Abstract
Background
Tumor infiltrating regulatory T (TITreg) cells are highly infiltrated in gastric cancer (GC) and associated with worse prognosis of GC patients. We aim to develop and validate a radiomics signature for evaluation of TITreg cells and outcome prediction of GC patients.
Methods
A total of 165 GC patients from three independent cohorts were enrolled in this retrospective study. The abundance of TITreg cells were evaluated by using multispectral immunohistochemical analysis and CIBERSORT algorithm. The radiomics features were extracted by using PyRadiomics software and the radiomics signature was generated by using the least absolute shrinkage and selection operator (LASSO) logistic regression model. The receiver operator characteristic (ROC) curves were applied to assess the performance of radiomics signature for estimating TITreg cells. Univariable and multivariable Cox regression analysis were used for identifying risk factor of overall survival (OS). The prognostic value of the radiomics signature and the TITreg cells were evaluated by using the Kaplan-Meier method and log-rank test.
Results
Six robust features were selected for building the radiomics signature. The radiomics signature showed good ability for estimating TITreg in the training, validation and testing cohort, with area under the curve (AUC) of 0.884, 0.869 and 0.847, respectively. Multivariable Cox regression analysis showed that the radiomics signature was an independent risk factor of unfavorable OS of GC patients.
Conclusions
The proposed CT-based radiomics signature is a promising non-invasive biomarker of TITreg cells and outcome prediction of GC patients.
Keywords: Gastric cancer (GC), radiomics, regulatory T cells (Treg cells), immunotherapy
Introduction
Regulatory T (Treg) cells are a subset of CD4+ T cells which distinguished by expression of the transcription factor forehead box protein P3 (FoxP3). Accumulating evidence demonstrated that a large number of Treg cells were infiltrated in tumor microenvironment (1). In addition, most of Treg cells within tumors had high proliferative activity and could lead to profound immunosuppression within tumors. A number of studies showed that higher fraction of Treg cells was associated with worse prognosis of breast cancer (2), non-small-cell lung cancer (3), pancreatic cancer (4), renal cell cancer (5), head and neck squamous cell cancer (6) and gastric cancer (GC) (7). Rapid progression referred to as hyper progression disease (HPD) had been observed in up to 10% of patients received immune checkpoint inhibitor (ICI) of several types of tumors (8,9). Recently, Kamada et al. reported that Treg cells may contribute to the HPD in proportion of GC patients treated with anti-PD-1 therapy (8). These results suggested a potential role for tumor infiltrating Treg (TITreg) cells in prediction of outcome and treatment response of immunotherapies.
Computational medical imaging, known as Radiomics, are processes that translate medical images into high-dimensional quantitative data and thus enable the characterization of microscopic features of tumor tissues such as cellular, molecular and even gene expression properties. In previous studies, radiomics had successful applicated in the differential diagnosis, prognosis or lymph node metastasis prediction of GC patients (10-12). Therefore, the radiomics-based biomarkers could be a novel non-invasive biomarker for diagnosis, prognosis and treatment response of cancer patients and promising complimentary to other gene-driven tumor markers or biopsies.
To our knowledge, there is no previous study that had investigated the value of radiomics for assessing TITreg cells. Thus, the aim of current study is to develop and validate a CT-based radiomics signature for evaluation of TITreg cells in GC. The prognostic value of the radiomics signature was also evaluated.
Methods
Patients and study design
This study was conducted in three independent cohorts. One hundred and thirty-five GC patients who had undergone radical gastrectomy at Tianjin Medical University Cancer Institute and Hospital between June 2012 and April 2016 were enrolled. The patients were randomly divided into training cohort (n=90) and validation cohort (n=45). The inclusion criteria: (I) patients received radical gastrectomy with D2 lymphadenectomy; (II) GC diagnosis was histologically confirmed; (III) contrast-enhanced abdominal CT images were acquired within 14 days before operation; (IV) Image quality was satisfactory for analysis. The exclusion criteria: (I) patients received chemotherapy or radiotherapy before surgery; (II) with other malignant tumors; (III) incomplete clinical information.
The Cancer Genome Atlas (TCGA) Stomach Adenocarcinoma (STAD) dataset was used as an external testing cohort. The CT images of TCGA-STAD dataset were obtained from The Cancer Imaging Archive (TCIA). The RNA-sequencing data and clinical information of patients were acquired from the TCGA portal. The abundance of TITreg cells of each patient was calculated using CIBERSORT web tools (13). All patients of TCGA dataset had undergone radical gastrectomy and the diagnosis of GC were also histologically confirmed.
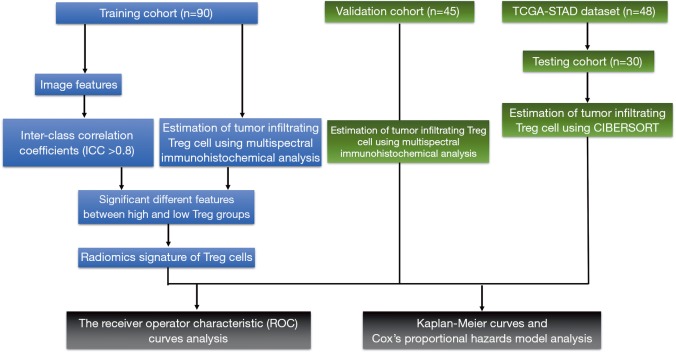
This study was approved by ethics committee of our institute and informed consent was obtained from all the patients. The flowchart of study design is showed in Figure 1.
Figure 1.
Flowchart of overall study design.
CT scanning protocol
Contrast-enhanced abdominal CT was performed by using Discovery CT750 HD (GE Medical Systems, Milwaukee, Wisconsin) or Somatom Sensation 64 scanner (Siemens Medical Solutions, Forchheim, Germany). Oral doses of water (500–1,000 mL) were administered to ensure stomach distension prior to CT examination. The parameters were as follows: tube voltage was 120 kVp, tube current was 150–200 mA; field of view, 350 mm × 350 mm; matrix, 512×512; Images reconstruction section thickness: 1.25 mm. Arterial phase images were obtained following delays of 20 seconds after intravenous injection of contrast media (2.5 mL/s, 1.2 mL/kg; Omnipaque 300, GE Healthcare, Chicago, Illinois) via a syringe pump. The portal venous phase was obtained after delays of 60 seconds.
Multispectral immunohistochemical analysis of TITreg cells
The abundance of TITreg cells was analyzed utilizing Opal 7-Colour Manual IHC Kit (PerkinElmer NEL811001KT) according to the manufacture’s protocol (14). In brief, the slides were incubated with Antibody Diluent blocking buffer (PerkinElmer) at room temperature (RT) for 10 min. The primary antibody for CD4 (Abcam, ab133616, 1:500) and FoxP3 (R&D, MAB8214, 1:400) were incubated at RT for 1 h. Then, a secondary HRP antibody were incubated at RT for 10 min. Signal amplification was performed using Opal 520 TSA (PerkinElmer) and incubated at RT for 10 min. Visualization of the slides was done using the Mantra Quantitative Pathology Imaging System (PerkinElmer) and analyzed using InForm Image Analysis software (PerkinElmer, version 2.1). The TITreg cell fraction in CD4+ cells were calculated and data were presented as mean ± SD.
Estimation of TITreg cells using CIBERSORT algorithm
CIBERSORT is a gene expression-based deconvolution algorithm for assessing immune cell composition. Normalized gene expression data of TCGA-STAD dataset was uploaded to CIBERSORT web portal (13). The proportion of 22 types of intratumoral immune cells, including Treg cells, were evaluated with the default algorithm at 1000 permutations (15).
Tumor segmentation and feature extraction
The portal venous phase of contrast-enhanced CT images was selected for the analysis because most of the lesions had significant enhancement in this phase (16).
The volume of interest (VOI) of tumors were manually drawn along the tumor boundaries by using 3D Slicer software (version 4.8.1). The VOIs were delineated by two radiologists (Reader 1, X Gao; Reader 2, T Ma) with 6 years and 9 years of experience, respectively. Reader 1 performed tumor segmentations for patients of all cohorts. Reader 2 independently segmented tumors of the training cohort. Inter-class correlation coefficients (ICCs) were calculated for evaluation of inter-observer reliability of the extracted features. Feature extraction was performed by using PyRadiomics software (version 2.2.0) (17). Eight hundred and fifty-nine radiomic features were extracted from VOIs. The features were divided into four catalogs including: first-order, shape, texture, and wavelet features. Details of radiomic features are showed in Table S1.
Feature selection and radiomics signature construction
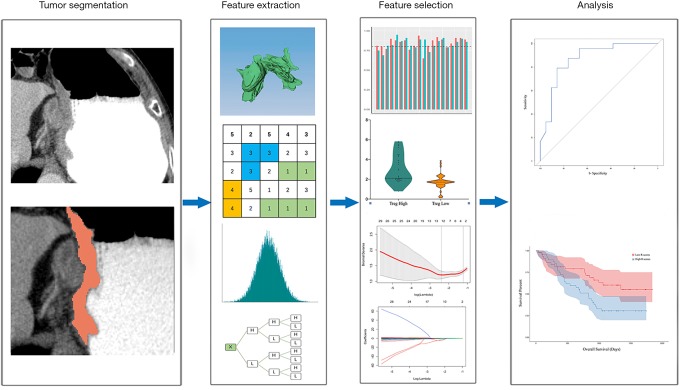
To evaluate the inter-observer reliability of the extracted features, tumors of the training cohort were independently segmented by two radiologists (X Gao and T Ma) and ICCs were calculated for each feature. Only features with ICC ≥0.8 were considered as highly reproducible and retained. Then, the patients were stratifying into high or low Treg cell group on the basis of median value of Treg cell fraction in CD4+ cells. The Mann-Whitney U test was utilized to compare the value of each radiomics feature between high and low TITreg cell groups. The features with P<0.05 were considered as significant different features between two groups and included for further analysis. Finally, the least absolute shrinkage and selection operator (Lasso) regression model was used for feature selection and signature construction (18). The regularization parameter λ was defined by use of 10-fold cross-validation. Lasso logistic regression analysis was done utilizing “glmnet” package of R software. The workflow of radiomics analysis is showed in Figure 2.
Figure 2.
Workflow of radiomics analysis.
Statistical analysis
The t-test, Mann-Whitney U test or one-way ANOVA test was used for numerical variables. χ2 test or Fisher’s exact test was used for categorical variables. The receiver operator characteristic (ROC) curves and the area under the curve (AUC) were utilized to assess the performance of radiomics signature for estimating TITreg cells. Univariable and multivariable Cox regression analysis were used for identifying risk factors of overall survival (OS). Factors with statistical significance at the univariable analysis were analyzed for the multivariable model with step-wise forward approach. The optimal cutoff value of radiomics score was decided with the maximized Youden index from the training cohort and patients were divided into high or low radiomics score group. OS was evaluated using the Kaplan-Meier curve along with log-rank test. P<0.05 was considered as statistical significance. Statistical analysis was performed by using R (version 3.4.2).
Results
Patients characteristics
The clinical characteristics of patients are showed in Table 1. The contrast-enhanced abdominal CT images, RNA-seq data and clinical information of TCGA-STAD dataset were downloaded from TCGA and TCIA data portal. 18 patients were excluded for lacking of RNA-seq data and 30 patients were finally included as the testing cohort. All patients from the three cohorts were pathological diagnosed with gastric adenocarcinoma. No significant difference in clinical covariates was observed among the three cohorts.
Table 1. Characteristics of the study population.
| Variable | Training cohort (n=90), n (%) | Validation cohort (n=45), n (%) | Testing cohort (n=30), n (%) | P |
|---|---|---|---|---|
| Age (years) | 0.346 | |||
| <60 | 36 (40.0) | 19 (42.2) | 8 (26.7) | |
| ≥60 | 54 (60.0) | 26 (57.8) | 22 (73.3) | |
| Gender | 0.261 | |||
| Male | 66 (73.3) | 32 (71.1) | 26 (86.7) | |
| Female | 24 (26.7) | 13 (28.9) | 4 (13.3) | |
| Vital status | 0.564 | |||
| Alive | 35 (38.9) | 19 (42.2) | 15 (50.0) | |
| Dead | 55 (61.1) | 26 (57.8) | 15 (50.0) | |
| Tumor site | 0.987 | |||
| Upper | 21 (23.3) | 12 (26.7) | 6 (20.0) | |
| Middle | 11 (12.2) | 6 (13.3) | 5 (16.7) | |
| Lower | 42 (46.7) | 20 (44.4) | 13 (43.3) | |
| Overlap | 16 (17.8) | 7 (15.6) | 6 (20.0) | |
| Differentiation | 0.881 | |||
| Moderate | 55 (61.1) | 26 (57.8) | 19 (63.3) | |
| Poorly | 35 (38.9) | 19 (42.2) | 11 (36.7) | |
| TNM stage | 0.625 | |||
| I | 6 (6.7) | 2 (4.4) | 1 (3.3) | |
| II | 22 (24.4) | 9 (20.0) | 4 (13.3) | |
| III | 62 (68.9) | 34 (75.6) | 25 (83.3) | |
| T stage | 0.860 | |||
| T1 | 5 (5.6) | 4 (8.9) | 1 (3.3) | |
| T2 | 12 (13.3) | 5 (11.1) | 3 (10.0) | |
| T3 | 14 (15.6) | 4 (8.9) | 5 (16.7) | |
| T4 | 59 (65.6) | 32 (71.1) | 21 (70.0) | |
| N stage | 0.994 | |||
| N0 | 23 (25.6) | 9 (20.0) | 7 (23.3) | |
| N1 | 16 (17.8) | 8 (17.8) | 6 (20.0) | |
| N2 | 25 (27.8) | 14 (31.1) | 9 (30.0) | |
| N3 | 26 (28.9) | 14 (31.1) | 8 (26.7) | |
| Adjuvant therapy | 0.990 | |||
| None | 55 (61.1) | 25 (55.6) | 16 (53.3) | |
| Chemotherapy | 15 (16.7) | 8 (17.8) | 6 (20.0) | |
| Radiotherapy | 2 (2.2) | 1 (2.2) | 1 (3.3) | |
| Chemoradiotherapy | 18 (20.0) | 11 (24.4) | 7 (23.3) | |
Evaluation of TITreg cells abundance in the training, validation and testing cohort
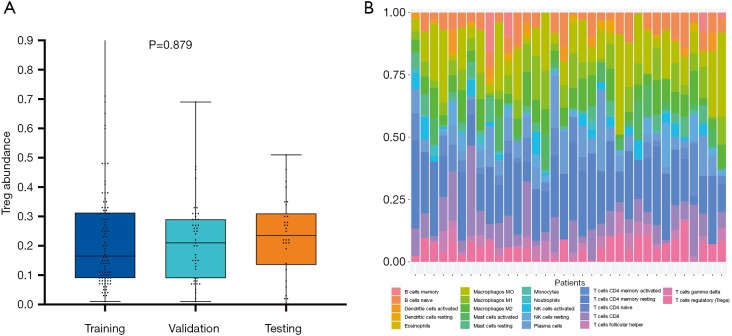
The abundance of TITreg cell of 135 patients of the training and the validation cohort were estimated using multispectral immunohistochemical analysis. The TITreg cell fraction in CD4+ cells of the training cohort and the validation cohort were 22.41%±17.53% and 21.38%±13.72%, respectively. As showed in Figure 3, the relative fraction of 22 tumor-infiltrating immune cell of 30 GC patients of the testing cohort were estimated utilizing CIBERSORT. The TITreg cell fraction in CD4+ cells of the testing cohort were 23.21%±13.28%. The fraction of TITreg cells were not significantly differed among three cohorts (P=0.879, Figure 3A).
Figure 3.
Estimation of tumor-infiltrating Treg cells. (A) The Treg cell fraction in CD4+ cells of training cohort, validation and the testing cohort. (B) The proportion of 22 types tumor infiltrating-immune cells in patients of the TCGA-STAD dataset using CIBERSORT.
Selection of features and establishment of radiomics signature
Of 859 features extracted from the selected ROIs of the training cohort, 70 features with ICC <0.8 were excluded. Then, 145 of the remained 789 features were identified as significant differed features between high and low Treg groups (P<0.05) and enrolled into LASSO regression analysis. Finally, the radiomics signature was built with 6 features. The calculation formula radiomics score is provided in supplementary materials.
The performance of radiomics signature for estimating TITreg cells
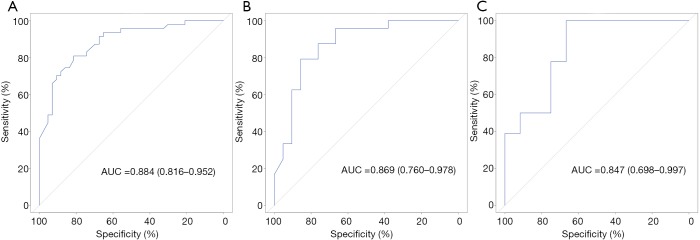
The ability of radiomics signature for estimating TITreg cells abundance was evaluated by using ROC curves. The AUCs of radiomics signature for the training and the validation cohorts were 0.884 (95% CI: 0.816–0.952, Figure 4A) and 0.869 (95% CI: 0.760–0.978, Figure 4B), respectively. To verify the predictive ability of the radiomics signature in different ethnic population, TCGA-STAD dataset which enrolled non-Asian patients was used as an external testing cohort. The radiomics signature also showed a good predictive performance in the testing cohort with an AUC of 0.847 (95% CI: 0.698–0.997, Figure 4C).
Figure 4.
ROC curves for the radiomics signature. (A) ROC curves of the radiomics signature in the training cohort. (B) ROC curves of the radiomics signature in the validation cohort. (C) ROC curves of the radiomics signature in the testing cohort.
Prognostic value of TITreg cells and radiomics signature
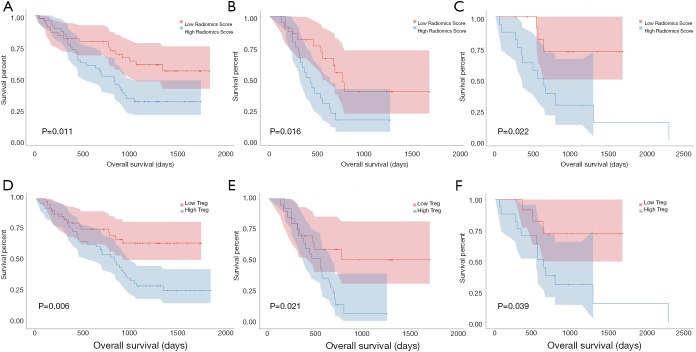
As shown in Table 2, univariable and multivariable Cox regression analysis showed that the radiomics signature [hazard ratio (HR) 2.018, 95% CI: 1.133–3.721, P=0.012], T-stage (HR 1.012, 95% CI: 1.008–1.875, P=0.042) and TNM stage (HR 1.226, 95% CI: 1.101–2.007, P=0.033) were independent predictors of OS. Kaplan-Meier analysis revealed that higher radiomics score were associated with poorer OS in the training (P=0.011, Figure 5A), validation (P=0.016, Figure 5B) and the testing cohort (P=0.022, Figure 5C). The higher abundance of TITreg cells were also related to unfavorable OS in the training (P=0.006, Figure 5D), validation (P=0.021, Figure 5E) and the testing cohort (P=0.039, Figure 5F).
Table 2. Univariate and multivariate Cox analyses of risk factors of overall survival.
| Variable | Univariate Cox regression | Multivariate Cox regression | |||
|---|---|---|---|---|---|
| OR (95% CI) | P value | OR (95% CI) | P value | ||
| Gender (male vs. female) | 1.261 (0.713–1.556) | 0.564 | – | – | |
| Age (<60 vs. ≥60 years) | 0.861 (0.637–1.213) | 0.340 | – | – | |
| Tumor site | 0.723 (0.556–1.013) | 0.452 | – | – | |
| Differentiation | 1.121 (0.872–1.334) | 0.513 | – | – | |
| TNM stage | 1.556 (1.221–2.314) | 0.021 | 1.226 (1.101–2.007) | 0.033 | |
| T stage (T1–2 vs. T3–4) | 1.176 (1.108–1.987) | 0.034 | 1.012 (1.007–1.875) | 0.042 | |
| N stage (N0 vs. N1–3) | 1.137 (0.891–1.732) | 0.071 | – | – | |
| Radiomics signature | 2.334 (1.210–3.116) | 0.009 | 2.018 (1.133–3.721) | 0.012 | |
Figure 5.
Kaplan-Meier curves for radiomics signature and Treg cells. (A) OS of patients relative to radiomics signature in the training cohort. (B) OS of patients relative to radiomics signature in the validation cohort. (C) OS of patients relative to radiomics signature in the testing cohort. (D) OS of patients relative to the abundance of Treg cells in the training cohort. (E) OS of patients relative to the abundance of Treg cells in the validation cohort. (F) OS of patients relative to the abundance of Treg cells in the testing cohort.
Discussion
In our study, six robust image features were identified and utilized to build a radiomics signature for assessing TITreg cells of GC patients. The AUCs of radiomics signature for the training and the validation cohort were 0.884 and 0.869, respectively. Then, by linking image features and gene expression signature of Treg cells, the performance of the radiomics signature was confirmed in an independent cohort with an AUC of 0.847. In addition, survival analysis showed that the radiomics signature could also sever as a predictor of OS in GC patients.
Treg cells are a subset of CD4+ T cells with immunosuppressive function. An increasing body of evidence demonstrated that Treg cells are highly infiltrated in various tumors. A recent meta-analysis showed that higher Treg cells was significantly correlated with poor outcome in most of solid tumors (19). Thus, TITreg cells play a vital role in shaping immunosuppressive tumor microenvironment, leading to the progression, invasion and metastasis of tumor (20,21).
ICIs represented by anti-CTLA-4 and anti-PD-1/L1 agents exhibited encouraging efficacy in various solid tumors and has deeply changed the strategy of cancer treatment. Some studies demonstrated that PD-1 also expressed on a subset of Treg cells (22-24). Furthermore, through interaction with PD-L1 on CD8+ T cells, upregulation of PD-1 on Treg cells could strengthen the suppression of immune response of CD8+ T cells (23). Recently, Kamada et al. reported that PD-1+ Treg cells significantly amplified in GC patients who received anti-PD-1 and contributed to the development of HPD (8). Thus, the radiomics signature established in our study maybe a potential non-invasive biomarker for predicting and monitoring the development of HPD induced by anti-PD-1/PD-L1 agents.
To date, studies about the relationship between radiomics features and intratumoral immune landscape are still limited. Tang et al. identified a radiomics signature consist of four features for prognosis prediction of lung cancer. The signature divided patients into four clusters which associated with OS. Specifically, a favorable outcome cluster with low CT intensity and high heterogeneity had the low level of PD-L1+ tumor cell and the high level of infiltrating CD3+ T cells (25). However, whether CD3+ T cells rather than specific subset of T cells could be a reliable biomarker for immune response is still a controversial problem (26). Sun and colleagues proposed and tested a radiomic signature for assessing tumor-infiltrating CD8+ cells in three datasets. Their results showed that the radiomic signature could be a useful biomarker for assessing CD8+ cell abundance and outcome predication for patients received PD-1/PD-L1 therapy (27). However, Sun et al.’s study enrolled more than 10 types of tumors. Since the patterns of intratumoral immune cell are tremendously different among different types of tumors, their results may still need to be confirmed in specific tumor type.
This study has some limitations. First, the retrospective nature of this study was inherently disadvantageous. Second, although the radiomics signature was established and tested with three independent cohorts, the sample size was relatively small. Third, no patients received anti-PD-1 treatment was enrolled for evaluation the ability of the radiomics signature for predicting treatment response. Fourth, although same CT scan parameters were used in different scanners and cohorts, images acquired with different scanners and at different centers may still introduce bias.
Conclusions
In conclusion, we proposed a CT-based radiomics signature for estimating TITreg cells and outcome prediction of GC. Multicenter studies with larger dataset are warranted to verify our results for clinical practice. GC Patients received anti-PD-1 are also required to further investigate the potential of the radiomics signature for predicting treatment response of anti-PD-1 agents.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by grants from the Fund of National Natural Science Foundation of China (grant number 81772620), and Tianjin major project for chronic disease prevention and control (grant number 17ZXMFSY0).
Supplementary
Radiomics score calculation formula
Radiomics score = (0.645 × GLSZM.Gray Level Non Uniformity Normalized) – (0.211×GLSZM.Gray Level Variance) – (0.216 × GLSZM.Small Area High Gray Level Emphasis) – (0.137 × Wavelet-WLW.ngtdm.Complexity) + (0.08 × GLCM.MCC − 0.221) × (Wavelet-LLL.firstorder.Maximum).
Table S1. Feature type and associated features.
| Feature type | Methods | Feature name |
|---|---|---|
| Shape-based | – | Maximum 3D diameter (M3D) |
| Maximum 2D Diameter Slice (M2DS) | ||
| Sphericity | ||
| Minor Axis (MA) | ||
| Elongation | ||
| Surface Volume Ratio (SVR) | ||
| Volume | ||
| Major Axis (MA1) | ||
| Surface Area (SA) | ||
| Flatness | ||
| Least Axis (LA) | ||
| Maximum 2D Diameter Column (M2DC) | ||
| Maximum 2D Diameter Row (M2DR) | ||
| First order-based | Histogram | Interquartile Range (IQR) |
| Skewness | ||
| Uniformity | ||
| Median | ||
| Energy | ||
| Robust Mean Absolute Deviation (RMAD) | ||
| Mean Absolute Deviation (MAD) | ||
| Total Energy (TE) | ||
| Maximum | ||
| Root Mean Squared (RMS) | ||
| 90 Percentile | ||
| Minimum | ||
| Entropy | ||
| Range | ||
| Variance | ||
| 10 Percentile | ||
| Kurtosis | ||
| Mean | ||
| Texture-based | GLCM | Joint Average (JA) |
| Sum Average (SA) | ||
| Joint Entropy (JE) | ||
| Cluster Shade (CS) | ||
| Maximum Probability (MP) | ||
| Idmn | ||
| Joint Energy (JE) | ||
| Contrast | ||
| Difference Entropy (DE) | ||
| Inverse Variance (IV) | ||
| Difference Variance (DV) | ||
| Idn | ||
| Idm | ||
| Correlation | ||
| Autocorrelation | ||
| Sum Entropy (SE) | ||
| Sum Squares (SS) | ||
| Cluster Prominence (CP) | ||
| Imc2 | ||
| Imc1 | ||
| Difference Average (DA) | ||
| Id | ||
| Cluster Tendency (CT) | ||
| GLSZM | Gray Level Variance (GLV) | |
| Zone Variance (ZV) | ||
| Gray Level Non-Uniformity Normalized (GLNUN) | ||
| Size Zone Non-Uniformity Normalized (SZNUN) | ||
| Size Zone Non-Uniformity (SZNU) | ||
| Gray Level Non-Uniformity (GLNU) | ||
| Large Area Emphasis (LAE) | ||
| Small Area High Gray Level Emphasis (SAHGLE) | ||
| Zone Percentage (ZP) | ||
| Large Area Low Gray Level Emphasis (LALGLE) | ||
| Large Area High Gray Level Emphasis (LAHGLE) | ||
| High Gray Level Zone Emphasis (HGLZE) | ||
| Small Area Emphasis (SAE) | ||
| Low Gray Level Zone Emphasis (LGLZE) | ||
| Zone Entropy (ZE) | ||
| Small Area Low Gray Level Emphasis (SALGLE) | ||
| GLRLM | Short Run Low Gray Level Emphasis (SRLGLE) | |
| Gray Level Variance (GLV) | ||
| Low Gray Level Run Emphasis (LGLRE) | ||
| Gray Level Non-Uniformity Normalized (GLNUN) | ||
| Run Variance (RV) | ||
| Gray Level Non-Uniformity (GLNU) | ||
| Long Run Emphasis (LRE) | ||
| Short Run High Gray Level Emphasis (SRHGLE) | ||
| Run Length Non-Uniformity (RLNU) | ||
| Short Run Emphasis (SRE) | ||
| Long Run High Gray Level Emphasis (LRHGLE) | ||
| Run Percentage (RP) | ||
| Long Run Low Gray Level Emphasis (LRLGLE) | ||
| Run Entropy (RE) | ||
| High Gray Level Run Emphasis (HGLRE) | ||
| Run Length Non-Uniformity Normalized (RLNUN) | ||
| NGTDM | Coarseness | |
| Complexity | ||
| Strength | ||
| Contrast | ||
| Busyness | ||
| GLDM | Gray Level Variance (GLV) | |
| High Gray Level Emphasis (HGLE) | ||
| Dependence Entropy (DE) | ||
| Dependence Non-Uniformity (DNU) | ||
| Gray Level Non-Uniformity (GLNU) | ||
| Small Dependence Emphasis (SDE) | ||
| Small Dependence High Gray Level Emphasis (SDHGLE) | ||
| Dependence Non-Uniformity Normalized (DNUN) | ||
| Large Dependence Emphasis (LDE) | ||
| Large Dependence Low Gray Level Emphasis (LDLGLE) | ||
| Dependence Variance (DV) | ||
| Large Dependence High Gray Level Emphasis (LDHGLE) | ||
| Small Dependence Low Gray Level Emphasis (SDLGLE) | ||
| Low Gray Level Emphasis (LGLE) | ||
| Wavelet-based | First-order statistic and texture of wavelet decomposition | First-order features |
| GLCM features | ||
| GLSZM features | ||
| Decomposition levels: LLL, LLH, LHL, LHH, HLL, HLH, HHL, HHH | GLRLM features | |
| NGTDM features | ||
| GLDM features |
GLCM, gray-level co-occurrence matrix, describe the second-order joint probability function of the voxel intensities within the contoured volume; GLSZM, gray-level size-zone matrix, quantify the number of connected voxels within the contoured volume that share the same gray level intensity; GLRLM, gray-level run-Length matrix, quantify the number of consecutive voxels that have the same gray level value; NGTDM, neighboring gray-tone difference matrix, quantify the difference between a gray value and the average gray value of its neighbors within 3×3×3 voxels neighborhood window; GLDM, gray-level dependence matrix, quantify the gray level dependencies in the contoured volume which is defined as the number of connected voxels within a specific distance that are dependent on the center voxel; Decomposition levels, i.e., LLH interpreted as the high-pass sub band, resulting from directional filtering of the volume with a low-pass filter along x-direction, a low pass filter along y-direction and a high-pass filter along z-direction
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (No. E2019082) and informed consent was obtained from all the patients.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.114). The authors have no conflicts of interest to declare.
References
- 1.Tanaka A, Sakaguchi S. Targeting Treg cells in cancer immunotherapy. Eur J Immunol 2019;49:1140-6. [DOI] [PubMed] [Google Scholar]
- 2.Li F, Zhao Y, Wei L, et al. Tumor-infiltrating Treg, MDSC, and IDO expression associated with outcomes of neoadjuvant chemotherapy of breast cancer. Cancer Biol Ther 2018;19:695-705. 10.1080/15384047.2018.1450116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barua S, Fang P, Sharma A, et al. Spatial interaction of tumor cells and regulatory T cells correlates with survival in non-small cell lung cancer. Lung Cancer 2018;117:73-9. 10.1016/j.lungcan.2018.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang X, Lang M, Zhao T, et al. Cancer-FOXP3 directly activated CCL5 to recruit FOXP3+ Treg cells in pancreatic ductal adenocarcinoma. Oncogene 2017;36:3048-58. 10.1038/onc.2016.458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ghatalia P, Gordetsky J, Kuo F, et al. Prognostic impact of immune gene expression signature and tumor infiltrating immune cells in localized clear cell renal cell carcinoma. J Immunother Cancer 2019;7:139. 10.1186/s40425-019-0621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seminerio I, Descamps G, Dupont S, et al. Infiltration of FoxP3+ Regulatory T Cells is a Strong and Independent Prognostic Factor in Head and Neck Squamous Cell Carcinoma. Cancers (Basel) 2019. doi: . 10.3390/cancers11020227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knegt P, de Jong PC, van der Schans EJ. Pharyngeal diverticula. Clin Otolaryngol Allied Sci 1987;12:319-20. [DOI] [PubMed] [Google Scholar]
- 8.Kamada T, Togashi Y, Tay C, et al. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc Natl Acad Sci U S A 2019;116:9999-10008. 10.1073/pnas.1822001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Togasaki K, Sukawa Y, Kanai T, et al. Clinical efficacy of immune checkpoint inhibitors in the treatment of unresectable advanced or recurrent gastric cancer: an evidence-based review of therapies. Onco Targets Ther 2018;11:8239-50. 10.2147/OTT.S152514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Zhang L, Tian C, et al. Prognostic value of computed tomography radiomics features in patients with gastric cancer following curative resection. Eur Radiol 2019;29:3079-89. 10.1007/s00330-018-5861-9 [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Liu W, Yu Y, et al. CT radiomics nomogram for the preoperative prediction of lymph node metastasis in gastric cancer. Eur Radiol 2020;30:976-86. 10.1007/s00330-019-06398-z [DOI] [PubMed] [Google Scholar]
- 12.Ma Z, Fang M, Huang Y, et al. CT-based radiomics signature for differentiating Borrmann type IV gastric cancer from primary gastric lymphoma. Eur J Radiol 2017;91:142-7. 10.1016/j.ejrad.2017.04.007 [DOI] [PubMed] [Google Scholar]
- 13.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nghiem PT, Bhatia S, Lipson EJ, et al. PD-1 Blockade with Pembrolizumab in Advanced Merkel-Cell Carcinoma. N Engl J Med 2016;374:2542-52. 10.1056/NEJMoa1603702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu H, Zhu Y, Wang Y, et al. Identification and Validation of Stromal Immunotype Predict Survival and Benefit from Adjuvant Chemotherapy in Patients with Muscle-Invasive Bladder Cancer. Clin Cancer Res 2018;24:3069-78. 10.1158/1078-0432.CCR-17-2687 [DOI] [PubMed] [Google Scholar]
- 16.Tsurumaru D, Miyasaka M, Nishimuta Y, et al. Differentiation of early gastric cancer with ulceration and resectable advanced gastric cancer using multiphasic dynamic multidetector CT. Eur Radiol 2016;26:1330-7. 10.1007/s00330-015-3938-2 [DOI] [PubMed] [Google Scholar]
- 17.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang YQ, Liang CH, He L, et al. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J Clin Oncol 2016;34:2157-64. 10.1200/JCO.2015.65.9128 [DOI] [PubMed] [Google Scholar]
- 19.Shang B, Liu Y, Jiang SJ, et al. Prognostic value of tumor-infiltrating FoxP3+ regulatory T cells in cancers: a systematic review and meta-analysis. Sci Rep 2015;5:15179. 10.1038/srep15179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi C, Chen Y, Chen Y, et al. CD4+ CD25+ regulatory T cells promote hepatocellular carcinoma invasion via TGF-beta1-induced epithelial-mesenchymal transition. Onco Targets Ther 2018;12:279-89. 10.2147/OTT.S172417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Ma K, Song S, et al. Tight correlation between FoxM1 and FoxP3+ Tregs in gastric cancer and their clinical significance. Clin Exp Med 2018;18:413-20. 10.1007/s10238-018-0505-6 [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Chikuma S, Hori S, et al. Nonoverlapping roles of PD-1 and FoxP3 in maintaining immune tolerance in a novel autoimmune pancreatitis mouse model. Proc Natl Acad Sci U S A 2016;113:8490-5. 10.1073/pnas.1608873113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park HJ, Park JS, Jeong YH, et al. PD-1 upregulated on regulatory T cells during chronic virus infection enhances the suppression of CD8+ T cell immune response via the interaction with PD-L1 expressed on CD8+ T cells. J Immunol 2015;194:5801-11. 10.4049/jimmunol.1401936 [DOI] [PubMed] [Google Scholar]
- 24.Asano T, Meguri Y, Yoshioka T, et al. PD-1 modulates regulatory T-cell homeostasis during low-dose interleukin-2 therapy. Blood 2017;129:2186-97. 10.1182/blood-2016-09-741629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang C, Hobbs B, Amer A, et al. Development of an Immune-Pathology Informed Radiomics Model for Non-Small Cell Lung Cancer. Sci Rep 2018;8:1922. 10.1038/s41598-018-20471-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin S, Xu B, Yu L, et al. The PD-1, PD-L1 expression and CD3+ T cell infiltration in relation to outcome in advanced gastric signet-ring cell carcinoma, representing a potential biomarker for immunotherapy. Oncotarget 2017;8:38850-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun R, Limkin EJ, Vakalopoulou M, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol 2018;19:1180-91. 10.1016/S1470-2045(18)30413-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as