Abstract
Triple-negative breast cancer (TNBC) is the most aggressive breast cancer subtype. Despite the progress made in precision treatment of cancer patients, targeted treatment is still at its early stage in TNBC, and chemotherapy remains the standard treatment. With the advances in next generation sequencing technology, genomic and transcriptomic analyses have provided deeper insight into the inter-tumoral heterogeneity of TNBC. Much effort has been made to classify TNBCs into different molecular subtypes according to genetic aberrations and expression signatures and to uncover novel treatment targets. In this review, we summarized the current knowledge regarding the molecular classification of TNBC and explore the future paradigm for using molecular classification to guide the development of precision treatment and clinical practice.
Keywords: Triple-negative breast cancer (TNBC), molecular subtypes, precision treatment
Introduction
Triple-negative breast cancer (TNBC) encompasses a subset of breast cancers that lack expression of the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBC accounts for 15% to 20% of newly diagnosed breast cancer cases (1,2), and is known for its aggressive biological behavior and poor patient outcomes compared with hormone receptor-positive breast cancer (2-4). It has also been of special interest to breast cancer researchers due to its varying response towards standard chemotherapy, and its lack of other effective treatment options.
Advances in sequencing technologies have allowed for comprehensive characterization of tumor molecular features. TNBC has been increasingly recognized as a heterogeneous disease that exhibits substantial differences in terms of genomic and transcriptomic profiles (5,6). The extreme heterogeneity of TNBC has led to difficulties in finding suitable molecular targets in preclinical studies and has been reflected in the limited benefit from targeted therapies observed in clinical trials for unselected TNBC patients. Therefore, TNBC subtyping based on the biologically and clinically relevant characteristics may contribute to the identification of therapeutic targets, optimization of clinical trial designs, and patient risk stratification. In this study, we discuss the current status of TNBC molecular subtyping and its relevance to prognostication and therapeutic strategies, including our own application of molecular subtyping to establish a treatment paradigm incorporating precision medicine for refractory TNBC patients.
Histological classification
TNBC encompasses a variety of histologic types. While invasive carcinoma of no special type (NST) comprises the vast majority of TNBC, other histologic types, which are referred to as special histologic types, account for approximately 10% of all TNBC (7). Invasive carcinoma NST represents a heterogeneous group of tumors that fail to exhibit sufficient histological features to be classified as any special type. The morphological features of this subtype vary vastly among different cases. The most common special histological types of TNBC include metaplastic carcinoma, medullary carcinoma, invasive lobular carcinoma, apocrine carcinoma and adenoid cystic carcinoma (8). Several studies have reported the prognostic implications of special histological types in TNBC. Compared with invasive carcinoma NST, metaplastic and invasive lobular carcinoma were associated with a poorer prognosis, while medullary, apocrine and adenoid cystic carcinoma indicated a better prognosis (8-10).
Classification of TNBC based on histopathological features has been well described in past literature. However, very little treatment recommendations are made in current guidelines according to the histological type of breast cancer. The prognostic implications of certain histologic types may be valuable in making clinical decisions regarding patient follow-up and therapeutic approaches, for example the dose intensity and the duration of adjuvant chemotherapy.
Molecular classification
Somatic genomic alterations
Cancer genomes harbor a large number of somatic alterations, but only a few of them play a role in driving carcinogenesis by conferring selective advantage to tumor cell growth (11-13). Genomic profiling studies of human TNBC, including whole-exome and whole-genome analyses, have identified numerous recurrent alterations in these cancer driver genes. Somatic mutations and copy number alterations (CNAs) account for most genomic alterations in TNBC. TP53 is the most frequently mutated gene, but at present no drugs targeting TP53 have been approved for clinical practice. Aside from TP53, a handful of genes which mutated at >5% prevalence in TNBC have been identified including PIK3CA, PTEN, KMT2C and RB1 (14-16). MYC amplification is the most frequent CNA event in TNBC. Other genes frequently affected by somatic CNAs include EGFR, PTEN, CCND1, RB1 and CCNE1 (14,15,17).
Due to the complexity of the genomic landscape, analysis of alterations in single genes is often insufficient for TNBC classification. Thus, effort has been made to group the aberrations of individual genes according to the molecular pathways, such as PDGF/VEGF signaling, PI3K/AKT/mTOR signaling, JAK/STAT3 signaling and cell cycle pathways to achieve better classification (14,18). Such grouping has helped us better understand the tumor biology and facilitated the development of drugs targeting these pathways.
Different mutational processes often generate different combinations of mutation types, which is termed “signature” (19). In the past decade, large-scale sequencing studies have revealed many mutational signatures across the spectrum of human cancer types (19-21). For example, Nik-Zainal et al. analyzed whole genome sequences of 560 breast cancers and identified twelve base substitution and six rearrangement signatures (17). Based on these established mutational signatures, Jiang et al. (15) classified TNBC into the following four mutation subtypes: (I) APOBEC, characterized by APOBEC-related signatures 2 and 13; (II) HRD, by homologous recombination deficiency (HRD)-related signature 3; (III) Clock-like: by clock-like signatures 1 and 5; and (IV) Mixed: no dominant signature. The mutational signature-based subtyping advances our understanding of the mutational processes underlying TNBC carcinogenesis.
HRD status in TNBCs
Defects in BRCA1/2 lead to HR DNA repair deficiency (22,23) and are associated with increased tumor sensitivity to DNA damaging agents, such as platinum salt and poly(ADP-ribose) polymerase (PARP) inhibitors (24,25). In breast cancer, several clinical trials have demonstrated the efficacy of DNA damaging agents in patients with germline mutations in BRCA1 or BRCA2 (BRCA1/2) (26-29).
Previous studies reported that 10–20% TNBCs have germline mutations in BRCA1/2 (30-33). These tumors typically exhibited HR deficiency, but some sporadic (BRCA1 and BRCA2 germline wild-type) TNBCs can also display functional BRCA1/2 deficiency and harbor DNA repair defects through, for instance, mutations of HR pathway genes (34,35). These tumors may be also sensitive to DNA-damaging agents (27). Thus, researchers sought to uncover additional biomarkers indicative of HR deficiency beyond germline BRCA1/2 mutations. Three quantitative metrics have been developed to measure the genomic instability that is the consequence of HR deficiency: loss of heterozygosity (LOH) score (36), telomeric allelic imbalance (TAI) score (37) and large-scale state transitions (LST) score (38). A combined HRD score is defined as the arithmetic mean of these three scores (39). Several studies defined a group of HR deficiency TNBCs as TNBCs with a high HRD score (>42) and/or the presence of BRCA1/2 mutations (26,40,41). In the neoadjuvant setting, either HR deficiency in all TNBCs or a high HRD score in the BRCA1/2 wild-type subgroup predicted better response towards platinum (26,40,41). However, in the metastatic setting, different studies reported conflicting data. For example, the TNT trial indicated that the HRD score assessed by Myriad HRD assay was not predictive of platinum response (42), while the TBCR009 trial found that HRD-LOH and HRD-LST assays can discriminate platinum responders and non-responders (43). The discrepancies between trials may be attributed to the different measures of HR deficiency status used in these studies, and further research or a unified method of measurement may remedy this problem.
By analyzing the whole genome sequencing data of 560 breast cancer samples, Davies et al. developed a mutational-signature-based predictor of BRCA1/2 deficiency called HRDetect (44). A followi6ng study demonstrated that in advanced-stage breast cancers, elevated HRDetect was associated with clinical benefit from platinum-based therapy independent of BRCA1/2 mutation status (45). More recently, Staaf et al. applied the HRDetect algorithm to 254 TNBC samples with whole genome sequencing data and classified them into HRDetect-high, HRDetect-intermediate and HRDetect-low subgroups (16). The HRDetect-high subgroup contained nearly 33% cases without germline/somatic mutations in BRCA1/BRCA2, BRCA1 promoter hypermethylation, RAD51C hypermethylation or biallelic loss of PALB2. Compared with the HRDetect-low subgroup, the HRDetect-high subgroup was associated with a better prognosis and a higher degree of sensitivity to standard adjuvant chemotherapy. The HRDetect-intermediate and HRDetect-low subgroups were characterized by the enrichment of CCNE1 amplifications and PIK3CA/AKT1 pathway abnormalities respectively. With the progress in sequencing technology and reduced sequencing costs, the HRDetect model may be used in the future to inform trial stratification and improve clinical decision-making.
Transcriptome-based classification
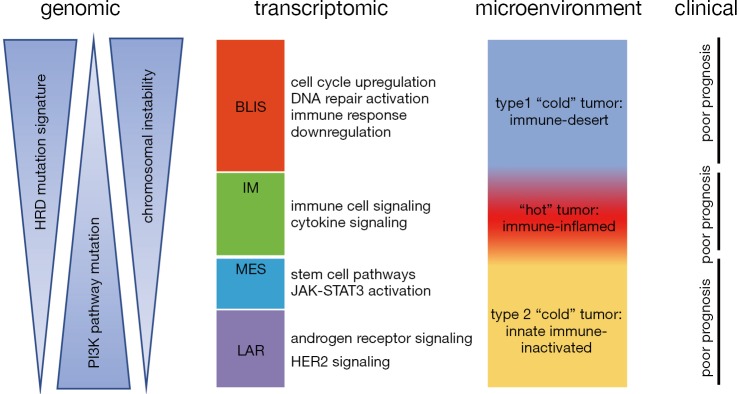
Transcriptome profiling analysis provided great insight into the molecular heterogeneity of TNBCs and enabled robust and unbiased classification. Lehmann et al. first initiated the molecular subtyping of TNBC based on transcriptome profiling data (5). They analyzed gene expression profiles of 587 TNBCs from 21 microarray data sets, performed k-means clustering on the most differentially expressed genes and classified TNBC tumors into 6 clusters: basal-like 1 (BL1), basal-like 2 (BL2), immunomodulatory (IM), mesenchymal (M), mesenchymal stem cell-like (MSL), and luminal androgen receptor (LAR). These six TNBC subtypes were characterized by distinct gene ontologies and gene expression patterns. The BL1 subtype was enriched for gene ontologies involving cell cycle and cell division. The BL2 subtype displayed gene ontologies involving growth factor signaling, glycolysis and gluconeogenesis. The IM subtype was enriched for gene ontologies in immune cell processes. Both MSL and M subtypes were enriched in gene expression for cell motility, but the MSL subtype can be distinguished by the low expression of proliferation and high expression of stem cell-related genes. The LAR subtype was characterized by androgen receptor signaling and showed luminal gene expression patterns. Another important contribution of Lehmann et al. was that they aligned representative TNBC cell lines to each of the 6 TNBC subtypes and uncovered that the cell lines displayed different sensitivities to therapeutic agents. Their results showed that identifying the unique expression of cellular components and presence of mutations in key oncogenes and tumor suppressors in each subtype may help understand the heterogeneity of TNBC and provide clues for the development of effective precision treatment. In a follow-up study, the same research team amended their classification from the previous six subtypes to four tumor-specific subtypes (BL1, BL2, M and LAR), because they found that the transcripts of the IM and MSL subtypes were contributed from tumor infiltrating lymphocytes (TILs) and tumor-associated stromal cells, respectively (46). They further pointed out that these four tumor-specific subtypes differed in response to standard neoadjuvant chemotherapy. Another highly important study on transcriptome-based TNBC subtyping was by Jiang et al. (15), where they comprehensively analyzed clinical, genomic and transcriptomic data of 465 primary TNBCs treated at Fudan University Shanghai Cancer Center (FUSCC). They classified TNBCs into four mRNA-based subtypes: (I) a LAR subtype characterized by androgen receptor signaling; (II) an IM subtype showing high immune cell signaling and cytokine signaling gene expression; (III) a basal-like and immune-suppressed (BLIS) subtype characterized by activation of DNA repair and downregulation of immune response genes; and 4) a mesenchymal-like (MES) subtype enriched in breast cancer stem cell pathways. Distinct genomic alterations and clinical features were identified in the four subtypes (Figure 1). Two aspects of this study distinguish it from the previous work. First, they analyzed TNBCs from an East Asian population, which demonstrated the similarity in molecular features among different ethnic groups and at the same time identified subtle difference, including a higher frequency of PIK3CA mutation and a higher proportion of LAR subtype in their Asian TNBC cohort. This large collection of comprehensively profiled TNBCs with well-documented clinical information will be an important supplement to the international compendium of molecular information regarding human breast cancer. Second, this study laid down the groundwork for subtype-specific treatment strategies for TNBC patients and a subsequent clinical trial is currently underway (NCT3805399) pending publication of preliminary results.
Figure 1.
Schematic representation of TNBC molecular subtypes. This figure shows the association of the transcriptomic TNBC subtypes with other tumor characteristics. The BLIS subtype is correlated with enrichment of HRD mutation signature and high chromosomal instability. The LAR subtype and MES subtype present high frequency of PI3K pathway mutations. The IM subtype is characterized by high immune infiltration and good prognosis. TNBC, triple-negative breast cancer; HRD, homologous recombination deficiency; HER2, human epidermal growth factor receptor 2.
Other works concerning the transcriptome-based classification of TNBCs include studies by Burstein et al. and Liu et al. Burstein et al. classified TNBCs into four subtypes using array-based RNA profiling data and summarized the highly expressed molecules in specific TNBC subtypes that can be targeted (47). Liu et al. developed a classification system characterized by the integration of transcriptome profiles of both messenger RNA and long noncoding RNA (48). Despite the differences in the classification methods and nomenclature used in the above studies, the classification results showed obvious correlations between each other. International effort to sharing and merging large-scale data and cross-comparison of different classification approaches may lead to a final consensus.
Tumor microenvironment (TME) classification
The TME has been increasingly recognized as an important indicator for patient prognosis and the efficacy of immunotherapy (49,50). The prevalence of TILs basically characterizes the phenotype of TME and can be easily assessed on haematoxylin and eosin stained sections. Several studies classified TNBCs according to the TILs levels. High levels of TILs were associated with a better prognosis in patients with early-stage TNBC, regardless of whether the patients received chemotherapy (51,52). TILs were also a predictor of response to neoadjuvant chemotherapy in TNBCs (53,54). In the metastatic setting, high levels of TILs correlated with a higher tumor response rate to immunotherapy and longer survival (55). Except TILs, one recent study concentrated on tumor-infiltrating myeloid (neutrophils and macrophages) and classified TNBCs into neutrophil-enriched and macrophage-enriched subtypes. While the macrophage-enriched subtype exhibited varying responses to immune checkpoint inhibitors (ICI), neutrophil-enriched subtype is resistant to immune checkpoint blockade (ICB) due to the enrichment of immunosuppressive neutrophils (56).
The transcriptome profiling of tumor tissue provides information on both tumor and microenvironment cells and allows for systematic analysis on the TME. Xiao et al. performed expression profile–based estimation of the abundance of microenvironment cells in tumor tissue and classified TNBCs into three microenvironment clusters: (I) the “immune-desert” cluster, with low microenvironment cell infiltration; (II) the “innate immune-inactivated” cluster, with resting innate immune cells and nonimmune stromal cells infiltration; and (III) the “immune-inflamed” cluster, with abundant adaptive and innate immune cells infiltration (57). Based on the multi-omics profiling data, they hypothesized that the MYC amplification, hyper-activated PI3K-AKT pathway and high expression of immune checkpoint molecules were the mechanisms of immune escape for each of the three clusters, respectively. He et al. classified TNBC into immunity high, immunity medium, and immunity low subgroups based on the expression of immune signatures, which may facilitate the optimal stratification of TNBC patients responsive to immunotherapy (58). Saleh et al. analyzed the transcriptome of tumor-associated stroma and characterized the TME of TNBC using four stromal axes: T cells, B cells, epithelial markers, and desmoplasia. The combination of these axes generated a novel TNBC classification approach and informed patient prognosis (59).
Recently, digital image analysis has been used to study the spatial structure of TME (60-63). Through multiplexed ion beam imaging, Keren et al. demonstrated the variability in the cellular composition and spatial organization of TNBC TME across individuals and divided TNBC tumors into cold (no infiltrate), mixed (immune cells mixed with tumor cells), and compartmentalized (immune cells spatially separated from tumor cells) subtypes. These three subtypes were associated with distinct tumor and immune populations expressing immunoregulatory proteins and with patient prognosis (64). The combination of transcriptomic analysis and digital image analysis on pathological phenotype may better reveal the TME heterogeneity of TNBC and contribute to realizing the personalized immunotherapy for patients with TNBC.
Therapeutic implications
Targeting deregulated signaling pathway in the context of transcriptomic subtypes
The genomic alterations in certain genes drive carcinogenesis by deregulating signaling pathways involving cellular differentiation, proliferation and apoptosis. The PI3K signaling pathway plays an important role in the regulation of cell growth and survival (65). Its activation is associated with tumor evolution and chemotherapy resistance, and is a common genomic abnormality detected in TNBCs (66,67). Multiple genomic aberrations contribute to the PI3K pathway activation, including PIK3CA activating mutations, AKT1 activating mutations, mTOR activating mutations, and PTEN loss. Taken together, the PI3K pathway is altered in approximately 25% of primary TNBCs, and the rate is even higher in metastatic TNBCs (67). Multiple clinical trials have been conducted to test the efficacy of different PI3K pathway inhibitors.
Buparlisib, a pan-PI3K inhibitor, has been demonstrated efficacy in PIK3CA-mutated HR+HER2- breast cancer (68). However, interim analysis of the BELLE-4 trial indicated that the addition of buparlisib to paclitaxel led to a worse progression free survival (PFS) in patients with advanced TNBC, and no benefit was found in patients with PIK3CA mutation or PTEN loss (69). Two phase II clinical trials suggested promising efficacy of AKT inhibitors in the first line treatment of metastatic TNBCs. In the LOTUS trial, median PFS in the intention-to-treat population was 6.2 months with ipatasertib plus paclitaxel versus 4·9 months with placebo plus paclitaxel. According to the preliminary analysis of overall survival (OS), the addition of ipatasertib to paclitaxel improved OS from 16.2 to 23.1 months (70). In the prespecified analysis of patients with PIK3CA/AKT1/PTEN-altered (genetic PIK3CA/AKT1-activating mutations or PTEN-inactivating alterations) tumors, PFS was improved from 4.9 to 9 months with the addition of ipatasertib to paclitaxel (71). The PAKT trial tested another AKT inhibitor, capivasertib. The capivasertib plus paclitaxel group showed a median PFS of 5.9 months, while the PFS in the placebo plus paclitaxel group was 4.2 months. At the same time, median OS was 19.1 months with capivasertib plus paclitaxel and 12.6 months with placebo plus paclitaxel. In patients with PIK3CA/AKT1/PTEN-altered tumors, an improvement in PFS was observed from 3.7 to 9.3 months with the addition of capivasertib to paclitaxel (72). The results of the PAKT trial were quite consistent with those of the LOTUS trial, which provided additional evidence supporting the use of AKT inhibitors in TNBC. In the neoadjuvant setting, the FAIRLNE trial indicated that the addition of ipatasertib to 12 weeks of paclitaxel improved the overall response rate but did not significantly increase pathological complete response (pCR) rate (73). Except for ipatasertib and capivasertib, MK-2206, a selective allosteric inhibitor of AKT, was also tested in patients with advanced breast cancer selected for PIK3CA/AKT1 or PTEN mutations or PTEN loss, but it showed limited clinical activity (74). Everolimus, a selective mTOR inhibitor, showed promising antitumor activity in hormone receptor positive and HER2 positive metastatic breast cancers (75,76). However, most clinical trials testing everolimus in combination with chemotherapy did not demonstrated clear efficacy in advanced HER2-negative or TNBCs (77-80).
Since the frequency of PI3K pathway alterations varied in TNBC subtypes (5,15), the efficacy of PI3K pathway inhibitors was examined in certain TNBC subtypes. For example, a phase I trial examined the combination of mTOR inhibitors, temsirolimus or everolimus, with liposomal doxorubicin and bevacizumab in advanced metaplastic TNBC, which was used as a surrogate of MES TNBC. A 21% objective response rate was observed and the PI3K pathway activation was found to be associated with an improved response rate (81). In addition, Lehmann et al. recently demonstrated in a phase II clinical trial that patients with LAR subtype metastatic TNBC tended to have better response to enzalutamide in combination with taselisib (a PI3K inhibitor) than those with non-LAR tumors (clinical benefit rate at 16 weeks, 75.0% vs. 12.5%) (82).
Aside from the PI3K signaling pathway, alterations in the EGFR, VEGF JAK/STAT and NOTCH pathways were also enriched in TNBCs. EGFR overexpression and high EGFR gene copy number were found in a substantial percentage of TNBCs (83). Several phase II trials evaluated the addition of monoclonal antibody against EGFR (cetuximab or panitumumab) to chemotherapy in patients with metastatic TNBC. Despite modest improvement on overall response rate or PFS, prespecified end points of these studies were not met (84-88). Nabholtz et al. carried out two phase II trials to investigate anti-EGFR monoclonal antibodies in combination with chemotherapy as neoadjuvant therapy for operable, stage II–III TNBC. The addition of panitumumab to anthracycline/taxane-based chemotherapy appeared efficacious (more than 25% absolute increase in pCR) (89), but cetuximab in combination with docetaxel showed modest activity (less than 15% increase in pCR) (90). TNBCs exhibit high levels of vascular endothelial growth factor (VEGF) and enhanced angiogenesis (91-93). Bevacizumab is a monoclonal antibody against VEGFA. The addition of bevacizumab to standard neoadjuvant chemotherapy increased the pCR rate in patients with TNBC (94-97). Patients with metastatic TNBC derived PFS and response benefits from the combination of bevacizumab with either first-line or second-line chemotherapy (98-101). However, the addition of bevacizumab to chemotherapy resulted in an increased rate of serious adverse events and bring no benefit in OS for breast cancer patients (102). In addition, in the adjuvant setting, the addition of bevacizumab to chemotherapy did not improve invasive disease-free survival (IDFS) or OS in unselected patients with TNBC (103). The mitogen-activated protein kinase (MAPK; also known as RAS-RAF-MEK-ERK) pathway plays a critical role in TNBC progression and chemoresistance. MEK is a key component of this pathway and MEK inhibitors have demonstrated anticancer efficacy against TNBC cells in preclinical studies (104-107). Nevertheless, clinical trials reported limited efficacy of MEK1/2 inhibitors in TNBC (108-110). Preclinical studies reported the activation of JAK/STAT signaling pathway in TNBC and the antitumor efficacy of JAK/STAT pathway inhibition (30,111,112). However, A phase II study observed no objective responses in metastatic TNBC patients treated with ruxolitinib, a selective JAK1/2 inhibitor, as a single agent (113). The activation of NOTCH signaling pathway has also been identified in TNBC, especially in the mesenchymal of mesenchymal stem-like TNBCs (5,114), but the treatment of PF-03084014, a selective gamma-secretase inhibitor, in combination with docetaxel demonstrated limited preliminary antitumor activity in patients with advanced TNBC (115).
Although researchers have made much effort to find genetically altered signaling pathways and to develop targeted therapies for TNBC, many clinical trials failed to demonstrate a statistically significant improvement in patient outcome by targeting these pathways. These limited benefits from targeted therapies observed in clinical trials for unselected TNBC patients further highlighted the molecular heterogeneity of TNBC and suggested a subtype-specific treatment paradigm for TNBC patients.
ICB immunotherapy in the context of TME clusters
Immunotherapy refers to any approach aimed at manipulating or amplifying the host immune system to better fight disease (116). ICB immunotherapy has become of special interest for cancer treatment in the past decades. Programmed cell death protein 1 (PD-1) and its ligand programmed death-ligand 1 (PD-L1) are the most studied targets of ICIs. Atezolizumab is a monoclonal antibody targeting PD-L1. The phase 3 study IMpassion130 evaluated atezolizumab plus nab-paclitaxel compared with placebo plus nab-paclitaxel as the first-line treatment for patients with unresectable locally advanced or metastatic TNBC. The analysis of PFS demonstrated that the addition of atezolizumab plus nab-paclitaxel prolonged PFS in both the intention-to-treat population (median PFS: 7.2 vs. 5.5 months) and the subgroup of patients with PD-L1 immune cell-positive tumors (median PFS: 7.5 vs. 5.0 months) (117). The recently updated data on OS suggested that although the difference between the treatment groups in the intention-to-treat population was not statistically significant, a clinically meaningful OS benefit with atezolizumab plus nab-paclitaxel was observed in patients with PD-L1 immune cell-positive tumors (median OS: 25.0 vs. 18.0 months) (118). Another phase I study PCD4989g investigated single-agent atezolizumab therapy in patients with metastatic TNBC and reported durable clinical activity and encouraging survival benefit, particularly in first-line patients or those with tumors of high levels of immune cells and PD-L1–positive immune cells (55). The GeparNuevo study examined another PD-L1 inhibitor, durvalumab, and suggested that the addition of durvalumab to anthracycline-/taxane-based neoadjuvant therapy increases pCR rate (119). Pembrolizumab is a highly selective, humanized monoclonal antibody against PD-1. The phase II KEYNOTE-086 study investigated pembrolizumab as a monotherapy in patients with metastatic TNBC. Pembrolizumab monotherapy demonstrated durable antitumor activity either in patients with previously untreated, PD-L1-positive mTNBC or in patients with previously treated metastatic TNBC regardless of the PD-L1 expression (120,121). The phase II TOPACIO study indicated that the combination of niraparib and pembrolizumab provided promising antitumor activity in patients with advanced or metastatic TNBC, especially in those with tumor BRCA mutations (122). JS001, another humanized monoclonal antibody for PD-1 was investigated in a phase I trial and showed a moderate response in advanced TNBC patients(123). The results of these studies suggested that targeting PD-1 and PD-L1 may be a promising therapeutic strategy for patients with TNBC.
Identification of robust biomarkers that can assist prediction of response to ICB is essential to further advance precision immunotherapy. The results of clinical trials highlighted the predictive value of immune cell PD-L1 expression (117). Besides PD-L1 expression, many other biomarkers have been suggested to predict tumor response to ICIs including TILs, tumor mutation burden and several single-gene biomarkers, such as EGFR amplification and IFN-γ expression (55,124-128). Due to the complexity of anti-cancer immunity, the use of a single marker may not be able to effectively identify tumors that respond well towards ICIs. The study by Xiao et al. initiated a new concept for guiding individualized immunotherapy. They performed a comprehensive analysis by integrating multi-omics profiles of TNBC with clinical and pathological information and classified TNBCs into three TME clusters. They suggested the selective application of ICIs to the “immune-inflamed” cluster, which is characterized by high infiltration of innate and adaptive immune cells, and high expression of immune checkpoint molecules (57). The classification by Xiao et al. simultaneously informed multiple aspects of TME and might be used to more effectively predict response and clinical benefits from ICIs.
Summary: subtyping-based TNBC precision medicine paradigm
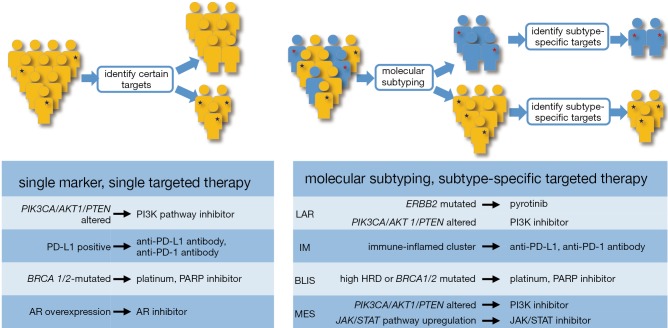
Combining all the analytical assumptions of preclinical studies and the results of clinical trials, we suggested the shifting of TNBC precision treatment paradigm from the biomarker-based paradigm to a subtyping-based paradigm (Figure 2). That is, first, TNBCs are classified into molecular subtypes with enrichment of certain actionable genomic alterations. Then, subtype-specific therapeutic targets and biomarkers are detected. The combination of these two methods would prompt a targeted treatment suitable for this individual disease.
Figure 2.
Transformation of precision medicine patterns for TNBC. This figure shows the evolution of precision treatment paradigm for TNBCs from biomarker-based targeted therapy to subtyping-based targeted therapy. The subtyping-based paradigm provides deeper insight to the molecular essence of patient tumors and contributes to more efficient target identification and drug development. TNBC, triple-negative breast cancer; HRD, homologous recombination deficiency; PD-1, programmed Cell Death 1; PD-1, programmed Cell Death ligand 1; AR, androgen receptor.
On the basis of such precision medicine paradigm, FUSCC has recently launched a study for precision treatment of refractory TNBC, called the FUSCC Refractory TNBC Umbrella (FUTURE) study (NCT 03805399). This study is a Phase Ib/II, open-label, umbrella study evaluating the efficacy and safety of multiple targeted treatment in patients with metastatic TNBC which had progressed during or following standard treatment with chemotherapy including anthracyclines, taxanes, platinums, vinorelbine, capecitabine, and gemcitabine. Refractory TNBC patients will be screened and tumor specimens of entered into different treatment arms according to their molecular subtype and the results of their gene panel testing. The FUTURE trial aims to provide refractory TNBC patients with precision treatment targeting their specific molecular characteristics to achieve better survival. Currently, treatment arms include: (I) pyrotinib with capecitabine, for the LAR subtype with ERBB2 activated mutation; (II) AR inhibitor with mTOR inhibitor, for the LAR subtype without ERBB2 activated mutation, but with a PIK3CA mutation; (III) AR inhibitor with CDK4/6 inhibitor, for the LAR subtype with neither ERBB2 activated mutation nor PIK3CA mutation; (IV) anti PD-1 with nab-paclitaxel for IM subtype (CD8 positive T cell more than 20%); (V) PARP inhibitor included therapy, for BLIS subtype with BRCA1/2 gene pathogenic mutation; (VI) anti-VEGFR included therapy, for the BLIS subtype without BRCA1/2 gene pathogenic mutation and for the MES subtype without PI3K/AKT pathway activation; (VII) mTOR inhibitor with nab-paclitaxel, for the MES subtype with PI3K/AKT pathway activation.
The FUTURE trial is still currently enrolling, with primary results yet to be reported (unpublished manuscript), but there is no doubt that this precision treatment paradigm will continue to evolve with increased understanding of this heterogeneous disease and increase in precision following our progress in biomarker and drug development.
Clinical transformation challenge
Molecular subtyping of TNBC via gene expression profiling is essential for deconstructing the molecular portraits of this heterogeneous disease and for developing individualized treatment. However, the high cost, complicated technological process and potential batch effects make its use in routine clinical practice less feasible. Thus, there is a need for a simplified approach to classify TNBCs into molecular subtypes for clinical use, and our research team has recently devised a surrogate IHC-based classification method on the basis of the transcriptome-based subtyping system proposed by Jiang et al. By taking full advantage of the gene expression profiling data and tumor tissue samples, we determined the markers that can be used to identify specific molecular subtypes through IHC test and developed an IHC-based classifier (unpublished manuscript). We demonstrated a substantial agreement (Cohen’s kappa coefficient >0.6) between the results of the IHC-based classification and the mRNA-based classification. In addition, most molecular features and therapeutic implications of different mRNA-based subtypes are preserved in the corresponding IHC-based subtypes. Our IHC-based classification allows for subgrouping of TNBC patients in large clinical trials and evaluating the efficacy of targeted therapies within certain subtypes. It will facilitate the translation of TNBC molecular classification into clinical practice.
Conclusions
TNBC has been increasingly recognized as a heterogeneous disease. Although progress in next generation sequencing technology has facilitated identifying potentially actionable targets, few of the findings have been translated into daily clinical practice due to limited benefit from targeted therapy observed in clinical trials for unselected TNBC patients. The molecular subtyping of TNBC enables the identification of molecularly homogeneous groups with enrichment of certain genomic alterations. The subtype-specific clinical investigations may be an effective method for drug development. With the increasing data accumulation from clinical trials, a precision medicine paradigm in the context of transcriptomic subtyping can be developed and fine-tuned for patients with TNBC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.194). The authors have no conflicts of interest to declare.
References
- 1.Bauer KR, Brown M, Cress RD, et al. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 2007;109:1721-8. 10.1002/cncr.22618 [DOI] [PubMed] [Google Scholar]
- 2.Criscitiello C, Azim HA, Jr, Schouten PC, et al. Understanding the biology of triple-negative breast cancer. Ann Oncol 2012;23 Suppl 6:vi13-8. 10.1093/annonc/mds188 [DOI] [PubMed] [Google Scholar]
- 3.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 2006;295:2492-502. 10.1001/jama.295.21.2492 [DOI] [PubMed] [Google Scholar]
- 4.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol 2008;26:1275-81. 10.1200/JCO.2007.14.4147 [DOI] [PubMed] [Google Scholar]
- 5.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011;121:2750-67. 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Metzger-Filho O, Tutt A, de Azambuja E, et al. Dissecting the heterogeneity of triple-negative breast cancer. J Clin Oncol 2012;30:1879-87. 10.1200/JCO.2011.38.2010 [DOI] [PubMed] [Google Scholar]
- 7.Gluz O, Liedtke C, Gottschalk N, et al. Triple-negative breast cancer--current status and future directions. Ann Oncol 2009;20:1913-27. 10.1093/annonc/mdp492 [DOI] [PubMed] [Google Scholar]
- 8.Zhao S, Ma D, Xiao Y, et al. Clinicopathologic features and prognoses of different histologic types of triple-negative breast cancer: A large population-based analysis. Eur J Surg Oncol 2018;44:420-8. 10.1016/j.ejso.2017.11.027 [DOI] [PubMed] [Google Scholar]
- 9.Montagna E, Maisonneuve P, Rotmensz N, et al. Heterogeneity of triple-negative breast cancer: histologic subtyping to inform the outcome. Clin Breast Cancer 2013;13:31-9. 10.1016/j.clbc.2012.09.002 [DOI] [PubMed] [Google Scholar]
- 10.Mills MN, Yang GQ, Oliver DE, et al. Histologic heterogeneity of triple negative breast cancer: A National Cancer Centre Database analysis. Eur J Cancer 2018;98:48-58. 10.1016/j.ejca.2018.04.011 [DOI] [PubMed] [Google Scholar]
- 11.Garraway LA, Lander ES. Lessons from the cancer genome. Cell 2013;153:17-37. 10.1016/j.cell.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 12.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature 2009;458:719-24. 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bareche Y, Venet D, Ignatiadis M, et al. Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann Oncol 2018;29:895-902. 10.1093/annonc/mdy024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang YZ, Ma D, Suo C, et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell. 2019;35:428-40.e5. 10.1016/j.ccell.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 16.Staaf J, Glodzik D, Bosch A, et al. Whole-genome sequencing of triple-negative breast cancers in a population-based clinical study. Nat Med 2019;25:1526-33. 10.1038/s41591-019-0582-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47-54. 10.1038/nature17676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balko JM, Giltnane JM, Wang K, et al. Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov 2014;4:232-45. 10.1158/2159-8290.CD-13-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. 10.1038/nature12477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexandrov LB, Stratton MR. Mutational signatures: the patterns of somatic mutations hidden in cancer genomes. Curr Opin Genet Dev 2014;24:52-60. 10.1016/j.gde.2013.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nik-Zainal S, Alexandrov LB, Wedge DC, et al. Mutational processes molding the genomes of 21 breast cancers. Cell 2012;149:979-93. 10.1016/j.cell.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moynahan ME, Chiu JW, Koller BH, et al. Brca1 controls homology-directed DNA repair. Mol Cell 1999;4:511-8. 10.1016/S1097-2765(00)80202-6 [DOI] [PubMed] [Google Scholar]
- 23.Tutt A, Bertwistle D, Valentine J, et al. Mutation in Brca2 stimulates error-prone homology-directed repair of DNA double-strand breaks occurring between repeated sequences. EMBO J 2001;20:4704-16. 10.1093/emboj/20.17.4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 2005;434:917-21. 10.1038/nature03445 [DOI] [PubMed] [Google Scholar]
- 25.Pommier Y, O'Connor MJ, de Bono J. Laying a trap to kill cancer cells: PARP inhibitors and their mechanisms of action. Sci Transl Med 2016;8:362ps17. 10.1126/scitranslmed.aaf9246 [DOI] [PubMed] [Google Scholar]
- 26.von Minckwitz G, Schneeweiss A, Loibl S, et al. Neoadjuvant carboplatin in patients with triple-negative and HER2-positive early breast cancer (GeparSixto; GBG 66): a randomised phase 2 trial. Lancet Oncol 2014;15:747-56. 10.1016/S1470-2045(14)70160-3 [DOI] [PubMed] [Google Scholar]
- 27.Silver DP, Richardson AL, Eklund AC, et al. Efficacy of neoadjuvant Cisplatin in triple-negative breast cancer. J Clin Oncol 2010;28:1145-53. 10.1200/JCO.2009.22.4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litton JK, Rugo HS, Ettl J, et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N Engl J Med 2018;379:753-63. 10.1056/NEJMoa1802905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robson M, Im SA, Senkus E, et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N Engl J Med 2017;377:523-33. 10.1056/NEJMoa1706450 [DOI] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Network . Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61-70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartman AR, Kaldate RR, Sailer LM, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer 2012;118:2787-95. 10.1002/cncr.26576 [DOI] [PubMed] [Google Scholar]
- 32.Greenup R, Buchanan A, Lorizio W, et al. Prevalence of BRCA mutations among women with triple-negative breast cancer (TNBC) in a genetic counseling cohort. Ann Surg Oncol 2013;20:3254-8. 10.1245/s10434-013-3205-1 [DOI] [PubMed] [Google Scholar]
- 33.Sharma P, Klemp JR, Kimler BF, et al. Germline BRCA mutation evaluation in a prospective triple-negative breast cancer registry: implications for hereditary breast and/or ovarian cancer syndrome testing. Breast Cancer Res Treat 2014;145:707-14. 10.1007/s10549-014-2980-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner N, Tutt A, Ashworth A. Hallmarks of 'BRCAness' in sporadic cancers. Nat Rev Cancer 2004;4:814-9. 10.1038/nrc1457 [DOI] [PubMed] [Google Scholar]
- 35.Ledermann JA, Drew Y, Kristeleit RS. Homologous recombination deficiency and ovarian cancer. Eur J Cancer 2016;60:49-58. 10.1016/j.ejca.2016.03.005 [DOI] [PubMed] [Google Scholar]
- 36.Abkevich V, Timms KM, Hennessy BT, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012;107:1776-82. 10.1038/bjc.2012.451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birkbak NJ, Wang ZC, Kim JY, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012;2:366-75. 10.1158/2159-8290.CD-11-0206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popova T, Manie E, Rieunier G, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454-62. 10.1158/0008-5472.CAN-12-1470 [DOI] [PubMed] [Google Scholar]
- 39.Timms KM, Abkevich V, Hughes E, et al. Association of BRCA1/2 defects with genomic scores predictive of DNA damage repair deficiency among breast cancer subtypes. Breast Cancer Res 2014;16:475. 10.1186/s13058-014-0475-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loibl S, Weber KE, Timms KM, et al. Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response-final results from GeparSixto. Ann Oncol 2018;29:2341-7. 10.1093/annonc/mdy460 [DOI] [PubMed] [Google Scholar]
- 41.Telli ML, Timms KM, Reid J, et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin Cancer Res 2016;22:3764-73. 10.1158/1078-0432.CCR-15-2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tutt A, Tovey H, Cheang MCU, et al. Carboplatin in BRCA1/2-mutated and triple-negative breast cancer BRCAness subgroups: the TNT Trial. Nat Med 2018;24:628-37. 10.1038/s41591-018-0009-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Isakoff SJ, Mayer EL, He L, et al. TBCRC009: A Multicenter Phase II Clinical Trial of Platinum Monotherapy With Biomarker Assessment in Metastatic Triple-Negative Breast Cancer. J Clin Oncol 2015;33:1902-9. 10.1200/JCO.2014.57.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies H, Glodzik D, Morganella S, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517-25. 10.1038/nm.4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao EY, Shen Y, Pleasance E, et al. Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin Cancer Res 2017;23:7521-30. 10.1158/1078-0432.CCR-17-1941 [DOI] [PubMed] [Google Scholar]
- 46.Lehmann BD, Jovanovic B, Chen X, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 2016;11:e0157368. 10.1371/journal.pone.0157368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Burstein MD, Tsimelzon A, Poage GM, et al. Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clin Cancer Res 2015;21:1688-98. 10.1158/1078-0432.CCR-14-0432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu YR, Jiang YZ, Xu XE, et al. Comprehensive transcriptome analysis identifies novel molecular subtypes and subtype-specific RNAs of triple-negative breast cancer. Breast Cancer Res 2016;18:33. 10.1186/s13058-016-0690-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature 2017;541:321-30. 10.1038/nature21349 [DOI] [PubMed] [Google Scholar]
- 50.Adams S, Gray RJ, Demaria S, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 2014;32:2959-66. 10.1200/JCO.2013.55.0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loi S, Drubay D, Adams S, et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J Clin Oncol 2019;37:559-69. 10.1200/JCO.18.01010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park JH, Jonas SF, Bataillon G, et al. Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Ann Oncol 2019;30:1941-9. 10.1093/annonc/mdz395 [DOI] [PubMed] [Google Scholar]
- 53.Denkert C, Loibl S, Noske A, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 2010;28:105-13. 10.1200/JCO.2009.23.7370 [DOI] [PubMed] [Google Scholar]
- 54.Russo L, Maltese A, Betancourt L, et al. Locally advanced breast cancer: Tumor-infiltrating lymphocytes as a predictive factor of response to neoadjuvant chemotherapy. Eur J Surg Oncol 2019;45:963-8. 10.1016/j.ejso.2019.01.222 [DOI] [PubMed] [Google Scholar]
- 55.Emens LA, Cruz C, Eder JP, et al. Long-term Clinical Outcomes and Biomarker Analyses of Atezolizumab Therapy for Patients With Metastatic Triple-Negative Breast Cancer: A Phase 1 Study. JAMA Oncol 2019;5:74-82. 10.1001/jamaoncol.2018.4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim IS, Gao Y, Welte T, et al. Immuno-subtyping of breast cancer reveals distinct myeloid cell profiles and immunotherapy resistance mechanisms. Nat Cell Biol 2019;21:1113-26. 10.1038/s41556-019-0373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao Y, Ma D, Zhao S, et al. Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer. Clin Cancer Res 2019;25:5002-14. 10.1158/1078-0432.CCR-18-3524 [DOI] [PubMed] [Google Scholar]
- 58.He Y, Jiang Z, Chen C, et al. Classification of triple-negative breast cancers based on Immunogenomic profiling. J Exp Clin Cancer Res 2018;37:327. 10.1186/s13046-018-1002-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Saleh SMI, Bertos N, Gruosso T, et al. Identification of Interacting Stromal Axes in Triple-Negative Breast Cancer. Cancer Res 2017;77:4673-83. 10.1158/0008-5472.CAN-16-3427 [DOI] [PubMed] [Google Scholar]
- 60.Jackson HW, Fischer JR, Zanotelli VRT, et al. The single-cell pathology landscape of breast cancer. Nature 2020;578:615-20. 10.1038/s41586-019-1876-x [DOI] [PubMed] [Google Scholar]
- 61.Saltz J, Gupta R, Hou L, et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep 2018;23:181-93.e7. 10.1016/j.celrep.2018.03.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kather JN, Krisam J, Charoentong P, et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PLoS Med 2019;16:e1002730. 10.1371/journal.pmed.1002730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ligorio M, Sil S, Malagon-Lopez J, et al. Stromal Microenvironment Shapes the Intratumoral Architecture of Pancreatic Cancer. Cell 2019;178:160-75.e27. 10.1016/j.cell.2019.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keren L, Bosse M, Marquez D, et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018;174:1373-87.e19. 10.1016/j.cell.2018.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cantley LC. The phosphoinositide 3-kinase pathway. Science 2002;296:1655-7. 10.1126/science.296.5573.1655 [DOI] [PubMed] [Google Scholar]
- 66.Wein L, Loi S. Mechanisms of resistance of chemotherapy in early-stage triple negative breast cancer (TNBC). Breast 2017;34 Suppl 1:S27-30. 10.1016/j.breast.2017.06.023 [DOI] [PubMed] [Google Scholar]
- 67.Pascual J, Turner NC. Targeting the PI3-kinase pathway in triple-negative breast cancer. Ann Oncol 2019;30:1051-60. 10.1093/annonc/mdz133 [DOI] [PubMed] [Google Scholar]
- 68.Baselga J, Im SA, Iwata H, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2017;18:904-16. 10.1016/S1470-2045(17)30376-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martín M, Chan A, Dirix L, et al. A randomized adaptive phase II/III study of buparlisib, a pan-class I PI3K inhibitor, combined with paclitaxel for the treatment of HER2- advanced breast cancer (BELLE-4). Ann Oncol 2017;28:313-20. 10.1093/annonc/mdw562 [DOI] [PubMed] [Google Scholar]
- 70.Dent R, Im SA, Espie M, et al. Overall survival update of the double-blind placebo-controlled randomized phase 2 LOTUS trial of first-line ipatasertib + paclitaxel for locally advanced/metastatic triple-negative breast cancer. J Clin Oncol 2018;36:1008. 10.1200/JCO.2018.36.15_suppl.1008 [DOI] [Google Scholar]
- 71.Kim SB, Dent R, Im SA, et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): a multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2017;18:1360-72. 10.1016/S1470-2045(17)30450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmid P, Abraham J, Chan S, et al. Capivasertib Plus Paclitaxel Versus Placebo Plus Paclitaxel As First-Line Therapy for Metastatic Triple-Negative Breast Cancer: The PAKT Trial. J Clin Oncol 2020;38:423-33. 10.1200/JCO.19.00368 [DOI] [PubMed] [Google Scholar]
- 73.Oliveira M, Saura C, Nuciforo P, et al. FAIRLANE, a double-blind placebo-controlled randomized phase II trial of neoadjuvant ipatasertib plus paclitaxel for early triple-negative breast cancer. Ann Oncol 2019;30:1289-97. 10.1093/annonc/mdz177 [DOI] [PubMed] [Google Scholar]
- 74.Xing Y, Lin NU, Maurer MA, et al. Phase II trial of AKT inhibitor MK-2206 in patients with advanced breast cancer who have tumors with PIK3CA or AKT mutations, and/or PTEN loss/PTEN mutation. Breast Cancer Res 2019;21:78. 10.1186/s13058-019-1154-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baselga J, Campone M, Piccart M, et al. Everolimus in postmenopausal hormone-receptor-positive advanced breast cancer. N Engl J Med 2012;366:520-9. 10.1056/NEJMoa1109653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morrow PK, Wulf GM, Ensor J, et al. Phase I/II study of trastuzumab in combination with everolimus (RAD001) in patients with HER2-overexpressing metastatic breast cancer who progressed on trastuzumab-based therapy. J Clin Oncol 2011;29:3126-32. 10.1200/JCO.2010.32.2321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Decker T, Marschner N, Muendlein A, et al. VicTORia: a randomised phase II study to compare vinorelbine in combination with the mTOR inhibitor everolimus versus vinorelbine monotherapy for second-line chemotherapy in advanced HER2-negative breast cancer. Breast Cancer Res Treat 2019;176:637-47. 10.1007/s10549-019-05280-2 [DOI] [PubMed] [Google Scholar]
- 78.Park IH, Kong SY, Kwon Y, et al. Phase I/II clinical trial of everolimus combined with gemcitabine/cisplatin for metastatic triple-negative breast cancer. J Cancer 2018;9:1145-51. 10.7150/jca.24035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yardley DA, Bosserman LD, O'Shaughnessy JA, et al. Paclitaxel, bevacizumab, and everolimus/placebo as first-line treatment for patients with metastatic HER2-negative breast cancer: a randomized placebo-controlled phase II trial of the Sarah Cannon Research Institute. Breast Cancer Res Treat 2015;154:89-97. 10.1007/s10549-015-3599-5 [DOI] [PubMed] [Google Scholar]
- 80.Jovanović B, Mayer IA, Mayer EL, et al. A Randomized Phase II Neoadjuvant Study of Cisplatin, Paclitaxel With or Without Everolimus in Patients with Stage II/III Triple-Negative Breast Cancer (TNBC): Responses and Long-term Outcome Correlated with Increased Frequency of DNA Damage Response Gene Mutations, TNBC Subtype, AR Status, and Ki67. Clin Cancer Res 2017;23:4035-45. 10.1158/1078-0432.CCR-16-3055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Basho RK, Gilcrease M, Murthy RK, et al. Targeting the PI3K/AKT/mTOR Pathway for the Treatment of Mesenchymal Triple-Negative Breast Cancer: Evidence From a Phase 1 Trial of mTOR Inhibition in Combination With Liposomal Doxorubicin and Bevacizumab. JAMA Oncol 2017;3:509-15. 10.1001/jamaoncol.2016.5281 [DOI] [PubMed] [Google Scholar]
- 82.Lehmann BD, Abramson VG, Sanders M, et al. TBCRC 032 IB/II Multicenter Study: Molecular insights to AR antagonist and PI3K inhibitor efficacy in patients with AR+ metastatic triple-negative breast cancer. Clin Cancer Res 2019. doi: . 10.1158/1078-0432.CCR-19-2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Park HS, Jang MH, Kim EJ, et al. High EGFR gene copy number predicts poor outcome in triple-negative breast cancer. Mod Pathol 2014;27:1212-22. 10.1038/modpathol.2013.251 [DOI] [PubMed] [Google Scholar]
- 84.Carey LA, Rugo HS, Marcom PK, et al. TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol 2012;30:2615-23. 10.1200/JCO.2010.34.5579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Cowherd S, Miller LD, Melin SA, et al. A phase II clinical trial of weekly paclitaxel and carboplatin in combination with panitumumab in metastatic triple negative breast cancer. Cancer Biol Ther 2015;16:678-83. 10.1080/15384047.2015.1026481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crozier JA, Advani PP, LaPlant B, et al. N0436 (Alliance): A Phase II Trial of Irinotecan With Cetuximab in Patients With Metastatic Breast Cancer Previously Exposed to Anthracycline and/or Taxane-Containing Therapy. Clin Breast Cancer 2016;16:23-30. 10.1016/j.clbc.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yardley DA, Ward PJ, Daniel BR, et al. Panitumumab, Gemcitabine, and Carboplatin as Treatment for Women With Metastatic Triple-Negative Breast Cancer: A Sarah Cannon Research Institute Phase II Trial. Clin Breast Cancer 2016;16:349-55. 10.1016/j.clbc.2016.05.006 [DOI] [PubMed] [Google Scholar]
- 88.Baselga J, Gómez P, Greil R, et al. Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol 2013;31:2586-92. 10.1200/JCO.2012.46.2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nabholtz JM, Abrial C, Mouret-Reynier MA, et al. Multicentric neoadjuvant phase II study of panitumumab combined with an anthracycline/taxane-based chemotherapy in operable triple-negative breast cancer: identification of biologically defined signatures predicting treatment impact. Ann Oncol 2014;25:1570-7. 10.1093/annonc/mdu183 [DOI] [PubMed] [Google Scholar]
- 90.Nabholtz JM, Chalabi N, Radosevic-Robin N, et al. Multicentric neoadjuvant pilot Phase II study of cetuximab combined with docetaxel in operable triple negative breast cancer. Int J Cancer 2016;138:2274-80. 10.1002/ijc.29952 [DOI] [PubMed] [Google Scholar]
- 91.Linderholm BK, Hellborg H, Johansson U, et al. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol 2009;20:1639-46. 10.1093/annonc/mdp062 [DOI] [PubMed] [Google Scholar]
- 92.Nalwoga H, Arnes JB, Stefansson IM, et al. Vascular proliferation is increased in basal-like breast cancer. Breast Cancer Res Treat 2011;130:1063-71. 10.1007/s10549-011-1740-7 [DOI] [PubMed] [Google Scholar]
- 93.Chang HY, Nuyten DS, Sneddon JB, et al. Robustness, scalability, and integration of a wound-response gene expression signature in predicting breast cancer survival. Proc Natl Acad Sci U S A 2005;102:3738-43. 10.1073/pnas.0409462102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gerber B, Loibl S, Eidtmann H, et al. Neoadjuvant bevacizumab and anthracycline-taxane-based chemotherapy in 678 triple-negative primary breast cancers; results from the geparquinto study (GBG 44). Ann Oncol 2013;24:2978-84. 10.1093/annonc/mdt361 [DOI] [PubMed] [Google Scholar]
- 95.Kim HR, Jung KH, Im SA, et al. Multicentre phase II trial of bevacizumab combined with docetaxel-carboplatin for the neoadjuvant treatment of triple-negative breast cancer (KCSG BR-0905). Ann Oncol 2013;24:1485-90. 10.1093/annonc/mds658 [DOI] [PubMed] [Google Scholar]
- 96.Sikov WM, Berry DA, Perou CM, et al. Impact of the addition of carboplatin and/or bevacizumab to neoadjuvant once-per-week paclitaxel followed by dose-dense doxorubicin and cyclophosphamide on pathologic complete response rates in stage II to III triple-negative breast cancer: CALGB 40603 (Alliance). J Clin Oncol 2015;33:13-21. 10.1200/JCO.2014.57.0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.von Minckwitz G, Eidtmann H, Rezai M, et al. Neoadjuvant chemotherapy and bevacizumab for HER2-negative breast cancer. N Engl J Med 2012;366:299-309. 10.1056/NEJMoa1111065 [DOI] [PubMed] [Google Scholar]
- 98.Brufsky A, Valero V, Tiangco B, et al. Second-line bevacizumab-containing therapy in patients with triple-negative breast cancer: subgroup analysis of the RIBBON-2 trial. Breast Cancer Res Treat 2012;133:1067-75. 10.1007/s10549-012-2008-6 [DOI] [PubMed] [Google Scholar]
- 99.Hamilton E, Kimmick G, Hopkins J, et al. Nab-paclitaxel/bevacizumab/carboplatin chemotherapy in first-line triple negative metastatic breast cancer. Clin Breast Cancer 2013;13:416-20. 10.1016/j.clbc.2013.08.003 [DOI] [PubMed] [Google Scholar]
- 100.Robert NJ, Dieras V, Glaspy J, et al. RIBBON-1: randomized, double-blind, placebo-controlled, phase III trial of chemotherapy with or without bevacizumab for first-line treatment of human epidermal growth factor receptor 2-negative, locally recurrent or metastatic breast cancer. J Clin Oncol 2011;29:1252-60. 10.1200/JCO.2010.28.0982 [DOI] [PubMed] [Google Scholar]
- 101.Thomssen C, Pierga JY, Pritchard KI, et al. First-line bevacizumab-containing therapy for triple-negative breast cancer: analysis of 585 patients treated in the ATHENA study. Oncology 2012;82:218-27. 10.1159/000336892 [DOI] [PubMed] [Google Scholar]
- 102.Sasich LD, Sukkari SR. The US FDAs withdrawal of the breast cancer indication for Avastin (bevacizumab). Saudi Pharm J 2012;20:381-5. 10.1016/j.jsps.2011.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cameron D, Brown J, Dent R, et al. Adjuvant bevacizumab-containing therapy in triple-negative breast cancer (BEATRICE): primary results of a randomised, phase 3 trial. Lancet Oncol 2013;14:933-42. 10.1016/S1470-2045(13)70335-8 [DOI] [PubMed] [Google Scholar]
- 104.Bartholomeusz C, Xie X, Pitner MK, et al. MEK Inhibitor Selumetinib (AZD6244; ARRY-142886) Prevents Lung Metastasis in a Triple-Negative Breast Cancer Xenograft Model. Mol Cancer Ther 2015;14:2773-81. 10.1158/1535-7163.MCT-15-0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee J, Galloway R, Grandjean G, et al. Comprehensive Two- and Three-Dimensional RNAi Screening Identifies PI3K Inhibition as a Complement to MEK Inhibitor AS703026 for Combination Treatment of Triple-Negative Breast Cancer. J Cancer 2015;6:1306-19. 10.7150/jca.13266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee J, Lim B, Pearson T, et al. Anti-tumor and anti-metastasis efficacy of E6201, a MEK1 inhibitor, in preclinical models of triple-negative breast cancer. Breast Cancer Res Treat 2019;175:339-51. 10.1007/s10549-019-05166-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawakami H, Huang S, Pal K, et al. Mutant BRAF Upregulates MCL-1 to Confer Apoptosis Resistance that Is Reversed by MCL-1 Antagonism and Cobimetinib in Colorectal Cancer. Mol Cancer Ther 2016;15:3015-27. 10.1158/1535-7163.MCT-16-0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schmid P, Forster MD, Summers YJ, et al. A study of vistusertib in combination with selumetinib in patients with advanced cancers: TORCMEK phase Ib results. J Clin Onco 2017;35:2548. 10.1200/JCO.2017.35.15_suppl.2548 [DOI] [Google Scholar]
- 109.Ramaswamy B, Mrozek E, Lustberg M, et al. Abstract LB-216: NCI 9455: Phase II study of trametinib followed by trametinib plus AKT inhibitor,GSK2141795 in patients with advanced triple negative breast cancer. Cancer Res 2016. doi: . 10.1158/1538-7445.AM2016-LB-216 [DOI] [Google Scholar]
- 110.Brufsky A, Miles D, Zvirbule Z, et al. Abstract P5-21-01: Cobimetinib combined with paclitaxel as firstline treatment for patients with advanced triple-negative breast cancer (COLET study): primary analysis of cohort I. Cancer Res 2018;78:P5-21. [Google Scholar]
- 111.Marotta LLC, Almendro V, Marusyk A, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+CD24− stem cell-like breast cancer cells in human tumors. J Clin Invest 2011;121:2723-35. 10.1172/JCI44745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Balko JM, Schwarz LJ, Luo N, et al. Triple-negative breast cancers with amplification of JAK2 at the 9p24 locus demonstrate JAK2-specific dependence. Sci Transl Med 2016;8:334ra53. 10.1126/scitranslmed.aad3001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stover DG, Gil Del Alcazar CR, Brock J, et al. Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast Cancer 2018;4:10. 10.1038/s41523-018-0060-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee CW, Raskett CM, Prudovsky I, et al. Molecular dependence of estrogen receptor-negative breast cancer on a notch-survivin signaling axis. Cancer Res 2008;68:5273-81. 10.1158/0008-5472.CAN-07-6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Locatelli MA, Aftimos P, Dees EC, et al. Phase I study of the gamma secretase inhibitor PF-03084014 in combination with docetaxel in patients with advanced triple-negative breast cancer. Oncotarget 2017;8:2320-8. 10.18632/oncotarget.13727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Waldmann TA. Immunotherapy: past, present and future. Nature Med 2003;9:269-77. 10.1038/nm0303-269 [DOI] [PubMed] [Google Scholar]
- 117.Schmid P, Adams S, Rugo HS, et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. The N Engl J Med 2018;379:2108-21. 10.1056/NEJMoa1809615 [DOI] [PubMed] [Google Scholar]
- 118.Schmid P, Rugo HS, Adams S, et al. Atezolizumab plus nab-paclitaxel as first-line treatment for unresectable, locally advanced or metastatic triple-negative breast cancer (IMpassion130): updated efficacy results from a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21:44-59. 10.1016/S1470-2045(19)30689-8 [DOI] [PubMed] [Google Scholar]
- 119.Loibl S, Untch M, Burchardi N, et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: clinical results and biomarker analysis of GeparNuevo study. Ann Oncol 2019;30:1279-88. 10.1093/annonc/mdz158 [DOI] [PubMed] [Google Scholar]
- 120.Adams S, Loi S, Toppmeyer D, et al. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann Oncol 2019;30:405-11. 10.1093/annonc/mdy518 [DOI] [PubMed] [Google Scholar]
- 121.Adams S, Schmid P, Rugo HS, et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: cohort A of the phase II KEYNOTE-086 study. Ann Oncol 2019;30:397-404. 10.1093/annonc/mdy517 [DOI] [PubMed] [Google Scholar]
- 122.Vinayak S, Tolaney SM, Schwartzberg L, et al. Open-Label Clinical Trial of Niraparib Combined With Pembrolizumab for Treatment of Advanced or Metastatic Triple-Negative Breast Cancer. JAMA Oncol 2019;5:1132-40. 10.1001/jamaoncol.2019.1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bian L, Zhang H, Wang T, et al. JS001, an anti-PD-1 mAb for advanced triple negative breast cancer patients after multi-line systemic therapy in a phase I trial. Ann Transl Med 2019;7:435. 10.21037/atm.2019.09.08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chabanon RM, Pedrero M, Lefebvre C, et al. Mutational Landscape and Sensitivity to Immune Checkpoint Blockers. Clin Cancer Res 2016;22:4309-21. 10.1158/1078-0432.CCR-16-0903 [DOI] [PubMed] [Google Scholar]
- 125.Kato S, Goodman A, Walavalkar V, et al. Hyperprogressors after Immunotherapy: Analysis of Genomic Alterations Associated with Accelerated Growth Rate. Clin Cancer Res 2017;23:4242-50. 10.1158/1078-0432.CCR-16-3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Loi S, Adams S, Schmid P, et al. Relationship between tumor infiltrating lymphocyte (TIL) levels and response to pembrolizumab (pembro) in metastatic triple-negative breast cancer (mTNBC): results from KEYNOTE-086. Ann Oncol 2017;28:mdx440.005.
- 127.Gao J, Shi LZ, Zhao H, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell 2016;167:397-404.e9. 10.1016/j.cell.2016.08.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Arora S, Velichinskii R, Lesh RW, et al. Existing and Emerging Biomarkers for Immune Checkpoint Immunotherapy in Solid Tumors. Adv Ther 2019;36:2638-78. 10.1007/s12325-019-01051-z [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as