Abstract
With the continuous development of RNA biology and massive genome-wide transcriptome analysis, more and more RNA molecules and their functions have been explored in the last decade. Increasing evidence has demonstrated that RNA-related regulatory networks play an important role in a variety of human diseases, including cardiovascular diseases. In this review, we focus on RNA regulatory networks in heart disease, most of which are devastating conditions with no known cure. We systemically summarize recent discoveries of important new components of RNA regulatory networks, including microRNAs, long non-coding RNAs, and circular RNAs, as well as multiple regulators that affect the activity of these networks in cardiac physiology and pathology. In addition, this review covers emerging micropeptides, which represent short open reading frames (sORFs) in long non-coding RNA transcripts that may modulate cardiac physiology. Based on the current knowledge of RNA regulatory networks, we think that ongoing discoveries will not only provide us a better understanding of the molecular mechanisms that underlie heart disease, but will also identify novel biomarkers and therapeutic targets for the diagnosis and treatment of cardiac disease.
Keywords: heart disease, diagnosis and therapy, RNA regulatory network, non-coding RNA, microRNA, micropeptide
Graphical Abstract
Heart disease is the leading cause of death around the world with no known cure in many cases. Recent discoveries of important new components of RNA regulatory networks in cardiac pathology reviewed herein identify potential novel biomarkers and therapeutic targets for the diagnosis and treatment of this devastating disease.
Main Text
Cardiac disease is the leading cause of death and disease around the world. In response to external stress or stimulus, the heart maintains homeostasis through dynamic remodeling. In the beginning of remodeling, these adaptations are an attempt to compensate for contractile dysfunction. As cardiac remodeling progresses, compensatory changes are gradually replaced by decompensatory changes. This transition leads to contractile and conduction dysfunction and progression toward heart failure.1 When the heart is confronted with serious pathological changes, such as the creation of collagenous, non-contractile scar tissue, thinning of the myocardial wall, or progressive enlargement and dilation of the ventricle, patients have very poor prognosis and an increased risk of death.2 Despite significant progress in the treatment of cardiac disease, including heart failure, in the past decade, there remains a lack of therapeutic options that can significantly alter the morbidity and mortality. Consequently, discovery of novel therapeutic targets is urgently required to develop effective treatments for heart disease.
Most research has focused on coding genes involved in the occurrence and progression of cardiac disease. However, the vast majority of the mammalian genome that is transcriptionally active (about 75%–90%) does not encode proteins, as only ∼2% of the DNA encodes proteins.3 Therefore, exploration of RNA regulatory networks is imperative, as increasing evidence indicates that non-coding RNAs (ncRNAs) participate in regulating the expression of protein-coding genes. ncRNAs include a variety of functional RNA species, and among all ncRNAs, microRNAs (miRNAs), long ncRNAs (lncRNAs), and circular RNAs (circRNAs) have received the most attention with respect to physiology and pathophysiology, including cardiovascular biology and disease. Recently, it has been discovered that previously “mislabeled” ncRNAs encode stable and functional peptides through short open reading frames (sORFs), and the micropeptides produced from these sORFs participate in the regulation of the physiological function of the heart.4 Alternatively, most RNAs undergo a series of modifications after transcription. There are more than 100 known modifications of RNA molecules,5 which affect processes such as splicing, nucleation, stabilization, and translation of mRNAs, thereby regulating gene expression. Among these, N6-methyladenosine (m6A) is one RNA modification that is closely associated with human disease. Although early research on m6A RNA modifications focused on tumor biology, it has recently been shown that m6A modifications are involved in heart disease.6,7
In this review, we summarize the latest research of ncRNA and RNA modifications in heart disease, including cardiac remodeling, fibrosis, and regeneration. We also discuss recent developments and challenges for the development of diagnostic and therapeutic applications of ncRNAs in cardiac disease.
ncRNAs Play Important Roles in Heart Disease
miRNAs Mediate Post-Transcriptional Regulation of Gene Expression in Heart Disease
miRNAs are small, single-stranded ncRNAs with a length of 20–22 nt, which play a crucial role in regulating posttranscriptional gene expression by guiding their target mRNAs to the RNA-induced silencing complex (RISC).8 Lin-4 of C. elegans was the first documented miRNA in the early 1990s. This molecule inhibited expression of target genes to regulate developmental timing in worm larvae.9 Subsequent studies showed that one-third of the genes in the human genome are regulated by miRNAs,10 which indicated that miRNAs play a critical role in various biological processes. Large amounts of data concluded that miRNAs are involved in virtually every cellular process, including proliferation, differentiation, apoptosis, and tumorigenesis.11, 12, 13 Furthermore, accumulating evidence reveals that miRNAs are closely connected to the regulation of cardiac physiology and pathology14,15 (Table 1).
Table 1.
List of miRNA-Mediated Regulation and Cardiac Function Summarized in This Review
| miRNA | Upregulated/Downregulated | Potential Target | Cardiac Function | Refs. |
|---|---|---|---|---|
| Hypertrophy and Fibrosis | ||||
| miR-208a | up | THRAP1, myostatin | cardiac hypertrophy | 17,18 |
| miR-208a | down | MED13/NcoR1 | accelerate the conversion from compensated RVH to decompensated heart failure | 19 |
| miR-1 | down | FBLN2, TWF1, CALM1, CALM2, MEF2A, MYLK3, RasGAP, Cdk9, Rheb | inhibition of cardiac hypertrophy | 22, 23, 24, 25, 26, 27, 28, 29, 30 |
| miR-21(CF) | up | SPRY1, Jagged1, mt-Cytb | cardiac fibrosis; myocardial fibroblast proliferation and fibroblast-to-myofibroblast transformation; reduce blood pressure and attenuate cardiac hypertrophy in SHRs | 39,43,159 |
| miR-133 | down | CTGF, RHOA, CDC42, NELF-a/WHSC2 | inhibition of cardiac hypertrophy |
160,161 |
| miR-155 | down | Jarid2, | cardiac hypertrophy and cardiac remodeling | 162 |
| miR-155 (CF) | up | TP53INP1 | collagen deposition and fibrosis | 163 |
| miR-222 | up | HMBOX1, p27, HIPK1/2 | inhibition of cardiac hypertrophy | 164 |
| miR-221/222 (CF) | down | JNK1, TGF-β1, TGF-β2, ETS-1 | inhibition of fibroblast activation and differentiation | 165 |
| miR-15 family | up | Sirt4, MO25, SIRT3, TGFβR1, p38, SMAD3, SMAD7 | inhibition of hypertrophy | 48,50,166,167 |
| Cardiac Ischemic Disease | ||||
| miR-1 | up | MYOCD, Bcl2, Hsp90aa1, LXRα | apoptosis | 31, 32, 33,168 |
| miR-208 | up | BAX, CHD9, QKI15 | apoptosis | 169, 170, 171 |
| miR-126 | up | ERRFI1 | anti-apoptosis | 172 |
| miR-499 | down | CnAα/β, PDCD4, PACS2 | anti-apoptosis | 173, 174, 175 |
| miR-195 | up | CHEK1 | inhabitation of proliferation | 52,176 |
| miR-15 family | up | SMAD7, Bcl2, β2-AR, c-myb, LC3BII, MFN2, ARL2, MAPK3, CIAPIN1 | apoptosis | 53, 54, 55, 56, 57, 58, 59, 60, 61, 62 |
| Cardiac Arrhythmias | ||||
| miR-208a | down | GATA4 | cardia conduction defect | 18 |
| miR-1/133 | up | GJA1,KCNJ2, HCN2,HCN4,NCX1,B56α,CACNA1C,IRX5 | ventricular arrhythmia; cardiac conduction slow | 34,36, 37, 38,177 |
| miR-328 | up | Cacna1c, Cacnb2 | atrial fibrillation | 178 |
| miR-499 | down | KCCN3, CACNB2 | atrial fibrillation | 179,180 |
THRAP1, thyroid hormone receptor-associated protein 1; MED13/NcoR1, mediator of transcription 13/nuclear receptor corepressor 1; FBLN2, fibulin-2; TWF1, twinfilin-1; CALM1/2, calmodulin 1/2; MYLK3, myosin light chain kinase 3; RasGAP, Ras GTPase-activating protein; MEF2A, myocyte enhancer factor 2; Cdk9, cyclin-dependent kinase 9; Rheb, Ras homolog enriched in brain; CF, cardiac fibroblast; SPRY1, sprouty homolog 1; mt-Cytb, mtDNA-encoded cytochrome b; CTGF, connective tissue growth factor; RHOA, a GDP-GTP exchange protein associated with cardiac growth; CDC42, a signaling kinase involved in pathological hypertrophy; NELF-A/WHSC2, a nuclear factor correlated with cardiogenesis; Jarid2, jumonji, AT-rich interactive domain 2; TP53INP1, tumor protein p53-inducible nuclear protein 1; HMBOX1, homeobox containing 1; HIPK1/2, homeodomain interacting protein kinase 1/2; JNK1, c-Jun N-terminal kinase 1; ETS-1, ETS proto-oncogene 1; SIRT3/4, sirtuin 3/4; MYOCD, myocardin; Bcl-2, B cell CLL/lymphoma 2; BAX, BCL2-associated X; QKI15, RNA-binding protein Quaking 15; CHD9, chromodomain helicase DNA-binding protein 9; ERRFI1, ERBB receptor feedback inhibitor 1; CnAα/β, calcineurin catalytic subunits; PDCD4, programmed cell death 4; PACS2, phosphofurin acidic cluster sorting protein 2; CHEK1, checkpoint kinase 1; β2-AR, β2 adrenergic receptor; MFN2, mitofusin 2; ARL2, ADP-ribosylation factor-like protein 2; MAPK3, mitogen-activated protein kinase 3; CIAPIN1, cytokine-induced apoptosis inhibitor 1; GJA1, gap junction protein α1; KCNJ2, potassium inwardly rectifying channel subfamily J member 2; HCN2/HCN4, hyperpolarization activated cyclic nucleotide gated potassium and sodium channel 2/4; NCX1, sodium/calcium exchanger protein; CACNA1C, calcium voltage-gated channel subunit α1C; IRX5, iroquois homeobox 5; CACNB2, voltage-dependent calcium channel β2 subunit; Cacna1c, calcium voltage-gated channel subunit 1C; Cacnb2, calcium voltage-gated channel auxiliary subunit 2; KCNN3, potassium calcium-activated channel subfamily N member 3.
Cardiac-Enriched miRNAs. A subset of miRNAs are enriched in the heart, such as miR-1, miR-133, miR-208, and miR-499.16 miR-208 was one of the first miRNAs reported to be involved in cardiac hypertrophy.17 Both gain- and loss-of function studies demonstrated that miR-208 was required for cardiac hypertrophy by targeting the thyroid hormone receptor-associated protein 1 (THRAP1). miR-208a, which is encoded within an intron of Myh6, and miR-208b, which is encoded within an intron of Myh7, are members of a miRNA family that is differentially expressed during cardiac development and pathology. Callis et al.18 demonstrated that overexpression of miR-208a is sufficient to induce cardiac hypertrophy, accompanied with increased β-myosin heavy chain (β-MHC) expression. miR-208a targets Thrap1 and myostatin, two important negative regulators of hypertrophic growth. In addition, miR-208a is required for normal cardiac conduction. Electrocardiograms (ECGs) showed that approximately 80% of Mir208a–/– mice lacked P waves and had prolonged PR intervals compared to wild-type mice. An additional study confirmed that miR-208a regulates expression of Cx40 and Hop through the transcriptional cofactor GATA4. Furthermore, a recent study reported that miR-208 is progressively downregulated as right ventricular hypertrophy progressed because of pulmonary hypertension. miR-208 also inhibited the expression of Mef2 through the Med13-NCoR1 axis, and therefore suppresses the disease transition from compensation to decompensation.19
miR-1 is another well-studied, cardiac-enriched miRNA. miR-1-1 and miR-1-2 are members of the miR-1 family and are located at separate chromosomal loci. miR-1 and miR-133a form a miRNA gene cluster and are co-expressed during cardiomyocyte differentiation and proliferation.20 Sayed et al.21 showed that several targets of miR-1 are involved in progressive myocardial hypertrophy and cardiac remodeling, including Ras GTPase-activating protein (RasGAP) and cyclin-dependent kinase 9 (Cdk9), activators of cardiac hypertrophy,22,23 Ras homolog enriched in brain (Rheb), an upstream activator of protein synthesis, and the cell growth-related mammalian target of rapamycin (mTOR)/S6 kinase pathway.24,25 Recent studies confirmed that miR-1 suppresses cardiac hypertrophy by inhibiting the expression of various downstream targets, including fibulin-2 (FBLN2),26 twinfilin-1 (TWF1),27 CALM1 and CALM2, MEF2A,28 MYLK3,29 and GATA4.30 In addition, the serum level of miR-1 and miR-133 is elevated in animal models and human patients with acute myocardial infarction (AMI). Inhibition of miR-1 with antisense oligonucleotides attenuates myocardial apoptosis by targeting Bcl2.31 Other studies reveal that miR-1 also represses expression of Hsp90aa1 and the liver X receptor α (LXRα), which affects cardiomyocyte apoptosis during myocardial infraction (MI).32,33 Similar to miR-208a, miR-1 is also required for normal cardiac electrophysiology. Widening of the QRS complex and a prolonged QT interval were observed in miR-1-transfected hearts.34 miR-1 repressed expression of its targets, GJA1 and KCNJ2, and led to a lower protein level of Cx43 and Kir2.1, resulting in a propensity for arrhythmia. In addition, it has been reported that miR-1 and miR-133 targeted several ion channel- and gap junction-associated genes, such as HCN2, HCN4,35 NCX1,36, B56α,37 CACNA1C, and IRX5.38
Therefore, these cardiac-enriched miRNAs seem to be housekeepers of cardiomyocytes. They maintain cardiomyocyte physiology, including assembly and function of the contractile apparatus as well as controlling electrophysiological function, to ensure efficient and coordinated pumping of blood to the circulation.
Ubiquitously Expressed miRNAs. Other than cardiac-enriched miRNAs, some ubiquitously expressed miRNAs also play important roles in cardiac pathology. Previous studies have shown that miR-21 is closely involved in the pathological progression of multiple cardiac abnormalities, including aberrant remodeling, arrhythmia, heart failure, and infarction. Thum et al.39 found that miR-21 activated the ERK/MAPK (extracellular signal-regulated kinase/mitogen-activated protein kinase) signaling pathway by inhibiting Spry1 expression, thereby promoting cardiac fibroblast activation and growth factor secretion. Interestingly, intravenous injection of antagomiR-21 suppresses myocardial fibrosis and preserves cardiac function; however, the precise mechanism remains poorly understood. It was suggested that fibroblast exosomal-derived miR-21_3p (miR-21∗) is a potent paracrine-acting RNA molecule that induces cardiomyocyte hypertrophy.40 A recent study showed that miR-21 plays a key role in myocardial fibroblast activation and myocardial fibrosis following MI by targeting the transforming growth factor β (TGF-β)1/Smad7 signaling pathway.41 Interestingly, phosphorylated Smad2 and Smad3, which are downstream effectors of TGF-β signaling, interact with DROSHA to promote processing of primary miR-21 under pressure overload through a feedback loop.42 Zhou et al.43 also showed that miR-21 promotes myocardial fibroblast proliferation and fibroblast-to-myofibroblast transformation by targeting Jagged1. Also note that loss of miR-21 through genetic engineering could not recapitulate the cardiac phenotype observed as a consequence of antagomiR interference,44 indicating that the transient interference with the function of miR-21 could be compensated for by other mechanisms in the long term.
miR-21 also participates in the regulation of cardiomyocyte apoptosis in ischemic cardiomyopathy. It was reported that miR-21 is downregulated in the infarcted region 6 h after AMI.45 Additional studies demonstrated that miR-21 inhibits hypoxia-induced apoptosis through the PDCD4/AP-1 (activator protein 1) pathway by targeting PDCD4. Therefore, miR-21 appears to play a protective role in reducing oxidative stress in cardiomyocytes due to ischemia/reperfusion (I/R) injury.46,47
The miR-15 family consists of six members, which possess a common seed sequence, including miR-15a, miR-15b, miR-16-1, miR-16-2, miR-195, and miR-497. Recently, several studies have shown that the miR-15 family plays crucial roles in the pathogenesis of cardiac disease. Tijsen et al.48 found that the miR-15 family was upregulated in the hypertrophic heart. Inhibition of miR-15b with locked nucleic acid (LNA)-based antimiRs leads to a significant increase in heart weight, excessive fibrosis, and collagen deposition during hypertrophy. The miR-15 family inhibits canonical and non-canonical TGF-β signaling, which constitutes a critical pathway for cardiac fibrosis and hypertrophy, by targeting multiple direct and indirect genes, including TGFβR1, P38, SMAD3, SMAD7, and endoglin. A previous study indicated that miR-195 plays an essential role in hypertrophic growth and chamber remodeling of the heart in response to pathological signaling.49 It was further demonstrated that the elevated expression of miR-195 in hypertrophic cardiomyocytes impedes the formation of LKB1/STRAD/MO25 complexes and activates the AMPK (AMP-activated protein kinase) pathway by suppressing MO25.50
Most mammalian cardiomyocytes lose the ability to regenerate shortly after birth. Once the heart is severely damaged by injuries such as those caused by MI, cardiomyocyte replenishment is insufficient to repair the damage.51 Porrello et al.52 found that multiple miR-15 family members, including miR-195, miR-497, miR-15a, and miR-16, are upregulated in the mouse ventricles between postnatal day 7 and 14. Inhibition of the miR-15 family prevents cardiomyocyte mitotic arrest and improves cardiac function after MI. Additional experiments showed that miR-195 regulates a number of mitotic genes in vivo by targeting Chek1. Other studies revealed that the miR-15 family not only regulates cardiomyocyte proliferation and cardiac regeneration, but it also modulates cardiomyocyte apoptosis. Loss of miR-15 family members in vitro or in vivo renders cardiomyocytes resistant to hypoxia-induced cell death, reduces infarct size, and suppresses cardiac remodeling.53 Recent studies further demonstrated that the miR-15 family targets other downstream genes involved in regulating cardiomyocyte apoptosis, such as SMAD7,54 Bcl2,55, 56, 57 β2 adrenergic receptor (β2-AR),58 c-myb,59 LC3B-II,57 mitofusin 2 (MFN2),60 ADP-ribosylation factor-like protein 2 (ARL2),53 MAPK3,61 and cytokine-induced apoptosis inhibitor 1 (CIAPIN1).62
In summary, a large effort has been expended on investigating these “tiny” miRNAs. Many of the miRNAs listed in Table 1, but not discussed here in detail, participate in the regulation of many aspects of cardiac physiology and pathology.
lncRNAs Have a Variety of Molecular Functions in Regulating Heart Disease
New technologies for genome-wide, massively parallel sequencing have led to the discovery that vast regions of the mammalian genome are actively transcribed into RNA. Surprisingly, all protein-coding sequences originate from about only 1.5% of the human genome sequence.63 As a result, numerous non-coding transcripts have been identified. lncRNAs belong to a class of ncRNAs with a length of more than 200 nt. Because of a huge effort, more and more lncRNAs are now known to have significant regulatory functions in cardiovascular biology.64,65 Herein, we have cataloged lncRNAs with important functions in cardiac remodeling, including those involved in hypertrophy, apoptosis, necrosis, and fibrosis66,67 (Table 2).
Table 2.
List of Cardiac Function of lncRNAs and Their Molecular Mechanisms Summarized in This Review
| lncRNA | Upregulated/Downregulated | Potential Mechanism | Effect | Refs. |
|---|---|---|---|---|
| Hypertrophy | ||||
| Mhrt | down | interacts with Brg1 | inhibits developing heart failure | 68,69,181 |
| Chaer | Up | interacts with PRC2 | promotes cardiac hypertrophy | 70,182 |
| Chrf | Up | sponge for miR-489 | promotes cardiac hypertrophy | 71 |
| Apoptosis and Autophagy | ||||
| APF | Up | sponge for miR-188-3p | promotes deregulated autophagy and cell death | 76 |
| CAIF | down | interacts with p53 | inhibits autophagy | 77 |
| MALAT1 | Up | sponge for miR-203 | worsens cardiomyocyte inflammation and apoptosis | 78 |
| Electrical Activity | ||||
| MALAT1 | up | sponge for miR-200c | regulates transient outward potassium current | 79 |
| Cardiac Fibrosis | ||||
| MALAT1 | Up | sponge for miR-145 | promotes cardiac fibrosis and deteriorates cardiac function after MI | 80 |
| Wisper | Up | interacts with TIAL1 | promotes cardiac fibrosis | 81 |
| MEG3 | down | interacts with p53 | promotes cardiac fibrosis and impaired diastolic performance | 82 |
| GAS5 | down | sponge for miR-21 | inhibits cardiac fibrosis | 83 |
Mhrt, Myheart; Brg1, also known as Smarca4 (SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4); Chaer, cardiac hypertrophy-associated epigenetic regulator; PRC2, polycomb repressive complex 2; Chrf, cardiac hypertrophy-related factor; APF, autophagy-promoting factor; CAIF, cardiac autophagy inhibitory factor; MALAT1, metastasis-associated lung adenocarcinoma transcript1; MI, myocardial infarction; Wisper, Wisp2 super-enhancer-associated RNA; TIAL1, TIA1 cytotoxic granule-associated RNA-binding protein-like 1; MEG3, maternally expressed gene 3; GAS5, growth arrest-specific 5.
Cardiac hypertrophy is an adaptive response by the heart to counteract cardiac overload to maintain output. However, sustained hypertrophy often leads to heart failure. Recently, lncRNA Myheart (Mhrt), which originates the from MYH7 locus and is enriched in adult hearts, was found to protect the adult heart from pathological hypertrophy by interacting with the helicase domain of Brg1 and inhibiting the function of Brg1, a chromatin-remodeling factor that is activated by stress and triggers aberrant gene expression and cardiomyopathy.68,69 Conversely, lncRNA Chaer (cardiac hypertrophy-associated epigenetic regulator) is required for the pathogenesis of cardiac hypertrophy. Chaer interacts with PRC2 and interferes with the targeting of the PRC2 complex to genomic loci, which inhibits PRC2-dependent histone H3 lysine 27 trimethylation at the promotor of prohypertrophic genes and the activation of their expression.70 The molecular mechanisms that underlie lncRNA regulation of cardiac hypertrophy are not limited to their action as a decoy for epigenetic regulators, as they also function as endogenous sponges for miRNAs. For example, lncRNA Chrf serves as a competing RNA by sequestering miR-489 and de-repressing the miR’s target, MYD88.71 Furthermore, ROR,72 H19,73 Plscr4,74 and MI-associated transcript (MIAT)75 regulate cardiac hypertrophy through a similar mechanism by inhibiting the function of different miRNAs.
lncRNAs, such as APF, CAIF, and MALAT1 (metastasis-associated lung adenocarcinoma transcript 1) were reported to regulate cardiomyocyte apoptosis and autophagy in heart disease. Under pathological conditions, upregulating the autophagy promoting factor (APF) de-represses the autophagy gene ATG7 by sequestering miR-188-3p. This leads to abnormal autophagy as well as cell death.76 In contrast, lncRNA CAIF (cardiac autophagy inhibitory factor) acts as a cardioprotective factor. CAIF inhibits p53-induced transcription of myocardin by directly binding to its promoter, which leads to the suppression of cardiac autophagy and protection of the heart during MI.77 During I/R injury, MALAT1 is highly expressed in heart, and it leads to a more severe cardiomyocyte inflammation and apoptosis by sequestering miR-203.78
Other than cardiac hypertrophy and apoptosis, lncRNAs have also been reported to regulate arrhythmia and fibrosis in heart disease. MALAT1 was reported to regulate electrical activity in an arrhythmic rat model by modulating expression of the miR-200c-HMGB1 axis in cardiomyocytes. Expression of transient outward potassium current and Kv4.2/Kv4.3 channel proteins are regulated via HMGB1 when MALAT1 is knocked down.79 In addition, knockdown of MALAT1 inhibits AngII-induced fibroblast proliferation and collagen synthesis, and then suppresses cardiac fibrosis following MI by suppressing TGF-β1 activity via miR-145.80 Other lncRNAs, such as Wisp2 super-enhancer-associated RNA (Wisper),81 MEG3,82 and GAS5,83 have also been reported to participate in the regulation of cardiac fibrosis through various molecular mechanisms.
As mentioned above, a large number of lncRNAs play important roles in cardiac remodeling during stress. It is also noteworthy that several lncRNAs, including Braveheart (Bvht)84 and Fendrr,85 are critical to cardiac lineage commitment and lead to developmental defects in the heart when these lncRNAs are deleted. Both of these lncRNAs interact with the PRC2 complex and epigenetically regulate the cardiac transcriptome during cardiac development.
circRNAs Primarily Function as a miRNA Sponge in Heart Disease
circRNAs are a class of ncRNA molecules shaped by a covalently closed continuous loop. Previous studies indicated that circRNAs play vital roles in the regulation of gene expression, including miRNA sponge effects, transcriptional and post-transcriptional gene expression regulation, alternative splicing, and protein coding and protein decoy activity. Some of these molecules are expressed in a tissue-specific manner.86, 87, 88, 89, 90 Recently, it has been shown that circRNAs are closely related to the pathological and physiological processes of various cardiac diseases, such as myocardial ischemia, myocardial fibrosis, cardiac hypertrophy, and heart failure.
An early study on the role of circRNAs in hypertrophy and heart failure revealed decreased expression of circRNA HRCR, which functions as a sponge to sequester cardiac miR-223, in the failing heart. In vivo overexpression of HRCR results in increased expression of miR-223’s downstream target ACR,91 which is an apoptosis repressor with a CARD domain. ACR plays a crucial role in cardiomyocyte hypertrophy and apoptosis92 and protects the heart from hypertrophy and failure. Another interesting study reported that expression of circRNA Foxo3 is significantly higher in aged hearts compared to young hearts. It induces cellular senescence and doxorubicin-induced heart failure by interacting with the anti-senescence proteins ID1 and E2F1, and the anti-stress proteins FAK and HIF-1α. These interactions block the nuclear translocation of these proteins and inhibit their function as transcription factors.93
Multiple circRNAs have been reported to regulate apoptosis and survival in heart disease. circRNA cerebellar degeneration-related protein 1 transcript (Cdr1) contains complementary binding sites for miR-7a that may function as miRNA sponges. This circRNA de-represses targets of miR-7a, PARP, and SP1, and it participates in the regulation of apoptosis after MI injury.94 In another study, it was shown that mitochondrial fission and apoptosis-related circRNA (MFCAR) plays an essential role in modulating mitochondrial fission and apoptosis by acting as a sponge for miR-652-3p. MFCAR prevents miR-652-3p from binding with mitochondrial membrane-associated protein 18 (MTP18). Knockdown of MFCAR decreases expression of MTP18 and attenuates mitochondrial fission and cardiomyocyte apoptosis in MI injury.95 Other than acting as miRNA sponges, circRNAs can interact with proteins and regulate their activities. A recent study reported that circ-Amotl1 binds to AKT and PDK1 and induces their nuclear translocation.96 In vivo, circ-Amotl1 overexpression enhances cardiomyocyte survival and, therefore, protects the heart in doxorubicin-induced cardiomyopathy.96 Furthermore, Zhou et al.97 report that a circRNA, autophagy-related circRNA (ACR), protects the heart from I/R injury and reduces the extent of the infarct. Mechanistically, ACR directly binds to Dnmt3B and blocks Dnmt3B-mediated DNA methylation of the promoter of Pink1, which suppresses autophagy via phosphorylating its downstream target, FAM65B.
Emerging evidence indicates that circRNAs also participate in the regulation of cardiac regeneration. Super-enhancer-associated circRNA circNfix was found to enhance expression in the adult heart.98 This study showed that circNfix regulates cardiomyocyte proliferation through diverse molecular mechanisms. circNfix functioned as a miRNA sponge to modulate Gsk3β signaling activity by sequestering miR-214. Alternatively, circNfix interacts with Ybx1 (Y-box-binding protein 1) and Nedd4l (an E3 ubiquitin ligase) and enhances the interaction of these two proteins, which induces Ybx1 degradation through ubiquitination. Knockdown of circNfix promotes cardiomyocyte proliferation and angiogenesis and, therefore, attenuates cardiac dysfunction and protects the heart after MI.
Aside from their function for RNA transcripts, a recent study showed that ribosome-associated cardiac circRNAs produce detectable peptides.4,99 The roles of these peptides in cardiac disease are currently unknown and, consequently, provide a new direction for future exploration.
Micropeptides Encoded by “Non-coding” RNAs in Heart Disease
Micropeptides are a group of protein molecules less than 100–150 aa in length.100 Micropeptides are significantly different from bioactive peptides, because the former originate from sORFs, which nest in transcripts identified as lncRNAs and TUFs (transcripts of unknown function), whereas the latter are derived from larger precursor proteins and contain N-terminal signal sequences.101 Because they are short, traditional computational prediction programs of protein-coding ORFs excluded these sORFs as false positives.102,103 Studies have shown that some of these sORF have non-classical start codons as well as low sequence conservation, which posed a challenge to uncover these micropeptides in the mammalian genome.100
Using emerging technologies and experimental approaches, researchers have begun to address this challenge. For example, Anderson et al.104 reported a group of micropeptides, named myoregulin (MLN), phospholamban (PLN), and sarcolipin (SLN). These peptides have similar conserved regions in their peptide sequence as well as a homologous function to inhibit SERCA activity by regulating cardiac calcium uptake in muscle. Another two micropeptides that have functions similar to MLN/PLN/SLN were subsequently identified and named endoregulin (ELN) and another-regulin (ALN).105 The search for SERCA-associated regulatory micropeptides did not end there, as the identification of the micropeptide dwarf ORFs (DWORFs) revealed enhanced SERCA activity by displacing the SERCA inhibitors PLN, SLN, and MLN in the mouse heart. So far, DWORF is the only endogenous peptide known to activate the SERCA pump by a physical interaction, resulting in enhanced muscle contraction.106
As more micropeptides are identified, the questions of how many micropeptides are present in the heart and whether they share any common features will eventually be answered. For instance, a genome-wide study recently identified micropeptides in diseased hearts. As a result, hundreds of micropeptides were found in human, mouse, and rat hearts. Interestingly, the overall coding sequence for these micropeptides were less conserved than that observed in traditional proteins. Furthermore, this study indicated that many microproteins are produced from sORFs located in lncRNAs identified with previously described cardiac functions, such as Myheart,69 chaer,70 UPPERHAND (also known as UPH or HAND2-AS1),107 ZFAS1,108 and TRDN-AS (also known as RP11-532N4.2).109 Although the subcellular location of micropeptides varied, most of these localized to mitochondria,4 which suggests that micropeptides could have important regulatory functions for mitochondrial biogenesis and function. Indeed, recent studies have shown a micropeptide named MOXI (micropeptide regulator of β-oxidation)110 or Mtln (mitoregulin)111 interacts with the mitochondrial trifunctional protein (MTP) and several mitochondrial complexes to regulate mitochondrial function, including fatty acid β-oxidation, respiratory (super)complex formation and activity, Ca2+ retention, and reactive oxygen species formation.
It is important to recognize that as more micropeptides are identified, it will be necessary to take this into account in future investigations of RNA regulatory networks, especially those determining the function of lncRNAs. To accurately define the function of a “non-coding” gene, the coding potential of a transcript needs to be carefully excluded when investigating the function of lncRNAs. For example, recent studies identified UPPERHAND as a critical lncRNA during cardiac development.107,112 However, a potential coding sORF was also identified in both human and murine UPPERHAND.4 Therefore, further studies need to be carried out to determine whether the discovered function of UPPERHAND was derived from the RNA transcript or the micropeptide. Alternatively, more effort should be expended on genome-wide discovery of sORFs to define the noncoding gene. Various approaches, such as computational analyses, ribosome profiling (Ribo-seq [ribosome sequencing]), mass spectrometry, and combinations of these procedures are recommended for accurately identifying protein-coding sORFs.
Dysregulation of RNA Modifications Is Associated with Heart Disease
RNA molecules often undergo various modifications post-transcriptionally. m6A methylation is one of the most widespread, internal, post-transcriptional modifications of eukaryotic mRNAs, involving the regulation of physiological and pathological activities by modifying mRNA or ncRNA.113 Although m6A was first discovered in 1974,114 its location in mRNAs and functions are not fully understood. Recently, the dynamics and function of m6A modifications of mRNAs in different biological processes have been intensively investigated. The m6A modifications were recently reported to facilitate cap-independent mRNA translation.115 The modification of m6A can be dynamically deposited, removed, and identified by a series of methyltransferases (METTL3/14, WTAP, RBM15/15B, ZC3H13, KIAA1429, and METTL16, termed “writers”), demethylases (FTO and ALKBH5, termed “erasers”), and m6A-binding proteins (YTHDF1/2/3, IGF2BP1 and HNRNPA2B1, termed “readers”).116,117 More and more studies have demonstrated that the abnormal dynamics of methylation of RNA on N6-adenosines are closely related to tumorigenesis.118
Until recently, the connection between m6A RNA modifications and heart disease was yet to be explored. Dorn et al.6 demonstrated that m6A modification of a subset of mRNAs was significantly increased in response to a hypertrophic stimulus in cardiomyocytes. As an important enzyme for N6-adenosine methylation, overexpression of METTL3 was sufficient to induce adaptive cardiac hypertrophy in the heart. Conversely, inhibition of METTL3 expression suppressed the hypertrophic growth of cardiomyocytes. Furthermore, the METTL3 knockout mouse showed gradual pathological changes during aging and stress. Interestingly, m6A modifications were found to occur specifically at MAPK mRNAs, which are important for hypertrophic growth of cardiomyocytes. In an ischemic heart mouse model, Song et al.119 have shown that increased Mettl3 activity promoted the association of HNRNPD with Tfeb pre-mRNA by regulating m6A modifications in the Tfeb 3′ UTR, and then decreased Tfeb mRNA stability in hypoxia/reoxygenation-treated cardiomyocytes, which inhibited the autophagic flux and promoted apoptosis of cardiomyocytes. In another recent study, decreased FTO expression and increased m6A RNA modifications were found in failing mammalian hearts and hypoxic cardiomyocytes. Importantly, myocardial overexpression of FTO showed a protective effect in ischemic hearts.7 It was shown that loss of FTO leads to abnormal calcium homeostasis and sarcomeric dynamics. In contrast, FTO overexpression selectively increases demethylation of contractile protein-related mRNAs, thereby inducing their expression. In addition, decreased cardiac fibrosis and enhanced angiogenesis were observed in the FTO-overexpressing ischemic myocardium through an unknown mechanism. Future studies may uncover the underlying mechanisms, which could lead to an identification of novel therapeutic strategy for MI. So far, while the m6A modification of mRNA was linked to heart disease, it will be interesting to find out whether m6A modifications of ncRNAs are involved in the pathogenesis of cardiac disease.
Increased mRNA translation is an essential step for cardiac remodeling, in which several key signaling pathways are involved, including AKT120 and AMPK.121,122 Similar to m6A modifications, which affect the translational activity of mRNA, the length of the poly(A) tail of the PABPC1 mRNA, coding a poly(A)-binding protein known to promote translation, was reported to be a key modification regulating the translation efficiency of its own mRNA.123 Pabpc1 poly(A) tail length was found to be significantly shorter in the adult heart compared to its length in the embryonic heart. This effect is correlated with the translational silencing of Pabpc1 in the adult heart under physiological conditions. The shortening of the poly(A) tail was reversed in the hypertrophic heart. It significantly enhanced the translation of Pabpc1 and triggered the subsequent global mRNA translational enhancement observed in cardiac hypertrophy. Unfortunately, the detailed mechanism of how this modification is regulated remains to be thoroughly explored.
RNA Molecules Are Potential Targets for Clinical Diagnosis and Gene Therapy for Cardiac Disease
One of the ultimate goals for investigating RNA regulatory networks in cardiac disease is to develop clinical applications with those RNA molecules, which can serve as biomarkers for disease diagnosis/prognosis and/or therapeutic targets.
Other than behaving as regulatory factors in the pathogenesis of cardiac disease, ncRNAs also function as paracrine factors by interacting with proteins to form RNA-protein complexes, as well as with lipids or high-density lipoproteins in the circulation.124 These complexes are stable and resistant to RNase degradation. Therefore, some ncRNAs with different expression levels in the serum of healthy and diseased people have the potential to act as biomarkers for the diagnosis of heart disease.
Effective biomarkers are important for assessing post-infarction risk and treatment responses in AMI. miR-1, miR-126, and cTnT expression levels in plasma from patients with AMI are significantly elevated, suggesting that miR-1 and miR-126 could be valuable indicators for AMI.125 miR-499 is specifically expressed in cardiomyocytes and only increases after AMI.126 Therefore, miR-499 could be an important biomarker for MI, especially NSTEMI.127 Other miRNAs, such as miR-208,128 miR-133,129 miR-1254,130 miR-99a,131 miR-122-5p,132 miR-874-3p,133 miR-19b, miR-223, and miR-483-5p,134 also have the potential to predict MI as biomarkers. The potential for lncRNAs to serve as biomarkers of cardiac disease has also been investigated. Vausort et al.135 demonstrated that levels of circulating lncRNAs aHIF, KCNQ1OT1, and MALAT1 were higher in patients with MI than in healthy volunteers, while levels of the circulating lncRNA ANRIL were lower. A recent study showed that high plasma ANRIL levels were correlated with a high risk of in-stent restenosis (ISR).136 Other evidence suggests that HOTAIR,137 UCA1,138 MHRT,139 MIAT,140 LIPCAR,141 CDR1AS, and ZFAS1142 could serve as potential markers for diagnosis and prognosis of AMI or CAD. circRNAs were found to be abundant in circulating blood and more stable than linear RNAs because of the closed-loop structure.143 These attributes allow the detection of these circRNAs using a convenient method.144 circRNA MICA was found to be downregulated in peripheral blood samples from MI patients.145 A study of 472 patients with AMI showed that circRNA MICRA improved the predictive value of a multivariable clinical model and it also improved the risk classification of patients after MI.146
The potential of ncRNAs as biomarkers for heart failure was also investigated. Similar to established diagnostic protein biomarkers, such as cTnI, circulating cardiac-enriched miRNAs (myomirs) increased up to 140-fold in advanced heart failure.147 In a study of chronic heart failure, miR-660-3p, miR-665, miR-1285-3p, and miR-4491, which were derived from cardiac fibroblasts, were significantly increased in heart and plasma.148 Recent studies also showed that many circulating miRNAs were differentially expressed in heart failure, including miR-18a-5p, miR-26b-5p, miR-27a-3p, miR-199a-3p,149 miR-499,150 miR-155-5p, and miR-595.151 Some of these were also demonstrated to be effective in the assessment of risk. For example, a decrease in plasma miR-18a-5p and miR-652-3p during early hospitalization was found to correlate with an increased risk of mortality within 180 days.149 Other circulating ncRNAs, such as lncRNAs, were also investigated for their potential as biomarkers in heart failure. Previous studies showed lncRNA UCA1 could predict a similar survival rate compared to BNP in patients with chronic heart failure.152 Similarly, quantitative analysis of lncRNAs in plasma revealed that NRON and MHRT have great potential as predictive biomarkers for heart failure.153
Other than biomarkers, ncRNAs are also attractive candidates for therapeutic targets in treating various human diseases.154 Some pioneering studies for cardiac regeneration with miRNAs have been carried out. Studies demonstrated that the miR-17-92 cluster plays a critical role in regulating cardiomyocyte proliferation in postnatal and adult hearts.155 A recent follow-up study explored the therapeutic potential of miR-19a/19b in protecting the heart in response to MI.156 In a MI mouse model, direct injection of miR-19a/19b mimics or AAV9-miR-19a/19b into infarcted hearts reduced scar formation, improved cardiac function, and promoted cardiomyocyte proliferation. Also note that transient overexpression of miR-19a/19b by injecting miR mimics has a long-term protective effect. Further investigation of the therapeutic effect of miR-19a/19b in a large animal model needs to be performed to demonstrate the therapeutic potential of this miR for MI in humans. Another miR, miR-199a, has also been shown to regulate cardiac regeneration. miR-199a promoted cardiomyocyte proliferation in both neonatal and adult rats.157 Excitingly, miR-199a also showed a therapeutic potential for MI in a large animal model. In an I/R injury pig model, overexpression of miR-199a in the myocardium using adenovirus-associated virus had a protective effect on the injured heart with better global cardiac function and regional/segmental contractility 28 days after injury.158 Further evidence demonstrated that morphological and functional improvements are associated with the role of miR-199a in promoting endogenous cardiomyocyte proliferation. However, it has been noticed that persistent and uncontrolled expression of miR-199a can cause sudden death due to arrhythmia. Therefore, several key factors, such as dosage, time window, and delivery approach, have to be carefully investigated before human trials can proceed.
A huge amount of effort has been spent on exploring targets and developing approaches for clinical applications in the diagnosis/treatment of cardiac diseases using proteins. Although great strides have been made, the clinical need has not yet been met. Numerous investigations of RNA regulatory networks, especially ncRNAs, will continue to provide new RNA targets with therapeutic potential. RNA targets have their own advantages as opposed to proteins, such as not relying on antibodies for their detection and their ease of synthesis and delivery. Therefore, in combination with protein targets, the discoveries of RNA regulatory networks will likely lead to a breakthrough in clinical applications for heart disease.
Conclusion and Perspective
After annotation of the human genome, people surprisingly found that the amount of protein-coding genes and the length of coding sequences were comparable to many other vertebrates and even invertebrates, such as C. elegans. However, humans have more abundant non-coding DNA sequences than other lower species. Until the last two decades, ncRNAs started to be explored, and the known regulatory networks in cardiac pathology, which mainly consist of proteins, are likely to be just a “the tip of iceberg” phenomenon. In this review, we summarize the main discoveries in RNA regulatory networks in cardiac disease, which are just the beginning of exploring the “dark matter” of the human genome. Clearly, RNA molecules are one of an indispensable component of these networks. Further work will help us better understand the underlying molecular mechanisms of cardiac disease (Figure 1). Perhaps more importantly, this knowledge may provide a roadmap to defeat heart disease.
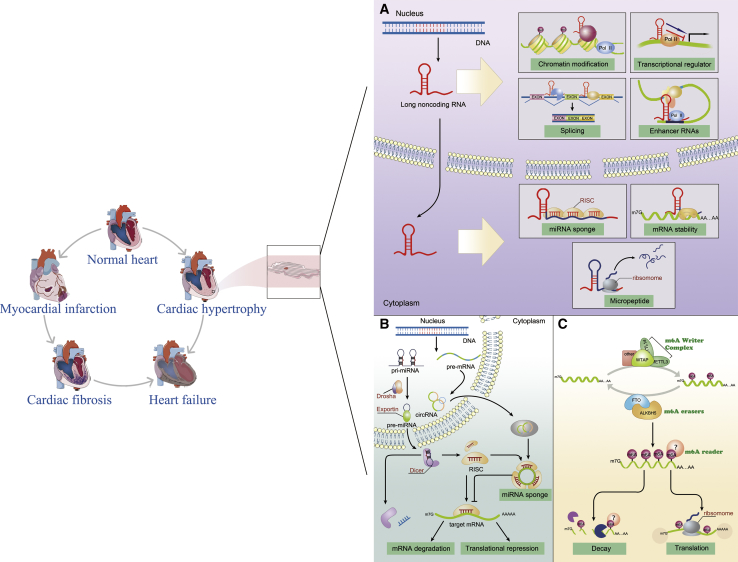
Figure 1.
Molecular Mechanisms of Components of the RNA Regulatory Networks in the Heart
(A) Molecular functions of lncRNAs in the heart. (B) Gene expression regulated by cardiac miRNAs and circRNAs. (C) m6A RNA modifications participate in the regulation of cardiac gene expression.
Author Contributions
R.T., T.L., and Z.-P.H. prepared the manuscript. R.T. and T.L. wrote the main parts of the article and produced graphics. K.O.L. and Y.C. reviewed and edited the manuscript. Z.-P.H drafted the final version of the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank members of the Huang laboratory for advice and support. We also thank Dr. Douglas Cowan for editing the manuscript. This work is supported by grants from the National Natural Science Foundation of China (81873463), the Guangdong Basic and Applied Basic Research Foundation (2019B151502003), the Guangdong Science and Technology Department (2018A050506026) and the Fundamental Research Funds for the Central Universities (20ykzd06) to Z.-P.H.
References
- 1.Hastings C.L., Roche E.T., Ruiz-Hernandez E., Schenke-Layland K., Walsh C.J., Duffy G.P. Drug and cell delivery for cardiac regeneration. Adv. Drug Deliv. Rev. 2015;84:85–106. doi: 10.1016/j.addr.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Hashimoto H., Olson E.N., Bassel-Duby R. Therapeutic approaches for cardiac regeneration and repair. Nat. Rev. Cardiol. 2018;15:585–600. doi: 10.1038/s41569-018-0036-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atianand M.K., Fitzgerald K.A. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol. Med. 2014;20:623–631. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.L. The translational landscape of the human heart. Cell. 2019;178:242–260.e29. doi: 10.1016/j.cell.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorn L.E., Lasman L., Chen J., Xu X., Hund T.J., Medvedovic M., Hanna J.H., van Berlo J.H., Accornero F. The N6-methyladenosine mRNA methylase METTL3 controls cardiac homeostasis and hypertrophy. Circulation. 2019;139:533–545. doi: 10.1161/CIRCULATIONAHA.118.036146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathiyalagan P., Adamiak M., Mayourian J., Sassi Y., Liang Y., Agarwal N., Jha D., Zhang S., Kohlbrenner E., Chepurko E. FTO-dependent N6-methyladenosine regulates cardiac function during remodeling and repair. Circulation. 2019;139:518–532. doi: 10.1161/CIRCULATIONAHA.118.033794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 10.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Shenoy A., Blelloch R.H. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat. Rev. Mol. Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomasetti M., Amati M., Santarelli L., Neuzil J. MicroRNA in metabolic re-programming and their role in tumorigenesis. Int. J. Mol. Sci. 2016;17:754. doi: 10.3390/ijms17050754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boon R.A., Iekushi K., Lechner S., Seeger T., Fischer A., Heydt S., Kaluza D., Tréguer K., Carmona G., Bonauer A. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. [DOI] [PubMed] [Google Scholar]
- 14.Wahlquist C., Jeong D., Rojas-Muñoz A., Kho C., Lee A., Mitsuyama S., van Mil A., Park W.J., Sluijter J.P., Doevendans P.A. Inhibition of miR-25 improves cardiac contractility in the failing heart. Nature. 2014;508:531–535. doi: 10.1038/nature13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small E.M., Frost R.J., Olson E.N. MicroRNAs add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan B., Wang H., Tan Y., Fu W. MicroRNAs in cardiovascular disease: small molecules but big roles. Curr. Top. Med. Chem. 2019;19:1918–1947. doi: 10.2174/1568026619666190808160241. [DOI] [PubMed] [Google Scholar]
- 17.van Rooij E., Sutherland L.B., Qi X., Richardson J.A., Hill J., Olson E.N. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 18.Callis T.E., Pandya K., Seok H.Y., Tang R.H., Tatsuguchi M., Huang Z.P., Chen J.F., Deng Z., Gunn B., Shumate J. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulin R., Sutendra G., Gurtu V., Dromparis P., Haromy A., Provencher S., Bonnet S., Michelakis E.D. A miR-208-Mef2 axis drives the decompensation of right ventricular function in pulmonary hypertension. Circ. Res. 2015;116:56–69. doi: 10.1161/CIRCRESAHA.115.303910. [DOI] [PubMed] [Google Scholar]
- 20.Ivey K.N., Muth A., Arnold J., King F.W., Yeh R.F., Fish J.E., Hsiao E.C., Schwartz R.J., Conklin B.R., Bernstein H.S., Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayed D., Hong C., Chen I.Y., Lypowy J., Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 22.Sano M., Abdellatif M., Oh H., Xie M., Bagella L., Giordano A., Michael L.H., DeMayo F.J., Schneider M.D. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 2002;8:1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- 23.Lypowy J., Chen I.Y., Abdellatif M. An alliance between Ras GTPase-activating protein, filamin C, and Ras GTPase-activating protein SH3 domain-binding protein regulates myocyte growth. J. Biol. Chem. 2005;280:25717–25728. doi: 10.1074/jbc.M414266200. [DOI] [PubMed] [Google Scholar]
- 24.Saucedo L.J., Gao X., Chiarelli D.A., Li L., Pan D., Edgar B.A. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 2003;5:566–571. doi: 10.1038/ncb996. [DOI] [PubMed] [Google Scholar]
- 25.Stocker H., Radimerski T., Schindelholz B., Wittwer F., Belawat P., Daram P., Breuer S., Thomas G., Hafen E. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 2003;5:559–565. doi: 10.1038/ncb995. [DOI] [PubMed] [Google Scholar]
- 26.Karakikes I., Chaanine A.H., Kang S., Mukete B.N., Jeong D., Zhang S., Hajjar R.J., Lebeche D. Therapeutic cardiac-targeted delivery of miR-1 reverses pressure overload-induced cardiac hypertrophy and attenuates pathological remodeling. J. Am. Heart Assoc. 2013;2:e000078. doi: 10.1161/JAHA.113.000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q., Song X.W., Zou J., Wang G.K., Kremneva E., Li X.Q., Zhu N., Sun T., Lappalainen P., Yuan W.J. Attenuation of microRNA-1 derepresses the cytoskeleton regulatory protein twinfilin-1 to provoke cardiac hypertrophy. J. Cell Sci. 2010;123:2444–2452. doi: 10.1242/jcs.067165. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda S., He A., Kong S.W., Lu J., Bejar R., Bodyak N., Lee K.H., Ma Q., Kang P.M., Golub T.R., Pu W.T. MicroRNA-1 negatively regulates expression of the hypertrophy-associated calmodulin and Mef2a genes. Mol. Cell. Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ai J., Zhang R., Gao X., Niu H.F., Wang N., Xu Y., Li Y., Ma N., Sun L.H., Pan Z.W. Overexpression of microRNA-1 impairs cardiac contractile function by damaging sarcomere assembly. Cardiovasc. Res. 2012;95:385–393. doi: 10.1093/cvr/cvs196. [DOI] [PubMed] [Google Scholar]
- 30.Hua Y., Zhang Y., Ren J. IGF-1 deficiency resists cardiac hypertrophy and myocardial contractile dysfunction: role of microRNA-1 and microRNA-133a. J. Cell. Mol. Med. 2012;16:83–95. doi: 10.1111/j.1582-4934.2011.01307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Y., Zheng J., Sun Y., Wu Z., Liu Z., Huang G. MicroRNA-1 regulates cardiomyocyte apoptosis by targeting Bcl-2. Int. Heart J. 2009;50:377–387. doi: 10.1536/ihj.50.377. [DOI] [PubMed] [Google Scholar]
- 32.Zhu W.S., Guo W., Zhu J.N., Tang C.M., Fu Y.H., Lin Q.X., Tan N., Shan Z.X. Hsp90aa1: a novel target gene of miR-1 in cardiac ischemia/reperfusion injury. Sci. Rep. 2016;6:24498. doi: 10.1038/srep24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng Y., Zhao W., Zhang X., Sun L., Yang H., Wang Y., Cao Y., Chu Y., Liu G. Downregulation of microRNA-1 attenuates glucose-induced apoptosis by regulating the liver X receptor α in cardiomyocytes. Exp. Ther. Med. 2018;16:1814–1824. doi: 10.3892/etm.2018.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B., Zhang Y., Xu C., Bai Y., Wang H. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 35.Luo X., Lin H., Pan Z., Xiao J., Zhang Y., Lu Y., Yang B., Wang Z. Retraction. J. Biol. Chem. 2011;286:28656. doi: 10.1074/jbc.A111.801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kumarswamy R., Lyon A.R., Volkmann I., Mills A.M., Bretthauer J., Pahuja A., Geers-Knörr C., Kraft T., Hajjar R.J., Macleod K.T. SERCA2a gene therapy restores microRNA-1 expression in heart failure via an Akt/FoxO3A-dependent pathway. Eur. Heart J. 2012;33:1067–1075. doi: 10.1093/eurheartj/ehs043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terentyev D., Belevych A.E., Terentyeva R., Martin M.M., Malana G.E., Kuhn D.E., Abdellatif M., Feldman D.S., Elton T.S., Györke S. miR-1 overexpression enhances Ca2+ release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56α and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ. Res. 2009;104:514–521. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers R., Timofeyev V., Li N., Kim C., Ledford H.A., Sirish P., Lau V., Zhang Y., Fayyaz K., Singapuri A. Feedback mechanisms for cardiac-specific microRNAs and cAMP signaling in electrical remodeling. Circ. Arrhythm. Electrophysiol. 2015;8:942–950. doi: 10.1161/CIRCEP.114.002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thum T., Gross C., Fiedler J., Fischer T., Kissler S., Bussen M., Galuppo P., Just S., Rottbauer W., Frantz S. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 40.Bang C., Batkai S., Dangwal S., Gupta S.K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan J., Chen H., Ge D., Xu Y., Xu H., Yang Y., Gu M., Zhou Y., Zhu J., Ge T. Mir-21 promotes cardiac fibrosis after myocardial infarction via targeting Smad7. Cell. Physiol. Biochem. 2017;42:2207–2219. doi: 10.1159/000479995. [DOI] [PubMed] [Google Scholar]
- 42.García R., Nistal J.F., Merino D., Price N.L., Fernández-Hernando C., Beaumont J., González A., Hurlé M.A., Villar A.V. p-SMAD2/3 and DICER promote pre-miR-21 processing during pressure overload-associated myocardial remodeling. Biochim. Biophys. Acta. 2015;1852:1520–1530. doi: 10.1016/j.bbadis.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Zhou X.L., Xu H., Liu Z.B., Wu Q.C., Zhu R.R., Liu J.C. miR-21 promotes cardiac fibroblast-to-myofibroblast transformation and myocardial fibrosis by targeting Jagged1. J. Cell. Mol. Med. 2018;22:3816–3824. doi: 10.1111/jcmm.13654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patrick D.M., Montgomery R.L., Qi X., Obad S., Kauppinen S., Hill J.A., van Rooij E., Olson E.N. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J. Clin. Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dong S., Cheng Y., Yang J., Li J., Liu X., Wang X., Wang D., Krall T.J., Delphin E.S., Zhang C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 2009;284:29514–29525. doi: 10.1074/jbc.M109.027896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng Y., Liu X., Zhang S., Lin Y., Yang J., Zhang C. MicroRNA-21 protects against the H2O2-induced injury on cardiac myocytes via its target gene PDCD4. J. Mol. Cell. Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng Y., Zhu P., Yang J., Liu X., Dong S., Wang X., Chun B., Zhuang J., Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc. Res. 2010;87:431–439. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tijsen A.J., van der Made I., van den Hoogenhof M.M., Wijnen W.J., van Deel E.D., de Groot N.E., Alekseev S., Fluiter K., Schroen B., Goumans M.J. The microRNA-15 family inhibits the TGFβ-pathway in the heart. Cardiovasc. Res. 2014;104:61–71. doi: 10.1093/cvr/cvu184. [DOI] [PubMed] [Google Scholar]
- 49.van Rooij E., Sutherland L.B., Liu N., Williams A.H., McAnally J., Gerard R.D., Richardson J.A., Olson E.N. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H., Untiveros G.M., McKee L.A., Perez J., Li J., Antin P.B., Konhilas J.P. Micro-RNA-195 and -451 regulate the LKB1/AMPK signaling axis by targeting MO25. PLoS ONE. 2012;7:e41574. doi: 10.1371/journal.pone.0041574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsieh P.C., Segers V.F., Davis M.E., MacGillivray C., Gannon J., Molkentin J.D., Robbins J., Lee R.T. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat. Med. 2007;13:970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Porrello E.R., Johnson B.A., Aurora A.B., Simpson E., Nam Y.J., Matkovich S.J., Dorn G.W., 2nd, van Rooij E., Olson E.N. miR-15 family regulates postnatal mitotic arrest of cardiomyocytes. Circ. Res. 2011;109:670–679. doi: 10.1161/CIRCRESAHA.111.248880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hullinger T.G., Montgomery R.L., Seto A.G., Dickinson B.A., Semus H.M., Lynch J.M., Dalby C.M., Robinson K., Stack C., Latimer P.A. Inhibition of miR-15 protects against cardiac ischemic injury. Circ. Res. 2012;110:71–81. doi: 10.1161/CIRCRESAHA.111.244442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y., Ding S., Xu G., Chen F., Ding F. MicroRNA-15a inhibition protects against hypoxia/reoxygenation-induced apoptosis of cardiomyocytes by targeting mothers against decapentaplegic homolog 7. Mol. Med. Rep. 2017;15:3699–3705. doi: 10.3892/mmr.2017.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu L., Zhang G., Liang Z., Liu X., Li T., Fan J., Bai J., Wang Y. MicroRNA-15b enhances hypoxia/reoxygenation-induced apoptosis of cardiomyocytes via a mitochondrial apoptotic pathway. Apoptosis. 2014;19:19–29. doi: 10.1007/s10495-013-0899-2. [DOI] [PubMed] [Google Scholar]
- 56.Hang P., Sun C., Guo J., Zhao J., Du Z. BDNF-mediates down-regulation of microRNA-195 inhibits ischemic cardiac apoptosis in rats. Int. J. Biol. Sci. 2016;12:979–989. doi: 10.7150/ijbs.15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li X., Zeng Z., Li Q., Xu Q., Xie J., Hao H., Luo G., Liao W., Bin J., Huang X., Liao Y. Inhibition of microRNA-497 ameliorates anoxia/reoxygenation injury in cardiomyocytes by suppressing cell apoptosis and enhancing autophagy. Oncotarget. 2015;6:18829–18844. doi: 10.18632/oncotarget.4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu J., Sun F., Wang Y., Yang W., Xiao H., Zhang Y., Lu R., Zhu H., Zhuang Y., Pan Z. Suppression of microRNA-16 protects against acute myocardial infarction by reversing beta2-adrenergic receptor down-regulation in rats. Oncotarget. 2017;8:20122–20132. doi: 10.18632/oncotarget.15391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen C., Jia K.Y., Zhang H.L., Fu J. miR-195 enhances cardiomyocyte apoptosis induced by hypoxia/reoxygenation injury via downregulating c-myb. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3410–3416. [PubMed] [Google Scholar]
- 60.Qin L., Yang W., Wang Y.X., Wang Z.J., Li C.C., Li M., Liu J.Y. MicroRNA-497 promotes proliferation and inhibits apoptosis of cardiomyocytes through the downregulation of Mfn2 in a mouse model of myocardial ischemia-reperfusion injury. Biomed. Pharmacother. 2018;105:103–114. doi: 10.1016/j.biopha.2018.04.181. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y., Yang L., Yin J., Su D., Pan Z., Li P., Wang X. MicroRNA-15b deteriorates hypoxia/reoxygenation-induced cardiomyocyte apoptosis by downregulating Bcl-2 and MAPK3. J. Investig. Med. 2018;66:39–45. doi: 10.1136/jim-2017-000485. [DOI] [PubMed] [Google Scholar]
- 62.Zhang H.J., Zhang Y.N., Teng Z.Y. Downregulation of miR-16 protects H9c2(2-1) cells against hypoxia/reoxygenation damage by targeting CIAPIN1 and regulating the NF-κB pathway. Mol. Med. Rep. 2019;20:3113–3122. doi: 10.3892/mmr.2019.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Clark A.L., Naya F.J. MicroRNAs in the myocyte enhancer factor 2 (MEF2)-regulated Gtl2-Dio3 noncoding RNA locus promote cardiomyocyte proliferation by targeting the transcriptional coactivator Cited2. J. Biol. Chem. 2015;290:23162–23172. doi: 10.1074/jbc.M115.672659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Naya F.J., Wang D.Z. (MYO)SLIDing our way into the vascular pool of long noncoding RNAs. Arterioscler. Thromb. Vasc. Biol. 2016;36:2033–2034. doi: 10.1161/ATVBAHA.116.308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pfeffer M.A., Braunwald E. Ventricular remodeling after myocardial infarction. experimental observations and clinical implications. Circulation. 1990;81:1161–1172. doi: 10.1161/01.cir.81.4.1161. [DOI] [PubMed] [Google Scholar]
- 67.Zhou H., Wang B., Yang Y.-X., Jia Q.-J., Zhang A., Qi Z.-W., Zhang J.P. Long noncoding RNAs in pathological cardiac remodeling: a review of the update literature. BioMed Res. Int. 2019;2019:7159592. doi: 10.1155/2019/7159592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hang C.T., Yang J., Han P., Cheng H.-L., Shang C., Ashley E., Zhou B., Chang C.P. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature. 2010;466:62–67. doi: 10.1038/nature09130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han P., Li W., Lin C.-H., Yang J., Shang C., Nuernberg S.T., Jin K.K., Xu W., Lin C.Y., Lin C.J. A long noncoding RNA protects the heart from pathological hypertrophy. Nature. 2014;514:102–106. doi: 10.1038/nature13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Z., Zhang X.-J., Ji Y.-X., Zhang P., Deng K.-Q., Gong J., Ren S., Wang X., Chen I., Wang H. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nat. Med. 2016;22:1131–1139. doi: 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang K., Liu F., Zhou L.-Y., Long B., Yuan S.-M., Wang Y., Liu C.Y., Sun T., Zhang X.J., Li P.F. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ. Res. 2014;114:1377–1388. doi: 10.1161/CIRCRESAHA.114.302476. [DOI] [PubMed] [Google Scholar]
- 72.Jiang F., Zhou X., Huang J. Long non-coding RNA-ROR mediates the reprogramming in cardiac hypertrophy. PLoS ONE. 2016;11:e0152767. doi: 10.1371/journal.pone.0152767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu L., An X., Li Z., Song Y., Li L., Zuo S., Liu N., Yang G., Wang H., Cheng X. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016;111:56–65. doi: 10.1093/cvr/cvw078. [DOI] [PubMed] [Google Scholar]
- 74.Lv L., Li T., Li X., Xu C., Liu Q., Jiang H., Li Y., Liu Y., Yan H., Huang Q. The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR-214. Mol. Ther. Nucleic Acids. 2018;10:387–397. doi: 10.1016/j.omtn.2017.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu X.H., Yuan Y.X., Rao S.L., Wang P. lncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016;20:3653–3660. [PubMed] [Google Scholar]
- 76.Wang K., Liu C.-Y., Zhou L.-Y., Wang J.-X., Wang M., Zhao B., Zhao W.K., Xu S.J., Fan L.H., Zhang X.J. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015;6:6779. doi: 10.1038/ncomms7779. [DOI] [PubMed] [Google Scholar]
- 77.Liu C.-Y., Zhang Y.-H., Li R.-B., Zhou L.-Y., An T., Zhang R.-C., Zhai M., Huang Y., Yan K.W., Dong Y.H. lncRNA CAIF inhibits autophagy and attenuates myocardial infarction by blocking p53-mediated myocardin transcription. Nat. Commun. 2018;9:29. doi: 10.1038/s41467-017-02280-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang S., Yu W., Chen J., Yao T., Deng F. lncRNA MALAT1 sponges miR-203 to promote inflammation in myocardial ischemia-reperfusion injury. Int. J. Cardiol. 2018;268:245. doi: 10.1016/j.ijcard.2018.03.085. [DOI] [PubMed] [Google Scholar]
- 79.Zhu P., Yang M., Ren H., Shen G., Chen J., Zhang J., Liu J., Sun C. Long noncoding RNA MALAT1 downregulates cardiac transient outward potassium current by regulating miR-200c/HMGB1 pathway. J. Cell. Biochem. 2018;119:10239–10249. doi: 10.1002/jcb.27366. [DOI] [PubMed] [Google Scholar]
- 80.Huang S., Zhang L., Song J., Wang Z., Huang X., Guo Z., Chen F., Zhao X. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J. Cell. Physiol. 2019;234:2997–3006. doi: 10.1002/jcp.27117. [DOI] [PubMed] [Google Scholar]
- 81.Micheletti R., Plaisance I., Abraham B.J., Sarre A., Ting C.-C., Alexanian M., Maric D., Maison D., Nemir M., Young R.A. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017;9:eaai9118. doi: 10.1126/scitranslmed.aai9118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Piccoli M.-T., Gupta S.K., Viereck J., Foinquinos A., Samolovac S., Kramer F.L., Garg A., Remke J., Zimmer K., Batkai S., Thum T. Inhibition of the cardiac fibroblast-enriched lncRNA Meg3 prevents cardiac fibrosis and diastolic dysfunction. Circ. Res. 2017;121:575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- 83.Tao H., Zhang J.-G., Qin R.-H., Dai C., Shi P., Yang J.-J., Deng Z.Y., Shi K.H. lncRNA GAS5 controls cardiac fibroblast activation and fibrosis by targeting miR-21 via PTEN/MMP-2 signaling pathway. Toxicology. 2017;386:11–18. doi: 10.1016/j.tox.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 84.Klattenhoff C.A., Scheuermann J.C., Surface L.E., Bradley R.K., Fields P.A., Steinhauser M.L., Ding H., Butty V.L., Torrey L., Haas S. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152:570–583. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Grote P., Wittler L., Hendrix D., Koch F., Währisch S., Beisaw A., Macura K., Bläss G., Kellis M., Werber M., Herrmann B.G. The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev. Cell. 2013;24:206–214. doi: 10.1016/j.devcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 87.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 88.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 89.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 92.Murtaza I., Wang H.X., Feng X., Alenina N., Bader M., Prabhakar B.S., Li P.F. Down-regulation of catalase and oxidative modification of protein kinase CK2 lead to the failure of apoptosis repressor with caspase recruitment domain to inhibit cardiomyocyte hypertrophy. J. Biol. Chem. 2008;283:5996–6004. doi: 10.1074/jbc.M706466200. [DOI] [PubMed] [Google Scholar]
- 93.Du W.W., Yang W., Chen Y., Wu Z.K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 94.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou L.Y., Zhai M., Huang Y., Xu S., An T., Wang Y.H., Zhang R.C., Liu C.Y., Dong Y.H., Wang M. The circular RNA ACR attenuates myocardial ischemia/reperfusion injury by suppressing autophagy via modulation of the Pink1/FAM65B pathway. Cell Death Differ. 2019;26:1299–1315. doi: 10.1038/s41418-018-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W. Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation. 2019;139:2857–2876. doi: 10.1161/CIRCULATIONAHA.118.038361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Mor H., Wyler E., Perez-Hernandez D., Ramberger E. Translation of circRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Makarewich C.A., Olson E.N. Mining for micropeptides. Trends Cell Biol. 2017;27:685–696. doi: 10.1016/j.tcb.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hashimoto Y., Kondo T., Kageyama Y. Lilliputians get into the limelight: novel class of small peptide genes in morphogenesis. Dev. Growth Differ. 2008;50(Suppl 1):S269–S276. doi: 10.1111/j.1440-169X.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- 102.Gish W., States D.J. Identification of protein coding regions by database similarity search. Nat. Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 103.Kochetov A.V. AUG codons at the beginning of protein coding sequences are frequent in eukaryotic mRNAs with a suboptimal start codon context. Bioinformatics. 2005;21:837–840. doi: 10.1093/bioinformatics/bti136. [DOI] [PubMed] [Google Scholar]
- 104.Anderson D.M., Anderson K.M., Chang C.-L., Makarewich C.A., Nelson B.R., McAnally J.R., Kasaragod P., Shelton J.M., Liou J., Bassel-Duby R., Olson E.N. A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell. 2015;160:595–606. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Anderson D.M., Makarewich C.A., Anderson K.M., Shelton J.M., Bezprozvannaya S., Bassel-Duby R., Olson E.N. Widespread control of calcium signaling by a family of SERCA-inhibiting micropeptides. Sci. Signal. 2016;9:ra119. doi: 10.1126/scisignal.aaj1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nelson B.R., Makarewich C.A., Anderson D.M., Winders B.R., Troupes C.D., Wu F., Reese A.L., McAnally J.R., Chen X., Kavalali E.T. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;351:271–275. doi: 10.1126/science.aad4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Anderson K.M., Anderson D.M., McAnally J.R., Shelton J.M., Bassel-Duby R., Olson E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature. 2016;539:433–436. doi: 10.1038/nature20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang Y., Jiao L., Sun L., Li Y., Gao Y., Xu C., Shao Y., Li M., Li C., Lu Y. lncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca2+ overload and contractile dysfunction in a mouse model of myocardial infarction. Circ. Res. 2018;122:1354–1368. doi: 10.1161/CIRCRESAHA.117.312117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang L., Salgado-Somoza A., Vausort M., Leszek P., Devaux Y., Cardiolinc™ network A heart-enriched antisense long non-coding RNA regulates the balance between cardiac and skeletal muscle triadin. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865:247–258. doi: 10.1016/j.bbamcr.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 110.Makarewich C.A., Baskin K.K., Munir A.Z., Bezprozvannaya S., Sharma G., Khemtong C., Shah A.M., McAnally J.R., Malloy C.R., Szweda L.I. MOXI is a mitochondrial micropeptide that enhances fatty acid β-oxidation. Cell Rep. 2018;23:3701–3709. doi: 10.1016/j.celrep.2018.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Stein C.S., Jadiya P., Zhang X., McLendon J.M., Abouassaly G.M., Witmer N.H., Anderson E.J., Elrod J.W., Boudreau R.L. Mitoregulin: a lncRNA-encoded microprotein that supports mitochondrial supercomplexes and respiratory efficiency. Cell Rep. 2018;23:3710–3720.e8. doi: 10.1016/j.celrep.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Han X., Zhang J., Liu Y., Fan X., Ai S., Luo Y., Li X., Jin H., Luo S., Zheng H. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development. 2019;146:dev176198. doi: 10.1242/dev.176198. [DOI] [PubMed] [Google Scholar]
- 113.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 114.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Coots R.A., Liu X.M., Mao Y., Dong L., Zhou J., Wan J., Zhang X., Qian S.B. m6A facilitates eIF4F-independent mRNA translation. Mol. Cell. 2017;68:504–514.e7. doi: 10.1016/j.molcel.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen X.Y., Zhang J., Zhu J.S. The role of m6A RNA methylation in human cancer. Mol. Cancer. 2019;18:103. doi: 10.1186/s12943-019-1033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang S., Chai P., Jia R., Jia R. Novel insights on m6A RNA methylation in tumorigenesis: a double-edged sword. Mol. Cancer. 2018;17:101. doi: 10.1186/s12943-018-0847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Song H., Feng X., Zhang H., Luo Y., Huang J., Lin M., Jin J., Ding X., Wu S., Huang H. METTL3 and ALKBH5 oppositely regulate m6A modification of TFEB mRNA, which dictates the fate of hypoxia/reoxygenation-treated cardiomyocytes. Autophagy. 2019;15:1419–1437. doi: 10.1080/15548627.2019.1586246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dorn G.W., 2nd, Force T. Protein kinase cascades in the regulation of cardiac hypertrophy. J. Clin. Invest. 2005;115:527–537. doi: 10.1172/JCI24178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chan A.Y., Soltys C.L., Young M.E., Proud C.G., Dyck J.R. Activation of AMP-activated protein kinase inhibits protein synthesis associated with hypertrophy in the cardiac myocyte. J. Biol. Chem. 2004;279:32771–32779. doi: 10.1074/jbc.M403528200. [DOI] [PubMed] [Google Scholar]
- 122.Li Y., Wang Y., Zou M., Chen C., Chen Y., Xue R., Dong Y., Liu C. AMPK blunts chronic heart failure by inhibiting autophagy. Biosci. Rep. 2018;38 doi: 10.1042/BSR20170982. BSR20170982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chorghade S., Seimetz J., Emmons R., Yang J., Bresson S.M., Lisio M., Parise G., Conrad N.K., Kalsotra A. Poly(A) tail length regulates PABPC1 expression to tune translation in the heart. eLife. 2017;6:e24139. doi: 10.7554/eLife.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Busch A., Eken S.M., Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann. Transl. Med. 2016;4:236. doi: 10.21037/atm.2016.06.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Long G., Wang F., Duan Q., Chen F., Yang S., Gong W., Wang Y., Chen C., Wang D.W. Human circulating microRNA-1 and microRNA-126 as potential novel indicators for acute myocardial infarction. Int. J. Biol. Sci. 2012;8:811–818. doi: 10.7150/ijbs.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Devaux Y., Vausort M., Goretti E., Nazarov P.V., Azuaje F., Gilson G., Corsten M.F., Schroen B., Lair M.L., Heymans S., Wagner D.R. Use of circulating microRNAs to diagnose acute myocardial infarction. Clin. Chem. 2012;58:559–567. doi: 10.1373/clinchem.2011.173823. [DOI] [PubMed] [Google Scholar]
- 127.Zhang L., Chen X., Su T., Li H., Huang Q., Wu D., Yang C., Han Z. Circulating miR-499 are novel and sensitive biomarker of acute myocardial infarction. J. Thorac. Dis. 2015;7:303–308. doi: 10.3978/j.issn.2072-1439.2015.02.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ji X., Takahashi R., Hiura Y., Hirokawa G., Fukushima Y., Iwai N. Plasma miR-208 as a biomarker of myocardial injury. Clin. Chem. 2009;55:1944–1949. doi: 10.1373/clinchem.2009.125310. [DOI] [PubMed] [Google Scholar]
- 129.Zhu L., Liu F., Xie H., Feng J. Diagnostic performance of microRNA-133a in acute myocardial infarction: a meta-analysis. Cardiol. J. 2018;25:260–267. doi: 10.5603/CJ.a2017.0126. [DOI] [PubMed] [Google Scholar]
- 130.de Gonzalo-Calvo D., Cediel G., Bär C., Núñez J., Revuelta-Lopez E., Gavara J., Ríos-Navarro C., Llorente-Cortes V., Bodí V., Thum T., Bayes-Genis A. Circulating miR-1254 predicts ventricular remodeling in patients with ST-segment-elevation myocardial infarction: a cardiovascular magnetic resonance study. Sci. Rep. 2018;8:15115. doi: 10.1038/s41598-018-33491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yang S.Y., Wang Y.Q., Gao H.M., Wang B., He Q. The clinical value of circulating miR-99a in plasma of patients with acute myocardial infarction. Eur. Rev. Med. Pharmacol. Sci. 2016;20:5193–5197. [PubMed] [Google Scholar]
- 132.Wang Y., Chang W., Zhang Y., Zhang L., Ding H., Qi H., Xue S., Yu H., Hu L., Liu D. Circulating miR-22-5p and miR-122-5p are promising novel biomarkers for diagnosis of acute myocardial infarction. J. Cell. Physiol. 2019;234:4778–4786. doi: 10.1002/jcp.27274. [DOI] [PubMed] [Google Scholar]
- 133.Yan Y., Song X., Li Z., Zhang J., Ren J., Wu J., Li Y., Guan Y., Wang J. Elevated levels of granzyme B correlated with miR-874-3p downregulation in patients with acute myocardial infarction. Biomarkers Med. 2017;11:761–767. doi: 10.2217/bmm-2017-0144. [DOI] [PubMed] [Google Scholar]
- 134.Li L., Li S., Wu M., Chi C., Hu D., Cui Y., Song J., Lee C., Chen H. Early diagnostic value of circulating microRNAs in patients with suspected acute myocardial infarction. J. Cell. Physiol. 2019;234:13649–13658. doi: 10.1002/jcp.28045. [DOI] [PubMed] [Google Scholar]
- 135.Vausort M., Wagner D.R., Devaux Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014;115:668–677. doi: 10.1161/CIRCRESAHA.115.303836. [DOI] [PubMed] [Google Scholar]
- 136.Wang F., Su X., Liu C., Wu M., Li B. Prognostic value of plasma long noncoding RNA ANRIL for in-stent restenosis. Med. Sci. Monit. 2017;23:4733–4739. doi: 10.12659/MSM.904352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao L., Liu Y., Guo S., Yao R., Wu L., Xiao L., Wang Z., Liu Y., Zhang Y. Circulating long noncoding RNA HOTAIR is an essential mediator of acute myocardial infarction. Cell. Physiol. Biochem. 2017;44:1497–1508. doi: 10.1159/000485588. [DOI] [PubMed] [Google Scholar]
- 138.Yan Y., Zhang B., Liu N., Qi C., Xiao Y., Tian X., Li T., Liu B. Circulating Long noncoding RNA UCA1 as a novel biomarker of acute myocardial infarction. BioMed Res. Int. 2016;2016:8079372. doi: 10.1155/2016/8079372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhang J., Gao C., Meng M., Tang H. Long noncoding RNA MHRT protects cardiomyocytes against H2O2-induced apoptosis. Biomol. Ther. (Seoul) 2016;24:19–24. doi: 10.4062/biomolther.2015.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Azat M., Huojiahemaiti X., Gao R., Peng P. Long noncoding RNA MIAT: a potential role in the diagnosis and mediation of acute myocardial infarction. Mol. Med. Rep. 2019;20:5216–5222. doi: 10.3892/mmr.2019.10768. [DOI] [PubMed] [Google Scholar]
- 141.Zhang Z., Gao W., Long Q.Q., Zhang J., Li Y.F., Liu D.C., Yan J.J., Yang Z.J., Wang L.S. Increased plasma levels of lncRNA H19 and LIPCAR are associated with increased risk of coronary artery disease in a Chinese population. Sci. Rep. 2017;7:7491. doi: 10.1038/s41598-017-07611-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang Y., Sun L., Xuan L., Pan Z., Li K., Liu S., Huang Y., Zhao X., Huang L., Wang Z. Reciprocal changes of circulating long non-coding RNAs ZFAS1 and CDR1AS predict acute myocardial infarction. Sci. Rep. 2016;6:22384. doi: 10.1038/srep22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shang Q., Yang Z., Jia R., Ge S. The novel roles of circRNAs in human cancer. Mol. Cancer. 2019;18:6. doi: 10.1186/s12943-018-0934-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Memczak S., Papavasileiou P., Peters O., Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE. 2015;10:e0141214. doi: 10.1371/journal.pone.0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vausort M., Salgado-Somoza A., Zhang L., Leszek P., Scholz M., Teren A., Burkhardt R., Thiery J., Wagner D.R., Devaux Y. Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J. Am. Coll. Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 146.Salgado-Somoza A., Zhang L., Vausort M., Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Akat K.M., Moore-McGriff D., Morozov P., Brown M., Gogakos T., Correa Da Rosa J., Mihailovic A., Sauer M., Ji R., Ramarathnam A. Comparative RNA-sequencing analysis of myocardial and circulating small RNAs in human heart failure and their utility as biomarkers. Proc. Natl. Acad. Sci. USA. 2014;111:11151–11156. doi: 10.1073/pnas.1401724111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li H., Fan J., Yin Z., Wang F., Chen C., Wang D.W. Identification of cardiac-related circulating microRNA profile in human chronic heart failure. Oncotarget. 2016;7:33–45. doi: 10.18632/oncotarget.6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ovchinnikova E.S., Schmitter D., Vegter E.L., Ter Maaten J.M., Valente M.A., Liu L.C., van der Harst P., Pinto Y.M., de Boer R.A., Meyer S. Signature of circulating microRNAs in patients with acute heart failure. Eur. J. Heart Fail. 2016;18:414–423. doi: 10.1002/ejhf.332. [DOI] [PubMed] [Google Scholar]
- 150.Corsten M.F., Dennert R., Jochems S., Kuznetsova T., Devaux Y., Hofstra L., Wagner D.R., Staessen J.A., Heymans S., Schroen B. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 151.Ikitimur B., Cakmak H.A., Coskunpinar E., Barman H.A., Vural V.A. The relationship between circulating microRNAs and left ventricular mass in symptomatic heart failure patients with systolic dysfunction. Kardiol. Pol. 2015;73:740–746. doi: 10.5603/KP.a2015.0082. [DOI] [PubMed] [Google Scholar]
- 152.Yu X., Zou T., Zou L., Jin J., Xiao F., Yang J. Plasma long noncoding RNA urothelial carcinoma associated 1 predicts poor prognosis in chronic heart failure patients. Med. Sci. Monit. 2017;23:2226–2231. doi: 10.12659/MSM.904113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Xuan L., Sun L., Zhang Y., Huang Y., Hou Y., Li Q., Guo Y., Feng B., Cui L., Wang X. Circulating long non-coding RNAs NRON and MHRT as novel predictive biomarkers of heart failure. J. Cell. Mol. Med. 2017;21:1803–1814. doi: 10.1111/jcmm.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 155.Chen J., Huang Z.P., Seok H.Y., Ding J., Kataoka M., Zhang Z., Hu X., Wang G., Lin Z., Wang S. mir-17-92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ. Res. 2013;112:1557–1566. doi: 10.1161/CIRCRESAHA.112.300658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gao F., Kataoka M., Liu N., Liang T., Huang Z.P., Gu F., Ding J., Liu J., Zhang F., Ma Q. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat. Commun. 2019;10:1802. doi: 10.1038/s41467-019-09530-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Eulalio A., Mano M., Dal Ferro M., Zentilin L., Sinagra G., Zacchigna S., Giacca M. Functional screening identifies miRNAs inducing cardiac regeneration. Nature. 2012;492:376–381. doi: 10.1038/nature11739. [DOI] [PubMed] [Google Scholar]
- 158.Gabisonia K., Prosdocimo G., Aquaro G.D., Carlucci L., Zentilin L., Secco I., Ali H., Braga L., Gorgodze N., Bernini F. MicroRNA therapy stimulates uncontrolled cardiac repair after myocardial infarction in pigs. Nature. 2019;569:418–422. doi: 10.1038/s41586-019-1191-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li H., Zhang X., Wang F., Zhou L., Yin Z., Fan J., Nie X., Wang P., Fu X.D., Chen C., Wang D.W. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation. 2016;134:734–751. doi: 10.1161/CIRCULATIONAHA.116.023926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Duisters R.F., Tijsen A.J., Schroen B., Leenders J.J., Lentink V., van der Made I., Herias V., van Leeuwen R.E., Schellings M.W., Barenbrug P. miR-133 and miR-30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ. Res. 2009;104:170–178. doi: 10.1161/CIRCRESAHA.108.182535. [DOI] [PubMed] [Google Scholar]
- 161.Carè A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 162.Seok H.Y., Chen J., Kataoka M., Huang Z.P., Ding J., Yan J., Hu X., Wang D.Z. Loss of microRNA-155 protects the heart from pathological cardiac hypertrophy. Circ. Res. 2014;114:1585–1595. doi: 10.1161/CIRCRESAHA.114.303784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.He W., Huang H., Xie Q., Wang Z., Fan Y., Kong B., Huang D., Xiao Y. miR-155 knockout in fibroblasts improves cardiac remodeling by targeting tumor protein p53-inducible nuclear protein 1. J. Cardiovasc. Pharmacol. Ther. 2016;21:423–435. doi: 10.1177/1074248415616188. [DOI] [PubMed] [Google Scholar]
- 164.Uchida S., Dimmeler S. Exercise controls non-coding RNAs. Cell Metab. 2015;21:511–512. doi: 10.1016/j.cmet.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 165.Verjans R., Peters T., Beaumont F.J., van Leeuwen R., van Herwaarden T., Verhesen W., Munts C., Bijnen M., Henkens M., Diez J. MicroRNA-221/222 family counteracts myocardial fibrosis in pressure overload-induced heart failure. Hypertension. 2018;71:280–288. doi: 10.1161/HYPERTENSIONAHA.117.10094. [DOI] [PubMed] [Google Scholar]
- 166.Xiao Y., Zhang X., Fan S., Cui G., Shen Z. MicroRNA-497 inhibits cardiac hypertrophy by targeting Sirt4. PLoS ONE. 2016;11:e0168078. doi: 10.1371/journal.pone.0168078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang X., Ji R., Liao X., Castillero E., Kennel P.J., Brunjes D.L., Franz M., Möbius-Winkler S., Drosatos K., George I. MicroRNA-195 regulates metabolism in failing myocardium via alterations in sirtuin 3 expression and mitochondrial protein acetylation. Circulation. 2018;137:2052–2067. doi: 10.1161/CIRCULATIONAHA.117.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lee S., Lim S., Ham O., Lee S.Y., Lee C.Y., Park J.H., Lee J., Seo H.H., Yun I., Han S.M. ROS-mediated bidirectional regulation of miRNA results in distinct pathologic heart conditions. Biochem. Biophys. Res. Commun. 2015;465:349–355. doi: 10.1016/j.bbrc.2015.07.160. [DOI] [PubMed] [Google Scholar]
- 169.Zhou Y.L., Sun Q., Zhang L., Li R. miR-208b targets Bax to protect H9c2 cells against hypoxia-induced apoptosis. Biomed. Pharmacother. 2018;106:1751–1759. doi: 10.1016/j.biopha.2018.07.141. [DOI] [PubMed] [Google Scholar]
- 170.Zhang S., Zhang R., Wu F., Li X. MicroRNA-208a regulates H9c2 cells simulated ischemia-reperfusion myocardial injury via targeting CHD9 through Notch/NF-kappa B signal pathways. Int. Heart J. 2018;59:580–588. doi: 10.1536/ihj.17-147. [DOI] [PubMed] [Google Scholar]
- 171.Wang F., Yuan Y., Yang P., Li X. Extracellular vesicles-mediated transfer of miR-208a/b exaggerate hypoxia/reoxygenation injury in cardiomyocytes by reducing QKI expression. Mol. Cell. Biochem. 2017;431:187–195. doi: 10.1007/s11010-017-2990-4. [DOI] [PubMed] [Google Scholar]
- 172.Wang W., Zheng Y., Wang M., Yan M., Jiang J., Li Z. Exosomes derived miR-126 attenuates oxidative stress and apoptosis from ischemia and reperfusion injury by targeting ERRFI1. Gene. 2019;690:75–80. doi: 10.1016/j.gene.2018.12.044. [DOI] [PubMed] [Google Scholar]
- 173.Wang J., Jia Z., Zhang C., Sun M., Wang W., Chen P., Ma K., Zhang Y., Li X., Zhou C. miR-499 protects cardiomyocytes from H2O2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 2014;11:339–350. doi: 10.4161/rna.28300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wang J.X., Jiao J.Q., Li Q., Long B., Wang K., Liu J.P., Li Y.R., Li P.F. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nat. Med. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 175.Zhu J., Yao K., Wang Q., Guo J., Shi H., Ma L., Liu H., Gao W., Zou Y., Ge J. Ischemic postconditioning-regulated miR-499 protects the rat heart against ischemia/reperfusion injury by inhibiting apoptosis through PDCD4. Cell. Physiol. Biochem. 2016;39:2364–2380. doi: 10.1159/000452506. [DOI] [PubMed] [Google Scholar]
- 176.Porrello E.R., Mahmoud A.I., Simpson E., Johnson B.A., Grinsfelder D., Canseco D., Mammen P.P., Rothermel B.A., Olson E.N., Sadek H.A. Regulation of neonatal and adult mammalian heart regeneration by the miR-15 family. Proc. Natl. Acad. Sci. USA. 2013;110:187–192. doi: 10.1073/pnas.1208863110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Luo X., Lin H., Pan Z., Xiao J., Zhang Y., Lu Y., Yang B., Wang Z. Retraction. J. Biol. Chem. 2011;286:28656. doi: 10.1074/jbc.A111.801035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lu Y., Zhang Y., Wang N., Pan Z., Gao X., Zhang F., Zhang Y., Shan H., Luo X., Bai Y. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 179.Ling T.Y., Wang X.L., Chai Q., Lau T.W., Koestler C.M., Park S.J., Daly R.C., Greason K.L., Jen J., Wu L.Q. Regulation of the SK3 channel by microRNA-499—potential role in atrial fibrillation. Heart Rhythm. 2013;10:1001–1009. doi: 10.1016/j.hrthm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ling T.Y., Wang X.L., Chai Q., Lu T., Stulak J.M., Joyce L.D., Daly R.C., Greason K.L., Wu L.Q., Shen W.K. Regulation of cardiac CACNB2 by microRNA-499: potential role in atrial fibrillation. BBA Clin. 2017;7:78–84. doi: 10.1016/j.bbacli.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liew C.-C., Dzau V.J. Molecular genetics and genomics of heart failure. Nat. Rev. Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 182.Viereck J., Thum T. Long noncoding RNAs in pathological cardiac remodeling. Circ. Res. 2017;120:262–264. doi: 10.1161/CIRCRESAHA.116.310174. [DOI] [PubMed] [Google Scholar]