine thorax without respiratory and cardiac motion artifact. Am J Pathol 2011;179:75–82. 10.1016/j.ajpath.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimura H et al. Real-time imaging of single cancer-cell dynamics of lung metastasis. J Cell Biochem 2010;109:58–64. 10.1002/jcb.22379. [DOI] [PubMed] [Google Scholar]
- 55.Sandison JC. A new method for the microscopic study of living growing tissues by the introduction of a transparent chamber in the rabbit’s ear. Anat Rec 1924;28: 281–287. [Google Scholar]
- 56.Algire GH An Adaptation of the Transparent-Chamber Technique to the Mouse 1943;4:1–11. [Google Scholar]
- 57.Leunig M, Yuan F, Menger MD, Boucher Y, Goetz AE, Messmer K, Jain RK. Angiogenesis, microvascular architecture, microhemodynamics, and interstitial fluid pressure during early growth of human adenocarcinoma LS174T in SCID mice. Cancer Res 1992;52:6553–6560. [PubMed] [Google Scholar]
- 58.Ring A et al. Induction of angiogenesis and neovascularization in adjacent tissue of plasma-collagen-coated silicone implants. Eplasty 2010;10:504–520. [PMC free article] [PubMed] [Google Scholar]
- 59.Laschke MW, Augustin V, Kleer S, Tschernig T, Menger MD. Locally applied macrophage-activating lipopeptide-2 (MALP-2) promotes early vascularization of implanted porous polyethylene (Medpor(R)). Acta Biomater 2014;10:4661–4669. 10.1016/j.actbio.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 60.Laschke MW et al. Vascularisation of porous scaffolds is improved by incorporation of adipose tissue-derived microvascular fragments. Eur Cell Mater 2012;24: 266–277. [DOI] [PubMed] [Google Scholar]
- 61.Sorg H, Krueger C, Vollmar B. Intravital insights in skin wound healing using the mouse dorsal skin fold chamber. J Anat 2007;211:810–818. 10.1111/j.1469-7580.2007.00822.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alexander S, Koehl GE, Hirschberg M, Geissler EK, Friedl P. Dynamic imaging of cancer growth and invasion: A modified skin-fold chamber model. Histochem Cell Biol 2008;130:1147–1154. 10.1007/s00418-008-0529-1. [DOI] [PubMed] [Google Scholar]
- 63.Greene GF, Kitadai Y, Pettaway CA, von Eschenbach A, Bucana CD, Fidler IJ. Correlation of metastasis-related gene expression with metastatic potential in human prostate carcinoma cells implanted in nude mice using an in situ messenger RNA hybridization technique. Am J Pathol 1997;150:1571–1582. [PMC free article] [PubMed] [Google Scholar]
- 64.Shan S, Sorg B, Dewhirst MW. A novel rodent mammary window of orthotopic breast cancer for intravital microscopy. Microvasc Res 2003;65:109–117. [DOI] [PubMed] [Google Scholar]
- 65.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat Biotechnol 2006;24: 461–465. [DOI] [PubMed] [Google Scholar]
- 66.Kedrin D et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Meth 2008;5:1019–1021. 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Entenberg D et al. Setup and use of a two-laser multiphoton microscope for multi-channel intravital fluorescence imaging. Nat Protoc 2011;6:1500–1520. 10.1038/nprot.2011.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jeong HS et al. Investigation of the lack of angiogenesis in the formation of lymph node metastases. J Nat Cancer Institute 2015;107:djv155 10.1093/jnci/djv155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritsma L et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci Transl Med 2012;4: 158ra145 10.1126/scitranslmed.3004394. [DOI] [PubMed] [Google Scholar]
- 70.Entenberg D et al. In vivo subcellular resolution optical imaging in the lung reveals early metastatic proliferation and motility. Intravital 2015;4:1–11. 10.1080/21659087.2015.1086613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Christie RH, Bacskai BJ, Zipfel WR, Williams RM, Kajdasz ST, Webb WW, Hyman BT. Growth arrest of individual senile plaques in a model of Alzheimer’s disease observed by in vivo multiphoton microscopy. J Neurosci 2001;21:858–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang G, Pan F, Parkhurst CN, Grutzendler J, Gan WB. Thinned-skull cranial window technique for long-term imaging of the cortex in live mice. Nat Protoc 2010;5: 201–208. 10.1038/nprot.2009.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shih AY, Mateo C, Drew PJ, Tsai PS, Kleinfeld D. A polished and reinforced thinned-skull window for long-term imaging of the mouse brain. J Visualiz Experimen: JoVE 2012:e3742 10.3791/3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sonntag WE, Lynch CD, Cooney PT, Hutchins PM. Decreases in cerebral microvasculature with age are associated with the decline in growth hormone and insulin-like growth factor 1. Endocrinology 1997;138:3515–3520. 10.1210/endo.138.8.5330. [DOI] [PubMed] [Google Scholar]
- 75.Xu HT, Pan F, Yang G, Gan WB. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci 2007;10:549–551. 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- 76.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron 2007; 56:43–57. 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Andermann ML, Kerlin AM, Reid RC. Chronic cellular imaging of mouse visual cortex during operant behavior and passive viewing. Front Cell Neurosci 2010;4:3 10.3389/fncel.2010.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldey GJ et al. Removable cranial windows for long-term imaging in awake mice. Nat Protoc 2014;9:2515–2538. 10.1038/nprot.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Askoxylakis V et al. A cerebellar window for intravital imaging of normal and disease states in mice. Nat Protoc 2017;12:2251–2262. 10.1038/nprot.2017.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suero-Abreu GA et al. In vivo Mn-enhanced MRI for early tumor detection and growth rate analysis in a mouse medulloblastoma model. Neoplasia 2014;16: 993–1006. 10.1016/j.neo.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Snuderl M et al. Targeting placental growth factor/neuropilin 1 pathway inhibits growth and spread of medulloblastoma. Cell 2013;152:1065–1076. 10.1016/j.cell.2013.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kerschensteiner M, Schwab ME, Lichtman JW, Misgeld T. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat Med 2005;11: 572–577. 10.1038/nm1229. [DOI] [PubMed] [Google Scholar]
- 83.Bareyre FM. Neuronal repair and replacement in spinal cord injury. J Neurol Sci 2008;265:63–72. 10.1016/j.jns.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 84.Dray C, Rougon G, Debarbieux F. Quantitative analysis by in vivo imaging of the dynamics of vascular and axonal networks in injured mouse spinal cord. Proc Natl Acad Sci U S A 2009;106:9459–9464. 10.1073/pnas.0900222106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kawakami N. Intravital imaging of T cells within the spinal cord. Methods Mol Biol 2018;1763:119–127. 10.1007/978-1-4939-7762-8_11. [DOI] [PubMed] [Google Scholar]
- 86.Figley SA et al. A spinal cord window chamber model for in vivo longitudinal multimodal optical and acoustic imaging in a murine model. PLoS One 2013;8:e58081 10.1371/journal.pone.0058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fenrich KK et al. Long-term in vivo imaging of normal and pathological mouse spinal cord with subcellular resolution using implanted glass windows. J Physiol 2012;590:3665–3675. 10.1113/jphysiol.2012.230532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fenrich KK, Weber P, Rougon G, Debarbieux F. Long-and short-term intravital imaging reveals differential spatiotemporal recruitment and function of myelomonocytic cells after spinal cord injury. J Physiol 2013;591:4895–4902. 10.1113/jphysiol.2013.256388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le VH et al. In vivo longitudinal visualization of bone marrow engraftment process in mouse calvaria using two-photon microscopy. Sci Rep 2017;7:44097 10.1038/srep44097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reismann D et al. Longitudinal intravital imaging of the femoral bone marrow reveals plasticity within marrow vasculature. Nat Commun 2017;8:2153 10.1038/s41467-017-01538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bochner F, Fellus-Alyagor L, Kalchenko V, Shinar S, Neeman M. A novel Intravital imaging window for longitudinal microscopy of the mouse ovary. Sci Rep 2015;5: 12446 10.1038/srep12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sipkins DA et al. In vivo imaging of specialized bone marrow endothelial micro-domains for tumour engraftment. Nature 2005;435:969–973. 10.1038/nature03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diez JA et al. Pancreatic islet blood flow dynamics in primates. Cell Rep 2017;20: 1490–1501. 10.1016/j.celrep.2017.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Svensson AM, Ostenson CG, Efendic S, Jansson L. Effects of glucagon-like peptide-1-(7–36)-amide on pancreatic islet and intestinal blood perfusion in Wistar rats and diabetic GK rats. Clin Sci (Lond) 2007;112:345–351. 10.1042/CS20060272. [DOI] [PubMed] [Google Scholar]
- 95.Harney AS et al. Real-time imaging reveals local, transient vascular permeability, and tumor cell intravasation stimulated by TIE2hi macrophage-derived VEGFA. Cancer Dis 2015;5:932–943. 10.1158/2159-8290.CD-15-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sparano JA et al. A metastasis biomarker (MetaSite breast score) is associated with distant recurrence in hormone receptor-positive, HER2-negative early-stage breast cancer. Nat PJ Breast Cancer 2017;3:42 10.1038/s41523-017-0043-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kienast Y et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med 2010;16:116–122. 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- 98.Fischer AH, Jacobson KA, Rose J, Zeller R. Cryosectioning tissues. CSH Protoc 2008;3:2008 10.1101/pdb.prot4991. [DOI] [PubMed] [Google Scholar]
- 99.Ramos-Vara JA. Principles and methods of immunohistochemistry. Methods Mol Biol 2017;1641:115–128. 10.1007/978-1-4939-7172-5_5. [DOI] [PubMed] [Google Scholar]
- 100.Fischer AH, Jacobson KA, Rose J, Zeller R. Paraffin embedding tissue samples for sectioning. CSH Protoc 2008;3:2008 10.1101/pdb.prot4989. [DOI] [PubMed] [Google Scholar]
- 101.Kurugol S et al. Automated delineation of dermal-epidermal junction in reflectance confocal microscopy image stacks of human skin. J Invest Dermatol 2015;135: 710–717. 10.1038/jid.2014.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thekkek N, Anandasabapathy S, Richards-Kortum R. Optical molecular imaging for detection of Barrett’s-associated neoplasia. World J Gastroenterol 2011;17: 53–62. 10.3748/wjg.v17.i1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neumann H, Kiesslich R, Wallace MB, Neurath MF. Confocal laser endomicroscopy: Technical advances and clinical applications. Gastroenterology 2010;139:388–392. 10.1053/j.gastro.2010.06.029. [DOI] [PubMed] [Google Scholar]
- 104.Paull PE, Hyatt BJ, Wassef W, Fischer AH. Confocal laser endomicroscopy: A primer for pathologists. Arch Pathol Lab Med 2011;135:1343–1348. 10.5858/arpa.2010-0264-RA. [DOI] [PubMed] [Google Scholar]
- 105.Nguyen VX, Nguyen CC, De Petris G, Sharma VK, Das A. Confocal endomicroscopy (CEM) improves efficiency of Barrett surveillance. J Interv Gastroenterol 2012;2:61–65. 10.4161/jig.22175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gaddam S et al. Novel probe-based confocal laser endomicroscopy criteria and interobserver agreement for the detection of dysplasia in Barrett’s esophagus. Am J Gastroenterol 2011;106:1961–1969. 10.1038/ajg.2011.294. [DOI] [PubMed] [Google Scholar]
- 107.Wang KK et al. Use of probe-based confocal laser endomicroscopy (pCLE) in gastrointestinal applications. A consensus report based on clinical evidence. United European Gastroenterol J 2015;3:230–254. 10.1177/2050640614566066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kiesslich R et al. In vivo diagnosis of collagenous colitis by confocal endomicroscopy. Gut 2006;55:591–592. 10.1136/gut.2005.084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kiesslich R, Goetz M, Neurath MF. Confocal laser endomicroscopy for gastrointestinal diseases. Gastrointest Endosc Clin N Am 2008;18:451–466. 10.1016/j.giec.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 110.Kitabatake S et al. Confocal endomicroscopy for the diagnosis of gastric cancer in vivo. Endoscopy 2006;38:1110–1114. 10.1055/s-2006-944855. [DOI] [PubMed] [Google Scholar]
- 111.Gunther U et al. Diagnostic value of confocal endomicroscopy in celiac disease.Endoscopy 2010;42:197–202. 10.1055/s-0029-1243937. [DOI] [PubMed] [Google Scholar]
- 112.Kiesslich R, Goetz M, Burg J, Stolte M, Siegel E, Maeurer MJ, Thomas S, Strand D,Galle PR, Neurath MF. Diagnosing helicobacter pylori in vivo by confocal laser endoscopy. Gastroenterology 2005;128:2119–2123. [DOI] [PubMed] [Google Scholar]
- 113.Pohl H et al. Endocytoscopy for the detection of microstructural features in adult patients with celiac sprue: A prospective, blinded endocytoscopy-conventional histology correlation study. Gastrointest Endosc 2009;70:933–941. 10.1016/j.gie.2009.04.043. [DOI] [PubMed] [Google Scholar]
- 114.Tontini GE et al. Prediction of clinical outcomes in Crohn’s disease by using confocal laser endomicroscopy: Results from a prospective multicenter study. Gastrointest Endosc 2018;87:1505–1514. 10.1016/j.gie.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 115.Kiesslich R et al. Chromoscopy-guided endomicroscopy increases the diagnostic yield of intraepithelial neoplasia in ulcerative colitis. Gastroenterology 2007;132: 874–882. 10.1053/j.gastro.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 116.Buchner AM et al. Comparison of probe-based confocal laser endomicroscopy with virtual chromoendoscopy for classification of colon polyps. Gastroenterology 2010; 138:834–842. 10.1053/j.gastro.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 117.Kuiper T et al. New classification for probe-based confocal laser endomicroscopy in the colon. Endoscopy 2011;43:1076–1081. 10.1055/s-0030-1256767. [DOI] [PubMed] [Google Scholar]
- 118.Goetz M et al. In vivo confocal laser endomicroscopy of the human liver: A novel method for assessing liver microarchitecture in real time. Endoscopy 2008;40: 554–562. 10.1055/s-2008-1077296. [DOI] [PubMed] [Google Scholar]
- 119.Meining A et al. Detection of cholangiocarcinoma in vivo using miniprobe-based confocal fluorescence microscopy. Clin Gastroenterol Hepatol 2008;6:1057–1060. 10.1016/j.cgh.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 120.Napoleon B et al. A novel approach to the diagnosis of pancreatic serous cystadenoma: Needle-based confocal laser endomicroscopy. Endoscopy 2015;47: 26–32. 10.1055/s-0034-1390693. [DOI] [PubMed] [Google Scholar]
- 121.Rajadhyaksha M, Menaker G, Flotte T, Dwyer PJ, Gonzalez S. Confocal examination of nonmelanoma cancers in thick skin excisions to potentially guide mohs micrographic surgery without frozen histopathology. J Invest Dermatol 2001;117: 1137–1143. 10.1046/j.0022-202x.2001.01524.x. [DOI] [PubMed] [Google Scholar]
- 122.Jain M, Rajadhyaksha M, Nehal K. Implementation of fluorescence confocal mosaicking microscopy by “early adopter” Mohs surgeons and dermatologists: Recent progress. J Biomed Opt 2017;22:24002 10.1117/1.JBO.22.2.024002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Krishnamurthy S et al. Confocal fluorescence microscopy platform suitable for rapid evaluation of small fragments of tissue in surgical pathology practice. Arch Pathol Lab Med 2019;143:305–313. 10.5858/arpa.2018-0352-OA. [DOI] [PubMed] [Google Scholar]


