Abstract
Background
The new coronavirus pneumonia (NCP) is now causing a severe public health emergency. The novel coronavirus 2019 (2019-nCoV) infected individuals by binding human angiotensin converting enzyme II (ACE2) receptor. ACE2 is widely expressed in multiple organs including respiratory, cardiovascular, digestive and urinary systems in healthy individuals. These tissues with high expression level of ACE2 seemed to be more vulnerable to SARS-CoV-2 infection. Recently, it has been reported that patients with tumors were likely to be more susceptible to SARS-CoV-2 infection and indicated poor prognosis.
Methods
The tissue atlas database and the blood atlas were used to analyze the distribution of ACE2 in human tissues or organs of cancers and normal samples. Starbase dataset was applied to predict the prognosis of cancers according to expression level of ACE2.
Results
In this study, we demonstrated a landscape profiling analysis on expression level of ACE2 in pan-cancers and showed the risky of different type of cancers to SARS-CoV-2 according to the expression level of ACE2. In addition, we found that ACE2 was both differential expression and related to the prognosis only in liver hepatocellular carcinoma (LIHC). Relative high expression of ACE2 indicated a favorable prognosis in LIHC, but they might be more susceptible to SARS-CoV-2.
Conclusions
We indeed emphasized that LIHC patients with high expression level of ACE2 should be more cautious of the virus infection. Our study might provide a potential clue for preventing infection of SARS-CoV-2 in cancers.
Keywords: SARS-CoV-2, cancer, angiotensin converting enzyme II (ACE2), liver hepatocellular carcinoma (LIHC)
Introduction
COVID-19, full name coronavirus disease 2019, is an infectious disease caused by a coronavirus called “SARS-CoV-2” (formerly known as “2019-nCoV”). At the end of 2019, the disease began in Wuhan, Hubei Province (1). So far, 72,530 confirmed cases (including medical staff) have been confirmed in China, including 59,989 in Wuhan, and similar cases have been confirmed in other provinces of China, German, Japan, South Korea, the United States and so on (2-6).
In general, the pathogenicity and transmissibility of most coronaviruses are slight (7), but there are two kinds of coronaviruses (severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) which have been in global outbreak (8-10). The mortality of SARS-CoV is about 10%, and that of MERS-CoV is nearly 37% (11,12). The related research points out that the clinical manifestations of COVID-19 are very similar to SARS-CoV, which can progress to acute respiratory distress syndrome and need intensive care (13-17). The median age of the patients was 49 years (IQR, 41.0–58.0 years), and 13 (32%) cases had underlying diseases such as diabetes, hypertension or cardiovascular disease (18,19). This suggests that our susceptible people may be older, which may be closely related to their own immune status.
At present, the pathogenic mechanism of SARS-CoV-2 has not been clear, and there is lack of targeted drugs (20,21). SARS-CoV-2 is considered to be similar to SARS-CoV in origin to bats HKU9-1 (15,16,22). It has been reported that angiotensin-converting enzyme 2 (ACE2) might have a strong binding ability to the SARS-CoV-2 (23). The cells or tissues expressed ACE2 may be more susceptible to be infected by SARS-CoV-2. In healthy individuals, the organs including lung, heart, kidney, bladder and oesophagus which expressed high level of ACE2 seemed to be more vulnerable to SARS-CoV-2 infection (23-25). Whereas, the gene expression in cancer patients is totally different from that in healthy people. Here, we demonstrate a landscape profiling analysis on expression level of ACE2 in pan-cancers and try to explore the risky of different type of cancers to SARS-CoV-2 infection. This might improve the vigilance of specific tumor patients to virus and provide a potential clue for preventing complications of SARS-CoV-2 infection.
Methods
The Tissue Atlas database
The Tissue Atlas database (https://www.proteinatlas.org/humanproteome/tissue) included numerous expression profiles of RNA and protein expressed in specific tissues and organs. The results defined the tissues-restricted gene is that its expression must significant elevate more than four-fold higher in special tissues compared to others. Deep sequencing of RNA (RNA-seq) was performed to acquire the mRNA expression data in thirty-seven different types of normal tissues and organs. This protein dataset included 15,313 available protein data. The expressions of proteins were validated in forty-four normal tissues by using immunohistochemistry and all images of immunohistochemistry were also available in this website.
The Blood Atlas analysis
The Blood Atlas (https://www.proteinatlas.org/humanproteome/blood) contains the information of more than 15 types of cells in blood including monocytes, NK-cells and dendritic cells etc. The RNA expression of ACE2 in these blood cells and the ACE2 protein detected in human plasma were analyzed in this database. The ACE2 protein concentration was estimated through mass spectrometry or immune assays.
Starbase dataset
Starbase database constructed the most comprehensive pan cancer expression map and interaction network including 14 cancer types (>6,000 samples). Differential expressed of ACE2 in pan-cancers were analyzed automatically when we enter the gene symbol into the search line. The gene expression of ACE2 in clinical cancer specimens was compared with that in paired normal controls using the Student’s t test. Next, Kaplan-Meier and log-rank methods were used to analyze the overall survival by Starbase dataset (http://starbase.sysu.edu.cn/panCancer.php).
Results
Expression profile of ACE2 in healthy human tissues
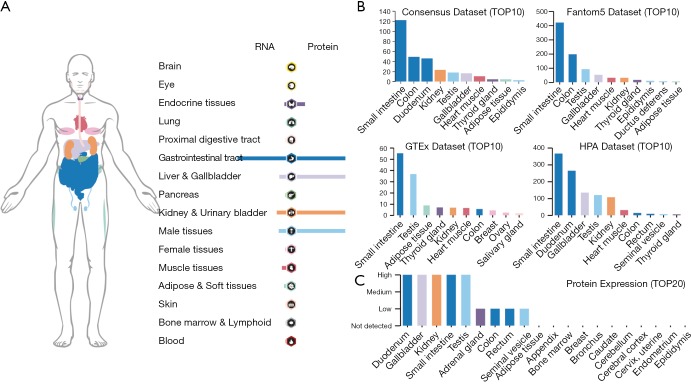
A human cell receptor ACE2 was reported to have strong binding affinity to SARS-CoV-2 (23). Here, we demonstrated an overview of RNA and protein expression data of ACE2 in healthy human tissues. These tissues were first divided into groups with different color-code according to their functional features. We found that ACE2 RNA is more enriched in endocrine tissues, gastrointestinal tract, liver and gallbladder, kidney and urinary bladder, male tissues and muscle tissues. The tendency of ACE2 protein expression level was similar to its RNA, but exhibited much more expression level in Liver and gallbladder, Kidney and urinary bladder and male tissues (Figure 1A). To further confirm the expression of ACE2 in these tissues, we analyzed the ACE2 RNA expression level in four datasets and listed the top 10 tissues expressed high ACE2 in Figure 1B. The result demonstrated that small intestine was the tissue with the highest expression of ACE2 in four datasets, the colon and kidney were exhibited a relative higher expression compared to other organs. In addition, these results showed that ACE2 was also high expressed in testis tissue, which wasn’t caught our attention before (Figure 1B). At protein level, the result showed that ACE2 was enriched in digestive and urinary system including duodenum, gallbladder, small intestine, colon, rectum and kidney (Figure 1C). Furthermore, we indicated that the male reproductive system (testis) was also high expressed ACE2 protein, which was a little different with some published reports.
Figure 1.
Expression of ACE2 in human tissues and organs. (A) The mRNA and protein expression profile of ACE2 in all tissues and organs; (B) the top 10 tissues or organs with ACE2 mRNA expression in four databases (Consensus dataset, Fantom5 dataset, GTEx dataset and HPA dataset); (C) TOP20 tissues or organs with ACE2 protein expression in related database. ACE2, angiotensin converting enzyme II.
ACE2 protein concentration in plasma
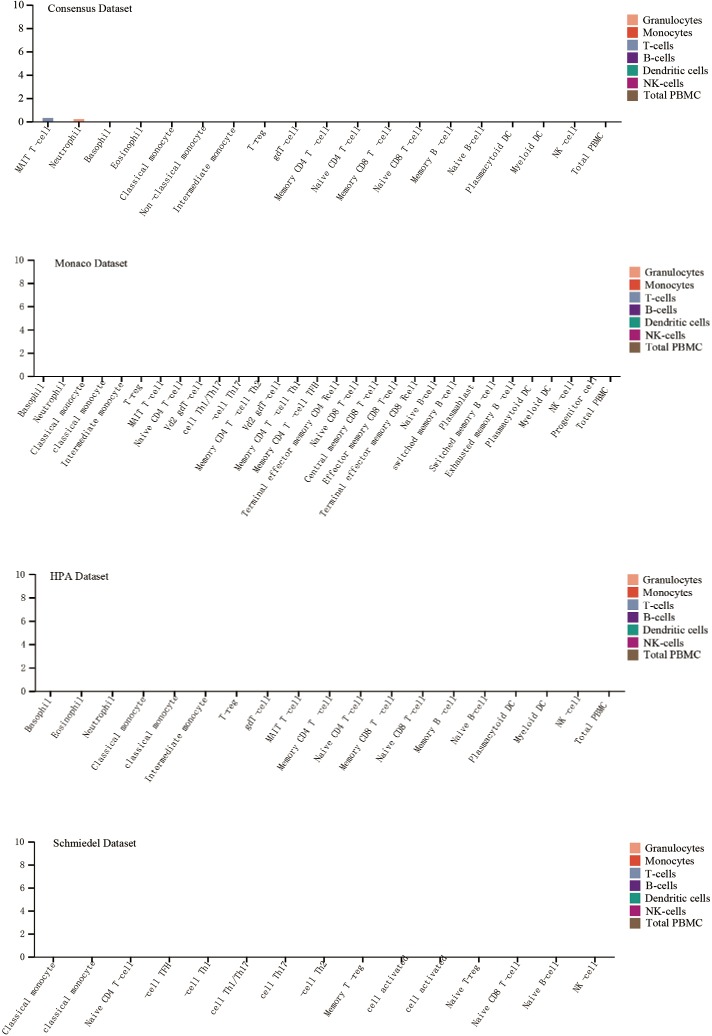
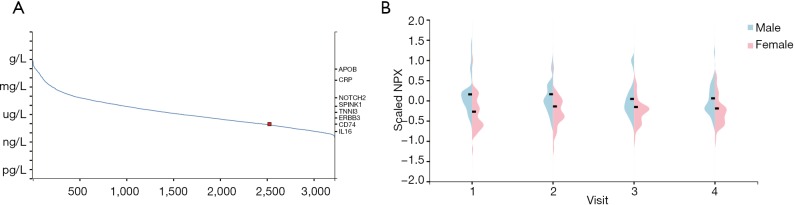
It has been reported that high concentrations of cytokines were recorded in the plasma of severe patients (19). We analyzed the ACE2 expression in immune cells and we found that it was hardly expressed in all type of immune cells in blood (Figure S1). Next, we wonder whether these abnormal cytokines were associated with the concentration of ACE2 protein in plasma. The ACE2 protein concentration was quantified by mass spectrometry and estimated from spectral counts in a publicly available data set obtained from the peptide atlas. The data showed that ACE2 protein was detected in plasma with about 85 ng/L (Figure 2A). Due to the infection rate of male is higher than that of female, we further compared the protein expression level in plasma between male and female by using the result of proximity extension assay (PEA). To our surprise, the protein concentration of ACE2 was a little higher in male plasma than that in female in four visits at three month intervals (Figure 2B). This might interpret the different susceptibility between male and female.
Figure S1.
Expression of ACE2 in human blood cells. ACE2, angiotensin converting enzyme II.
Figure 2.
ACE2 protein concentration in plasma. (A) The concentration of ACE2 protein estimated in plasma; (B) the difference of ACE2 protein concentration between male and female. ACE2, angiotensin converting enzyme II.
Expression profile and prognosis of ACE2 in pan-cancers
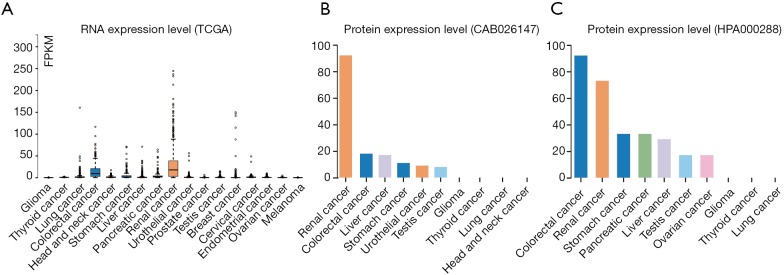
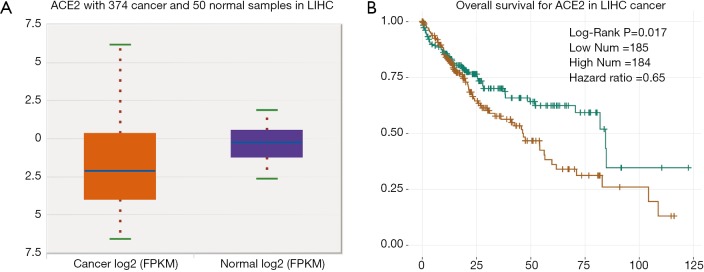
Cancer patients are suffering novel coronavirus pneumonia and risk of poor prognosis after infection due to malignant tumor itself, chemotherapy and surgery (26). We next estimated the expression profile of ACE2 in Pan-cancers. In TCGA dataset, we found that ACE2 RNA was majority highly expressed in renal cancer and colorectal cancer, which was consistent with its expression in normal tissues (Figure 3A). In addition, the protein expression level was a little different from that of RNA. This protein was not only expressed in renal and colorectal cancers but also expressed in liver and testis cancers both in CAB026147 and HP000288 datasets (Figure 3B,C). Next, we filtered the differentially expression of ACE2 between tumor and normal tissues in pan-cancer by ENCORI. In total, we found that it was significant differentially expressed in six type of cancers including kidney chromophobe (KICH), breast invasive carcinoma (BRCA), prostate adenocarcinoma (PRAD), thyroid carcinoma (THCA), liver hepatocellular carcinoma (LIHC) and stomach adenocarcinoma (STAD) with P value <0.05 and FDR <0.05 (Table 1). The expression of ACE2 was much lower in 374 patients with liver cancer compared with that in 50 normal samples (P=0.0012, FDR =0.0038). We then examined the efficiency of ACE2 in the survival of pan-cancer patients. The result revealed that decreased ACE2 expression was significantly associated with poor prognosis in overall survival (OS) of patients with kidney renal clear cell carcinoma (KIRC, P=0.00016, HR =0.55), mesothelioma (MESO, P=0.0028, HR =0.48), ovarian serous cystadenocarcinoma (OV, P=0.0073, HR =0.7) and LIHC (P=0.017, HR =0.65) (Table 2). To our attention, we found that ACE2 was both differential expression and related to the prognosis only in LIHC (Figure 4A,B).
Figure 3.
Expression Profile and Prognosis of ACE2 in Pan-cancers. (A) The RNA expression level of ACE2 in pan-cancers in TCGA dataset. The protein expression level of ACE2 in pan-cancer in CAB026147 (B) and HPA000288 (C). ACE2, angiotensin converting enzyme II.
Table 1. ACE2 significant differentially expressed in six type of cancers compared with normal samples.
| Cancer | Cancer full name | Cancer number | Normal number | Cancer Exp | Normal Exp | Fold change | P value | FDR |
|---|---|---|---|---|---|---|---|---|
| KICH | Kidney chromophobe | 65 | 24 | 0.1 | 9.49 | 0.01 | 1.10E-27 | 1.50E-25 |
| BRCA | Breast invasive carcinoma | 1,104 | 113 | 1.09 | 0.48 | 2.27 | 9.10E-22 | 8.20E-21 |
| PRAD | Prostate adenocarcinoma | 499 | 52 | 0.19 | 1.8 | 0.11 | 1.60E-13 | 3.40E-12 |
| THCA | Thyroid carcinoma | 510 | 58 | 0.6 | 0.72 | 0.83 | 0.00074 | 0.0023 |
| LIHC | Liver hepatocellular carcinoma | 374 | 50 | 2.34 | 1.07 | 2.19 | 0.0012 | 0.0038 |
| STAD | Stomach adenocarcinoma | 375 | 32 | 4.24 | 35.62 | 0.12 | 0.0048 | 0.013 |
| LUAD | Lung adenocarcinoma | 526 | 59 | 2.79 | 0.82 | 3.38 | 0.047 | 0.097 |
| UCEC | Uterine corpus endometrial carcinoma | 548 | 35 | 0.66 | 0.38 | 1.75 | 0.039 | 0.1 |
| KIRP | Kidney renal papillary cell carcinoma | 289 | 32 | 40.82 | 20.93 | 1.95 | 0.057 | 0.14 |
| COAD | Colon adenocarcinoma | 471 | 41 | 13.62 | 16.42 | 0.83 | 0.073 | 0.17 |
| PAAD | Pancreatic adenocarcinoma | 178 | 4 | 4.83 | 0.78 | 6.18 | 0.046 | 0.38 |
| ESCA | Esophageal carcinoma | 162 | 11 | 2.62 | 0.91 | 2.87 | 0.21 | 0.5 |
| HNSC | Head and neck squamous cell carcinoma | 502 | 44 | 1.41 | 1.1 | 1.17 | 0.47 | 0.62 |
| CHOL | Cholangiocarcinoma | 36 | 9 | 10.94 | 1.22 | 8.97 | 0.56 | 0.67 |
| BLCA | Bladder urothelial carcinoma | 411 | 19 | 1.28 | 1.01 | 1.26 | 0.48 | 0.73 |
| KIRC | Kidney renal clear cell carcinoma | 535 | 72 | 22.66 | 17.24 | 1.31 | 0.72 | 0.76 |
| LUSC | Lung squamous cell carcinoma | 501 | 49 | 1.56 | 0.84 | 1.85 | 0.67 | 0.77 |
ACE2, angiotensin converting enzyme II; Exp, expression level.
Table 2. The efficiency of ACE2 expression in the survival of pan-cancer patients.
| Cancer | Cancer full name | Cancer number | Median | coef. | HR | P value |
|---|---|---|---|---|---|---|
| KIRC | Kidney renal clear cell carcinoma | 517 | 15.45 | −0.59 | 0.55 | 0.00016 |
| MESO | Mesothelioma | 85 | 0.03 | −0.73 | 0.48 | 0.0028 |
| OV | Ovarian serous cystadenocarcinoma | 374 | 0.15 | −0.36 | 0.7 | 0.0073 |
| LIHC | Liver hepatocellular carcinoma | 369 | 0.22 | −0.43 | 0.65 | 0.017 |
| LUSC | Lung squamous cell carcinoma | 469 | 0.8 | −0.28 | 0.76 | 0.054 |
| STAD | Stomach adenocarcinoma | 365 | 1.28 | −0.06 | 0.94 | 0.7 |
| KICH | Kidney chromophobe | 64 | 0.07 | −0.16 | 0.86 | 0.82 |
| LUAD | Lung adenocarcinoma | 503 | 1.02 | 0.03 | 1.03 | 0.84 |
| BRCA | Breast invasive carcinoma | 1,082 | 0.05 | −0.03 | 0.97 | 0.85 |
| PRAD | Prostate adenocarcinoma | 495 | 0.1 | −0.02 | 0.98 | 0.97 |
ACE2, angiotensin converting enzyme II; coef., co-efficiency.
Figure 4.
Prognosis of ACE2 in LIHC. (A) The expression level of ACE2 in LIHC compared with normal samples; (B) overall survival for ACE2 in LIHC cancer with P=0.017. ACE2, angiotensin converting enzyme II; LIHC, liver hepatocellular carcinoma.
Discussion
Novel coronavirus (2019-nCoV) outbreak has been reported around the world with characteristic of multiple organ dysfunction syndrome (MODS) (18,27). Recently, it has been reported that tumor patients are likely to have higher risk of SARS-CoV-2 infection and poor prognosis compared with non-tumor individuals (26). Functionally, a receptor angiotensin-converting enzyme ACE2 which is widely distributed in multiple organs was considered as the potential target protein of COVID-19 (24). Consistent with other reports, we found that ACE2 was most highly expressed in digestive system especially in small intestine and colon (28). These results might interpret why SARS-CoV-2 could be isolated from excrement and urine and be transmitted via the fecal-oral route. Majority of patients infected SARS-CoV-2 also had renal dysfunction and this might be related to the high expression of ACE2 protein (23,29). In addition, ACE2 was also detected high expressed in reproductive system. Some paper had pointed out that there was no evidence that SARS-CoV-2 could pass through the placental barrier to transmit from mother to infant (30,31). Therefore, we wondered whether this virus could penetrate the reproductive barrier to affect the function of sperm. ACE2 was reported to play an important role in myocardial infarction (32). In our dataset, we didn’t detect the high expression of ACE2 in heart system.
ACE2 is located on the X chromosome, which means that the number of ACE2 in men is half that in women, but the risk of infection in men is not lower than in women. Our data showed that the protein concentration of ACE2 in plasma was a little higher in male than that in female in healthy individuals. This might be the reason how the different susceptibility between men and women. To the limitation of the public data, we couldn’t analyze the relationship between age and COVID-19. Because the cytokine storm was detected in severe infected patients, we hypothesized that this might be closely related to the immune status of patients.
In this study, we also showed the risky of different type of cancers to SARS-CoV-2 infection according the expression level of ACE2. In accordance with the result that the incidence of severe events in lung cancer patients was not significantly higher than that in other cancers, our data also demonstrated that ACE2 was not highly expressed in lung cancer cells. Furthermore, we found the unique cancer type LIHC was associated with ACE2 expression and prognosis. Relative high expression of ACE2 indicated a favorable prognosis in LIHC, but they might be more susceptible to SARS-CoV-2. Therefore, we indeed emphasized that LIHC patients with high expression level of ACE2 should be more aware of the virus infection. Our study might provide a potential clue for preventing infection of SARS-CoV-2 in cancers.
Supplementary
The article’s supplementary files as
Acknowledgments
We thank all of our colleagues at the Sun Yat-sen University Cancer Center for constructive discussions and technical help.
Funding: This work was supported by the National Natural Science Foundation of China [Grant number 81873428].
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm.2020.03.61). The authors have no conflicts of interest to declare.
References
- 1.Zhu N, Zhang D, Wang W, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med 2020;382:727-33. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rothe C, Schunk M, Sothmann P, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med 2020;382:970-1. 10.1056/NEJMc2001468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mowbray H. In Beijing, coronavirus 2019-nCoV has created a siege mentality. BMJ 2020;368:m516. 10.1136/bmj.m516 [DOI] [PubMed] [Google Scholar]
- 4.Holshue ML, DeBolt C, Lindquist S, et al. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med 2020;382:929-36. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishiura H, Kobayashi T, Yang Y, et al. The Rate of Under ascertainment of Novel Coronavirus (2019-nCoV) Infection: Estimation Using Japanese Passengers Data on Evacuation Flights. J Clin Med 2020. doi: . 10.3390/jcm9020419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin X, Gong Z, Xiao Z, et al. Novel Coronavirus Pneumonia Outbreak in 2019: Computed Tomographic Findings in Two Cases. Korean J Radiol 2020;21:365-8. 10.3348/kjr.2020.0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol 2019;17:181-92. 10.1038/s41579-018-0118-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ksiazek TG, Erdman D, Goldsmith CS, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 2003;348:1953-66. 10.1056/NEJMoa030781 [DOI] [PubMed] [Google Scholar]
- 9.Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 2003;362:263-70. 10.1016/S0140-6736(03)13967-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaki AM, van Boheemen S, Bestebroer TM, et al. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 2012;367:1814-20. 10.1056/NEJMoa1211721 [DOI] [PubMed] [Google Scholar]
- 11.Perlman S, Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat Rev Microbiol 2009;7:439-50. 10.1038/nrmicro2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hui DS, Azhar EI, Kim YJ, et al. Middle East respiratory syndrome coronavirus: risk factors and determinants of primary, household, and nosocomial transmission. Lancet Infect Dis 2018;18:e217-e227. 10.1016/S1473-3099(18)30127-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Zheng X, Tong Q, et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J Med Virol 2020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan JF, Kok KH, Zhu Z, et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect 2020;9:221-36. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coutard B, Valle C, de Lamballerie X, et al. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 2020;176:104742. 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020. [Epub ahead of print]. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, Gayle AA, Wilder-Smith A, et al. The reproductive number of COVID-19 is higher compared to SARS coronavirus. J Travel Med 2020. [Epub ahead of print]. 10.1093/jtm/taaa021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020;395:507-13. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richardson P, Griffin I, Tucker C, et al. Baricitinib as potential treatment for 2019-nCoV acute respiratory disease. Lancet 2020;395:e30-e31. 10.1016/S0140-6736(20)30304-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russell CD, Millar JE, Baillie JK. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. Lancet 2020;395:473-5. 10.1016/S0140-6736(20)30317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xia S, Zhu Y, Liu M, et al. Fusion mechanism of 2019-nCoV and fusion inhibitors targeting HR1 domain in spike protein. Cell Mol Immunol 2020. [Epub ahead of print]. 10.1038/s41423-020-0374-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wan Y, Shang J, Graham R, et al. Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol 2020. [Epub ahead of print]. 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 2013;503:535-8. 10.1038/nature12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu A, Peng Y, Huang B, et al. Genome Composition and Divergence of the Novel Coronavirus (2019-nCoV) Originating in China. Cell Host Microbe 2020. [Epub ahead of print]. 10.1016/j.chom.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang W, Guan W, Chen R, et al. Cancer patients in SARS-CoV-2 infection: a nationwide analysis in China. Lancet Oncol 2020;21:335-7. 10.1016/S1470-2045(20)30096-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou X, Chen K, Zou J, et al. The single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to Wuhan 2019-nCoV infection. Front Med 2020. [Epub ahead of print]. 10.1007/s11684-020-0754-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. 2020;395:565-74. [DOI] [PMC free article] [PubMed]
- 30.Favre G, Pomar L, Musso D, et al. 2019-nCoV epidemic: what about pregnancies? Lancet 2020;395:e40. 10.1016/S0140-6736(20)30311-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen H, Guo J, Wang C, et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet 2020;395:809-15. 10.1016/S0140-6736(20)30360-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ferrario CM. Myocardial infarction increases ACE2 expression in rat and humans. Eur Heart J 2005;26:1141; author reply 1141-3. 10.1093/eurheartj/ehi256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as